Summary
This protocol describes an in vitro fluorogenic assay to measure the proteolytic activity and identify inhibitors of Mpro, the main protease produced by SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2). Studies to identify potential inhibitors of Mpro mainly rely on in silico molecular dynamics simulations or on FRET (Fluorescence Resonance Energy Transfer) substrates. The protocol is based on an aminomethyl coumarin substrate. High sensitivity, specificity, and an easily detectable fluorescent read-out are the advantages offered by this rapid assay, which allows high throughput screening of new Mpro inhibitors.
Subject areas: Biotechnology and bioengineering, Chemistry, Clinical Protocol, Health Sciences, High Throughput Screening, Molecular/Chemical Probes, Protein Biochemistry
Graphical abstract
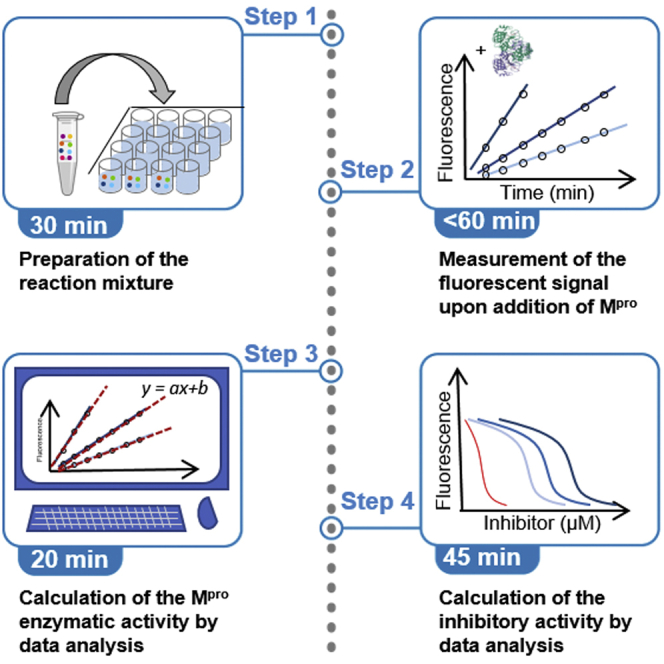
Highlights
-
•
Mpro is the main protease produced by SARS-CoV-2
-
•
An in vitro fluorogenic assay for the measurement of the proteolytic activity of Mpro
-
•
An in vitro fluorogenic assay for the inhibitor screening of Mpro
-
•
High sensitivity, specificity, and an easily detectable fluorescent read-out
This protocol describes an in vitro fluorogenic assay to measure the proteolytic activity and identify inhibitors of Mpro, the main protease produced by SARS-CoV-2. Studies to identify potential inhibitors of Mpro mainly rely on in silico molecular dynamics simulations or on FRET substrates. The protocol is based on an aminomethyl coumarin substrate. High sensitivity, specificity, and an easily detectable fluorescent read-out are the advantages offered by this rapid assay, which allows high throughput screening of new Mpro inhibitors.
Before you begin
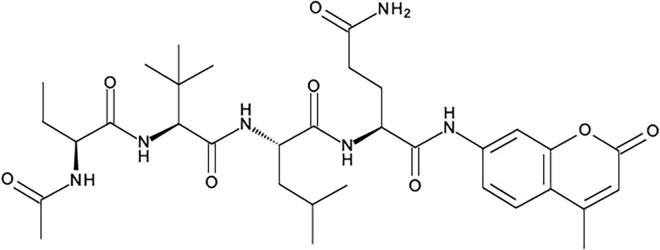
This protocol details an in vitro fluorogenic assay to measure the activity of the main protease produced by SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2) and its application to the evaluation of Mpro inhibitory compounds. The protocol assays the Mpro protease activity using a specific fluorogenic probe Ac-Abu-Tle-Leu-Gln-AMC which has been described by Rut et al. (2020) as the preferred synthetic substrate for Mpro (KM = 207 μM for analog substrate Ac-Abu-Tle-Leu-Gln-ACC). The fluorogenic substrate molecule contains the two non-natural amino acids L-2-aminobutyric acid (Abu) and L-tert-leucine (Tle) and the two natural amino acids L-leucine (Leu) and L-glutamine (Gln). The oligopeptide carries the fluorophore 7-Amino-4-methylcoumarin (AMC) at the C-terminal end of glutamine, i.e., linked via a peptide bond (Figure 1). Upon cleavage by Mpro, a fluorogenic moiety is released and a fluorescent signal is produced. The assay can be used for, e.g., the analysis of MPro activity in infected cell cultures and assessment of potential Mpro inhibitors.
Figure 1.
Chemical structure of the synthetic fluorogenic substrate Ac-Abu-Tle-Leu-Gln-AMC for the main protease Mpro of SARS-CoV-2.
Prior to the experiment, the assay buffer and the stock solutions of substrate, enzyme, and inhibitors should be prepared to ensure a straightforward assembly of the reaction mix in the multiwell plate before measurement.
Before you start, familiarize yourself with the material safety data sheets (MSDS) for all the reagents.
Note: Fluorescence can be expressed in arbitrary units (AU), relative fluorescence units (RFU) or sometimes simply abbreviated as FL and its values are device-dependent (Beal et al., 2018).
Prepare fluorogenic substrate solution
Timing: 10 min
A stock solution (5 mM or 3.28 mg/mL) of the fluorogenic substrate Ac-Abu-Tle-Leu-Gln-AMC (M.W. 656.77 g/mol) should be prepared and can be stored as described for future use.
-
1.
Weigh 1–2 mg of Ac-Abu-Tle-Leu-Gln-AMC in a microcentrifuge tube using an analytical balance.
-
2.Add an exact volume of dimethyl sulfoxide (DMSO, calculated from weighing) to reach a final concentration of 3.28 mg/mL Ac-Abu-Tle-Leu-Gln-AMC, e.g., add 0.5 mL of DMSO to 1.74 mg of substrate.
-
a.Vortex briefly to dissolve.
-
a.
-
3.
Store stock solution at −20°C, protected from light. The solution is stable under these conditions for at least one year.
Preparation of the Mpro stock solution
Timing: 10 min
Stock solutions of SARS-CoV-2 main protease (final concentration 1 mg/mL or 28 μM) can be prepared as follows:
-
4.Reconstitute the Mpro lyophilisate directly in the vial of the supplier, i.e., containing 1 mg, by adding 1 mL of 50% v/v glycerol in water.
-
a.Dissolve any solid completely by gently pipetting up and down avoiding the formation of bubbles (Troubleshooting 2).
-
a.
-
5.
Divide in 0.1 mL aliquots of stock solution.
-
6.
Store at a temperature between −15°C and −25°C and avoid freeze/thaw cycles, i.e., prepare aliquots for a single use. The solution is stable under these conditions for at least one year.
Preparation of the assay buffer
Timing: 30 min
The assay buffer contains 20 mM (2.42 g/l) TRIS, 1 mM EDTA, 150 mM NaCl, 1 mM DTT and has a pH of 7.3. It can be prepared as follows for a 100 mL final volume:
-
7.Weigh 242 mg tris(hydroxymethyl)aminomethane (TRIS, M.W. 121.14 g/mol) and place it in a beaker containing approximately 80 mL of ultrapure water and a magnetic stirrer.
-
a.Dissolve by stirring.
-
a.
-
8.Weigh 37.2 mg of ethylenediaminetetraacetic acid tetrasodium salt tetrahydrate (EDTA, tetrasodium salt, M.W. 452.23 g/mol).
-
a.Add it to the TRIS-containing solution and stir to dissolve.
-
a.
-
9.Weigh 876 mg of NaCl.
-
a.Add it to the TRIS-containing solution and stir to dissolve.
-
b.Adjust pH to 7.3 by adding drop-wise 1 N HCl.
-
a.
-
10.Add ultrapure water to reach a volume of 100 mL in a measuring cylinder.
-
a.Stir with a magnetic bar for 1 min.
-
a.
-
11.In a separate tube, weigh an amount between 200 and 400 mg of dithiothreitol (DTT, M.W. 154.253 g/mol).
-
a.Dissolve it in an exact volume of ultrapure water (calculated from weighing) to achieve a 1 M stock solution (154.25 mg/mL).
-
a.
-
12.
To prepare the buffer for activity measurement, add 1 μL DTT stock solution per mL buffer, e.g., add 5 μL of DTT stock solution to 5 mL of assay buffer.
| Reagent | Final concentration | Amount |
|---|---|---|
| TRIS | 20 mM | 242 mg |
| EDTA | 1 mM | 37.2 mg |
| NaCl | 150 mM | 876 mg |
| DTT | 1 mM | 100 μL (1 μL of 1M DTT stock solution per mL buffer) |
| Ultrapure water | - | Up to 100 mL |
CRITICAL: For best results, use freshly prepared buffer in your experiments. DTT has a half-life of less than one hour under certain conditions in aqueous solution, i.e. 40°C and pH 8.5, and should always be added freshly to solutions. DTT stock solution can be stored in aliquots for up to one year at −80°C, but multiple freezing and thawing cycles should be avoided. The addition of EDTA to the DTT stock solution at a 0.1 mM concentration improves its stability (Stevens et al. 1983).
Alternatives: Buffers alternative to TRIS that have been used with Mpro include 20 mM HEPES, pH 6.0, 0.4 mM EDTA, 1 mM DTT, 1% glycerol that has been used when assaying the activity with a FRET (fluorescence resonance energy transfer) substrate (Hung et al. 2020).
CRITICAL: DTT is often used in inhibition assays with proteases, but may interfere with the efficacy of certain inhibitors, e.g. ebselen, disulfiram, tideglusib, carmofur, shikonin, and PX-12 (Ma et al. 2020).example, ebselen. In the initial screenings of potential inhibitors and in control assays with ebselen, it is recommended to omit DTT from the assay buffer or test different reducing agents (Lee et al. 2012). DTT-free assay buffer can be stored at +4°C for several months. Similarly, EDTA prevents the possible interference of metal ions with the protease and thus compromises its activity (Kozak et al. 2020).
Preparation of the stock solutions of the inhibitor ebselen
Timing: 10 min
Ebselen (M.W. 274.2 g/mol) is used here as a positive inhibition control and solutions of additional potential inhibitors to be tested should be prepared similarly and ahead of time. For the preparation of the ebselen stock solution:
-
13.
Weigh an amount of ebselen between 5 and 7 mg in a 2 mL microcentrifuge tube.
-
14.From the weight, calculate the exact volume of DMSO required for a final 20 mM concentration (5.48 mg/mL).
-
a.Add the calculated volume of DMSO.
-
b.Dissolve by vortexing (Troubleshooting 4).
-
a.
-
15.
Stock solution can be stored for one year at −20°C.
CRITICAL: Concentration of inhibitor stock solution should be at least 200-fold higher than highest final concentration to be tested in the in assay to avoid any potential interference of the solvent.
Preparation of the dilution series of inhibitor
Timing: 10 min
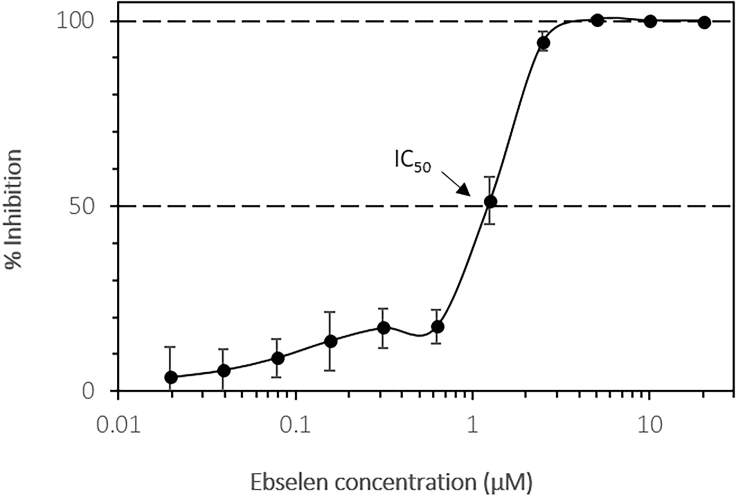
By supplementing the assay buffer with an inhibitory molecule, the assay measurement can be used to assess the impact of the compound tested on the Mpro protease activity. A high concentration of ebselen can be used as a positive control for activity inhibition. Here, suggested concentrations are presented. Mpro inhibition can be presented as a relative value to the signal obtained without ebselen that will be considered as 0% inhibition (100% activity) reference value. The IC50 value is the inhibitor concentration where the enzyme activity is inhibited by 50%. An IC50 of 0.67 μM for ebselen has been experimentally determined towards SARS-CoV-2 Mpro (Jin et al. 2020). The inhibitory activity of ebselen is tested in a 0.02–20 μM concentration range, with solutions dispensed in triplicate wells. For each novel inhibitor the optimal concentration range, covering the IC50, needs to be determined individually. In order to account for assay variability, we recommend to prepare and measure three replicate dilution series for each inhibitor.
The serial dilution of the inhibitors should be prepared right before measurement, possibly while the instrument is equilibrating to the desired temperature of 37°C.
The stock solution of inhibitor, i.e., ebselen, prepared in advance, can be diluted in a time-efficient way directly in the multiwell plate as follows:
-
16.In three 1.5 mL microcentrifuge tubes, combine 0.998 mL of assay buffer without DTT (in the case of ebselen) and 2 μL of 200× inhibitor stock solution (20 mM ebselen). This 2× ebselen working solution has a concentration of 40 μM.
-
a.Mix well by vortexing.
-
a.
-
17.Prepare wells with decreasing ebselen concentrations as follows (Table 1):
-
a.Add 0.2 mL of each 2× ebselen working solution (40 μM) to wells A1, B1, C1 of a black multiwell plate with black bottom.
-
b.Add 0.1 mL assay buffer without DTT to Wells A2-A12, B2-B12, and C2-C12 using a multi-channel pipette.
-
c.Transfer 0.1 mL of solution from wells A1, B1, C1 to wells A2, B2, C2.
-
d.Mix with new pipette tips by pipetting up and down 5 times, then transfer 0.1 mL to Wells A3, B3, and C3.
-
e.Mix with new pipette tips, then transfer 0.1 mL to adjacent wells to the right.
-
f.Continue this procedure until wells A11, B11, C11 are reached.
-
g.Remove 0.1 mL from wells A11, B11, C11 after mixing.
-
a.
-
18.Prepare control wells without Mpro (background fluorescence).
-
a.Add 0.1 mL 2× ebselen working solution (40 μM) to well D1.
-
b.Add 0.1 mL assay buffer without DTT to well D2.
-
a.
CRITICAL: use new pipette tips for each step to avoid cross contamination.
Table 1.
Plate layout for the serial dilutions of ebselen; final concentration in assay is indicated (μM)
| Row/Column | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 20 | 10 | 5 | 2.5 | 1.25 | 0.625 | 0.313 | 0.156 | 0.078 | 0.039 | 0.02 | 0 |
| B | 20 | 10 | 5 | 2.5 | 1.25 | 0.625 | 0.313 | 0.156 | 0.078 | 0.039 | 0.02 | 0 |
| C | 20 | 10 | 5 | 2.5 | 1.25 | 0.625 | 0.313 | 0.156 | 0.078 | 0.039 | 0.02 | 0 |
| D | 20 | 0 |
Preparation of the stock solution of 7-amino-4-methylcoumarin (AMC)
Timing: 10 min
AMC is the fluorogenic moiety released by the Mpro proteolytic activity and it is here used to obtain a conversion factor between the fluorescence values measured by the fluorimeter and the concentration of substrate consumed by the enzymatic reaction.
To prepare a 10 mM stock solution of 7-Amino-4-methylcoumarin (AMC, M.W. 175.18 g/mol):
-
19.
Weigh between 10 and 20 mg, i.e., approximately 17.5 mg, of AMC in a 10 mL measuring flask, note the exact weighed amount.
-
20.Add assay buffer to a final volume of 10 mL.
-
a.Vortex to dissolve (Troubleshooting 2).
-
b.Calculate the exact concentration of stock solution from the weight
-
a.
-
21.
Use freshly prepared during the day and store on ice when not in use.
Experimental setup
Timing: 15 min
Use a plate reader with fluorescence function, e.g., SpectraMax M5 plate reader (black multiwell plate, PMT setting “low”, 0.2 mL assay volume). In general, ensure that the reading settings are optimal by measuring a solution of AMC with the same concentration as the maximum final concentration achievable during the assay; measurement conditions under which this solution does not saturate the detector of the plate reader are suitable. Assays can be done in plate readers with or without temperature control. If a plate reader with temperature control is used, assays are best performed at 37°C, mimicking conditions of viral replication in human hosts.
Once the plate reader has reached temperature set point, place the plate in the reader and pre-incubate for 10 min in order to equilibrate to the desired assay temperature.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Ac-Abu-Tle-Leu-Gln-AMC | Biosynth Carbosynth | FA178674 |
| AMC | Biosynth Carbosynth | FA00826 |
| DTT | Biosynth Carbosynth | FD02370 |
| EDTA, tetrasodium salt | Biosynth Carbosynth | FE04031 |
| Ebselen | Biosynth Carbosynth | FE75162 |
| Glycerol | Merck Millipore | 137028 |
| HCl | Sigma-Aldrich | H9892 |
| NaCl | Sigma-Aldrich | S7653 |
| SARS-CoV-2 main protease | Biosynth Carbosynth | BS178678 |
| TRIS | Biosynth Carbosynth | FT15751 |
| Other | ||
| Black 96-well plate | Thermo Fisher, Nunc | 137101 |
| SpectraMax M5 plate reader | Molecular Devices | 0310-5625 |
Materials and equipment
Alternatives: In our study, we used a SpectraMax M5 plate reader (Molecular Devices) but other models are also suitable for the assay to be performed. Software able to fit the data obtained and produce slope values indicative of the protease activity, i.e. variation of fluorescence per min, such as Microsoft Excel are useful and time-savers.
Alternatives: Measurements should be performed in black multiwell plates with black bottom to avoid crosstalk among wells. The use of black plates is recommended as it helps to reduce the background signal noise; opaque white plates are an alternative but conditions might need to be tuned as the background signal will be more evident.
Step-by-step method details
Assembly of the assay mixture for Mpro activity measurement
Timing: 10 min
Depending on the extent of your experiment and the number of samples to measure, plan the number of wells of the multiwell plate you will need and the volume of reagents required, i.e., considering a single reaction volume of 200 μL but also a possible 10% disposable volume. To measure Mpro activity in three replicate measurements, we have routinely used a working solution containing the substrate at a concentration of 100 μM. This working solution is prepared as follows:
-
1.
Prepare the 1.025× substrate working solution by adding 20.5 μL of substrate stock solution to 979.5 μL of assay buffer with DTT.
-
2.
After adding the assay buffer and the substrate but before the addition of the enzyme, pre-warm the plate for 5 min at the desired assay temperature in the plate reader, e.g., 22°C or 37°C.
-
3.
Before measurement, let all solutions equilibrate to room temperature (20°C–22°C).
-
4.
Dispense 195 μL of the 1.025× working solution in each of 5 wells of a black multiwell plate.
After adding the assay buffer and the substrate but before the addition of the enzyme, pre-warm the plate for 5 min at the desired assay temperature in the plate reader, e.g., 22°C or 37°C.
Measurement of the enzymatic activity of the Mpro protease
Timing: 10–60 min
Enzymatic activity is defined as enzyme units and one enzyme unit corresponds to the amount of enzyme that will release one micromole of 7-amino-4-methylcoumarin (AMC) in one minute in assays with 0.1 mM Ac-Abu-Tle-Leu-Gln-AMC at 37°C and pH 7.3. In this case, the enzymatic activity can be expressed as increase of fluorescence (arbitrary units, RFU) over time. Most microplate readers allow the analysis of the curves produced by the measurements and a slope value expressed in variation of fluorescent signal per minute can be obtained. These values can be used to compare results obtained within the same measurement or by the same instrument, i.e., in RFU/min.
For routine activity assays and inhibition studies, the comparison of fluorescence values as RFU is sufficient. In this respect, a positive control reaction is set up and can be used as a reference of 100% relative to the other reactions.
In the procedure, the enzyme mixture is added last to initiate the reaction, the measurement should start immediately (Figure 2) with settings enabling fluorescence measurement from the top with an excitation wavelength of 380 nm and an emission wavelength of 455 nm every 30 s for at least 10 min.
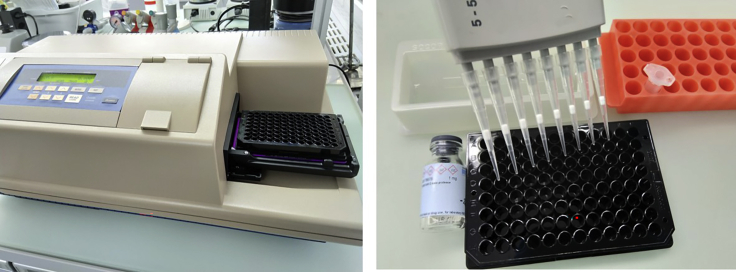
Figure 2.
Required Equipment
Image of the SpectraMax M5 plate reader (left) and photos taken during the activity measurement (right), i.e., multi-channel pipette, black multiwell plate, vial with protease stock solution, polypropylene tube for preparation of working solution and assay buffer (in the reservoir). The use of pipette tips with filter is facultative.
Upon preparation of the working solution, the fluorogenic reaction is started by the addition of the protease.
-
5.To each well containing the substrate working solution as previously described:
-
a.Add 5 μL of sample containing active 25 μg/mL MPro for a final concentration of 0.025 μg/mL.
-
b.Mix by pipetting up and down several times avoiding bubble formation.
-
a.
-
6.Proceed to measure the fluorescence immediately.
-
a.Place the microplate in the plate reader and measure the fluorescence from the top with an excitation wavelength of 380 nm and an emission wavelength of 455 nm every 30 s over a 10–60 min period until sufficient data points for a reliable data fitting are gathered.
-
b.Shaking during measurement is an option some equipment offer and is beneficial to ensure a more homogenous distribution of both substrate and enzyme.
-
a.
-
7.
Export the data obtained as fluorescence intensity (RFU or AU) vs. time (Troubleshooting 1).
Calculation of the enzymatic activity by data analysis
Timing: 20 min
After collecting the data on the development of fluorescence (Figure 3) upon addition of Mpro to the substrate working solution, analysis of the curves allows for the calculation of the enzymatic activity in relative or absolute values. The spectrophotometer proprietary software might allow a first-degree polynomial fitting (straight line) of the curves obtained and the direct export of the slope values. Otherwise, this can be performed in Microsoft Excel as follows:
-
8.Plot fluorescence intensity values versus time using a scatter plot.
-
a.Set the time scale to minutes.
-
a.
-
9.
Select at least 5 adjacent points covering more than 1 min in the initial part of the curve for the fitting and plot them separately.
- 10.
-
11.
Record the slope value as the activity value in RFU/min (Troubleshooting 3).
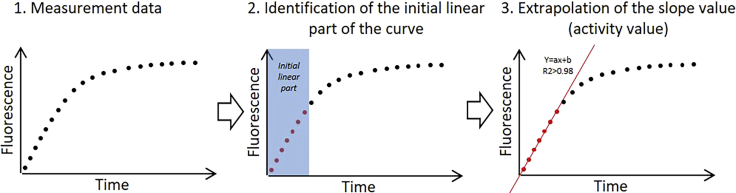
Figure 3.
Example of scatter plot, identification of the linear part of the curve to obtain the slope value corresponding ot the enzymatic activity value as variation of fluorescence over time
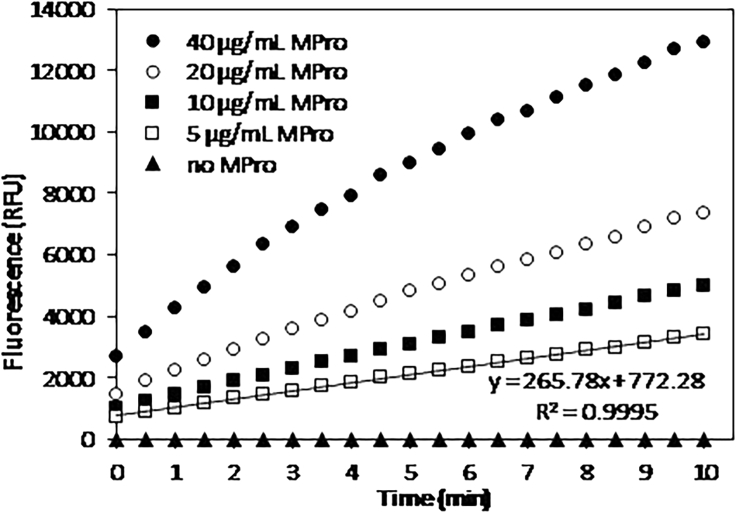
Figure 4.
Development of the fluorescent signal over time upon incubation of 0.1 mM fluorogeneic substrate Ac-Abu-Tle-Leu-Gln-AMC with Mpro at different concentrations.
The protease-free control sample is reported as filled triangles.
Measurement of Mpro inhibition
Timing: 10–60 min
In order to save reagent while maintaining a substrate concentration below the reported KM, the inhibitor screening assays are performed at 25 μM fluorogenic substrate concentration and 5 μg/mL Mpro concentration by diluting the stock solution 200×, i.e., 100 μL of respective stock solutions to 1.9 mL of buffer. The assay is performed in a black multiwell plate and in a preheated plate reader at 37°C to simulate human body conditions. When scouting for novel inhibitors, the step here presented with ebselen should be adapted to include a wider inhibitor concentration range between 1 nM and 1 mM and through log10 dilution steps.
-
12.
Add 80 μL of working solution MPro to the previously described setup of diluted inhibitor in wells A1-A12, B1-B12 and C1-C12 using a multichannel pipette.
-
13.
Add 80 μL assay buffer to negative control wells D1 and D2 for protease-free control reactions.
-
14.
Pre-warm the plate for 5 min at 37°C by placing it in the plate reader.
-
15.
Add 20 μL of the 10× fluorogenic substrate to all wells using a multichannel pipette and mix by pipetting up-and down 3 times. Use new pipette tips for every well.
-
16.Measure fluorescence immediately from the top with excitation wavelength at 380 nm and emission wavelength of 455 nm for at least 10 min in a kinetic mode.
-
a.Highest concentration of inhibitor ebselen in wells A1, B1, C1 is 20 μM, concentration decreases serially by factor two from wells A/B/C2 to A/B/C11.
-
b.Wells A12, B12 and C12 correspond to uninhibited MPro activity.
-
a.
Data analysis of Mpro inhibition study
Timing: 45 min
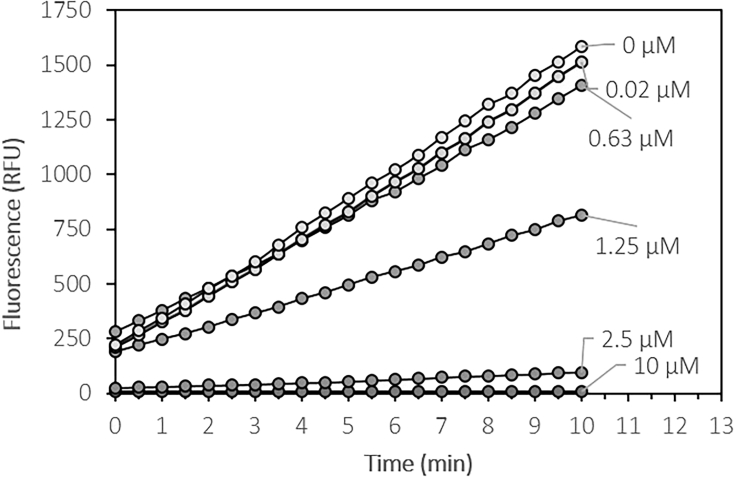
The spectrophotometer proprietary software might allow a first degree polynomial fitting (straight line) of the curves obtained and the direct export of the slope values (Figure 5). These values are activity values, e.g., 265.78 RFU/min in Figure 5. Otherwise, the data can be analyzed in Microsoft Excel as previously presented (section above) to obtain the variation of fluorescence per minute (enzymatic activity value).
Figure 5.
Fluorescence development curves over time in the presence of different ebselen concentrations.
After collecting the data on the development of fluorescence upon addition of Mpro in the presence of both the substrate and the inhibitor, the calculation of the inhibitory activity is carried out in software such as Microsoft Excel.
-
17.By plotting the fluorescence values (RFU) over time and fitting the curve to a straight line, extract the rate of RFU increase over time (RFU/min) for each well using the trend line fitting in Microsoft Excel (Figure 3).
-
a.Show the trend line equation and R2 fitting value.
-
b.In all uninhibited and partially inhibited wells, RFU increase should be linear over the whole measurement period. The correlation coefficient for these wells should be above 0.98.
-
a.
-
18.Control wells (D1, D2) without MPro should show low RFU values which do not increase over time.
-
a.Wells at inhibitor concentrations higher than a 10-fold IC50 concentration or higher should show no or only marginal RFU increase.
-
b.Calculate the average activity values and corresponding standard deviations of all wells with similar inhibitor concentration using the respective functions in Microsoft Excel, e.g., average values of results from wells A1-B1-C1.
-
a.
-
19.Divide the average activity, i.e., slope of the curve in RFU/min, of wells with inhibitor by the calculated average slope of samples without inhibitor.
-
a.Calculate the relative activity as percentage (%) by multiplying by 100. The relative inhibition value can be obtained by subtracting this activity from 100, i.e., a calculated 10% relative activity in the presence of the inhibitor corresponds to a 90% relative inhibition.
-
b.Calculate the relative standard deviation by dividing the standard deviation of the slopes of replicate wells by the corresponding standard deviation of the average of the corresponding uninhibited wells and multiplying by 100.
-
a.
-
20.Draw inhibition curve using a scatter graph in Microsoft Excel, i.e., the x-axis presents the inhibitor concentration in a μM concentration and the y-axis presents the relative inhibition as percentage (%).
-
a.Rescale to a logarithmic scale on the x-axis.
-
a.
-
21.
Calculate a rough estimation of the IC50 value by interpolation between data points above and below the 50%; for more precise values fit with a sigmoidal curve (Troubleshooting 5).
Note: For the determination of IC50 with the highest accuracy, it is recommended to use multiple dilution steps with smaller changes (IC50 ± 10–30%) in a second experiment, i.e. to achieve a higher resolution.
Absolute quantification of the amount of substrate transformed
Timing: 60 min
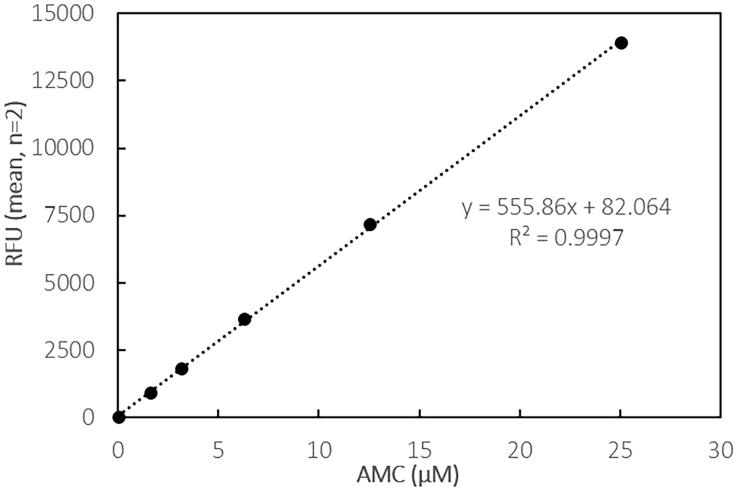
When the enzymatic activity needs to be expressed in enzyme units, i.e., U/mL or U/mg, a conversion needs to be carried out. The conversion factor is specific for each fluorimeter and can be obtained by preparing a calibration curve of AMC, the fluorophore produced by the enzymatic reaction (Table 2, Figure 6).
-
22.The stock solution of ±10 mM AMC is first diluted in assay buffer to reach a 100 μM concentration.
-
a.Calculate exact volume (±100 μL) of AMC stock solution to reach a final concentration of 100 μM in 10 mL total volume
-
b.Add calculated volume of AMC stock solution to a 10 mL measuring flask and fill to exactly 10 mL with assay buffer.
-
c.Store in the dark and use fresh.
-
a.
-
23.
In a black microplate, prepare a series of solutions with varying 7-Amino-4-methylcoumarin (AMC, M.W. 5.18 g/mol) concentration ranging from 1.5 μM) to 25 μM, see Table 2 as example. Include a negative control well with assay buffer only. Each measurement should be performed at least in triplicates, e.g., three replicate wells per concentration.
-
24.Measure the fluorescence intensity of the solutions containing different amounts of AMC with the same settings used for the activity assay.
-
a.Export the values in fluorescence units, e.g., RFU or AU.
-
a.
-
25.Plot them against the corresponding concentration of AMC.
-
a.Fit the data points in the linear part of the curve and obtain the equation of the fitting curve (Figure 6).
-
a.
-
26.
This equation, i.e., the slope coefficient 555.86 RFU/μM in Figure 7, can be used to convert the instrument-specific fluorescence values to the concentration of AMC in the assay, resulting from conversion of substrate.
CRITICAL: Each measurement should be performed at least in duplicate, e.g. two identical wells per condition and an average value calculated and used for the curve fitting. Aim for an R2 value above 0.98.
Table 2.
Example of dilutions used for the preparation of the calibration curve
| AMC final concentration (μM) | Dilution factor | Volume of 100 μM AMC solution per well (μl) | Volume of assay solution per well (μl) | Total volume (μl) |
|---|---|---|---|---|
| 25 | 4 | 50.0 | 150.0 | 200 |
| 12.5 | 8 | 25.0 | 175.0 | 200 |
| 6.25 | 16 | 12.5 | 187.5 | 200 |
| 3.125 | 32 | 6.2 | 193.7 | 200 |
| 1.5625 | 64 | 3.1 | 196.9 | 200 |
| 0 | - | 0 | 200.0 | 200 |
Figure 6.
Inhibition curve of ebselen and Mpro protease obtained by plotting the enzymatic activity in fluorescence variation over time vs. inhibitor concentration. Measurements were carried out in triplicate and the average is reposted with the corresponding standard deviation (error bars).
Figure 7.
Example of standard curve of the fluorescent moiety AMC alone in assay buffer and data measured with the SpectraMax M5 plate reader (black multiwell plate, PMT setting “low”, 0.2 mL assay volume)
This result is device-dependent.
Expected outcomes
Successfully carried out, this protocol allows the detection and measurement of the proteolytic activity of the previously referred to as main protease Mpro of SARS-CoV-2 as a fluorescent signal (Figure 3). By supplementing potential inhibitors to the activity assay, their inhibitory activity on Mpro protease can be assessed. The number of natural molecules acting as inhibitors is growing (Bharadwaj et al. 2021; Ghahremanpouret al. 2020, Qiao et al. 2021) and the assay can serve as a tool for the high throughput screening of new inhibitors to the clinically relevant protease Mpro (Figure 4) and the determination of their characteristic IC50 values (Table 3, Figure 6). Moreover, it presents how it is possible to convert device-specific fluorescence values into values reflecting the substrate concentration (Figure 3, Table 4).
Table 3.
Example of raw data obtained from the testing of the inhibitory action of ebselen
| Well (ebselen, μM) |
A1 (20) |
A2 (10) |
A3 (5) |
A4 (2.5) |
A5 (1.25) |
A6 (0.63) |
A7 (0.31) |
A8 (0.16) |
A9 (0.08) |
A10 (0.04) |
A11 (0.02) |
A12 (0, control) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (min) | Fluorescence (RFU) | |||||||||||
| 0.0 | 11 | 10 | 13 | 25 | 189 | 282 | 299 | 308 | 197 | 211 | 211 | 221 |
| 0.5 | 11 | 9 | 13 | 28 | 219 | 331 | 350 | 371 | 246 | 267 | 266 | 285 |
| 1.0 | 11 | 10 | 13 | 30 | 248 | 379 | 398 | 419 | 301 | 319 | 325 | 344 |
| 1.5 | 11 | 10 | 13 | 34 | 273 | 432 | 453 | 481 | 355 | 372 | 379 | 408 |
| 2.0 | 11 | 10 | 12 | 36 | 303 | 482 | 505 | 531 | 414 | 432 | 444 | 477 |
| 2.5 | 10 | 10 | 13 | 38 | 337 | 536 | 561 | 582 | 474 | 491 | 507 | 537 |
| 3.0 | 11 | 10 | 12 | 41 | 365 | 584 | 614 | 641 | 528 | 551 | 567 | 600 |
| 3.5 | 11 | 10 | 12 | 45 | 395 | 639 | 670 | 705 | 587 | 609 | 635 | 679 |
| 4.0 | 11 | 10 | 12 | 48 | 431 | 700 | 734 | 763 | 647 | 678 | 704 | 756 |
| 4.5 | 11 | 10 | 12 | 50 | 461 | 758 | 790 | 819 | 724 | 751 | 768 | 822 |
| 5.0 | 11 | 10 | 11 | 54 | 496 | 814 | 846 | 871 | 770 | 808 | 827 | 891 |
| 5.5 | 11 | 9 | 11 | 59 | 530 | 881 | 906 | 937 | 833 | 865 | 900 | 961 |
| 6.0 | 12 | 10 | 11 | 63 | 558 | 922 | 961 | 989 | 886 | 930 | 964 | 1021 |
| 6.5 | 11 | 9 | 11 | 66 | 585 | 982 | 1024 | 1042 | 945 | 1001 | 1025 | 1090 |
| 7.0 | 12 | 10 | 11 | 72 | 622 | 1040 | 1086 | 1113 | 1016 | 1089 | 1100 | 1171 |
| 7.5 | 12 | 10 | 10 | 77 | 647 | 1111 | 1153 | 1183 | 1089 | 1156 | 1162 | 1242 |
| 8.0 | 11 | 10 | 11 | 80 | 683 | 1159 | 1209 | 1247 | 1140 | 1224 | 1239 | 1319 |
| 8.5 | 11 | 10 | 11 | 85 | 720 | 1216 | 1262 | 1293 | 1198 | 1303 | 1297 | 1370 |
| 9.0 | 11 | 9 | 10 | 87 | 748 | 1280 | 1329 | 1350 | 1257 | 1356 | 1371 | 1453 |
| 9.5 | 11 | 9 | 10 | 93 | 786 | 1346 | 1398 | 1428 | 1323 | 1432 | 1446 | 1515 |
| 10.0 | 11 | 10 | 11 | 96 | 814 | 1408 | 1479 | 1484 | 1387 | 1503 | 1512 | 1585 |
| Calculated slope (RFU/min) | 0.02 | −0.02 | −0.28 | 7.23 | 63 | 112 | 116 | 117 | 120 | 130 | 130 | 137 |
| R2 | 0.201 | −0.24 | −0.90 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 |
Table 4.
Example of fluorescence data obtained for the preparation of the standard curve correlating AMC concentrations and fluorescence values
| AMC concentration (μM) | Replicate | Fluorescence (RFU) | Mean fluorescence (RFU) |
|---|---|---|---|
| 25 | A | 13917 | 13903 |
| B | 13889 | ||
| 12.5 | A | 7070 | 7172 |
| B | 7273 | ||
| 6.25 | A | 3620 | 3648 |
| B | 3675 | ||
| 3.125 | A | 1798 | 1825 |
| B | 1852 | ||
| 1.5625 | A | 9123 | 911 |
| B | 909 | ||
| 0 | A | 11 | 8.8 |
| B | 5 |
Limitations
This protocol relies on the high specificity of the fluorogenic substrate used for Mpro. It has however been tested only with the purified form of the Mpro protease and thus results obtained from complex Mpro-containing samples might differ. In studies analyzing activity of Mpro in infected cells, an antibody-based assay specific for Mpro might be used to further support the evidence obtained with the fluorescence-based assay presented here.
Troubleshooting
Problem 1
No fluorescent signal is detected upon addition of the enzyme to the substrate solution during the measurement of the enzymatic activity (step 6).
Potential solution
Ensure to be using a fresh Mpro solution, dispense a higher concentration of Mpro in the assay mixture, increase the detection sensitivity of the reader, or monitor the reaction for longer time. Additionally, you might want to test different concentrations of protease and of substrates at first to assess the sensitivity of the detection of your instrument, e.g., being able to detect the signal of a solution containing an AMC concentration as expected to be produced by the reaction without saturating the detector. For example, to obtain concentration of MPro main protease lower than 1 mg/mL, pre-dilute MPro solution in assay buffer, e.g., 0.1 mL of sock solution added of 0.9 mL of assay buffer for a 10-fold dilution.
Problem 2
The substrate, inhibitor, or enzyme are not soluble during the preparation of the stock solutions (steps 1–15 in “before you begin”).
Potential solution
Control the pH of the buffer for protease dilution and check if a suitable organic solvent such as DMSO has been used for the substrate
Problem 3
No linearity is found in the development of fluorescence during the measurement of the enzymatic activity (step 10).
Potential solution
Repeat the measurement by adjusting the reading settings of the instrument or by using higher or lower concentration of Mpro protease, e.g., 10-fold higher or lower, or measure for a longer time, e.g., 1 h.
Problem 4
The inhibitor is not soluble in DMSO and precipitates giving a cloudy solution during the preparation of the stock solutions (step 14 in “before you begin”).
Potential solution
Depending on the chemical properties of investigated inhibitors use other solvents, e.g., water, ethanol, methanol, or dimethyl formamide, and mild heating may be better suited.
Problem 5
No S-shaped curve is visible after plotting the activity data obtained in the presence of an inhibitor during the inhibition study (step 20).
Potential solution
Ensure that the dilution series encompasses the IC50 value, if known, for the inhibitor used as reference, e.g., ebselen to be used as positive control. As previously mentioned referring to unknown inhibitors, the IC50 might lay outside the presented range. In this case, an initial step to test inhibition and encompassing a concentration between 1 nM and 1 mM of the inhibitor through log10 dilution steps should be performed.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Greta Faccio, greta.faccio@biosynth-carbosynth.com.
Materials availability
This study did not generate new unique reagents, strains or plasmids. All described materials are commercially available without use restrictions.
Acknowledgments
The Biosynth Carbosynth group has provided financial support in the form of authors’ salaries and research materials. The funder provided support in the form of salaries for the authors but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the “author contributions” section.
Author contributions
I.J. curated the conceptualization, data analysis, investigation, writing, review, and editing; Y.C. contributed to assay development; F.G. and S.T. curated the writing, original draft, writing, review, and editing; S.U. curated the conceptualization, resources, writing, review, and editing.
Declaration of interests
I.J., F.G., Y.C., and S.U. are employed by Biosynth AG, a company part of the Biosynth Carbosynth group, a corporation that markets the enzyme and substrate used in this protocol. This does not alter our adherence to the journal’s policies on sharing data and materials. No further conflicts of interest are declared.
Contributor Information
Julien Ihssen, Email: julian.ihssen@biosynth-carbosynth.com.
Greta Faccio, Email: greta.faccio@biosynth-carbosynth.com.
Data and code availability
This study did not generate code and datasets.
References
- Bharadwaj S., Dubey A., Yadava U., Mishra S.K., Kang S.G., Dwivedi V.D. Exploration of natural compounds with anti-SARS-CoV-2 activity via inhibition of SARS-CoV-2 Mpro. Brief. Bioinform. 2021;22:1361–1377. doi: 10.1093/bib/bbaa382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal J., Haddock-Angelli T., Baldwin G., Gershater M., Dwijayanti A., Storch M., de Mora K., Lizarazo M., Rettberg R., the iGEM Interlab Study Contributors Quantification of bacterial fluorescence using independent calibrants. PLoS One. 2018;13:e0199432. doi: 10.1371/journal.pone.0199432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahremanpour M.M., Tirado-Rives J., Deshmukh M., Ippolito J.A., Zhang C.H., de Vaca I.C., Liosi M.E., Anderson K.S., Jorgensen W.L. Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. ACS Med. Chem. Lett. 2020;11:2526–2533. doi: 10.1021/acsmedchemlett.0c00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, Ke Y.Y., Huang S.Y., Huang P.N., Kung Y.A., Chang T.Y., Yen K.J., Peng T.T., Chang S.E., Huang C.T. Discovery of M protease inhibitors encoded by SARS-CoV-2. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00872-20. e00872–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Kozak J.J., Gray H.B., Garza-López R.A. Structural stability of the SARS-CoV-2 main protease: Can metal ions affect function? J. Inorg. Biochem. 2020;211:111179. doi: 10.1016/j.jinorgbio.2020.111179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Torres J., Truong L., Chaudhuri R., Mittal A., Johnson M.E. Reducing agents affect inhibitory activities of compounds: Results from multiple drug targets. Anal. Biochem. 2012;423:46–53. doi: 10.1016/j.ab.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Hu Y., Townsend J.A., Lagarias P.I., Marty M.T., Kolocouris A., Wang J. Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol. Transl. Sci. 2020;3:1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Li Y.S., Zeng R., Liu F.L., Luo R.H., Huang C., Wang Y.F., Zhang J., Quan B., Shen C. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371:1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Hilgenfeld R., Drag M. Substrate specificity profiling of SARS-CoV-2 Mpro protease provides basis for anti-COVID-19 drug design. bioRxiv. 2020 doi: 10.1101/2020.03.07.981928. [DOI] [Google Scholar]
- Stevens R., Stevens L., Price N.C. The stabilities of various thiol compounds used in protein purifications. Biochem. Educ. 1983;11:70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate code and datasets.