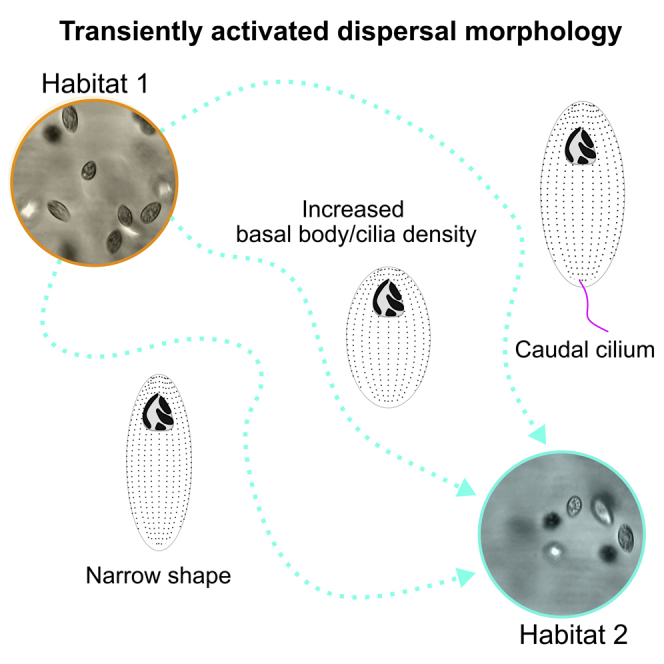
Summary
Dispersal is the movement of organisms from one habitat to another that potentially results in gene flow. It is often plastic, allowing organisms to adjust dispersal movements depending on environmental conditions. A fundamental aim in ecology is to understand the determinants underlying dispersal and its plasticity. We utilized 22 strains of the ciliate Tetrahymena thermophila to determine if different phenotypic dispersal strategies co-exist within a species and which mechanisms underlie this variability. We quantified the cell morphologies impacting cell motility and dispersal. Distinct differences in innate cellular morphology and dispersal rates were detected, but no universally utilized combinations of morphological parameters correlate with dispersal. Rather, multiple distinct and plastic morphological changes impact cilia-dependent motility during dispersal, especially in proficient dispersing strains facing challenging environmental conditions. Combining ecology and cell biology experiments, we show that dispersal can be promoted through plastic motility-associated changes to cell morphology and motile cilia.
Subject areas: Microorganism, Evolutionary biology, Cell biology
Graphical abstract
Highlights
-
•
Tetrahymena thermophila exhibits intra-specific diversity in morphology and dispersal
-
•
Cell motility behavior during dispersal changes with cilia length and cell shape
-
•
Proficient dispersing strains transiently change basal body and cilia position
-
•
Starvation triggers increased cilia density and caudal cilium formation
Microorganism; Evolutionary biology; Cell biology
Introduction
Organisms move within or between habitats in the search for food or partners, or to avoid unsuitable local conditions (Clobert et al., 2012). Routine movements occur mostly within habitats, while a movement between habitat patches influencing gene flow from one population to another is defined as dispersal (Ronce, 2007; Van Dyck and Baguette, 2005). During dispersal, an organism leaves a local habitat patch (emigration), moves through a matrix separating habitats (transit), and finally colonizes or settles in a new habitat (immigration). Dispersal movements often depend upon both the environmental conditions encountered at each of its three phases (e.g., density- or resource-dependent dispersal), and upon the phenotype of the individuals (Stevens et al., 2013). Dispersing individuals can be phenotypically different from residents for a suite of correlated traits. This correlation between the dispersal rate and phenotypic traits defines a dispersal syndrome (Clobert et al., 2009; Stevens et al., 2013). Traits involved in dispersal syndromes can be diverse, linked to morphology, behavior, physiology, and/or life history (Clobert et al., 2009). Moreover, dispersal syndromes have been described in a variety of taxa including plants, fish, rodents, and arthropods (Snell et al., 2019; Comte and Olden, 2018; Le Galliard et al., 2012; Bonte and Saastamoinen, 2012; Stevens et al., 2010, 2013).
The acquisition of a repertoire of dispersal strategies may enhance a species' ability to cope with the spatiotemporal heterogeneity of environmental conditions (Cheptou et al., 2008). However, in most organisms, neither the extent of intra-specific variability in dispersal syndromes (Snell et al., 2019; Comte and Olden, 2018), nor the underlying mechanisms behind this diversity (genetic variation or phenotypic plasticity) are deeply understood (Ronce and Clobert, 2012). Depending on these underlying mechanisms, the direction and speed of response to environmental changes diverges. Plastic strategies can facilitate the rapid fine-tuning of dispersal-related traits of a single genotype to environmental conditions but correspond with the costs of expressing such phenotypes (Murren et al., 2015). Plasticity (including that which is linked to dispersal) can also limit local adaptation to new environmental conditions in cases where it buffers the phenotypic variation of genetically-diversified populations because it hinders the diversity of existing genotypes to the effects of natural selection (Fox et al., 2019). Conversely, genetically driven dispersal strategies may result from a long adaptive history of the species to its environmental landscape without paying the cost of environment-dependent strategies. But genetic diversity might not be sufficient for a population to face rapid and drastic environmental shifts. Evaluating the extent of phenotypic variation in dispersal syndromes that rely on both genetic and plastic mechanisms is important to capture the consequences of dispersal strategies on ecological and evolutionary dynamics, especially in the face of current global change (Clobert et al., 2012; Cote et al., 2017).
Understanding the morphological and behavioral changes during dispersal requires their quantitative observation at corresponding spatial and temporal scales. Microbes are tractable for dispersal behavior and morphology studies because of their small sizes, short generation times, and morphologies and motilities that can now be readily observed (Bayless et al., 2015). The ciliate, Tetrahymena thermophila, lives in freshwater ecosystems and undergoes environmentally stimulated morphological and motility dynamics (Nanney et al., 1978; Nanney, 1966; Doerder and Brunk, 2012). T. thermophila isolates exhibit context-dependent dispersal behaviors in response to crowding, nutrient availability, presence of kin, and temperature (Chaine et al., 2010; Jacob et al., 2016, 2017, 2018; Fjerdingstad et al., 2007; Schtickzelle et al., 2009). T. thermophila cells swim by the coordinated beating of hundreds of motile cilia that cover the approximately 20 × 40 μm rugby ball shaped cell. Cell shape is dictated, in part, by the aspect ratio of length relative to width. Ciliary beating produces hydrodynamic flow along the cell shape and is essential for both directed cell swimming and tortuous navigation through the cell's environment (Pennekamp et al., 2019). Cilia are nucleated and positioned by basal bodies (BBs). Importantly, changes to cilia length and their positioning directly influence both ciliary motility and cell morphology (Galati et al., 2014; Jiang et al., 2019; Elgeti and Gompper, 2013; Omori et al., 2020; Bottier et al., 2019). In the case of cilia length, intraflagellar transport defects cause cilia to shorten, reducing cell motility, and eventually paralyzing cells (Nanney and McCoy, 1976; Jiang et al., 2019; Wloga et al., 2006). In the case of BB and cilia position, loss of BB stability factors reduces BB and cilia number in ciliary rows and also slows cellular motility (Pearson et al., 2009; Bayless et al., 2012, 2016). Furthermore, disrupting BB anchoring structures causes BBs to be mispositioned, which alters cell morphology and reduces the speed and linearity of cell swimming (Galati et al., 2014; Junker et al., 2019; Soh et al., 2020). In contrast, starvation of T. thermophila results in highly motile cells with behaviors (fast and directed swimming) suggested to be associated with dispersal. Upon starvation, cells halt the cell division cycle, narrow in cell shape, increase ciliary density, and increase their ciliary beating rate (Nelsen, 1978; Nelsen and Debault, 1978; Fjerdingstad et al., 2007). These changes increase the rate of cell motility. Thus, morphology changes to T. thermophila cells are likely associated with dispersal behaviors.
In T. thermophila, dispersing cells generally take the form of smaller, more elongated, and fast-swimming cells as a result of plastic mechanisms (Jacob et al., 2020), although the morphologies of dispersers can vary among genotypes (e.g., large, round cells (Jacob et al., 2016), see also (Fjerdingstad et al., 2007)). This variability in gross morphology associated with cell shape (aspect ratio) and ciliary spacing influences the dynamics of spatially structured cell population but has not been studied in detail (Jacob et al., 2016, 2019, 2020; Pennekamp et al., 2014; Fjerdingstad et al., 2007). We hypothesize that the described dispersal syndromes and their variability result from mechanistic changes at BBs and cilia that impact cell morphology and motility. Here, we define the link between fine-scale cellular processes and dispersal using both innate (genetic and stable non-genetic factors) and plastic (non-genetic factors sensitive to the environment) variation in behavior and cell morphology between strains of T. thermophila.
We used an experimental approach to relate the dispersal rates of 22 clonal strains of T. thermophila with both innate and plastic cell morphologies impacting cell motility. Given the diversity of documented dispersal responses of this species when exposed to either standard or stressful conditions, we hypothesize that several morphological, especially cilia-dependent, and behavioral dispersal strategies co-exist in T. thermophila (Fjerdingstad et al., 2007; Murren et al., 2015; Jacob et al., 2017, 2018). We first used two-patch systems commonly used to study dispersal under non-challenging environmental conditions to decipher the basics of dispersal phenotypic determinants (Pennekamp et al., 2014; Jacob et al., 2016, 2019, 2020). We then exposed the most and least dispersive strains to starvation, a stressful environmental condition described as a driver of strong dispersal response, and measured the same phenotypic metrics as in standard conditions (Fjerdingstad et al., 2007; Nelsen, 1978). By bridging the usually unrelated evolutionary ecology and cell biology fields, we reveal an unexpected repertoire of dispersal strategies that rely on both innate and plastic phenotypes, opening new perspectives in the extent of intra-specific dispersal syndrome variability.
Results
Tetrahymena thermophila isolates exhibit unique dispersal propensities
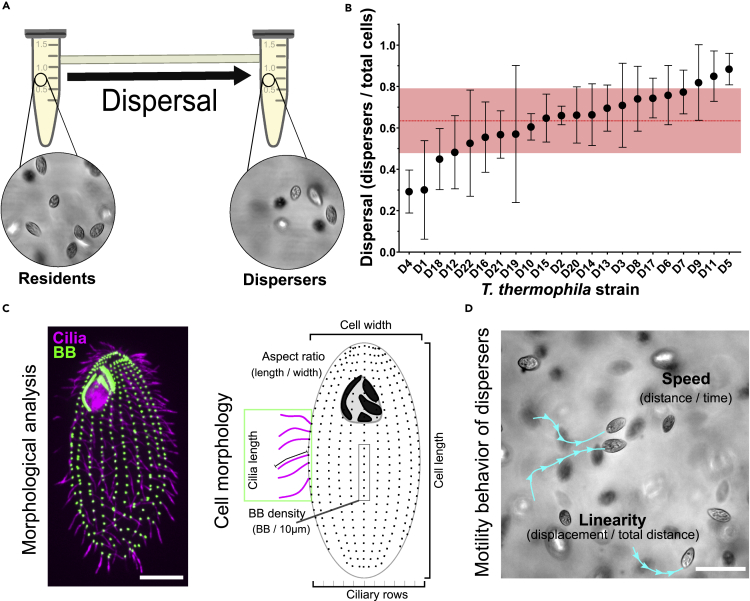
We first leveraged dispersal rate variation in 22 clonal T. thermophila strains under standard conditions (D1 to D22, Figure S1A). Using a system of two habitat patches connected by a corridor tube (Figure 1A), cells were seeded into one habitat patch and allowed to disperse to the second patch through the corridor over 4 hr (less than one asexual generation; time interval allows a representative estimate of a strain dispersal rate, V. Thuillier and N. Schtickzelle, unpublished). This routinely used setup allows for discrimination between T. thermophila “resident” cells that remain in the original patch and “dispersing” cells that move to the second patch (Altermatt et al., 2015). Cell movement might lead to a homogeneous distribution across the system within a few minutes after inoculation (Laurent et al., 2020). However, this does not occur, meaning that movements between patches clearly deviate from random cell diffusion and instead result from dispersal decisions (Laurent et al., 2020). The proportion of dispersing cells was used to define the dispersal rate. Most of the strains fell within one standard deviation of the mean dispersal fraction (Figure 1B). However, strains harboring both low and high dispersal propensities were observed. D4 is the lowest dispersing strain with 29% of the cells dispersed to the secondary chamber. Conversely, D5 is the highest dispersing strain with 88% of the cells dispersed to the secondary chamber. This amounts to a threefold range in dispersal rates across the 22 strains. Thus, T. thermophila show differing dispersal propensities, which may inform the rates at which strains move between habitat patches in nature.
Figure 1.
Differential dispersal propensities in T. thermophila strains
(A) Schematic of the two-patch dispersal system composed of 1.5 mL conical tubes connected by a narrow passageway. Cells in dispersal assays are seeded in the left patch and allowed to disperse to the right patch over 4 hr.
(B) Dispersal propensity is different between the 22 T. thermophila isolates. Dispersal rate is measured as a ratio between number of dispersers and total number of cells in 5 separate biological replicates. Graph shows the mean and standard deviation in dispersal rate for each strain. Red line and transparent red band denote the inter-strain mean and inter-strain standard deviation for all strains.
(C) Fluorescence image of T. thermophila cell with labeled BBs (α-TtCen1; green) and cilia (α-α-tubulin; magenta) (scale bar, 10 μm). Schematic of cell morphologies that were measured in the 22 T. thermophila isolates.
(D) Schematic for T. thermophila cell behavior measurements (scale bar, 10 μm).
Diverse innate morphologies and motilities of Tetrahymena thermophila
To determine if innate cell morphologies associated with cell motility could underlie the variability in dispersal propensity observed among strains, we quantified the distribution of cell and ciliary morphologies in the 22 strains under uniform growth conditions (Figures 1C, S1, and S2) and compared them with the laboratory strain B2086. Mean ± standard deviation measurements from 19.7 ± 1.7 cells were generated for each strain and compared with the cumulative inter-strain mean for each parameter. Each strain was then plotted relative to the inter-strain average to describe their cell morphology. The length of all 22 strains was 42.0 ± 5.0 μm and the width was 20.0 ± 2.9 μm. The aspect ratio (length/width) was 2.1 ± 0.4. The BB, and therefore cilia density, was 7.1 ± 0.4 BBs/10 μm. The number of ciliary rows was 20.0 ± 1.9 rows. The cilia length was 5.5 ± 0.4 μm. Finally, the swim speed was 244 ± 96 μm/s. The laboratory strain B2086 closely resembled the inter-strain means but exhibited 97% less variability compared to the 22 strains (Figures S2B and S2C). Overall, the 22 isolates exhibited a broad spectrum of morphological variation from the inter-strain mean.
Innate cell shape and cilia length inform swimming linearity in dispersers
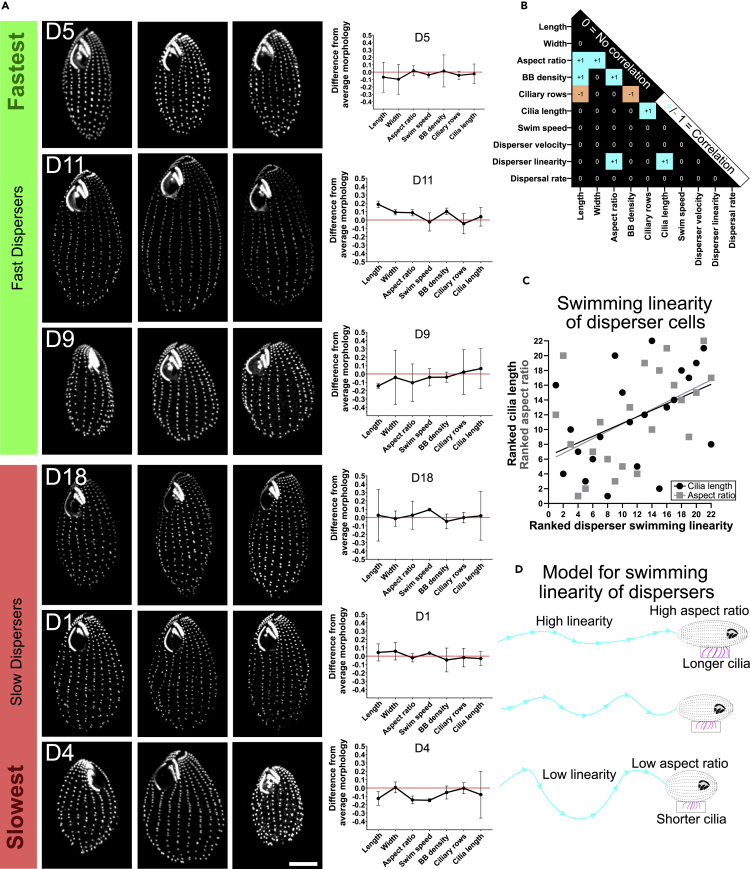
Motility of organisms from one habitat to another is an essential component of dispersal, and the BB and cilia parameters that promote cilia-dependent cell propulsion is important for cell motility (Galati et al., 2014; Junker et al., 2019; Omori et al., 2020; Rode et al., 2021; Ito et al., 2019). First, we tried to identify innate patterns of trait associations across the 22 strains that could be linked to dispersal propensity based on the set of traits described in the previous section. Second, we added dispersers' swimming behaviors (swim speed and linearity of cell having moved to the second patch, Figure 1D) to the analysis in the aim to find correlations between innate morphological traits and dispersers' behavior. The morphological analysis of the three most and three least proficient dispersing strains are shown as examples (Figure 2A). Each measured parameter was ranked among the 22 strains and simple linear regression was used to compare the parameters (Figure 2B).
Figure 2.
Increased linearity of swimming in high disperser cells correlates with narrow cell shape and longer cilia
(A) Fluorescence images of BBs (α-TtCen1; greyscale) and cell morphology from the three highest and three lowest dispersing strains among the 22 T. thermophila strains (scale bar, 10 μm) (n = 19.7 ± 1.7 cells per strain). Graphs indicate the individual mean and standard deviation for each morphology parameter relative to the mean morphology for all strains (red line).
(B) Plot of correlations between the ranked morphologies and the dispersal rate of the 22 T. thermophila strains (n = 19.7 ± 1.7 cells per strain).Correlation was calculated using linear regression between morphologies. Correlation is defined as F-test non-zero slopes of p=<0.05 denoted by a boxed blue “+1” (positive correlation) or orange “-1” (negative correlation), and no correlation is defined as F-test non-zero slopes of p=>0.05 denoted by a black boxed by “0” (see STAR Methods).
(C) Plot of the ranked innate aspect ratio (cell length/width) and innate cilia length relative to ranked swimming linearity (displacement/total distance) in disperser cells. Linear regression shows slopes are non-zero for aspect ratio (p = 0.04) and cilia length (p = 0.03). Higher ranked numbers (0–22) denote increased ranked linearity, increased ranked aspect ratio, and longer ranked cilia.
(D) Model of T. thermophila cell morphology relative to swimming behavior during dispersal.
Innate cell aspect ratio significantly correlated with cell length and width (R2 = 0.47, p = <0.01 and R2 = 0.28, p = 0.01, respectively), as expected. Tetrahymena lab strains exhibit a counting mechanism that maintains a constant number of BBs per cell (Nanney et al., 1978; Galati et al., 2015). Changes to BB density and the number of ciliary rows were observed to impact cell length and width. Consistently, we showed in the 22 strains that BB density predicted cell length and the number of ciliary rows such that increased BBs density correlated with increased cell length and reduced number of total rows (Figure 2B). However, the number of ciliary rows did not correlate with cell width as previously observed with T. thermophila lab strains (Nanney et al., 1978; Galati et al., 2015). This is because several strains exhibited unique morphologies when comparing ciliary row number and cell width. D9 for instance had many ciliary rows (20.2) relative to its narrow width (18.9 μm) and D11 a low frequency of ciliary rows (18.6) but was very wide (22.0 μm). This suggests that parameters beyond BB and ciliary placement control cell size and shape. In addition, cilia length increases in cells with more ciliary rows (Figure 2B).
The linearity of swimming in cells that have moved to the second patch (dispersers) separately correlated with innate cell aspect ratio and innate cilia length (Figure 2C; p < 0.04). Dispersers that swim more linearly have longer cilia and/or have narrow cell shapes (high aspect ratio, Figure 2D). In contrast, dispersers that swim less linearly have shorter cilia and/or have ‘fatter’ cell shapes (low aspect ratio). Cilia length and cell shape do not independently correlate with each other (Figure 2B). Thus, strains with differing innate morphologies employ differing behavioral strategies during dispersal.
Transient increase in BB density and cell shortening correspond with dispersal
In addition to innate factors as shown in the previous section, dispersal can be modulated by plastic mechanisms. We tested if dispersal propensity was impacted by intra-generational plasticity in cell morphology during the dispersal process by comparing resident and disperser morphologies. Such cell morphology differences after dispersal assays in isogenic strains can indeed be explained by cells plastically expressing new morphologies when confronted with dispersal (increase in global phenotypic variance). However, it can also be explained by morphological subpopulations in the starting population that sort into residents and dispersers during dispersal (equal global phenotypic variances), or by selective mortality of morphological subpopulations during dispersal (decrease in global phenotypic variance). We thus additionally compared trait distributions in starting populations to residents and dispersers to exclude these two last scenarios.
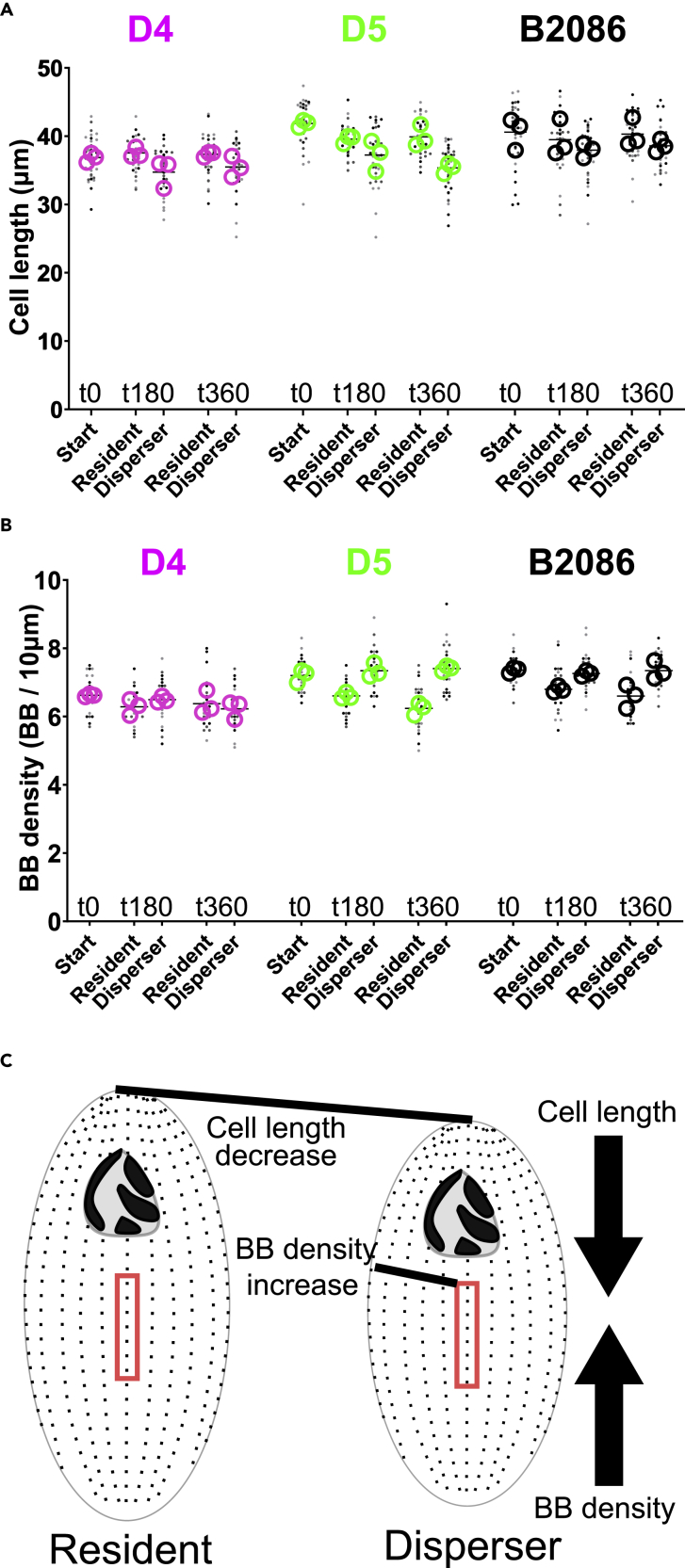
We detailed changes in cell morphologies between dispersers and residents 0, 180, and 360 min after dispersal induction in three strains. We leveraged the spectrum of low (D4), high (D5) and very high (B2086) dispersing strains to ask how plastic changes to morphology is linked to dispersal propensity (Figures 3 and S3). Clear plastic changes between residents and dispersers were not observed for most morphological parameters (Figures S3J and S3K). However, consistent with the relationship between cell shape and BB spacing found above, we identified a relationship between cell length and BB density in disperser cells. While there was a general decrease in cell length at 180 and 360 min amongst all cells (Figure 3A), dispersers showed a more pronounced decrease compared to residents. Dispersers from D4, a low dispersing strain, showed a 7% and 5% decrease in cell length, respectively after 180 and 360 min compared to residents. D4 BB density remained unchanged at each time point (Figure 3B). Dispersers from D5, a highly dispersing strain, had a more pronounced decrease in cell length at 180 (13%) and 360 (14%) min compared to resident cells. B2086 disperser cell length was unchanged at 180 min but shortened at 360 min (4%). Consistent with the shorter cells, BB density in disperser cells increased at 180 (10% and 6% for D5 and B2086, respectively) and 360 (16% and 10%) min compared with residents. The distribution of morphologies in cell length and BB density of residents and dispersers extend outside the range of morphology displayed in the starting population (Figures 3A and 3B). The overlap of the start morphologies to the respective means in the dispersal systems are lowest for cell length in the D5 (6% overlap) and B2086 (8% overlap) t360 dispersers, as compared with the D4 (19% overlap) t360 dispersers (Figures S3G and S3H). The overlap of the start conditions to the respective means in simulations are lowest for BB density in the D5 (4% overlap) and B2086 (10% overlap) t360 residents, as compared with the D4 (21% overlap) t360 residents. Thus, the innate morphologies do not appreciably contribute to the mean morphology observed in resident and dispersal populations. Moreover, the standard deviation is increased in residents and dispersers in higher dispersing cells compared to their respective start conditions (D5 and B2086 cells (Figures S3E and S3F). This suggests morphological plasticity in residents and dispersers, rather than sorting of innate starting population morphologies or selective mortality, best explains the decreased in cell length and increased BB density in cells with higher dispersal. This highlights the importance of transient morphological changes to support motility during dispersal.
Figure 3.
BB and cilia density and cell length increase in higher dispersing cells
(A) Cell length in D4 disperser cells relative to resident cells is reduced at 180 (p < 0.0001) and 360 (p = 0.02) min, cell length decreases in D5 disperser cells at 180 and 360 (p < 0.0001) min. Cell length remains the same in B2086 cells at 180 (p = 0.09) min and decreases at 360 (p = 0.03) min. Cell length measurements were taken from 3 biological replicates with n = 35.6 ± 2.1 cells per condition and time point (total of 534 cells measured). Data are represented as strain mean (black bar), the experimental means (hollow circles), and individual cell length (dots).
(B) BB density in D4 disperser cells relative to resident cells is unchanged at 180 min (p = 0.15) and 360 min (p = 0.33). BB density is increased in D5 disperser cells at 180 and 360 min (p < 0.0001). BB density is increased in B2086 disperser cells at 180 and 360 min (p < 0.0001). The measurements for BB density were taken from 3 biological replicates n = 28.9 ± 1.1 cells per condition and time point (total of 434 cells). Data are represented as strain mean (black bar), the experimental means (hollow circles), and mean cell BB density (dots).
(C) Schematic for how a decrease in cell length can increase BB (and ciliary) density.
Starvation promotes plastic changes to cell swimming and shape that trigger dispersal
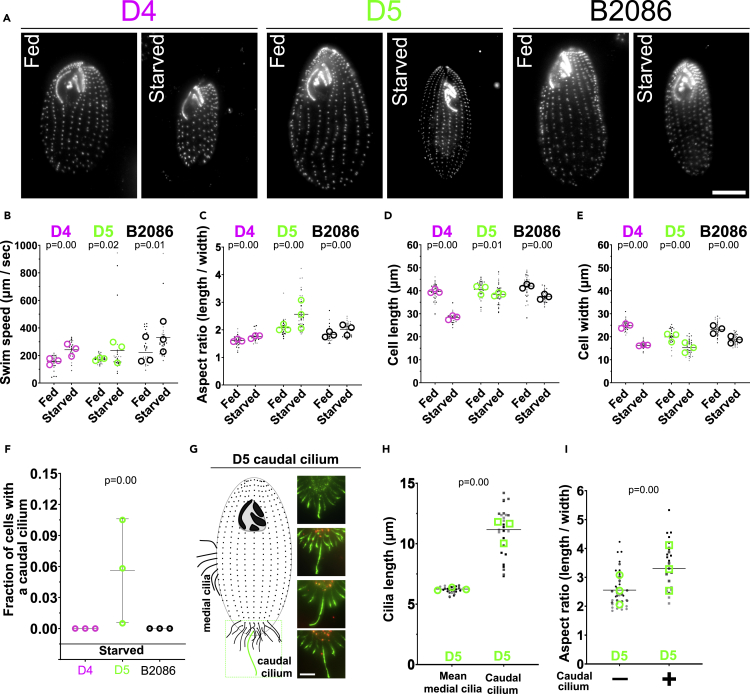
Starvation is predicted to enhance dispersal in some strains and triggers T. thermophila cells to become narrower and swim 2-3-fold faster (Nelsen, 1978; Fjerdingstad et al., 2007). We next tested whether dispersal promoting cell changes are induced by the environmental signal of starvation, and whether this response depends on the innate dispersal propensity of each strain. To test for the effect of starvation on plastic dispersal changes according to innate dispersal propensity, cells from D4 (low dispersal), D5 (high dispersal), and B2086 (very high dispersal) were starved by replacing growth media (1X SPP) with 10 mM Tris-HCl to induce “extreme disperser” phenotypes (Nelsen, 1978; Nelsen and Debault, 1978).
A peak of fast cell swimming was observed for D5 and B2086 240 min after starvation (Figure S4A). Analyses were thus conducted to compare fed cultures with those that were starved for 240 min (Figure 4A). The rate of swimming increased for each strain (Figure 4B). The maximum swim speed of D5 (944 μm/s) and B2086 (941 μm/s) increased more than D4 (332 μm/s). Moreover, the variation in swim speed between cells was greater in D5 and B2086 compared with D4. The standard deviation and coefficient of variation (%CV) for D4, D5 and B2086 was 60 μm/s (25 %CV), 199 μm/s (87 %CV) and 154 μm/s (49 %CV), respectively. In summary, starvation caused all strains to increase their swimming speed, but D5 and B2086 exhibited a greater maximum change (2.8-fold) and cell-to-cell variation in swim speed compared with D4 cells.
Figure 4.
Starvation-induced dispersal increases BB density and formation of a caudal cilium
(A) BB localization (α-TtCen1; greyscale) and cell morphology in D4, D5, and B2086 cells from cycling (fed) and starved conditions (scale bar, 10 μm).
(B) Starved D4, D5, and B2086 cells swim faster than fed cells (p < 0.0001, 0.02, and 0.01, respectively). Maximum speeds of both B2086 and D5 are 2.8-fold faster than the maximum speed of D4. The measurements for swim speeds were taken from 3 biological replicates n = 30.2 ± 2.4 cells per condition (total of 181 cells). Data are represented as strain mean (black bar), the experimental means (hollow circles), and individual cell speed (dots).
(C) Starved cells increase in cell aspect ratio after starvation for all strains (p < 0.0001 for D4, D5, and B2086). The measurements for aspect ratio were taken from 3 biological replicates n = 36.6 ± 4.7 cells per condition (total of 220 cells). Data are represented as strain mean (black bar), the experimental means (hollow circles), and individual cell aspect ratio (dots).
(D) Cell length decreases in all strains after starvation (D4, p < 0.0001; D5, p = 0.01; B2086, p < 0.0001). The measurements for cell length were taken from 3 biological replicates n = 36.6 ± 4.7 cells per condition (total of 220 cells). Data are represented as strain mean (black bar), the experimental means (hollow circles), and individual cell length (dots).
(E) Cell width decreases after starvation (p < 0.0001 for D4, D5, and B2086). The measurements for cell width were taken from 3 biological replicates n = 36.6 ± 4.7 cells per condition (total of 220 cells). Data are represented as strain mean (black bar), the experimental means (hollow circles), and individual cell width (dots).
(F) Starved D4 and B2086 (D4 n = 604 cells, B2086 n = 555 cells) cells do not generate a caudal cilium while starved D5 cell (D5 n = 600 cells) exhibit a subpopulation of cells with a caudal cilium (p < 0.0001). Data are represented as strain mean and standard deviation (black bars), and the experimental means (hollow circles).
(G) Model of D5 cell and cilia location (left). Fluorescence images of the caudal or cell posterior end in starved D5 cells. BBs are stained for BBs (α-TtCen1, red) and cilia (α -glutamylated and α-α-tubulin; green) (scale bar, 5 μm).
(H) The caudal cilium is longer than the average cortical medial cilium (p < 0.0001) (n = 26 caudal cilia from 26 cells, n = 300 cilia from 30 cells). Data are represented as a morphotype mean (black bar), the experimental means (hollow circles/squares), and mean cilia length of individual cells (dots).
(I) Starved D5 cells with a caudal cilium have an increased aspect ratio (narrow cell shape) compared with starved D5 cells without a caudal cilium (p < 0.0001) (n = 26 cells with caudal cilia, n = 42 cells without caudal cilia). Data are represented as the morphotype mean (black bar), the experimental means (hollow circles/squares), and the aspect ratio of individual cells (dots).
We next analyzed the cell size and shape differences between D4, D5, and B2086 cells (Figure 4A). Narrow cell shape is associated with increased dispersal and motility (Fjerdingstad et al., 2007; Stevens et al., 2013; Pennekamp et al., 2019; Ito et al., 2019; Omori et al., 2020). Consistently, in response to starvation, the three strains increased their aspect ratio (8%, 19%, and 11%, respectively, for D4, D5 and B2086, Figure 4C). This occurred in D4 cells by a dramatic reduction of both cell length (29%) and width (34%), in D5 cells by a small reduction in cell length (5%) and a large reduction of cell width (23%), and in B2086 cells by a small reduction in cell length (11%), and a greater reduction in cell width (20%) (Figures 4D and 4E). Thus, general cell morphology changes in response to starvation are different between each strain.
Increased BB density and caudal cilia are associated with dispersal propensity
To determine the relationship between cell swim speed and cilia, we quantified differences in BBs and cilia between the D4, D5, and B2086 cells after starvation for 240 min. No difference in the number of ciliary rows between fed and starved cells was observed. Combined with the decreases in cell width (D4 = 34%, D5 = 23%, B2086 = 20%), the lateral spacing between rows of BBs and cilia correspondingly decreased resulting in an increase in lateral BB density (Figures 4E and S4B). This is consistent with original starvation analyses (Nelsen, 1978). The BB density within ciliary rows in each strain also uniquely responded to starvation (Figure S4C): D4 cells exhibited a 10% reduction in BB density, D5 cells had no change, and B2086 cells increased BB density by 18%. This demonstrates that BB density in starvation increases as strain dispersal propensity increases.
No differences in cilia length were detected between fed and starved cells (Figure S4D). This suggests that cilia length is not responsive to starvation, exactly as cilia length does not change between residents and dispersers (see above). Interestingly, a small fraction of starved D5 cells, but not D4 or B2086 cells, possess a single caudal cilium (Figures 4F and 4G) that is 1.2–2.3-fold longer than the average medial cilium length (Figure 4H). The subpopulation of starved D5 cells with a caudal cilium are longer (16%, Figure S4E) and narrower (23%, Figures 4I, S4F, S4J, and S4K). These caudal cilia, only described once before in T. thermophila, do not beat and may serve as a rudder for steering motility (Video S1 (Nelsen, 1978)). The presence of a caudal cilium in a fraction of the starved D5 cells and their associated distinct morphology further supports the importance of plasticity in the dispersal process.
Discussion
We have used an experimental approach linking ecological and cell biology competencies to determine how innate and plastic changes to fine-scale cellular activities can impact the dispersal process. Deciphering the mechanisms underlying a repertoire of intra-specific dispersal strategies is a critical step to understand dispersal evolution (e.g., how species modulate their dispersal response when facing environmental changes, or how variability in dispersal syndromes impacts ecosystem function or metapopulation dynamics) (Jacob et al., 2019; Little et al., 2019).
Innate and plastic mechanisms underlie variation in dispersal propensity
T. thermophila have distinct intra-specific diversity in dispersal propensity and motility associated morphologies. Cell aspect ratio and cilia length predict the linearity of dispersers' motility. Because no innate cell or cilia morphology directly correlates with the rate of dispersal, we suggest that different T. thermophila isolates implement different strategies to disperse. This diversity of strategies might result from tradeoffs between dispersal behaviors (e.g., linearity in motility) and efficiency in motility involving cell shape, cilia length and BB density, and/or colonization ability (Jacob et al., 2016). Interestingly, innate swim speed does not correlate with dispersal rate, nor with dispersers' swimming characteristics. This absence of correlation is expected when dispersers' movement dynamics result from plastic variability during dispersal, as was previously described in T. thermophila (Jacob et al., 2020; Pennekamp et al., 2019). Thus, we suggest that dispersal rates as measured in two-patch systems are not a simple by-product of random movements of each strain within a patch, but rather result at least in part from phenotypic characteristics specifically expressed during the dispersal process. Accordingly, during the dispersal and starvation experiments, cells develop new morphologies that are not present in the original population including in narrow cell shape, increased BB and cilia density, and a caudal cilium. Both cell shape and increased BB and cilia density are associated with increased phenotypic variance after the dispersal assays. For instance, when dispersal associated behavior is triggered by starvation, higher dispersing strains in standard conditions display fast but variable swim speeds, narrow cell shape, increased BB density, and/or a caudal cilium. Thus, our study highlights that phenotypic plasticity plays a major role in the morphological determinants of T. thermophila dispersal.
Dispersal is an inherently costly process involving movement through a matrix that separates habitats (Bonte et al., 2012). A morphology that improves the dispersal propensity of an individual may be detrimental to other fundamental processes like nutrient acquisition. To manage these tradeoffs, one strategy is to plastically activate a costly morphology in a context-dependent manner (Cote et al., 2017; Benard and McCauley, 2008). Another strategy is to use existing morphological variation in a population, where only a fraction of individuals exhibits potentially costly morphologies that are beneficial to dispersal. Both models can produce populations of individuals with unique dispersal capacity, consistent with the observations in this study. Moreover, this provides a morphological basis for dispersal propensity independent from a generalized innate morphology. We have shown in T. thermophila that plasticity mainly explains the variability in dispersal strategies observed (cells in resident, dispersing, or starved populations have morphologies that are not present in the original starting population). However, variation could also be generated through asymmetric cell division events that can produce unique cellular morphologies. In this study, we minimize this source of variation by restricting the dispersal time to four hours (less than one generation), six hours (less than two generations), or by pausing the cell cycle through starvation. While innate population variation may play a slight role in dispersal, our studies suggest that plastic changes of cell morphology that are transiently activated before or during dispersal is the main promotor of dispersal morphologies.
Starvation is generally a strong environmental cue that activates dispersal behavior, especially in microorganisms (McDougald et al., 2012). We observe that strains that proficiently disperse in fed conditions activate highly variable swimming behavior in response to starvation. Rather than an entire population synchronizing its motility rate when exposed to starvation, a spectrum of individuals with high motility is produced in strains with high dispersal propensity. The observed variation in swim speed could rely on multiple changes including gross cell morphology, BB and cilia spacing, ciliary dynamics, or metabolic output. We suspect that combinatorial effects, with differences in BB and ciliary morphology could explain the high variation in swim speeds measured for strains D5 and B2086. These strains increased their phenotypic heterogeneity in response to starvation stress through phenotypic plasticity. Such a diversifying phenotypic plasticity could be evolutionarily viewed as a risk spreading strategy in fluctuating and discontinuous environments allowing for highly heterogeneous dispersal behaviors in cells with the same genotype.
Plasticity in cilia density and cell aspect ratio occurs during dispersal
Motility is an essential component of active dispersal. In most aquatic eukaryotic microorganisms, including T. thermophila, motility is driven by cilia-dependent fluid flow (Mitchell, 2007). For multiple cilia to efficiently produce fluid flow, they are properly spaced relative to one another (Nanney, 1966; Elgeti and Gompper, 2013; Omori et al., 2020; Wolff et al., 1992; Rode et al., 2021; Ito et al., 2019). In our dispersal assays, decreases in cell length and increases in BB and cilia density in highly dispersive strains were observed in cells that have successfully dispersed. This is consistent with T. thermophila's BB counting mechanism which ensures the number of BBs in each cell remains nearly constant (Nanney, 1966). Here, a reduction in cell length will increase BB density. An increase in BB density is expected to increase cilia-dependent fluid flow and motility (Omori et al., 2020; Elgeti and Gompper, 2013; Rode et al., 2021; Ito et al., 2019). For comparison, T. thermophila genetic mutants that cause BB instability often display 15–35% decreases in BB density and exhibit reduced cell swimming (Bayless et al., 2012, 2016; Pearson et al., 2009). In this study, highly dispersive D5 and B2086 strains have increased BB density and we suggest this to be what improves their cilia-dependent cell swimming during dispersal. We suggest that the plastic increase in BB and cilia density in ciliated microorganisms that control BB frequency increases effective cell motility during dispersal.
The cell's global geometry is also likely to impact cell motility (Omori et al., 2020; Ito et al., 2019). Elevated cell aspect ratio creates cells that are elongated relative to their width. The increased aspect ratio that is triggered by nutrient removal is consistent with strain dispersal rates and the general finding of associations between dispersal and narrow cell shape. However, passive plastic changes in cell volume could also occur because of changes to osmotic pressure during starvation. We suspect that combinatorial effects, with differences in BB and ciliary morphology could explain the high variation in swim speeds measured for the efficient dispersing strains D5 and B2086. It is plausible that narrow cell shape is an environment-dependent change that either directly impacts motility through effects on ciliary motility or reflects a metabolic state of the cell that promotes dispersal behaviors.
The caudal cilium is a determinant of rapid cell motility
The caudal cilium is a transient structure that we observed in this study upon exposure to starvation. It is present only in a sub-population of starved D5 cells and is not found in fed cells. Moreover, the caudal cilium is associated with variable fast motility of cells. This is a distinct example of a transiently emerging morphology that is not present before dispersal behavior is initiated. Caudal cilia do not appear to beat and may serve as a rudder for steering rapid cell motility (Video S1 (Nelsen, 1978)). Caudal cilia have also been detected on other Tetrahymena species and ciliates including Ichthyophthirius, Uronema, Paramecium, and Coleps/Levicoleps (Corliss, 1957; Foissner et al., 2008; Holz and Corliss, 1956; Lu et al., 2016; Nelsen and Debault, 1978; Tamm, 1978; Kozel, 1986). Ichthyophthirius, a parasitic ciliate closely related to T. thermophila, has distinct morphologies during its life cycle and possesses a caudal cilium only during its free-swimming dispersal stage prior to host invasion (Kozel, 1986). The transient development of a caudal cilium further supports the importance of morphological plasticity and variation in dispersal. We posit that plasticity and variation in morphology is critical for managing the costs and risks surrounding the dispersal process.
Conclusion
By decrypting the morphological determinants of dispersal at a previously unseen cellular level, we show that the dispersal process does not rely on innate phenotypic stereotypes. T. thermophila rather employs differential transient cell and ciliary morphologies to promote motility for dispersal, which could be important in managing energy cost trade-offs between morphologies that promote feeding and the transition to morphologies employed for efficient dispersal (Omori et al., 2020; Gilpin et al., 2017; Bonte et al., 2012; Ito et al., 2019). To determine if this link between cilia-dependent motility and dispersal is ubiquitous, future studies should now be developed in other taxa inhabiting ponds and lakes, but also river, ocean, and terrestrial biotas. Future studies should also elaborate on the molecular determinants, costs, and benefits of this panel of dispersal strategies, as well as their ecological and evolutionary consequences in real landscapes both when individuals colonize new patch (as in our study) and settle in an already occupied patch.
Limitations of the study
We studied dispersal within highly simplified microcosms. The limitations of artificial systems, especially the generalization of the corresponding observations to nature, is a long-standing matter of concern in scientific research, but their obvious advantages lead to their massive use (Bestion et al., 2019; Benton et al., 2007). In the context of dispersal, behaviors appear to be similar in nature and in artificial systems for some species (Legrand et al., 2012, 2015; Benton et al., 2007; Le Galliard et al., 2005). In Tetrahymena, prior studies from our groups and others have shown that dispersal behavior studied in the simple two-patch systems are congruent with what is known for other species in natura. For instance, cells adjust dispersal decisions relative to kinship, population density, resources, and temperature within these simplified microcosms (Chaine et al., 2010; Jacob et al., 2016, 2017, 2018, 2019; Pennekamp et al., 2014). However, techniques are not available to study the T. thermophila dispersal behaviors in natura, as for most microorganisms. Yet, our system is extremely simplified (axenic media, only two patches, absence of sex, only clonemates with a likely unreasonable cell density) and therefore far from natural conditions. The severity of some of these limitations could be easily evaluated, e.g., feeding T. thermophila with a single or a diversity of bacteria, introducing top predators and/or competing species, working on a heterogeneous populations of T. thermophila, or having multiple patches of different sizes and in physical different conditions. This would test whether dispersal and its related morphologies are dependent on specific environmental conditions. Nonetheless, in the long run, studies in natura will be necessary.
The morphological analysis of Tetrahymena cells, BBs and cilia requires cell fixation and preparation for light microscopy. This can impact natural morphologies and subtle changes to BB and cilia morphology are to be expected. Moreover, this study measured a limited number of morphological parameters that can be expanded upon to further explore the diversity in changes with dispersal. These include, but are not limited to, cortical rigidity, oral apparatus positioning, ciliary beat frequency, waveform and metachronicity, mechanical coupling between BBs, and number and volume of vacuoles. In addition, our studies highlight a subset of the population and examples like the caudal cilium suggest that we may not accurately estimate the variance of morphological diversity in these systems. The low frequency and high variation of caudal cilia appearance in starved D5 cells make it challenging to study the functional role of this transient cilium. To image live cells with a caudal cilium, cells were slowed using viscous media and imaged when cells swam just above the cover glass. While the caudal cilium appears to be immotile compared with neighboring cilia, subtle beating may be difficult to visualize. Future quantitative studies will use automated approaches analyzing cell morphologies, and cilia dynamics to describe morphological and behavioral variance.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α-TtCen1 | (Stemm-Wolf et al., 2005) | NA |
| α-glutamylation GT335 | Adipogen | Cat# AG-20B-0020-C100 |
| α-α-tubulin 12G10 | DSHB | Cat# AB_1157911 |
| Alexa Fluor 488 goat α–rabbit IgG | Invitrogen | Cat# A11034 |
| Alexa Fluor 594 goat α–rabbit IgG | Invitrogen | Cat# A32740 |
| Alexa Fluor 647 goat α–rabbit IgG | Invitrogen | Cat# A21244 |
| Alexa Fluor 488 goat α–mouse IgG | Invitrogen | Cat# A11029 |
| Alexa Fluor 594 goat α–mouse IgG | Invitrogen | Cat# A32742 |
| Alexa Fluor 647 goat α–mouse IgG | Invitrogen | Cat# A21236 |
| Chemicals, peptides, and recombinant proteins | ||
| Polyethelene oxide | ACROS Organics™www.fishersci.com | Cat # AC183220050 |
| Citifluor™ Mountant solution AF1 | Ted Pella Inc. | Cat # 17,970-25 |
| Experimental models: organisms/strains | ||
| T. thermophila: Doerder strains 1-22 | tetrahymena.vet.cornell.edu | Figure S1 |
| T. thermophila: B2086 | tetrahymena.vet.cornell.edu | Cat # SD01625 |
| Software and algorithms | ||
| Prism8 | GraphPad Software | San Diego, California USA, www.graphpad.com |
Resource availability
Lead contact
Further information and requests for resources and reagents used in this study should be directed to and will be fulfilled by the lead contact, Chad Pearson (chad.pearson@cuanschutz.edu).
Materials availability
This study did not generate any new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This study does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subjects details
Tetrahymena thermophila D1-D22 strains (wild type natural isolates) and wild type B2086 (laboratory strain) were used in experiments for this study. Each strain is verified as T. thermophila through successful conjugation/mating events with known T. thermophila strains and genome sequencing (Arslanyolu and Doerder, 2000). The sex of each Tetrahymena thermophila strain were not accounted for because T. thermophila has 7 mating types (sexes) and the exact mating types were unknown for most strains. All experiments were performed in accordance with governmental and institutional guidelines.
Method details
Tetrahymena cell culture
Tetrahymena thermophila D1-D22 strains were kindly provided by Paul Doerder (Cleveland State University, see Figure S1H) and B2086 was obtained from the Tetrahymena Stock Center (tetrahymena.vet.cornell.edu/index). Cultures for D1-D22 were maintained in 24-well culture plates by passaging approximately every 10 days, with two periods of cryopreservation. Each strain was grown independently and cultured in 0.6% SPP media (0.6% protease peptone, 0.03% yeast extract, 0.06% glucose, and 0.0009% Fe-EDTA) at 23°C. Cells collected for analysis were grown to mid-log phase (approximately 3×105 cells/mL). Cell counts were determined using a Coulter Counter Z1 (Beckman Coulter), except for the quantification of dispersal rates (see below).
Quantitative analyses of tetrahymena cell swimming and morphology
Swim speed was calculated from 5 s videos acquired at 20 frames per sec, individual cell swim paths were followed (for 4–10 frames) to determine average speed in μm/second (Figure S1B). The swim speed of cells is defined as the total distance traveled by cells divided by the duration of the trajectory, and linearity is the ratio between the net distance traveled (Euclidian distance between start and end positions), and the total distance traveled, such that higher values indicate straighter trajectories (Figure S1B). Cell length was measured as the distance from the anterior tip to the posterior end of the cell (Figure S1C). Cell width was measured as the distance across the cell's dorsal-ventral axis perpendicular to the anterior-posterior cell axis, 1 μm posterior to the cell's oral apparatus (Figure S1D). Aspect ratio was calculated as the length/width of the cell (Figure S1E). BB density was measured as the number of BBs (α-TtCen1) in a 10 μm distance of a ciliary row within the medial region of the cell (middle 50% of anterior-posterior axis) (Figure S1F). Ciliary rows per cell were counted through full cell z-stacks as the total number of individual rows of BBs around the cell's lateral circumference (Figure S1G). Cilia length was measured using a segmented line as the distance from the distal end of the BB (α-TtCen1) to the tip of the cilium (α-–α-tubulin and α-glutamylated tubulin) (Figure S1H). Each of these measure parameters were used to generate an average morphology for all strains analyzed (Figure S1I).
Dispersal
We performed dispersal assays to characterize the 22 T. thermophila strains using two-patch dispersal systems consisting of 1.5 mL tubes connected by a corridor made of a 2.5 cm long silicone tube with a 4 mm internal diameter (Chaine et al., 2010; Jacob et al., 2015, 2016). In T. thermophila, cells adjust dispersal decisions relative to kinship, population density, resources, and temperature within these simplified microcosms (Chaine et al., 2010; Pennekamp et al., 2014; Jacob et al., 2016, 2017, 2018, 2019). Such results can be observed because two-patch systems accurately mimic simplified metapopulations, i.e., suitable patches separated by a corridor (over 1000 times the size of a T. thermophila cell), with each patch being of sufficient size to host very large population sizes (up to ∼500,000 cells per patch (Jacob et al., 2019)). Importantly, dispersal movements in these experimental microcosms influence T. thermophila local adaptation by generating gene flow among patches and affecting population differentiation (Jacob et al., 2017). The movements we quantify in these microcosms therefore match the classical definition of dispersal movements: movements between populations than can lead to gene flow (Clobert et al., 2012; Ronce, 2007). The dispersal system was filled with 4 mL of 0.6% SPP media and the central corridor was clamped. 100,000 cells were added to the resident patch. After 30 min of acclimatization, the corridor clamp was removed, and cells were allowed to disperse to the disperser patch for 4 hr at 23°C. After the 4 hr, the corridor was clamped again and cells from each patch were collected, counted, and/or analyzed for swimming parameters described above. Dispersal rate was measured as the fraction of dispersers/total number of cells, as measured from 2 x 10 μL pipetted from each tube into chambers of a multichambered counting slide (Kima precision cell 301890). We immediately acquired 20 s live cell movies from each chamber under dark-field microscopy to count cells and quantify movement characteristics using BEMOVI R-package (Pennekamp et al., 2015).
To quantify swimming and morphology changes associated with dispersal in D4, D5 and B2086, large two-patch systems with more cells were required to perform immunofluorescence experiments. Dispersal assays were conducted using a two-patch system consisting of 5 mL habitat microtubes connected by an 8 cm long silicone tube corridor with a 4 mm internal diameter. This dispersal system was optimized for the increase in habitat size and corridor length (Figures S3A–S3D). This dispersal system was filled with 11 mL of 0.6% SPP media and the central corridor was clamped. 400,000 cells were added to the resident patch. As above, after acclimatization, the corridor clamp was removed, and cells were allowed to disperse to the disperser patch for 6 hr at 30°C.
Starvation
Starvation experiments were conducted by growing cells in 5 mL cultures with 2% SPP growth media at 30°C to mid log (approximately 3×105 cells/mL). Cells were centrifuged in 10 mM Tris HCl pH 7.4, then washed in 10 mM Tris HCl pH 7.4 again before resuspending cells (3×105 cells/mL) in 5 mL of 10 mM Tris HCl pH 7.4. Cells were incubated 240 min at 30°C before collection and analysis. Analysis of swim speed was conducted as described above, standard deviation and percent coefficient of variation (% CV) was calculated in Graphpad Prism on the mean speeds of individual cells.
Visualization of the caudal cilium in live D5 cells was performed by pelleting cells at 0.6 xg in a microfuge and resuspending them in 2% polyethylene oxide (PEO, molecular weight 900,000, Acros Organics) dissolved in 10 mM Tris pH 7.4 (Galati et al., 2014). The PEO solution increases viscosity and slows mobility for visualization. DIC movies were acquired at 100 frames per sec (10 ms exposure).
Sample preparation for immunofluorescence
Immunofluorescence imaging procedures were modified from prior studies to maximize the quality of BB, cilia staining and retention of cell shape (Junker et al., 2019). For BB and cilia visualization, cells were washed in 10 mM Tris HCl pH 7.4, followed by PHEM buffer (60 mM 1,4-piperazinediethanesulfonic acid, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mM EGTA, and 2 mM MgCl2, pH 6.9), and then fixed in a Triton X-100/paraformaldehyde solution (0.25% Triton X-100 and 3.2% paraformaldehyde in PHEM buffer) for 5 min. Cells were then incubated in Triton X-100 solution (0.25% Triton X-100 in PHEM buffer) for 10 min on ice. They were washed three times in 0.5% BSA in PBS (BSA-PBS) before a 24 hr incubation in primary antibody diluted in 0.5% BSA-PBS at 4°C. They were then washed three times in 0.5% BSA-PBS before incubation in secondary antibody diluted in 0.5% BSA-PBS at 23°C for 2 hr. They were again washed three times in 0.5% BSA-PBS and pelleted. 2 μL of cells from the pellet were added to a coverslip and mounted in 6.5 μL Citifluor mounting media (Ted Pella). Samples were then sealed using clear nail polish.
The primary antibodies used in this study were α-TtCen1 (1:1000 dilution (Stemm-Wolf et al., 2005)), α-glutamylation (1:1000 dilution, GT335, Adipogen AG-20B-0020-C100 (Wolff et al., 1992)), and α-α-tubulin (1:200 dilution, 12G10, DSHB AB_1157911 (Thazhath et al., 2002)). Secondary antibodies (Alexa Fluor 488 (Invitrogen A11034), Alexa Fluor 594 (Invitrogen A32740), or Alexa Fluor 647 (Invitrogen A21244) goat α–rabbit IgG, and Alexa Fluor 488 (Invitrogen A11029), Alexa Fluor 594 (Invitrogen A32742), or Alexa Fluor 647 (Invitrogen A21236) goat α–mouse IgG) were used at 1:1000 dilution.
Microscopy
Fluorescence imaging was performed using either a Nikon wide-field TiE fluorescence microscope stand equipped with a 100x NA 1.4 Plan Apo objective and an Andor Xyla 4.2 CMOS camera or a Nikon X1 confocal TiE fluorescence microscope stand equipped with a 100x NA 1.4 Plan Apo objective and an Andor iXon EMCCD camera were used depending upon the experiment. Images were acquired using NIS Elements software. All fluorescence imaging was conducted at approximately 25°C and exposure times were between 50 and 500 msec, depending on the experiment. Only T. thermophila cells in Stage 1 (G1) of cell cycle and cells that were not visibly damaged by the fixation and staining process were imaged and included in the analyses.
Data representation
Each data point (representing a single cell) is indicated as a single dot on the figure plots. Replicates for morphological data are denoted by different shades (light gray, dark gray and black), the mean of each replicate is denoted by hollow colored circles. Lines denote the mean for the entire experimental condition. All lines indicate the mean, and all error bars indicate standard deviation.
Quantification and statistical analysis
All experimental datasets represent a minimum of three biological replicates. For morphological analysis, each replicate has approximately 10 cells (at least 30 cells per experimental condition) unless otherwise indicated in the Figure Legends. Statistical p values are included in each dataset in the corresponding Figure Legend. Statistical tests were run in Prism8 (GraphPad Software). Normally distributed continuous datasets were analyzed using unpaired, two-tailed Student's t-test. Non-normally distributed continuous datasets were analyzed using unpaired, two-tailed Mann-Whitney test. Data sets with multiple comparisons were analyzed using one-way ANOVA. All p values are numerically presented to two decimal places. Lines and bars on all plots indicate the mean and standard deviation, respectively. For correlations between traits, each measured parameter was ranked among the isolates and simple linear regression was used to compare the parameters (Figure 2B). Parameters that correlated had an F-test p value for a non-zero slope of <0.05 and were assigned a value of 1. Parameters that did not correlate had p value for a non-zero slope of >0.05 and were assigned a value of 0. Standard deviation of residents and dispersers from the start populations was calculated as the standard deviation of each strain's resident or disperser measurements to their start population's mean (Figures S3E and S3F). One-way ANOVA for each strain with Dunett's multiple comparisons to the start condition mean was conducted and reported in the figure legend. The fraction of start overlap to simulation means was calculated using the fraction of start condition measurements that fell within the range of their corresponding resident or disperser 95% confidence intervals (Figures S3G and S3H). This is used as an approximation of morphological contribution of innate start population morphology to the mean morphology observed in corresponding residents and dispersers.
Acknowledgments
A.D.J. and C.G.P. were supported by NIH/NHLBI #F31HL1474495 and NIH/NIGMS #R01GM099820. S.J., H.P., and D.L. are part of the French Laboratory of Excellence project TULIP (ANR-10-LABX-41). C.G.P. was supported by the 2018 Visiting Scientist program of French Laboratory of Excellence project TULIP (ANR-10-LABX-41). We thank Alex Stemm-Wolf for critical comments on the manuscript. S.J., H.P., and D.L. warmly thank Michèle Huet for the breeding of strains and her precious experimental advice.
Author contributions
A.D.J., S.J., D.L., and C.G.P. designed the study and analyzed experimental data. A.D.J., S.J., and D.L. performed the experiments and collected the data. A.D.J. and S.J. conducted the computational analyses. H.P. provided conceptual and experimental assistance. All authors contributed substantially to the writing and revisions of this manuscript.
Declaration of interests
The authors declare no competing interests.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102915.
Contributor Information
Delphine Legrand, Email: delphine.legrand@sete.cnrs.fr.
Chad G. Pearson, Email: chad.pearson@cuanschutz.edu.
Supplemental information
Caudal cilium at the posterior end of D5 cell does not exhibit ciliary beating as observed in neighboring cortical cilia.
References
- Altermatt F., Fronhofer E.A., Garnier A., Giometto A., Hammes F., Klecka J., Legrand D., Mächler E., Massie T.M., Pennekamp F. Big answers from small worlds: a user's guide for protist microcosms as a model system in ecology and evolution. Methods Ecol. Evol. 2015;6:218–231. [Google Scholar]
- Arslanyolu M., Doerder F.P. Genetic and environmental factors affecting mating type frequency in natural isolates of Tetrahymena thermophila. J. Eukaryot. Microbiol. 2000;47:412–418. doi: 10.1111/j.1550-7408.2000.tb00068.x. [DOI] [PubMed] [Google Scholar]
- Bayless B.A., Galati D.F., Junker A.D., Backer C.B., Gaertig J., Pearson C.G. Asymmetrically localized proteins stabilize basal bodies against ciliary beating forces. J. Cell Biol. 2016;215:457–466. doi: 10.1083/jcb.201604135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B.A., Galati D.F., Pearson C.G. Tetrahymena basal bodies. Cilia. 2015;5:1. doi: 10.1186/s13630-016-0022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B.A., Giddings T.H., Jr., Winey M., Pearson C.G. Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol. Biol. Cell. 2012;23:4820–4832. doi: 10.1091/mbc.E12-08-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard M.F., McCauley S.J. Integrating across life-history stages: consequences of natal habitat effects on dispersal. Am. Nat. 2008;171:553–567. doi: 10.1086/587072. [DOI] [PubMed] [Google Scholar]
- Benton T.G., Solan M., Travis J.M., Sait S.M. Microcosm experiments can inform global ecological problems. Trends Ecol. Evol. 2007;22:516–521. doi: 10.1016/j.tree.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Bestion E., Cote J., Jacob S., Winandy L., Legrand D. Habitat fragmentation experiments on arthropods: what to do next? Curr. Opin. Insect Sci. 2019;35:117–122. doi: 10.1016/j.cois.2019.07.011. [DOI] [PubMed] [Google Scholar]
- Bonte D., Saastamoinen M. Oxford University Press; 2012. Dispersal Syndromes in Spiders and Butterflies. Dispersal Ecology and Evolution. [Google Scholar]
- Bonte D., Van Dyck H., Bullock J.M., Coulon A., Delgado M., Gibbs M., Lehouck V., Matthysen E., Mustin K., Saastamoinen M. Costs of dispersal. Biol. Rev. Camb Philos. Soc. 2012;87:290–312. doi: 10.1111/j.1469-185X.2011.00201.x. [DOI] [PubMed] [Google Scholar]
- Bottier M., Thomas K.A., Dutcher S.K., Bayly P.V. How does cilium length Affect beating? Biophys. J. 2019;116:1292–1304. doi: 10.1016/j.bpj.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaine A.S., Schtickzelle N., Polard T., Huet M., Clobert J. Kin-based recognition and social aggregation in a ciliate. Evol. Int. J. Org. Evol. 2010;64:1290–1300. doi: 10.1111/j.1558-5646.2009.00902.x. [DOI] [PubMed] [Google Scholar]
- Cheptou P.-O., Carrue O., Rouifed S., Cantarel A. Rapid evolution of seed dispersal in an urban environment in the weed <em>Crepis sancta</em>. Proc. Natl. Acad. Sci. U S A. 2008;105:3796–3799. doi: 10.1073/pnas.0708446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clobert J., Baguette M., Benton T.G., Bullock J.M. OUP Oxford; 2012. Dispersal Ecology and Evolution. [Google Scholar]
- Clobert J., Le Galliard J.F., Cote J., Meylan S., Massot M. Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecol. Lett. 2009;12:197–209. doi: 10.1111/j.1461-0248.2008.01267.x. [DOI] [PubMed] [Google Scholar]
- Comte L., Olden J.D. Evidence for dispersal syndromes in freshwater fishes. Proc. R. Soc. B Biol. Sci. 2018;285:20172214. doi: 10.1098/rspb.2017.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corliss J. Tetrahymena paravorax n. sp., the first caudal-ciliated member of the genus referrable to the vorax-patula complex. J. Protozool. 1957;4:13. [Google Scholar]
- Cote J., Bestion E., Jacob S., Travis J., Legrand D., Baguette M. Evolution of dispersal strategies and dispersal syndromes in fragmented landscapes. Ecography. 2017;40:56–73. [Google Scholar]
- Doerder F.P., Brunk C. Natural populations and inbred strains of Tetrahymena. Methods Cell Biol. 2012;109:277–300. doi: 10.1016/B978-0-12-385967-9.00009-8. [DOI] [PubMed] [Google Scholar]
- Elgeti J., Gompper G. Emergence of metachronal waves in cilia arrays. Proc. Natl. Acad. Sci. U S A. 2013;110:4470–4475. doi: 10.1073/pnas.1218869110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjerdingstad E.J., Schtickzelle N., Manhes P., Gutierrez A., Clobert J. Evolution of dispersal and life history strategies--Tetrahymena ciliates. BMC Evol. Biol. 2007;7:133. doi: 10.1186/1471-2148-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner W., Kusuoka Y., Shimano S. Morphology and gene sequence of Levicoleps biwae n. gen., n. sp. (Ciliophora, Prostomatida), a proposed endemic from the ancient Lake Biwa, Japan. J. Eukaryot. Microbiol. 2008;55:185–200. doi: 10.1111/j.1550-7408.2008.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R.J., Donelson J.M., Schunter C., Ravasi T., Gaitán-Espitia J.D. The Royal Society; 2019. Beyond Buying Time: The Role of Plasticity in Phenotypic Adaptation to Rapid Environmental Change. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati D.F., Abuin D.S., Tauber G.A., Pham A.T., Pearson C.G. Automated image analysis reveals the dynamic 3-dimensional organization of multi-ciliary arrays. Biol. Open. 2015;5:20–31. doi: 10.1242/bio.014951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati D.F., Bonney S., Kronenberg Z., Clarissa C., Yandell M., Elde N.C., Jerka-Dziadosz M., Giddings T.H., Frankel J., Pearson C.G. DisAp-dependent striated fiber elongation is required to organize ciliary arrays. J. Cell Biol. 2014;207:705–715. doi: 10.1083/jcb.201409123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin W., Prakash V.N., Prakash M. Vortex arrays and ciliary tangles underlie the feeding–swimming trade-off in starfish larvae. Nat. Phys. 2017;13:380–386. [Google Scholar]
- Holz G.G., Jr., Corliss J.O. Tetrahymena setifera n. sp., a member of the genus Tetrahymena with a caudal cilium. J. Protozool. 1956;3:112–118. [Google Scholar]
- Ito H., Omori T., Ishikawa T. Swimming mediated by ciliary beating: comparison with a squirmer model. J. Fluid Mech. 2019;874:774–796. [Google Scholar]
- Jacob S., Chaine A.S., Huet M., Clobert J., Legrand D. Variability in dispersal syndromes is a key driver of metapopulation dynamics in experimental microcosms. Am. Nat. 2019;194:613–626. doi: 10.1086/705410. [DOI] [PubMed] [Google Scholar]
- Jacob S., Chaine A.S., Schtickzelle N., Huet M., Clobert J. Social information from immigrants: multiple immigrant-based sources of information for dispersal decisions in a ciliate. J. Anim. Ecol. 2015;84:1373–1383. doi: 10.1111/1365-2656.12380. [DOI] [PubMed] [Google Scholar]
- Jacob S., Laurent E., Haegeman B., Bertrand R., Prunier J.G., Legrand D., Cote J., Chaine A.S., Loreau M., Clobert J., Schtickzelle N. Habitat choice meets thermal specialization: competition with specialists may drive suboptimal habitat preferences in generalists. Proc. Natl. Acad. Sci. U S A. 2018;115:11988–11993. doi: 10.1073/pnas.1805574115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob S., Laurent E., Morel-Journel T., Schtickzelle N. Fragmentation and the context-dependence of dispersal syndromes: matrix harshness modifies resident-disperser phenotypic differences in microcosms. Oikos. 2020;129:158–169. [Google Scholar]
- Jacob S., Legrand D., Chaine A.S., Bonte D., Schtickzelle N., Huet M., Clobert J. Gene flow favours local adaptation under habitat choice in ciliate microcosms. Nat. Ecol. Evol. 2017;1:1407–1410. doi: 10.1038/s41559-017-0269-5. [DOI] [PubMed] [Google Scholar]
- Jacob S., Wehi P., Clobert J., Legrand D., Schtickzelle N., Huet M., Chaine A. Cooperation-mediated plasticity in dispersal and colonization. Evolution. 2016;70:2336–2345. doi: 10.1111/evo.13028. [DOI] [PubMed] [Google Scholar]
- Jiang Y.Y., Maier W., Baumeister R., Minevich G., Joachimiak E., Wloga D., Ruan Z., Kannan N., Bocarro S., Bahraini A. LF4/MOK and a CDK-related kinase regulate the number and length of cilia in Tetrahymena. PLoS Genet. 2019;15:e1008099. doi: 10.1371/journal.pgen.1008099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A.D., Soh A.W.J., O'toole E.T., Meehl J.B., Guha M., Winey M., Honts J.E., Gaertig J., Pearson C.G. Microtubule glycylation promotes attachment of basal bodies to the cell cortex. J. Cell Sci. 2019;132:jcs233726. doi: 10.1242/jcs.233726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozel T.R. Scanning electron microscopy of theronts of Ichthyophthirius multifiliis: their penetration into host tissues. Trans. Am. Microsc. Soc. 1986:357–364. [Google Scholar]
- Laurent E., Schtickzelle N., Jacob S. Fragmentation mediates thermal habitat choice in ciliate microcosms. Proc. Biol. Sci. 2020;287:20192818. doi: 10.1098/rspb.2019.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Galliard J.F., Ferrière R., Clobert J. Effect of patch occupancy on immigration in the common lizard. J. Anim. Ecol. 2005;74:241–249. [Google Scholar]
- Le Galliard J.F., Remy A., Ims R.A., Lambin X. Patterns and processes of dispersal behaviour in arvicoline rodents. Mol. Ecol. 2012;21:505–523. doi: 10.1111/j.1365-294X.2011.05410.x. [DOI] [PubMed] [Google Scholar]
- Legrand D., Guillaume O., Baguette M., Cote J., Trochet A., Calvez O., Zajitschek S., Zajitschek F., Lecomte J., Bénard Q. The Metatron: an experimental system to study dispersal and metaecosystems for terrestrial organisms. Nat. Methods. 2012;9:828–833. doi: 10.1038/nmeth.2104. [DOI] [PubMed] [Google Scholar]
- Legrand D., Trochet A., Moulherat S., Calvez O., Stevens V.M., Ducatez S., Clobert J., Baguette M. Ranking the ecological causes of dispersal in a butterfly. Ecography. 2015;38:822–831. [Google Scholar]
- Little C.J., Fronhofer E.A., Altermatt F. Dispersal syndromes can impact ecosystem functioning in spatially structured freshwater populations. Biol. Lett. 2019;15:20180865. doi: 10.1098/rsbl.2018.0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B.R., Ma M.Z., Gao F., Shi Y.H., Chen X.R. Morphology and molecular phylogeny of two colepid species from China, Coleps amphacanthus Ehrenberg, 1833 and Levicoleps biwae jejuensis Chen et al., 2016 (Ciliophora, Prostomatida) Dongwuxue Yanjiu. 2016;37:176–185. doi: 10.13918/j.issn.2095-8137.2016.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougald D., Rice S.A., Barraud N., Steinberg P.D., Kjelleberg S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 2012;10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- Mitchell D.R. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv. Exp. Med. Biol. 2007;607:130–140. doi: 10.1007/978-0-387-74021-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murren C.J., Auld J.R., Callahan H., Ghalambor C.K., Handelsman C.A., Heskel M.A., Kingsolver J.G., Maclean H.J., Masel J., Maughan H. Constraints on the evolution of phenotypic plasticity: limits and costs of phenotype and plasticity. Heredity. 2015;115:293–301. doi: 10.1038/hdy.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney D., Chen S., Meyer E. Scalar constraints in Tetrahymena evolution. Quantitative basal body variations within and between species. J. Cell Biol. 1978;79:727–736. doi: 10.1083/jcb.79.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney D.L. Corticotype transmission in Tetrahymena. Genetics. 1966;54:955–968. doi: 10.1093/genetics/54.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney D.L., McCoy J.W. Characterization of the species of the Tetrahymena pyriformis complex. Trans. Am. Microsc. Soc. 1976;95:664–682. [PubMed] [Google Scholar]
- Nelsen E.M. Transformation in Tetrahymena thermophila. Development of an inducible phenotype. Dev. Biol. 1978;66:17–31. doi: 10.1016/0012-1606(78)90270-1. [DOI] [PubMed] [Google Scholar]
- Nelsen E.M., Debault L.E. Transformation in Tetrahymena pyriformis: description of an inducible phenotype. J. Protozool. 1978;25:113–119. doi: 10.1111/j.1550-7408.1978.tb03880.x. [DOI] [PubMed] [Google Scholar]
- Omori T., Ito H., Ishikawa T. Swimming microorganisms acquire optimal efficiency with multiple cilia. Proc. Natl. Acad. Sci. U S A. 2020;117:30201–30207. doi: 10.1073/pnas.2011146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Osborn D.P., Giddings T.H., Jr., Beales P.L., Winey M. Basal body stability and ciliogenesis requires the conserved component Poc1. J. Cell Biol. 2009;187:905–920. doi: 10.1083/jcb.200908019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennekamp F., Clobert J., Schtickzelle N. The interplay between movement, morphology and dispersal in Tetrahymena ciliates. PeerJ. 2019;7:e8197. doi: 10.7717/peerj.8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennekamp F., Mitchell K.A., Chaine A., Schtickzelle N. Dispersal propensity in Tetrahymena thermophila ciliates - a reaction norm perspective. Evolution. 2014;68:2319–2330. doi: 10.1111/evo.12428. [DOI] [PubMed] [Google Scholar]
- Pennekamp F., Schtickzelle N., Petchey O.L. BEMOVI, software for extracting behavior and morphology from videos, illustrated with analyses of microbes. Ecol. Evol. 2015;5:2584–2595. doi: 10.1002/ece3.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode S., Elgeti J., Gompper G. Multi-ciliated microswimmers-metachronal coordination and helical swimming. Eur. Phys. J. E Soft Matter. 2021;44:76. doi: 10.1140/epje/s10189-021-00078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronce O. How does it Feel to Be like a Rolling Stone? Ten questions about dispersal evolution. Annu. Rev. Ecol. Evol. Syst. 2007;38:231–253. [Google Scholar]
- Ronce O., Clobert J. Dispersal syndromes. Dispersal Ecol. Evol. 2012;155:119–138. [Google Scholar]
- Schtickzelle N., Fjerdingstad E.J., Chaine A., Clobert J. Cooperative social clusters are not destroyed by dispersal in a ciliate. BMC Evol. Biol. 2009;9:251. doi: 10.1186/1471-2148-9-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell R.S., Beckman N.G., Fricke E., Loiselle B.A., Carvalho C.S., Jones L.R., Lichti N.I., Lustenhouwer N., Schreiber S.J., Strickland C. Consequences of intraspecific variation in seed dispersal for plant demography, communities, evolution and global change. AoB Plants. 2019;11:plz016. doi: 10.1093/aobpla/plz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh A.W.J., Van Dam T.J.P., Stemm-Wolf A.J., Pham A.T., Morgan G.P., O'toole E.T., Pearson C.G. Ciliary force-responsive striated fibers promote basal body connections and cortical interactions. J. Cell Biol. 2020;219:e201904091. doi: 10.1083/jcb.201904091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A.J., Morgan G., Giddings T.H., Jr., White E.A., Marchione R., Mcdonald H.B., Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell. 2005;16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens V.M., Trochet A., Blanchet S., Moulherat S., Clobert J., Baguette M. Dispersal syndromes and the use of life-histories to predict dispersal. Evol. Appl. 2013;6:630–642. doi: 10.1111/eva.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens V.M., Turlure C., Baguette M. A meta-analysis of dispersal in butterflies. Biol. Rev. 2010;85:625–642. doi: 10.1111/j.1469-185X.2009.00119.x. [DOI] [PubMed] [Google Scholar]
- Tamm S.L. Laser microbeam study of a rotary motor in termite flagellates. Evidence that the axostyle complex generates torque. J. Cell Biol. 1978;78:76–92. doi: 10.1083/jcb.78.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thazhath R., Liu C., Gaertig J. Polyglycylation domain of beta-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat. Cell Biol. 2002;4:256–259. doi: 10.1038/ncb764. [DOI] [PubMed] [Google Scholar]
- Van Dyck H., Baguette M. Dispersal behaviour in fragmented landscapes: routine or special movements? Basic Appl. Ecol. 2005;6:535–545. [Google Scholar]
- Wloga D., Camba A., Rogowski K., Manning G., Jerka-Dziadosz M., Gaertig J. Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Mol. Biol. Cell. 2006;17:2799–2810. doi: 10.1091/mbc.E05-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A., De Néchaud B., Chillet D., Mazarguil H., Desbruyères E., Audebert S., Eddé B., Gros F., Denoulet P. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 1992;59:425–432. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Caudal cilium at the posterior end of D5 cell does not exhibit ciliary beating as observed in neighboring cortical cilia.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This study does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.