Abstract
During our survey of the diversity of woody litter fungi in China and Thailand, three Hermatomyces species were collected from dead woody twigs of Dipterocarpus sp. (Dipterocarpaceae) and Ehretiaacuminata (Boraginaceae). Both morphology and multigene analyses revealed two taxa as new species (Hermatomycesturbinatus and H.jinghaensis) and the remaining collections as new records of H.sphaericus. Hermatomycesturbinatus is characterized by 1) dimorphic conidia, having circular to oval lenticular conidia and 2) turbinate conidia consisting of two columns with two septa composed of 2–3 cells in each column. Hermatomycesjinghaensis is characterized by dimorphic conidia, having circular to oval lenticular conidia and clavate or subcylindrical to cylindrical conidia and consisting of one or two columns with 6–8 cells in each column. Phylogenetic analyses of combined LSU, ITS, tub2, tef1-α and rpb2 sequence data supports the placement of these new taxa within Hermatomycetaceae with high statistical support.
Keywords: 2 new species, hyphomycetes, phylogeny, taxonomy, woody litter fungi
Introduction
Over the past few decades, the number of studies using a molecular-based approach to study microfungal diversity in the greater Mekong subregion (GMS) has increased rapidly, especially on freshwater and woody litter fungi from China (Yunnan Province) and Thailand (Hapuarachchi et al. 2019; Dong et al. 2020; Li et al. 2020; Monkai et al. 2020; Wanasinghe et al. 2020, 2021; Mortimer et al. 2021). Hyde et al. (2018) reported that about 96% of fungi from Thailand are new to science. Feng and Yang (2018) estimated 104,000 fungal species currently exist in Yunnan Province, China; however, only about 6,000 are extant. Therefore, further studies need to be conducted to fill gaps in knowledge regarding the diversity, taxonomy and phylogeny of microfungi in the GMS. Supporting this obligation, we have begun to study plant-based ascomycetes in GMS. The current study accounts for hermatomyces-like ascomycetes recovered from the woody litter in China (Yunnan Province) and Thailand.
Hermatomyces was introduced by Spegazzini (1911) with H.tucumanensis as the type species. Doilom et al. (2017) accommodated Hermatomyces in Lophiotremataceae based on combined LSU, SSU, tef1-α and rpb2 sequence data. Later, Hashimoto et al. (2017) validated Hermatomycetaceae (Hermatomycetaceae Locq. 1984 was not validly published, Art. 39.1) to accommodate the genus Hermatomyces. This genus is known only by its asexual morph that is characterized by sporodochial conidiomata and dimorphic (lenticular or cylindrical) conidia of one or two types. The lenticular conidia are globose to subglobose, hyaline to pale brown peripheral cells with dark brown central cells, and the cylindrical conidia is hyaline, cylindrical to subcylindrical or turbinate and consisting of 1–4 columns of 2–12 cells (Spegazzini 1911; Tibpromma et al. 2016; Hashimoto et al. 2017; Hyde et al. 2019; Pem et al. 2019; Phukhamsakda et al. 2020).
Based on morphological comparisons and phylogenetic affinities, Koukol et al. (2018) revised Hermatomyces species and described five new species (viz. H.bifurcatus, H.constrictus, H.megasporus, H.sphaericoides and H.verrucosus) and one new combination, H.reticulatus, from Panama. Accordingly, H.chromolaenae, H.saikhuensis, H.tectonae were treated as H.sphaericus and H.subiculosus, H.chiangmaiensis, H.thailandicus were synonymized with H.reticulatus, H.krabiensis and H.indicus, respectively (Koukol et al. 2018). These are probably species complexes that need more detailed study. Subsequent studies introduced H.bauhiniae, H.biconisporus, H.clematidis, H.trangensis and H.truncates into Hermatomyces (Tibpromma et al. 2018; Hyde et al. 2019; Koukol et al. 2019; Nuankaew et al. 2019; Phukhamsakda et al. 2020). Currently, 24 species are recognized in Hermatomyces (Koukol et al. 2018, 2019; Nuankaew et al. 2019; Delgado et al. 2020; Phukhamsakda et al. 2020; Table 2).
Table 2.
Synopsis of the morphological characteristics of Hermatomyces species.
| Species | Lenticular conidia size (μm) | Cylindrical / turbinate conidia feature | Host | Country | Reference | ||
|---|---|---|---|---|---|---|---|
| Shape | Length × width (μm) | Number of columns (cells) | |||||
| Hermatomyces amphisporus | 27–36(–38) × 18–29(–31) | Cylindrical, pyriform or turbinate | 30‒38 × 20‒26 | 2(–4) (6–12 cells) | Cyathea sp., Sabalminor | Mexico, USA | Castañeda-Ruiz and Heredia (2000); Delgado et al. (2020) |
| H. bauhiniae | 25–36 × 15–20 | Cylindrical | 20–28 × 8–11 | 1 (2–3-septate) | Bauhinia variegata | Thailand | Hyde et al. (2019) |
| H. biconisporus | 28–34 × 15–25 | Cylindrical | 32–39 × 14.5–26 | 1–2 (3–4 cells) | Pandanus sp. | China | Tibpromma et al. (2018) |
| H. bifurcatus | (24–)30–36.5(–41) × (18–)21.5–26(–28) | Cylindrical | Apex: 7–16 × 7–12 Basal: 9–14 × 13–18.5 | 2 (2–3 cells) | Unknown | Panama | Koukol et al. (2018) |
| H. chromolaenae | 9.2–10.4 × 10.2–11.5 | NA | NA | NA | Chromolaena odorata | Thailand | Tibpromma et al. (2017) |
| H. clematidis | 30–45 × 24–31 | Cylindrical | 29–35 × 12–14 | 1–2 (5–6 cells) | Clematis sikkimensis | Thailand | Phukhamsakda et al. (2020) |
| H. constrictus | (22–)25.5–29.5(–32) × 19–23.5(–27.5) | Cylindrical | Lower cells: (20–)24–30.5(–37) × 12–17 Upper cells: (16–)20–26(–30) × 8–14 | 1 (2 cells) | Bauhinia cumanensis | Panama | Koukol et al. (2018) |
| H. dimorphus | 35‒55 × 15‒20 | Cylindrical | 15‒40 × 10‒15 | 4 (7 cells) | Unknown | India | Rao and de Hoog (1986) |
| H. indicus | 18‒30 × 11.5‒19 | Turbinate | 22.4‒35.4 × 11.4‒21.6 | 2 (6–7 cells) | Phoenix rupicola | India | Prasher and Prasher (2014) |
| H. iriomotensis | 30–36 × 20–27 | Cylindrical | 20.5–33 × 7–12.5 | 1–2 (3–7 cells) | Unknown | Japan | Hashimoto et al. (2017) |
| H. jinghaensis | 30–40 × 25–30 | Clavate, subcylindrical | 33–43 × 11–13 | 1–2 (6–8 cells) | Unknown | China | This study |
| H. krabiensis | 24.3–32.5 × 12.1–21.3 | Cylindrical | 20.4–26.4 × 8.6–19.7 | 1–2 (2–3 cells) | Pandanus odorifer | Thailand | Tibpromma et al. (2016) |
| H. megasporus | (45–)49–56(–59) × (31–)37–46 | Cylindrical | (37–)49.5–60.5(67–) × 18–28(–32) | 2 ((5–)6–7(–10) cells) | Unknown | Panama | Koukol et al. (2018) |
| H. nabanheensis | 20.2–25.1 × 16.6–20.7 | Cylindrical | 15.3–26.8 × 12.1–18.2 | 1–2 (2–3 cells) | Pandanus sp. | China | Hyde et al. (2017) |
| H. pandanicola | 12–15.7 × 20–30.1 | Cylindrical | 13.2–20.6 × 8.9–11.9 | 2 (2 cells) | Pandanus odorifer | Thailand | Tibpromma et al. (2016) |
| H. reticulatus | 3–40(–45) × 25–34(–41) | NA | NA | NA | Unknown | Thailand, Panama | Hyde et al. (2016); Koukol et al. (2018) |
| H. saikhuensis | 14.2–21.4 × 11.2–19.3 | NA | NA | NA | Pandanus odorifer | Thailand | Tibpromma et al. (2016) |
| H. sphaericoides | (20.5–)24.5–28(–31) × (20–)23–26(–29) | NA | NA | NA | Unknown | Panama | Koukol et al. (2018) |
| H.sphaericus (PMA 116080) | (21–)24–29(–32.5) × (18–)21–27(–31.5) | NA | NA | NA | Various host plants | Tropical or subtropical | Koukol et al. (2018) |
| H. sphaericus | 27–29 × 26–28 | NA | NA | NA | Dipterocarpus sp., Ehretiaacuminata | China, Thailand | This study |
| H. tectonae | (23–)26–29(–33) × (19–)23–25(–28) | Cylindrical | (27–)31–32(–35) ×(21–)23 | 2 (6 cells) | Tectona grandis | Thailand | Doilom et al. (2017) |
| H. trangensis | 27.5‒35 × 25‒32.5 | NA | NA | NA | Arenga pinnata | Thailand | Nuankaew et al. (2019) |
| H. truncates | (26–)31.5–36.5(–37) × 22–27(–30) | Cylindrical | Lower cells: 14–22.5(–28) × 8.5–14.5 | 1 (2–3 cells) | Averrhoa carambola | Ghana, Panama | Koukol et al. (2019) |
| Upper cells: 12–18(–30) × (6–)8–12.5 | |||||||
| H. tucumanensis | (22–)27–35 × 18–25 | Obclavate or subcylindrical | (21–)23–26(–28.5) × 7–14 | 2 (3–6 cells) | Unknown | Panama | Koukol et al. (2018) |
| H. turbinatus | 24–30 × 17–21 | Turbinate | 27–36 × 19–28 | 2 (2–3 cells) | Dipterocarpus sp. | Thailand | This study |
| H. uniseriatus | 27–36 × 15.5–24 | Cylindrical | 19–34 × 10–12.5 | 1 (3–4 cells) | Smilax campestris | Argentina | Leão-Ferreira et al. (2013) |
| H. verrucosus | 23–30(–39) × 21–29.5 | NA | NA | NA | Unknown | Panama | Koukol et al. (2018) |
NA: absent
Our investigation led to the discovery of three Hermatomyces species, including two novel species, on dead woody-based substrates. Morphological illustrations and multi-gene phylogenetic analyses with ML, MP and BI of combined LSU, ITS, tub2, tef1-α and rpb2 sequence data are used to confirm the phylogenetic placement of the novel species within Hermatomyces.
Materials and methods
Sample collection, examination and isolation
Woody litter samples were collected from China (Yunnan Province) during the dry season (December 2019) and Thailand (Tak Province) during the wet season (August 2019). Samples were brought to the laboratory using plastic Ziploc bags. Fungal specimens were then examined using a stereomicroscope (Olympus SZ61, China). Pure cultures were obtained via single spore isolation on potato dextrose agar (PDA) following the methods described in Senanayake et al. (2020). Cultures were incubated at 25 °C for three weeks. Micro-morphological structures were photographed using a Nikon compound microscope (Nikon ECLIPSE Ni) fitted with a Canon (EOS 600D) digital camera. Measurements were taken using the Tarosoft (R) Image Frame Work program. Figures were processed using Adobe Photoshop CS6. Type specimens were deposited in the herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS). Ex-type living cultures were deposited at the Culture Collection of Mae Fah Luang University (MFLUCC) and Kunming Institute of Botany Culture Collection (KUMCC).
DNA extraction, amplification and sequencing
DNA extraction, amplification, sequencing, sequence alignment and phylogenetic analyses follow the methods of Dissanayake et al. (2020) with the following details. Two partial rDNA genes and three protein coding genes were used in our study, including internal transcribed spacer region (ITS) using primer pair ITS5/ITS4 (White et al. 1990), 28S large subunit nuclear ribosomal (LSU) using primer pair LR0R/LR5 (Vilgalys and Hester 1990), translation elongation factor 1-alpha gene (tef1-α) using primer pair EF1-983F/EF1-2218R (Rehner and Buckley 2005), RNA polymerase II second largest subunit (rpb2) using primer pair fRPB2-5F/fRPB2-7cR (Liu et al. 1999) and β-tubulin (tub2) using primer pair T1/T22 (O’Donnell and Cigelnik 1997). Amplification reactions were performed in a total volume of 25 μL of PCR mixtures containing 8.5 μL ddH2O, 12.5 μL 2× PCR MasterMix (TIANGEN Co., China), 2 μL DNA template and 1 μL of each primer. The PCR thermal cycle program for LSU, ITS, tef1-α and rpb2 were set as described in Tibpromma et al. (2018). The PCR amplification condition of tub2 was set as denaturation at 94 °C for 3 minutes, followed by 35 cycles of denaturation at 94 °C for 45 seconds, annealing at 56 °C for 50 seconds and extension at 72 °C for 1 minute, with a final extension step at 72 °C for 10 minutes. PCR products were sent to the Qingke Company, Kunming City, Yunnan Province, China, for sequencing. Sequences were deposited in GenBank (Table 1).
Table 1.
GenBank accession numbers of sequences used for the phylogenetic analyses.
Sequence alignment and phylogenetic analyses
Representative species used in the phylogenetic analyses were selected based on previous publications (Koukol et al. 2018; Nuankaew et al. 2019; Delgado et al. 2020; Phukhamsakda et al. 2020). Sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/), and their accession numbers are listed in Table 1. The newly generated sequences in this study were assembled by BioEdit 7.0.9.0 (Hall 1999). Individual gene regions were separately aligned in MAFFT v.7 web server (http://mafft.cbrc.jp/alignment/server/) (Katoh et al. 2019). The alignments of each gene were improved by manually deleting the ambiguous regions plus gaps and combined using BioEdit 7.2.3. Final alignments containing LSU, ITS, tub2, tef1-α and rpb2 were converted to NEXUS format (.nxs) using CLUSTAL X (2.0) (Thompson et al. 1997) and processed for Bayesian and maximum parsimony analysis. The FASTA format was changed into PHYLIP format via the Alignment Transformation Environment (ALTER) online program (http://www.sing-group.org/ALTER/) and used for maximum likelihood analysis (ML).
ML was carried out in CIPRES Science Gateway v.3.3 (http://www.phylo.org/portal2/; Miller et al. 2010) using RAxML-HPC2 on XSEDE (8.2.12) (Stamatakis 2014) with the GTRGAMMA substitution model and 1,000 bootstrap iterations. Maximum parsimony analysis (MP) was performed in PAUP v. 4.0b10 (Swofford 2002) with the heuristic search option and Tree-Bisection-Reconnection (TBR) of branch-swapping algorithm for 1,000 random replicates. Branches with a minimum branch length of zero were collapsed, and gaps were treated as missing data (Hillis and Bull 1993).
Bayesian analysis was executed in MrBayes v.3.2.2 (Ronquist et al. 2012). The model of evolution was estimated using MrModeltest v. 2.3 (Nylander et al. 2008) via PAUP v. 4.0b10 (Ronquist and Huelsenbeck 2003). The SYM+I+G for LSU and ITS; HKY+I for tub2; GTR+I+G for tef1-α and rpb2 were used in the final command. Markov chain Monte Carlo sampling (MCMC) in MrBayes v.3.2.2 (Ronquist et al. 2012) was used to determine posterior probabilities (PP) (Rannala and Yang 1996; Zhaxybayeva and Gogarten 2002). Bayesian analyses of six simultaneous Markov chains were run for 2,000,000 generations and trees were sampled and printed to output at every 200 generations (resulting in 10,001 total trees). The first 25% of sampled trees were discarded as part of the burn-in procedure, the remaining 7,501 trees were used to create the consensus tree and the average standard deviation of split frequencies was set as 0.01.
Phylogenetic trees were visualized in FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/; Rambaut 2012), the tree was edited using Microsoft PowerPoint before being saved in PDF format and finally converted to JPG format using Adobe Illustrator CS6 (Adobe Systems, USA). The finalized alignments and trees were deposited in TreeBASE, submission ID: TB2:S28514 (http://purl.org/phylo/treebase/phylows/study/TB2:S28514).
Ex-type strains are indicated with superscript “T”, and newly generated sequence is shown in bold. NA represents sequences that are unavailable in GenBank. Abbreviations:
ANM A.N. Miller;
BCCBIOTEC Culture Collection, Bangkok, Thailand;
CBSCentraal Bureau voor Schimmel cultures, Utrecht, The Netherlands;
CCF Culture Collection of Fungi, Charles University, Prague, Czech Republic;
HKAS The herbarium of Cryptogams Kunming Institute of Botany Academia Sinica;
KH K. Hirayama;
KUMCC Culture Collection of Kunming Institute of Botany, Kunming, China;
KZP O. Koukol;
MFLUCC Mae Fah Luang University Culture Collection, Chiang Rai, Thailand;
PMAHerbarium of the University of Panama, Panama City, Panama;
PRCHerbarium of the Charles University, Prague, Czech Republic;
PRM Herbarium of the National Museum, Prague, Czech Republic.
T1 Type of Hermatomycesthailandicus;
T2 Type of H.chiangmaiensis;
T3 Type of H.subiculosus;
T4 Type of H.chromolaenae;
T5 Type of H.saikhuensis;
T6 Type of H.tectonae.
Results
Phylogenetic analysis
The phylogenetic analysis was conducted using 57 strains in Hermatomycetaceae, and two outgroup taxa Anteagloniumglobosum (ANM 925.2) and A.parvulum (MFLUCC 14-0821) in Pleosporales (Table 1). The aligned sequence matrix comprised five gene regions (LSU: 887 bp, ITS: 530 bp, tub2: 606 bp, tef1-α: 952 bp and rpb2: 1,028 bp) and a total of 4,003 characters (including gaps), of which 3,207 characters were constant, 174 variable characters were parsimony-uninformative and 622 characters were parsimony-informative. The Kishino-Hasegawa test shows length = 1,388 steps with CI = 0.671, RI = 0.884, RC = 0.593 and HI = 0.329. The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of -13406.555506. Estimated base frequencies were as follows: A = 0.241874, C = 0.266701, G = 0.257552, T = 0.233873; substitution rates AC = 1.188604, AG = 4.826453, AT = 1.273226, CG = 0.855218, CT = 11.409386, GT = 1.00; gamma distribution shape parameter α = 0.16102.
In the phylogenetic tree obtained from ML, MP and BI analysis (Fig. 1) the maximum likelihood analysis resulted in trees largely with similar topology and clades as in the maximum parsimony and Bayesian analyses. The new species, Hermatomycesturbinatus, is sister to H.nabanheensis (KUMCC 16-0149) with high support (94% ML, 91% MP and 1.00 BYPP, Fig. 1). Hermatomycesjinghaensis is nested between H.trangensis and H.clematidis with a strongly supported monophyletic group (98% ML, 92% MP, 1.00 PP; Fig. 1). New isolates of H.sphaericus (KUMCC 20-0231; MFLUCC 21-0036) clustered with remaining H.sphaericus strains as a monophyletic group (Fig. 1). The topology of the phylogenetic tree is in accordance with recent phylogenetic studies discussing species in Hermatomycetaceae (Nuankaew et al. 2019; Phukhamsakda et al. 2020).
Figure 1.
Phylogenetic RAxML tree based on analysis of a combined LSU, ITS, tub2, tef1-α and rpb2 and dataset. Bootstrap support values for ML and MP equal to or higher than 75% and Bayesian PP equal to or greater than 0.95 are shown at nodes. Hyphens (--) represent support values less than 75% / 0.95 BYPP. The ex-type strains are in bold and the new isolate in this study is in blue bold. The tree is rooted with Anteagloniumglobosum (ANM 925.2) and A.parvulum (MFLUCC 14-0821). The scale bar represents the expected number of nucleotide substitutions per site.
Taxonomy
Hermatomyces turbinatus
G.C. Ren & K.D. Hyde sp. nov.
479CDBD8-11CB-58C0-93ED-BED46E6A7908
558166
Facesoffungi Number No: FoF09735
Figure 2.
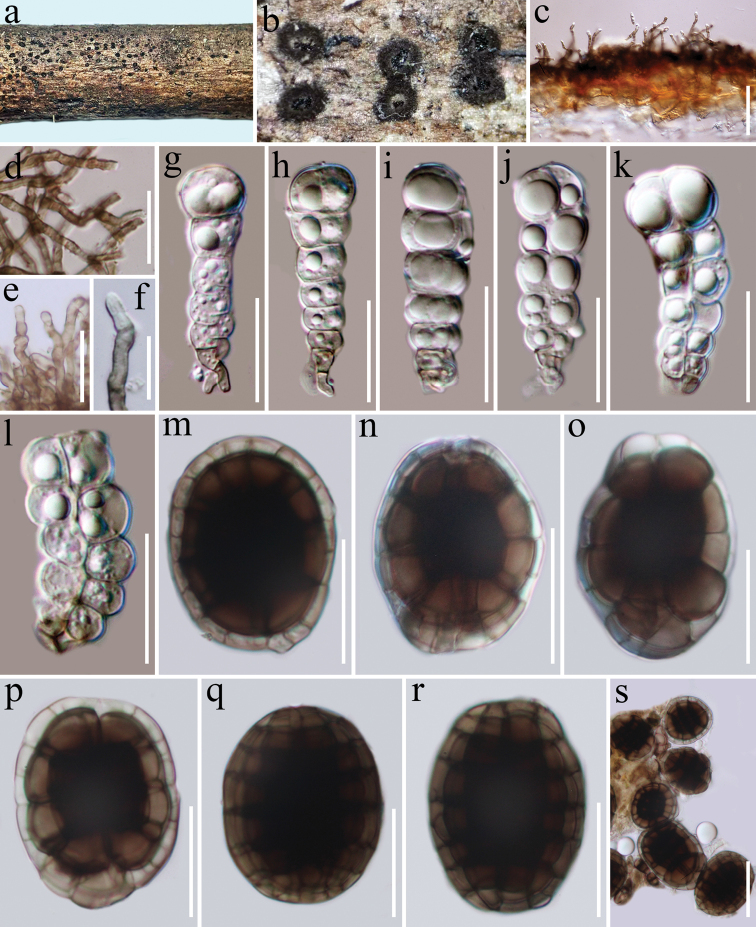
Hermatomycesturbinatus (HKAS 112724, holotype) a, b sporodochia on natural substrate c vertical section of sporodochium d conidiophores and conidiogenous cells e–h turbinate conidia i turbinate and mature lenticular conidia j–m mature lenticular conidia n germinated conidium o, p culture characters on PDA. Scale bars: 30 μm (c); 20 μm (d–n); 30 mm (o, p).
Etymology.
Referring to the turbinate shape of the conidia.
Holotype.
HKAS 112724.
Description.
Saprobic on woody litter of Dipterocarpus sp. (Dipterocarpaceae) Sexual morph Undetermined. Asexual morphColonies on natural substrate forming sporodochial conidiomata, superficial, scattered, small groups, circular or oval, sterile mycelial outer zone enclosing a black-brown velvety margin, sparse, black sporulating center, shiny, glistening, circular or oval, conidia readily liberated when agitated. Mycelium superficial, branched, septate, hyaline to pale brown, 2–3 μm wide. Conidiophores 6–8 × 2–3 μm, micronematous, straight or flexuous, smooth, short, pale brown. Conidiogenous cells 3–5 × 2–3 μm, monoblastic, integrated, terminal, determinate, often arising directly on the superficial mycelium, subsphaerical, ovoid or ampulliform, hyaline to pale brown, smooth finely verruculose. Conidia dimorphic, solitary, smooth-walled. Lenticular conidia 24–30 × 17–21 μm (x = 27 × 20 μm, n = 20), 12–15 μm thick, thick-walled, circular to oval in front view, smooth, solitary, muriform, central cells dark brown to black, peripheral cells hyaline to pale brown, forming a weakly ring, sometimes slightly constricted at septa, obovoid or oblong in lateral view, arranged in 2 rows, a row of composed of 4–6 cells, end cells pale brown to hyaline, middle cells dark brown. Turbinate conidia turbinate, pyriform, 27–36 μm in length, 19–28 µm wide in broadest part of lower cells, (x = 32 × 23 μm, n = 20), asymmetrical with the upper cells smaller than lower cells, thick-walled, smooth, septate, constricted distinct at septa, consisting of two columns with two septa composed of 2–3 rectangular to globose cells in each column, usually upper part of terminal cells dark brown, becoming hyaline towards the lower side, two cells hyaline in the lower cells swollen with oil globules.
Known host and distribution.
Dipterocarpus sp. (Thailand).
Culture characteristics.
Colonies on PDA, reaching 30–40 mm diam., after 3 weeks at 25–30 °C, circular, convex with papillate and radially furrowed at the center, rough, labate, crenate edge, fluffy, dense, gray black, in reverse darkens at the center, pale brown to gray at edge.
Material examined.
Thailand, Tak Province. Ban Na Sam Ngao District, on woody litter of Dipterocarpus sp. (Dipterocarpaceae), 22 August 2019, G. C. Ren, TSY04 (HKAS 112724, holotype), ex-type living culture, MFLUCC 21-0038.
Notes.
Hermatomycesturbinatus is introduced as a new species based on its distinct morphology, which is supported by phylogenetic analyses. In the phylogenetic analyses, H.turbinatus is distinct from extant species in this genus and formed a sister clade to H.nabanheensis with strong support (94% ML, 91% MP, 1.00 PP; Fig. 1). Hermatomycesturbinatus differs from H.nabanheensis in having turbinate conidia with two columns, while H.nabanheensis has cylindrical conidia with one or two columns. Hermatomycesturbinatus has two conidial types, and its lenticular conidia are similar to H.tectonae in shape and size. However, the turbinate conidia of H.turbinatus have 2 columns of 2–3 cells in each column, while the turbinate conidia of H.tectonae have 2 columns of 3 cells in each column. We also compared the morphological characters of H.turbinatus to other species of Hermatomyces (Table 2). Despite no molecular data being available for the three species viz. H.dimorphus, H.uniseriatus and H.truncates, H.turbinatus nonetheless differs from these species in conidial characteristics (Table 2).
Hermatomyces jinghaensis
G.C. Ren & K.D. Hyde sp. nov.
586F538B-54BB-5D95-A10A-50FE2A43AEE0
558165
Facesoffungi Number No: FoF09736
Figure 3.
Hermatomycesjinghaensis (HKAS 112167, holotype) a, b sporodochia on natural substrate c vertical section of sporodochium d conidiophores e, f conidiogenous cells g–l cylindrical conidia m–s mature lenticular conidia. Scale bars: 50 μm (c); 30 μm (d); 20 μm (e–r); 30 μm (s).
Etymology.
The species epithet “jinghaensis” refers to the location where the species was collected.
Holotype.
HKAS 112167.
Description.
Saprobic on unidentified woody litter. Sexual morph Undetermined. Asexual morphColonies on natural substrate forming sporodochial conidiomata, superficial, scattered, small groups, circular, sterile mycelial outer zone enclosing a black velvety margin, dense, thick, black sporulating center, shiny, glistening, circular or oval, conidia readily liberated when agitated. Mycelium superficial, branched, septate, hyaline to pale brown, 2–3 μm wide. Conidiophores 30–45 × 2–3 μm, mononematous, cylindrical, straight or flexuous, smooth, pale brown. Conidiogenous cells 4–6 × 2–3 μm, monoblastic, integrated, terminal, determinate, often arising directly on the superficial mycelium, cylindrical, ampulliform, hyaline to pale brown, smooth finely verruculose. Conidia dimorphic solitary, smooth-walled. Lenticular conidia 30–40 × 25–30 μm (x = 37 × 28 μm, n = 20), 21–25 μm thick, thick-walled, circular to oval in front view, smooth, solitary, muriform, central cells brown to dark brown, peripheral cells hyaline to subhyaline, forming a wide and distinct ring, sometimes slightly constricted at septa, obovoid or oblong in lateral view, central cells brown to dark brown, peripheral cells pale brown to brown. Cylindrical conidia 33–43 μm in length, 11–13 µm wide in broadest part of lower cells (x = 39 × 12 μm, n = 20), clavate or subcylindrical, straight or flexuous, septate, constricted distinct at the septa, with large guttules, consisting of one or two columns, each column with 6–8 cells, apical cell rectangular to globose, smooth, hyaline, smooth, basal cells acute, rectangular to cylindrical, pale brown.
Known host and distribution.
Unidentified woody litter (China)
Material examined.
China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Jinghong, Jingha (21°78.06'N, 101°05.61'E), on unidentified woody litter, 19 December 2019, D.N. Wanasinghe, DW57 (HKAS 112167, holotype), no living culture.
Notes.
Hermatomycesjinghaensis is introduced as a new species based on its distinct morphology and the phylogenetic results of a combined LSU, ITS, tub2, tef1-α and rpb2 dataset. Hermatomycesjinghaensis nested with H.clematidis and H.trangensis in a strongly supported monophyletic group (99% ML, 100% MP, 1.00 PP; Fig. 1). Hermatomycesjinghaensis is characterized by both lenticular and cylindrical conidia. Hermatomycesjinghaensis differs from H.clematidis in having cylindrical conidia with one or two columns, each of which has 6–8 cells with large guttules, while the latter has 5–6 cells for each column conidia. Hermatomycestrangensis differs from H.jinghaensis in having only lenticular conidia.
Hermatomyces sphaericus
(Sacc.) S. Hughes 1953.
272AC737-BC12-5F42-AA42-C0262ADA5FD2
298410
Facesoffungi Number No: FoF05259
Figure 4.
Hermatomycessphaericus (HKAS 112725) a, b colonies on the natural substrate c mycelia d–g young conidia h–k mature conidia (h–j surface view k thickness view) l germinated conidium m, n culture characters on PDA. Scale bars: 1000 μm (a); 200 μm (b); 20 μm (c–i, l); 30 μm (j, k); 3 cm (m, n).
Description.
Saprobic on woody litter of Dipterocarpus sp. (Dipterocarpaceae) and Ehretiaacuminata (Boraginaceae). Sexual morph Undetermined. Asexual morphColonies on natural substrate forming sporodochial conidiomata, superficial, circular or irregular, scattered or crowded, consisting of a velvety, dense, annular, gray brown, sterile mycelial outer zone and a black, glistening, abundantly sporulating granulose center, with conidia readily liberated when agitated. Mycelium 2–2.5 μm wide, superficial, composed of a tightly network of branched, septate, smooth or finely verruculose, hyaline or pale brown hyphae. Conidiophores 10–13 × 2–4 μm (x = 12 × 3 μm, n = 10) micronematous, cylindrical or forked, smooth, hyaline or pale brown, often corresponding to conidiogenous cells. Conidiogenous cells 5–8 × 3–5 μm (x = 7 × 4 μm, n = 20), monoblastic, integrated, terminal, cylindrical, hyaline to pale brown, smooth or finely verruculose. Conidia of one type, 27–29 × 26–28 μm (x = 28 × 27 μm, n = 30) μm, 19–24 μm thick, solitary, lenticular, globose, subglobose in front view, muriform, smooth, central cells brown, dark brown, outer ring of peripheral cells narrow, pale brown to brown, often constricted at septa, disk-shaped in lateral view, consisting of two rows, each row with 4–6 cells, hyaline to light brown at lower and upper cells, middle cells brown to black brown.
Known host and distribution.
Tropical and subtropical regions of Central and South America, Africa, Asia, Oceania and North America. The species were found as saprobes on Acanthaceae, Apocynaceae, Arecaceae, Asteraceae, Dipterocarpaceae, Euphorbiaceae, Fabaceae, Lamiaceae, Leguminosae, Mimosaceae, Nyctaginaceae, Oxalidaceae, Pandanaceae, Pinaceae, Rhamnaceae, and Sterculiaceae (Zhang et al 2009; Koukol et al. 2018, 2019).
Culture characteristics.
Colonies on PDA, reaching 35–40 mm diam., after 3 weeks at 25–30 °C, with circular, umbonate, fluffy, velvety, entire edge, a circular raised band, gray white, in reverse dark gray, black toward the center.
Material examined.
Thailand, Tak Province, Tha Song Yang District, on woody litter of Dipterocarpus sp. (Dipterocarpaceae), 22 August 2019, G. C. Ren, T903 (HKAS 112725), living culture, MFLUCC 21-0036; China, Yunnan Province, Xishuangbanna (21°55.19'N, 101°15.24'E), on woody litter of Ehretiaacuminata (Boraginaceae), 4 August 2020, G. C. Ren, JH39 (HKAS 112166), living culture, KUMCC 20-0231.
Notes.
The characters of our new strain of Hermatomycessphaericus (KUMCC 20-0231, MFLUCC 21-0036) are similar to the type collection (K(M)–IMI 37763) in having gray black to black sporodochia, mononematous, pale brown, smooth, monoblastic, integrated, terminal, cylindrical, hyaline to pale brown conidiogenous cells and globose to subglobose conidia (Hughes 1953). A multigene phylogeny indicates that novel strains clustered within the H.sphaericus clade (Fig. 1). We name our strain (KUMCC 20-0231, MFLUCC 21-0036) as H.sphaericus, which has been reported from different plant families and genera (Koukol et al. 2018). However, we consider this might be a species complex that need further detailed studies. Our study provides the new host records of H.sphaericus on Dipterocarpus sp. (Dipterocarpaceae) and Ehretiaacuminata (Boraginaceae), and updates sequence data for the new collections of H.sphaericus.
Discussion
This study introduces two new species of woody-based litter fungi; Hermatomycesjinghaensis from Yunnan, China and Hermatomycesturbinatus on Dipterocarpus sp. from Thailand. We also report for the first time two new records of H.sphaericus on Dipterocarpus sp. and Ehretiaacuminata in China and Thailand.
Hermatomyces (Hermatomycetaceae) is different from other similar genera in its sporodochial conidiomata and in having one to two (lenticular and cylindrical conidia) unusual conidial types (Spegazzini 1911). All species of Hermatomyces have lenticular conidia with similar characteristics, whereas some species have cylindrical and turbinate conidia, which have greater variance in shape, size, number of columns and cells. Koukol et al. (2018, 2019) have reported that multiple species may occur together on a single sample, a phenomenon we observed, which may complicate morphological identification and separation for culturing. Therefore, molecular sequence data are more reliable for the identification of Hermatomyces species (Tibpromma et al. 2016, 2017, 2018; Nuankaew et al. 2019; Phukhamsakda et al. 2020).
Hermatomycessphaericus was introduced by Hughes (1953), which may be the most widespread of species in Hermatomyces distributed across many subtropical and tropical regions worldwide (Wijayawardene et al. 2014; Doilom et al. 2017; Koukol et al. 2018, 2019; Hyde et al. 2019; Jayasiri et al. 2019; Nuankaew et al. 2019; Phukhamsakda et al. 2020). This species has been reported as saprobic on dead plant tissues of several host families (Tibpromma et al. 2016, 2017; Doilom et al. 2017; Jayasiri et al. 2019). In addition, Koukol et al. (2018) reported that H.sphaericus (ARIZ: PS0053) was isolated from seeds of Apeibamembranacea (Malvaceae), suggesting this species could be an endophyte. Previous studies have indicated that H.sphaericus is not restricted to any single host (Koukol et al. 2018, 2019; Jayasiri et al. 2019), whereas other species of Hermatomyces are saprobic on a limited number of hosts and are limited to specific regions (Rao and de Hoog 1986; Leão-Ferreira et al. 2013; Prasher and Prasher 2014; Hyde et al. 2016, 2017, 2019; Tibpromma et al. 2016, 2017, 2018; Doilom et al. 2017; Hashimoto et al. 2017; Koukol et al. 2018, 2019; Nuankaew et al. 2019; Delgado et al. 2020; Phukhamsakda et al. 2020; Table 2). In this study, our new strains of H.sphaericus had slight morphological differences in lenticular conidia size compared to the type strains and other strains of H.sphaericus (Hughes 1953, Table 2). As reported by Koukol et al. (2018), H.sphaericus is a plurivorous species, and accordingly the phenotypic variation among strains could be influenced by environmental factors and culture conditions or it could have speciated in isolated polulations (Hyde et al. 2020).
Species delineation in Hermatomyces, especially in the H.sphaericus clade, is subject to much controversy due to species inconsistency in morphological and phylogenetic status. Koukol et al. (2018) synonymized H.chromolaenae, H.saikhuensis and H.tectonae under H.sphaericus based on morphological and molecular comparisons and suspected that H.pandanicola could either be a hybrid species or incorrect sequences were used in the analysis. Koukol et al. (2019) considered that during isolation of H.biconisporus, a conidium of H.sphaericus might have been taken instead, leading to contamination when extracting DNA and the misinterpretation of its taxonomic placement. Phukhamsakda et al. (2020) further confirmed that H.biconisporus, H.pandanicola and H.sphaericus should be treated as the same species based on Genealogical Concordance Phylogenetic Species Recognition (GCPSR) analysis.
Hermatomyces had long been treated as “incertae sedis” within Ascomycota (Wijayawardene et al. 2012). Doilom et al. (2017) placed Hermatomyces in Lophiotremataceae baed on phylogenetic analyses, and consequently, Hashimoto et al. (2017) revised the family Lophiotremataceae based on morphological observations and phylogenetic analyses, and Hermatomyces was accepted in the family Hermatomycetaceae, as monophyletic. Recent studies and our study indicate Hermatomyces to be highly polyphyletic, and Hermatomyces morphology has evolved, which is mainly characterized by lenticular and cylindrical conidia (Fig. 1; Koukol et al. 2018, 2019; Hyde et al. 2019; Phukhamsakda et al. 2020). Support for a single H.sphaericus species (Fig. 1) lacks internal statistical support and includes H.biconisporus, H.chromolaenae, H.pandanicola, H.saikhuensis and H.tectonae and we suspect that this is a species complex. Tibpromma et al. (2018) also noted that H.sphaericus could be a species complex including several species and did not accept the synonymy of H.saikhuensis and H.tectonae in H.sphaericus owing to their significant base-pair differences.
In this study, we combined two non-translated loci (LSU, ITS) and three protein-coding regions (tub2, tef1-α and rpb2) to carry out phylogenetic analysis for Hermatomyces species in order to validate phylogenetic placement of the taxa within Hermatomyces. In our phylogenetic analyses, H.tectonae, H.chromolaenae, H.biconisporus, H.pandanicola and H.saikhuensis grouped together with strains of H.sphaericus (PRC 4100, PRC 4104, PMA 116081). Hermatomycessaikhuensis and H.chromolaenae are characterized by one conidium type (lenticular) similar to H.sphaericus, however, they differ in the shape, color and size of conidia (Tibpromma et al. 2016, 2017; Table 2). Hermatomycestectonae, H.biconisporus and H.pandanicola are characterized by dimorphic conidia which differ from H.sphaericus (Tibpromma et al. 2016, 2018; Doilom et al. 2017; Koukol et al. 2018; Table 2). Hermatomycessphaericus (PRC 4100, PRC 4104, PMA 116081) did not have a morphological description for inter-species comparison (Koukol et al. 2018). Further taxon sampling and more sequence data are needed to elucidate this clade.
Supplementary Material
Acknowledgements
This work was supported by Open Research Fund Program of Science and Technology on Aerospace Chemical Power Laboratory (STACPL320181B04). We thank the support from the National Natural Science Foundation of China (NSFC21975066, NSFC21875061). We also would like to thank the Thailand Research Fund for the grant entitled Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (No. RDG6130001). Dhanushka Wanasinghe thanks CAS President’s International Fellowship Initiative (PIFI) for funding his postdoctoral research (number 2021FYB0005), the Postdoctoral Fund from Human Resources and Social Security Bureau of Yunnan Province and the National Science Foundation of China and Chinese Academy of Sciences (grant no. 41761144055) for financial support.
Citation
Ren G-C, Wanasinghe DN, Monkai J, Mortimer PE, Hyde KD, Xu J-C, Pang A, Gui H (2021) Novel saprobic Hermatomyces species (Hermatomycetaceae, Pleosporales) from China (Yunnan Province) and Thailand. MycoKeys 82: 57–79. https://doi.org/10.3897/mycokeys.82.67973
Funding Statement
Open Research Fund Program of Science and Technology on Aerospace Chemical Power Laboratory (STACPL320181B04). The National Natural Science Foundation of China (NSFC21975066, NSFC21875061). The Thailand Research Fund for the grant entitled Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (No. RDG6130001).
Contributor Information
Aimin Pang, Email: ppam@tom.com.
Heng Gui, Email: guiheng@mail.kib.ac.cn.
References
- Castañeda-Ruiz RF, Heredia G. (2000) Two new dematiaceous hyphomycetes on Cyathea from Mexico. Cryptogamie Mycologie 21: 221–228. 10.1016/S0181-1584(00)01047-2 [DOI] [Google Scholar]
- Delgado G, Koukol O, Heredia G, Piepenbring M. (2020) Texas microfungi: Hermatomycesamphisporus (Pleosporales, Dothideomycetes) revisited. Czech Mycology 72: 95–107. 10.33585/cmy.72107 [DOI] [Google Scholar]
- Dissanayake AJ, Bhunjun CS, Maharachchikumbura SSN, Liu JK. (2020) Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 11: 2652–2676. 10.5943/mycosphere/11/1/18 [DOI] [Google Scholar]
- Doilom M, Dissanayake AJ, Wanasinghe DN, Boonmee S, Liu JK, Bhat DJ, Taylor JE, Bahkali AH, McKenzie EHC, Hyde KD. (2017) Microfungi on Tectonagrandis (teak) in Northern Thailand. Fungal Diversity 82: 107–182. 10.1007/s13225-016-0368-7 [DOI] [Google Scholar]
- Dong W, Wang B, Hyde KD, McKenzie EHC, Raja HA, Tanaka K, Abdel-Wahab MA, Abdel-Aziz FA, Doilom M, Phookamsak R, Hongsanan S, Wanasinghe DN, Yu XD, Wang GN, Yang H, Yang J, Thambugala KM, Tian Q, Luo ZL, Yang JB, Miller AN, Fournier J, Boonmee S, Hu DM, Nalumpang S, Zhang H. (2020) Freshwater Dothideomycetes. Fungal Diversity 105: 319–575. 10.1007/s13225-020-00463-5 [DOI] [Google Scholar]
- Feng B, Yang Z. (2018) Studies on diversity of higher fungi in Yunnan, southwestern China: A review. Plant Diversity 40: 165–171. 10.1016/j.pld.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hapuarachchi KK, Karunarathna SC, Phengsintham P, Yang HD, Kakumyan P, Hyde KD, Wen TC. (2019) Ganodermataceae (Polyporales): Diversity in Greater Mekong Subregion countries (China, Laos, Myanmar, Thailand and Vietnam). Mycosphere 10: 221–309. 10.5943/mycosphere/10/1/6 [DOI] [Google Scholar]
- Hashimoto A, Matsumura M, Hirayama K, Tanaka K. (2017) Revision of Lophiotremataceae (Pleosporales, Dothideomycetes): Aquasubmersaceae, Cryptocoryneaceae, and Hermatomycetaceae fam. nov. Persoonia 39: 51–73. 10.3767/persoonia.2017.39.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Hughes SJ. (1953) Fungi from the Gold Coast. II. Mycological Papers 50: 1–104. [Google Scholar]
- Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Abdel-Aziz FA, Abdel–Wahab MA, Banmai S, Chomnunti P, Cui BK, Daranagama DA, Das K, Dayarathne MC, de Silva NI, Dissanayake AJ, Doilom M, Ekanayaka AH, Gibertoni TB, Góes-Neto A, Huang SK, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li WJ, Lin CG, Liu JK, Lu YZ, Luo ZL, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, de Azevedo Santiago ALCM, Drechsler-Santos ER, Senanayake IC, Tanaka K. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. 10.1007/s13225-016-0373-x [DOI] [Google Scholar]
- Hyde KD, Norphanphoun C, Abreu VP, Bazzicalupo A, Chethana KWT, Clericuzio M, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, He MQ, Hongsanan S, Huang SK, Jayasiri SC, Jayawardena RS, Karunarathna A, Konta S, Kusan I, Lee H, Li J, Lin CG, Liu NG, Lu YZ, Luo ZL, Manawasinghe IS, Mapook A, Perera RH, Phookamsak R, Phukhamsakda C, Siedlecki I, Soares AM, Tennakoon DS, Tian Q, Tibpromma S, Wanasinghe DN, Xiao YP, Yang J, Zeng XY, Abdel-Aziz FA, Li WJ, Senanayake IC, Shang QJ, Daranagama DA, de Silva NI, Thambugala KM, Abdel-Wahab MA, Bahkali AH, Berbee ML, Boonmee S, Bhat DJ, Bulgakov TS, Buyck B, Camporesi E, Castañeda-Ruiz RF, Chomnunti P, Doilom M, Dovana F, Gibertoni TB, Jadan M, Jeewon R, Jones EBG, Kang JC, Karunarathna SC, Lim YW, Liu JK, Liu ZY, Plautz Jr HL, Lumyong S, Maharachchikumbura SSN, Matočec N, McKenzie EHC, Mešic A, Miller D, Pawlowska J, Pereira OL, Promputtha I, Romero AI, Ryvarden L, Su HY, Suetrong S, Tkalčec Z, Vizzini A, Wen TC, Wisitrassameewong K, Wrzosek M, Xu JC, Zhao Q, Zhao RL, Mortimer PE. (2017) Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Diversity 87: 1–235. 10.1007/s13225-017-0391-3 [DOI] [Google Scholar]
- Hyde KD, Norphanphoun C, Chen J, Dissanayake AJ, Doilom M, Hongsanan S, Jayawardena RS, Jeewon R, Perera RH, Thongbai B, Wanasinghe DN, Wisitrassameewong K, Tibpromma S, Stadler M. (2018) Thailand’s amazing diversity – up to 96% of fungi in northern Thailand are novel. Fungal Diversity 93: 215–239. 10.1007/s13225-018-0415-7 [DOI] [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R, Bhat DJ, Maharachchikumbura SSN, Rossi W, Leonardi M, Lee HB, Mun HY, Houbraken J, Nguyen TTT, Jeon SJ, Frisvad JC, Wanasinghe DN, Luücking R, Aptroot A, Cáceres MES, Karunarathna SC, Hongsanan S, Phookamsak R, de Silva NI, Thambugala KM, Jayawardena RS, Senanayake IC, Boonmee S, Chen J, Luo ZL, Phukhamsakda C, Pereira OL, Abreu VP, Rosado AWC, Buyck B, Randrianjohany E, Hofstetter V, Gibertoni TB, da Silva Soares AM, Plautz Jr HL, Sotão HMP, Xavier WKS, Bezerra JDP, de Oliveira TGL, de Souza-Motta CM, Magalhães OMC, Bundhun D, Harishchandra D, Manawasinghe IS, Dong W, Zhang SN, Bao DF, Samarakoon MC, Pem D, Karunarathna A, Lin CG, Yang J, Perera RH, Kumar V, Huang SK, Dayarathne MC, Ekanayaka AH, Jayasiri SC, Xiao YP, Konta S, Niskanen T, Liimatainen K, Dai YC, Ji XH, Tian XM, Mešić A, Singh SK, Phutthacharoen K, Cai L, Sorvongxay T, Thiyagaraja V, Norphanphoun C, Chaiwan N, Lu YZ, Jiang HB, Zhang JF, Abeywickrama PD, Aluthmuhandiram JVS, Brahmanage RS, Zeng M, Chethana T, Wei DP, Réblová M, Fournier J, Nekvindová J, do Nascimento Barbosa R, dos Santos JEF, de Oliveira NT, Li GJ, Ertz D, Shang QJ, Phillips AJL, Kuo CH, Camporesi E, Bulgakov TS, Lumyong S, Jones EBG, Chomnunti P, Gentekaki E, Bungartz F, Zeng XY, Fryar S, Tkalčec Z, Liang J, Li GS, Wen TC, Singh PN, Gafforov Y, Promputtha I, Yasanthika E, Goonasekara ID, Zhao RL, Zhao Q, Kirk PM, Liu JK, Yan JY, Mortimer PE, Xu JC. (2019) Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. 10.1007/s13225-019-00429-2 [DOI] [Google Scholar]
- Hyde KD, Jeewon R, Chen YJ, Bhunjun CS, Calabon MS, Jiang HB, Lin CG, Norphanphoun C, Sysouphanthong P, Pem D, Tibpromma S, Zhang Q, Doilom M, Jayawardena RS, Liu JK, Maharachchikumbura SSN, Phukhamsakda C, Phookamsak R, Al-Sadi AM, Naritsada Thongklang N, Wang Y, Gafforov Y, Jones EBG, Lumyong S. (2020) The numbers of fungi: is the descriptive curve flattening? Fungal Diversity 103: 219–271. 10.1007/s13225-020-00458-2 [DOI]
- Jayasiri SC, Hyde KD, Jones EBG, McKenzie EHC, Jeewon R, Phillips AJL, Bhat DJ, Wanasinghe DN, Liu JK, Lu YZ, Kang JC, Xu J, Karunarathna SC. (2019) Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10: 1–186. 10.5943/mycosphere/10/1/1 [DOI] [Google Scholar]
- Jayasiri SC, Jones EBG, Kang JC, Promputtha I, Bahkali AH, Hyde KD. (2016) A new species of genus Anteaglonium (Anteagloniaceae, Pleosporales) with its asexual morph. Phytotaxa 263: 233–244. 10.11646/phytotaxa.263.3.4 [DOI] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in bioinformatics 20: 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukol O, Delgado G, Hofmann TA, Piepenbring M. (2018) Panama, a hot spot for Hermatomyces (Hermatomycetaceae, Pleosporales) with five new species, and a critical synopsis of the genus. International Mycological Association 9: 107–141. 10.5598/imafungus.2018.09.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukol O, Delgado G. (2019) Do not forget Africa – revision of fungarium collections at Kew revealed a new species of Hermatomyces (Hermatomycetaceae, Pleosporales). Nova Hedwigia 109: 413–423. 10.1127/nova_hedwigia/2019/0559 [DOI] [Google Scholar]
- Leão-Ferreira SM, Gusmão LFP, Castañeda RF. (2013) Conidial fungi from the semi-arid Caatinga biome of Brazil. Three new species and new records. Nova Hedwigia 96: 479–494. 10.1127/0029-5035/2013/0084 [DOI] [Google Scholar]
- Li G, Slippers B, Wingfield MJ, Chen S. (2020) Variation in Botryosphaeriaceae from Eucalyptus plantations in Yunnan Province in southwestern China across a climatic gradient. IMA Fungus 11: e22. 10.1186/s43008-020-00043-x [DOI] [PMC free article] [PubMed]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Monkai J, Boonmee S, Ren GC, Wei DP, Phookamsak R, Mortimer PE. (2020) Distoseptisporahydei sp. nov. (Distoseptisporaceae), a novel lignicolous fungus on decaying bamboo in Thailand. Phytotaxa 459: 093–107. 10.11646/phytotaxa.459.2.1 [DOI] [Google Scholar]
- Mortimer PE, Jeewon R, Xu JC, Lumyong S, Wanasinghe DN. (2021) Morpho-Phylo Taxonomy of novel dothideomycetous fungi associated with dead woody twigs in Yunnan Province, China. Front Microbiol 12: e654683. 10.3389/fmicb.2021.654683 [DOI] [PMC free article] [PubMed]
- Mugambi GK, Huhndorf SM. (2009) Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi). Systematics and Biodiversity 7: 453–464. 10.1017/S147720000999020X [DOI] [Google Scholar]
- Nuankaew S, Suetrong S, Wutikhun T, Pinruan U. (2019) Hermatomycestrangensis sp. nov., a new dematiaceous hyphomycete (Hermatomycetaceae, Pleosporales) on sugar palm in Thailand. Phytotaxa 391: 277–288. 10.11646/phytotaxa.391.5.1 [DOI] [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. (2008) AWTY: a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24: 581–583. 10.1093/bioinformatics/btm388 [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Pem D, Hongsanan S, Doilom M, Tibpromma S, Wanasinghe DN, Dong W, Ningguo L, Phookamsak R, Phillips AJL, Jeewon R, Hyde KD. (2019) https://www.dothideomycetes.org: An online taxonomic resource for the classification, identification, and nomenclature of Dothideomycetes. Asian Journal of Mycology 2: 287–297. 10.5943/ajom/2/1/19 [DOI] [Google Scholar]
- Prasher IB, Prasher S. (2014) Hermatomycesindicus sp. nov. (Hyphomycetes) from India. Nova Hedwigia 99: 551–556. 10.1127/0029-5035/2014/0177 [DOI] [Google Scholar]
- Phukhamsakda C, McKenzie EHC, Phillips AJL, Jones EBG, Bhat DJ, Marc S, Bhunjun CS, Wanasinghe DN, Thongbai B, Camporesi E, Ertz D, Jayawardena RS, Perera RH, Ekanayake AH, Tibpromma S, Doilom M, Xu JC, Hyde KD. (2020) Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Diversity 102: 1–203. 10.1007/s13225-020-00448-4 [DOI] [Google Scholar]
- Rambaut A. (2012) FigTree version 1.4.0. http://tree.bio.ed.ac.uk/software/figtree [accessed 1 May 2020]
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Rao V, de Hoog GS. (1986) New or critical hyphomycetes from India. Studies in Mycology 28: 1–84. [Google Scholar]
- Rehner SA, Buckley E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Rathnayaka AR, Marasinghe DS, Calabon MS, Gentekaki E, Lee HB, Hurdeal VG, Pem D, Dissanayake LS, Wijesinghe SN, Bundhun D, Nguyen TTT, Goonasekara ID, Abeywickrama PD, Bhunjun CS, Jayawardena RS, Wanasinghe DN, Jeewon R, Bhat DJ, Xiang MM. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11: 2678–2754. 10.5943/mycosphere/11/1/20 [DOI] [Google Scholar]
- Spegazzini CL. (1911) Mycetes Argentinenses. Series V. Anales Museo Nacional de Historia Natural Buenos Aires 3: 446.
- Stamatakis A. (2014) RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP: phylogenetic analysis using parsimony, version 4.0 b10. Sinauer Associates, Sunderland. 10.1111/j.0014-3820.2002.tb00191.x [DOI]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24: 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibpromma S, Bhat DJ, Doilom M, Lumyong S, Nontachaiyapoom S, Yang JB, Hyde KD. (2016) Three new Hermatomyces species (Lophiotremataceae) on Pandanusodorifer from Southern Thailand. Phytotaxa 275: 127–139. 10.11646/phytotaxa.275.2.4 [DOI] [Google Scholar]
- Tibpromma S, Hyde KD, Jeewon R, Maharachchikumbura SSN, Liu JK, Bhat DJ, Jones EBG, McKenzie EHC, Camporesi E, Bulgakov TS, Doilom M, Santiago A, Das K, Manimohan P, Gibertoni TB, Lim YW, Ekanayaka AH, Thongbai B, Lee HB, Yang JB, Kirk PM, Sysouphanthong P, Singh SK, Boonmee S, Dong W, Raj KNA, Latha KPD, Phookamsak R, Phukhamsakda C, Konta S, Jayasiri SC, Norphanphoun C, Tennakoon DS, Li JF, Dayarathne MC, Perera RH, Xiao YP, Wanasinghe DN, Senanayake IC, Goonasekara ID, de Silva NI, Mapook A, Jayawardena RS, Dissanayake AJ, Manawasinghe IS, Chethana KWT, Luo ZL, Hapuarachchi KK, Baghela A, Soares AM, Vizzini A, Meiras-Ottoni A, Mesic A, Dutta AK, de Souza CAF, Richter C, Lin CG, Chakrabarty D, Daranagama DA, Lima DX, Chakraborty D, Ercole E, Wu F, Simonini G, Vasquez G, da Silva GA, Plautz HL, Ariyawansa HA, Lee H, Kusan I, Song J, Sun J, Karmakar J, Hu K, Semwal KC, Thambugala KM, Voigt K, Acharya K, Rajeshkumar KC, Ryvarden L, Jadan M, Hosen MI, MiksiK M, Samarakoon MC, Wijayawardene NN, Kim NK, Matocec N, Singh PN, Tian Q, Bhatt RP, de Oliveira RJV, Tulloss RE, Aamir S, Kaewchai S, Marathe SD, Khan S, Hongsanan S, Adhikari S, Mehmood T, Bandyopadhyay TK, Svetasheva TY, Nguyen TTT, Antonin V, Li WJ, Wang Y, Indoliya Y, Tkalcec Z, Elgorban AM, Bahkali AH, Tang AMC, Su HY, Zhang H, Promputtha I, Luangsa-Ard J, Xu JC, Yan JY, Ji-Chuan K, Stadler M, Mortimer PE, Chomnunti P, Zhao Q, Phillips AJL, Nontachaiyapoom S, Wen TC, Karunarathna SC. (2017) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 83: 1–261. 10.1007/s13225-017-0378-0 [DOI] [Google Scholar]
- Tibpromma S, Hyde KD, McKenzie EHC, Bhat DJ, Phillips AJL, Wanasinghe DN, Samarakoon MC, Jayawardena RS, Dissanayake AJ, Tennakoon DS, Doilom M, Phookamsak R, Tang AMC, Xu JC, Mortimer PE, Promputtha I, Maharachchikumbura SSN, Khan S, Karunarathna SC. (2018) Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 93: 1–160. 10.1007/s13225-018-0408-6 [DOI] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Wijayawardene NN, Xu JC, Cheewangkoon R, Mortimer PE. (2020) Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China). PLoS ONE 15: e0235855. 10.1371/journal.pone.0235855 [DOI] [PMC free article] [PubMed]
- Wanasinghe DN, Mortimer PE, Xu J. (2021) Insight into the systematics of microfungi colonizing dead woody twigs of Dodonaeaviscosa in Honghe (China). Journal of Fungi 7: e180. 10.3390/jof7030180 [DOI] [PMC free article] [PubMed]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: A Guide to Methods and Applications. Academic Press, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Crous PW, Kirk PM, Hawksworth DL, Boonmee S, Braun U, Dai DQ, D’souza MJ, Diederich P, Dissanayake A, Doilom M, Hongsanan S, Jones EB, Groenewald JZ, Jayawardena R, Lawrey JD, Liu JK, Lucking R, Madrid H, Manamgoda DS, Muggia L, Nelsen MP, Phookamsak R, Suetrong S, Tanaka K, Thambugala KM, Wanasinghe DN, Wikee S, Zhang Y, Aptroot A, Ariyawansa HA, Bahkali AH, Bhat DJ, Gueidan C, Chomnunti P, De Hoog GS, Knudsen K, Li WJ, McKenzie EHC, Miller AN, Phillips AJ, Piatek M, Raja HA, Shivas RS, Slippers B, Taylor JE, Tian Q, Wang Y, Woudenberg JH, Cai L, Jaklitsch WM, Hyde KD. (2014) Naming and outline of Dothideomycetes 2014 including proposals for the protection or suppression of generic names. Fungal Diversity 69: 1–55. 10.1007/s13225-014-0309-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene NN, McKenzie EHC, Hyde KD. (2012) Towards incorporating anamorphic fungi in a natural classification – checklist and notes for 2011. Mycosphere 3: 157–228. 10.5943/mycosphere/3/2/5 [DOI] [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3: 1–4. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhao G, Zhang X, Liu H, Wu Y. (2009) 26 Genera of Dematiaceous Dictyosporous Hyphomycetes excluding Alternaria. [Flora Fungorum Sinicorum no. 31.] Science Press, Beijing.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.