Abstract
GATA transcription factors (TFs) are widespread eukaryotic regulators whose DNA-binding domain is a class IV zinc finger motif (CX2CX17–20CX2C) followed by a basic region. We identified 262 GATA genes (389 GATA TFs) from seven Populus genomes using the pipeline of GATA-TFDB. Alternative splicing forms of Populus GATA genes exhibit dynamics of GATA gene structures including partial or full loss of GATA domain and additional domains. Subfamily III of Populus GATA genes display lack CCT and/or TIFY domains. 21 Populus GATA gene clusters (PCs) were defined in the phylogenetic tree of GATA domains, suggesting the possibility of subfunctionalization and neofunctionalization. Expression analysis of Populus GATA genes identified the five PCs displaying tissue-specific expression, providing the clues of their biological functions. Amino acid patterns of Populus GATA motifs display well conserved manner of Populus GATA genes. The five Populus GATA genes were predicted as membrane-bound GATA TFs. Biased chromosomal distributions of GATA genes of three Populus species. Our comparative analysis approaches of the Populus GATA genes will be a cornerstone to understand various plant TF characteristics including evolutionary insights.
Subject terms: Protein analysis, Gene regulatory networks, Sequence annotation
Introduction
A transcription factor (TF) is a protein that controls the rate of transcription by binding to specific DNA sequences, including promoter regions. TF can also combine and interact with cis-acting elements in the promoter region as well as interact with other proteins to regulate the start site of transcription1. In plant, TF plays important roles such as controlling flower developments2, 3, circadian clock4, carbon and nitrogen regulatory networks5, protein–protein interaction6, cell differentiation7, pathogen and hormone responses8, and disease resistance9.
Due to a large number of plant genomes available (2,220 genomes from 725 species; Plant Genome Database Release 2.75; http://www.plantgenome.info/; Park et al., in preparation), many genome-wide analyses of plant TFs have been conducted10–15. One of the genome-wide TF databases is the plantTFDB which identifies 58 TF families from 165 plant species11. Some of these TF families are plant-specific, including AP2/ERF16, NAC17, WRKY18, and GRAS19, 20, and some are general in eukaryotic such as bHLH (basic helix-loop-helix)21, 22, bZIP (basic leucine-zipper)23, and GATA24–27. With the published plant genomes, various genome-wide analyses of TF families have been conducted; AP2/ERF, NAC28–31, bHLH32–34, bZIP23, 35–37, GRAS20, 38, 39, and GATA25, 40, 41 TF families displaying their features in various aspects including evolutionary aspect. Genome-wide analyses of TF families in Arabidopsis thaliana have also been studied, presenting 122 AP2/ERF genes42, 105 NAC genes28, 162 bHLH genes34, 75 bZIP genes23, 32 GRAS genes20, 29 GATA genes25 as well as 566 GATA genes from 19 A. thaliana genomes43.
GATA TFs contain more than one highly conserved type IV zinc finger motifs (CX2X17–20CX2C) followed by a basic region that can bind to a consensus DNA sequence, WGATAR (W means T or A; R indicates G or A)25, 44, 45. Most plant GATA TFs contain a single GATA domain of which pattern is CX2CX18CX2C (type IVb) or CX2CX20CX2C (type IVc)27. Except for these known types, additional patterns were also identified: e.g., CX4CX18CX2X, which have four amino acids in the first Cysteine-Cysteine, named as type IV443.
Plant GATA TFs have various roles like the chloroplast development46, photosynthesis and growth47, epithelial innate immune responses48, seed germination49, hypocotyl and petiole elongation50, and cryptochrome1-dependent response51. Genome-wide analyses and/or expression analyses of GATA TFs have been reported in 21 plant species25, 40, 41, 43, 52–66 (Table S1), including P. trichocarpa of which genome-wide identification of GATA genes was conducted based on the old gene model (Version 3.0; Table S1).
Populus genus is a model system for investigating the wood development, crown formation, and disease resistance in perennial plants67, which has several advantages including rapid growth, ease of cloning, and small genome68. Owing to it, its genome was sequenced as a first wood plant genome69, and then additional genome sequences of Populus species have been sequenced and analyzed55, 70–75 (Table 1), which is an excellent resource to identify genus-wide analyses of Populus GATA TFs. These species were classified into independent three clades based on phylogenetic studies using single-copy76 nuclear genes and whole chloroplast genome sequences77, 78: (i) P. tremuloides, P. tremula, and P. tremula x alba, (ii) P. pruinosa and P. euphratica, and (iii) P. trichocarpa and P. deltoides. In spite of abundant resources of Salicaceae genomes including Salix purpurea79, there is only one study for characterizing the biological function of Populus GATA gene (PdGNC), which regulates chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis80, suggesting genome-wide identification of Populus GATA genes are strongly required.
Table 1.
Characteristics of identified GATA TFs from seven Populus genomes.
| Populus species name | Version | # of GATA genes (A) | # of GATA TFs | # of GATA genes having alternative splicing forms (B) | # of GATA TFs having alternative splicing forms | # of genes | # of proteins | Ratio (B/A) (%) |
|---|---|---|---|---|---|---|---|---|
| Populus trichocarpa | 3.1 | 39 | 67 | 13 | 41 | 42,950 | 63,498 | 33.33 |
| Populus pruinosa | 1 | 37 | 37 | 0 | 0 | 35,131 | 35,131 | 0.00 |
| Populus euphratica | 1 | 40 | 55 | 9 | 24 | 30,688 | 49,676 | 22.50 |
| Populus deltoides | 2.1 | 38 | 55 | 7 | 24 | 44,853 | 57,249 | 18.42 |
| Populus tremuloides | 1.1 | 37 | 44 | 7 | 14 | 36,830 | 48,320 | 18.92 |
| Populus tremula | 1.1 | 33 | 60 | 16 | 43 | 35,309 | 83,720 | 48.48 |
| Populus tremula x alba | 1.1 | 38 | 71 | 16 | 49 | 41,335 | 73,013 | 42.11 |
| Total | 262 | 389 | 68 | 195 | 267,096 | 410,607 | 30.22* |
*It shows that total ratio (B/A) of Populus genomes except P. pruinosa.
Here, we conducted genome-wide identification, phylogenetic analyses, expression level analysis, identification of amino acid patterns and transmembrane helix of GATA TFs in seven Populus genomes with the GATA-TFDB (http://gata.genefamily.info/; Park et al., in preparation). Our comparative and comprehensive analyses conducted with the integrated bioinformatic pipeline provided by the GATA-TFDB will be a cornerstone to understand various plant TF characteristics including evolutionary insights.
Results and discussions
Identification of GATA TFs from seven Populus genomes
We identified 262 GATA genes (389 GATA TFs) from seven Populus genomes available in public using the pipeline of the GATA-TFDB (http://gata.genefamily.info/; Table S2). The number of GATA genes for each Populus genome ranges from 33 to 40 (Table 1), which is larger than those of A. thaliana (29 to 30 GATA genes)25, 43, V. vinifera (19 GATA genes), and O. sativa (28 GATA genes)81; while is smaller than that of G. max (64 GATA genes)40. The phylogenetic relationship of the seven Populus species inferred from the complete chloroplast genomes (Fig. 1a), congruent to the previous studies76, 78, shows no correlation with the number of GATA genes. It can be explained that the number of GATA genes is rather affected by the accuracy of the gene model (e.g., the largest number of GATA TFs is not from P. trichocarpa, which is the model Populus species; Fig. 1b). The proportion of Populus GATA genes against whole genes ranges from 0.08% (P. deltoides) to 0.13% (P. euphratica; Table 1), which is similar to that of A. thaliana (0.11%) and is slightly higher than those of V. vinifera (0.07%), G. max (0.06%), and O. sativa (0.05%).
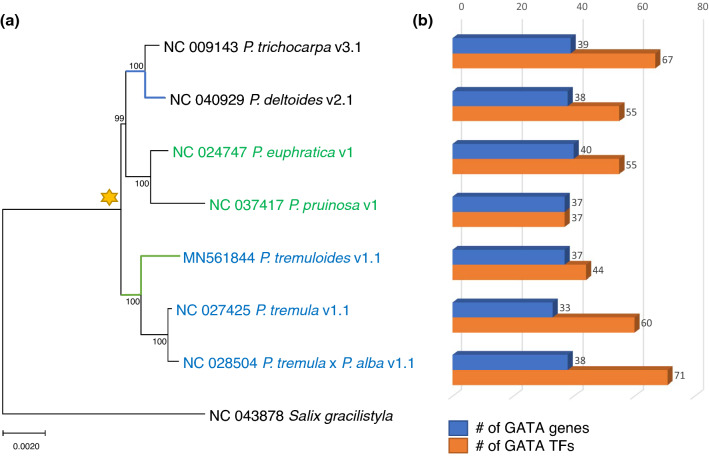
Figure 1.
Phylogenetic tree of seven Populus species in complete chloroplast genomes. (a) shows a phylogenetic tree of seven Populus species in complete chloroplast genomes. Bootstrap analyses with 1000 pseudo-replicates were conducted with the same options. S. gracilistyla is aligned to the seven Populus genomes as an outgroup. Light blue and green lines indicate the gene loss of GATA TFs in P. deltoides and P. tremuloides lineage, respectively. Yellow star means gene duplication events of the 10 paralogous pairs. Black, green, blue letters of 7 Populus genomes mean independent three clades based on phylogenetic studies. (b) presents the number of GATA genes and GATA TFs for each genome.
Among Populus genomes, P. euphratica has the largest number of GATA genes (40); while P. tremula contains the smallest (33; Fig. 1b): difference of the number of GATA genes between the largest and the smallest is seven. Similarity, three Arabidopsis genomes, Arabidopsis halleri, Arabidopsis lyrata, and A. thaliana, contain 22, 28, and 30 GATA genes, respectively (Fig. S1) and four Oryza genomes, O. sativa, Oryza glaberrima, Oryza brachyantha, and Oryza rufipogon, showed 28, 25, 24, and 28 GATA genes, respectively25 (Fig. S1), displaying similar interspecies differences. While, Gossypium raimondii, Gossypium arboretum, and Gossypium hirsutum presented 46, 46, and 87, respectively because G. hirsutum is a tetraploid species52. These interspecific differences of GATA genes indicate that many evolutionary events including the gain and loss of GATA genes were occurred in three genera.
Except P. pruinosa genome not containing alternative splicing forms, numbers of GATA TFs are larger than those of GATA genes (Table 1). Numbers of GATA genes which have alternative splicing forms range from 7 to 16 (Table 1 and Table S3), accounting for 30.22% of Populus GATA genes, which is similar that of A. thaliana (9 out of 30 GATA genes; 30.00%). PdGATA6 from P. deltoides contains nine alternative splicing forms, which is the largest number. In addition, P. tremula (PtaGATA26) and P. trichocarpa (PtrGATA12) show seven, P. euphratica (PeGATA35) displays six, P. tremula x alba (PtaaGATA36) presents five, and P. tremuloides (PtsGATA3, 17, 22, 27, 30, 35, and 36) has two. Average numbers of alternative splicing forms of GATA genes range from 2.00 (P. tremuloides) to 3.43 (P. deltoides). These differences can be partially explained by that alternative splicing forms are controlled via multilayered regulatory network82, however, further studies are required.
Differences in the number of alternative splicing forms of GATA genes in Populus genus can be caused by (i) different gene prediction programs83–85 and (ii) amount of evidence transcript sequences, covering fully characterized genes, expressed sequences tags (ESTs), and/or RNA-Seq data. In the gene prediction process, evidence sequences are essential to achieve accurate prediction of genes as well as alternative splicing forms. More available EST or RNA-Seq sequences will bring plentiful alternative splicing forms of GATA genes: e.g., the human genome contains around averagely of 10 alternative splicing forms per one gene, predicted from a large amount of transcript sequences86. Available RNA-Seq data of Populus genus (As of 2018 Jun) deposited in NCBI Short Read Archive show that P. trichocarpa and P. tremula presenting a large proportion of alternative splicing forms of GATA genes contain a large amount of RNA-Seq data (Table S4).
In-depth investigations of alternative splicing forms of Populus GATA genes
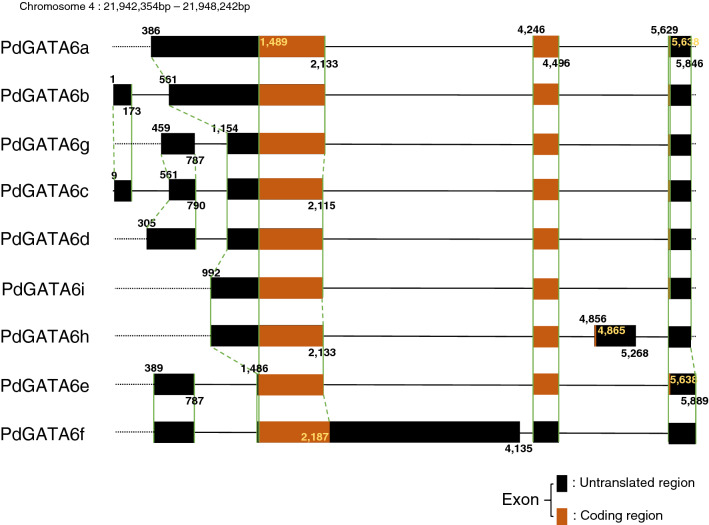
We identified the phenomena that some alternative splicing forms originated from one Populus GATA gene encode the same amino acids. Each of the nine alternative splicing forms of PdGATA6, a typical example of this phenomenon, is composed of 232 aa, 295 aa, and 301 aa in protein length and exon is composed of between 3 and 5. Among the nine alternative splicing forms of PdGATA6, all except PdGATA6f and 6h present the same start and end positions of ORFs. The first ORF exons of the eight alternative splicing forms except PdGATA6f contain start methionine without stop codon are classified into two types: one is 627 bp and the other is 645 bp. It results in two types of amino acid sequences from the eight alternative splicing forms, indicating that most of alternative splicing events are occurred in 5’ and 3’ UTR regions (Fig. 2). In addition, GATA domain sequences of eight alternative splicing forms of PdGATA6 are identical, suggesting that the GATA domain is important to bind DNA.
Figure 2.
Diagram of alternative splicing forms of PdGATA6. It shows the gene structure of the PdGATA6 gene (P. deltoides). Orange thick boxes indicate translated regions and black thick boxes display untranslated regions. Black thin lines mean intron and black dotted lines are intergenic regions. Green dotted and solid lines indicate the conserved and different structure of GATA genes including exon, intron, and, untranslated regions, respectively. The number around the boxes display relative positions of translated, untranslated, and exons based on the start position of PdGATA6b. Names of alternative splicing forms of the PdGATA6 gene are displayed in the left part of each gene diagram.
Interestingly, some of alternative splicing forms of Populus GATA genes display the same amino acids: one GATA gene from P. tremuloides, four from P. euphratica and P. deltoides, six from P. trichocarpa and P. tremula x alba, and eight from P. tremula. One of known roles of untranslated regions of messenger RNA is changing the amount of translated proteins87. The number of TFs will increase or decrease the transcription amount of target genes, so that these alternative splicing forms may be important to the regulatory network of GATA TFs.
We also identified that some alternative splicing forms of the twelve GATA genes of three Populus species (P. tremula x alba, P. tremula, and P. tremuloides; Table S5) missed GATA domain which was not included in the Populus GATA TFs list. Interestingly, GATA TFs without GATA domain can negatively regulate the target genes by competing with normal GATA TFs88, indicating that these twelve GATA genes containing alternative splicing forms without domain can also play a role of negative regulators. In addition, one Populus GATA gene, PtaGATA28 (from P. tremula), has five out of six alternative splicing forms that missed GATA domain, suggesting that this gene might have a dominant role of negative regulation in contrast to the normal GATA genes even though additional research such as expression level of each alternative splicing forms in various conditions are required. PtaGATA33 from P. tremula has two out of three, and the rest ten GATA genes have one.
Identification and investigation characteristics of Populus GATA subfamilies
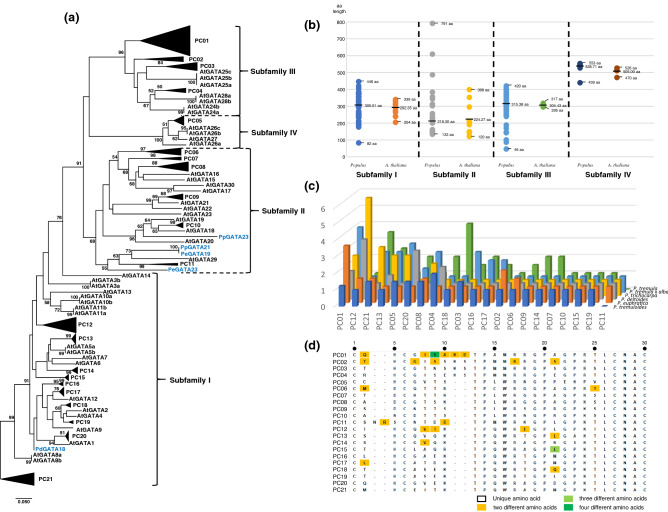
We constructed a neighbor-joining phylogenetic tree based on amino acid sequences of GATA domains of 389 Populus and 41 Arabidopsis GATA TFs together to identify subfamilies (Fig. 3a), resulting those four subfamilies (I to IV; see Material and Methods; Table S2) were successfully identified. Subfamily I has the largest number of Populus GATA genes; while subfamily IV contains the smallest number (Table S6) as same as A. thaliana25, V. vinifera89, and G. max40 except O. sativa81, monocot species. Subfamily III presents the largest average number of alternative splicing forms (1.81) and subfamily II displays the lowest (1.15; Table S6). GATA domains belonging to subfamilies I, II, and III are located adjacent to the C-terminal; however, those in subfamily IV are at the N-terminal as same as A. thaliana25, V. vinifera89, G. max40, and O. sativa81.
Figure 3.
Sequence characteristics of Populus GATA TFs. (a) shows that the phylogenetic tree of GATA domain sequences constructed by the neighbor-joining method. Black triangles on the tree indicate a group of Populus GATA domains. Names of the Populus GATA genes not condensed were displayed with blue colors. Bootstrap values calculated from 10,000 replicates are shown on the node except that those are lower than 50. The scale bar corresponds to 0.050 estimated amino acid substitutions per site. Ranges of subfamilies were presented with lines on the right side. (b) displays distribution of amino acid length of Populus and A. thaliana GATA TFs. The length distribution of GATA genes in each subfamily was plotted separated with the dotted lines. The Y-axis represents an amino acid length of GATA TF. Bold lines mean average length of GATA genes. Thin lines present maximum or minimum the length of GATA genes. (c) provides the information of conserved and diversity splicing forms of GATA genes among six Populus. The X-axis displays a list of PCs. Y-axis indicates Populus species name. Z-axis shows the ratio of the number of GATA TF per GATA gene. (d) presents the pattern of amino acid sequence of GATA motif (CX2-4CX18-20CX2C) along with Populus gene clusters (PCs). Yellow, light green, and dark green shaded alphabets mean two, three, and four different amino acids in that position, respectively.
Amino acid lengths of Populus GATA TFs in each subfamily present a wider range than those of A. thaliana (Fig. 3b). However, three Populus GATA TFs display extremely short lengths: PdGATA18 belonging to subfamily I is 82 aa, PtsGATA29 and PdGATA36 from subfamily III are 46 and 86 aa, respectively (Table S2). Interestingly, some of GATA TFs of the other plant species including A. thaliana also display the short GATA TFs: 120 aa (AtGATA23) in A. thaliana25, 109 aa (VvGATA13) in V. vinifera89, 80 aa (GmGATA10) in G. max40, and 101 aa (OsGATA8b) in O. sativa81. It indicates that the three shortest Populus GATA TFs may be functional GATA TFs, suspecting that the gene prediction program can miss some of exons nearby the exon containing the GATA domain.
Two Populus GATA TFs in subfamily II have unique domains in comparison to those of A. thaliana; PpGATA21 contains NIR domain (IPR005343) found in the Noc2 gene family in Arabidopsis. This domain seems to be involved in protein–protein interaction, indicating that PpGATA21 may have partner protein for forming protein complex to regulate target genes. In addition, PpGATA23 covers two HMA domains (IPR006121), which can bind heavy metal ions90. These two Populus GATA genes will have unknown additional functions like WC1, which is involved in circadian clock mechanism of Neurospora crassa with light-sensing domain91.
A. thaliana GATA TFs in subfamily III contain two known additional domains: CCT domain (IPR010402) found in circadian clock and a flowering control gene (CONSTANS92) and TIFY domain (IPR010399) to mediate homo- and heteromeric interactions between TIFY proteins and other specific TFs93, 94. In contrast to A. thaliana25 and G. max40, fourteen GATA TFs in six Populus species except P. euphratica lack CCT and/or TIFY domains. Some of GATA TFs of O. sativa also presented the same phenomenon81. In detail, nine of the 14 GATA TFs are the unique transcript, indicating that they completely lost CCT and/or TIFY domains during evolution, similar to those of V. vinifera89. The remaining five GATA TFs have other alternative splicing forms, presenting the selective loss of these domains.
Comparisons of the principal component analysis of seven Populus species
In a previous study, six Populus species except P. tremula x alba were used for conducting the phylogenetic analysis based on a total of 76 morphological properties for buds, leaves, inflorescences, flowers, and fruits95, which is congruent to the chloroplast phylogenetic tree (Fig. 1), except P. trichocarpa and P. deltoides. The incongruency of the two species is caused by limited species in Fig. 1. However, the principal component analysis of Populus GATA genes shows one cluster covering P. euphratica, P. tremula x alba, P. tremuloides, and P. trichocarpa (Fig. S2), which is incongruent to the aforementioned two phylogenetic trees. It reflects that GATA TFs may not evolve in the similar to species evolution due to their widely regulatory roles96.
Identification of Populus GATA gene clusters (PCs) on phylogenetic tree of Populus GATA genes
To understand the phylogenetic relationship of Populus GATA genes clearly, we clustered them in the phylogenetic tree (Fig. 3a), resulting in 21 distinct Populus GATA gene clusters (PCs; Fig. 3a and Table S7). All PCs except PC11 containing five species except P. tremuloides and P. tremula covers all seven Populus species, displaying conserveness of Populus GATA genes. Eleven of the 21 PCs (52.38%) contain the same amount of GATA genes for each Populus species, while the rest 9 PCs (42.86%) show different numbers (Table S8). PC03, PC05, and PC14 lack of only one GATA gene from P. pruinosa, P. tremula, and P. deltoides, respectively; while PC12 and PC20 have additional GATA gene of P. deltoides and P. euphratica, respectively. The remaining 4 PCs display a complex pattern of the number of GATA genes for each Populus species (Table S8), indicating complex history of gain and loss of GATA genes in the Populus genus.
Subfamily I, containing the largest Populus GATA genes, displays the most complex structure with the largest number of PCs (Fig. 3a). Interestingly, AtGATA3, AtGATA10, AtGATA11, AtGATA13, AtGATA14, and PC12 do not show neighbor GATA genes like an independent clade (Fig. 3a). Subfamily II shows the largest ratio of the number of PCs to the number of GATA genes, implying faster evolution might be occurred. In addition, four Populus GATA genes (PpGATA21, PpGATA23, PeGATA19, and PeGATA23) are not clustered into PCs (Fig. 3a), presenting species-specific GATA genes. In subfamily III, PC01, containing four Populus GATA genes per species, might be experienced a gene duplication event in comparison to the other PCs. In addition, PC01 and PC02 seem to be independent of three Arabidopsis GATA genes (Fig. 3a), suggesting Populus-specific GATA genes, while PC03 and PC04 have their partner Arabidopsis GATA genes (Fig. 3a). Subfamily IV covers only one PC and two Arabidopsis GATA genes, the smallest subfamily (Fig. 3a).
Among six Populus species except P. pruinosa, PCs that have a relatively high average of the ratio of the number of GATA TF per GATA gene are PC01 (2.58), PC12 (2.36), and PC21 (2.17) (Fig. S3), suggesting that GATA TFs in these PCs may have diverse biological functions, similar to the case of OsGATA2381. In the species level, PC01 in P. euphratica (3.50) and P. tremula x alba (4.00), PC12 in P. deltoides (3.67) and P. trichocarpa (6.00), PC04 in P. tremuloides (2.00), and PC18 in P. tremula (4.00) display the high ratio, while five PCs (PC07, PC10, PC11, PC15, and PC19) have one GATA TF per GATA gene (Fig. 3c). We suspected that GATA TFs of each Populus species might have dynamic features of their functional diversification, including subfunctionalization and neofunctionalization97–99.
In total, 23 out of 556 (4.14%) amino acid positions in the CX2-4CX18-20CX2C region (inter-species variations) show more than one amino acid (Fig. 3d), which is larger than that of 19 A. thaliana genomes43 (intraspecific variations; 0.93%). Positions of variable amino acids in the CX2-4CX18-20CX2C region are scattered throughout this region along with PCs (Fig. 3d). Most variable positions are 9th in PC01 (four amino acids) and 21st in PC15 (three amino acids; Fig. 3d); while the maximum number of different amino acids in Arabidopsis GATA genes is 243.
Genome-wide inference of GATA TF functions based on characterized A. thaliana GATA TFs
Till now, biological functions of one Populus80 and 15 A. thaliana GATA TFs have been characterized43. Nine of 15 characterized Arabidopsis GATA genes were also successfully mapped based on the PCs (Fig. 3a and Table 2) and similarity of amino acids, resulting in that seven PCs are related to the characterized Arabidopsis GATA genes. 129 Populus GATA TFs in the seven PCs are candidates for deducing their functional roles in Populus. In addition, PdGNC from P. nigra x (P. deltoides x P. nigra), a uniquely characterized GATA TF, known to regulate chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis80 is successfully mapped to PtrGATA19 (PC09) with 98.02% amino acid similarity. It supports this inference method because both GNC in Arabidopsis and PdGNC are in the same PC with the similar functions even though Arabidopsis and Populus belong to Brassicaceae and Salicaceae and are an herb and a tree species, respectively. Moreover, OsGATA12 involved in the seedling stage based on expression profile, is similar to that of BME3 in A. thaliana81. Based on this result, researchers can efficiently and systematically identify the biological functions of Populus GATA genes in the near future.
Table 2.
Function inference of Populus GATA gene clusters (PCs) based on characterized A. thaliana GATA TFs.
| PC name | GATA gene | Biological functions | Subfamily | References |
|---|---|---|---|---|
| PC03 | AtGATA25 | Hypocotyl and petiole elongation | III | 50 |
| PC04 | AtGATA28 (ZML2) | Mediation of cryptochrome1-dependent response | 133 | |
| PC08 | AtGATA15 (GATA15) | Cytokinin-regulated development, including greening, hypocotyl elongation, phyllotaxy, floral organ initiation, accessory meristem formation, flowering time, and senescence | II | 135 |
| AtGATA16 (GATA16) | ||||
| PC09 | AtGATA21 (GNC) | a nitrate‐inducible member important for chlorophyll synthesis and glucose sensitivity | 136 | |
| Modulation of chlorophyll biosynthesis (greening) and glutamate synthase (GLU1/Fd-GOGAT) expression | 137, 138 | |||
| Downstream effectors of floral homeotic gene action by controlling two MADS-box TFs | 139 | |||
| Control of convergence of auxin and gibberellin signaling | 140, 141 | |||
| Control of greening, cold tolerance, and flowering time | 142 | |||
| Regulation of chloroplast development, growth, and division as well as photosynthetic activities | 143, 144 | |||
| Cytokinin-regulated development, including greening, hypocotyl elongation, phyllotaxy, floral organ initiation, accessory meristem formation, flowering time, and senescence | 135 | |||
| PIF- and light-regulated stomata formation in hypocotyls | 145 | |||
| PC10 | AtGATA18 (HAN) | Regulation of shoot apical meristem and flower development | 145–149 | |
| Stable establishment of cotyledon identity during embryogenesis | 148 | |||
| Position the proembryo boundary in the early Arabidopsis embryo | 149 | |||
| PC19 | AtGATA2 (GATA-2) | Regulation of light-responsive genes | I | 45 |
| AtGATA4 (GATA-4) | ||||
| PC20 | AtGATA1 (GATA-1) |
Expression level analysis of GATA genes in P. deltoides and P. pruinosa
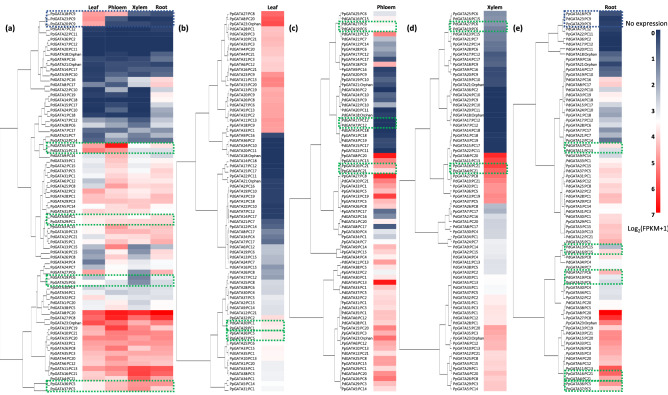
Based on available RNA-Seq raw reads obtained from leaf, phloem, xylem, and root tissues of P. deltoides and P. pruinosa (Table S9), expression levels of Populus GATA TFs were calculated (Fig. 4). In the four tissues, three GATA genes in the PC9 were well clustered displaying leaf and phloem specific expressions (Fig. 4a), which is congruent to the putative functions of GATA genes in PC9, such as regulation of chloroplast development, growth, and divisions (Table 2). In addition, four clusters covering GATA genes in PC13, PC1, PC6, and PC5, also presented similar expression patterns across the tissues, but these clusters did not cover all members in each PC. PC1 and PC5 showed high expression in all four tissues; while PC13 was leaf and phloem specific and PC6 was expressed lowly, especially in xylem, which can be a clue to understand their biological functions due to lack of homologous genes of which biological functions were characterized. Expression profiles of each tissue exhibited that some clustered GATA genes from the same PC were same as those in the four tissues and the rest were not (Fig. 4b–e), showing that expression level of Populus GATA genes partially reflects their conserveness across the species.
Figure 4.
Heatmap of GATA genes in four tissues of P. deltoides and P. pruinosa. Dendrograms at the left side of heatmaps are the result of hierarchical clustering of each expression data using hclust function in R package stats version 4.0.3. Heatmaps present expression levels based on FPKM values calculated by cufflink with gradient colors from blue to red displayed in the legend on the right side. Dendrograms at the left side of heatmaps are the result of hierarchical clustering of each expression data. Labels consist of GATA gene and PC names separated with ‘:’. Green dotted boxes indicate the case that some of members in the same PC were clustered. Blue dotted boxes mean that all members in the PC were clustered together. (a) displays heatmap of GATA genes of two Populus species in the four tissues, (b)–(e) are for heatmaps in leaf, phloem, xylem, root tissues.
Domain types of Populus GATA genes
The DNA-binding motif of GATA TFs was classified into three types designated as type IVa (CX2CX17CX2C), IVb (CX2CX18CX2C), and IVc (CX2CX20CX2C) among which Type IVb and IVc are common in plants25, 40, 81, 89. Additional types, including type IVp (p indicates partial; mentioned in Arabidopsis43 and G. max GATA TF analyses25) and type IV4 (four amino acids between the first two cysteines, which seems to be functionally active43), were also found.
In Populus GATA TFs, type IVb is the most abundant (272 of 389; 69.92%), and type IVc is the second (102; 26.22%). Type IVb, a common type of DNA-binding motifs of plant GATA TFs, occupies the largest proportion in Populus GATA TFs and is found in subfamilies I, II, and IV and Type IVc, the second largest, is a common in subfamily III of Populus GATA TFs, which is similar to those of A. thaliana, G. max, V. vinifera, and O. sativa. Type IV4 is found only in eight Populus GATA TFs (2.06%) belonging to subfamily II. It was also identified in some species including A. thaliana and G. max, suggesting that this type was independently occurred during evolution by adding two amino acids only in the first two cysteines of subfamily II. In contrast, type IVp is found in all subfamilies of Populus GATA TFs as well as those of A. thaliana, G. max, and O. sativa, suggesting that random modifications of GATA domains of all subfamilies have been occurred during evolution. Interestingly, type IVp can be generated by alternative splicing forms: PtaaGATA36d is GATA TF displaying type IVp; while the rest four alternative splicing forms of PtaaGATA36 show type IVc domain, which is similar to the cases of OsGATA881 and AtGATA26. Type IVp was also considered as an ancestral form of GATA zinc finger40, requiring more research of Type IVp with additional GATA genes from many plant genomes.
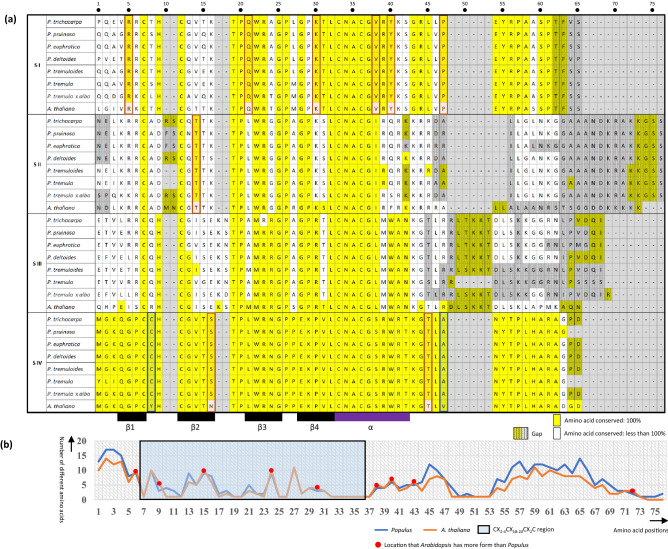
Amino acid patterns of GATA domains in seven Populus species and A. thaliana
Amino acid sequences of 422 GATA domain from 382 Populus and 40 A. thaliana GATA TFs excluding seven type IVp domains of Populus and one Arabidopsis were used for multiple sequence alignment (Fig. 5a). Subfamily IV of Populus displays the most conserved manner (49 of 55 conserved amino acids are identical.) in their domains, while subfamily II of Populus shows the least conserveness (18 of 66 conserved amino acids). Considering with the number of Populus GATA genes in each subfamily, subfamily I, the largest subfamily, is more conserved than subfamilies II and III. Some of dominant amino acids in GATA domain are different among Populus species (e.g., the 5’ region of GATA domains; Fig. 5a).
Figure 5.
Conserved amino acids of GATA domain along with subfamilies and genera (A. thaliana and Populus). (a) shows the conserved amino acid in each position of the GATA domain. Yellow shaded characters mean 100% conserved amino acids and gray background color in amino acid indicate that there are gaps in the position. S I, S II, S III, and S IV are shortened forms of subfamily I, II, III, and IV, respectively. Blue-colored transparent boxes show incongruent of conserved amino acids between the two species. Red-colored transparent boxes present amino acids which are 100% conserved in the Populus genus but not in A. thaliana. (b) The X-axis indicates each amino acid position of the aligned amino acids of GATA domain and Y-axis displays the number of different amino acids in the specific position of aligned GATA domain of Populus (blue line) and A. thaliana (orange line). Red dots on the graph mean position where A. thaliana has more different amino acids than that of Populus. The blue-colored transparent box presents the CX2CX18-20CX2C region.
Subfamily IV displays incongruent of conserved amino acids between Populus and A. thaliana at 8th and 47th (Fig. 5a; blue-colored transparent boxes), indicating that subfamily IV was evolved and stabilized in early stage. In addition, six, one, and two amino acids which are 100% conserved in Populus genus but not in A. thaliana are found in subfamilies I, II, and IV, respectively (Fig. 5a; red-colored transparent boxes), suggesting that subfamily I has been most diversified in the lineage of A. thaliana.
Twelve conserved amino acids of Populus and A. thaliana belonging to the zinc finger motif (CX2-4CX18-20CX2C) are identical in all subfamilies (Fig. 5a) suggesting that the zinc finger motif is the most conserved and important region in the GATA domain. 22nd and 28th conserved amino acids in subfamilies I, II, and IV are Tryptophan and Glycine, respectively; while subfamily III displays methionine and glutamic acid (Fig. 5a). These differences can be key factors to classify four subfamilies.
As expected, the zinc finger motif, which can bind to DNA and is the most important region in GATA domain, contains a smaller number of different amino acids (Fig. 5b). Despite of large number of species in Populus genus used in this study, four positions in this region show a high number of amino acids in A. thaliana (Fig. 5b), suggesting that selection pressures have been differently applied in the two lineages. This phenomenon is also found outside this region (Fig. 5b). It is congruent to the findings described in the previous paragraph. Once more plant genomes including the large number of resequencing data of A. thaliana and P. trichocarpa are analyzed, the detailed evolutionary history of the GATA domain will be uncovered.
Identification of transmembrane helix (TMH) of GATA gene family in seven Populus genomes
Membrane-bound transcription factors (MTFs) are docked in cellular membranes using their transmembrane domains100. MTFs have usually been found in plant species101 of which are related to seed germination102, cell division101, heat stress103, and salt stress104. Mechanisms of MTFs are well known in two major plant TF families: NAC TF family and bZIP TF family105 (Fig. S4). NTL6 (NAC TF) of A. thaliana is localized in the plasma membrane under normal conditions; while under stress conditions, NTL6 is processed by an as-yet-unidentified intramembrane protease and SnRK2.8 kinase phosphorylates NTL6 and facilitates its nuclear import105. (ii) Intracellular movement of Arabidopsis bZIP60 and bZIP28 was characterized105. bZIP60 and bZIP28, which are other MTFs in Arabidopsis, were localized on the membrane of the endoplasmic reticulum and then transported to nucleus by cleaving TMH.
Five Populus GATA TFs were identified with TMHs predicted by TMHMM106. PtrGATA14b, 14c (P. trichocarpa) in subfamily I, PpGATA21, 25 (P. pruinosa), and PtaaGATA23 (P. tremula x alba; Table S10), belonging to subfamily II. These putative GATA MTFs have one TMH, which is the same as the previously characterized Arabidopsis MTFs107. As far as we know, this is the first time to report putative GATA MTFs in plant species; additional putative GATA MTFs were also identified in other plant species using our pipeline: VvGATA19 (V. vinifera; subfamily IV)89, GmGATA39 (G. max; subfamily I)40, OsGATA14 (O. sativa; subfamily II)81 with one TMH, while no GATA MTF was found in A. thaliana25. It is interesting that there are no common subfamilies containing putative GATA MTFs along with different species. In addition, some alternative splicing forms of PtrGATA14 (P. trichocarpa) have TMH, indicating that truncation of TMH region by alternative splicing may switch their functions by changing subcellular localization of GATA TFs, similar to the bZIP60 in A. thaliana105. With accumulating more data including expression profiles and subcellular localization, roles of these putative GATA MTFs can be uncovered. Moreover, these results can be a corner stone to understand plant GATA MTFs together with a large number of plant genomes available now108–110 in a broad taxonomic range of plant species.
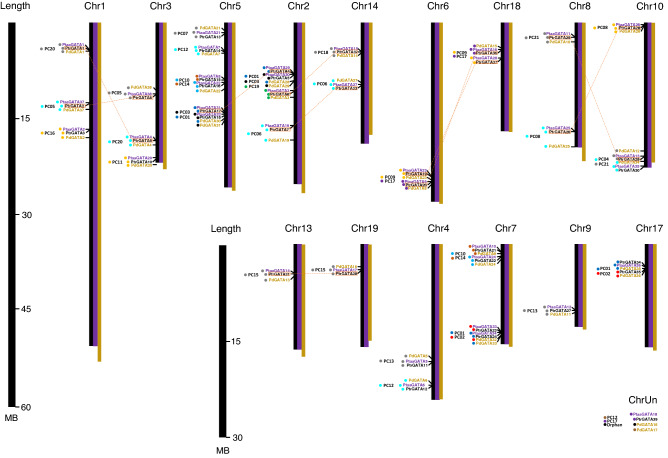
Chromosomal distribution of P. trichocarpa, P. tremula x alba, and P. deltoides GATA gene family
Chromosomal distribution of Populus GATA genes from the three species, P. trichocarpa, P. tremula x alba, and P. deltoides belonging to the same clade (Fig. 6), presents several important features: (i) 38 of 39 GATA genes in P. trichocarpa, 37 out of 38 in P. tremula x alba, and 36 of 38 in P. deltoides were distributed on 15 of 19 chromosomes (Fig. 6), presenting similar chromosomal distribution among three species. (ii) Chromosome 5 in both species contains the largest number of GATA genes; while chromosomes 9, 13, and 19 in both species contain the smallest (Fig. 6). This biased chromosomal distribution was also found in many plant species including A. thaliana25, V. vinifera89, G. max40, O. sativa81. (iii) In the three species, chromosome 7 shows the highest density of GATA genes in both species; and chromosome 5 is a second rank. (iv) Most of GATA genes of three species are in the same PCs and in similar chromosomal position (Fig. 6) except the four genes of which chromosomal positions are not assigned (See ChrUn in Fig. 6). It indicates that there might be no chromosomal rearrangement events and biological functions of GATA genes may have similar functions among the three species. Interestingly, three of the four genes are additional copy of GATA TFs in PC12 and PC17, which is the result of independent gene gain events.
Figure 6.
Chromosomal distribution of P. trichocarpa, P. tremula x alba, and P. deltoides GATA genes. Black, purple, and yellow bars indicate P. trichocarpa, P. tremula x alba, and P. deltoides chromosomes, respectively. Black, purple, yellow letters mean GATA gene names of P. trichocarpa, P. tremula x alba, and P. deltoides. ChrUn present scaffold sequences which are not assigned to any chromosomes. Yellow-colored transparent boxes indicate 10 paralogous pairs.
Based on paralogous GATA TFs of P. trichocarpa identified in the previous study111, 10 paralogous pairs were successfully mapped to GATA TFs from three Populus species (Fig. 6), displaying that all paralogous pairs contain three GATA TF from each species in the similar chromosomal positions, indicating that gene duplication events of the 10 paralogous pairs were occurred before speciation of three Populus species (see yellow star in Fig. 1a).
In addition, three PCs, PC14 (PtrGATA15 and PtaaGATA8 in chromosome 5), PC02 (PtrGATA35 and PdGATA36 in chromosome 17), and PC09 (PtrGATA37 and PtaaGATA28 in chromosome 18), have not complete set of Populus GATA TFs. PC14 and PC09 suggest the loss event of GATA genes in the lineage of P. deltoides (see the light blue line in Fig. 1). PC02 indicates another loss event occurred in the lineage of P. tremuloides (See green lines in Fig. 1). Taken together with the incongruency inferred from the PCA, GATA TFs were evolved with the events occurred in various lineages in Populus species, which is independent to their species evolution.
Conclusion
Using the identification pipeline of GATA TFs in the GATA-TFDB, we successfully identified 262 GATA genes (389 GATA TFs) from seven Populus species. Alternative splicing forms of Populus GATA genes display the high number of alternative splicing forms (nine is maximum) with only changes in untranslated regions and loss of DNA-binding motif or additional domains. Populus GATA genes were classified into the four subfamilies, same to Arabidopsis GATA genes, except that some genes in subfamily III lack CCT and/or TIFY domains. 21 Populus GATA gene clusters (PCs) were identified from the phylogenetic tree of GATA domain sequences and 20 of 21 PCs cover the seven Populus species, displaying the conserveness of Populus GATA genes. Distribution of alternative splicing forms in the PCs exhibits the possibility of subfunctionalization and neofunctionalization of Populus GATA genes. Through the expression analysis of GATA genes of two Populus species, the five PCs which display similar expression patterns across the four tissues were identified for predicting their biological functions. Populus-specific conserved amino acids in the GATA domain were discovered in comparison to A. thaliana, suggesting a complex evolutionary history of the GATA domain. Five Populus GATA TFs contain one transmembrane helix (TMH), which is the first report of membrane-bound GATA TFs. Together with the biased distribution of GATA genes across the chromosomes, paralogous pairs of GATA genes suggested several gene duplication events in the lineages of Populus genus. Taken together, our first comprehensive analyses of genus-wide GATA TFs in plants successfully provide characteristics of Populus GATA genes across the seven species as well as their putative functions and evolutionary traits of Populus GATA genes.
Materials and methods
Collecting Populus genome sequences from various sources
We utilized the seven Populus genomes sequences deposited from the Populus Comparative Genome Database108–110 (http://www.populusgenome.info/), which adopted-whole genome sequences from the Plant Genome Database (http://www.plantgenome.info/; Park et al., in preparation). These genomes were originated from the NCBI genome database (http://genome.ncbi.nlm.nih.gov/) and Phytozome (http://www.phyotozome.info/)112 in the standardized form of genome sequences provided by the GenomeArchive® (http://www.genomearchive.info/)113.
Identifying GATA TFs from whole Populus genome sequences
Amino acid sequences from seven Populus genomes were subjected to InterProScan114 to identify GATA TFs. The pipeline for identifying Populus GATA TFs implemented at the GATA-TFDB (http://gata.genefamily.info/; Park et al., in preparation), which is an automated pipeline for identifying GATA TFs with GATA DNA-binding motif InterPro term (IPR000679) and post-process to filter-out false positive results and for analyzing various analyses including domain sequence analysis, gene family analysis, as well as phylogenetic analysis. GATA-TFDB was constructed and maintained as one of members of the Gene Family Database (http://www.genefamily.info/; Park et al., in preparation).
Exon structure and alternative splicing forms of Populus GATA TFs
Based on the Populus Comparative Genome Database (http://www.populusgenome.info/; Park et al., in preparation), exon structure and alternative splicing forms of GATA TFs were retrieved. Diagrams of exon structure and alternative splicing forms of GATA TFs were drawn primarily based on the diagram generated by the GATA-TFDB (http://gata.genefamily.info; Park et al., in preparation) with adding additional information manually.
Construction of phylogenetic tree of Populus GATA TFs
Phylogenetic trees were constructed with a Neighbor-joining method with bootstrap option (10,000 repeats) by ClustalW 2.1115 based on the alignment of amino acids of GATA domains obtained from the GATA-TFDB (http://gata.genefamily.info; Park et al., in preparation) also by ClustalW 2.1115.
Chromosomal distribution of Populus GATA TFs
We drew the chromosomal distribution map of Populus GATA genes from three species based on the chromosomal coordination from their pseudo-molecule level assemblies deposited in the Plant Genome Database (http://www.plantgenome.info/).
Prediction of transmembrane helixes on Populus GATA TFs
Transmembrane helixes on Populus GATA TFs were predicted by TMHMM106 under the environment of the Plant Genome Database (http://www.plantgenome.info/).
Principal component analysis of Populus GATA TFs
Principal component analysis (PCA) was conducted based on 19 characteristics of GATA genes using prcomp function in R package stats version 4.0.3116. The result was visualized into a scatterplot including variance, with the first two principal components.
Expression analysis of GATA TFs based on Populus RNA-seq data
Raw reads of RNA-Seq experiments of P. pruinosa and P. deltoides were downloaded from NCBI (Table S9). RNA-Seq raw reads were aligned against the whole genome of P. pruinosa and P. deltoides with hisat2 v2.2.0117 after generating datasets of each Populus genome. After generating bam file for each SRA raw reads, bam files from the same experiments were merged using samtools v1.9118. Expression levels of the merged bam files were calculated by cufflink v2.2.1119.
Hierarchical clustering was conducted for the five datasets: one covers four different conditions (four tissues) and the last four contain each condition (Fig. 4) using hclust function in R package stats version 4.0.3116.
Construction of phylogenetic tree of seven Populus species based on complete chloroplast genomes
Complete chloroplast genomes of seven Populus chloroplast genomes69, 120–123 and Salix gracilistyla78, used as an outgroup, were aligned using MAFFT v7.450124. All chloroplast genome sequences were retrieved from the PCD (http://www.cp-genome.net; Park et al., in preparation). The maximum-likelihood trees were reconstructed in MEGA X125. During the ML analysis, a heuristic search was used with nearest-neighbor interchange branch swapping, the Tamura-Nei model, and uniform rates among sites. All other options were set to their default values. Bootstrap analyses with 1,000 pseudoreplicates were conducted with the same options. All bioinformatic processes were conducted under the environment of the Genome Information System (GeIS) used in the various previous studies43, 126–134.
Supplementary Information
Acknowledgements
This study was supported by InfoBoss Research Grant (IBB-001) to JP.
Author contributions
J.P. designed this manuscript and M.K. and H.X. identified and GATA TFs and analyzed them. M.K. curated GATA TFs. M.K., S.P., and Y.Y. visualized the analyzed data. M.K. and J.P. wrote the original manuscript and all authors improved the manuscript. All authors read and approved the final draft of the manuscript.
Data availability
All GATA TFs identified in this study can be accessed at the Populus Comparative Genome Database (http://www.populusgenome.info/).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-95940-5.
References
- 1.Singh KB, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002;5:430–436. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, et al. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell. 2013;112:107854. doi: 10.1105/tpc.112.107854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics. 1995;140:345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gendron JM, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. 2012;109:3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos LA, de Souza SR, Fernandes MS. OsDof25 expression alters carbon and nitrogen metabolism in Arabidopsis under high N-supply. Plant Biotechnol. Rep. 2012;6:327–337. doi: 10.1007/s11816-012-0227-2. [DOI] [Google Scholar]
- 6.Jensen MK, Skriver K. NAC transcription factor gene regulatory and protein–protein interaction networks in plant stress responses and senescence. IUBMB Life. 2014;66:156–166. doi: 10.1002/iub.1256. [DOI] [PubMed] [Google Scholar]
- 7.Du J, Miura E, Robischon M, Martinez C, Groover A. The Populus Class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS ONE. 2011;6:e17458. doi: 10.1371/journal.pone.0017458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang B, Jiang Y, Rahman MH, Deyholos MK, Kav NN. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 2009;9:1–19. doi: 10.1186/1471-2229-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramírez V, et al. Drought tolerance in Arabidopsis is controlled by the OCP3 disease resistance regulator. Plant J. 2009;58:578–591. doi: 10.1111/j.1365-313X.2009.03804.x. [DOI] [PubMed] [Google Scholar]
- 10.Riechmann JL, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 11.Jin, J. et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic acids research, gkw982 (2016). [DOI] [PMC free article] [PubMed]
- 12.Xiong Y, et al. Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol. Biol. 2005;59:191–203. doi: 10.1007/s11103-005-6503-6. [DOI] [PubMed] [Google Scholar]
- 13.Pérez-Rodríguez P, et al. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010;38:D822–D827. doi: 10.1093/nar/gkp805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mochida K, et al. In silico analysis of transcription factor repertoire and prediction of stress responsive transcription factors in soybean. DNA Res. 2009;16:353–369. doi: 10.1093/dnares/dsp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao W, et al. Transcriptome analysis of transcription factor genes under multiple abiotic stresses in Populus simonii× P. nigra. Gene. 2019;707:189–197. doi: 10.1016/j.gene.2019.04.071. [DOI] [PubMed] [Google Scholar]
- 16.Saleh A. Plant AP2/ERF transcription factors. Genetika. 2003;35:37–50. doi: 10.2298/GENSR0301037S. [DOI] [Google Scholar]
- 17.Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA)-Gene Regulat. Mech. 2012;1819:97–103. doi: 10.1016/j.bbagrm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 19.Xu K, et al. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 2015;15:141. doi: 10.1186/s12870-015-0532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian C, Wan P, Sun S, Li J, Chen M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 2004;54:519–532. doi: 10.1023/B:PLAN.0000038256.89809.57. [DOI] [PubMed] [Google Scholar]
- 21.Cowles, M. W. et al. Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development, dev. 098616 (2013). [DOI] [PubMed]
- 22.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakoby M, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 24.Scazzocchio C. The fungal GATA factors. Curr. Opin. Microbiol. 2000;3:126–131. doi: 10.1016/S1369-5274(00)00063-1. [DOI] [PubMed] [Google Scholar]
- 25.Reyes JC, Muro-Pastor MI, Florencio FJ. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004;134:1718–1732. doi: 10.1104/pp.103.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon MC. Gotta have GATA. Nat. Genet. 1995;11:9. doi: 10.1038/ng0995-9. [DOI] [PubMed] [Google Scholar]
- 27.Park J-S, et al. A comparative genome-wide analysis of GATA transcription factors in fungi. Genom. Inf. 2006;4:147–160. [Google Scholar]
- 28.Ooka H, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 29.Nuruzzaman M, et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Moyano E, et al. Genome-wide analysis of the NAC transcription factor family and their expression during the development and ripening of the Fragaria× ananassa fruits. PLoS ONE. 2018;13:e0196953. doi: 10.1371/journal.pone.0196953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J, et al. Genome wide analysis of the NAC transcription factor family in Chinese cabbage to elucidate responses to temperature stress. Sci. Hortic. 2014;165:82–90. doi: 10.1016/j.scienta.2013.11.005. [DOI] [Google Scholar]
- 32.Carretero-Paulet L, et al. Genome wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss and algae. Plant Physiol. 2010;110:153593. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song X-M, et al. Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis) Mol. Genet. Genom. 2014;289:77–91. doi: 10.1007/s00438-013-0791-3. [DOI] [PubMed] [Google Scholar]
- 34.Li X, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei K, et al. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012;19:463–476. doi: 10.1093/dnares/dss026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baloglu MC, Eldem V, Hajyzadeh M, Unver T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE. 2014;9:e96014. doi: 10.1371/journal.pone.0096014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu W, et al. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016;6:22783. doi: 10.1038/srep22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song X-M, et al. Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica rapa ssp. pekinensis) Genomics. 2014;103:135–146. doi: 10.1016/j.ygeno.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Lu J, Wang T, Xu Z, Sun L, Zhang Q. Genome-wide analysis of the GRAS gene family in Prunus mume. Mol. Genet. Genom. 2015;290:303–317. doi: 10.1007/s00438-014-0918-1. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C, et al. Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress. PLoS ONE. 2015;10:e0125174. doi: 10.1371/journal.pone.0125174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H, et al. Genome-wide identification, evolution, and expression analysis of GATA transcription factors in apple (Malus× domestica Borkh) Gene. 2017;627:460–472. doi: 10.1016/j.gene.2017.06.049. [DOI] [PubMed] [Google Scholar]
- 42.Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim M, Xi H, Park J. Genome-wide comparative analyses of GATA transcription factors among 19 Arabidopsis ecotype genomes: Intraspecific characteristics of GATA transcription factors. PLoS ONE. 2021;16:e0252181. doi: 10.1371/journal.pone.0252181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teakle GR, Manfield IW, Graham JF, Gilmartin PM. Arabidopsis thaliana GATA factors: organisation, expression and DNA-binding characteristics. Plant Mol. Biol. 2002;50:43–56. doi: 10.1023/A:1016062325584. [DOI] [PubMed] [Google Scholar]
- 46.Hudson D, et al. Rice cytokinin GATA transcription Factor1 regulates chloroplast development and plant architecture. Plant Physiol. 2013;162:132–144. doi: 10.1104/pp.113.217265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An, Y. et al. A GATA transcription factor PdGNC plays an important role in photosynthesis and growth in Populus. J. Exp. Botany (2020). [DOI] [PMC free article] [PubMed]
- 48.Shapira M, et al. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu PP, Koizuka N, Martin RC, Nonogaki H. The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. 2005;44:960–971. doi: 10.1111/j.1365-313X.2005.02588.x. [DOI] [PubMed] [Google Scholar]
- 50.Shikata M, et al. Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 2004;55:631–639. doi: 10.1093/jxb/erh078. [DOI] [PubMed] [Google Scholar]
- 51.Shaikhali J, et al. The Cryptochrome1-dependent response to excess light is mediated through the transcriptional activators Zinc Finger protein expressed in inflorescence meristem like1 and ZML2 in Arabidopsis. Plant Cell. 2012;24:3009–3025. doi: 10.1105/tpc.112.100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, Z. et al. Genome-wide identification and analysis of the evolution and expression patterns of the GATA transcription factors in three species of Gossypium genus. Gene (2018). [DOI] [PubMed]
- 53.Ao T, Liao X, Xu W, Liu A. Identification and characterization of GATA gene family in Castor Bean (Ricinus communis) Plant Diver. Resour. 2015;37:453–462. [Google Scholar]
- 54.Yuan Q, Zhang C, Zhao T, Yao M, Xu X. A genome-wide analysis of GATA transcription factor family in tomato and analysis of expression patterns. Int. J. Agric. Biol. 2018;20:1274–1282. [Google Scholar]
- 55.Apuli R-P, et al. Inferring the genomic landscape of recombination rate variation in European aspen (Populus tremula) G3: Genes Genom. Genet. 2020;10:299–309. doi: 10.1534/g3.119.400504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, W., Guo, Y., Chen, Y., Wu, D. & Jiang, L. Genome-Wide Identification and Characterization of GATA Family Genes in Brassica Napus. (2020). [DOI] [PMC free article] [PubMed]
- 57.Huang, Q., Shi, M., Wang, C., Hu, J. & Kai, G. Genome-wide Survey of the GATA Gene Family in Camptothecin-producing Plant Ophiorrhiza Pumila. (2021). [DOI] [PMC free article] [PubMed]
- 58.Zhang Z, et al. Characterization of the GATA gene family in Vitis vinifera: genome-wide analysis, expression profiles, and involvement in light and phytohormone response. Genome. 2018;61:713–723. doi: 10.1139/gen-2018-0042. [DOI] [PubMed] [Google Scholar]
- 59.Liu, H. et al. TaZIM‐A1 negatively regulates flowering time in common wheat (Triticum aestivum L.). Journal of integrative plant biology (2018). [DOI] [PubMed]
- 60.Wang, T. et al. Genome-wide analysis of GATA factors in moso bamboo (Phyllostachys edulis) unveils that PeGATAs regulate shoot rapid-growth and rhizome development. bioRxiv, 744003 (2019).
- 61.Qi Y, Chunli Z, Tingting Z, Xiangyang X. Bioinformatics analysis of GATA transcription factor in pepper. Chin. Agric. Sci. Bull. 2017;2017:5. [Google Scholar]
- 62.Jiang L, Yu X, Chen D, Feng H, Li J. Identification, phylogenetic evolution and expression analysis of GATA transcription factor family in maize (Zea mays) Int. J. Agric. Biol. 2020;23:637–643. [Google Scholar]
- 63.Yu, R. et al. Genome-wide identification of the GATA gene family in potato (Solanum tuberosum L.) and expression analysis. J. Plant Biochem. Biotechnol. pp. 1–12 (2021).
- 64.Yu C, et al. Genome-wide identification and function characterization of GATA transcription factors during development and in response to abiotic stresses and hormone treatments in pepper. J. Appl. Genet. 2021;62:265–280. doi: 10.1007/s13353-021-00618-3. [DOI] [PubMed] [Google Scholar]
- 65.Peng W, et al. Genome-wide characterization, evolution, and expression profile analysis of GATA transcription factors in Brachypodium distachyon. Int. J. Mol. Sci. 2021;22:2026. doi: 10.3390/ijms22042026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niu L, et al. The GATA gene family in chickpea: structure analysis and transcriptional responses to abscisic acid and dehydration treatments revealed potential genes involved in drought adaptation. J. Plant Growth Regul. 2020;39:1647–1660. doi: 10.1007/s00344-020-10201-5. [DOI] [Google Scholar]
- 67.Zhuang J, et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008;371:468–474. doi: 10.1016/j.bbrc.2008.04.087. [DOI] [PubMed] [Google Scholar]
- 68.Bradshaw H, Ceulemans R, Davis J, Stettler R. Emerging model systems in plant biology: poplar (Populus) as a model forest tree. J. Plant Growth Regul. 2000;19:306–313. doi: 10.1007/s003440000030. [DOI] [Google Scholar]
- 69.Tuskan GA, et al. The genome of black cottonwood Populus trichocarpa (Torr & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 70.Yang W, et al. The draft genome sequence of a desert tree Populus pruinosa. Gigascience. 2017;6:gix75. doi: 10.1093/gigascience/gix075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma T, et al. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013;4:1–9. doi: 10.1038/ncomms3797. [DOI] [PubMed] [Google Scholar]
- 72.Tuskan GA, et al. Hardwood tree genomics: Unlocking woody plant biology. Front. Plant Sci. 2018;9:1799. doi: 10.3389/fpls.2018.01799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin Y-C, et al. Functional and evolutionary genomic inferences in Populus through genome and population sequencing of American and European aspen. Proc. Natl. Acad. Sci. 2018;115:E10970–E10978. doi: 10.1073/pnas.1801437115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schiffthaler, B. et al. An improved genome assembly of the European aspen Populus tremula. bioRxiv, 805614 (2019).
- 75.Xue L-J, Alabady MS, Mohebbi M, Tsai C-J. Exploiting genome variation to improve next-generation sequencing data analysis and genome editing efficiency in Populus tremula× alba 717–1B4. Tree Genet. Genom. 2015;11:1–8. doi: 10.1007/s11295-015-0907-5. [DOI] [Google Scholar]
- 76.Wang M, et al. Phylogenomics of the genus Populus reveals extensive interspecific gene flow and balancing selection. New Phytol. 2020;225:1370–1382. doi: 10.1111/nph.16215. [DOI] [PubMed] [Google Scholar]
- 77.Wang Z, et al. Phylogeny reconstruction and hybrid analysis of Populus (Salicaceae) based on nucleotide sequences of multiple single-copy nuclear genes and plastid fragments. PLoS ONE. 2014;9:103645. doi: 10.1371/journal.pone.0103645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zong D, et al. Comparative analysis of the complete chloroplast genomes of seven Populus species: Insights into alternative female parents of Populus tomentosa. PLoS ONE. 2019;14:0218455. doi: 10.1371/journal.pone.0218455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen J-H, et al. Genome-wide analysis of Cushion willow provides insights into alpine plant divergence in a biodiversity hotspot. Nat. Commun. 2019;10:1–12. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.An Y, Han X, Tang S, Xia X, Yin W. Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ Culture (PCTOC) 2014;119:313–327. doi: 10.1007/s11240-014-0536-y. [DOI] [Google Scholar]
- 81.Gupta P, Nutan KK, Singla-Pareek SL, Pareek A. Abiotic stresses cause differential regulation of alternative splice forms of GATA transcription factor in rice. Front. Plant Sci. 2017;8:1944. doi: 10.3389/fpls.2017.01944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han H, et al. Multilayered control of alternative splicing regulatory networks by transcription factors. Mol. Cell. 2017;65:539–553. doi: 10.1016/j.molcel.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 83.Alioto, T., Picardi, E., Guigó, R. & Pesole, G. ASPic-GeneID: A lightweight pipeline for gene prediction and alternative isoforms detection. BioMed Res. Int.2013 (2013). [DOI] [PMC free article] [PubMed]
- 84.Foissac S, Schiex T. Integrating alternative splicing detection into gene prediction. BMC Bioinf. 2005;6:25. doi: 10.1186/1471-2105-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keller O, Kollmar M, Stanke M, Waack S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics. 2011;27:757–763. doi: 10.1093/bioinformatics/btr010. [DOI] [PubMed] [Google Scholar]
- 86.Flicek P, et al. Ensembl 2011. Nucleic Acids Res. 2010;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zwicky R, Müntener K, Csucs G, Goldring MB, Baici A. Exploring the role of 5’alternative splicing and of the 3’-untranslated region of cathepsin B mRNA. Biol. Chem. 2003;384:1007–1018. doi: 10.1515/BC.2003.113. [DOI] [PubMed] [Google Scholar]
- 88.Kelemen O, et al. Function of alternative splicing. Gene. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang, Z. et al. Characterization of GATA gene family in Vitis vinifera: genome-wide analysis, expression profiles, and involvement in light and phytohormone response. Genome (2018). [DOI] [PubMed]
- 90.Bull PC, Cox DW. Wilson disease and Menkes disease: new handles on heavy-metal transport. Trends Genet. 1994;10:246–252. doi: 10.1016/0168-9525(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 91.Ballario P, et al. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 1996;15:1650–1657. doi: 10.1002/j.1460-2075.1996.tb00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suárez-López P, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 93.Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R. The ZIM domain mediates homo-and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009;59:77–87. doi: 10.1111/j.1365-313X.2009.03852.x. [DOI] [PubMed] [Google Scholar]
- 94.Melotto M, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine-and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55:979–988. doi: 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eckenwalder JE. Systematics and evolution of Populus. Biol. Populus Implications Manage. Conservation. 1996;7:32. [Google Scholar]
- 96.Xu Z, Casaretto JA, Bi YM, Rothstein SJ. Genome-wide binding analysis of AtGNC and AtCGA1 demonstrates their cross-regulation and common and specific functions. Plant Direct. 2017;1:00016. doi: 10.1002/pld3.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Higo A, et al. Transcription factor DUO1 generated by neo-functionalization is associated with evolution of sperm differentiation in plants. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-018-07728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vandenbussche M, Theissen G, Van de Peer Y, Gerats T. Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 2003;31:4401–4409. doi: 10.1093/nar/gkg642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yao S, Deng L, Zeng K. Genome-wide in silico identification of membrane-bound transcription factors in plant species. PeerJ. 2017;5:e4051. doi: 10.7717/peerj.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim Y-S, et al. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell. 2006;18:3132–3144. doi: 10.1105/tpc.106.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park J, et al. Integration of auxin and salt signals by a NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 2011;111:177071. doi: 10.1104/pp.111.177071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gao H, Brandizzi F, Benning C, Larkin RM. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 2008;105:16398–16403. doi: 10.1073/pnas.0808463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim SG, Lee AK, Yoon HK, Park CM. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 2008;55:77–88. doi: 10.1111/j.1365-313X.2008.03493.x. [DOI] [PubMed] [Google Scholar]
- 105.Seo PJ. Recent advances in plant membrane-bound transcription factor research: Emphasis on intracellular movement. J. Integr. Plant Biol. 2014;56:334–342. doi: 10.1111/jipb.12139. [DOI] [PubMed] [Google Scholar]
- 106.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 107.Tajima H, Iwata Y, Iwano M, Takayama S, Koizumi N. Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem. Biophys. Res. Commun. 2008;374:242–247. doi: 10.1016/j.bbrc.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 108.Park J, Xi H, Yongsung K. Plant genome database release 2.5: A standardized plant genome repository for 233 species. Plant Animal Genome. 2018 doi: 10.13140/RG.2.2.22162.68801. [DOI] [Google Scholar]
- 109.Park, J., Yongsung, K. & Xi, H. Plant Genome Database: An integrated platform for plant genomes. doi:10.13140/RG.2.2.18807.24484 (2017).
- 110.Park, J., Min, J., Kim, Y. & Chung, Y. The Comparative Analyses of Six Complete Chloroplast Genomes of Morphologically Diverse Chenopodium album L.(Amaranthaceae) Collected in Korea. International Journal of Genomics2021 (2021). [DOI] [PMC free article] [PubMed]
- 111.An Y, et al. The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 2020;71:1969–1984. doi: 10.1093/jxb/erz564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goodstein DM, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Park, J. & Xi, H. GenomeArchive(R): A standardized whole genome database. 10.13140/RG.2.2.27092.22408 (2018).
- 114.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thompson, J. D., Gibson, T. J. & Higgins, D. G. Multiple sequence alignment using ClustalW and ClustalX. Current Protocols Bioinf., 2.3. 1–2.3. 22 (2003). [DOI] [PubMed]
- 116.Team, R. C. R: A language and environment for statistical computing. (2020).
- 117.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zong D, et al. Plastome sequences help to resolve deep-level relationships of Populus in the family Salicaceae. Front. Plant Sci. 2019;10:5. doi: 10.3389/fpls.2019.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang Q-J, Gao L-Z. The complete chloroplast genome sequence of desert poplar (Populus euphratica) Mitochondrial DNA Part A. 2016;27:721–723. doi: 10.3109/19401736.2014.913159. [DOI] [PubMed] [Google Scholar]
- 122.Gai Z-S, et al. Complete chloroplast genome sequence of Populus pruinosa Schrenk from PacBio Sequel II Platform. Mitochondrial DNA Part B. 2020;5:3452–3454. doi: 10.1080/23802359.2020.1824593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kersten B, et al. Genome sequences of Populus tremula chloroplast and mitochondrion: implications for holistic poplar breeding. PLoS ONE. 2016;11:e0147209. doi: 10.1371/journal.pone.0147209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Park J, Xi H, Oh S-H. Comparative chloroplast genomics and phylogenetic analysis of the Viburnum dilatatum complex (Adoxaceae) in Korea. Korean J. Plant Taxonomy. 2020;50:8–16. doi: 10.11110/kjpt.2020.50.1.8. [DOI] [Google Scholar]
- 127.Park J, Xi H, Kim Y. The complete mitochondrial genome of Arabidopsis thaliana (Brassicaceae) isolated in Korea. Korean J. Plant Taxonomy. 2021;51:176–180. doi: 10.11110/kjpt.2021.51.2.176. [DOI] [Google Scholar]
- 128.Yoo S-C, Oh S-H, Park J. Phylogenetic position of Daphne genkwa (Thymelaeaceae) inferred from complete chloroplast data. Korean J. Plant Taxonomy. 2021;51:171–175. doi: 10.11110/kjpt.2021.51.2.171. [DOI] [Google Scholar]
- 129.Choi, N. J., Xi, H. & Park, J. A Comparative Analyses of the Complete Mitochondrial Genomes of Fungal Endosymbionts in Sogatella furcifera, White-Backed Planthoppers. Int. J. Genom.2021 (2021). [DOI] [PMC free article] [PubMed]
- 130.Park, J., Min, J., Kim, Y. & Chung, Y. The comparative analyses of six complete chloroplast genomes of morphologically diverse Chenopodium album L. (Amaranthaceae) collected in Korea. Int. J. Genom.2021 (2021). [DOI] [PMC free article] [PubMed]
- 131.Park, J., Xi, H. & Kim, Y. The complete chloroplast genome of Arabidopsis thaliana isolated in Korea (Brassicaceae): an investigation of intraspecific variations of the chloroplast genome of Korean A. thaliana. Int. J. Genom.2020 (2020). [DOI] [PMC free article] [PubMed]
- 132.Lee J, Park J, Xi H, Park J. Comprehensive analyses of the complete mitochondrial genome of Figulus binodulus (Coleoptera: Lucanidae) J. Insect Sci. 2020;20:10. doi: 10.1093/jisesa/ieaa090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Park, J, & Xi, H. Investigation of nucleotide diversity based on 17 sea cucumber mitochondrial genomes and assessment of sea cucumber mitochondrial gene markers. Ad Oceanogr & Marine Biol.2(5). 10.13140/AOMB.MS.ID.000547 (2021).
- 134.Park, J., Xi, H., Kim, Y. & Kim, M. Complete genome sequence of lentilactobacillus parabuchneri strain KEM. Microbiol. Resour. Announc., 10(20), e01208-20 (2021). [DOI] [PMC free article] [PubMed]
- 135.Ranftl QL, Bastakis E, Klermund C, Schwechheimer C. LLM-domain containing B-GATA factors control different aspects of cytokinin-regulated development in Arabidopsis thaliana. Plant Physiol. 2016;170:2295–2311. doi: 10.1104/pp.15.01556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bi YM, et al. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005;44:680–692. doi: 10.1111/j.1365-313X.2005.02568.x. [DOI] [PubMed] [Google Scholar]
- 137.Hudson D, et al. GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE. 2011;6:e26765. doi: 10.1371/journal.pone.0026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bastakis E, Hedtke B, Klermund C, Grimm B, Schwechheimer C. LLM-domain B-GATA transcription factors play multifaceted roles in controlling greening in Arabidopsis. Plant Cell. 2018;30:582–599. doi: 10.1105/tpc.17.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mara CD, Irish VF. Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol. 2008;147:707–718. doi: 10.1104/pp.107.115634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Richter R, Behringer C, Zourelidou M, Schwechheimer C. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 2013;110:13192–13197. doi: 10.1073/pnas.1304250110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Richter R, Behringer C, Müller IK, Schwechheimer C. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from della proteins and phytochrome-interacting factors. Genes Dev. 2010;24:2093–2104. doi: 10.1101/gad.594910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Richter R, Bastakis E, Schwechheimer C. Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol. 2013;162:1992–2004. doi: 10.1104/pp.113.219238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chiang Y-H, et al. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012;160:332–348. doi: 10.1104/pp.112.198705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zubo YO, et al. Coordination of chloroplast development through the action of the GNC and GLK transcription factor families. Plant Physiol. 2018;178:130–147. doi: 10.1104/pp.18.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Klermund C, et al. LLM-domain B-GATA transcription factors promote stomatal development downstream of light signaling pathways in Arabidopsis thaliana hypocotyls. Plant Cell. 2016;28:646–660. doi: 10.1105/tpc.15.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao Y, et al. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell. 2004;16:2586–2600. doi: 10.1105/tpc.104.024869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang X, et al. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell. 2013;25:83–101. doi: 10.1105/tpc.112.107854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kanei M, Horiguchi G, Tsukaya H. Stable establishment of cotyledon identity during embryogenesis in Arabidopsis by ANGUSTIFOLIA3 and HANABA TARANU. Development. 2012;139:2436–2446. doi: 10.1242/dev.081547. [DOI] [PubMed] [Google Scholar]
- 149.Nawy T, et al. The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Dev. Cell. 2010;19:103–113. doi: 10.1016/j.devcel.2010.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All GATA TFs identified in this study can be accessed at the Populus Comparative Genome Database (http://www.populusgenome.info/).