Abstract
Alien invasive plants pose a threat to global biodiversity and the cost of control continues to rise. Early detection and prediction of potential risk areas are essential to minimize ecological and socio-economic costs. In this study, the Maxent model was used to predict current and future climatic conditions to estimate the potential global distribution of the invasive plant Xanthium italicum. The model consists of 366 occurrence records (10 repeats, 75% for calibration and 25% for verification) and 10 climate prediction variables. According to the model forecast, the distribution of X. italicum was expected to shrink in future climate scenarios with human intervention, which may be mainly caused by the rise in global average annual temperature. The ROC curve showed that the AUC values of the training set and the test set are 0.965 and 0.906, respectively, indicating that the prediction result of this model was excellent. The contribution rates of annual mean temperature, monthly mean diurnal temperature range, standard deviation of temperature seasonal change and annual average precipitation to the geographical distribution of X. italicum were 65.3%, 11.2%, 9.0%, and 7.7%, respectively, and the total contribution rate was 93.2%. These four variables are the dominant environmental factors affecting the potential distribution of X. italicum, and the influence of temperature is greater than that of precipitation. Through our study on the potential distribution prediction of X. italicum under the future climatic conditions, it has contribution for all countries to strengthen its monitoring, prevention and control, including early warning.
Subject terms: Biological techniques, Environmental sciences
Introduction
With the rapid development of economic globalization, international trade exchanges and human activities are becoming more and more frequent, alien invasive species are expanding at an unprecedented rate1–3. The resulting ecological problems and increasing risks of biological invasion have caused huge eco-environmental hazards and economic losses all over the world4,5. The biological invasion has become a great challenge for human beings. Xanthium italicum Moretti is one of the most widely distributed invasive plants in the world, and its biological characteristics of easily carrying seeds make it more likely to spread widely6. X. italicum is an annual herbaceous plant of the genus Xanthium in the Compositae family, propagated by seeds7, and native to North America but has been spread to many countries and regions in Europe, Asia, and Oceania8,9. Previous studies shown that X. italicum seeds have been found in old crawler excavators imported from Japan, rapeseed seeds from Mongolia, and soybeans imported from Brazil, Argentina and the United States10–12. Because of its strong ecological adaptability, X. italicum can grow in wasteland, fields, river beaches and ditch roadsides, and plants grow more luxuriantly and taller in wetland, irrigated land and ditches. X. italicum has high plant coverage and strong competitiveness, it can compete water, nutrition, light and growth space with native species, to form a dominant population in the habitats13. Moreover, X. italicum also has strong communicative power, and the involucre bears dense barbs, which can easily be attached to livestock and poultry, wild animals, agricultural machinery, seeds and packaging of agricultural by-products for long-distance transmission9. It caused serious harm to agriculture, animal husbandry and biodiversity in the invaded areas14. It has been reported that the X. italicum caused 43% and 90% of the yield loss after introducing sunflower and corn crops to agricultural production areas, respectively15,16. In some farming and pastoral areas, the large and prickly fruits of X. italicum are often carried by the fur of some livestock, which not only caused the further spread of X. italicum, but also reduced the quality of products such as wool for sale17. X. italicum has strong phenotypic plasticity to different environmental conditions, showing great invasive potential18. A clear understanding of the potential distribution area of X. italicum under global natural conditions has great significance for its monitoring, prevention and control, and early warning.
The niche model uses the well-known distribution data of species and relevant environmental variables, constructs the model in accordance with a deep learning algorithm, induces or simulates the ecological needs of species, and projects the calculation results at different time and space to predict the potential distribution of species19,20. At present, the niche models to predict species potential distribution areas mainly include the Genetic Algorithm for the Rule Set Production (GARP), Maximum Entropy (MAXENT), Match Climates Regional Algorithm (CLIMEX), Climatic Envelope (BIOCLIM)21–24. Each model has a different theoretical basis, data requirements and analysis methods. Most studies show that, compared with other niche models, the Maxent model not only has good prediction effect and stability but also has the advantages of simple and fast operation, and small sample demand, so it has become an ideal prediction tool for many scholars25–28. It builds a prediction model based on the actual distribution points and environmental variables of the distribution area stored in GIS, and to simulate the potential distribution of species in the target space. The output is a thematic map reflecting the suitability of the relative distribution of species29,30. Maxent model has been widely used in the field of species distribution research31,32, such as the potential distribution of Haloxylon persicum in Central Asia under global warming33, the prediction of the suitable growth zone of Rhinopithecus roxellana due to the sharp reduction of human disturbance34, and the distribution of five economic tree species in the Amazon River basin combined with remote sensing technology35. Besides, Maxent niche models have also been used to study the potential distribution of invasive species in recent years, such as Solidago canadensis, Ageratina adenophora, Mimosa pigra, Flaveria bidentis, Solenopsis invicta and Ambrosia artemisiifolia36–41.
As the prevention and control of X. italicum is an extremely time-consuming and costly project, the prediction and simulation of the potential distribution area of X. italicum is highly important for the future prevention and control management. In this paper, the distribution record points of X. italicum were determined by consulting the literature and GBIF website. In combination with the relevant environmental variables of X. italicum invaded area, taking the SSP245 scenario with human intervention as forecasting background, the global potential distribution area of X. italicum in current and future was predicted with the help of Maxent model tools to provide theoretical support for decision-makers to formulate corresponding prevention and control management measures.
Materials and methods
Distribution data
The foreign distribution data of X. italicum were downloaded from the Global Biodiversity Information Facility (GBIF) http://data.gbif.org/welcome.htm, while the data of China were mainly collected from literature and publications7,13. We queried the specific coordinates of the place in Geonames website (http://www.geonames.org/), deleted the duplicate and invalid distribution points, and finally got 336 effective distribution points. Detail coordinate point information was included in the supplementary material Table S1, and the species names and longitude and latitude of X. italicum distribution data were recorded in the Excel file as .csv format, in which the east longitude and north latitude were marked as positive values, and the west longitude and south latitude were marked as negative values.
Selection and treatment of environmental variables
The sixth IPCC assessment report publishes four climate change scenarios, namely, SSP126 scenario, SSP245 scenario, SSP370 scenario, and SSP585 scenario. We chose the SSP245 scenario, where greenhouse gas emissions are about the same as current condition (1970–2000) and the global average temperature tends to reduce with human intervention. 19 environmental-climate factors derived from the WorldClim environmental database (http://www.worldclim.org/), with a spatial resolution of 5 km (Table 1).
Table 1.
Environmental variables extracted from current X. italicum habitats.
| Code | Variables description | Code | Variables description |
|---|---|---|---|
| Bio1 | Annual mean temperature | Bio11 | Mean temperature of coldest quarter |
| Bio2 | Mean diurnal range | Bio12 | Annual precipitation |
| Bio3 | Isothermality | Bio13 | Precipitation of wettest month |
| Bio4 | Temperature seasonality | Bio14 | Precipitation of driest month |
| Bio5 | Max temperature of warmest month | Bio15 | Precipitation seasonality |
| Bio6 | Min temperature of coldest month | Bio16 | Precipitation of wettest quarter |
| Bio7 | Temperature annual range | Bio17 | Precipitation of driest quarter |
| Bio8 | Mean temperature of wettest quarter | Bio18 | Precipitation of warmest quarter |
| Bio9 | Mean temperature of driest quarter | Bio19 | Precipitation of coldest quarter |
| Bio10 | Mean temperature of warmest quarter |
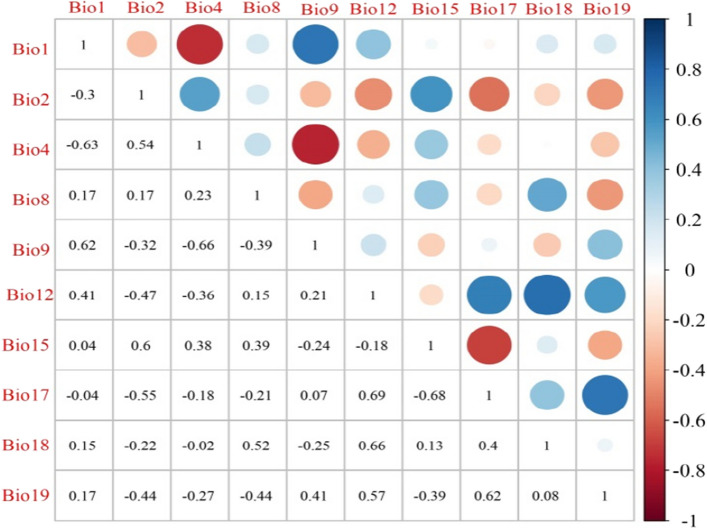
Environmental variable data is an important parameter for constructing a niche model, and using too many environmental variables to construct the model will enhance the spatial correlation between variables, cause over-fitting, and then reduce the transferability of the model. On the contrary, choosing the moderate and reasonable environmental variables can significantly improve the prediction ability of the model42,43. Therefore, it is necessary to calculate the correlation of environmental variables and exclude the high correlation environmental variables.Therefore, we used Pearson correlation analysis of R v4.0.2 software44 to screen 10 environmental variables whose correlation is less than 0.8 (Fig. 1, Table 2) and set them as environmental parameters into Maxent software45.
Figure 1.
Pearson correlation analysis of various environment variables (r < 0.8). Created in R 4.0.2 (https://www.r-project.org/).
Table 2.
Retained environmental variables used in this study.
| Code | Variables description |
|---|---|
| Bio1 | Annual mean temperature |
| Bio2 | Mean diurnal range |
| Bio4 | Temperature seasonality |
| Bio8 | Mean temperature of wettest quarter |
| Bio9 | Mean temperature of driest quarter |
| Bio12 | Annual precipitation |
| Bio15 | Precipitation seasonality |
| Bio17 | Precipitation of driest quarter |
| Bio18 | Precipitation of warmest quarter |
| Bio19 | Precipitation of coldest quarter |
Map data and software sources
The global vector map comes from the website of the National Fundamental Geographic Information System (http://nfgis.nsdi.gov.en); the Maxent software version 3.3.3 k (http://www.Cs.princeton.edu/-schapire/maxent/); ArcGIS spatial technology platform is a complete set of GIS products developed by ESRI company in the United States, and the ArcGIS software version is 10.2.246. Species current distribution was again overlaid on the Köppen–Geiger climatic classification system (1976–2000)47.
Data processing
Import distribution data and environment data into Maxent v3.3.3, randomly select 25% of the distribution points as testing data, the remaining 75% as training data, and the model was trained for 10 repetition48. In the environment parameter settings, we used the Jackknife method, and other parameter settings followed software default values. The LHQPT feature classes were selected in this model and the regularization multiplier was set to 1.049. The ENMeval package was used to calculate the corrected Akaike information criterion correction value (AICc value) under these parameters, and the “checkerboard2” method was used to calculate the AICc value50. The output was a grid layer of ASCII format, and the value of each grid in the layer represented the adaptation of X. italicum to the environment in that area, with a range of 0 to 1. We loaded the calculation result of Maxent in ArcGIS10.2.2 software for visual expression51.
Model accuracy test
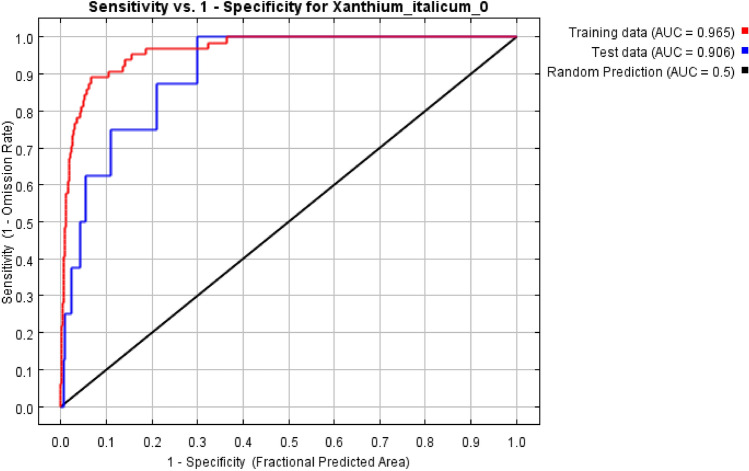
The receiver operating characteristic curve (ROC) analysis method was used to test the accuracy of this model. The ROC curve takes the true positive rate as the ordinate (the ratio that exists and is predicted to exist) and the false positive rate (the ratio that does not exist but is predicted to exist) as the abscissa. The AUC value is the area enclosed by the abscissa and ROC curve, and the range is 0–1. The larger AUC value is, the farther the distance from the random distribution is, the greater correlation between environmental variables and predicted geographical distribution of species is, and the better prediction effect of this model is. On the contrary, it means that the prediction effect of the model is worse. The AUC value of 0.5–0.6 means the simulation effect of this model is failed; 0.6–0.7 means the simulation effect is poor; 0.7–0.8 means the simulation effect is average; 0.8–0.9 means the simulation effect is good; 0.9–1 means the simulation effect is perfect52,53. Moreover, three other approaches also have performed in the supplementary table S2 to test the performance of Maxent model, which includes kappa coefficient (K), Normalized Mutual Information (NMI) n(s) and True Skill Statistic (TSS)30,54,55.
Ethics approval
This paper evaluates published data and does not need specific ethics approvals.
Consent to participate
Human subjects were not involved in this study.
Consent for publication
Human subjects were not involved in this study.
Results
Global potential distribution prediction of X. italicum
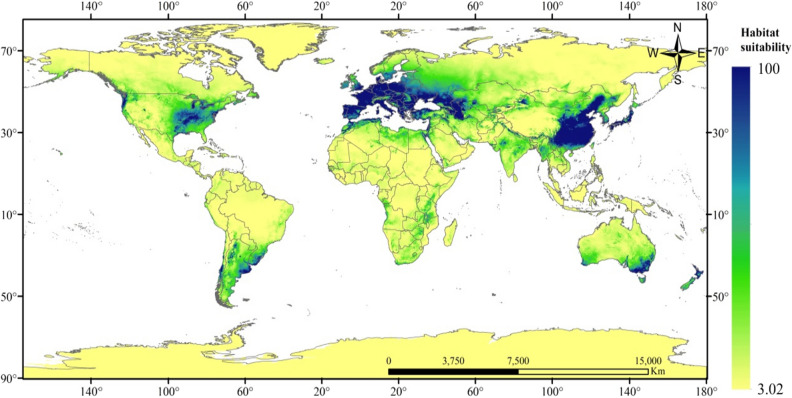
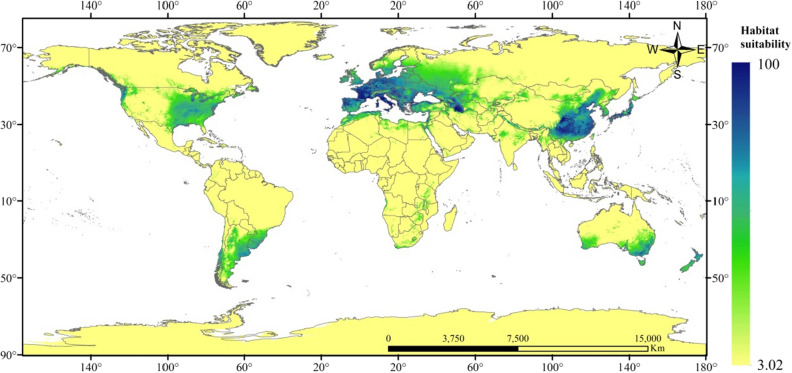
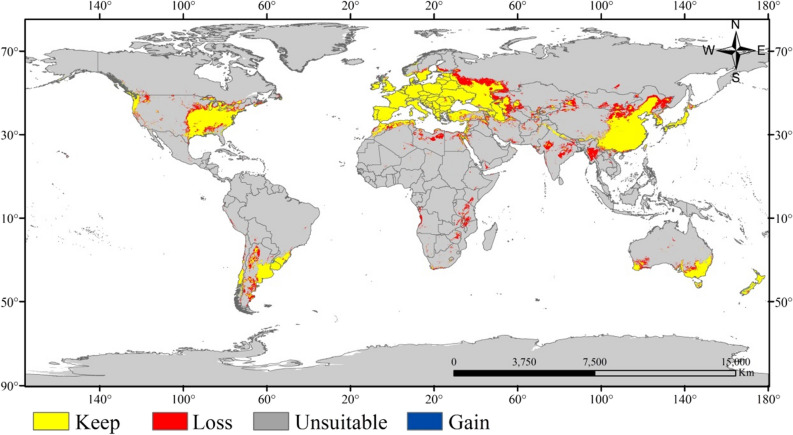
In the base climate (1970–2000), the suitable growth area prediction result of X. italicum was shown in Fig. 2. In the SSP245 scenario (2050), the potential distribution prediction of X. italicum was shown in Figs. 3 and 4 showed the change of suitable growth area between these two maps. As it can be seen from Fig. 4, the global suitable area of X. italicum in the future (17.8650 million km2) is less than that the present (19.4315 million km2), which relatively reduces the pressure on the ecological environment in some areas, but its invasion potential cannot be ignored. Therefore, it is extremely unwise to relax the early warning of X. italicum. Except for Antarctica, there are suitable zones for X. italicum on all continents.
Figure 2.
Current potential binary distribution of X. italicum under normal climate (1970–2000) according to the simulations. Created in ESRI ArcMap 10.2.2 (https://support.esri.com/en/Products/Desktop/arcgis-desktop/arcmap/10-2-2#downloads).
Figure 3.
Future potential binary distribution of X. italicum under SSP245 scenarios (2050) according to the simulations. Created in ESRI ArcMap 10.2.2 (https://support.esri.com/en/Products/Desktop/arcgis-desktop/arcmap/10-2-2#downloads).
Figure 4.
Changes in the potential habitat suitability of X. italicum. Created in ESRI ArcMap 10.2.2 (https://support.esri.com/en/Products/Desktop/arcgis-desktop/arcmap/10-2-2#downloads).
The suitable growth area of X. italicum is mostly concentrated in the north–south temperate zone, rarely distributed in the tropics and no distribution in frigid zones. From the potential distribution range of X. italicum, it prefers temperate marine climate, Mediterranean climate, and temperate continental monsoon climate, while the distribution area in temperate continental climate is obviously reduced. So moisture may be a key factor affecting the potential distribution of X. italicum.
Accuracy evaluation of Maxent model
The final ROC curve of this model was shown in Fig. 5. In this study, the AUC values of training data and verification data are 0.965 and 0.906 respectively, indicating that the Maxent model is very effective in predicting the global potential distribution of X. italicum. The predicted geographical distribution results and the actual distribution have a high degree of overlap between the regions of X. italicum, which means the results can be applied to the suitable regionalization of this species.
Figure 5.
Receiver operating characteristic (ROC) curve and AUC values of the MaxEnt model. The red curve indicates training data, the blue curve indicates test data, and the black line indicates random prediction.
Weight analysis of dominant environmental variables
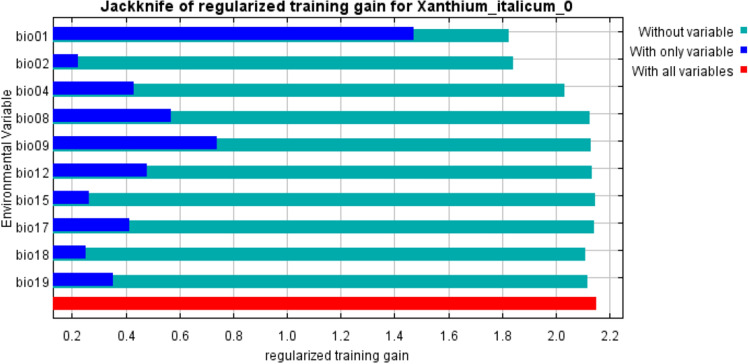
Jackknife method was used in the Maxent model, and the results can show the weight of different environmental factors affecting the habitat suitability of X. italicum (Fig. 6). The vertical axis represents the screened environmental variables, and the horizontal axis represents the score of each environmental variable. The dark blue column represents the model score with only this environmental factor exists, and the light blue column represents the sum of the scores of other variables except for this variable, and red represents the sum of all variables scores. As can be seen from Table 3, the main environmental factors affecting the potential distribution of X. italicum are annual mean temperature, monthly mean diurnal temperature range, the standard deviation of temperature seasonal change and annual average precipitation. The contribution rates are 65.3%, 11.2%, 9.0% and 7.7% respectively, and the total contribution rate is 93.2%. The annual mean temperature has the highest contribution rate, which indicates that the annual mean temperature is the most important factor affecting the potential distribution of X. italicum.
Figure 6.
Important analysis of environmental variables based on Jackknife tests.
Table 3.
The percentage contribution of ten climatic variables for X. italicum based on Jackknife tests.
| Variable | Contribution (%) |
|---|---|
| (Bio1) Annual mean temperature | 65.3 |
| (Bio2) Mean diurnal range | 11.2 |
| (Bio4) Temperature seasonality | 9.0 |
| (Bio12) Annual precipitation | 7.7 |
| (Bio19) Precipitation of coldest quarter | 2.7 |
| (Bio9) Mean temperature of driest quarter | 1.7 |
| (Bio18) Precipitation of warmest quarter | 1.5 |
| (Bio17) Precipitation of driest quarter | 0.5 |
| (Bio8) Mean temperature of wettest quarter | 0.4 |
| (Bio15) Precipitation seasonality | 0 |
Discussion
The Global Invasive Species Program (GISP) has proposed that preventing the invasion of alien species is more feasible and economical than controlling its outbreak56. Therefore, it is quite importance to understand the climate-driven changes on the potential distribution and range shifts of this plant for better planning and decision-making of control and management actions. Using all available global occurrence data and reliable species responsive environmental predictors, we demonstrated that the overall potential suitability for X. italicum will contract in the future under SSP245 scenarios relative to the current climate (Fig. 4). The simulation results showed that the suitable growth range of X. italicum is very wide in the world, and it is mostly concentrated in the north–south temperate zone. Although the potentially suitable zone of X. italicum in 2050 is less than the current predicted distribution range, it still has great potential for invasion. Some countries, such as China, South Korea, Japan, the United States, Australia, Angola, Namibia, the Republic of Mozambique, Kazakhstan and European countries all have high suitable zones of X. italicum, which has a very high risk of invasion. This was consistent with the information that has been reported and various countries government should attach great importance to this species7,57.
Distribution and modelling results can also be influenced by other intrinsic factor (dispersal distance and rate of the species, and its generation time) and extrinsic factor (human activity and natural enemy)50. According to the analysis of the variable contribution and the Jackknife test (Table 3; Fig. 6), the temperature and precipitation were found to be a significantly important variable in determining the distribution of this plant. Climate, particularly temperature, is the main factor that controls the distribution of biological invaders on earth58. Among bioclimatic variables, the main environmental factors that have a great influence on the distribution of X. italicum are annual mean temperature (65.3%), monthly mean diurnal temperature range (11.2%), the standard deviation of temperature seasonal change (9.0%) and annual average precipitation (7.7%). The first three were all environmental factors related to temperature, and the contribution percentage of annual average precipitation is only the fourth, which showed that the contribution rate of temperature factor to the geographical distribution of X. italicum was greater than that of precipitation factor. This was consistent with the results of literature reports and field investigation, X. italicum can grow in wetlands, farmland, deserts and other environments with great differences in soil moisture, its relative anti-waterlogging, and drought-resistant growth characteristics lead to the contribution rate7,9,18. Whether temperature can be used to slow down or stop the trend of rapid invasion of X. italicum remains to be verified by further research.
Based on the SSP245 scenario, we predicted the global suitable growth region of X. italicum in 2050. Among these four climate scenarios, the SSP245 scenario is the closest scenario to reality. The predicted reduction of potential geographical distribution may be due to the increase of global annual mean temperature with human intervention because the annual mean temperature accounts for a very high percentage of all ecological factors, and the temperature factor has a significant effect on the growth of X. italicum. Although the global total suitable area of X. italicum has decreased, the suitable area of temperate maritime climate, Mediterranean climate, and temperate continental monsoon climate have not increased. It indicated that the contribution percentage of annual average precipitation to X. italicum distribution is not high, but it still has a certain impact on X. italicum growth. A possible explanation is that the regions with a temperate maritime climate, Mediterranean climate, and temperate continental monsoon climate are relatively humid, and the temperature of humid air is more stable than that of dry air59. Therefore, the annual mean temperature does not decrease significantly in the above-mentioned areas, so did not cause a change in the suitable area of X. italicum. Our habitat suitability maps indicate a highly conducive environment for X. italicum, these areas are predicted to be found rather steady under future climate scenarios as well (Fig. 5).
In recent years, ROC curve analysis has been widely used in the evaluation of species potential distribution prediction models, especially in invasive species60–63. In this paper, the performance of the MaxEnt model was revealed by the ROC curve, whereas the accuracy of the prediction mapping (current and future) was determined by the percentage contribution and jackknife test. Additionally, classification accuracy measures, such as K, NMI and TSS, were in agreement with Bhandari30. Further, it concurs about the sample size of the studied species to determine the training and testing of geo-coordinates used for prediction mapping (Supplementary material Table S2). Thus, Our results showed that the simulation effect of this model is very good, and can accurately simulate the global distribution of X. italicum. However, the premise of applying a niche model is to assume that species niche demand is conservative, but the niche drift of invasive species sometimes occurs, there may produce a deviation in the potential distribution of X. italicum after niche drift64,65. Moreover, because alien species usually spread from one or several locations of the earliest invasion, in a specific time, the ecological characteristics of alien invasive species in the introduction area are difficult to reflect the complete ecological needs of species. Using the non-equilibrium distribution data of the intrusion site, especially the early distribution data, there will be an error in the prediction of suitable growth areas4,66. It may be more accurate to use the equilibrium distribution data of invasive regions to predict other suitable growth areas. So, the demography of this species requires us to follow up the invasion area, but it may not be allowed in some countries. At present, many countries pay less attention to X. italicum, and the report of this species is not detailed enough. Therefore, it is important to strengthen the quarantine control on the importation of commodities, especially of transport vehicles and goods at potential donor spots (i.e., border control/border biosecurity system), to decrease further risks of this biological invader.
Invasive plants respond to the changing climate i.e., increases in temperatures and CO2 levels, changes in precipitation67. Thus, long-term changes in the climate can have potential influences on the distribution of X. italicum habitats due to the limitation of suitable climate conditions. At present, this plant need immediate and ongoing control and management measures as the current predicted potential ranges are very high. This study provides insights for decision-makers that climate change influences on potential distribution of invasive species should be considered for long-term effective management of this species. Our results also provide detail information relevant to potentially suitable areas of this invasive plant in current and future under climate change. Defining likely spread areas and recognizing the pattern of invasion in the future are important components of climate change-integrated short-term and long-term conservation management strategies. The study has mapped the areas potentially suitable for the distribution of X. italicum across the worldwide under current and future climate scenarios. Administrative managers can use these maps for identifying high-risk areas and thus to prioritize conservation actions to those areas. Further, this information is useful to them for future surveying and monitoring efforts, and designing conservation strategies and management plans.
Supplementary Information
Author contributions
G.R., K.Z. and X.W. downloaded and analyzed the data, Y.Z. wrote the manuscript and J.T. submitted and revised the article.
Funding
This work was supported by the National Natural Science Foundation of China (no. 62072157) and the Natural Science Foundation of Henan Province (no. 202300410102).
Data availability
Data from the current study are available from the corresponding author upon reasonable request.
Code availability
R script is available from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yang Zhang and Jieshi Tang.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-96041-z.
References
- 1.Jane AC, Peter AV, David MR, Petr P. Quantifying levels of biological invasion: Towards the objective classification of invaded and invasible ecosystems. Glob. Change Biol. 2012;18:44–62. doi: 10.1111/j.1365-2486.2011.02549.x. [DOI] [Google Scholar]
- 2.Peter S, Linda P, Heikki H, Colin L, Claudia AOT, Celia VH. The hidden faces of a biological invasion: Parasite dynamics of invaders and natives. Int. J. Parasitol. 2020;50:111–123. doi: 10.1016/j.ijpara.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Ren GQ, et al. Resource conservation strategy helps explain patterns of biological invasion in a low-N environment. Biochem. Sys. Ecol. 2021;94:104–205. [Google Scholar]
- 4.Dufour CMS, Clark DL, Herrel A, Losos JB. Recent biological invasion shapes species recognition and aggressive behaviour in a native species: A behavioural experiment using robots in the field. J. Anim. Ecol. 2020;89:1604–1614. doi: 10.1111/1365-2656.13223. [DOI] [PubMed] [Google Scholar]
- 5.Wendlandt CE, et al. Decreased coevolutionary potential and increased symbiont fecundity during the biological invasion of a legume-rhizobium mutualism. Evolution. 2021;75:731–747. doi: 10.1111/evo.14164. [DOI] [PubMed] [Google Scholar]
- 6.Tang JS, Huang L, Liu Y, Toshmatov Z, Zhang C, Shao H. Two phytotoxins isolated from the pathogenic fungus of the invasive weed Xanthium italicum. Chem. Biodivers. 2020;17:23–34. doi: 10.1002/cbdv.202000043. [DOI] [PubMed] [Google Scholar]
- 7.Shao H, Shi K, Han CX, Li WJ, Zhang C. Allelopathic plants 27: Xanthium italicum Moretti. Allelopathy J. 2019;48:139–153. [Google Scholar]
- 8.Che JD, Sun GQ. Two new weeds, Xanthium sibiricum and Xanthium italicum, found in Beijing. Pest Predict. 1992;12:39–40. [Google Scholar]
- 9.Tang JS, Ma M. Genetic diversity and genetic differentiation of invasive weed Xanthium italicum in China. CR. Biol. 2020;343:63–72. doi: 10.5802/crbiol.7. [DOI] [PubMed] [Google Scholar]
- 10.Wang ZG, Guan W, Chen DH, Zhang JW. Interception of quarantine weeds in Zhongshan Bureau—Xanthium italicum. Plant Quarantine. 2008;1:59. [Google Scholar]
- 11.Yuan SZ, et al. Investigation on harmful weeds carried by rapeseed imported from Mongolia. China Plant Protect. 2016;36:72–75. [Google Scholar]
- 12.Li LJ, Xiang ZC, Yu F, Song T, Wang LG, Wang K. Interception and identification of Xanthium italicum L. in imported soybean from Shandong port. J. Anhui Agric. Sci. 2018;46:171–175. [Google Scholar]
- 13.Shi TJ, Long ZZ, Miao M. Glomus mosseae promotes Xanthium italicum invasion. Sains Malays. 2020;49:2425–2432. doi: 10.17576/jsm-2020-4910-08. [DOI] [Google Scholar]
- 14.Liu HY, Ming GH. Distribution and control measures of Xanthium italicum. Biol. Bull. 2008;43:15–16. [Google Scholar]
- 15.Hocking PJ, Liddle MJ. The biology of Australian weeds: 15. Xanthium occidentale Bertol. and Xanthium spinosum L. J. Agric. Sci. 1986;50:191–221. [Google Scholar]
- 16.Kazinczi G, Torma M, Béres I. Competition between Xanthium italicum and crops under field conditions. Cereal. Res. Commun. 2009;37:77–80. [Google Scholar]
- 17.Baldoni G, Viggiani P, Bonetti A, Dinelli G, Catizone P. Classification of Italian Xanthium strumarium complex based on biological traits, electrophoretic analysis and response to maize interference. Weed Res. 2010;40:191–204. doi: 10.1046/j.1365-3180.2000.00181.x. [DOI] [Google Scholar]
- 18.Li, J. Phenotypic plasticity and adaptability of alien invasive plant Xanthium italicum Moretti. (Dissertation, University of Shihezi, China 2019).
- 19.Zhao P, Qu JJ, Xu XY, Yu QS, Jiang SX, Zhao HR. Desert vegetation distribution and species-environment relationships in an oasis-desert ecotone of northwestern China. J. Arid Land. 2019;11:461–476. doi: 10.1007/s40333-019-0055-0. [DOI] [Google Scholar]
- 20.Hu WJ, et al. Predicting potential mangrove distributions at the global northern distribution margin using an ecological niche model: Determining conservation and reforestation involvement. Forest Ecol. Manag. 2020;478:118–127. doi: 10.1016/j.foreco.2020.118517. [DOI] [Google Scholar]
- 21.Adineh F, Motamedvaziri B, Ahmadi H, Moeini A. Landslide susceptibility mapping using genetic algorithm for the rule set production(GARP) model. J. MT. Sci. Engl. 2018;15:2013–2026. doi: 10.1007/s11629-018-4833-5. [DOI] [Google Scholar]
- 22.Zhang JL, Liu F. Predictive modeling of suitable habitats for Cinnamomum camphora (L.) Presl using Maxent model under climate change in China. Int. J. Environ. Res. Public Health. 2019;16:3185. doi: 10.3390/ijerph16173185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubos, N. et al. Assessing the effect of sample bias correction in species distribution models. arXiv:2103.07107v1 (2021).
- 24.Booth TH. Why understanding the pioneering and continuing contributions of BIOCLIM to species distribution modelling is important. Austral. Ecol. 2018;43:852–860. doi: 10.1111/aec.12628. [DOI] [Google Scholar]
- 25.Yang ZX, Zhou GS, Yin XJ, Jia BR. Geographic distribution of Larix gmelinii natural forest in China and its climatic suitability. Chin. J. Ecol. 2014;33:1429–1436. [Google Scholar]
- 26.Beck HE, Zimmermann NE, McVicar TR, Vergopolan N, Berg A, Wood EF. Data descriptor: Present and future Köppen–Geiger climate classification maps at 1-km resolution. Sci. Data. 2018;5:180214. doi: 10.1038/sdata.2018.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song XY, et al. Blow to the Northeast? Intraspecific differentiation of Populus davidiana suggests a north-eastward skew of a phylogeographic break in East Asia. J. Biogeogr. 2020;48:187–201. doi: 10.1111/jbi.13992. [DOI] [Google Scholar]
- 28.Anand V, Oinam B, Singh IH. Predicting the current and future potential spatial distribution of endangered Rucervus eldii eldii (Sangai) using Maxent model. Environ. Monit. Assess. 2021;193:147. doi: 10.1007/s10661-021-08950-1. [DOI] [PubMed] [Google Scholar]
- 29.Phillips SJ, Anderson RP, Schapire RE. Maximun entropy modeling of species geographic distributions. Ecol. Model. 2006;190:231–259. doi: 10.1016/j.ecolmodel.2005.03.026. [DOI] [Google Scholar]
- 30.Bhandari MS, Shankhwar R, Maikhuri S. Prediction of ecological and geological niches of Salvadora oleoides in arid zones of India: Causes and consequences of global warming. Arab. J. Geosci. 2021;14:524–542. doi: 10.1007/s12517-020-06384-6. [DOI] [Google Scholar]
- 31.Elith J, Geaham HC, Anderson PR. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29:129–151. doi: 10.1111/j.2006.0906-7590.04596.x. [DOI] [Google Scholar]
- 32.Gebrewahid Y, et al. Current and future predicting potential areas of Oxytenanthera abyssinica (A. Richard) using Maxent model under climate change in Northern Ethiopia. Ecol. Process. 2020;9:1–15. doi: 10.1186/s13717-019-0210-8. [DOI] [Google Scholar]
- 33.Emad K, Francis G. Assessment of the extinction risks of medicinal plants in Egypt under climate change by integrating species distribution models and IUCN Red List criteria. J. Arid Environ. 2019;170:1–9. [Google Scholar]
- 34.Dong X, Chu YMR, Gu X, Huang Q, Zhang J, Bai W. Suitable habitat prediction of sichuan snub-nosed monkeys (Rhinopithecus roxellana) and its implications for conservation in baihe nature reserve, Sichuan, China. Environ. Sci. Pollut. Res. 2019;26:32374–32384. doi: 10.1007/s11356-019-06369-3. [DOI] [PubMed] [Google Scholar]
- 35.Saatchi S, Buermann W, Steege HT, Mori S, Smith TB. Modeling distribution of Amazonian tree species and diversity using remote sensing measurements. Remote Sens. Environ. 2008;112:2000–2017. doi: 10.1016/j.rse.2008.01.008. [DOI] [Google Scholar]
- 36.Wang R, Wang YZ. Invasion dynamics and potential spread of the invasive alient plant species Ageratina adenophora (Asteraceae) in China. Divers. Distrib. 2006;12:397–408. doi: 10.1111/j.1366-9516.2006.00250.x. [DOI] [Google Scholar]
- 37.Morrison LW, Korzukhin MD, Porter SD. Predicted range expansion of the invasive fire ant, Solenopsis invicta, in the eastern United States based on the VEMAP global warming scenario. Divers. Distrib. 2005;11:199–204. doi: 10.1111/j.1366-9516.2005.00142.x. [DOI] [Google Scholar]
- 38.Cao XF, Qian GL, Hu BS, Liu FQ. Prediction of potential suitable distribution area of Flaveria bidentis in China based on niche models. Chin. J. Appl. Ecol. 2010;21:3063–3069. [PubMed] [Google Scholar]
- 39.Lie JC, Xu HG. Maxent-based prediction of potential distribution of Solidago canadensis in China. J. Ecol. Rural Envir. 2010;26:137–141. [Google Scholar]
- 40.Yue MF, Feng L, Tian XS, Yang CH. Maxent-based prediction of potential distribution areas of Mimosa pigra L. J. Biosaf. 2013;22:173–180. [Google Scholar]
- 41.Shao Y, Cao W. Prediction of the potential distribution area of invasive alien plant Ambrosia artemisiifolia in Northeast China. J. Arid Land Resour. Environ. 2017;31:172–176. [Google Scholar]
- 42.Zhu GP, Petersen MJ, Bu MJ. Selecting biological meaningful environmental dimensions of low discrepancy among ranges to predict potential distribution of bean plataspid invasion. PLoS One. 2012;7:e46247. doi: 10.1371/journal.pone.0046247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saupe EE, Barve V, Myers CE, Barve N, Hensz CM. Variation in niche and distribution model performance: The need for a priori assessment of key causal factors. Ecol. Model. 2012;237:11–22. doi: 10.1016/j.ecolmodel.2012.04.001. [DOI] [Google Scholar]
- 44.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R foundation for statistical computing. https://www.R-project.org/ (2020).
- 45.Bay RA, Harrigan RJ, Buermann W, Underwood VL, Gibbs HL, Smith TB, Ruegg K. Response to comment on "Genomic signals of selection predict climate-driven population declines in a migratory bird". Science. 2018;361:eaat7279. doi: 10.1126/science.aat7956. [DOI] [PubMed] [Google Scholar]
- 46.Li JY, Chang H, Liu T, Zhang C. The potential geographical distribution of Haloxylon across Central Asia under climate change in the 21st century. Agric. Forest Meteorol. 2019;275:243–254. doi: 10.1016/j.agrformet.2019.05.027. [DOI] [Google Scholar]
- 47.Kottek MJ, Grieser C, Beck B, Rudolf RF. World map of the Köppen–Geiger climate classification updated. Meteorol. Z. 2006;15:259–263. doi: 10.1127/0941-2948/2006/0130. [DOI] [Google Scholar]
- 48.Zaidi F, Fatima SH, Jan T, Fatima M, Ali A, Khisroon M, Adnan M, Rasheed SB. Environmental risk modeling and potential sand-fly vectors of Cutaneous leishmaniasis in Chitral District: A leishmanial focal point of mount Tirchmir, Pakistan. Trop. Med. Int. Health. 2017;22:1130–1140. doi: 10.1111/tmi.12916. [DOI] [PubMed] [Google Scholar]
- 49.Lissovsky AA, Dudov SV. Species-distribution modeling: Advantages and limitations of its application. 2. MaxEnt. Biol. Bull. Rev. 2021;11:265–275. doi: 10.1134/S2079086421030087. [DOI] [Google Scholar]
- 50.Gao T, Xu Q, Liu Y, Zhao JQ, Shi J. Predicting the potential geographic distribution of Sirex nitobei in China under climate change using Maximum Entropy model. Forests. 2021;12:151–168. doi: 10.3390/f12020151. [DOI] [Google Scholar]
- 51.Williams RAJ, Owens HL, Clamp J, Peterson AT, Warren A, Martin-Cereceda M. Endemicity and climatic niche differentiation in three marine ciliated protists. Limnol. Oceanogr. 2018;63:2727–2736. doi: 10.1002/lno.11003. [DOI] [Google Scholar]
- 52.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 53.Kong F, Tang L, He H, Yang FX, Tao J, Wang WC. Assessing the impact of climate change on the distribution of Osmanthus fragrans using Maxent. Environ. Sci. Pollut. Res. 2021;28:34655–34663. doi: 10.1007/s11356-021-13121-3. [DOI] [PubMed] [Google Scholar]
- 54.Fielding AH, Bell JF. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 1997;24:38–49. doi: 10.1017/S0376892997000088. [DOI] [Google Scholar]
- 55.Allouche O, Tsoar A, Kadmon R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS) J. Appl. Ecol. 2006;43:1223–1232. doi: 10.1111/j.1365-2664.2006.01214.x. [DOI] [Google Scholar]
- 56.Waage JK, Reaser JK. A global strategy to defeat invasive species. Science. 2001;292:1477–1486. doi: 10.1126/science.292.5521.1486a. [DOI] [PubMed] [Google Scholar]
- 57.Lee S, Ka JO, Song HG. Growth promotion of Xanthium italicum by application of rhizobacterial isolates of Bacillus aryabhattai in microcosm soil. J. Microbiol. 2012;50:45–49. doi: 10.1007/s12275-012-1415-z. [DOI] [PubMed] [Google Scholar]
- 58.Gallardo B, Zieritz A, Aldridge DC. The importance of the human footprint in shaping the global distribution of terrestrial, freshwater and marine invaders. PLoS One. 2015;10:e0125801. doi: 10.1371/journal.pone.0125801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soler-Méndez M, Parras-Burgos D, Mas-Espinosa E, et al. Standardization of the dimensions of a portable weighing lysimeterdesigned to be applied to vegetable crops in mediterranean climates. Sustainability. 2021;13:2210. doi: 10.3390/su13042210. [DOI] [Google Scholar]
- 60.Mamun M, Kim S, An KG. Distribution pattern prediction of an invasive alien species largemouth bass using a maximum entropy model (Maxent) in the Korean peninsula. J. Asia-Pac. Biodivers. 2018;11:516–524. doi: 10.1016/j.japb.2018.09.007. [DOI] [Google Scholar]
- 61.Yan HY, et al. Predicting the potential distribution of an invasive species, Erigeron canadensis L. in China with a maximum entropy model. Glob. Ecol. Conserv. 2019;21:00822. [Google Scholar]
- 62.Park JU, Lee T, Dong GK, Shin S. Prediction of potential habitats and distribution of the marine invasive sea squirt, Herdmania momus. Environ. Biol. Res. 2020;38:179–188. doi: 10.11626/KJEB.2020.38.1.179. [DOI] [Google Scholar]
- 63.Balceniuk MD, et al. Association between the atherosclerotic disease risk score and carotid artery stenosis. J. Surg. Res. 2021;257:189–194. doi: 10.1016/j.jss.2020.06.041. [DOI] [PubMed] [Google Scholar]
- 64.Svensson EI, Gómez-Llano MA, Torres AR, Bensch HM. Frequency dependence and ecological drift shape coexistence of species with similar niches. Am. Nat. 2018;191:691–703. doi: 10.1086/697201. [DOI] [PubMed] [Google Scholar]
- 65.Peterson AT. Predicting potential geographic distributions of invading species. Curr. Sci. India. 2005;89:9. [Google Scholar]
- 66.Broennimann O, Treier UA, Thuiller W, Peterson AT, Guisan A. Evidence of climatic niche shift during biological invasion. Ecol. Lett. 2007;10:701–709. doi: 10.1111/j.1461-0248.2007.01060.x. [DOI] [PubMed] [Google Scholar]
- 67.Kariyawasam CS, Kumar L, Ratnayake SS. Potential distribution of aquatic invasive alien plants, Eichhornia crassipes and Salvinia molesta under climate change in Sri Lanka. Wetl. Ecol. Manag. 2021;1:1–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the current study are available from the corresponding author upon reasonable request.
R script is available from the corresponding author.