Abstract
Standard tests for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detect the presence of viral RNA using real-time reverse transcription (rRT)-PCR. Recently, convenient, rapid, and relatively inexpensive SARS-CoV-2 antigen (Ag) detection methods have been developed. The STANDARD Q COVID-19 Ag test (SD Biosensor, Inc., Suwon, Korea) is a rapid immunochromatography test that qualitatively detects the nucleocapsid protein of SARS-CoV-2 using gold conjugated antibodies. We evaluated its performance in comparison with that of Allplex 2019-nCoV Assay (Seegene, Seoul, Korea) in a retrospective case-control study using residual samples. The sensitivity and specificity of the STANDARD Q COVID-19 Ag test were 89.2% (58/65) and 96.0% (96/100), respectively. Cycle threshold (Ct) values for the three target SARS-CoV-2 genes (envelope, RNA-dependent RNA polymerase, and nucleocapsid genes) included in Allplex 2019-nCoV Assay were significantly lower in Ag test-positive patients than in Ag test-negative patients (P<0.001). The Ag test sensitivity was higher in samples with Ct≤30 and those collected one to five days post symptom onset. In conclusion, the STANDARD Q COVID-19 Ag test can serve as an alternative in high-prevalence settings, when the low sensitivity is compensated or when rRT-PCR tests are limited.
Keywords: Severe Acute Respiratory Syndrome Coronavirus 2, STANDARD Q COVID-19 Ag test, Allplex 2019-nCoV Assay, Antigen, Immunochromatography, Nucleocapsid protein, Performance
In December 2019, a case of unexplained, atypical pneumonia was reported in Wuhan, Hubei Province, China. This atypical pneumonia was caused by a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. The World Health Organization (WHO) named the disease caused by SARS-CoV-2 as coronavirus disease 2019 (COVID-19) [2]. As of July 14, 2021, SARS-CoV-2 had infected 187,296,646 patients and caused 4,046,470 deaths in 210 countries, and COVID-19 remained a global pandemic [3].
Current standard tests for SARS-CoV-2 detect the presence of viral RNA using real-time reverse transcription (rRT)-PCR [4]. As COVID-19 exponentially spreads across the globe, a rapid method that allows on-field testing and yields immediate results is urgently needed. Compared with rRT-PCR, rapid diagnostic tests (RDTs) do not require complex technology and equipment and are convenient and relatively inexpensive. Therefore, an RDT capable of diagnosing SARS-CoV-2 with excellent clinical performance may serve as an alternative when rRT-PCR testing is limited, though currently the testing capacity in Korea is sufficient [5-7].
Tests for detecting the SARS-CoV-2 antigen (Ag) are commercially available [6], and their performance needs to be validated. The STANDARD Q COVID-19 Ag test (SD Biosensor, Inc., Suwon, Korea) is an RDT that detects the nucleocapsid protein (NP) of SARS-CoV-2 via immunochromatography, and it is the first SARS-CoV-2 Ag-based RDT approved by the Korea Ministry of Food and Drug Safety, as an alternative when PCR testing is limited. NP is the predominantly expressed structural protein of SARS-CoV-2, and an NP Ag-based test has proven useful in the early diagnosis of COVID-19 [8]. We evaluated the clinical usefulness of the STANDARD Q COVID-19 Ag test, and compared its performance with that of Allplex 2019-nCoV Assay (Seegene, Seoul, Korea), a molecular diagnostic test approved by the Korea Ministry of Food and Drug Safety and the United States Food and Drug Administration for emergency use. Performance evaluation studies of the STANDARD Q COVID-19 Ag test have been conducted both abroad [9-11] and in Korea [12]. This study was conducted using residual samples after COVID-19 testing was performed during hospitalizations and drive-through screenings at a single tertiary hospital at the time of the outbreak in Daegu. The Institutional Review Board (IRB) of Yeungnam University Hospital, Daegu, Korea, approved this study (IRB No. YUMC 2020-03-070). The IRB approved the collection of residual samples under a waiver of consent.
We conducted a retrospective diagnostic case-control study with a reversed-flow design [13]. Patients who visited Yeungnam University Hospital for COVID-19 drive-through screening or who were hospitalized between February 28, 2020 and April 30, 2020 were included. All samples were collected using nasopharyngeal swabs, transferred to the laboratory in viral transport medium (VTM), and stored at 4°C until and after testing with Allplex 2019-nCoV Assay in accordance with the guidelines reported by Hong, et al. [14]. All tests of the Allplex 2019-nCoV Assay were performed on the day of sample collection. The next day, residual samples were stored at -80°C. Samples collected in eNAT (Copan Diagnostics Inc., Brescia, Italy) were excluded, whereas those collected in REST UTM (Noble Biosciences, Inc., Hwaseoung, Korea) or ESwab (Copan Diagnostics) were included for the STANDARD Q COVID-19 Ag test as per the manufacturer’s recommendation. Among serial samples of patients with a positive Allplex 2019-nCoV Assay result, the ones tested at initial diagnosis (N=206) were included in this study. Sixty-five samples collected in eNAT and 25 samples with insufficient residual volume for the STANDARD Q COVID-19 Ag test were excluded. Finally, 65 positive samples were selected consecutively (Table 1). In addition, 100 negative samples were collected during the same period. Overall, the study included 75 male and 90 female patients (age range, 12–95 years). STANDARD Q COVID-19 Ag tests were conducted using residual samples in May 2020, in a randomized order, by a laboratory technician who was blinded to the Allplex 2019-nCoV Assay results.
Table 1.
Basic characteristics of COVID-19 patients
| Characteristic | Total (N = 65) | Ag (+) (N = 58) | Ag (–) (N = 7) |
|---|---|---|---|
| Age (yr) | 56 (48–63) | 58 (48–64) | 51 (49–56) |
| Sex | |||
| Male | 24 (36.9) | 22 (37.9) | 2 (28.6) |
| Female | 41 (63.1) | 36 (62.1) | 5 (71.4) |
| Patient location | |||
| Hospitalization | 1 (1.5) | 1 (1.8) | 0 (0.0) |
| Drive-through screening | 64 (98.5) | 57 (98.2) | 7 (100.0) |
| Clinical symptom | |||
| Cough | 28 (21.1) | 23 (19.6) | 5 (31.3) |
| Productive cough | 24 (18.1) | 22 (18.8) | 2 (12.5) |
| Fever | 22 (16.5) | 21 (18.0) | 1 (6.3) |
| Sore throat | 19 (14.3) | 17 (14.4) | 2 (12.5) |
| Myalgia | 11 (8.3) | 7 (6.0) | 4 (25.0) |
| Chills | 5 (3.8) | 5 (4.3) | 0 (0.0) |
| Rhinorrhea | 4 (3.0) | 4 (3.4) | 0 (0.0) |
| Headache | 3 (2.3) | 3 (2.6) | 0 (0.0) |
| Dyspnea | 3 (2.3) | 3 (2.6) | 0 (0.0) |
| Nasal obstruction | 3 (1.5) | 3 (2.6) | 0 (0.0) |
| Diarrhea | 2 (0.8) | 2 (1.7) | 0 (0.0) |
| Chest discomfort | 1 (0.8) | 0 (0.0) | 1 (6.3) |
| Cold sweating | 1 (0.8) | 0 (0.0) | 1 (6.3) |
| Anorexia | 1 (0.8) | 1 (0.9) | 0 (0.0) |
| Parosmia | 1 (0.8) | 1 (0.9) | 0 (0.0) |
| Fatigue | 1 (0.8) | 1 (0.9) | 0 (0.0) |
| Asymptomatic | 4 (3.0) | 4 (3.4) | 0 (0.0) |
| Symptom onset to diagnosis | |||
| Asymptomatic | 4 (6.1) | 4 (6.9) | 0 (0.0) |
| 1–5 days | 41 (63.1) | 39 (67.2) | 2 (28.6) |
| 6–10 days | 15 (23.1) | 11 (19.0) | 4 (57.1) |
| 11–21 days | 5 (7.7) | 4 (6.9) | 1 (14.3) |
Values are expressed as median (interquartile range) or number (%).
Abbreviations: COVID-19, coronavirus disease 2019; Ag, antigen.
Allplex 2019-nCoV Assay is a multiplex rRT-PCR test that simultaneously detects three target SARS-CoV-2 genes in a single tube. The test is designed to detect the RNA-dependent RNA polymerase (RdRP) and nucleocapsid (N) genes specific to SARS-CoV-2 and the envelope (E) gene that is common to all sarbecoviruses, including SARS-CoV-2. According to the manufacturer’s recommendations, the result was interpreted as positive only if the cycle threshold (CT) values of all three target genes were within the cutoff (<40) and negative if all were outside the cutoff or if there was no amplification; otherwise, the result was interpreted as inconclusive [15, 16].
The STANDARD Q COVID-19 Ag test qualitatively detects SARS-CoV-2 NP using gold-conjugated antibodies. It comprises a nitrocellulose membrane with control (“C”) and test (“T”) lines that are coated with mouse monoclonal anti-chicken IgY antibodies and mouse monoclonal anti-SARS-CoV-2 antibodies, respectively. The extraction buffer containing mouse monoclonal anti-SARS-CoV-2 antibodies conjugated with gold interact with SARS-CoV-2 antigens in the samples to form antigen–antibody complexes. These complexes migrate on the membrane until they reach the T line, where they are captured by the mouse monoclonal anti-SARS-CoV-2 antibodies. In this study, nasopharyngeal swab samples in VTM (100 μL) were mixed with the extraction buffer (100 μL) provided with the test kit.
The sensitivity and specificity of the STANDARD Q COVID-19 Ag test were evaluated and compared with those of Allplex 2019-nCoV Assay. Confidence intervals (CIs) for sensitivity and specificity were “exact” Clopper-Pearson CIs. Ct values for the target SARS-CoV-2 genes were compared between Ag test-positive and Ag-negative samples using the Mann–Whitney test. All statistical analyses were performed using MedCalc Statistical Software version 19.5.3 (MedCalc Software Ltd., Ostend, Belgium).
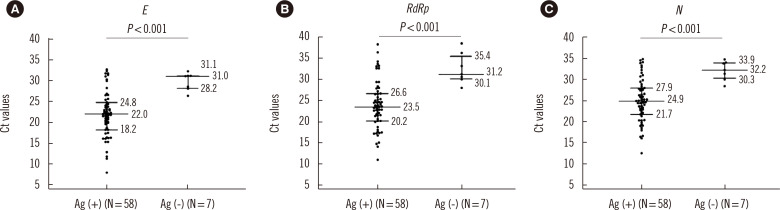
Based on the analysis of the 65 positive samples, the sensitivity and specificity of the STANDARD Q COVID-19 Ag test were 89.2% (95% CI, 79.1%–95.6%) and 96.0% (95% CI, 90.1%–98.9%), respectively (Table 2). Ct values for the target SARS-CoV-2 genes were significantly lower in Ag test-positive samples than in Ag test-negative samples (Fig. 1, P<0.001).
Table 2.
Performance of the STANDARD Q COVID-19 Ag test for the detection of SARS-CoV-2
| Groups | Allplex 2019-nCoV Assay | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| rRT-PCR† (+) (N = 65) | rRT-PCR† (–) (N = 100) | Sensitivity % (95% CI) | Specificity % (95% CI) | Positive predictive value % (95% CI) | Negative predictive value % (95% CI) | |||
|
|
|
|||||||
| Ag (+), N (%) | Ag (–), N (%) | Ag (+), N (%) | Ag (–), N (%) | |||||
| Overall | 58 (89.2) | 7 (10.8) | 4 (4.0) | 96 (96.0) | 89.2 (79.1–95.6) | 96.0 (90.1–98.9) | 93.6 (84.7–97.4) | 93.2 (87.2–96.5) |
| Ct* ≤ 30 | 51 (78.4) | 3 (4.6) | - | - | 94.4 (84.6–98.8) | - | - | - |
| Ct* > 30 | 7 (10.8) | 4 (6.2) | - | - | 63.6 (30.8–89.0) | - | - | - |
| Asymptomatic | 4 (6.9) | 0 (0.0) | - | - | 100 (39.8–100.0) | - | - | - |
| 1–5 days | 39 (67.2) | 2 (28.6) | - | - | 95.1 (83.5–99.4) | - | - | - |
| 6–10 days | 11 (19.0) | 4 (57.1) | - | - | 73.3 (44.9–92.2) | - | - | - |
| 11–21 days | 4 (6.9) | 1 (14.3) | - | - | 80.0 (28.4–99.5) | - | - | - |
*Ct value for the envelope gene in Allplex 2019-nCoV Assay; †Interpreted as described in the instruction of Allplex 2019-nCoV Assay.
Abbreviations: Ag, antigen; COVID-19, coronavirus disease 2019; CI, confidence interval; Ct, cycle threshold; rRT-PCR, real-time reverse transcription PCR; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Fig. 1.
Comparison of Ct values for the target SARS-CoV-2 genes (A) E, (B) RdRp, and (C) N in Ag test-positive and Ag test-negative samples. Values are expressed as median (interquartile range).
Abbreviations: Ct, cycle threshold; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; Ag, antigen; E, envelope gene; RdRp, RNA-dependent RNA polymerase gene; N, nucleocapsid gene.
The performance of the STANDARD Q COVID-19 Ag test was inferior to that of rRT-PCR (Table 2). However, its sensitivity was higher for samples with a Ct value ≤30 and samples collected one to five days post symptom onset than for those with a Ct value >30 and those collected after six days post symptom onset; the former possibly accounts for a significant proportion of transmission cases. This finding is consistent with the recommendation to use rapid Ag tests by the European Center for Disease Prevention and Control and WHO [17, 18].
Improvement in Ag test performance is required because of the potentially large number of false negatives due to low sensitivity, and false positives despite the high specificity. However, the STANDARD Q COVID-19 Ag test has several benefits over rRT-PCR for SARS-CoV-2 detection, such as simplicity of use, easy availability, low cost, and the short time needed to obtain the results. Therefore, the STANDARD Q COVID-19 Ag test, when used considering the minimal performance requirements set by the WHO, can serve as an effective alternative in high-prevalence settings, when the low sensitivity is compensated or when rRT-PCR tests are limited.
Our study had several limitations. First, the number of samples used in this study was small. More than a half the samples were collected one to five days post symptom onset and yielded a Ct value for E of<30 (36/65). Moreover, since only four asymptomatic patients were included, we cannot definitively conclude that the STANDARD Q COVID-19 Ag test shows good performance in asymptomatic patients, despite the 100% sensitivity in this group. Overall, the patient distribution may not reflect the general population with low-prevalence settings, and results of the performance evaluation may vary greatly depending on the patient group [12]. Second, as this was a retrospective study conducted with residual samples, we mixed nasopharyngeal swab samples in VTM with the extraction buffer, while directly mixing nasopharyngeal swab samples with extraction buffer is the primary recommendation. Sample dilution with VTM may have decreased the test sensitivity for samples with high Ct values. Furthermore, the C line of the STANDARD Q COVID-19 Ag test did not appear in preliminary tests of samples collected in eNAT, suggesting that guanidine thiocyanate in this transport medium may have denatured proteins [19]. Therefore, directly mixing the nasopharyngeal swab with the extraction buffer provided with STANDARD Q COVID-19 Ag test kit is required to accurately assess the clinical performance of the test. Third, Ct values of Ag test-positive samples may not accurately indicate the patient status. Ct values can vary depending on the sample quality and may not correlate with the presence of SARS-CoV-2 Ag.
In conclusion, the STANDARD Q COVID-19 Ag test can act as an alternative in high-prevalence settings, when the low sensitivity is compensated or when rRT-PCR tests are limited. It is necessary to evaluate the method further by directly mixing nasopharyngeal swabs with the extraction buffer provided with STANDARD Q COVID-19 Ag test kit. Moreover, analysis of a larger number of samples is needed to evaluate the performance in low-prevalence settings in the general population.
ACKNOWLEDGMENTS
We thank SD Biosensor for providing STANDARD Q COVID-19 Ag kits.
Footnotes
AUTHOR CONTRIBUTIONS
Lee JH conceived and designed the study and performed the STANDARD Q COVID-19 Ag tests. Kim HW performed data analysis and wrote the manuscript. Lee JH and Park MK reviewed the manuscript.
CONFLICTS OF INTEREST
None declared.
RESEARCH FUNDING
This work was supported by the 2020 Yeungnam University Research Grant.
REFERENCES
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization, author. [Updated on Feb 2020];Naming the coronavirus disease (COVID-19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it#:~:text=Official%20names%20have%20been%20announced,%2DCoV%2D2)-(covid-2019)-and-the-virus-that-causes-it .
- 3.World Health Organization, author. [Updated on July 2021];WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/
- 4.World Health Organization, author. [Updated on Sep 2020];Diagnostic testing for SARS-CoV-2. https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2 .
- 5.Kim YJ, Sung H, Ki CS, Hur M. COVID-19 Testing in South Korea: current status and the need for faster diagnostics. Ann Lab Med. 2020;40:349–50. doi: 10.3343/alm.2020.40.5.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foundation for Innovative New Diagnostics, author. [Updated on July 2021];SARS-CoV-2 diagnostic pipeline (continuously updated) 2020 https://www.finddx.org/covid-19/pipeline .
- 7.Huh HJ, Hong KH, Kim TS, Song SH, Roh KH, Lee H, et al. Surveillance of Coronavirus Disease 2019 (COVID-19) testing in clinical laboratories in Korea. Ann Lab Med. 2021;41:225–9. doi: 10.3343/alm.2021.41.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diao B, Wen K, Zhang J, Chen J, Han C, Chen Y, et al. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect. 2020;27:289.e1–4. doi: 10.1016/j.cmi.2020.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerutti F, Burdino E, Milia MG, Allice T, Gregori G, Bruzzone B, et al. Urgent need of rapid tests for SARS CoV-2 antigen detection: Evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J Clin Virol. 2020;132:104654. doi: 10.1016/j.jcv.2020.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jääskeläinen AE, Ahava MJ, Jokela P, Szirovicza L, Pohjala S, Vapalahti O, et al. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. J Clin Virol. 2021;137:104785. doi: 10.1016/j.jcv.2021.104785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mak GCK, Lau SSY, Wong KKY, Chow NLS, Lau CS, Lam ETK, et al. Evaluation of rapid antigen detection kit from the WHO Emergency Use List for detecting SARS-CoV-2. J Clin Virol. 2021;134:104712. doi: 10.1016/j.jcv.2020.104712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Kim SY, Huh HJ, Kim N, Sung H, Lee H, et al. Clinical performance of the Standard Q COVID-19 Rapid Antigen Test and simulation of its real-world application in Korea. Ann Lab Med. 2021;41:588–92. doi: 10.3343/alm.2021.41.6.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutjes AW, Reitsma JB, Vandenbroucke JP, Glas AS, Bossuyt PM. Case-control and two-gate designs in diagnostic accuracy studies. Clin Chem. 2005;51:1335–41. doi: 10.1373/clinchem.2005.048595. [DOI] [PubMed] [Google Scholar]
- 14.Hong KH, Lee SW, Kim TS, Huh HJ, Lee J, Kim SY, et al. Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID-19) in Korea. Ann Lab Med. 2020;40:351–60. doi: 10.3343/alm.2020.40.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung H, Roh KH, Hong KH, Seong MW, Ryoo N, Kim HS, et al. COVID-19 molecular testing in Korea: practical essentials and answers from experts based on experiences of emergency use authorization assays. Ann Lab Med. 2020;40:439–47. doi: 10.3343/alm.2020.40.6.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hur KH, Park K, Lim Y, Jeong YS, Sung H, Kim MN. Evaluation of four commercial kits for SARS-CoV-2 real-time reverse-transcription polymerase chain reaction approved by emergency-use-authorization in Korea. Front Med (Lausanne) 2020;7:521. doi: 10.3389/fmed.2020.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.European Centre for Disease Prevention and Control, author. [Updated on Nov 2020];Options for the use of rapid antigen tests for COVID-19 in the EU/EEA and the UK. https://www.ecdc.europa.eu/en/publications-data/options-use-rapid-antigen-tests-covid-19-eueea-and-uk#copy-to-clipboard .
- 18.World Health Organization, author. [Updated on Sep 2020];Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays. https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays .
- 19.Mason PE, Neilson GW, Dempsey CE, Barnes AC, Cruickshank JM. The hydration structure of guanidinium and thiocyanate ions: implications for protein stability in aqueous solution. Proc Natl Acad Sci U S A. 2003;100:4557–61. doi: 10.1073/pnas.0735920100. [DOI] [PMC free article] [PubMed] [Google Scholar]