Abstract
Background
The emergence of carbapenemase-producing Enterobacteriaceae (CPE) represents a major clinical problem. Recently, the occurrence of CPE has increased globally, but epidemiological patterns vary across region. We report the trends in the genotypic distribution and antimicrobial susceptibility of CPE isolated from rectal and clinical samples during a four-year period.
Methods
Between January 2016 and December 2019, 1,254 nonduplicated CPE isolates were obtained from four university hospitals in Korea. Carbapenemase genotypes were determined by multiplex real-time PCR. Antimicrobial susceptibility was profiled using the Vitek 2 system (bioMérieux, Hazelwood, MO, USA) or MicroScan Walkaway-96 system (Siemens West Sacramento, CA, USA). The proportions of carbapenemase genotypes and nonsusceptibility were analyzed using Pearson’s chi-square test.
Results
Among the 1,254 CPE isolates, 486 (38.8%), 371 (29.6%), 357 (28.5%), 8 (0.6%), 8 (0.6%), and 24 (1.9%) were Klebsiella pneumoniae carbapenemase (KPC), oxacillinase (OXA)-48-like, New Delhi metallo-β-lactamase (NDM), imipenemase (IMP), Verona integron-encoded metallo-β-lactamase (VIM), and multiple producers, respectively. The predominant species was K. pneumoniae (72.6%), followed by Escherichia coli (6.5%). More than 90% of the isolates harboring KPC, NDM, and OXA-48-like were nonsusceptible to cephalosporins, aztreonam, and carbapenems.
Conclusions
The impact of CPE is primarily due to KPC-, NDM-, and OXA-48-like-producing K. pneumoniae isolates. Isolates carrying these carbapenemase are mostly multidrug-resistant. Control strategies based on these genotypic distributions and antimicrobial susceptibilities of CPE isolates are required.
Keywords: Carbapenemase-producing Enterobacteriaceae, Klebsiella pneumoniae, Klebsiella pneumoniae carbapenemase, New Delhi metallo-β-lactamase, oxacillinase-48-like
INTRODUCTION
The emergence of carbapenemase-producing Enterobacteriaceae (CPE) is a major clinical concern because these bacteria are nonsusceptible to various classes of antimicrobials, which can lead to therapeutic failure and worse outcomes [1]. CPE mainly harbor three major classes of β-lactamases according to the Ambler classification: class A β-lactamases (Klebsiella pneumoniae carbapenemase [KPC]), class B metallo-β-lactamases (New Delhi metallo-β-lactamase [NDM], imipenemase [IMP], and Verona integron-encoded metallo-β-lactamase [VIM]), and class D β-lactamases (oxacillinase [OXA]-48) [2]. In CPE that cause infection outbreaks, the carbapenemase genes are located on mobile genetic elements, such as plasmids or transposons [3, 4].
The occurrence of CPE has increased globally, but their epidemiological patterns vary across region. KPC-producing K. pneumoniae was first isolated in North Carolina, USA, in 1996 [5]. Numerous outbreaks related to these strains have been reported in the USA, South America, Greece, Italy, Israel, and China [1]. NDM producers were identified in early 2008 from an Indian patient hospitalized previously in New Delhi, India; since then, they have been detected in most countries related with India [6]. Recently, isolates from the Balkan states and the Middle East have served as reservoirs of NDM producers [7]. The first OXA-48-producing K. pneumoniae strain was isolated in Istanbul, Turkey, in 2001 [8]. Various strains, including OXA-48 strains, have caused several outbreaks in Mediterranean and Southern Europe, the Indian subcontinent, and Northern Africa [2, 7].
Multicenter studies on the prevalence and molecular epidemiology or antimicrobial susceptibilities of CPE have been conducted in Korea [9, 10]. Reporting the recent status of genotypic distribution of CPE in relation to antimicrobial susceptibilities based on a multicenter study is necessary to increase the awareness about and control of these isolates. We report the genotypic distribution and antimicrobial susceptibilities of CPE isolated from rectal and clinical samples in four Korean university hospitals during a 4-year period.
MATERIALS AND METHODS
Study design and CPE isolate identification
Between January 2016 and December 2019, CPE isolates were prospectively collected from four university hospitals in a metropolitan area of Korea (two hospitals, Kangnam Sacred Heart Hospital with 572 beds [A] and Hangang Sacred Heart Hospital with 158 beds [B] in Seoul, and two, Hallym University Sacred Heart Hospital with 834 beds [C] and Dongtan Sacred Heart Hospital with 660 beds [D] in Gyeonggi-do). In total, 1,254 nonduplicated CPE isolates harboring carbapenemase genes were collected (N=308, hospital A; N=55, hospital B; N=856, hospital C; N=35, hospital D). The isolates had been obtained from rectal samples taken for epidemiological and infection control purposes and clinical samples taken during the work-up of patients with suspected infections. The following clinical samples were provided: sputum (N=158, 12.6%), urine (N=130, 10.3%), blood (N=58, 4.6%), body fluid (N=36, 2.9%), wound or pus (N=22, 1.8%), and swabs (N=10, 0.8%). The isolates from patients in the intensive care unit (ICU) were more than a half of the included CPE isolates (N=792, 63.2%). Bacteria were identified using a Vitek 2 system (bioMérieux, Hazelwood, MO, USA) at hospitals B and D, amatrix-assisted laser desorption ionization-time-of-flight mass spectrometry on a Vitek-MS instrument (bioMérieux) at hospital A, and a MicroScan Walkaway-96 system (Siemens, West Sacramento, CA, USA) at hospital C. After collecting carbapenem-nonsusceptible Enterobacteriaceae isolates (based on a minimal inhibitory concentration [MIC]>0.5 μg/mL for ertapenem or MIC>1 μg/mL for imipenem or meropenem) from the four hospitals, the modified Hodge test (MHT) and carbapenemase inhibition test (CIT) were used for CPE detection at hospital A [11]. Clinical information, such as hospital of origin, sampling date, isolation site, department, and hospitalization ward, was collected by reviewing the medical records. The study protocol was approved by the Institutional Review Board of Kangnam Sacred Heart Hospital, Seoul (HKS 2020-03-020), which waived the need for informed consent. The data used in this study have been deposited in Harvard Dataverse and are accessible through https://doi.org/10.7910/DVN/QR9DR6.
Genotypic detection of carbapenemase genes
PANA Real Typer CRE kits (PANAGENE Inc., Daejeon, Korea) utilizing peptide nucleic acid-mediated multiplex real-time PCR were used to type all CPE isolates that tested carbapenemase-positive using MHT and CIT. The primers and probes, which were designed to target six carbapenemase genes (blaKPC, blaGES, blaNDM, blaVIM, blaIMP, and blaOXA-48-like) and an internal control, were provided by PANAGENE Inc. The sequences have been previously published [12]. The multiplex real-time PCR assay was conducted in a 96-well plate on a CFX-96 Real-Time PCR detection system (Bio-Rad Laboratories Inc., Hercules, CA, USA) using the PANA Real Typer CRE kit per the manufacturer’s instructions. Reaction mixtures contained 5 μL of genomic DNA, 1μL of Taq DNA polymerase, and 19 μL of the primer/probe and multiplex real-time PCR master mixtures. Thermal cycles were as follows: 50°C for 2 minutes and 95°C for 15 minutes; 45 cycles of 95°C for 15 seconds, 58°C for 45 seconds, and 72°C for 15 seconds; 95°C for 5 minutes; 35°C for 5 minutes; and a melting-curve step (35°C to 80°C in 0.5°C increments for 5 seconds). Fluorescence was acquired on all four channels (Blue/FAM for blaKPC and blaIMP, Green/HEX for blaGES and blaNDM, Yellow/ROX for blaVIM and blaOXA-48, and Violet/Cy5 for the internal control). After each run, the threshold cycle (Ct) was measured based on the signal strength, at which the fluorescence exceeded the threshold. Rectal and clinical samples with a Ct value <35 were regarded positive. RNase-free water was used as a negative control in each run.
Antimicrobial susceptibility testing
The MICs of the isolates were determined using the Vitek 2 system at hospitals A, B, and D and the MicroScan Walkaway-96 system at hospital C. Nonsusceptibility rates were calculated by dividing the number of isolates showing nonsusceptibility by the total number of isolates. The analyzed antimicrobial agents were cefotaxime, ceftazidime, cefepime, aztreonam, ertapenem, imipenem (or meropenem), amikacin, gentamicin, ciprofloxacin (or levofloxacin), trimethoprim-sulfamethoxazole, and tigecycline. MIC breakpoints were applied according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (M100S) [13]. For tigecycline, MIC results were interpreted according to the European Committee for Antimicrobial Susceptibility Testing criteria [14]. Intermediate susceptibility was determined according to the criteria set in the CLSI guidelines and per the European Committee for Antimicrobial Susceptibility Testing [13, 14].
Statistical analysis
Statistical analyses were performed using PASW version 18.0 (SPSS Inc., Chicago, IL, USA), Analyse-it Method Evaluation Edition software (version 2.26; Analyse-it Software Ltd., Leeds, UK), and R statistical software (version 3.6.3; R Foundation for Statistical Computing, Vienna, Austria). Categorical variables were compared using Pearson’s chi-square test or Fisher’s exact test. All tests were two-sided, and differences with P<0.05 were considered significant.
RESULTS
Distribution of CPE isolates
The CPE isolates comprised 910 K. pneumoniae (72.6%), 81 Escherichia coli (6.5%), 79 Enterobacter cloacae (6.3%), 47 Citrobacter freundii (3.7%), 45 Klebsiella oxytoca (3.6%), 28 Serratia marcescens (2.2%), 20 Klebsiella aerogenes (1.6%), 13 Citrobacter koseri (1.0%), 12 Providencia rettgeri (1.0%), 3 Enterobacter hormaechei (0.2%), 3 Enterobacter kobei (0.2%), 3 Pantoea agglomerans (0.2%), 2 Citrobacter braakii (0.2%), 2 Enterobacter asubriae (0.2%), 2 Kluyvera ascorbate (0.2%), 2 Raoutella planticola (0.2%), 1 Klebsiella ozaenae (0.1%), and 1 Klebsiella variicola (0.1%) isolates.
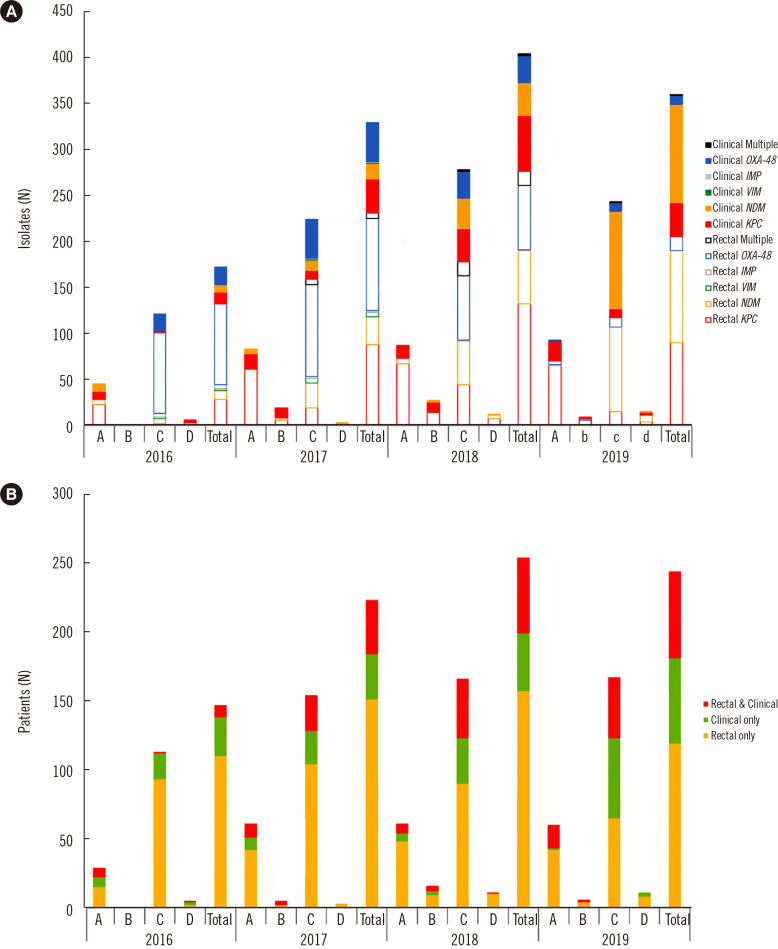
The distribution of carbapenemase genes identified among the CPE isolates is illustrated in Fig. 1A. Among the 1,254 CPE isolates, 486 (38.8%), 371 (29.6%), 357 (28.5%), 8 (0.6%), 8 (0.6%), and 24 (1.9%) were KPC, OXA-48-like, NDM, IMP, VIM, and multiple producers harboring more than two types of genes, respectively. Twenty-four CPE (1.9%) isolates harbored more than two carbapenemase genes, and most of these isolates (N=19, 1.5%) contained both OXA-48-like and NDM. The samples comprised rectal (N=829, 66.9%) and clinical (N=415, 33.1%) samples. The number of CPE isolates from clinical samples alone (P=0.018) and from rectal plus clinical samples (P<0.001) increased yearly (Fig. 1B).
Fig. 1.
Distributions of genotypes and patients according to the hospital. (A) Carbapenemase genotypes and (B) patients with carbapenemase-producing Enterobacteriaceae isolates according to year and sample type.
Abbreviations: A, Kangnam Sacred Heart Hospital; B, Hangang Sacred Heart Hospital; C, Hallym University Sacred Heart Hospital; D, Dongtan Sacred Heart Hospital; KPC, K. pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA-48, oxacillinase-48; IMP, imipenemase; VIM, Verona integron-encoded metallo-β-lactamase.
The distribution of carbapenemase genotypes according to species is presented in Table 1. The most prevalent species among the KPC, NDM, and OXA-48-like producers was K. pneumoniae (72.8% of KPC, 66.7% of NDM, and 83.6% of OXA-48-like producers), followed by E. coli isolates (7.6% of KPC, 5.9% for each of NDM and OXA-48-like producers). E. cloacae (8.7%) and C. freundii (8.4%) isolates were more frequently NDM producers than KPC or OXA-48-like producers. The distribution of CPE isolates according to the four hospitals is shown in Table 2. In hospital C, the proportion of NDM (19.3%) and OXA-48-like (22.4%) was larger than that of KPC (8.3%), unlike in the other hospitals (P<0.001), reflecting local differences.
Table 1.
Distribution of the most prevalent carbapenemase genotypes according to carbapenemase-producing Enterobacteriaceae species
| KPC | NDM | OXA-48-like | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Rectal | Clinical | Subtotal | Rectal | Clinical | Subtotal | Rectal | Clinical | Subtotal | |
| Klebsiella pneumoniae | 239 (49.2) | 115 (23.7) | 354 (72.8) | 114 (31.9) | 124 (34.7) | 238 (66.7) | 217 (58.5) | 93 (25.1) | 310 (83.6) |
| Escherichia coli | 29 (6.0) | 8 (1.6) | 37 (7.6) | 16 (4.5) | 5 (1.4) | 21 (5.9) | 18 (4.9) | 4 (1.1) | 22 (5.9) |
| Enterobacter cloacae | 16 (3.3) | 4 (0.8) | 20 (4.1) | 26 (7.3) | 5 (1.4) | 31 (8.7) | 4 (1.1) | 1 (0.3) | 5 (1.3) |
| Citrobacter freundii | 3 (0.6) | 2 (0.4) | 5 (1.0) | 27 (7.6) | 3 (0.8) | 30 (8.4) | 10 (2.7) | 0 (0.0) | 10 (2.7) |
| Klebsiella oxytoca | 11 (2.3) | 5 (1.0) | 16 (3.3) | 5 (1.4) | 3 (0.8) | 8 (2.2) | 17 (4.6) | 1 (0.3) | 18 (4.9) |
| Serratia marcescens | 10 (2.1) | 9 (1.9) | 19 (3.9) | 1 (0.3) | 8 (2.2) | 9 (2.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Klebsiella aerogenes | 12 (2.5) | 3 (0.6) | 15 (3.1) | 2 (0.6) | 0 (0.0) | 2 (0.6) | 2 (0.5) | 1 (0.3) | 3 (0.8) |
| Citrobacter koseri | 11 (2.3) | 2 (0.4) | 13 (2.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Providencia rettgeri | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 12 (3.4) | 12 (3.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Enterobacter hormaechei | 1 (0.2) | 1 (0.2) | 2 (0.4) | 1 (0.3) | 0 (0) | 1 (0.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Enterobacter kobei | 2 (0.4) | 0 (0.0) | 2 (0.4) | 1 (0.3) | 0 (0.0) | 1 (0.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Pantoea agglomerans | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.6) | 0 (0.0) | 2 (0.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Citrobacter braakii | 1 (0.2) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| Enterobacter asubriae | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Kluyvera ascorbate | 0 (0.0) | 1 (0.2) | 1 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| Raoutella planticola | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.6) | 0 (0.0) | 2 (0.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Klebsiella ozaenae | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.3) | 0 (0.0) | 1 (0.3) |
| Klebsiella variicola | 1 (0.2) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Total | 336 (69.1) | 150 (30.9) | 486 (100.0) | 197 (55.2) | 160 (44.8) | 357 (100) | 271 (73.0) | 100 (27.0) | 371 (100) |
Data are expressed as number (%).
Abbreviations: KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA-48, oxacillinase-48.
Table 2.
Distribution of carbapenemase-producing Enterobacteriaceae
| Hospital A | Hospital B | Hospital C | Hospital D | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| Rectal | Clinical | Subtotal | Rectal | Clinical | Subtotal | Rectal | Clinical | Subtotal | Rectal | Clinical | Subtotal | |
| Klebsiella pneumoniae | 133 (21.6) | 42 (6.8) | 175 (28.5) | 19 (17.3) | 18 (16.4) | 37 (33.6) | 72 (4.4) | 49 (3.0) | 121 (7.4) | 15 (21.4) | 6 (8.6) | 21 (30.0) |
| Escherichia coli | 22 (3.6) | 3 (0.5) | 25 (4.1) | 3 (2.7) | 2 (1.8) | 5 (4.5) | 4 (0.2) | 3 (0.2) | 7 (0.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Enterobacter cloacae | 16 (2.6) | 4 (0.7) | 20 (3.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Serratia marcescens | 10 (1.6) | 9 (1.5) | 19 (3.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Klebsiella oxytoca | 8 (1.3) | 0 (0.0) | 8 (1.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.1) | 5 (0.3) | 7 (0.4) | 1 (1.4) | 0 (0.0) | 1 (1.4) |
| Klebsiella aerogens | 8 (1.3) | 1 (0.2) | 9 (1.5) | 3 (2.7) | 2 (1.8) | 5 (4.5) | 1 (0.1) | 0 (0.0) | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Citrobacter koseri | 10 (1.6) | 2 (0.3) | 12 (2.0) | 1 (0.9) | 0 (0.0) | 1 (0.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Citrobacter freundii | 3 (0.5) | 0 (0.0) | 3 (0.5) | 0 (0.0) | 2 (1.8) | 2 (1.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Enterobacter hormaechei | 1 (0.2) | 1 (0.2) | 2 (0.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Enterobacter kobei | 2 (0.3) | 0 (0.0) | 2 (0.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Citrobacter braakii | 1 (0.2) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Klebsiella variicola | 1 (0.2) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| KPC | 215 (35.0) | 63 (10.2) | 278 (45.2) | 26 (23.6) | 24 (21.8) | 50 (45.5) | 79 (4.8) | 57 (3.5) | 136 (8.3) | 16 (22.9) | 6 (8.6) | 22 (31.4) |
| Klebsiella pneumoniae | 4 (0.7) | 0 (0.0) | 4 (0.7) | 2 (1.8) | 0 (0) | 2 (1.8) | 106 (6.5) | 124 (7.6) | 230 (14.1) | 1 (1.4) | 0 (0.0) | 2 (2.9) |
| Enterobacter cloacae | 2 (0.3) | 0 (0.0) | 2 (0.3) | 0 (0.0) | 2 (1.8) | 2 (1.8) | 24 (1.5) | 3 (0.2) | 27 (1.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Citrobacter freundii | 0 (0.0) | 1 (0.2) | 1 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 26 (1.6) | 2 (0.1) | 28 (1.7) | 1 (1.4) | 0 (0.0) | 1 (1.4) |
| Escherichia coli | 5 (0.8) | 0 (0.0) | 5 (0.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 5 (0.3) | 3 (0.2) | 8 (0.5) | 6 (8.6) | 2 (2.9) | 8 (11.4) |
| Providencia rettgeri | 0 (0.0) | 11 (1.8) | 11 (1.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.1) | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Serratia marcescens | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.1) | 8 (0.5) | 9 (0.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Klebsiella oxytoca | 1 (0.2) | 1 (0.2) | 2 (0.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (0.2) | 2 (0.1) | 5 (0.3) | 1 (1.4) | 0 (0.0) | 1 (1.4) |
| Klebsiella aerogenes | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.1) | 0 (0.0) | 2 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Pantoea agglomerans | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.1) | 0 (0.0) | 2 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Raoutella planticola | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.1) | 0 (0.0) | 2 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Enterobacter hormaechei | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.4) | 0 (0.0) | 1 (1.4) |
| Enterobacter kobei | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.1) | 0 (0.0) | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| NDM | 12 (2.0) | 13 (2.1) | 25 (4.1) | 2 (1.8) | 2 (1.8) | 4 (3.6) | 172 (10.5) | 143 (8.8) | 315 (19.3) | 11 (15.7) | 2 (2.9) | 13 (18.6) |
| Klebsiella pneumoniae | 2 (0.3) | 0 (0.0) | 2 (0.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 215 (13.2) | 93 (5.7) | 308 (18.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Escherichia coli | 1 (0.2) | 1 (0.2) | 2 (0.3) | 1 (0.9) | 0 (0.0) | 1 (0.9) | 16 (1.0) | 3 (0.2) | 19 (1.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Klebsiella oxytoca | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 17 (1.0) | 1 (0.1) | 18 (1.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Citrobacter freundii | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 10 (0.6) | 0 (0.0) | 10 (0.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Enterobacter cloacae | 1 (0.2) | 0 (0.0) | 1 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (0.2) | 1 (0.1) | 4 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Klebsiella aerogenes | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.1) | 1 (0.1) | 3 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Citrobacter braakii | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.1) | 0 (0.0) | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Kluyvera ascorbate | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.1) | 0 (0.0) | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Klebsiella ozaenae | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.1) | 0 (0.0) | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| OXA-48-like | 4 (0.7) | 1 (0.2) | 5 (0.8) | 1 (0.9) | 0 (0.0) | 1 (0.9) | 266 (16.3) | 99 (6.1) | 365 (22.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Total | 462 (75.1) | 153 (24.9) | 615 (100.0) | 58 (52.7) | 52 (47.3) | 110 (100) | 1,034 (63.4) | 598 (36.6) | 1,632 (100.0) | 53 (75.7) | 16 (22.9) | 70 (100.0) |
Data are expressed as number (%).
Abbreviations: KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA-48, oxacillinase-48; Hospitals A, Kangnam Sacred Heart Hospital; B, Hangang Sacred Heart Hospital; C, Hallym University Sacred Heart Hospital; D, Dongtan Sacred Heart Hospital.
Antimicrobial susceptibility profile
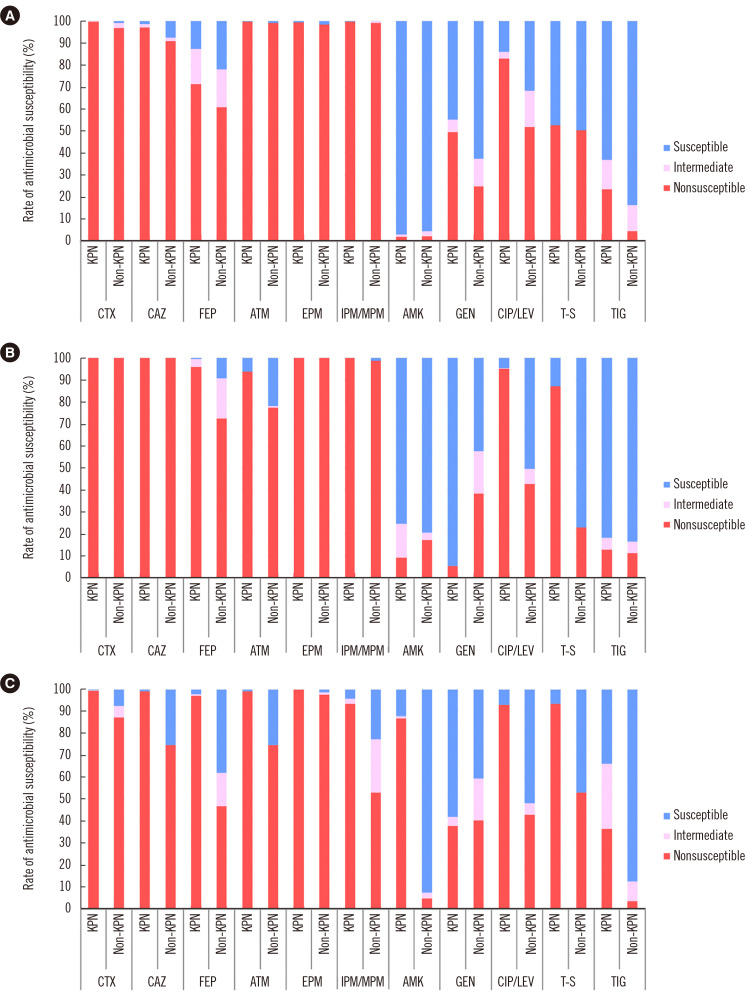
More than 90% of KPC-harboring Enterobacteriaceae isolates were nonsusceptible to cefotaxime, ceftazidime, aztreonam, ertapenem, imipenem, and meropenem (Fig. 2A). The isolates harboring NDM showed similar nonsusceptibility patterns; however, more than 90% of K. pneumoniae isolates (95.8% of NDM vs. 71.5% of KPC producers) were nonsusceptible to cefepime (Fig. 2B). As for Enterobacteriaceae harboring OXA-48-like, K. pneumoniae isolates were highly nonsusceptible (nonsusceptibility rate >90%) to cefotaxime, ceftazidime, cefepime, aztreonam, ertapenem, imipenem, meropenem, ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole when compared with non-K. pneumoniae isolates (Fig. 2C). All K. pneumoniae isolates harboring KPC, NDM, and OXA-48-like genes showed higher tigecycline nonsusceptibility rates (12.9%–36.5%) than non-K. pneumoniae isolates (3.8%–11.3%) (P<0.001).
Fig. 2.
Antimicrobial susceptibility profiles of Enterobacteriaceae isolates producing (A) KPC, (B) NDM, and (C) OXA-48-like.
Abbreviations: CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; EPM, ertapenem; IMP/MPM, imipenem (or meropenem); AMK, amikacin; GEN, gentamicin; CIP/LEV, ciprofloxacin (or levofloxacin); T-S, trimethoprim-sulfamethoxazole; TIG, tigecycline; KPN, K. pneumoniae; KPC, K. pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA-48, oxacillinase-48.
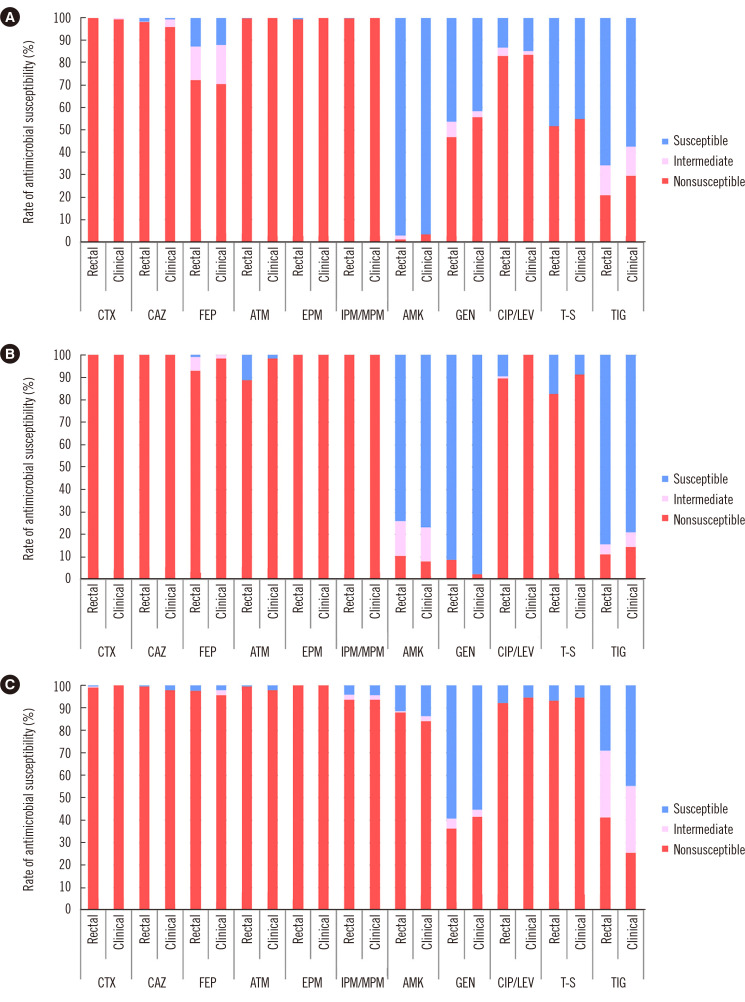
Regarding K. pneumoniae, representing most of the CPE isolates, more than 90% of the isolates with KPC, NDM, and OXA-48-like were nonsusceptible to cefotaxime, ceftazidime, cefepime, aztreonam, ertapenem, imipenem, and meropenem. KPC-harboring K. pneumoniae isolates and NDM-harboring K. pneumoniae from rectal samples showed lesser nonsusceptibility rates (<90%) to cefepime and aztreonam, respectively (Fig. 3). K. pneumoniae harboring OXA-48-like in both rectal and clinical samples presented more than 90% of nonsusceptibility to ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole.
Fig. 3.
Antimicrobial susceptibility profiles of K. pneumoniae isolates producing (A) KPC, (B) NDM, and (C) OXA-48-like by sample types.
Abbreviations: CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; EPM, ertapenem; IMP/MPM, imipenem (or meropenem); AMK, amikacin; GEN, gentamicin; CIP/LEV, ciprofloxacin (or levofloxacin); T-S, trimethoprim-sulfamethoxazole; TIG, tigecycline; KPC, K. pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA-48, oxacillinase-48.
DISCUSSION
Increasing trends in CPE prevalence and spread have been identified worldwide [7, 15]. Concordant with previous studies, we mainly identified KPC (38.8%), OXA-48-like (29.6%), and NDM (28.5%) producers [9, 16]. The predominantly isolated species (K. pneumoniae and E. coli) were also similar to those reported in other studies [9, 16].
In the USA and Europe, K. pneumoniae is the main pathogen harboring KPC, and the first case of infection with a KPC-producing extremely nonsusceptible K. pneumoniae isolate in Korea was reported in 2010 [17]. The first outbreak of KPC-producing K. pneumoniae in Korea in 2013 was likely due to clonal spread [18].
The Indian subcontinent is recognized as an NDM endemic zone, and the associated species are mostly K. pneumoniae and E. coli. NDM-producing K. pneumoniae isolates producing nearly identical XbaI pulsed-field gel electrophoresis patterns were first detected in Korea in 2010 [19]. Nosocomial transmission of genetically related NDM-producing E. coli from four elderly patients was reported in a Korean hospital in 2011 [20]. In this study, E. cloacae and C. freundii were frequently detected NDM producers in rectal samples. These species were also more commonly detected among NDM rather than KPC and OXA-48-like producers in Canada and major strains in Korea in a study conducted in 2015 [9, 21].
OXA-48-like producers are often detected in Northern Africa and the Mediterranean area and is increasingly being reported in Enterobacteriaceae, such as K. pneumoniae and E. coli [7, 16]. Korea is known for outbreaks of K. pneumoniae and E. coli that produce OXA-232, which was identified as an OXA-48 variant differing in a few amino acids [22, 23]. Clonal and horizontal spread of these OXA-232-harboring species has caused persistent outbreaks in Korea [9, 22].
Although the proportions of IMP (0.6%) and VIM (0.6%) producers among CPE were relatively low in this study, strains with these genes cause sporadic outbreaks mainly in Southeast Asia and the Americas, respectively [24]. In Korea, an outbreak of IMP-producing carbapenem-nonsusceptible K. pneumoniae strains from four ICU patients was reported in 2017 [25]. After the first identification of VIM in 1999, it has spread mainly in Pseudomonas aeruginosa, with several outbreaks [26, 27].
The predominant CPE genotypes in Korea showed hospital-specific differences. In particular, hospital C had more NDM and OXA-48-like producers than KPC compared with the other hospitals. Thus, the control of nosocomial spread of CPE may require different measures depending on the local circumstances.
KPC-producing Enterobacteriaceae isolates were highly nonsusceptible to cefotaxime, ceftazidime, aztreonam, ertapenem, imipenem, and meropenem. Treatment options for infections caused by KPC producers remain limited because these strains are mostly multidrug-nonsusceptible [7]. Among cephalosporins, nonsusceptibility rates for cefepime (71.5% of K. pneumoniae isolates and 60.9% of non-K. pneumoniae isolates) were lower than those for cefotaxime and ceftazidime (>90% of both K. pneumoniae and non-K. pneumoniae isolates). Cefepime is a fourth-generation cephalosporin proven to be an extended-spectrum antimicrobial against gram-negative pathogens. Compared with other cephalosporins, cefepime exhibits high affinity for KPC, which has led to high susceptibility rates in previous studies [28 29]. The use of cefepime in combination with amoxicillin-clavulanate has been recommended for the treatment of KPC-producing K. pneumoniae infections [30].
In line with our results, NDM-producing Enterobacteriaceae were nonsusceptible to nearly all antimicrobials, except tigecycline [2, 31]. Typically, metallo-β-lactamases, such as NDM, weakly hydrolyze aztreonam [32]. However, the NDM-producing K. pneumoniae isolates exhibited a high nonsusceptibility rate (>90%) to aztreonam in clinical samples. The isolates likely had extended-spectrum β-lactamase (ESBL) or permeability defects simultaneously [31]. Plasmids containing NDM are diverse and harbor numerous nonsusceptible genes related to carbapenemases, such as OXA-48, VIM types, ESBLs, AmpCs, and sulfamethoxazole-nonsusceptible genes, and thus frequently provide multidrug or extensive drug nonsusceptibility [7, 31].
In general, OXA-48-like producers effectively hydrolyze penicillins, whereas they weakly hydrolyze carbapenems and cephalosporins [23]. Non-K. pneumoniae isolates harboring OXA-48-like in our study showed similar trends. However, OXA-48-like-producing K. pneumoniae isolates exhibited high nonsusceptibility (>90%) to all cephalosporins, aztreonam, carbapenems, fluoroquinolones, and even trimethoprim-sulfamethoxazole. When combining ESBL or permeability defects, OXA-48 producers showed a high level of nonsusceptibility to broad-spectrum cephalosporins, aminoglycosides, fluoroquinolones, and carbapenems [23]. In a previous study in Korea, all OXA-232-producing K. pneumoniae isolates co-produced ESBL, such as CTX-M-15 [9]. Therefore, only tigecycline remains effective against these OXA-48-producing K. pneumoniae isolates [7, 9, 23]. The dissemination of the OXA-48-like gene to various Enterobacteriaceae has been worrisome, because it is unusually located on a small and self-transferable plasmid [23, 33].
This study has some limitations. We only evaluated carbapenemases, not ESBLs and AmpCs. However, most CPE isolated in Korea also harbor an ESBL [9]. Differences in the commercial platforms utilized for the identification and antimicrobial susceptibility testing in the different hospitals might have affected the results. In addition, hospital size likely influenced the distribution results.
In summary, the impact of CPE in Korea is primarily due to KPC-, NDM-, and OXA-48-like-producing K. pneumoniae isolates. Enterobacteriaceae isolates harboring these carbapenemase genes were mostly multidrug-nonsusceptible or extensively drug-nonsusceptible. Moreover, a stepwise and significant increase in CPE was noted in recent years. These genetic trends have affected the antimicrobial susceptibility patterns. These worsening antimicrobial nonsusceptibility trends should be reflected in precise and aggressive control strategies to combat CPE.
ACKNOWLEDGEMENTS
The authors appreciate the support and cooperation of clinical microbiology technicians in Hallym University Medical Centers.
Footnotes
AUTHOR CONTRIBUTIONS
Song W designed the study; Jeong S and Lee N analyzed the data and wrote the manuscript; and Kim HS, Kim HS, Park MJ, and Kim JS edited the manuscript. All authors reviewed and approved the manuscript.
CONFLICTS OF INTEREST
None declared.
RESEARCH FUNDING
This work was supported by a National Research Foundation of Korea (NRF) grant, funded by the Korean Government (Ministry of Science and ICT) (NRF-2017R1C1B2004597). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Doi Y, Paterson DL. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med. 2015;36:74–84. doi: 10.1055/s-0035-1544208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P. Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Med Mal Infect. 2014;44:51–6. doi: 10.1016/j.medmal.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–81. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 4.Lee M, Choi TJ. Antimicrobial resistance caused by KPC-2 encoded by promiscuous plasmids of the Klebsiella pneumoniae ST307 strain. Ann Lab Med. 2021;41:86–94. doi: 10.3343/alm.2021.41.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–54. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–8. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong SH, Kim HS, Kim JS, Shin DH, Kim HS, Park MJ, et al. Prevalence and molecular characteristics of carbapenemase-producing Enterobacteriaceae from five hospitals in Korea. Ann Lab Med. 2016;36:529–35. doi: 10.3343/alm.2016.36.6.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee H, Yoon EJ, Kim D, Jeong SH, Won EJ, Shin JH, et al. Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 to April 2017: first one-year report from Kor-GLASS. Euro Surveill. 2018;23:1800047. doi: 10.2807/1560-7917.ES.2018.23.42.1800047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song W, Hong SG, Yong D, Jeong SH, Kim HS, Kim HS, et al. Combined use of the modified Hodge test and carbapenemase inhibition test for detection of carbapenemase-producing Enterobacteriaceae and metallo-beta-lactamase-producing Pseudomonas spp. Ann Lab Med. 2015;35:212–9. doi: 10.3343/alm.2015.35.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong S, Kim JO, Jeong SH, Bae IK, Song W. Evaluation of peptide nucleic acid-mediated multiplex real-time PCR kits for rapid detection of carbapenemase genes in gram-negative clinical isolates. J Microbiol Methods. 2015;113:4–9. doi: 10.1016/j.mimet.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 13.CLSI, author. CLSI M100S. 27th ed. Clinical and Laboratory Standards Institute; Wayne, PA: 2016. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 14.European Committee on Antimicrobial Susceptibility Testing, author. [Updated on Jan 2021];Clinical breakpoints - breakpoints and guidance. http://www.eucast.org/clinical_breakpoints .
- 15.Song W, Park MJ, Jeong S, Shin DH, Kim JS, Kim HS, et al. Rapid identification of OXA-48-like, KPC, NDM, and VIM carbapenemase-producing Enterobacteriaceae from culture: evaluation of the RESIST-4 O.K.N.V. multiplex lateral flow assay. Ann Lab Med. 2020;40:259–63. doi: 10.3343/alm.2020.40.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20:821–30. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 17.Rhee JY, Park YK, Shin JY, Choi JY, Lee MY, Peck KR, et al. KPC-producing extreme drug-resistant Klebsiella pneumoniae isolate from a patient with diabetes mellitus and chronic renal failure on hemodialysis in South Korea. Antimicrob Agents Chemother. 2010;54:2278–9. doi: 10.1128/AAC.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong SK, Yong D, Kim K, Hong SS, Hong SG, Khosbayar T, et al. First outbreak of KPC-2-producing Klebsiella pneumoniae sequence type 258 in a hospital in South Korea. J Clin Microbiol. 2013;51:3877–9. doi: 10.1128/JCM.01730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim MN, Yong D, An D, Chung HS, Woo JH, Lee K, et al. Nosocomial clustering of NDM-1-producing Klebsiella pneumoniae sequence type 340 strains in four patients at a South Korean tertiary care hospital. J Clin Microbiol. 2012;50:1433–6. doi: 10.1128/JCM.06855-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo JS, Kim HM, Koo HS, Yang JW, Yoo JI, Kim HS, et al. Nosocomial transmission of NDM-1-producing Escherichia coli ST101 in a Korean hospital. J Antimicrob Chemother. 2013;68:2170–2. doi: 10.1093/jac/dkt126. [DOI] [PubMed] [Google Scholar]
- 21.Sekirov I, Croxen MA, Ng C, Azana R, Chang Y, Mataseje L, et al. Epidemiologic and genotypic review of carbapenemase-producing organisms in British Columbia, Canada, between 2008 and 2014. J Clin Microbiol. 2016;54:317–27. doi: 10.1128/JCM.02289-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong SH, Lee KM, Lee J, Bae IK, Kim JS, Kim HS, et al. Clonal and horizontal spread of the blaOXA-232 gene among Enterobacteriaceae in a Korean hospital. Diagn Microbiol Infect Dis. 2015;82:70–2. doi: 10.1016/j.diagmicrobio.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67:1597–606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 24.Walsh TR. Emerging carbapenemases: a global perspective. Int J Antimicrob Agents. 2010;36:S8–14. doi: 10.1016/S0924-8579(10)70004-2. [DOI] [PubMed] [Google Scholar]
- 25.Lee JY, Park JY, Kim JH, Lee YH, Yang HY, Yoo JS. Outbreak of imipenemase-1-producing carbapenem-resistant Klebsiella pneumoniae in an intensive care unit. Korean J Crit Care Med. 2017;32:29–38. doi: 10.4266/kjccm.2016.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, et al. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–90. doi: 10.1128/AAC.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K. Epidemiology and characteristics of metallo-beta-lactamase-producing Pseudomonas aeruginosa. Infect Chemother. 2015;47:81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tseng IL, Liu YM, Wang SJ, Yeh HY, Hsieh CL, Lu HL, et al. Emergence of carbapenemase producing Klebsiella pneumonia and spread of KPC-2 and KPC-17 in Taiwan: A nationwide study from 2011 to 2013. PLoS One. 2015;10:e0138471. doi: 10.1371/journal.pone.0138471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endimiani A, Perez F, Bonomo RA. Cefepime: a reappraisal in an era of increasing antimicrobial resistance. Expert Rev Anti Infect Ther. 2008;6:805–24. doi: 10.1586/14787210.6.6.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sader HS, Fritsche TR, Jones RN. Potency and spectrum trends for cefepime tested against 65746 clinical bacterial isolates collected in North American medical centers: results from the SENTRY Antimicrobial Surveillance Program (1998-2003) Diagn Microbiol Infect Dis. 2005;52:265–73. doi: 10.1016/j.diagmicrobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Ji S, Lv F, Du X, Wei Z, Fu Y, Mu X, et al. Cefepime combined with amoxicillin/clavulanic acid: a new choice for the KPC-producing K. pneumoniae infection. Int J Infect Dis. 2015;38:108–14. doi: 10.1016/j.ijid.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Bartolini A, Basso M, Franchin E, Menegotto N, Ferrari A, De Canale E, et al. Prevalence, molecular epidemiology and intra-hospital acquisition of Klebsiella pneumoniae strains producing carbapenemases in an Italian teaching hospital from January 2015 to September 2016. Int J Infect Dis. 2017;59:103–9. doi: 10.1016/j.ijid.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Fuster B, Tormo N, Salvador C, Gimeno C. Detection of two simultaneous outbreaks of Klebsiella pneumoniae coproducing OXA-48 and NDM-1 carbapenemases in a tertiary-care hospital in Valencia, Spain. New Microbes New Infect. 2020;34:100660. doi: 10.1016/j.nmni.2020.100660. [DOI] [PMC free article] [PubMed] [Google Scholar]