Abstract
In this study, the characteristic patterns of ferroptosis in clear cell renal cell carcinoma (ccRCC) were systematically investigated with the interactions between ferroptosis and the tumor microenvironment (TME). On the mRNA expression profiles of 57 ferroptosis-related genes (FRGs), three ferroptosis patterns were constructed, with distinct prognosis and immune cell infiltrations (especially T cells and dendritic cells). The high ferroptosis scores were characterized by poorer prognosis, increased T cell infiltration, higher immune and stromal scores, elevated tumor mutation burden, and enhanced response to anti-CTLA4 immunotherapy. Meanwhile, the low ferroptosis scores were distinctly associated with enhanced tumor purity and amino acid and fatty acid metabolism pathways. Following validation, the ferroptosis score was an independent and effective prognostic factor. Collectively, ferroptosis could be involved in the diverse and complex TME. Evaluation of the ferroptosis patterns may heighten the comprehension about immune infiltrations in the TME, assisting oncologists to generate individualized immunotherapeutic strategies.
Keywords: ferroptosis, clear cell renal cell carcinoma, tumor microenvironment, immunotherapy, prognosis
Graphical abstract
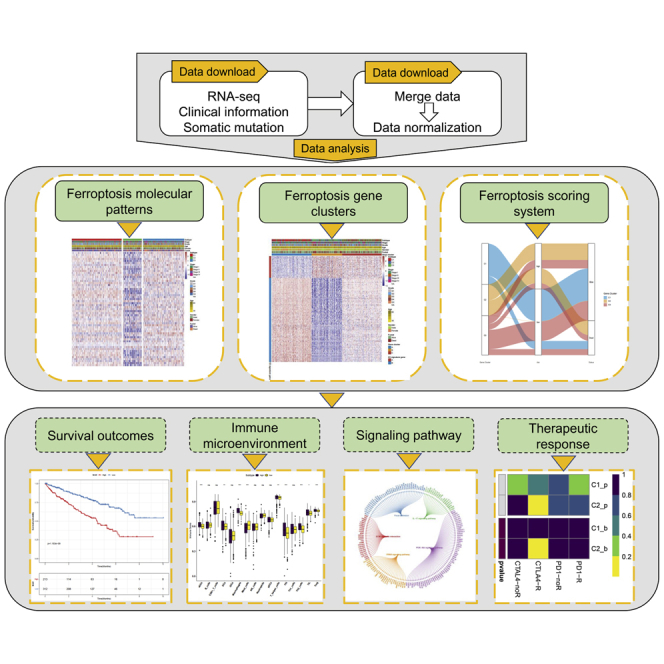
Characteristics of ferroptosis in clear cell renal cell carcinoma (ccRCC) were systematically investigated. Ferroptosis scores associated with increased T cell infiltration, higher immune and stromal scores, elevated tumor mutation burden, and enhanced anti-CTLA4 response involved in tumor microenvironment therefore are likely to affect the immunotherapy cohorts (GEO: GSE78220 and IMvigor210) in anti-PD-1 treatment.
Introduction
Renal cell carcinoma (RCC) is the second most lethal malignant tumor in the urinary system; it mainly consists of the subtypes including clear cell renal cell carcinoma (ccRCC), papillary renal cell carcinoma (pRCC), and chromophobe renal cell carcinoma (chRCC).1 ccRCC is the main histological subtype, occupying approximately 85% of all RCC cases.2 According to the statistics, about ∼74,000 new cases were diagnosed as ccRCC in the United States in 2019.3 Presently, laparoscopic partial nephrectomy and radical nephrectomy are the major therapeutic strategies, especially for the localized RCC patients.4 It is noteworthy that approximately 30% of patients with ccRCC develop distant metastases during the diagnosis.5 Considering that the ccRCC patients are insensitive to the radiation, hormone, and cytotoxic therapies, several targeted agents, including sunitinib, sorafenib, lenvatinib, and nivolumab, have been approved to treat metastatic ccRCC.6 However, the therapeutic effects are still limited. Moreover, despite that an increasing number of PD-1/PD-L1 blocking immunotherapy drugs have been approved for the treatment of ccRCC, not all ccRCC patients respond to the immunotherapy.7 Hence it is of clinical significance to identify which patients will benefit from immunotherapy.
Ferroptosis, as a cell death regulating process, is characterized by iron-dependent lipid peroxidation.8 Ferroptosis is tightly associated with various metabolism processes, such as iron, amino acids, NADH, phospholipids, and glutathione.9 Moreover, lipid peroxidation inhibitors, iron chelators, and reduction of intracellular polyunsaturated fatty acids can inhibit the progression of ferroptosis.10 Currently, the induction of ferroptosis has become a promising therapeutic approach to efficiently trigger cancer cell death, especially in those tumors resistant to the traditional treatment.11 Recent studies have revealed that CD8+ T cells suppress tumor growth through inducing the ferroptosis.12 Moreover, Liu et al.13 have developed the ferroptosis potential index (FPI) to explore the functional roles of ferroptosis in a variety of cancers and revealed that ferroptosis is associated with the cancer hallmarks tumor microenvironment (TME), drug resistance, and patient survival. These findings suggest that ferroptosis plays an essential role in the TME.
In the present study, the three ferroptosis patterns were constructed, associated with distinct prognosis and TME features. We first proposed using the ferroptosis scores to quantify the ferroptosis patterns for each ccRCC patient based on the mRNA expression profiles of ferroptosis-related genes (FRGs). The scoring system may assist oncologists to make more efficient and individualized immunotherapeutic strategies.
Results
Ferroptosis-related molecular patterns with distinct survival and TME features in ccRCC
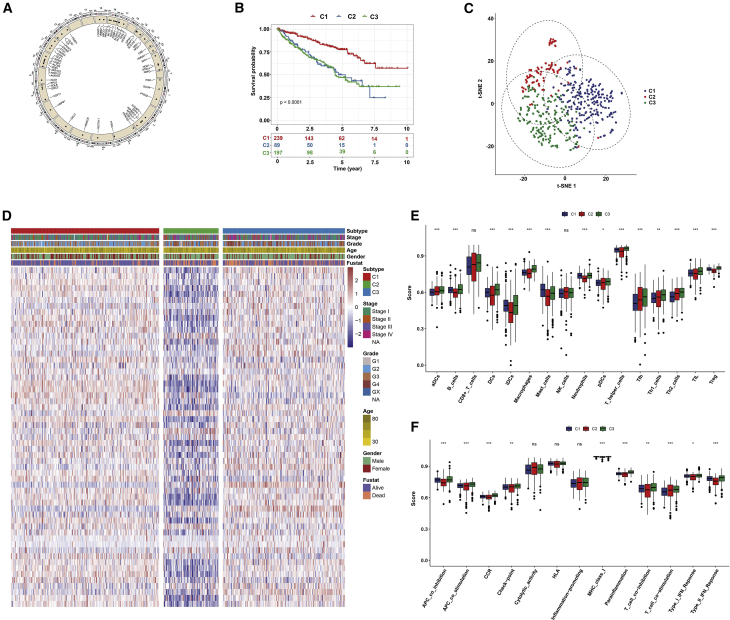
Totally, 57 FRGs were included in this study. Figure 1A showed the location of copy number variation (CNV) of these FRGs on chromosomes. Based on the mRNA expression profiles of 57 FRGs in ccRCC samples from The Cancer Genome Atlas (TCGA) database, ccRCC patients were classified into the three molecular patterns (C1: n = 239; C2: n = 89; C3: n = 197) by unsupervised clustering analysis (Figures S1A–S1C). The three ferroptosis patterns with distinct clinical outcomes were established (p < 0.0001; Figure 1B). Patients in C1 witnessed a significant increase in the survival time. t-Distributed stochastic neighbor embedding (t-SNE) confirmed that the three subtypes can be completely distinguished (Figure 1C). Figure 1D visualized the three patterns with different clinicopathological features of ccRCC patients. Furthermore, we evaluated the correlation between the patterns and tumor immune microenvironment features. Our data showed significant differences in immune cell infiltration (Figure 1E) and immune function (Figure 1F) among the three molecular patterns, especially for dendritic cells (DCs) and T cells.
Figure 1.
Ferroptosis-related molecular patterns with distinct prognosis and immune cell infiltrations and functions in ccRCC
(A) The location of CNV of 57 FRGs on chromosomes. (B) Kaplan-Meier curves for the three molecular patterns of ccRCC patients. (C) t-SNE of the mRNA expression profiles of FRGs from the ccRCC samples in the TCGA dataset confirmed the three clusters: C1 (purple), C2 (red), and C3 (green). (D) Heatmap depicted the correlation between the patterns and different clinicopathological features. (E and F) Boxplots showed the scores of immune infiltrations (E) and functions (F) among the three patterns. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Ns, not significant.
Prognosis and TME characteristics in three ferroptosis gene clusters for ccRCC
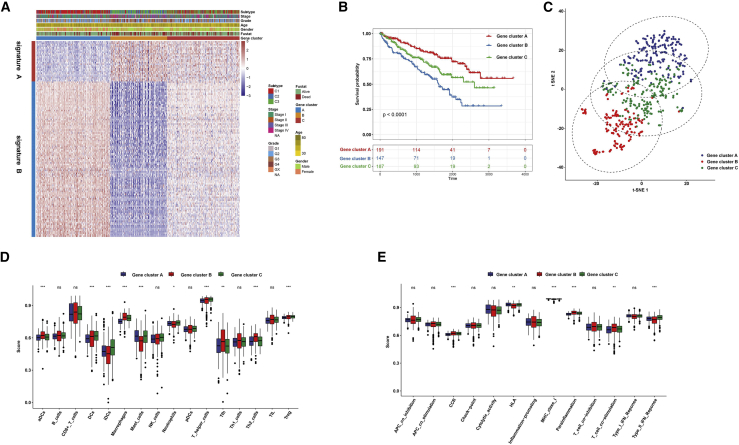
We further probed into the biological behaviors among the three ferroptosis molecular patterns. Differential expression analysis was constructed among these three patterns. Based on the common differentially expressed genes (DEGs), the unsupervised clustering was presented, and the TCGA-ccRCC cohort was classified into the three gene clusters (A–C; Figures S2A–S2C). The dimension reduction was presented by the Boruta algorithm based on the ferroptosis gene signatures A and B. The heatmap transcriptomic profile depicted the 276 most abundant DEGs identified across the genomic clusters (Figure 2A). These DEGs in the gene signatures A and B separately exhibited various biological functions (Figures S3A and S3B). We further probed into the prognostic implications of the ferroptosis gene clusters by overall survival (OS). The data showed that the subjects in gene cluster A exhibited longer OS time, while those in gene cluster B possessed a pessimistic prognosis (p < 0.0001; Figure 2B). Two-dimensional t-SNE confirmed the cluster assignments (Figure 2C). Then, we explored whether the three genes clusters had distinct TME features. As a result, gene cluster B showed the highest scores of T cells (including follicular helper T [Tfh] cells, type 2 T helper [Th2] cells, and regulatory T [Treg] cells), macrophages, and activated dendritic cells (aDCs) compared with gene clusters A and C (Figure 2D). Moreover, the highest CCR, parainflammation, and T cell stimulation scores and the lowest human leukocyte antigen (HLA) and type II IFN response scores were in gene cluster B rather than gene clusters A and C (Figure 2E). Collectively, the coherence between the prognostic and TME features in the three gene clusters indicated that this classification was reliable and reasonable.
Figure 2.
Prognosis and TME characteristics in three ferroptosis gene clusters for ccRCC patients
(A) Clinical features of the three ferroptosis gene clusters. (B) Kaplan-Meier survival analysis for patients in the three gene clusters. (C) t-SNE confirmed the classification. (D) Boxplots showed immune cell infiltration scores differed in the three clusters. (E) Boxplots depicted the differences in immune scores in the three clusters. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Development of the ferroptosis scoring system for ccRCC
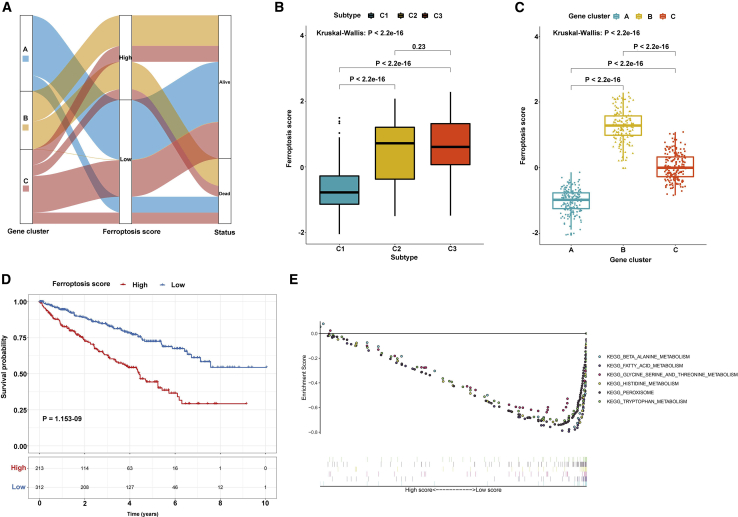
The ferroptosis score for each ccRCC sample was calculated based on principal-component analysis (PCA). All patients were then divided into the high and low ferroptosis score groups. Figure 3A showed the ferroptosis score distribution and survival outcomes of patients in the three gene clusters. Patients in C2 had the highest ferroptosis score compared with C1 and C3 (p < 2.2e−16; Figure 3B). Meanwhile, there was the highest ferroptosis score in gene cluster B (p < 2.2e−16; Figure 3C). Subjects with high ferroptosis score usually implied an unfavorable prognosis compared with those with low ferroptosis score (p = 1.153−9; Figure 3D), consistent with our previous studies. Gene set enrichment analysis (GSEA) results showed that β-alanine, fatty acid, glycine serine and threonine, histidine, and tryptophan metabolism pathways were significantly enriched in the low ferroptosis score group (Figure 3E). However, there were no significant pathways enriched in the high ferroptosis score group.
Figure 3.
Development of the ferroptosis scoring system for ccRCC
(A) Alluvial diagram of three gene clusters, ferroptosis scores, and survival status. (B) Boxplot depicted the differences in ferroptosis scores among the three molecular patterns. (C) Boxplot showed the differences in ferroptosis scores among the three gene clusters. (D) Kaplan-Meier survival analysis for patients with high and low ferroptosis scores. (E) GSEA identified metabolism-related pathways enriched in the low ferroptosis score group.
Ferroptosis score as an independent prognostic factor for ccRCC
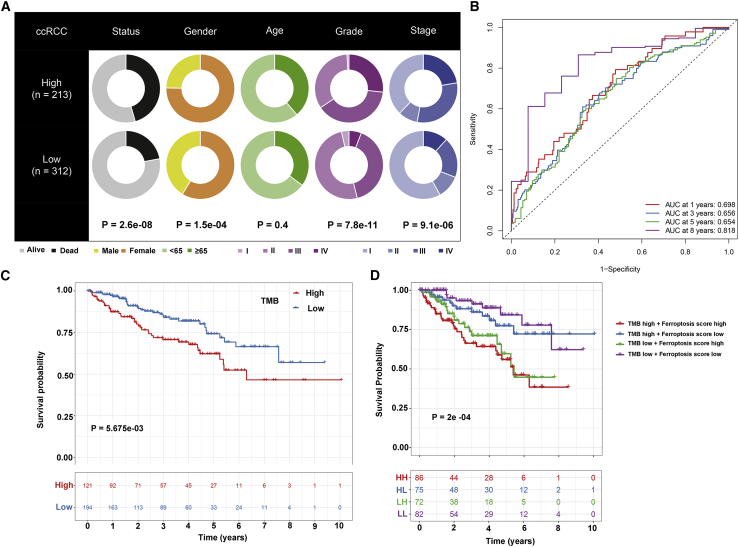
The prognostic values of ferroptosis scores were analyzed in depth. Our results showed that in the TCGA-ccRCC cohort, the ferroptosis score was significantly correlated to survival outcomes (p = 2.6e−8), gender (p = 1.5e−4), grade (p = 7.8e−11), and stage (p = 9.1e−6) in Figure 4A. Following multivariate cox regression analysis, ferroptosis score was confirmed as an independent risk factor for ccRCC (hazard ratio [HR]: 1.749, 95% confidence interval [CI]: 1.336–2.289, p = 4.66e−5; Table 1). Area under the curves (AUCs) for 1-, 3-, 5-, and 8-year survival time were 0.698, 0.656, 0.654, and 0.818, suggesting that the ferroptosis score possessed a robust and reliable capacity for predicting the prognosis for ccRCC patients (Figure 4B). The somatic mutations were frequently detected in ccRCC by the MutSigCV algorithm. Among ccRCC samples, VHL showed the most frequent mutations (49%), followed by PBRM1 (42%) and SETD2 (12%) in Figure S4. Our further analysis showed that patients with the high tumor mutation burden (TMB) score usually indicated shorter survival time than those with the low TMB score (p = 5.675e−3; Figure 4C). The survival advantage in the low ferroptosis score group was evident in the patients with the low TMB score (p = 2e−4; Figure 4D). Collectively, ferroptosis score may serve as an independent prognostic factor for ccRCC patients.
Figure 4.
The prognostic values of ferroptosis score for ccRCC patients
(A) Clinical features for the high and low ferroptosis score groups. (B) ROCs for 1-, 3-, 5-, and 8-year survival time based on the ferroptosis score. (C) Kaplan-Meier survival analysis for the high and low TMB score groups. (D) Kaplan-Meier survival analysis for patients stratified by ferroptosis and TMB scores.
Table 1.
Multivariate Cox regression analysis results of clinicopathological characteristics and ferroptosis score
| Variable | HR | Low 95% CI | Upper 95% CI | p value |
|---|---|---|---|---|
| OS | ||||
| Male gender | 0.826 | 0.59 | 1.157 | 2.06E |
| Age | 1.031 | 1.016 | 1.047 | 5.81E−5 |
| Grade | 1.289 | 1.01 | 1.646 | 4.15E−2 |
| Stage II | 1.076 | 0.534 | 2.17 | 8.38E−1 |
| Stage III | 2.147 | 1.382 | 3.335 | 6.77E−4 |
| Stage IV | 4.856 | 3.061 | 7.703 | 1.91E−11 |
| Score | 1.418 | 1.199 | 1.676 | 4.51E−5 |
| DFS | ||||
| Male gender | 0.746 | 0.436 | 1.276 | 2.84E−1 |
| Age | 1.012 | 0.99 | 1.035 | 2.74E−1 |
| Grade | 1.268 | 0.883 | 1.821 | 1.99E−1 |
| Stage II | 2.956 | 0.918 | 9.523 | 6.94E−2 |
| Stage III | 5.821 | 2.449 | 13.840 | 6.70E−5 |
| Stage IV | 18.507 | 7.564 | 45.277 | 1.63E−10 |
| Score | 1.749 | 1.336 | 2.289 | 4.66E−5 |
Ferroptosis score is associated with TME features of ccRCC
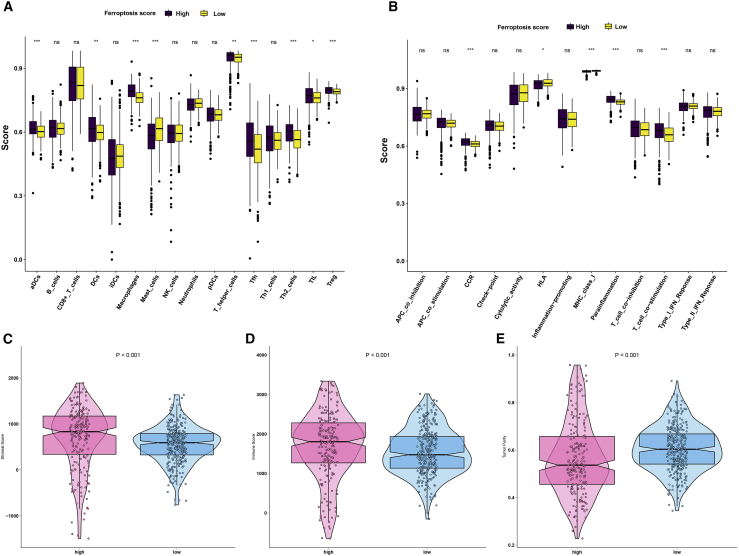
The correlation between ferroptosis score and TME features was further explored in this study. The data showed that the high ferroptosis score group had significantly higher scores of aDCs, DCs, plasmacytoid dendritic cells (pDCs), macrophages, Tfh cells, Th2 cells, tumor-infiltrating lymphocytes (TILs), and Treg cells than the low ferroptosis score group (Figure 5A). Meanwhile, there were lower scores of mast cells in the high ferroptosis score group compared with the low ferroptosis score group (Figure 5A). Patients with high ferroptosis score exhibited significantly higher scores of CCR, parainflammation, and T cell co-stimulation, and lower scores of HLA compared with those with low ferroptosis score (Figure 5B). Furthermore, patients in the high ferroptosis score group had distinctly higher stromal (p < 0.001; Figure 5C) and immune (p < 0.001; Figure 5D) scores in comparison with those in the low ferroptosis score group. On the contrary, lower tumor purity was detected in the high ferroptosis score group than in the low ferroptosis score group (p < 0.001; Figure 5E). Taken together, ferroptosis score exhibited a close association with TME for ccRCC.
Figure 5.
Ferroptosis score is associated with TME of ccRCC
(A) Boxplots depicted the differences in immune cell infiltrations between high and low ferroptosis score patients. (B) The differences in immune cell functions between the high and low ferroptosis score groups. (C–E) Violin diagram showed the differences in (C) stromal score, (D) immune score, and (E) tumor purity between the high and low ferroptosis score groups. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
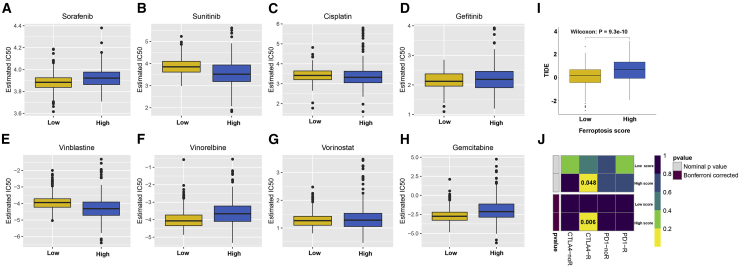
High ferroptosis score is more sensitive to vinorelbine chemotherapy, immune checkpoint blockade (ICB) therapy, and anti-CTLA4 immunotherapy
We compared the differences in the estimated half maximal inhibitory concentration (IC50) levels of eight chemotherapy drugs, including sorafenib (Figure 6A), sunitinib (Figure 6B), cisplatin (Figure 6C), gefitinib (Figure 6D), vinblastine (Figure 6E), vinorelbine (Figure 6F), vorinostat (Figure 6G), and gemcitabine (Figure 6H). Our data showed that there were distinctly higher estimated IC50 levels of sorafenib (p = 6.95e−7) and gemcitabine (p = 1.70e−10) in the high ferroptosis score group compared with the low ferroptosis score group. Oppositely, the estimated IC50 levels of vinorelbine (p = 2.98e−12) in the high ferroptosis score group were significantly lower than in the low ferroptosis score group, indicating that high ferroptosis patients were more sensitive to vinorelbine. The responses of ICB therapies between the two groups were assessed via Tumor Immune Dysfunction and Exclusion (TIDE) algorithm. As a result, subjects with high ferroptosis score exhibited higher TIDE score than those with low ferroptosis score (p = 9.3e−10; Figure 6I). In Figure 6J, patients in high ferroptosis score significantly responded to anti-CTLA4 therapy. Taken together, high ferroptosis score could be more sensitive to vinorelbine chemotherapy, ICB therapy, and anti-CTLA4 immunotherapy.
Figure 6.
High ferroptosis score is more sensitive to vinorelbine chemotherapy, ICB therapy, and anti-CTLA4 immunotherapy
Boxplots depicted the differences in the estimated IC50 levels of (A) sorafenib, (B) sunitinib, (C) cisplatin, (D) gefitinib, (E) vinblastine, (F) vinorelbine, (G) vorinostat, and (H) gemcitabine between the high and low ferroptosis score groups. (I) Boxplots showed the differences in TIDE scores between the two groups. (J) Heatmap visualized the response to anti-CTLA4 and anti-PD1 therapies between the two groups.
Potential small-molecule compounds based on ferroptosis score
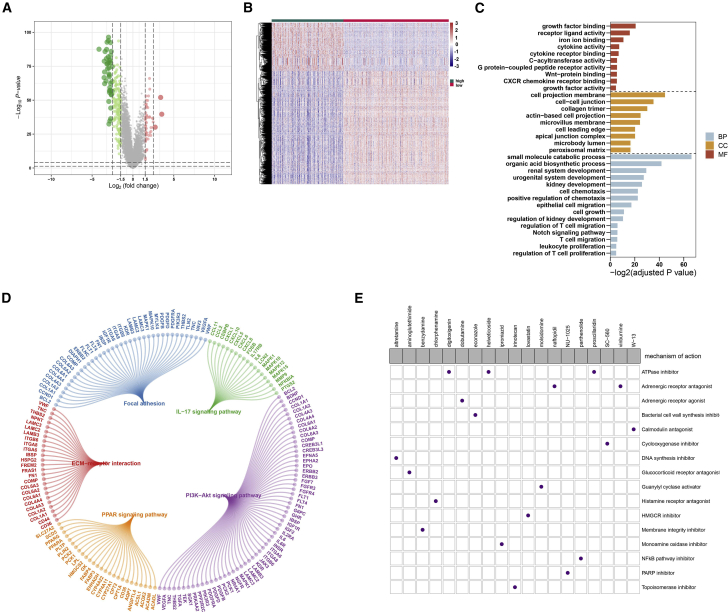
To screen out potential small-molecule compounds for the treatment of ccRCC patients, we further analyzed the DEGs between high and low ferroptosis scores. Consequently, 2,425 DEGs with corrected p < 0.05 and |fold change [FC]| > 1.5 were identified between the two groups, including 725 upregulated and 1,700 downregulated genes (Figures 7A and 7B). Gene Ontology (GO) enrichment analysis results revealed that DEGs were distinctly enriched in iron ion binding, cytokine receptor binding, cell projection membrane, small-molecule catabolic process, renal system development, kidney development, and T cell migration (Figure 7C). Furthermore, these DEGs were significantly associated with focal adhesion, ECM-receptor interaction, interleukin-17 (IL-17), PI3K-Akt, and PPAR signaling pathways (Figure 7D). The upregulated and downregulated genes were uploaded to the Connectivity map (CMap; https://portals.broadinstitute.org/cmap/) small-molecule drug database, and the underlying drug mechanisms were analyzed. A total of 19 potential small-molecule drugs (such as digitoxigenin, helveticoside, and proscillaridin) and 16 drug mechanisms (such as ATPase inhibitor, DNA synthesis inhibitor, PARP inhibitor, and nuclear factor κB [NF-κB] pathway inhibitor) were identified, as shown in Figure 7E.
Figure 7.
Potential small-molecule compounds based on ferroptosis score
(A) Volcano plot depicted the down- (green) and upregulated (red) genes between the high and low ferroptosis score groups. (B) Heatmap visualized the differences in expression patterns of DEGs between the two groups. (C) GO enrichment analysis of biological process (BP), cellular component (CC), and molecular function (MF) results ranked by adjusted p value. (D) KEGG pathway enrichment analysis results showed these pathways and enriched genes. (E) Heatmap showed small-molecule compounds (perturbagen) and their shared drug mechanisms of action (rows) through the CMap database.
The roles of ferroptosis patterns on response to anti-PD-1 immunotherapy
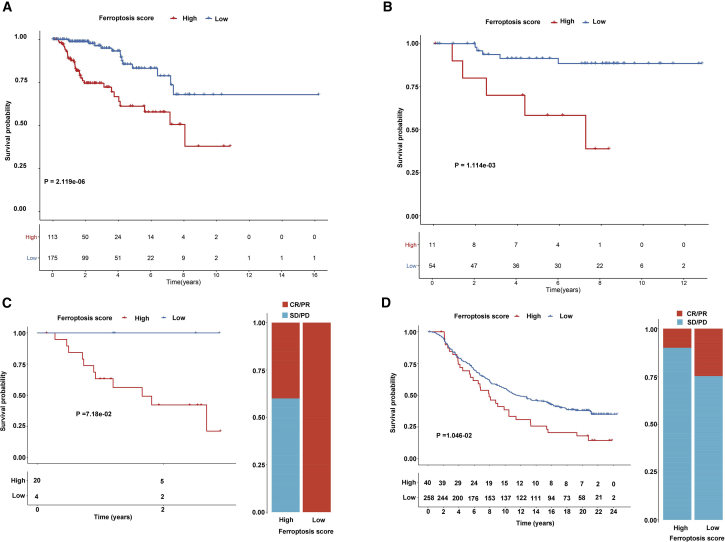
We further validated the predictive effectiveness of ferroptosis score for prediction of prognosis in the pRCC and chRCC datasets from TCGA database. Consistent with ccRCC, high ferroptosis score distinctly indicated a poorer prognosis than low ferroptosis score for both pRCC (p = 2.119e−6; Figure 8A) and chRCC (p = 1.114e−3; Figure 8B) patients. Furthermore, the practicability of the ferroptosis score was further evaluated for speculation of the therapeutic benefit for ccRCC patients. The patients who received anti-PD-L1 immunotherapy in the Gene Expression Omnibus (GEO): GSE78220 (Figure 8C) and IMvigor210 (Figure 8D) cohorts were assigned based on high and low ferroptosis scores. These patients in the high ferroptosis score group distinctly exhibited shorter survival time compared with the low ferroptosis score group in the two cohorts (Figures 8C and 8D). The response rate of anti-PD-L1 therapy in the low ferroptosis score group was distinctly higher than that in the high ferroptosis score group. Collectively, these findings indicated that ferroptosis score could be associated with response to anti-PD-L1 immunotherapy.
Figure 8.
Ferroptosis score in prediction of immunotherapeutic benefits
(A and B) Kaplan-Meier curves for high and low ferroptosis score in pRCC (A) and chRCC (B) patients. (C and D) Kaplan-Meier curves and clinical response to anti-PD-1 therapy for patients with high and low ferroptosis scores in the GEO: GSE78220 and IMvigor210 cohorts. CR, complete response; PD, progressive disease; PR, partial response, SD, stable disease.
Discussion
In this study, we comprehensively analyzed the clinical implications and TME features of ferroptosis patterns in ccRCC. Furthermore, the ferroptosis scoring system was proposed to assess ferroptosis for individuals, which could improve the comprehension about TME immune infiltrations and assist oncologists to make more efficient immunotherapeutic strategies.
Based on the mRNA expression profiles of ferroptosis genes, we developed three molecular patterns for ccRCC. There were distinct differences in prognosis, immune infiltrations, and functions among the three patterns. Genomic alterations in ccRCC may affect the responsiveness to immunotherapy.14 According to the DEGs correlated with the clusters signature, three gene clusters with distinct clinical outcomes and immune cell infiltrations and functions were constructed for ccRCC. The C2 and gene cluster B with the worst prognosis had the highest ferroptosis score among three gene patterns and clusters. By the Boruta algorithm, ferroptosis score was constructed for quantification of the ferroptosis patterns. Intriguingly, patients with high ferroptosis scores usually experienced shorter survival time, indicating that high ferroptosis score could contribute to poor prognosis for ccRCC patients. Ferroptosis is initiated and executed by amino acid, lipid, and iron metabolisms.15 Consistently, our GSEA results showed that amino acid and fatty acid metabolism pathways were significantly enriched, indicating that ferroptosis sensitivity can be regulated by these metabolic pathways.
Ferroptosis score was significantly correlated to clinicopathological characteristics of ccRCC. After adjusting these factors, our data suggested that ferroptosis score was an independent risk factor for ccRCC patients’ prognosis. Operating characteristic curves (ROCs) confirmed its high predictive efficacy for 1-, 3-, 5-, and 8-year survival time. Recently, a five-FRG model has been conducted for ccRCC prognosis.16 Thus, ferroptosis score has the potential to robustly predict the clinical outcomes of patients. The accumulation of genetic mutations leads to the occurrence of cancers, which is driven by exposure to excess iron.17 Our data confirmed that there was a distinct difference in genetic mutations between high and low ferroptosis scores. Higher TMB has been confirmed to be associated with worse survival outcome for ccRCC patients, consistent with our results.18 Patients had the survival advantage in the low ferroptosis score group, indicating that ferroptosis score possessed the potential to predict the responsiveness to immunotherapy, which could be independent of TMB.
Immune responses are key features of carcinogenesis, as well as treatment effect for ccRCC.19 Immune cells and stromal cells are the main components of the TME. High immune and stromal scores have correlations with clinicopathological features and unfavorable prognosis in ccRCC.16 Herein, the estimation of stromal and immune cells in malignant tumor tissues using expression data (ESTIMATE) algorithm was utilized to quantify immune and stromal scores for ccRCC patients. High ferroptosis score markedly exhibited lower tumor purity and higher immune and stromal scores compared with low ferroptosis score. This indicated that ferroptosis could be related to the modulation of the TME, thereby affecting tumor growth and progression. We found that elevated infiltration levels of T cells (T helper, Tfh, Th2, TIL, and Treg) and DCs were related to high ferroptosis score. As in a previous study, T cell-induced ferroptosis promotes the anti-tumor effectiveness for immunotherapy.12 Hence the immune system mediates tumorigenesis partly by inducing ferroptosis.20 In turn, vaccination with tumor cells in the early stage of ferroptosis can efficiently induce anti-tumor immunogenicity.21 Hence targeting ferroptosis could be a potential therapeutic strategy.
ccRCC is usually resistant to chemotherapy.22 Our data showed that patients with high ferroptosis were more sensitive to vinorelbine. Combination vinorelbine and targeting ferroptosis might alleviate resistance mechanisms. Despite favorable clinical benefits of ICB, only a set of ccRCC patients show response to the therapy.14 TIDE model has been developed to predict ICB response. Herein, we found that ccRCC patients with high ferroptosis scores exhibited higher TIDE scores than those with low ferroptosis scores. Furthermore, patients with high scores significantly responded to anti-CTLA4 therapy. Consistently, high levels of T cell infiltration and T cell co-stimulation were detected in high ferroptosis scores. This indicated that ferroptosis score could affect the response to ICB therapy.23 Ferroptosis score could have the potency for assisting oncologists to decide which patients may have response to ICB.
A total of 725 up- and 1,700 downregulated genes were identified between high and low ferroptosis score groups. These DEGs were distinctly enriched in iron ion binding, renal system development, and immunity. Moreover, these DEGs were significantly associated with ferroptosis-related pathways, including focal adhesion, ECM-receptor interaction, IL-17, PI3K-Akt, and PPAR signaling pathways. For example, ferroptosis could regulate IL-17 signaling pathway in hepatocellular carcinoma cells.24 Although iron therapeutics in cancers have been put forward in recent years, clinical applicability is dissatisfactory due to modest results from clinical trials or only tests in experimental settings.25 Abnormal proliferation of tumor cells commands much iron for maintaining DNA synthesis and repair, as well as cellular energy, which is the process upon iron addicted to ferroptosis resistance.26 In this study, we screened 19 pharmacological and genetic inhibitors for ferroptosis, involving 16 mechanisms of action, such as PARP and NF-κB inhibitors. MDM2/X facilitate ferroptosis via PPARα-induced lipid remodeling.27 Moreover, ferroptosis involves the NF-κB signaling pathway.28, 29, 30 Novel determinants of ferroptosis should be further validated by experiments.
The prognostic values of ferroptosis score have been validated in two independent datasets. Our data confirmed that pRCC and chRCC patients with high ferroptosis score were indicative of poorer prognosis than those with low ferroptosis score. Using the IMvigor210 and GEO: GSE78220 datasets, we confirmed the predictive efficacy of ferroptosis score for response to anti-PD-1 immunotherapy. Patients with higher ferroptosis score were more likely to benefit from anti-PD-1 immunotherapy. However, the current findings require validation in a larger ccRCC cohort experiencing immunotherapy.
Conclusions
Taken together, this study systematically analyzed the ferroptosis landscape in ccRCC, which provided the distinct interpretation about the clinical features and implications of ferroptosis. There were significant differences in immune microenvironment and response to immunotherapy between high and low ferroptosis score patients. Hence it is of clinical significance to comprehensively evaluate ferroptosis scores for each ccRCC patient, which may assist oncologists to make first-rank immunotherapy strategies.
Materials and methods
Patients and specimens
RNA sequencing (RNA-seq) and matched complete clinical information (age, gender, survival status, grade, and stage) for ccRCC (n = 525) were retrieved from TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) on October 19, 2020. Fragments per kilobase million (FPKMs) values were normalized as transcripts per kilobase million (TPMs). The specific information of 525 patients was listed in Table S1. CNV and somatic mutation data were obtained from TCGA database.
Ferroptosis-based consensus clustering analysis
The number of unsupervised classes in the TCGA-ccRCC dataset was estimated based on the mRNA expression profiles of 57 FRGs (Table S2) with the consensus clustering method via the ConsensusClusterPlus package in R.31 t-SNE was applied to verify the subtype assignments according to the expression profiles of the above genes.
Single sample gene set enrichment analysis (ssGSEA)
A set of marker genes for tumor-infiltrating immune cells (TIICs) was acquired from Bindea et al.32 By applying the ssGSEA, the enrichment scores of 16 immune cells and 13 immune functions for each ccRCC sample were quantified based on expressed gene signatures using the gene set variation analysis (GSVA) package.33 The immune infiltration levels and functions among different groups were compared by Kruskal-Wallis test.
Dimension reduction and ferroptosis score
ccRCC patients from TCGA database were clustered into the ferroptosis subtypes, and DEGs were screened among these subtypes by the linear models for microarray data (limma) package in R.34 The cutoff values were set as adjusted p < 0.05 and |L2FC| >1. DEGs that had positive and negative correlations with the clusters signature were separately named as the ferroptosis gene signatures A and B. The clusterProfiler in R package was employed to implement GO enrichment analysis for gene signatures A and B,35 including three terms: biological process (BP), cellular component (CC), and molecular function (MF). Based on the mRNA expression profiles of ferroptosis gene signatures, the patients were categorized into the gene clusters by employing unsupervised clustering analysis. The categorization was validated via t-SNE.
The dimension reduction of the ferroptosis gene signatures A and B was implemented by the Boruta algorithm. PCA was implemented to extract principal component 1 as the signature score. A method similar to gene expression grade36,37 was then applied to calculate the index of the ferroptosis score for each sample as follows: ferroptosis score = −, where PC1A represents the first component of signature A, and PC1B indicates the first component of signature B. The ferroptosis scores among the molecular subtypes or gene clusters were assessed via Kruskal-Wallis test.
GSEA
GSEA was presented between the high and low ferroptosis score groups. Pathways with nominal p < 0.05 and false discovery rate (FDR) < 0.05 were considered significantly enriched. The “c2.cp.kegg.v7.1.symbols” was chosen as the reference.
ESTIMATE
By implementing the ESTIMATE algorithm,38 stromal and immune scores were generated to estimate the levels of infiltrating stromal and immune cells in ccRCC tissues and tumor purity through the expression profiles. Then, we compared the differences in tumor purity and stromal and immune scores between the high and low ferroptosis score groups by Wilcoxon rank-sum test.
Drug sensitivity prediction
The sensitivity of each patient to chemotherapy drugs was estimated using the Genomics of Drug Sensitivity in Cancer (GDSC; https://www.cancerrxgene.org/) database.39 The IC50 was quantified via the pRRophetic package in R.40
Assessment of TMB and response to ICB
Somatic mutation data were visualized based on the Mutation Annotation Format (MAF) file using the Maftools package.41 The TMB for each patient was calculated as follows: TMB = (total count of variants)/(the whole length of exons). The response to ICB was predicted by the TIDE (http://tide.dfci.harvard.edu/login/) website. The differences in TIDE scores were compared between the high and low ferroptosis score groups by Wilcoxon test. Furthermore, the response to the anti-PD1 or anti-CTLA4 was predicted for melanoma patients between the two groups.
Screening small-molecule drugs
DEGs with FDR < 0.05 and |FC| ≥ 1.5 were identified between the high and low ferroptosis score groups using the limma package, which were visualized into volcano plots and heatmaps. GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were then presented by the clusterProfiler package. Adjusted p < 0.05 was significantly enriched. The up- and downregulated genes were uploaded into the CMap database.42 Candidate small-molecular drugs and mechanisms of action were discovered by CMap mode-of-action (MoA) analysis.
External dataset validation
RNA-seq and clinical information of pRCC (n = 325) and chRCC (n = 93) samples were required from TCGA database. The prognostic values of the ferroptosis scores were validated in the two datasets. Furthermore, two anti-PD-L1 immunotherapy cohorts were included to determine the ferroptosis scores as follows: GEO: GSE78220 dataset from the GEO (https://www.ncbi.nlm.nih.gov/gds/) repository on the GPL11154 platform14 and IMvigor210 cohort based on the Creative Commons 3.0 License.15
Statistical analysis
Statistical analysis was conducted through R (version 4.0.2). Comparisons between two groups were presented via Wilcoxon rank-sum test, while multiple comparisons were assessed via Kruskal-Wallis test. The cutoff point of each subgroup was identified by the survminer package in R. Kaplan-Meier curves for OS analysis were presented between different subgroups, followed by log rank test. Multivariate Cox regression analyses were utilized to evaluate the association between OS and disease-free survival (DFS) and clinicopathological characteristics and ferroptosis scores, which were visualized by the forestplot package in R. ROCs for 1-, 3-, 5-, and 8-year survival were delineated for evaluation of the predictive efficacy of the ferroptosis score. p value was corrected by Bonferroni’s test. Two-sided p < 0.05 was considered statistically significant.
Availability of data and material
Data underlying this work are available upon request for general research use whose access was approved by the dbGaP Data Access Committee (phs000178.v9.p8).
Acknowledgments
The Frontiers Science Center for Flexible Electronics (FSCFE) and the Research and Development Institute of Northwestern Polytechnical University in Shenzhen are acknowledged in providing space, computational facility, and services. We appreciated TCGA database for providing the original study data. This work is supported by the Science Technology and Innovation Commission of Shenzhen Municipality (JCYJ20190806153018791), Natural Science Foundation of Zhejiang Province (LGF19H200005), Natural Science Foundation of China (81601553), and Japan China Medical Association (2018920). The roles of the above funders are advocacy work, monitoring implementation outcomes, and dissemination of knowledge.
Author contributions
D.B.: conceptualization, methodology, investigation, writing – reviewing and editing, and funding acquisition. H.F.: investigation and formal analysis. J.Y.: investigation and data curation. A.Y.: resources and reviewing. X.L.: data curation and visualization. A.Q.: validation, resources, reviewing, and funding acquisition. H.S.: validation, editing, resources, and funding acquisition. All authors have read and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.05.009.
Contributor Information
Dan Bai, Email: danbai@xjtu.edu.cn.
Hiroshi Sugiyama, Email: sugiyama.hiroshi.3s@kyoto-u.ac.jp.
Supplemental information
References
- 1.Linehan W.M., Ricketts C.J. The Cancer Genome Atlas of renal cell carcinoma: findings and clinical implications. Nat. Rev. Urol. 2019;16:539–552. doi: 10.1038/s41585-019-0211-5. [DOI] [PubMed] [Google Scholar]
- 2.Carril-Ajuria L., Santos M., Roldán-Romero J.M., Rodriguez-Antona C., de Velasco G. Prognostic and Predictive Value of PBRM1 in Clear Cell Renal Cell Carcinoma. Cancers (Basel) 2019;12:16. doi: 10.3390/cancers12010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 4.Yang F., Zhou Q., Xing N. Comparison of survival and renal function between partial and radical laparoscopic nephrectomy for T1b renal cell carcinoma. J. Cancer Res. Clin. Oncol. 2020;146:261–272. doi: 10.1007/s00432-019-03058-z. [DOI] [PubMed] [Google Scholar]
- 5.Xue D., Wang H., Chen Y., Shen D., Lu J., Wang M., Zebibula A., Xu L., Wu H., Li G., Xia L. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol. Cancer. 2019;18:151. doi: 10.1186/s12943-019-1072-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh J.J., Purdue M.P., Signoretti S., Swanton C., Albiges L., Schmidinger M., Heng D.Y., Larkin J., Ficarra V. Renal cell carcinoma. Nat. Rev. Dis. Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKay R.R., Bossé D., Xie W., Wankowicz S.A.M., Flaifel A., Brandao R., Lalani A.A., Martini D.J., Wei X.X., Braun D.A. The Clinical Activity of PD-1/PD-L1 Inhibitors in Metastatic Non-Clear Cell Renal Cell Carcinoma. Cancer Immunol. Res. 2018;6:758–765. doi: 10.1158/2326-6066.CIR-17-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mou Y., Wang J., Wu J., He D., Zhang C., Duan C., Li B. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J. Hematol. Oncol. 2019;12:34. doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang C., Zhang X., Yang M., Dong X. Recent Progress in Ferroptosis Inducers for Cancer Therapy. Adv. Mater. 2019;31:e1904197. doi: 10.1002/adma.201904197. [DOI] [PubMed] [Google Scholar]
- 10.Hassannia B., Vandenabeele P., Vanden Berghe T. Targeting Ferroptosis to Iron Out Cancer. Cancer Cell. 2019;35:830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Friedmann Angeli J.P., Krysko D.V., Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Cancer. 2019;19:405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 12.Wang W., Green M., Choi J.E., Gijón M., Kennedy P.D., Johnson J.K., Liao P., Lang X., Kryczek I., Sell A. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z., Zhao Q., Zuo Z.X., Yuan S.Q., Yu K., Zhang Q., Zhang X., Sheng H., Ju H.Q., Cheng H. Systematic Analysis of the Aberrances and Functional Implications of Ferroptosis in Cancer. iScience. 2020;23:101302. doi: 10.1016/j.isci.2020.101302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mariathasan S., Turley S.J., Nickles D., Castiglioni A., Yuen K., Wang Y., Kadel E.E., III, Koeppen H., Astarita J.L., Cubas R. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGregor B.A., Lalani A.A., Xie W., Steinharter J.A., E Bakouny Z., Martini D.J., Fleischer J.H., Abou-Alaiwi S., Nassar A., Nuzzo P.V. Activity of cabozantinib after immune checkpoint blockade in metastatic clear-cell renal cell carcinoma. Eur. J. Cancer. 2020;135:203–210. doi: 10.1016/j.ejca.2020.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascón S., Hatzios S.K., Kagan V.E. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu G., Wang Q., Xu Y., Li Q., Cheng L. A new survival model based on ferroptosis-related genes for prognostic prediction in clear cell renal cell carcinoma. Aging (Albany NY) 2020;12:14933–14948. doi: 10.18632/aging.103553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torti S.V., Torti F.M. Iron: The cancer connection. Mol. Aspects Med. 2020;75:100860. doi: 10.1016/j.mam.2020.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C., Li Z., Qi F., Hu X., Luo J. Exploration of the relationships between tumor mutation burden with immune infiltrates in clear cell renal cell carcinoma. Ann. Transl. Med. 2019;7:648. doi: 10.21037/atm.2019.10.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu W.H., Xu Y., Wang J., Wan F.N., Wang H.K., Cao D.L., Shi G.H., Qu Y.Y., Zhang H.L., Ye D.W. Prognostic value and immune infiltration of novel signatures in clear cell renal cell carcinoma microenvironment. Aging (Albany NY) 2019;11:6999–7020. doi: 10.18632/aging.102233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stockwell B.R., Jiang X. A Physiological Function for Ferroptosis in Tumor Suppression by the Immune System. Cell Metab. 2019;30:14–15. doi: 10.1016/j.cmet.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efimova I., Catanzaro E., Van der Meeren L., Turubanova V.D., Hammad H., Mishchenko T.A., Vedunova M.V., Fimognari C., Bachert C., Coppieters F. Vaccination with early ferroptotic cancer cells induces efficient antitumor immunity. J. Immunother. Cancer. 2020;8:e001369. doi: 10.1136/jitc-2020-001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pal K., Madamsetty V.S., Dutta S.K., Mukhopadhyay D. Co-delivery of everolimus and vinorelbine via a tumor-targeted liposomal formulation inhibits tumor growth and metastasis in RCC. Int. J. Nanomedicine. 2019;14:5109–5123. doi: 10.2147/IJN.S204221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang B., Zhu J., Li J., Fan K., Gao Y., Cheng S., Kong C., Zheng L., Wu F., Weng Q. The ferroptosis and iron-metabolism signature robustly predicts clinical diagnosis, prognosis and immune microenvironment for hepatocellular carcinoma. Cell Commun. Signal. 2020;18:174. doi: 10.1186/s12964-020-00663-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vela D. Iron in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020;1259:39–51. doi: 10.1007/978-3-030-43093-1_3. [DOI] [PubMed] [Google Scholar]
- 28.Toyokuni S., Yanatori I., Kong Y., Zheng H., Motooka Y., Jiang L. Ferroptosis at the crossroads of infection, aging and cancer. Cancer Sci. 2020;111:2665–2671. doi: 10.1111/cas.14496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesh D., O’Brien N.A., Zandkarimi F., Tong D.R., Stokes M.E., Dunn D.E., Kengmana E.S., Aron A.T., Klein A.M., Csuka J.M. MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling. Genes Dev. 2020;34:526–543. doi: 10.1101/gad.334219.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen P.H., Wu J., Ding C.C., Lin C.C., Pan S., Bossa N., Xu Y., Yang W.H., Mathey-Prevot B., Chi J.T. Kinome screen of ferroptosis reveals a novel role of ATM in regulating iron metabolism. Cell Death Differ. 2020;27:1008–1022. doi: 10.1038/s41418-019-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., Angell H., Fredriksen T., Lafontaine L., Berger A. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., Nordgren H., Farmer P., Praz V., Haibe-Kains B. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X., Shi M., Chen T., Zhang B. Characterization of the Immune Cell Infiltration Landscape in Head and Neck Squamous Cell Carcinoma to Aid Immunotherapy. Mol. Ther. Nucleic Acids. 2020;22:298–309. doi: 10.1016/j.omtn.2020.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson I.R. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9:e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.P., Subramanian A., Ross K.N. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data underlying this work are available upon request for general research use whose access was approved by the dbGaP Data Access Committee (phs000178.v9.p8).