Abstract
Serotonin (5-hydroxytryptamine [5-HT]), a metabolite of tryptophan, acts on the components of the hypothalamus-hypophysis-gonad axis and induces puberty delay in mammals via 5-HT receptor 1A (HTR1A). However, the roles of HTR1A in the hypothalamus in pubertal regulation of gene expression are not fully understood. In the current study, the upregulated gonadotropin-releasing hormone (GnRH) expression in GT1-7 GnRH neuronal cells induced by the HTR1A antagonist WAY-100635 maleate was observed in vitro. Furthermore, RNA sequencing (RNA-seq) showed decreased expression of chromobox 4 (CBX4), a member of the polycomb-repressive complex 1 (PRC1), and the loss of RING2 and YY1 interaction with CBX4, suggesting the degradation of the PRC1 in GT1-7 cells treated with maleate. Chromatin immunoprecipitation sequencing (ChIP-seq) showed that the genome-wide occupancy of CBX4 and histone H2A lysine-119 ubiquitination (H2AK119ub) was compromised, especially on the promoter of GnRH. Finally, we determined that inactivation of phosphatidylinositol 3-kinase (PI3K)/Akt and mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) contributed to CBX4 downregulation. Taken together, we concluded that HTR1A antagonists could enhance GnRH transcription via PRC1 degradation and H2AK119ub loss driven by reduced CBX4 expression through PI3K/Akt and MAPK/ERK pathway suppression in GT1-7 cells and provided a potential epigenetic mechanism of action of HTR1A on GnRH gene expression for mammalian puberty onset.
Keywords: serotonin, HTR1A, GnRH, CBX4, H2AK119ub, PRC1, PI3K/Akt, puberty
Graphical abstract
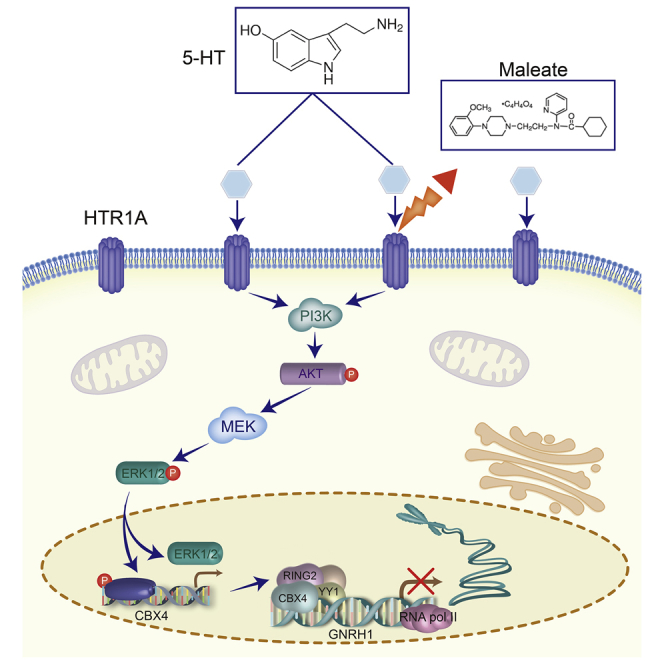
The roles of 5-HT in GnRH expression for puberty onset via HTRs are not fully understood. We highlight a novel regulatory mechanism that 5-HT enhances the activity of PI3K/Akt and MAPK/ERK pathways and upregulates CBX4 expression, maintains PRC1 formation, and represses GnRH transcription in vitro and in vivo.
Introduction
Adolescent sexual maturation is an important part of the individual ontogeny in mammalian development. The active and high hormone levels in this juvenile period promote fertility attainment, body growth, increased metabolism, and acceleration of psychological development. The initiation of puberty is governed by the hypothalamic–pituitary–gonadal axis and begins with hypothalamic gonadotropin-releasing hormone (GnRH) neurons.1 The pulsatile secretion of GnRH stimulates the gonadotroph cells of the pituitary gland to secrete gonadotropins, luteinizing hormone, and follicle-stimulating hormone, which further stimulate the production of estrogen from the ovaries in females and testosterone from the testes in males.2
Secretion of GnRH is regulated by many neurotransmitters, among which the serotoninergic system plays an important role in neuroendocrine control of reproductive hormone secretion.3 Serotonin (5-hydroxytryptamine [5-HT]), a metabolite of tryptophan, is converted from tryptophan within the central nervous system (CNS), particularly in the hypothalamus, via the enzymatic processes of hydroxylation and decarboxylation. Light and electron microscopic observations have both suggested that 5-HT neurons project directly to GnRH neurons in rodents.4 It is known that 5-HT, as an inhibitory neurotransmitter, exerts a repressive influence on puberty in sheep5 and rats,6 and 5-HT receptor (HTR) inhibitors have been used to enhance the excitability of GnRH neurons.4,7 Previous studies have identified seven HTR subtypes and 14 HTR members in mammals that are distributed in a variety of tissues, particularly in the CNS.4 The different potential properties of HTR subtypes usually result in a dual effect of 5-HT, as HTR1 and HTR5 are involved in the inhibition of neuronal discharge, whereas others are excitatory receptor subtypes. However, the role of HTRs in response to 5-HT to control GnRH secretion is complicated and obscure.
In the present study, we detected the dynamic changes in HTR expression during the pubertal process in female mouse hypothalamus in vivo and further studied the regulatory mechanism of HTR1A on GnRH transcription in mouse hypothalamic neuron cell GT1-7, a cell line that stably produces GnRH in vitro.8 Our study revealed a novel connection between the expression of HTR1A and GnRH, emphasizing the importance of epigenetic alterations in regulating GnRH transcription during the pubertal process.
Results
Close connection between the dynamic changes of HTR1A and GnRH release
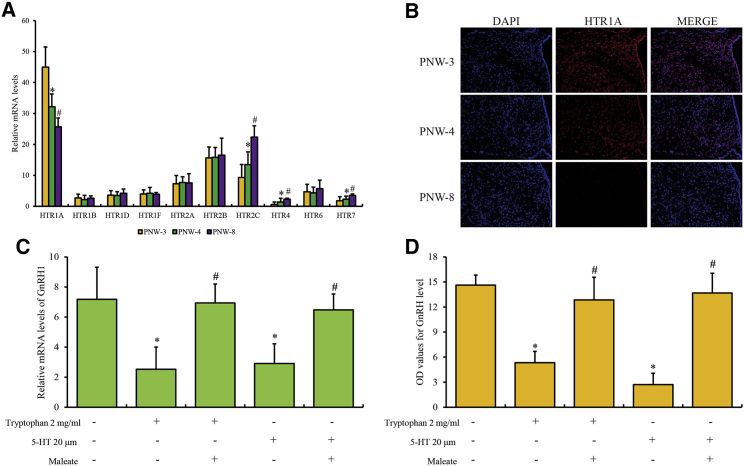
Initially, we investigated the profiling of serotonergic receptors in the hypothalamus of mice during different stages of puberty. Postnatal week (PNW)-3, -4, and -8 indicate the stages of prepuberty, puberty, and late puberty, as previously described.9 The mRNA levels of 5-HTRs (HTR1A, HTR1B, HTR1D, HTR1F, HTR2A, HTR2B, HTR2C, HTR4, HTR6, and HTR7) showed that HTR1A had the highest transcription abundance among all HTRs in the hypothalamus, and HTR1A was significantly downregulated, whereas HTR2C, HTR4, and HTR7 were upregulated from PNW-3, -4, and -8 (Figure 1A). Next, we focused on the hypothalamic arcuate nucleus (ARC), which contains the neuronal nucleus associated with pubertal development and GnRH secretion. Consistent with the mRNA levels, HTR1A protein was significantly downregulated in the hypothalamic ARC of PNW-4 and PNW-8 compared to PNW-3 (Figure 1B). In turn, the expression of HTR2C, HTR4, and HTR7 had no obvious changes compared to PNW-3, -4, and -8 (Figures S1A–S1C).
Figure 1.
Expression profile of HTR1A in ARC
(A) The mRNA levels of 5-HTRs in mouse hypothalamus of PNW-3, -4, and -8, detected by qPCR assay. (B) The expression of HTR1A in hypothalamic ARC (bregma: anterior-posterior, –1.40 mm; dorsal-ventral, –5.80 mm; lateral, +/–0.30 mm) of PNW-3, -4, and -8, detected by IF assay with 200× magnification. Third ventricle is located at the right side of all images. (C) The mRNA levels and (D) extracellular protein levels of GnRH in GT1-7 cells induced by HTR1A inhibitor WAY-100635 maleate and tryptophan or 5-HT treatment by qPCR and ELISA assay. The qPCR and ELISA data are presented as the mean ± SEM of three separate experiments. “∗” or “#” represents the significant difference with a p value less than 0.05 compared to control or the previous group by one-way ANOVA.
To further study the role of HTR1A in the regulatory mechanism of pubertal development, mouse hypothalamic neuron cell line GT1-7 was used for in vitro study. First, GT1-7 cells treated with tryptophan and 5-HT could both induce lower GnRH level in the medium supernatant accompanied with increased expression of HTR1A, with no significant changes in the expression of HTR2C, HTR4, and HTR7 compared to the control (Figures S2A–S2D). Furthermore, the transcription and extracellular secretion of GnRH in GT1-7 cells following stimulation with tryptophan or 5-HT treatment were rescued by pretreatment with WAY-100635 maleate (HTR1A inhibitor) (Figures 1C and 1D). Taken together, we determined that HTR1A contributed to the repression of GnRH release by tryptophan metabolism in pubertal regulation in vivo and in vitro.
Overview of transcription profiling by HTR1A antagonist maleate in GT1-7 cells
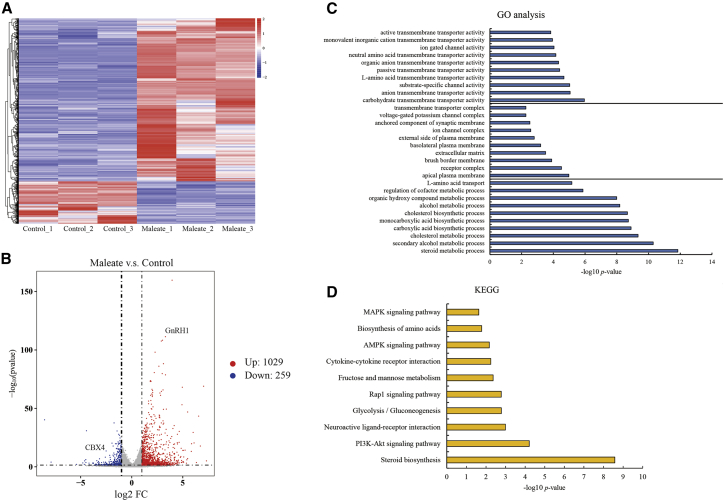
RNA sequencing (RNA-seq) was conducted to decipher the mRNA profiles of GT1-7 cells affected by maleate (Figure 2A). Compared to the negative control, 1,287 differentially expressed genes (DEGs) were observed in GT1-7 cells (Table S1). A total of 1,029 coding genes were upregulated (log2 fold change [FC] > 1, p < 0.05), whereas 259 genes were downregulated (log2 FC < −1, p < 0.05) in maleate-treated GT1-7 cells (Figure 2B). GNRH1 was upregulated (log2 FC = 3.83, p < 0.05), whereas no significant change in HTRs was observed except for HTR3B (FC = 2.66, p = 0.005) and HTR5A (FC = 1.33, p = 0.003), which indicated that maleate did not affect the transcription of HTR1A or most other HTRs. Gene ontology (GO) analysis showed that most of the proteins encoded by DEGs were distributed on the plasma membrane or extracellular matrix and contributed to signal transmission and organic exchange through multiple ion channels, ultimately affecting the metabolic processes and biosynthesis of a variety of substances, such as steroids, alcohols, cholesterol, and acids (Figure 2C). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that the phosphatidylinositol 3-kinase (PI3K)/Akt, Rap1, AMP-activated protein kinase (AMPK), and mitogen-activated protein kinase (MAPK) signaling pathways were involved in maleate treatment (Figure 2D). The RNA profiling of GT1-7 cells provided an overview of the potential mechanism behind the regulation of GnRH gene expression induced by maleate.
Figure 2.
Transcription profiling of GT1-7 cells treated with maleate
(A) Heatmap and (B) volcano plots of differentially expressed genes (DEGs) between maleate treatment and negative control. Color bars above the heatmap represent sample groups: red is for upregulated genes, and blue is for downregulated genes. Top ten of involved biological processes, molecular functions, and cellular components in (C) GO and (D) KEGG pathway analysis of DEGs.
Deterioration of polycomb-repressive complex 1 (PRC1) through chromobox 4 (CBX4) silencing affected by maleate
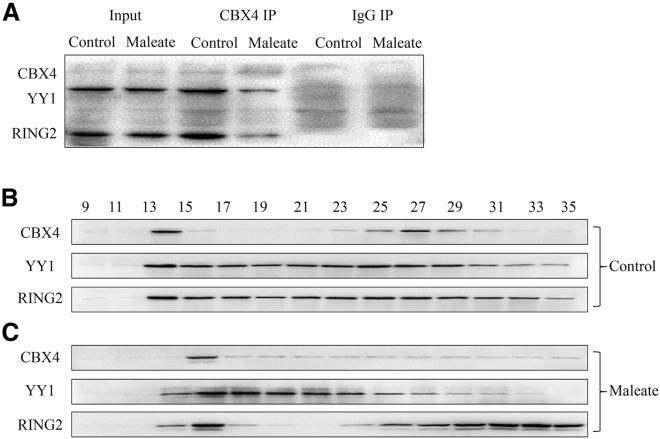
Here, CBX4 was found in the list of downregulated genes by comparing maleate-treated cells and control (log2 FC = −1.01, p < 0.05) in RNA-seq data. CBX4, as a member of the CBX protein family, participates in PRC1 formation10 and recognizes H3K9me3 to recruit the PRC1 subunit of ring finger protein 2 (RING2) via its C-terminal polycomb repressor box,11 whereas RING2 is known as an E3 ubiquitin ligase responsible for histone H2A lysine-119 ubiquitination (H2AK119ub).12,13 We hypothesized that the reduction of CBX4 expression by maleate was likely to cause PRC1 destruction. To this end, the integrity of the PRC1 was detected by coimmunoprecipitation (coIP). We determined that the interactions of CBX4 with the YY1 transcription factor (YY1) and RING2 expression were compromised in maleate-treated GT1-7 cells compared to the control (Figure 3A). Consistently, the gel filtration assay showed that the peaks of YY1 and RING2 protein were not located within the same fraction in maleate-treated GT1-7 cells compared to the control (Figures 3B and 3C), indicating that the PRC1 deteriorated due to the loss of CBX4 expression.
Figure 3.
Expression of CBX4 in GT1-7 cells treated with maleate
(A) The interactions between CBX4 and YY1 or RING2 in maleate-treated GT1-7 cells detected by IP-WB. The integrity of the PRC1 detected by gel filtration in (B) negative control and (C) maleate treatment.
Enhanced GnRH transcription regulated by maleate via the loss of CBX4 expression and H2AK119ub binding to the GnRH promoter
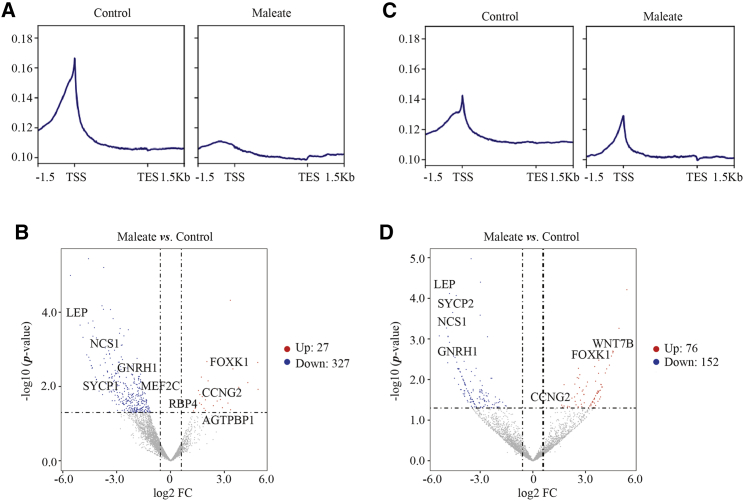
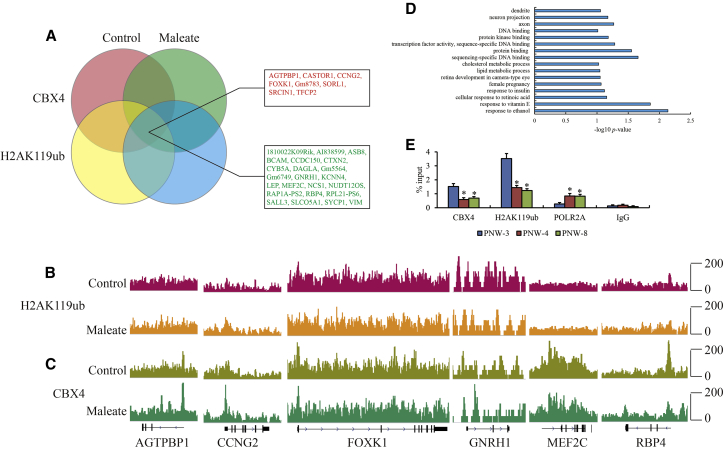
Subsequently, chromatin immunoprecipitation sequencing (ChIP-seq) was conducted to investigate the maleate-induced effects on the genome-wide enrichment of CBX4. The genomic affinity of CBX4 was obviously compromised in maleate-treated GT1-7 cells compared to the control (Figure 4A). A total of 354 genes with significantly differential peaks were annotated (log2 FC > 0.585 or < −0.585, p < 0.05) (Figure 4B; Table S2), and 326 genes had lost the CBX4 occupancy, including GNRH1, LEP, NCS1, and SYCP1, as well as multiple solute carrier family members that had an intimate connection with GnRH secretion and neuronal electrophysiology. To study the potential epigenetic function of PRC1 regulated by CBX4, genomic H2AK119ub modification was explored in parallel by ChIP-seq. Similar to CBX4, H2AK119ub modification also decreased in maleate-treated GT1-7 cells compared to control (Figure 4C), with a total of 228 genes with 76 upregulated and 152 downregulated H2AK119ub modifications (log2 FC > 0.585 or < −0.585, p < 0.05) (Figure 4D; Table S3). The intersected genes with significantly differential enrichment of CBX4 and H2AK119ub were analyzed (Figure 5A), and 31 genes containing AGTPBP1, CCNG2, FOXK1, GNRH1, MEF2C, and RBP4 were included (Figures 5B and 5C). The presence of weakening CBX4 and H2AK119ub enrichment in the GNRH1 promoter region was visualized for maleate treatment. GO analysis showed that certain terms such as ethanol, lipid, and cholesterol metabolic processes overlapped with RNA-seq data, which implied that blocking the tryptophan metabolic process might affect the homeostasis of other essential metabolites (Figure 5D). To confirm the transcriptional activity of GnRH affected by CBX4 expression, the enrichment of CBX4, H2AK119ub, and RNA polymerase II subunit A (POLR2A), as the largest subunit of POLR2 on the GnRH promoter, was studied by ChIP-quantitative PCR (qPCR) in the ARC of PNW-3, -4, and -8. Surprisingly, reduced CBX4 and H2AK119ub occupancies and elevated POLR2 enrichment at the GNRH1 promoter in PNW-4 and -8 compared to PNW-3 were observed (Figure 5E), indicating that the epigenetic regulation of GnRH gene expression affected by CBX4 expression was also tenable in normal pubertal development in vivo. Overall, the effect of the tryptophan metabolic axis on GnRH gene expression was substantially based on CBX4-mediated epigenetic regulation.
Figure 4.
Genome-wide enrichments of CBX4 and H2AK119ub in GT1-7 cells treated with maleate
The genome-wide occupancy representation of (A) CBX4 and (C) H2AK119ub at all annotated gene promoters determined by ChIP-seq. Average CBX4 enrichment measured by log2 (peak p values) in 200 bp bins is shown within genomic regions covering 1.5 kb up- and downstream of coding genes. Volcano plots of genes with significantly different peaks called from (B) CBX4 and (D) H2AK119ub enrichments.
Figure 5.
CBX4 occupancy and H2AK119ub modification of the GnRH promoter in GT1-7 cells treated with maleate
(A) Venn diagram view of genes with significantly different enrichments of CBX4 and H2AK119ub. Red and green fonts indicate up- and downregulation of CBX4 and H2AK119ub enrichment in maleate versus control. Gene browser views of (B) H2AK119ub and (C) CBX4 enrichment on AGTPBP1, CCNG2, FOXK1, GNRH1, MEF2C, and RBP4 in maleate-treated GT1-7 cells. (D) KEGG pathway analysis of intersected genes in Venn diagram. (E) Enrichments of CBX4, H2AK119ub, and POLR2A in the GnRH promoter from ARC of PNW-3, -4, and -8 by ChIP-qPCR. The qPCR data are presented as the mean ± SEM of three separate experiments. ∗ represents the significant difference with a p value less than 0.05 compared to PNW-3 by one-way ANOVA.
Contribution of PI3K/Akt suppression by maleate to CBX4 silencing
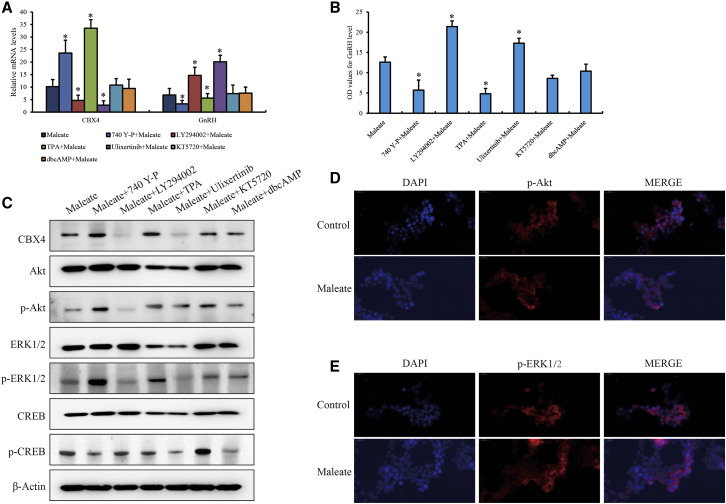
Finally, the regulatory network responding to maleate was studied to bridge the contacts between HTR1A and CBX4 in GT1-7 cells. Given the signaling pathways including PI3K/Akt, cyclic AMP (cAMP)/protein kinase A (PKA), and MAPK, which are responsible for the signals from HTR1A in transcriptomic analysis and recently reported studies,14, 15, 16 small-molecule inhibitors or agonists of these pathways were used to pretreat GT1-7 cells, followed by maleate. We observed that CBX4 expression could be rescued by 740 Y-P (PI3K agonist) and 12-O-tetradecanoylphorbol-13-acetate (TPA; extracellular signal-regulated kinase [ERK]1/2 agonist) or further suppressed by LY294002 (PI3K inhibitor) and ulixertinib (ERK1/2 inhibitor) compared to cAMP/PKA (in)activation and maleate-only treatment, which was opposite to GnRH (Figures 6A and 6B). Interestingly, we also observed that the PI3K/Akt block also weakened the phosphorylation of ERK1/2, whereas MAPK/ERK inhibition failed to affect PI3K/Akt (Figure 6C), implying that PI3K/Akt and MAPK/ERK are the essential pathways connecting HTR1A and CBX4 expression and that MAPK/ERK activity was likely dependent on PI3K/Akt in our system. Consistently, the phosphorylation of Akt and ERK1/2 in the nucleus was substantially decreased in maleate-treated GT1-7 cells, confirming that PI3K/Akt and MAPK/ERK were both blocked by maleate (Figures 6D and 6E).
Figure 6.
Activities of PI3K/Akt and MAPK/ERK pathways induced by maleate for regulating CBX4 expression
The mRNA levels of (A) CBX4 and (B) GnRH and the extracellular GnRH secretion impacted with the inhibitors or agonists of PI3K/Akt, MAPK/ERK, and cAMP/PKA in GT1-7 cells. The activities of PI3K/Akt and MAPK/ERK pathways affected by maleate detected by (C) WB and (D and E) IF assay with 200× magnification.
Taken together, these results show that the negative signals from HTR1A blocked PI3K/Akt and MAPK/ERK pathways to silence CBX4 transcription, and the latter led to the degradation of the PRC1, weakening H2AK119ub modification at the promoter of GnRH, and ultimately enhanced GnRH transcription. Our data revealed a novel regulatory mechanism of tryptophan metabolism for GnRH regulation in the mammalian pubertal process and provided a potential therapeutic strategy for CNS-derived disorders of sexual dysplasia.
Discussion
5-HT acts as an inhibitory neurotransmitter at the synapses of nerve cells and plays a role in the etiology and development of multiple neurological diseases through the corresponding receptor types and subtypes. A previous study showed the controversial discoveries that no HTRs were detected in GnRH-positive neurons in vivo17, whereas 5-HT1A, 5-HT2C, 5-HT4, and 5-HT7 were identified in GT1-7 cells in vitro.18 In our results, we focused on HTR1A because of its high expression compared to 5-HT2C, 5-HT4, and 5-HT7 in the hypothalamus in vivo. Additionally, in GT1-7 cells, only the expression of HTR1A could respond to tryptophan and 5-HT induction, and the antagonist of HTR1A could rescue the inhibitory effect of GnRH gene expression. We can conclude that HTR1A is enriched in GnRH neurons and is the main contributor to GnRH gene expression, forming a bridge between tryptophan and 5-HT. HTR1A exerts a unique inhibitory effect, whereas the other three HTRs may play an excitatory role in GnRH secretion, which needs to be investigated in future studies.
For ion channel activity, HTR1A reduces ionotropic glutamate receptor signaling through cAMP and Ca2+/calmodulin-dependent protein kinase II (CAMKII) inhibition. Multiple studies have revealed that HTR1A is a heptahelical G protein-coupled receptor that couples via inhibitory G proteins (Gi/Go) to inhibit adenylyl cyclase and reduce cAMP levels in non-neuronal cells.19 Although the 5-HT1A/Gi/phospholipase C (PLC) pathway has also been described in hippocampal and raphe cells, it is limited to non-differentiated cells or newborn tissues, indicating that HTR1A coupling to stimulate PLC may be restricted to development.20 Additionally, HTR1A can couple Gβγ and Gαo subunits to activate G-protein inward-rectifying potassium and calcium channels to hyperpolarize the membrane potential.21 Furthermore, HTR1A appears to be pivotal in the antidepressant response via PI3K/Akt and MAPK/ERK activation. HTR1A inhibition can block ketamine-induced Akt and ERK1/2 phosphorylation in the hippocampal response to depression.22,23 However, the downstream signaling pathways linking with HTR1A in GnRH neurons are poorly understood. In our RNA-seq data, we observed that most of the pathways mentioned above were also involved in HTR1A inhibitor-induced GT1-7 cells. Unlike in other brain tissues or neuron cells, GT1-7 cells displaying altered GnRH gene expression by maleate through the PI3K/Akt/MAPK/ERK axis were determined in this study. Our data show that GnRH can also be induced by inhibitors or suppressed by the agonists of PI3K/Akt and MAPK/ERK, which may be beneficial for application in central precocious puberty or delayed puberty in clinical trials.
To date, most studies show that HTR1A-mediated regulatory signals primarily appear in the cytoplasm and plasma membrane. Genome-wide transcriptional regulation in the nucleus in response to tryptophan and the 5-HT metabolic axis has seldom been investigated. We show that CBX4, a subunit of PRC1 regulators, is transcriptionally suppressed through the PI3K/Akt and MAPK/ERK pathways. However, we have still not determined all of the transcription factors activated by phosphorylated ERK1/2 that regulate CBX4 transcription. Nevertheless, we noticed that the number of DEGs as well as differentially binding target genes of CBX4 and H2AK119ub responding to maleate were few, suggesting that only a proportion of genes involved in synapse formation and metabolite production appear to be impacted. The fundamental functions of GT1-7 cells, such as cell proliferation, cell cycle, and cell fate, were not affected by maleate. However, with the consideration of the extensive distribution of HTR1A in brain tissues, the cross effects resulting from HTR1A inhibitors or agonists may cause other psychogenic changes in GnRH secretion in in vivo trials.
Overall, our study highlights the 5-HT-mediated regulatory mechanism of GnRH gene expression in mammalian pubertal development.
Materials and methods
Animal study
C57BL/6 mice purchased from SLACCAS (Shanghai, China) were housed in clean cages and maintained at 22°C ± 2°C with a constant 12-h light/dark schedule. The animals were allowed free access to food and water. PNW-3, -4, and -8 mice (n = 10 per group) were used in this study. Initially, a preliminary experiment for dye injection was used to target the location of the ARC using initial orientation (bregma: anterior-posterior, –1.40 mm; dorsal-ventral, –5.80 mm; lateral, +/–0.30 mm) as previously described.24,25 Mice were sacrificed via cervical dislocation, and the whole brains were isolated immediately. The hypothalamic ARC tissues in each group were harvested and gathered for subsequent experiments as previously described.9 All procedures were performed in accordance with the Institutional Animal Care and Use Committee of Shanghai Jiao Tong University.
Cell culture
GT1-7 cells were cultured in DMEM containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA). Drug treatment conditions of cells are listed below: 100 μg/mL HTR1A antagonist WAY-100635 maleate (Sigma-Aldrich, St. Louis, MO, USA) for 6 h; 0.5–4 mg/mL L-tryptophan (Sinopharm Chemical Reagent, Shanghai, China) and 2.5–40 μM 5-HT (Sigma-Aldrich) for 1 h, as referred to in previous literature;4,26 20 μM 740 Y-P (APExBIO, Houston, TX, USA) and 10 μM LY294002 (APExBIO) for 30 min;27 5 nM TPA (APExBIO)28 and 30 μM ulixertinib (APExBIO) for 6 h;29 and 0.5 μg/mL KT5720 (APExBIO) and 1 mM dibutyryl cAMP (dbcAMP; APExBIO) for 24 h.30
RNA-seq
Brain tissues or 1 × 106 GT1-7 cells under different conditions were stored in 1 mL TRIzol (Thermo Fisher Scientific) and ground in liquid nitrogen. Next, 100 μL chloroform was added, and the cells were fully mixed and centrifuged at the highest speed at 4°C for 10 min. The supernatant was transferred into a new tube, and isopropanol was added to the same volume and centrifuged at the highest speed at 4°C for 10 min. The precipitate was washed with 75% cold ethanol and dissolved in the appropriate diethyl pyrocarbonate (DEPC) water. The concentration and quality of RNA were measured using a Nanodrop 2000 (Thermo Fisher Scientific) and an Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). A total of 4 μg of RNA from each group was used for library preparation using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) following the manufacturer’s instructions and sequenced on an Illumina HiSeq platform.
The raw data were trimmed for adaptors, and low-quality reads were filtered out using Trimmomatic,31 and the quality of clean reads was checked using FastQC.32 Next, clean reads were aligned to the latest human genome assembly hg38 using HISAT2 (hierarchical indexing for spliced alignment of transcripts 2).33 The transcripts were assembled, and the expression levels were estimated with fragments per kilobase of exon per million (FPKM) values using the StringTie algorithm with default parameters.34 Differential mRNA and long non-coding RNA (lncRNA) expression among the groups were evaluated using the R package ballgown,35 and the significance of differences was computed using the Benjamini and Hochberg (BH) p value adjustment method. Gene annotation was performed using Database: Ensembl. The R package clusterProfiler was used to annotate the DEGs using GO terms and KEGG pathways.36 The raw sequencing documents were deposited in Database: ArrayExpress (assigned as ArrayExpress: E-MTAB-10171).
ChIP-seq
In brief, whole cell lysates of 1 × 107 GT1-7 cells were sonicated to break up the genomic DNA into 200–500 bp fragments. Next, 10% lysates were saved as input, and the remaining were incubated with 1 μg immunoprecipitation (IP)-grade antibodies of CBX4 (#30559; Cell Signaling Technology [CST], Beverly, MA, USA) and H2AK119ub (ABE569; Sigma-Aldrich) at 4°C overnight. This was followed by incubation for 2 h with Protein A beads (Thermo Fisher Scientific) at 37°C to pull down the bound DNA fragments.
For high-throughput sequencing, we added 3′-dA overhangs to the CBX4- or H2AK119ub-enriched or input DNA and ligated them to the adaptor to build a DNA library. DNA libraries with ligated adapters were isolated based on the appropriate size for sequencing using the Illumina HiSeq 2000 platform. The raw sequence reads of input and IP were trimmed based on adaptors, and low-quality reads were filtered out using Cutadapt (version [v.]1.9.1) and Trimmomatic (v.0.35). The quality of the clean reads was checked using FastQC. The clean reads were mapped to the human genome (assembly hg38) using the bowtie 2 (v.2.2.6) algorithm,37 and peak calling (p < 0.01) was performed using MACS 2 (v.2.1.1).38 The differentially bound genes were analyzed based on p values less than 0.05 and annotated using DiffBind.39 The relevant peaks on the genomic loci were visualized using the Integrative Genomics Viewer (IGV). GO analysis was used to determine the biological functions of genes associated with the differential peaks.40 The raw ChIP sequencing data were submitted to Database: ArrayExpress and registered as ArrayExpress: E-MTAB-10173.
For the in vivo study, genomic DNA fragments from ARC tissues of PNW-3, -4, and -8 were pulled down by 1 μg CBX4, H2AK119ub, and POLR2A (#2629; CST) and then purified by ethanol precipitation, and qPCR was performed to detect the enrichment of target binding proteins on the GNRH1 promoter (see qPCR assay for details).
qPCR assay
DNA or cDNA templates were detected using Fast Universal SYBR Green Real-time PCR Master Mix (Roche, Basel, Switzerland) under the following conditions: 95°C for 2 min, 40 cycles of 95°C for 5 s, 60°C for 10 s, 72°C for 30 s, and 72°C for 10 min. β-actin was used as the loading control. The primers used in this study are listed in Table S4.
IP
The initial procedures were similar to those of ChIP until bead purification. The CBX4 antibody was used to incubate cell lysates. After washing, proteins were extracted using loading buffer (0.28 M Tris-HCl [pH 6.8], 30% [V/V] glycerol, 1% [m/V] sodium dodecyl sulfate [SDS]-polyacrylamide gel, 0.5 M dithiothreitol, 0.0012% [m/V] bromophenol blue) at 100°C for 10 min, and YY1 and RING2 were detected by western blot (WB) assay.
Gel filtration assay
The gel filtration assay was performed as previously described.41 In brief, hypotonic buffer with Nonidet P-40 (NP-40) was used to separate the cytoplasm and nuclei of 1 × 108 GT1-7 cells, and the nuclear proteins were isolated using high salt extraction buffer (20 mM HEPES [pH 7.9], 420 mM NaCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, and protease inhibitors). The nuclear extracts (4 mg) were directly applied to a Sepharose 6B column (Sigma-Aldrich) equilibrated with column running buffer containing 20 mM HEPES (pH 7.9), 200 mM NaCl, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, and 10% glycerol. Fractions of 1 mL each were collected, and CBX4, YY1, and RING2 were detected by WB.
WB
A total of 1 × 106 GT1-7 cells were added to radioimmunoprecipitation assay (RIPA) buffer (Solarbio, Beijing, China). Total proteins (30 μg) were separated by SDS gel electrophoresis and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Primary antibodies, including HTR1A (1:2,000; SAB Biotech, College Park, MD, USA), HTR2C (1:2,000; SAB Biotech), HTR4 (1:2,000; SAB Biotech), HTR7 (1:2,000; SAB Biotech), CBX4 (1:1,000), YY1 (1:2,000; CST), RING2 (1:2,000; CST), Akt (1:2,000; CST), phosphorylated Akt (1:500; CST), ERK1/2 (1:2,000; CST), p-ERK1/2 (1:500; CST), CREB; 1:2,000; CST), p-CREB (1:500; CST), and β-actin (1:5,000; CST), were used for incubation with the PVDF membranes at 4°C overnight. The PVDF membranes were then incubated with secondary antibody for 1 h at room temperature. The membrane was developed using enhanced chemiluminescence (ECL) Plus reagents (Thermo Fisher Scientific) and exposed using the ChemiDoc XRS system (Bio-Rad Laboratories, Hercules, CA, USA).
Enzyme-linked immunosorbent assay (ELISA)
GnRH levels in the medium supernatant were assayed using commercial ELISA kits (E-EL0071c; Elabscience Biotechnology, Wuhan, Hubei, China) in accordance with the manufacturer’s instructions.
Immunofluorescence (IF) assay
Hypothalamus were sectioned into 25 μm samples (at a range of bregma −1.22 mm to −2.76 mm). GT1-7 cells were transplanted into 8 mm Teflon-printed black diagnostic slides (Immuno-Cell, Mechelen, Belgium) overnight, fixed with 3.7% formaldehyde for 15 min at 4°C the next day, and quenched with 0.125 M glycine for 10 min. After washing with prechilled PBS three times, 5 min each, tissues/cells were permeabilized using 0.2% Triton-100 for 5 min at room temperature and incubated with 3% horse serum for 30 min and then with primary antibodies for HTR1A, p-Akt, and p-ERK1/2 (diluted appropriately in 0.5% horse serum) overnight at 4°C. After washing with prechilled PBS, 3 times for 5 min each, sections were incubated with goat anti-rabbit secondary antibody (Alexa Fluor 488) or rabbit anti-mouse secondary antibody (Alexa Fluor 647; CST) for 30 min in a cool, dark place. After washing three times with prechilled PBS for 5 min, 800 mM 4′,6-diamidino-2-phenylindole (DAPI) was added to the sections, mounting medium was added, and the slides were covered with glass, sealed with nail polish, and observed using a microscope.
Statistical analysis
Data are presented as the mean ± standard deviation for three independent experiments. The differences in values were analyzed using one-way ANOVA. Statistical significance was set at p < 0.05.
Acknowledgments
We are grateful for the assistance provided by Shanghai Genefund Biotech Co., Ltd., for the generation of high-throughput sequencing data. We are also grateful to Coweldgen Scientific Co., Ltd., for its suggestions on histological sections and GnRH detection. The datasets, supporting materials, and analysis generated are available from the corresponding author upon reasonable request. This work was supported by the National Natural Science Foundation of China (81871131), sponsored by the Interdisciplinary Program of Shanghai Jiao Tong University (YG2021ZD25), Hospital Level Project of Shanghai Children’s Hospital (2018YQN002), Cultivation Plan for Excellent Young Talents of Shanghai Children’s Hospital (2017YYQ05), and Shanghai Sailing Program (20YF1440700).
Author contributions
S. Zhou and Y.S. performed all cellular and molecular experiments and analyzed the data. S. Zhou and S. Zang performed the animal experiments. X.Y. helped design the research route. P.L. designed and supported the entire project and drafted and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.05.014.
Supplemental information
References
- 1.Toro C.A., Aylwin C.F., Lomniczi A. Hypothalamic epigenetics driving female puberty. J. Neuroendocrinol. 2018;30:e12589. doi: 10.1111/jne.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chulani V.L., Gordon L.P. Adolescent growth and development. Prim. Care. 2014;41:465–487. doi: 10.1016/j.pop.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Comai S., Bertazzo A., Carretti N., Podfigurna-Stopa A., Luisi S., Costa C.V. Serum levels of tryptophan, 5-hydroxytryptophan and serotonin in patients affected with different forms of amenorrhea. Int. J. Tryptophan Res. 2010;3:69–75. doi: 10.4137/ijtr.s3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattarai J.P., Roa J., Herbison A.E., Han S.K. Serotonin acts through 5-HT1 and 5-HT2 receptors to exert biphasic actions on GnRH neuron excitability in the mouse. Endocrinology. 2014;155:513–524. doi: 10.1210/en.2013-1692. [DOI] [PubMed] [Google Scholar]
- 5.Domański E., Przekop F., Skubiszewski B., Wolińska E. The effect and site of action of indoleamines on the hypothalamic centers involved in the control of LH release and ovulation in sheep. Neuroendocrinology. 1975;17:265–273. doi: 10.1159/000122362. [DOI] [PubMed] [Google Scholar]
- 6.Arendash G.W., Gallo R.V. Serotonin involvement in the inhibition of episodic luteinizing hormone release during electrical stimulation of the midbrain dorsal raphe nucleus in ovariectomized rats. Endocrinology. 1978;102:1199–1206. doi: 10.1210/endo-102-4-1199. [DOI] [PubMed] [Google Scholar]
- 7.Prasad P., Ogawa S., Parhar I.S. Serotonin reuptake inhibitor citalopram inhibits GnRH synthesis and spermatogenesis in the male zebrafish. Biol. Reprod. 2015;93:102. doi: 10.1095/biolreprod.115.129965. [DOI] [PubMed] [Google Scholar]
- 8.Kanasaki H., Oride A., Mijiddorj T., Sukhbaatar U., Kyo S. How is GnRH regulated in GnRH-producing neurons? Studies using GT1-7 cells as a GnRH-producing cell model. Gen. Comp. Endocrinol. 2017;247:138–142. doi: 10.1016/j.ygcen.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Shen Y., Zhou S., Zhao X., Li H., Sun J. Characterization of Genome-Wide DNA Methylation and Hydroxymethylation in Mouse Arcuate Nucleus of Hypothalamus During Puberty Process. Front. Genet. 2020;11:626536. doi: 10.3389/fgene.2020.626536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen I., Ezhkova E. Cbx4: A new guardian of p63's domain of epidermal control. J. Cell Biol. 2016;212:9–11. doi: 10.1083/jcb.201512032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein E., Duncan E.M., Masui O., Gil J., Heard E., Allis C.D. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 13.Gao Z., Zhang J., Bonasio R., Strino F., Sawai A., Parisi F., Kluger Y., Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukumoto K., Iijima M., Funakoshi T., Chaki S. 5-HT1A receptor stimulation in the medial prefrontal cortex mediates the antidepressant effects of mGlu2/3 receptor antagonist in mice. Neuropharmacology. 2018;137:96–103. doi: 10.1016/j.neuropharm.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Prasad S., Ponimaskin E., Zeug A. Serotonin receptor oligomerization regulates cAMP-based signaling. J. Cell Sci. 2019;132:jcs230334. doi: 10.1242/jcs.230334. [DOI] [PubMed] [Google Scholar]
- 16.Zhou L., Cai M., Ren Y., Wu H., Liu M., Chen H., Shang J. The different roles of 5-HT1A/2A receptors in fluoxetine ameliorated pigmentation of C57BL/6 mouse skin in response to stress. J. Dermatol. Sci. 2018;92:222–229. doi: 10.1016/j.jdermsci.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Wright D.E., Jennes L. Lack of expression of serotonin receptor subtype -1a, 1c, and -2 mRNAs in gonadotropin-releasing hormone producing neurons of the rat. Neurosci. Lett. 1993;163:1–4. doi: 10.1016/0304-3940(93)90214-6. [DOI] [PubMed] [Google Scholar]
- 18.Wada K., Hu L., Mores N., Navarro C.E., Fuda H., Krsmanovic L.Z., Catt K.J. Serotonin (5-HT) receptor subtypes mediate specific modes of 5-HT-induced signaling and regulation of neurosecretion in gonadotropin-releasing hormone neurons. Mol. Endocrinol. 2006;20:125–135. doi: 10.1210/me.2005-0109. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y.F., Ghahremani M.H., Rasenick M.M., Jakobs K.H., Albert P.R. Stimulation of cAMP synthesis by Gi-coupled receptors upon ablation of distinct Galphai protein expression. Gi subtype specificity of the 5-HT1A receptor. J. Biol. Chem. 1999;274:16444–16450. doi: 10.1074/jbc.274.23.16444. [DOI] [PubMed] [Google Scholar]
- 20.Mogha A., Guariglia S.R., Debata P.R., Wen G.Y., Banerjee P. Serotonin 1A receptor-mediated signaling through ERK and PKCα is essential for normal synaptogenesis in neonatal mouse hippocampus. Transl. Psychiatry. 2012;2:e66. doi: 10.1038/tp.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albert P.R., Vahid-Ansari F. The 5-HT1A receptor: Signaling to behavior. Biochimie. 2019;161:34–45. doi: 10.1016/j.biochi.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Fukumoto K., Iijima M., Funakoshi T., Chaki S. Role of 5-HT1A Receptor Stimulation in the Medial Prefrontal Cortex in the Sustained Antidepressant Effects of Ketamine. Int. J. Neuropsychopharmacol. 2018;21:371–381. doi: 10.1093/ijnp/pyx116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiung S.C., Adlersberg M., Arango V., Mann J.J., Tamir H., Liu K.P. Attenuated 5-HT1A receptor signaling in brains of suicide victims: involvement of adenylyl cyclase, phosphatidylinositol 3-kinase, Akt and mitogen-activated protein kinase. J. Neurochem. 2003;87:182–194. doi: 10.1046/j.1471-4159.2003.01987.x. [DOI] [PubMed] [Google Scholar]
- 24.Kumar D., Candlish M., Periasamy V., Avcu N., Mayer C., Boehm U. Specialized subpopulations of kisspeptin neurons communicate with GnRH neurons in female mice. Endocrinology. 2015;156:32–38. doi: 10.1210/en.2014-1671. [DOI] [PubMed] [Google Scholar]
- 25.Krashes M.J., Koda S., Ye C., Rogan S.C., Adams A.C., Cusher D.S., Maratos-Flier E., Roth B.L., Lowell B.B. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal P., Singh D., Raisuddin S., Kumar R. Amelioration of ochratoxin-A induced cytotoxicity by prophylactic treatment of N-Acetyl-L-Tryptophan in human embryonic kidney cells. Toxicology. 2020;429:152324. doi: 10.1016/j.tox.2019.152324. [DOI] [PubMed] [Google Scholar]
- 27.Qiu Y., Chen D., Huang X., Huang L., Tang L., Jiang J., Chen L., Li S. Neuroprotective effects of HTR1A antagonist WAY-100635 on scopolamine-induced delirium in rats and underlying molecular mechanisms. BMC Neurosci. 2016;17:66. doi: 10.1186/s12868-016-0300-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Traore K., Sharma R., Thimmulappa R.K., Watson W.H., Biswal S., Trush M.A. Redox-regulation of Erk1/2-directed phosphatase by reactive oxygen species: role in signaling TPA-induced growth arrest in ML-1 cells. J. Cell. Physiol. 2008;216:276–285. doi: 10.1002/jcp.21403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward R.A., Colclough N., Challinor M., Debreczeni J.E., Eckersley K., Fairley G., Feron L., Flemington V., Graham M.A., Greenwood R. Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of ERK1/2. J. Med. Chem. 2015;58:4790–4801. doi: 10.1021/acs.jmedchem.5b00466. [DOI] [PubMed] [Google Scholar]
- 30.Mustafa S.B., Castro R., Falck A.J., Petershack J.A., Henson B.M., Mendoza Y.M., Choudary A., Seidner S.R. Protein kinase A and mitogen-activated protein kinase pathways mediate cAMP induction of alpha-epithelial Na+ channels (alpha-ENaC) J. Cell. Physiol. 2008;215:101–110. doi: 10.1002/jcp.21291. [DOI] [PubMed] [Google Scholar]
- 31.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guilherme B., Andrew S. Falco: high-speed FastQC emulation for quality control of sequenceing data. F1000Res. 2019;8:1874. doi: 10.12688/f1000research.21142.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frazee A.C., Pertea G., Jaffe A.E., Langmead B., Salzberg S.L., Leek J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015;33:243–246. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kołodziej-Wojnar P., Borkowska J., Wicik Z., Domaszewska-Szostek A., Połosak J., Cąkała-Jakimowicz M., Bujanowska O., Puzianowska-Kuznicka M. Alterations in the Genomic Distribution of 5hmC in In Vivo Aged Human Skin Fibroblasts. Int. J. Mol. Sci. 2020;22:78. doi: 10.3390/ijms22010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Sun Q., Liang J., Li H., Czajkowsky D.M., Shao Z. Q-Nuc: a bioinformatics pipeline for the quantitative analysis of nucleosomal profiles. Interdiscip. Sci. 2020;12:69–81. doi: 10.1007/s12539-019-00354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han B., Park H.K., Ching T., Panneerselvam J., Wang H., Shen Y., Zhang J., Li L., Che R., Garmire L., Fei P. Human DBR1 modulates the recycling of snRNPs to affect alternative RNA splicing and contributes to the suppression of cancer development. Oncogene. 2017;36:5382–5391. doi: 10.1038/onc.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.