Abstract
Like most enveloped viruses, HIV must acquire a lipid membrane as it assembles and buds through the plasma membrane of infected cells to spread infection. Several sets of host cell machinery facilitate this process, including proteins of the endosomal sorting complexes required for transport pathway, which mediates the membrane fission reaction required to complete viral budding, as well as angiomotin (AMOT) and NEDD4L, which bind one another and promote virion membrane envelopment. AMOT and NEDD4L interact through the four NEDD4L WW domains and three different AMOT Pro-Pro-x (any amino acid)-Tyr (PPxY) motifs, but these interactions are not yet well defined. Here, we report that individual AMOT PPxY and NEDD4L WW domains interact with the following general affinity hierarchies: AMOT PPxY1>PPxY2>PPxY3 and NEDD4L WW3>WW2>WW1∼WW4. The unusually high-affinity of the AMOT PPxY1–NEDD4L WW3 interaction accounts for most of the AMOT–NEDD4L binding and is critical for stimulating HIV-1 release. Comparative structural, binding, and virological analyses reveal that complementary ionic and hydrophobic contacts on both sides of the WW–PPxY core interaction account for the unusually high affinity of the AMOT PPxY1–NEDD4L WW3 interaction. Taken together, our studies reveal how the first AMOT PPxY1 motif binds the third NEDD4L WW domain to stimulate HIV-1 viral envelopment and promote infectivity.
Keywords: HIV, AMOT, Nedd4L, WW domains, PPxY
Abbreviations: AMOT, angiomotin; ESCRT, endosomal sorting complexes required for transport; FP, fluorescence polarization; HSQC, heteronuclear single-quantum correlation; RMSD, root mean square deviation; NOE, nuclear Overhauser effect; PPxY, Pro-Pro-x (any amino acid)-Tyr
Like other enveloped viruses, the assembling HIV-1 virion must be wrapped in a host-derived membrane (termed envelopment) and the membrane must then be severed to release the infectious particle from the producer cell (termed budding). HIV-1 assembly is organized by the viral Gag protein (1, 2), and budding is mediated by the machinery of the cellular endosomal sorting complexes required for transport (ESCRT) pathway (3, 4). To initiate the budding process, PTAP and YPxL peptide motifs located within the C-terminal p6 region of Gag recruit two early-acting ESCRT factors, TSG101 (a component of the ESCRT-I complex) and ALIX, respectively. These factors then recruit downstream ESCRT complexes that constrict the viral bud neck and perform the membrane fission reaction that releases the virion from the host plasma membrane.
HIV-1 release also requires ubiquitin transfer (5, 6, 7). Precisely how ubiquitin promotes HIV-1 release is not yet fully understood, but other retroviral Gag proteins recruit members of the NEDD4 family of HECT-containing ubiquitin E3 ligases and promote budding (8). Recruitment of NEDD4 is typically facilitated by a PPxY motif (where P = proline, x = any amino acid, and Y = tyrosine). The PTAP, YPxL, and PPxY motifs are collectively termed “late domains” because their mutation arrests virion morphogenesis at late stages of assembly. Different late domains can act autonomously and interchangeably, implying that they likely perform analogous functions in recruiting ESCRT factors. In the case of PPxY–NEDD4 family E3 ligase interactions, this could occur through local ubiquitination of proteins at sites of virus assembly because at least three different ESCRT factors, TSG101/ESCRT-I, ALIX, and ESCRT-II, have ubiquitin-binding activities (8, 9, 10, 11, 12, 13, 14, 15).
Although HIV-1 Gag does not contain an identifiable PPxY late domain, the NEDD4 family member NEDD4L can nevertheless stimulate virion release (16, 17, 18). This stimulation is particularly pronounced when a naturally occurring isoform of NEDD4L, isoform 2 (also called NEDD4-2s or NEDD4LΔC2), is overexpressed, and when TSG101/ESCRT-I and ALIX recruitment are impaired. HIV-1 constructs lacking the TSG101/ESCRT-I and ALIX recruiting late domains within p6Gag (HIV- 1ΔPTAP,ΔYP) are therefore useful for assaying the NEDD4L elements and activities that stimulate virion release. Loss of endogenous NEDD4L also impairs HIV-1ΔPTAP,ΔYP release, however, indicating that even at native levels, this enzyme can participate in a ubiquitin-dependent step of virus assembly (17).
We have also shown that the NEDD4L-binding partner angiomotin (AMOT) can associate with HIV-1 Gag and function together with NEDD4L to promote HIV-1 release (19). Importantly, AMOT is required for NEDD4L stimulation of HIV-1ΔPTAP,ΔYP release, and the two proteins act synergistically. Stimulatory activity is specific to the p130 isoform of AMOT, whereas shorter isoforms such as AMOT p80 that cannot bind NEDD4L do not stimulate virion release. These observations imply that AMOT and NEDD4L function together during HIV-1 assembly and release. In the absence of AMOT, assembling wt virions fail to form fully spherical enveloped particles, indicating that AMOT acts upstream of the ESCRT machinery, during the envelopment phase of virion assembly (19). AMOT p130 contains a Bin/amphiphysin/Rvs-like domain that can remodel and tubulate membranes in other contexts (20, 21), suggesting that this domain could mediate AMOT p130 recruitment when the assembling virion reaches the proper stage of membrane curvature and/or help complete virion envelopment.
In addition to HIV-1, other enveloped viruses may utilize similar host complexes to help drive membrane envelopment and release. For example, another motin family member, angiomotin-like 1, links the structural M protein of paramyxoviruses with NEDD4 family members to promote virion release in an ESCRT- and ubiquitin-dependent pathway (22, 23). AMOT also enhances the release of filoviruses (24, 25), whose structural proteins contain PPxY late domains that recruit NEDD4 directly. Thus, these very different enveloped viruses appear to utilize similar egress machineries, and learning how the AMOT–NEDD4L complex functions in HIV-1 release will therefore likely have broader implications for understanding enveloped virus assembly.
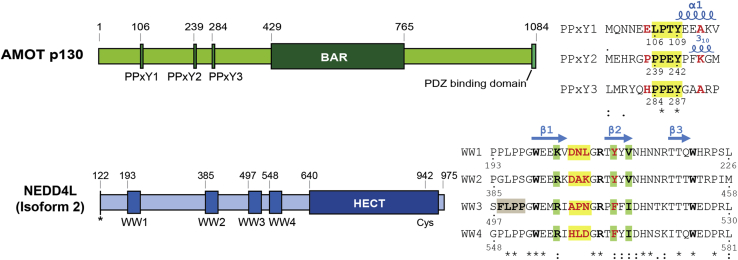
Here, we have investigated how NEDD4L and AMOT interact with one another to stimulate HIV-1 release. The two proteins have been shown to bind one another through interactions that are mediated by PPxY motifs in AMOT and WW domains in NEDD4L (19, 26). PPxY–WW domain interactions mediate protein–protein interactions in many different cell signaling and trafficking pathways (27, 28), and their recognition modes and structures have been defined in many cases (27, 29, 30). Nevertheless, it is not yet clear precisely how AMOT and NEDD4L interact because the N-terminal region of AMOT contains three different PPxY motifs and the central region of NEDD4L contains four different WW domains (Fig. 1). We undertook this study with the goals of determining which of the 12 different possible pairwise AMOT PPxY–NEDD4L WW domain interactions are most energetically and functionally important and determining the structural basis for these more favorable interaction(s).
Figure 1.
Domain organization and motif sequences of AMOT p130 and NEDD4L. PPxY color coding shows the core motif (yellow highlight), key variable residues (red font), and secondary structures (i + 4 α-helix in PPxY1 and i + 3 310-helix in PPxY2). WW domain color coding shows the two eponymous Trp residues (bold W), loop 1 residues (yellow highlights), hydrophobic pocket residues (green highlights), key variable residues (red), and a cryptic motif that binds in the binding site of an adjacent subunit in the WW3 crystal lattice (tan highlight, Fig. S2). Positions of the three canonical β-strands are shown. Residue conservation in the PPxY and WW domain alignments is denoted with asterisks (full conservation), colons (strong conservation), and dots (weak conservation). Experiments were carried out using the naturally occurring isoform 2 of NEDD4L (also referred to as NEDD4-2s or NEDD4DC2). Isoform 2 lacks the 121 N-terminal residues of the C2 domain present in NEDD4L isoform 1 (asterisk). See Refs. (17, 18, 19) for a full explanation of NEDD4L isoforms and activities. PPxY, Pro-Pro-x (any amino acid)-Tyr.
Results and discussion
AMOT PPxY1 is required for efficient HIV-1 release and infectivity
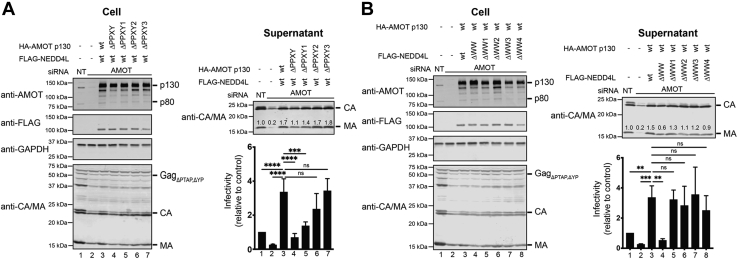
We began by testing the functional requirements for each of the three AMOT PPxY motifs on release and infectivity, using an HIV-1ΔPTAP,ΔYP construct that lacked functional PTAP and YPXL late domains and is therefore highly dependent on NEDD4L and AMOT (16, 17, 19). As expected (19), siRNA depletion of AMOT reduced HIV-1ΔPTAP,ΔYP virion release into the supernatants of HEK293T producer cells, and correspondingly decreased viral titers, as measured in single-cycle infectivity assays (Fig. 2A, compare lanes 1 and 2). Conversely, virion release and infectivity were strongly stimulated by coexpression of wt NEDD4L and an siRNA-resistant AMOT p130 expression construct (lane 3), but not by an AMOT construct with AAxY mutations in all three PPxY motifs (ΔPPxY, lane 4). Mutations in each of the three individual PPxY motifs altered virus release and infectivity to different degrees. Specifically, mutation of PPxY1 strongly impaired virus release (ΔPPxY1, lane 5), whereas mutations in the second and third PPxY motifs had moderate (ΔPPxY2, lane 6) or no (ΔPPxY3, lane 7) effects. Thus, the AMOT PPxY motifs are required for efficient HIV- 1ΔPTAP,ΔYP release and infectivity, and PPxY1 is the most important of the three motifs.
Figure 2.
Roles of AMOT–NEDD4L in HIV-1 release and infectivity.A, importance of AMOT PPxY motifs for NEDD4L-dependent release of HIV-1ΔPTAP,ΔYP. Left panels are Western blots showing HEK293T cellular levels of endogenous AMOT and exogenous HA-AMOT p130 or indicated mutants (panel 1, anti-AMOT), exogenous FLAG-NEDD4L (panel 2, anti-FLAG), endogenous GAPDH (panel 3, anti-GAPDH, loading control), and HIV-1 GagΔPTAP,ΔYP and the MA and CA proteolytic processing products (panel 4, anti-MA and anti-CA). Cells were cotransfected with a nontargeting (NT) siRNA (lane 1) or an siRNA-targeting endogenous AMOT (lanes 2–7) and expression vectors for HIV-1ΔPTAP,ΔYP, wt FLAG-NEDD4L (lanes 3–7), wt or mutant, siRNA-resistant HA-AMOT p130 (lanes 3–7), or an empty vector control (lanes 1 and 2). Right panels show corresponding levels of extracellular, virion-associated CAGag and MAGag proteins (panel 1, anti-MA and anti-CA) and viral titers (panel 2), relative to the value in the control experiment, set to 1.0. Titers for control experiment were 1–5 × 103 infectious units per milliliter. Numbers within the blots show integrated intensities of the MA band (relative to the value in the control experiment, set to 1.0; average of three independent repeats). Here and in panel B, error bars denote SD from three independent replicates. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, and ∗∗p < 0.01 by one-way ANOVA followed by Dunnett's multiple comparisons test. B, importance of WW domains for NEDD4L-dependent release of HIV-1ΔPTAP,ΔYP. Panels are equivalent to those in panel A, except that cells were cotransfected with a nontargeting (NT) siRNA (lane 1) or an siRNA-targeting endogenous AMOT (lanes 2–8), expression vectors for HIV- 1ΔPTAP,ΔYP (lanes 1–8), a wt, siRNA-resistant HA-AMOT p130 construct (lanes 3–8), wt or mutant FLAG-NEDD4L (lanes 3–8), or an empty vector control (lanes 1 and 2). ns, not significant; PPxY, Pro-Pro-x (any amino acid)-Tyr.
The importance of the individual NEDD4L WW domains was tested by coexpressing wt AMOT p130 together with wt and mutant NEDD4L constructs that carried inactivating Ala mutations in the essential second conserved Trp residue of each of the four different NEDD4L WW domains (Fig. 2B). A construct carrying mutations in all four WW domains failed to rescue HIV-1ΔPTAP,ΔYP release and infectivity, confirming the importance of the PPxY–WW domain for virus release (ΔWW, lane 4). However, all four NEDD4L constructs with mutations in single WW domains (ΔWW1-4, lanes 5–8) stimulated virion release and infectivity to near wt levels, demonstrating that retention of three functional WW domains can compensate for the loss of any single other WW domain.
AMOT PPxY1 binds with high affinity to the NEDD4L WW3 domain
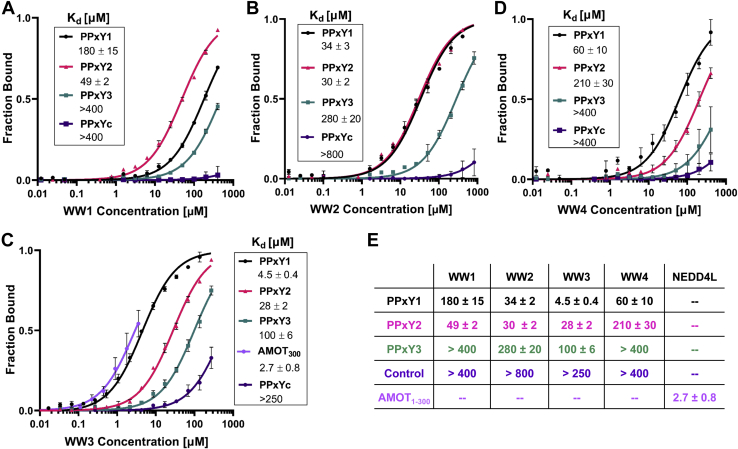
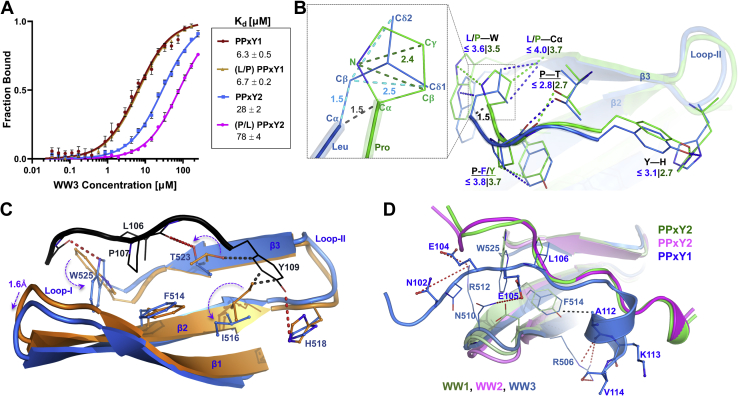
We used fluorescence polarization (FP) anisotropy binding assays to determine the affinities of labeled peptides corresponding to each of the three AMOT PPxY motifs for each of the four different recombinant NEDD4L WW domains (Fig. 3, A–D). The 12 different pairwise interactions spanned more than two orders of magnitude in binding affinity (Fig. 3E), with the following trends: (1) NEDD4L WW3 was consistently the strongest PPxY binder, (2) NEDD4L WW4 was the weakest PPxY binder in two of three cases, and (3) AMOT PPxY3 was consistently the weakest WW domain binder. The AMOT PPxY1–NEDD4L WW3 interaction was of particular note because it was more than 5-fold tighter than any other pairwise interaction (Kd = 4.5 ± 0.4 μM).
Figure 3.
Fluorescence polarization binding isotherms showing NEDD4L WW domains interacting with individual AMOT PPxY motifs. A–D, binding isotherms of synthetic AMOT PPxY peptides to the four different recombinant NEDD4L WW domains. Binding was measured by changes in the fluorescence polarization (FP) of PPxY peptides with C-terminal Oregon Green fluorophores (pH 7.5 and 25 °C). Color coding: PPxY1 (black), PPxY2 (pink), PPxY3 (green), and the control peptide lacking a PPxY motif (purple). Note that the FP binding isotherm for AMOT1-300 and full-length NEDD4L (violet) is also shown in panel C. Data points report the mean of three independent titrations, and fitted curves follow the equation Y=(maxFP∗[protein]/(Kd + [protein])). Error bars denote SD. E, fitted Kd mean values (μM) ± mean SEM. PPxY, Pro-Pro-x (any amino acid)-Tyr.
In some contexts, tandem WW domains can bind cooperatively to tandem PPxY motifs, and this can even occur when one of the PPxY motifs is cryptic. An example is the tandem WW1/WW2 domains of the KIBRA protein, which form a well-ordered multidomain module that binds tightly (Kd = 96 nM) to AMOT PPxY3 (284PPEY287) and to a cryptic 278LMRY281 motif located immediately upstream (31). To test whether the NEDD4–AMOT complex makes any such synergistic interactions, we quantified the binding of full-length recombinant NEDD4L to an Alexa Fluor–tagged recombinant AMOT construct that spanned all three PPxY motifs (AMOT1-300, Fig. 3, C and E). Although the limited solubility of full-length NEDD4L prevented sampling of the full isotherm, binding was observed with an apparent dissociation constant of 2.7 ± 0.8 μM. This interaction is only 1.5-fold tighter than that of the isolated AMOT PPxY1–NEDD4L WW3 complex, and we therefore conclude that this single interaction likely provides most of the binding energy in the AMOT1-300–NEDD4L complex. However, other PPxY–WW interactions also probably contribute to creating the slightly tighter binding observed in the AMOT1-300–NEDD4L complex, and we also cannot rule out the possibility of compensatory effects in the full-length protein. For example, autoinhibitory intramolecular WW domain interactions have been postulated to regulate NEDD4L and prevent self-ubiquitination (31). Nevertheless, our data indicate that the AMOT1-300–NEDD4L complex does not contain special high-affinity or highly cooperative interactions but rather that the different NEDD4L WW domains and AMOT PPxY motifs instead bind relatively independently, as has been seen in other cases where multiple WW domains can bind to multiple PPxY motifs (32, 33). This conclusion is consistent with the unique functional importance of the AMOT PPxY1 motif in HIV-1 release (Fig. 2A) and with a recent report that the homologous interaction also dominates binding of NEDD4 to angiomotin-like 1 (23).
To understand why the KIBRA WW1/WW2 element binds tightly to AMOT, whereas the similarly spaced, tandem NEDD4L WW3/WW4 element does not, we modeled NEDD4L WW3/WW4 based on the KIBRA WW1/WW2 structure (34) (Fig. S1). The model suggests that formation of a stable NEDD4L WW3/WW4 interdomain interface may be disfavored by the anionic Glu526 residue located between the WW3 and WW4 domains (replacing Ile35 in the equivalent position of KIBRA WW1/WW2). Consistent with this idea, the affinity of the KIBRA–AMOT complex is reduced 50-fold when Ile35 is mutated to Asp (34). Cooperative binding may also be disfavored by substitution of two other large hydrophobic KIBRA WW1/WW2 interface residues (Phe47 and Leu57) with smaller hydrophobics at equivalent positions in NEDD4L WW3/WW4 (Leu542 and Pro552, respectively).
Structural studies of AMOT PPxY–NEDD4L WW complexes
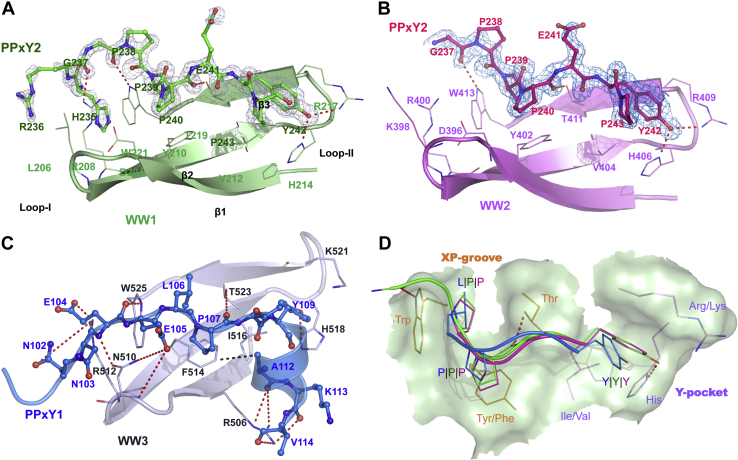
Having established that the different NEDD4L WW domains and AMOT PPxY motifs bind independently, we next investigated the structural basis for the observed affinity differences in the different pairwise complexes. This was done by determining and comparing the structures of three different NEDD4L WW domains bound to AMOT PPxY motifs, including the high-affinity AMOT PPxY1–NEDD4L WW3 complex.
To obtain structures, we screened a series of different high- and moderate-affinity AMOT PPxY–NEDD4L WW complexes for crystallization. Two such complexes, AMOT PPxY2–NEDD4L WW1 and AMOT PPxY2–NEDD4L WW2, yielded high-resolution crystal structures (1.52 Å and 1.74 Å, respectively) (Table S1 and Fig. 4, A and B).
Figure 4.
Structures of AMOT PPxY–NEDD4L WW complexes. A, crystal structure of AMOT PPxY2 (green sticks, chain b) bound to NEDD4L WW1 (pale green, chain a; ribbon diagram with key residue side chains shown explicitly). The three WW domain strands are labeled in this panel. B, crystal structure of AMOT PPxY2 (magenta, chain d) bound to NEDD4L WW2 (pink, chain c). Red dashed lines in panels A and B denote observed hydrogen bonds. Mesh renderings show the FO-FC map (3σ), calculated with the PPxY peptide omitted. C, solution structure of AMOT PPxY3 (blue) bound to NEDD4L WW3 (gray). Dashed red lines indicate observed hydrogen bonds or salt bridge interactions. The hydrophobic pocket interaction between WW3 Phe514 and PPxY1 Ala112 (∼3.9 Å) is shown as black dashed lines. Glu105 interacts with Asn510 (2.7–4.5 Å) in ∼50% of ensemble structures. Each of the residues Glu104 and Asn102 individually interacts with Arg512 (2.6–4.5 Å) in ∼40% of ensemble structures. Carbonyl oxygen of Glu105 hydrogen bonds to side-chain nitrogen of Asn510 (not shown for clarity). Arg506 interacts with the carbonyls of Ala112, Lys113, and Val114 (3.0–4.5 Å) in ∼25% of ensemble structures. D, surface rendering of the conserved WW core domain showing the characteristic XP groove (residue labels in orange) and Y-pocket (residue labels in purple). PPxY peptides are overlaid and colored following panels A-C. Red dashed lines denote canonical WW–PPxY hydrogen bonds (shown only for WW1–PPxY2 for clarity). PPxY, Pro-Pro-x (any amino acid)-Tyr.
Attempts to crystallize the high-affinity AMOT PPxY1–NEDD4L WW3 complex repeatedly yielded crystals of NEDD4L WW3 alone. The explanation for this became evident when we determined the apo-NEDD4L WW3 structure and observed that in the preferred crystal lattice, a proline-rich element from the N terminus of one NEDD4L WW3 domain (cryptic peptide, residues 498FLPP501, Fig. 1) bound in the PPxY-binding site of a neighboring molecule (Fig. S2A). The cryptic peptide binds in the opposite orientation of AMOT PPxY peptides (Fig. S2B) and is therefore not discussed further.
In the absence of suitable crystals, we used NMR spectroscopy to determine the solution structure of the AMOT PPxY1–NEDD4L WW3 complex. To drive complex formation and simplify NMR assignment/refinement strategies, we determined the structure of a recombinant construct in which the AMOT PPxY1 motif was fused to the NEDD4L WW3 N terminus through a short Gly–Ser linker. The intramolecular PPxY1 element bound in a native conformation within this construct, as evidenced by the nearly perfect overlap of 1H/15N heteronuclear single-quantum correlation (HSQC) signals for WW3 residues of the fusion construct and a nonfused NEDD4L WW3 titrated with AMOT PPxY1 peptide (Fig. S3, A and B). NMR spectra of the PPxY1-WW3 fusion protein were of high quality, and the resulting structure (Fig. 4C) was well ordered for the residues corresponding to AMOT residues 101QNNEELPTYEEAKV114 and NEDD4L WW3 residues 498 to 526 (0.5 Å root mean square deviation (RMSD) for 20 lowest energy structures) (Fig. S3C and Table S2). Finally, to identify conformational changes that occur upon AMOT PPxY1 binding, we also determined a high-resolution NMR solution structure of the unliganded NEDD4L WW3 protein (Table S2).
All five of our new structures exhibit canonical WW domains, with three-stranded β-sheets and canonical positioning of the two eponymous Trp residues (27, 29, 30). The three WW–PPxY complexes and the WW3 cryptic peptide complex are superimposable (<1.0 Å RMSD), whereas the apo WW3 WW domain structure (∼3.0 Å RMSD) differed in significant ways, as discussed below.
In all three WW–PPxY complexes, the PPxY core elements bind in an extended polyproline II helical conformation across the concave WW sheet (Fig. 4, A–C). As illustrated in Figure 4D, each conserved residue in the PPxY motif makes canonical binding interactions: the first Pro/Leu residue binds in the “XP-groove,” where it stacks against the second conserved WW Trp residue (Trp525 in WW3). The indole nitrogen of this Trp residue also hydrogen bonds to the backbone carbonyl located two residues before the PPxY motif. The second PPxY Pro packs against a Tyr/Phe residue from the second β-strand (β2), and its carbonyl oxygen forms a hydrogen bond to the hydroxyl of a conserved Thr (in β3). The fourth Tyr residue of PPxY binds in the characteristic “Y-pocket”, forming a hydrogen bond with conserved His (in loop-II between β2 and β3). Thus, the core WW–PPxY interactions are very similar in every case and apparently do not explain why AMOT PPxY1 binds WW3 with unusually high affinity.
The unique Leu106 residue in AMOT PPxY1
The AMOT PPxY1 motif is unique in having a Leu106 residue in place of the canonical Pro in the first position (Fig. 1). We therefore investigated whether this residue might underlie the uniquely high affinity of the PPxY1–WW3 interaction. This was not the case, however, because substitution of Leu106 by Pro did not measurably alter NEDD4L WW3 binding affinity (Fig. 5A). Similarly, mutation of Leu106 to Pro did not alter the ability of AMOT to stimulate HIVΔPTAP,ΔYP release and infectivity to the same extent as NEDD4L wt (Fig. 6A, compare lanes 2 and 3). These experiments were performed in the context of NEDD4L constructs in which the WW1, WW2, and WW4 elements were inactivated by mutation and only the WW3 domain was active (denoted NEDD4LWW3). These experiments showed that the NEDD4LWW3 construct alone could rescue HIV release and infectivity to wt levels (Fig. 6A, compare lanes 1 and 2), and this construct was used to ensure that any effects observed were restricted to the PPxY1–WW3 interaction. Thus, the unique Leu106 residue does not play a key binding or functional role although it could, in principle, contribute to specificity by helping to discriminate between different WW domains in NEDD4L and other WW proteins.
Figure 5.
Energetic and structural consequences of NEDD4L WW3 binding to wt and mutant AMOT PPxY motifs.A, fluorescence polarization (FP) binding isotherms and fitted Kd values (μM) for NEDD4L WW3 binding to peptides corresponding to wt AMOT PPxY1 (red), an PPxY1 L106P mutant (orange, denoted (L/P)PPxY1), wt PPxY2 (blue), and a PPxY2 P239L mutant (magenta, denoted (P/L)PPxY2). Error bars denote SD from four independent replicates. B, superpositions of PPxY1–WW3 (blue) and PPxY2–WW1 (green) complexes illustrating how the noncanonical Leu106 residue of the PPxY1 occupies the same position as the canonical Pro239 of PPxY2. Key conserved residues are shown explicitly; AMOT PPxY–WW distances (dashed lines, Å) are shown. WW3–PPxY1 distances are from ∼75% of ensemble structures. Inset: close-up view of Leu/Pro residues within the boxed region. Note that the Pro pyrrolidine ring and the Leu isopropyl group occupy the same WW binding site (Trp, Thr, Tyr/Phe, and His) and bury similar accessible surface areas (∼90 Å2). Distances (Å, dashed lines) are shown for scale and to illustrate the 1.5 Å shift between the PPxY1 Leu106 and PPxY2 Pro239 Cα positions. Substitution of Leu for Pro in the first position of the PPxY motif should reduce the propensity to form a PPII helix (37), but this unfavorable change may be compensated by burial of the larger Leu side chain. C, superposition of WW3 structures in the apo/unliganded (orange) and PPxY1 (blue) states. Displacements of the loop 1 and key side chains upon peptide (black) binding are shown as purple arrows and dashed lines. Red dashed lines depict canonical hydrogen bonds, and black dashed lines show steric clashes that require WW3 Ile516 (1.5 Å to Tyr109) and Thr523 (3.0 Å to Tyr109) to change conformation. D, superposition of PPxY2–WW1 (green), PPxY2–WW2 (magenta), and PPxY1–WW3 (blue), emphasizing the interactions unique to WW3–PPxY1. PPII, polyproline II; PPxY, Pro-Pro-x (any amino acid)-Tyr.
Figure 6.
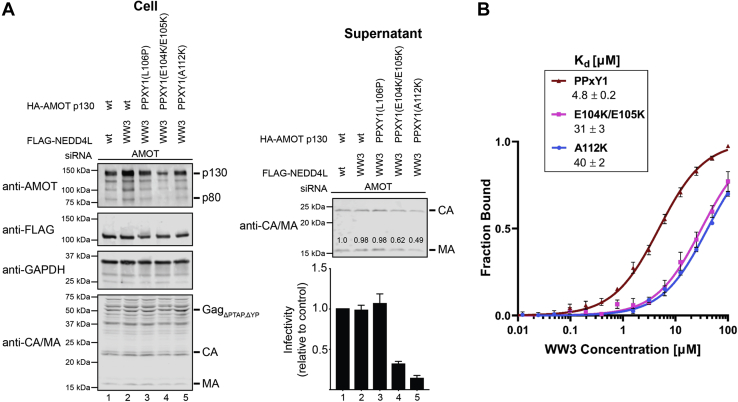
Impact of unique PPxY1 N- and C-terminal extensions on NEDD4L WW3 binding and HIV budding and infectivity.A, importance of AMOT PPxY1 residues for NEDD4LWW3-dependent release of HIV-1ΔPTAP,ΔYP. Left panels are Western blots showing HEK293T cellular levels of endogenous AMOT and exogenous HA-AMOT p130 or indicated mutants (panel 1, anti-AMOT), exogenous FLAG-NEDD4L (panel 2, anti-FLAG) or FLAG- NEDD4LWW3, endogenous GAPDH (panel 3, anti-GAPDH, loading control), and HIV-1 GagΔPTAP,ΔYP and the MA and CA proteolytic processing products (panel 4, anti-MA and anti-CA). Cells were cotransfected with an siRNA-targeting endogenous AMOT and expression vectors for HIV-1ΔPTAP,ΔYP, wt FLAG-NEDD4L (lane 1), FLAG-NEDD4LWW3 (lanes 2–5), and wt or mutant, siRNA-resistant HA-AMOT p130. Right panels show corresponding levels of extracellular, virion-associated CAGag and MAGag proteins (panel 1, anti-MA and anti-CA) and viral titers (panel 2), relative to the value in lane 1, set to 1.0. Numbers within the blots show integrated intensities of the MA band intensities (relative to the value in the control experiment, set to 1.0; average of two independent repeats). Error bars denote the SD from two independent replicates. B, importance of AMOT PPxY1 residues for NEDD4LWW3 binding. Fluorescence polarization binding isotherms and Kd values (μM) for the interaction of WW3 with wt PPxY1 (red), and the E104K/E105K (pink) and A112K (blue) mutants. Error bars denote the SD from three independent replicates. PPxY, Pro-Pro-x (any amino acid)-Tyr.
Our structural analyses reveal that the isopropyl Cβ-Cδ element of AMOT PPxY1 Leu106 occupies the same WW pocket as the pyrrolidine ring of the canonical first Pro residue (e.g., Pro239 in PPxY2, Fig. 5B inset). To accommodate the longer Leu side chain, the PPxY1 backbone shifts back by ∼1.5 Å as compared with PPxY2 backbone. We found, however, that equivalent Pro to Leu substitutions were not isoenergetic in all PPxY motifs, because substitution of Pro239 to Leu reduced PPxY2 binding to WW3 by 2.8-fold (Fig. 5A).
Conformational changes that accompany AMOT PPxY1 binding to NEDD4L WW3
Comparison of the peptide-bound and -unbound AMOT WW3 structures revealed that peptide engagement expands the central binding groove and creates the different subpockets that accommodate conserved PPxY residues (Fig. 5C). The groove slightly expands (<2.0 Å) by the movement of loop I away from loop II/β3. Five WW domain sidechains also rotate significantly: Trp525 rotates to stack against PPxY1 Leu106 and to align the indole nitrogen to hydrogen bond to a carbonyl upstream of PPxY motif; His518 rotates to stack and hydrogen bond with the PPxY1 Tyr109 hydroxyl; Phe514 slightly moves upward to stack against PPxY1 Pro107; Ile516 and Thr523 rotate to accommodate the stacking of PPxY1 Tyr109 in the Y-pocket and align the Thr523 hydroxyl to hydrogen bond with the PPxY1-Pro107 carbonyl (Fig. 5C). Analogous “induced fit” peptide-binding mechanisms have been described for other WW–PPxY interactions (35).
Structural basis for the high-affinity PPxY1–WW3 interaction
Comparison of the structure of PPxY1–WW3 to the lower affinity PPxY2–WW1 and PPxY2–WW2 complexes revealed that the high-affinity binding of PPxY1 to WW3 reflects a series of favorable contacts made uniquely in this complex by the N- and C-terminal extensions that flank the conserved PPxY core (Figs. 4C and 5D). In contrast, the PPxY2–WW1 and PPxY2–WW2 complexes make few favorable contacts outside the core (Figs. 4, A and B and 5D).
On the N-terminal side, the PPxY1 backbone approaches WW3 loop-I more closely (∼6 Å) than PPxY2 in the WW1 or WW2 complexes (11 Å and 14 Å, respectively) (Fig. 5D). Favorable side-chain interactions include hydrogen bonds between the PPxY1 Glu104 backbone nitrogen with the unique WW3 Asn510 side chain (a Leu in WW1 and Lys in WW2), and PPxY1 Glu105 Oε with the WW3 Asn510 backbone and side-chain nitrogen. The anionic and hydrophilic side chains of PPxY1 Asn102 and Glu104 also interact favorably with the WW3 Arg512 guanidinium (Figs. 4C and S4).
To assess the importance of the N-terminal WW3–PPxY1 interaction, we mutated AMOT PPxY1 Glu104 and Glu105 to Lys. This mutation reduced the affinity of the WW3–PPxY1 interaction 6-fold (from 4.8 to 31 μM) (Fig. 6B) and also reduced the ability of AMOT to stimulate virus release and infectivity in concert with the NEDD4LWW3 construct (Fig. 6A, compare lanes 2 and 4). Thus, the WW3 loop I composition contributes to PPxY1 binding, in good agreement with other studies (32, 33, 36, 37).
On the C-terminal side of the core, PPxY1 residues 109YEEAKV114 form a short α-helix, capped by Thr108, that sits in a hydrophobic groove created by WW3 Phe514, Ile516 and the aliphatic atoms of Lys521 and Thr523 (Figs. 4C and 5D) Two complementary residues enable this interaction: the aromatic WW3 Phe514 side chain (as opposed to the larger, hydroxylated Tyr residue in WW1/WW2) and a matching small PPxY1 α-helix packing interface residue, Ala112, that fits well in the hydrophobic pocket (as opposed to a larger Lys residue in the same position in the 310-helix of PPxY2). Tight PPxY1–WW3 packing also allows the conserved WW3 Arg506 residue to interact with the carbonyl oxygens of PPxY1 Ala112, Lys113, and Val114 (Fig. 4C). Analogous helix–hydrophobic groove interactions have also been observed in other PPxY–WW domain complexes (36, 37, 38).
To test the energetic contribution of the C-terminal PPxY1 helix–WW3 interaction, we mutated AMOT PPxY1 Ala112 to Lys. This mutation reduced WW3 binding affinity 8-fold (from 4.8 to 40 μM) (Fig. 6B). This mutation also reduced the ability of AMOT to stimulate virus release and infectivity in concert with the NEDD4LWW3 construct (Fig. 6A, compare lanes 2 and 5). Thus, the AMOT PPxY1 Ala112 residue contributes to the structure, affinity, and function of WW3 binding.
Summary
The cellular host proteins AMOT and NEDD4L can cooperate to stimulate HIV-1 release, particularly when ESCRT factor recruitment is inefficient (19). The two proteins bind one another through interactions between the three PPxY motifs in AMOT and the four WW domains in NEDD4L, and these interactions are essential for AMOT- and NEDD4L-dependent HIV-1 release and infectivity (Fig. 2). We find that the AMOT PPxY1 element plays an essential role in this process and binds the NEDD4L WW3 domain with unusually high affinity (Figs. 2 and 3). This affinity reflects mutual compatibility between the unique PPxY1 N- and C-terminal extensions and the complementary WW3 Loop-I and Phe514/Ile516/Arg506 hydrophobic pocket elements, respectively (Figs. 4C and 5D). Sequence variations prevent all other possible AMOT PPxY–NEDD4L WW combinations from making this full set of interactions, thereby explaining the uniquely favorable affinity and specificity of the PPxY1–WW3 interaction. In the context of an HIV infection, the AMOT–NEDD4L interaction helps drive virion envelopment (19). We speculate that AMOT also helps recruit NEDD4L ubiquitin ligase activity to the nascent virion at the proper time. This could occur if the AMOT Bin/amphiphysin/Rvs domain senses the extent of virus assembly by assessing the degree of membrane curvature. Once the proper degree of membrane curvature is sensed, AMOT might then recruit and activate NEDD4L, thereby promoting recruitment of the early-acting, ubiquitin-binding ESCRT factors required to initiate bud neck closure and membrane fission.
Experimental procedures
Plasmids
Constructs for expressing wt and mutant NEDD4L and AMOT proteins in Escherichia coli and human HEK293T cells were created by standard cloning and mutagenesis methods, with detailed methods available upon request. Expression constructs are described in detail in Table S3. All of the new expression constructs used in our studies have been submitted to the Addgene plasmid repository (https://www.addgene.org/)
Cell culture
HEK293T and HeLa-TZM-bl cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Thermo Fisher Scientific) containing 10% fetal bovine serum at 37 °C and 5% CO2. HEK293T cells were obtained from the American Type Culture Collection, and HeLa–TZM-bl cells were obtained from the HIV Reagent Program. Cells were tested for Mycoplasma contamination every 3 months using a PCR Mycoplasma Detection Kit (Applied Biological Materials).
Budding assays
HIV-1 budding assays were performed as follows: HEK293T cells (3 × 105 cells/well, 6-well plate) were transfected with 20 nM siRNA-targeting AMOT (19) using 7.5-μl Lipofectamine RNAiMax (Thermo Fisher Scientific). 24 h later, the medium was changed and cells were cotransfected with 20 nM siRNA-targeting AMOT, 0.5-μg HIV-1NL4-3 R9ΔPTAP,ΔYP, 0.5 μg of pCI-FLAG-NEDD4L, or indicated NEDD4L mutants and 1-μg pcDNA3 HA–AMOT p130 or indicated AMOT mutants. The medium was changed to fresh DMEM 6 to 8 h after transfection. Forty-eight hours after the second transfection, cells were harvested for Western blot analyses and supernatants harvested for titer measurements and virus purification.
Cells were washed off the plate in 1 ml PBS, pelleted by centrifugation, and lysed for 15 min at 4 °C in 150-μl RIPA buffer (10 mM Tris, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and 140 mM NaCl, pH 8.0) supplemented with mammalian protease inhibitor (Sigma-Aldrich). Insoluble material was removed by centrifugation (10 min, 15,000g, 4 °C). 150 μl of 2 × Laemmli SDS-PAGE loading buffer supplemented with 10% 2-mercaptoethanol (Sigma-Aldrich) were added and samples were boiled for 5 min. Virions from 1-ml supernatant were pelleted by centrifugation through a 200-μl 20% sucrose cushion (90 min, 15,000g, 4 °C) and denatured by adding 100 μl 1× Laemmli SDS-PAGE loading buffer and boiling for 5 min. Proteins were separated by SDS-PAGE, transferred onto PVDF membranes, and probed with antibodies. Primary antibodies were as follows: anti-AMOT (1;3000, UT691, Covance), anti-FLAG (1:10,000, F1804, Sigma-Aldrich), anti-HIV CA (1:1000, UT415, Covance), anti-HIV MA (1:1000, UT556, Covance), and anti-GAPDH (1:10,000, MAB374, EMD Millipore, Burlington, VT). Antibodies with UT designations and numbers were raised against recombinant proteins purified in the Sundquist laboratory. Bands were visualized by probing the membrane with fluorescently labeled secondary antibodies (LI-COR Biosciences) and scanning with an Odyssey Imager (LI-COR Biosciences).
HIV-1 titers were assayed in HeLa-TZM-bl indicator cells using a β-galactosidase assay. HeLa-TZM-bl cells (5000 cells per well, 96-well plate) were infected with three different dilutions of virus-containing culture media, each in triplicate, harvested after 48 h, washed once with 200-μl PBS, and lysed in 50-μl 25 mM Tris phosphate (pH 7.8), 2 mM DTT, 2 mM 1,2-diaminocyclohexane N,N,N′,N′-tetraacetic acid, 10% glycerol, and 1% Triton X-100 (10 min, 23 °C). Cell lysates were incubated with 50-μl assay buffer (200 mM sodium phosphate (pH 7.3), 2 mM MgCl2, 100 mM β-mercaptoethanol, 1.33 mg/ml ortho-nitrophenyl-β-galactoside) for 60 min at 37 °C. Reactions were terminated by addition of 200-μl 1 M Tris base, and absorbance at 420 nm was read in a plate reader.
Protein expression, purification, and labeling
Bacterial expression plasmids containing either GST-PP-WW or His-SUMO-WW domains, His-AMOT(1–300), or His-sortase (Staphylococcus aureus) were expressed in the Rosetta-pLysS bacterial strain grown in autoinduction media ZYP-5052 (39). Lysis buffer contained 50 mM Tris, 150 to 500 mM NaCl, 5% glycerol, 2 mM MgCl2, 1 mM TCEP, 0.5% NP-40, pH 7.5, supplemented with Roche cOmplete EDTA-free protease inhibitors, and 5 mM imidozole for His-tagged proteins. Lysate viscosity was reduced by addition of DNase (12.5 μg/ml, Roche) followed by ultrasonic disruption. Proteins captured on Ni-NTA cOmplete resin (Roche), or Glutathione Superose resin (GE Healthcare Bio-Sciences), were washed with the lysis buffer and eluted with the lysis buffer supplemented with 250 mM imidazole for His-tagged proteins, or GST-fusion proteins were separated from the GST tag by on-column cleavage with PreScission protease (1 nmol protease/30 nmol purified protein, GE Healthcare Bio-Sciences). The His-SUMO-Fusion proteins were further cleaved with ULP1 (1 nmol protease/30 nmol purified substrate protein). Further ion-exchange (pH 7.5, SP-column, HiTrap HP-SP, GE Healthcare Bio-Sciences) and S75 sizing column (150 mM NaCl, pH 7.5) purifications were performed, and the samples were snap-frozen for storage at −80 °C. Typical yields per liter culture were 60 nmol for WW domains, and 8 nmol for sortase and AMOT1-300. Protein identities were confirmed by electrospray mass spectrometry, which confirmed the expected full protein masses.
A labeled peptide (GGG-ALEXA488) was added to AMOT1-300 (which carried a sortase tag) by transpeptidation with sortase. Typical reactions contained 10 to 50 μM AMOT1-300, 30 to 150 μM GGG-ALEXA488 peptide and 20 μM sortase enzyme in the reaction buffer (50 mM Tris [pH 7.5], 150 mM NaCl and 10 mM CaCl2). Reactions were allowed to proceed at either room temperature or 4 °C until the reaction was complete (typically 1–4 h). AMOT1-300–ALEXA488 was then purified by ion-exchange chromatography (SP HiTrap) as described above.
FP binding experiments
Peptide preparation
Three AMOT PPxY peptides (residues 100–114, 233–247, and 278–292, see Fig. 1) and one AMOT control peptide (residues 196–209), each containing a non-native, N-terminal cysteine, were synthesized, purified, and fluorescently labeled in the University of Utah Peptide Synthesis Core.
Briefly, peptides were synthesized with a cysteine at the N terminus using Fmoc solid-phase technology, common protecting groups, and HBTU chemistry on an ABI 433 synthesizer (Applied Biosystems). Synthesized peptides were purified by reversed-phase chromatography using acetonitrile/water gradients with 0.1% TFA in both solvents. Peptide-containing fractions were pooled, dried, and redissolved in DMSO. Fluorescent labeling was performed in DMSO at 4 °C with approximately 1.3-fold molar excess of Oregon Green 488 maleimide (Life Technologies/Molecular Probes 06034) dissolved in a 1:1 solution of acetonitrile:DMSO. Reaction progress was monitored by HPLC, and labeled peptides were separated from free dye and residual unlabeled peptides using the same reversed-phase conditions described above. Labeled peptide masses were determined by MALDI-TOF-MS at the University of Utah Mass Spectrometry Core facility: dye-labeled PPxY1 was 2358.9 Da (theoretical 2360.5 Da); dye-labeled PPxY2 was 2336.9 Da (theoretical 2338.6 Da); dye-labeled PPxY3 was 2350.0 Da (theoretical 2351.6 Da); and dye-labeled PPxY-control was 2090.8 Da (theoretical 2092.2 Da). Confirmed peptide fractions were dried under vacuum and redissolved and neutralized in water. Concentrations were calculated using the absorbance of Oregon Green 488 at 491 nm (extinction coefficient 83,000 cm−1 M−1 in 50 mM potassium phosphate, pH 9.0) before diluting in FP binding buffer (see below).
FP binding assay
FP experiments were performed in 100-μl reaction volumes in the binding buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 0.5 mM TCEP) using 1 nM fluor-labeled PPxY peptides and 2-fold dilutions of WW domains (or associated mutants; WW1: 4 × 102–4.8 × 10−5 μM; WW2: 8.2 × 102−9.8 × 10−5 μM; WW3: 1.9 × 102–2.3 × 10−5 μM; WW4: 4 × 102–4.8 × 10−5 μM). Each dilution series was performed at least in triplicate. Each dilution (70 μl) was placed in a Corning 384 flat-bottom plate and allowed to equilibrate (26 °C, 15 min) in the plate reader. Measurements were taken in the BioTek Synergy Neo Hybrid Multi-Mode Microplate Reader. The instrument measured 50 data points for each well, with a 200-millisecond delay between wells. The lamp was positioned 9.0 mm away from the plate with excitation at 498 nm and emission monitored at 528 nm. The instrument gain was set on the control peptide sample, which consistently had a lower fluorescent intensity than the other peptides. Each binding isotherm was measured at least four independent times, with a one-minute delay between each read. Data were exported into Microsoft Excel and input into Graph Prism-6 (GraphPad, San Diego, CA). Curve fitting and Kd values were created using a pre-existing protocol in Graph Prism 6. The data were fit to the equation Y = (maxFP∗[protein])/(Kd+[protein]). The mean Kd values are reported as mean ± SEM.
Crystallization, data collection, and structure determination
The PPxY peptides and native WW domains were mixed (1:1 M ratio) at a final concentration of 10 to 30 mg/ml (20 mM Tris, pH 7.5, 150 mM NaCl). Crystals were initially screened using kits from Hampton Research. The cryoprotectant solution contained precipitant supplemented with 25 to 30% glycerol.
Apo-WW3 crystals were grown in a solution containing 3.6 M ammonium nitrate, 5% glycerol, and 0.1 M sodium acetate at pH 4.6 (20 °C). WW2–PPxY2 crystals were grown in a solution containing 2.0 M ammonium sulfate and 0.1 M Bis-Tris at pH 6.5 (20 °C). WW1–PPxY2 crystals were grown in a solution containing 1.6 M (NH4)2SO4, 0.1 M potassium sodium tartrate, 5% glycerol, and 0.1 M sodium acetate (37 °C).
Diffraction data were collected at SSRL beamline BL7-1 and processed using AutoXDS at SSRL http://smb.slac.stanford.edu/facilities/software/xds/#autoxds_script (40). Structures were determined by molecular replacement (Phaser in PHENIX (41)), using a solution structure of the NEDD4L WW3 domain (Protein Data Bank ID: 2MPT (31)) as a search model. Building was completed in Coot (42) and structures refined in PHENIX (43). Structure alignments, distance measurements, and figures were made using PyMol Molecular Graphics System (Schrödinger, LLC). Surface area accessibility was calculated using CoCoMaps (44).
NMR data collection and structure refinement
NMR data collection and assignments
NMR samples were typically 1 mM of 13C/15N-labeled protein in the NMR buffer (20 mM sodium phosphate, 50 mM NaCl, and 10% 2H2O). All NMR spectra were collected at 25 °C on a Varian Inova 600 MHz spectrometer equipped with a triple-resonance 1H/13C/15N cryo-probe with z-axis pulsed field gradient capability. Backbone NMR chemical shift assignments were made using a suite of triple resonance experiments as described previously (45), using 3D versions of the HNCACB, CBCACONH, HNCO, and HA(CA)CO. Side-chain assignments were completed using 3D-H(CCO)NH-TOCSY, 3D-(H)C(CCO)NH-TOCSY, and 3D HCCH-TOCSY experiments. Aromatic resonances were assigned using a combination of 1H/13C HSQC, 13C-edited NOESY experiments centered on the aromatic carbon resonances (125 ppm), a 13C-edited NOESY experiment centered on the aliphatic region, and heteronuclear correlation experiments that correlate the Cβ carbons to the Hδ and Hε protons of the aromatic rings. Stereospecific assignments for β-methylene protons (and estimation of χ1 dihedral angles) were obtained using a combination of HNHB, HN(CO)HB, 15N-edited TOCSY, and NOESY data. Stereospecific assignments of side-chain methyl groups and qualitative determination of χ1 and χ2 dihedral propensities were obtained using long-range carbon–carbon and carbon–proton couplings observed in long-range carbon-carbon and long-range carbon-hydrogen couplings experiments. Three-dimensional 15N-edited NOESY-HSQC and 3D 13C-edited NOESY-HSQC’s (100 msec mixing times) were used to generate distance restraints for refinement (46, 47, 48). Dihedral restraints were derived from backbone Ha, Ca, CO, and Cb chemical shifts using TALOS. All spectra were processed with FELIX 2007 (Accelrys) or NMRPipe (49) and referenced indirectly to sodium trimethylsilylpropanesulfonate.
Structure determinations
Backbone and side-chain correlations were assigned, and nuclear Overhauser effect (NOE) intensities were integrated using tools in SPARKY https://www.cgl.ucsf.edu/home/sparky/ (50). The solution structure for WW3–PPxY1 and WW3 were refined using automated NOE assignments and torsion angle dynamics as implemented in CYANA (version 3.9) (51). Initial refinements used NOE data alone to define the overall fold. The final refinements added dihedral restraints, hydrogen bonds, and stereospecific assignments. A total of 100 randomized conformers were “folded” into 3D structures by iteratively including NOE constraints in a stepwise manner using criteria defined by CYANA. Of the 100 CYANA structures calculated, the 20 conformers with the lowest target function were subjected to a molecular dynamics protocol in the explicit solvent (52) using XPLOR-NIH (53).
Structures were validated using PROCHECK-NMR and AQUA and the programs supplied at the Protein Data Bank deposition site. All figures were created using PyMOL Molecular Graphics System (Schrödinger, LLC).
NMR chemical shift mapping
Chemical shift mapping of the interaction interface between the WW3 and PPxY1-3 was accomplished by making independent NMR samples of 15N-labeled WW3 (50 μM) supplemented with final concentrations of PPxY peptides over the range 0 to 150 μM in a titration NMR buffer (20 mM sodium phosphate [pH 6.0], 50 mM NaCl, and 10% 2H2O). 1H/15N -HSQC spectra were collected for each titration point. Normalized shift changes for 1H and 15N were calculated at the final titration point for each amide pair using the expression: Δn = [25∗((Δ1H)2+(Δ15N/5)2)]0.5.
Data availability
Coordinates, structure factor, and NMR restraint and chemical shift files have been deposited in the Protein Data Bank (PDB) with the following codes: apo-WW3 (crystal, 7LP1), PPxY2-WW1 (7LP3), PPxY2-WW2 (7LP2), apo-WW3 (solution, 7LP4), and PPxY1-WW3 (7LP5).
Supporting information
This article contains supporting information (54, 55, 56).
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Peptide synthesis, sequencing of constructs, and validation of proteins were performed at the peptide synthesis, DNA sequencing, and mass spectrometry core facilities at the University of Utah, which are supported by NCI, National Institutes of Health (NIH), Grant P30CA042014. Mass spectrometry equipment in the Core was obtained through an NIH Shared Instrumentation Grant S10 OD018210. We thank Stanford Synchrotron Radiation Lightsource (SSRL) for access to beamline BL7-1 that contributed to the results presented here. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and the NIH and National Institute of General Medical Sciences (P30GM133894). NMR spectra were collected in the University of Utah HSC Core NMR Facility. Crystallographic support from Chris Hill and Frank Whitby from the Department of Biochemistry, University of Utah, is gratefully acknowledged. We also thank John McCullough and Chris Stringham for help with supporting biochemistry work.
Author contributions
L. R., T. T., E. L. P., S. L. A., and A. A. formal analysis; L. R., T. T., G. M., E. L. P., and F. C. P. investigation; L. R., T. T., F. C. P., S. L. A., and A. A. visualization; L. R., T. T., G. M., E. L. P., G. P., F. C. P., B. F. V., S. L. A., and A. A. writing–review and editing; B. F. V., S. L. A., A. A., and W. I. S. supervision; S. L. A., A. A., and W. I. S. conceptualization; S. L. A. and A. A. data curation; A. A. and W. I. S. writing–original draft; W. I. S. Funding acquisition; W. I. S. writing–review and editing.
Funding and additional information
This work was supported by NIH R37 AI051174-21 to W. I. S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Edited by Craig Cameron
Contributor Information
Steven L. Alam, Email: Steve.Alam@biofiredx.com.
Akram Alian, Email: akram.alian@biochem.utah.edu.
Wesley I. Sundquist, Email: wes@biochem.utah.edu.
Supporting information
References
- 1.Sundquist W.I., Krausslich H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tedbury P.R., Freed E.O. HIV-1 gag: An emerging target for antiretroviral therapy. Curr. Top. Microbiol. Immunol. 2015;389:171–201. doi: 10.1007/82_2015_436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurley J.H., Cada A.K. Inside job: How the ESCRTs release HIV-1 from infected cells. Biochem. Soc. Trans. 2018;46:1029–1036. doi: 10.1042/BST20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lippincott-Schwartz J., Freed E.O., van Engelenburg S.B. A consensus view of ESCRT-mediated human immunodeficiency virus type 1 abscission. Annu. Rev. Virol. 2017;4:309–325. doi: 10.1146/annurev-virology-101416-041840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogt V.M. Ubiquitin in retrovirus assembly: Actor or bystander? Proc. Natl. Acad. Sci. U. S. A. 2000;97:12945–12947. doi: 10.1073/pnas.97.24.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strack B., Calistri A., Accola M.A., Palu G., Gottlinger H.G. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubert U., Ott D.E., Chertova E.N., Welker R., Tessmer U., Princiotta M.F., Bennink J.R., Krausslich H.G., Yewdell J.W. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13057–13062. doi: 10.1073/pnas.97.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Serrano J. The role of ubiquitin in retroviral egress. Traffic. 2007;8:1297–1303. doi: 10.1111/j.1600-0854.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- 9.Sundquist W.I., Schubert H.L., Kelly B.N., Hill G.C., Holton J.M., Hill C.P. Ubiquitin recognition by the human TSG101 protein. Mol. Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 10.Agromayor M., Soler N., Caballe A., Kueck T., Freund S.M., Allen M.D., Bycroft M., Perisic O., Ye Y., McDonald B., Scheel H., Hofmann K., Neil S.J., Martin-Serrano J., Williams R.L. The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain. Structure. 2012;20:414–428. doi: 10.1016/j.str.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alam S.L., Langelier C., Whitby F.G., Koirala S., Robinson H., Hill C.P., Sundquist W.I. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat. Struct. Mol. Biol. 2006;13:1029–1030. doi: 10.1038/nsmb1160. [DOI] [PubMed] [Google Scholar]
- 12.Slagsvold T., Aasland R., Hirano S., Bache K.G., Raiborg C., Trambaiolo D., Wakatsuki S., Stenmark H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J. Biol. Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- 13.Dowlatshahi D.P., Sandrin V., Vivona S., Shaler T.A., Kaiser S.E., Melandri F., Sundquist W.I., Kopito R.R. ALIX is a Lys63-specific polyubiquitin binding protein that functions in retrovirus budding. Dev. Cell. 2012;23:1247–1254. doi: 10.1016/j.devcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keren-Kaplan T., Attali I., Estrin M., Kuo L.S., Farkash E., Jerabek-Willemsen M., Blutraich N., Artzi S., Peri A., Freed E.O., Wolfson H.J., Prag G. Structure-based in silico identification of ubiquitin-binding domains provides insights into the ALIX-V:ubiquitin complex and retrovirus budding. EMBO J. 2013;32:538–551. doi: 10.1038/emboj.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pashkova N., Gakhar L., Winistorfer S.C., Sunshine A.B., Rich M., Dunham M.J., Yu L., Piper R.C. The yeast Alix homolog Bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes. Dev. Cell. 2013;25:520–533. doi: 10.1016/j.devcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Usami Y., Popov S., Popova E., Gottlinger H.G. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J. Virol. 2008;82:4898–4907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung H.Y., Morita E., von Schwedler U., Muller B., Krausslich H.G., Sundquist W.I. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss E.R., Popova E., Yamanaka H., Kim H.C., Huibregtse J.M., Gottlinger H. Rescue of HIV-1 release by targeting widely divergent NEDD4-type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercenne G., Alam S.L., Arii J., Lalonde M.S., Sundquist W.I. Angiomotin functions in HIV-1 assembly and budding. Elife. 2015;4 doi: 10.7554/eLife.03778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heller B., Adu-Gyamfi E., Smith-Kinnaman W., Babbey C., Vora M., Xue Y., Bittman R., Stahelin R.V., Wells C.D. Amot recognizes a juxtanuclear endocytic recycling compartment via a novel lipid binding domain. J. Biol. Chem. 2010;285:12308–12320. doi: 10.1074/jbc.M109.096230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimura T., Morone N., Suetsugu S. Membrane re-modelling by BAR domain superfamily proteins via molecular and non-molecular factors. Biochem. Soc. Trans. 2018;46:379–389. doi: 10.1042/BST20170322. [DOI] [PubMed] [Google Scholar]
- 22.Pei Z., Bai Y., Schmitt A.P. PIV5 M protein interaction with host protein angiomotin-like 1. Virology. 2010;397:155–166. doi: 10.1016/j.virol.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray G., Schmitt P.T., Schmitt A.P. Angiomotin-like 1 links Paramyxovirus M proteins to NEDD4 family ubiquitin ligases. Viruses. 2019;11 doi: 10.3390/v11020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Z., Dash S., Sagum C.A., Ruthel G., Jaladanki C.K., Berry C.T., Schwoerer M.P., Harty N.M., Freedman B.D., Bedford M.T., Fan H., Sidhu S.S., Sudol M., Shtanko O., Harty R.N. Modular mimicry and engagement of the Hippo pathway by Marburg virus VP40: Implications for filovirus biology and budding. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Z., Ruthel G., Dash S., Berry C.T., Freedman B.D., Harty R.N., Shtanko O. Angiomotin regulates budding and spread of Ebola virus. J. Biol. Chem. 2020;295:8596–8601. doi: 10.1074/jbc.AC120.013171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C., An J., Zhang P., Xu C., Gao K., Wu D., Wang D., Yu H., Liu J.O., Yu L. The Nedd4-like ubiquitin E3 ligases target angiomotin/p130 to ubiquitin-dependent degradation. Biochem. J. 2012;444:279–289. doi: 10.1042/BJ20111983. [DOI] [PubMed] [Google Scholar]
- 27.Ingham R.J., Gish G., Pawson T. The Nedd4 family of E3 ubiquitin ligases: Functional diversity within a common modular architecture. Oncogene. 2004;23:1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- 28.Ilsley J.L., Sudol M., Winder S.J. The WW domain: Linking cell signalling to the membrane cytoskeleton. Cell Signal. 2002;14:183–189. doi: 10.1016/s0898-6568(01)00236-4. [DOI] [PubMed] [Google Scholar]
- 29.Zarrinpar A., Bhattacharyya R.P., Lim W.A. The structure and function of proline recognition domains. Sci. STKE. 2003;2003:RE8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- 30.Zarrinpar A., Lim W.A. Converging on proline: The mechanism of WW domain peptide recognition. Nat. Struct. Biol. 2000;7:611–613. doi: 10.1038/77891. [DOI] [PubMed] [Google Scholar]
- 31.Escobedo A., Gomes T., Aragon E., Martin-Malpartida P., Ruiz L., Macias M.J. Structural basis of the activation and degradation mechanisms of the E3 ubiquitin ligase Nedd4L. Structure. 2014;22:1446–1457. doi: 10.1016/j.str.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Aragon E., Goerner N., Xi Q., Gomes T., Gao S., Massague J., Macias M.J. Structural basis for the versatile interactions of Smad7 with regulator WW domains in TGF-beta Pathways. Structure. 2012;20:1726–1736. doi: 10.1016/j.str.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aragon E., Goerner N., Zaromytidou A.I., Xi Q., Escobedo A., Massague J., Macias M.J. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25:1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Z., Yang Z., Xie R., Ji Z., Guan K., Zhang M. Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity. Elife. 2019;8 doi: 10.7554/eLife.49439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi S., O'Hayre M., Gutkind J.S., Hurley J.H. Structural and biochemical basis for ubiquitin ligase recruitment by arrestin-related domain-containing protein-3 (ARRDC3) J. Biol. Chem. 2014;289:4743–4752. doi: 10.1074/jbc.M113.527473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panwalkar V., Neudecker P., Schmitz M., Lecher J., Schulte M., Medini K., Stoldt M., Brimble M.A., Willbold D., Dingley A.J. The Nedd4-1 WW domain recognizes the PY motif peptide through coupled folding and binding equilibria. Biochemistry. 2016;55:659–674. doi: 10.1021/acs.biochem.5b01028. [DOI] [PubMed] [Google Scholar]
- 37.Kanelis V., Bruce M.C., Skrynnikov N.R., Rotin D., Forman-Kay J.D. Structural determinants for high-affinity binding in a Nedd4 WW3∗ domain-Comm PY motif complex. Structure. 2006;14:543–553. doi: 10.1016/j.str.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 38.Kanelis V., Rotin D., Forman-Kay J.D. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat. Struct. Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 39.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Kabsch W. Integration, scalling, space-group assignment and post refinement. Acta Cryst. 2010;D66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bunkoczi G., Echols N., McCoy A.J., Oeffner R.D., Adams P.D., Read R.J. Phaser.MRage: Automated molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2013;69:2276–2286. doi: 10.1107/S0907444913022750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., Moriarty N.W., Oeffner R.D., Poon B.K., Prisant M.G., Read R.J. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vangone A., Spinelli R., Scarano V., Cavallo L., Oliva R. COCOMAPS: A web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics. 2011;27:2915–2916. doi: 10.1093/bioinformatics/btr484. [DOI] [PubMed] [Google Scholar]
- 45.Alam S.L., Sun J., Payne M., Welch B.D., Blake B.K., Davis D.R., Meyer H.H., Emr S.D., Sundquist W.I. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marion D., Driscoll P.C., Kay L.E., Wingfield P.T., Bax A., Gronenborn A.M., Clore G.M. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear overhauser-multiple quantum coherence spectroscopy: Application to interleukin 1 beta. Biochemistry. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 47.Marion D., Kay L.E., Sparks S.W., Torchia D.A., Bax A. Three-dimensional heteronuclear NMR of nitrogen-15 labeled proteins. J. Am. Chem. Soc. 1989;111:1515–1517. [Google Scholar]
- 48.Zuiderweg E.R., Fesik S.W. Heteronuclear three-dimensional NMR spectroscopy of the inflammatory protein C5a. Biochemistry. 1989;28:2387–2391. doi: 10.1021/bi00432a008. [DOI] [PubMed] [Google Scholar]
- 49.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 50.Goddard T.D., Kneller D.G. University of California; San Francisco, CA: 2001. SPARKY 3. [Google Scholar]
- 51.Gottstein D., Kirchner D.K., Guntert P. Simultaneous single-structure and bundle representation of protein NMR structures in torsion angle space. J. Biomol. NMR. 2012;52:351–364. doi: 10.1007/s10858-012-9615-8. [DOI] [PubMed] [Google Scholar]
- 52.Linge J.P., Williams M.A., Spronk C.A., Bonvin A.M., Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 53.Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 54.Fisher R.D., Chung H.Y., Zhai Q., Robinson H., Sundquist W.I., Hill C.P. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 55.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laskowski R.A., Swindells M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates, structure factor, and NMR restraint and chemical shift files have been deposited in the Protein Data Bank (PDB) with the following codes: apo-WW3 (crystal, 7LP1), PPxY2-WW1 (7LP3), PPxY2-WW2 (7LP2), apo-WW3 (solution, 7LP4), and PPxY1-WW3 (7LP5).