Abstract
Objectives
In the era of increasing antimicrobial resistance, the need for early identification and prompt treatment of multi-drug-resistant infections is crucial for achieving favorable outcomes in critically ill patients. As traditional microbiological susceptibility testing requires at least 24 hours, automated machine learning (AutoML) techniques could be used as clinical decision support tools to predict antimicrobial resistance and select appropriate empirical antibiotic treatment.
Methods
An antimicrobial susceptibility dataset of 11,496 instances from 499 patients admitted to the internal medicine wards of a public hospital in Greece was processed by using Microsoft Azure AutoML to evaluate antibiotic susceptibility predictions using patients’ simple demographic characteristics, as well as previous antibiotic susceptibility testing, without any concomitant clinical data. Furthermore, the balanced dataset was also processed using the same procedure. The datasets contained the attributes of sex, age, sample type, Gram stain, 44 antimicrobial substances, and the antibiotic susceptibility results.
Results
The stack ensemble technique achieved the best results in the original and balanced dataset with an area under the curve-weighted metric of 0.822 and 0.850, respectively.
Conclusions
Implementation of AutoML for antimicrobial susceptibility data can provide clinicians useful information regarding possible antibiotic resistance and aid them in selecting appropriate empirical antibiotic therapy by taking into consideration the local antimicrobial resistance ecosystem.
Keywords: Drug Resistance, Anti-Bacterial Agents, Machine Learning, Artificial Intelligence, Supervised Machine Learning
I. Introduction
As the spread of antibiotic-resistant infections has increased in recent years, a global health crisis has emerged with severe health and economic implications [1]. A study from the UN Interagency Coordinating Group on Antimicrobial Resistance cautioned that if no new major developments are made by 2050, mortality could rise to 10 million deaths globally each year [2]. A critical threat is posed in healthcare facilities from rising drug-resistant infections, as effective treatments are lacking [3].
A recent study [4] presented the resistance levels of multidrug-resistant isolates between the intensive care unit and other hospital wards in a public tertiary hospital in Greece. Feretzakis et al. [5] proposed a methodology that enables clinicians to select the most appropriate antibiotic therapy based on statistically significant sensitivity results from data available in the laboratory information system (LIS). The use of unit-specific local antibiograms within hospitals is highly recommended by the Infectious Disease Society of America (IDSA) as a principal guideline for the prescription of effective empiric treatment [6].
Artificial intelligence (AI) tools are increasingly applied in healthcare, potentially changing many aspects of patient care as well as administrative processes within hospitals. Several researchers suggest that AI can work in key healthcare activities, such as diagnosis and treatment, with equal or even greater accuracy than clinicians. A recent study [7] reported that AI outperformed experts by identifying cancers that radiologists missed in the images while ignoring features they falsely identified as possible tumors. The potential for AI in healthcare has been described in detail [8]. A recent review article [9] presented the opportunities offered by automated machine learning (AutoML) platforms for healthcare.
The current literature shows great interest in implementing machine learning (ML) techniques as clinical decision support tools for the prediction of antimicrobial resistance (AMR) [10–12] and the selection of appropriate empirical antibiotic treatment. Still, ML techniques are not widely implemented in clinical practice in this particular domain since clinicians take into account several patient-specific factors to choose an empirical therapy. Predictive models for antibiotic susceptibility can be an additional tool for decision support regarding early empirical therapy [13].
In this study, we assessed the effectiveness of AutoML-trained models to predict AMR based only on data available in the LIS of the clinical microbiology laboratory, such as the type of sample, the Gram stain, and the antibiotic susceptibility results together with simple patient demographics (age/sex). Age and sex have been reported in research studies [14,15] as factors influencing AMR.
II. Methods
We retrospectively analyzed the antimicrobial susceptibility data of the biopathological laboratory from 499 patients admitted to the internal medicine wards of a public hospital in Greece from January until December 2018. This study was approved by the Institutional Review Board of Sismanogleio General Hospital (No. 6682/2020). The dataset consisted of 11,496 instances and contained the attributes of sex (binary), age (numerical), sample type (categorical), Gram stain (positive or negative; binary), 44 antimicrobial substances (categorical), and the antibiotic susceptibility result (sensitive or resistant; binary). The different types of clinical samples that were taken into consideration for the antibiotic susceptibility analysis, together with simple summary statistics of the dataset, are presented in Table 1.
Table 1.
Summary statistics of the dataset
| Proportion (%) | |
|---|---|
| Agea) (yr) | 78.65 ± 14.94 |
| 82 (19–101) | |
|
| |
| Sex | |
| Male | 44 |
| Female | 56 |
|
| |
| Gram stain | |
| Positive | 20.13 |
| Negative | 79.87 |
|
| |
| Class | |
| Resistant | 33.32 |
| Sensitive | 66.68 |
|
| |
| Type of samples | |
| Blood | 19.05 |
| Tissue | 16.08 |
| Catheters | 2.30 |
| Sputum | 2.41 |
| Tracheobronchial | 9.86 |
| Urine | 50.30 |
Data are expressed as mean ± standard deviation and median (range).
AutoML automates the application of various ML techniques and enables researchers to develop large-scale and effective predictive models. Traditional ML model development is resource-intensive, as it needs significant domain knowledge and time to build and compare the performance of many models. For our experiments, we used the AutoML of the Microsoft Azure ML platform, and the ML algorithms during the automation and tuning process have been fully described [16]. In AutoML experiments, automatic scaling and normalization techniques are applied to all data by default. Since the purpose of our research paper is to present an easy-to-apply procedure that can be communicated to and even used by non-technical experts, we kept all the default parameters and avoided applying custom settings at all stages, including the feature selection process. Our proposed approach is summarized in Figure 1.
Figure 1.
The three-step proposed process. AutoML: automated machine learning, LIS: laboratory information system.
For the same reason, we chose to use a 10-fold cross-validation approach to evaluate the performance of the deduced models instead of using custom settings to split the data into training and testing sets. Cross-validation is an ML technique that uses all available instances for training and testing. It mimics the use of training and test sets by repeatedly training the algorithm K times with a fraction 1/K of training examples left out for testing purposes [17].
Due to the imbalance of our dataset, we also examined the performance of AutoML algorithms by applying an oversampling method since we wanted to keep all the intrinsic value of our dataset and avoided dropping any possible valuable instances by using an undersampling method. Synthetic minority oversampling technique (SMOTE) is a statistical method for uniformly increasing the number of cases in a dataset in order to make it balanced. The new instances are not just duplicates of existing minority instances. Instead, this method takes feature space samples for each target class and its nearest neighbors. After that, new instances are produced that combine features of the target case with those from its neighbors [18].
III. Results
The performance metrics [19] we used in our analysis are briefly presented below:
A receiver operating characteristic (ROC) [20] curve is a plot of the correctly classified classes versus the incorrectly classified classes for a particular model. The area under the curve-weighted (AUCW) is the arithmetic mean of the score for each class, weighted by the number of true instances in each class. The AUCW was set as the target metric for the AutoML procedure.
The average precision score-weighted (APSW) summarizes a precision-recall curve as the weighted mean of precisions achieved at each threshold, with the increase in recall from the previous threshold used as the weight.
The F1 score-weighted (F1W) is the harmonic mean of precision and recall.
Accuracy is the percentage of predicted classes that exactly match the true class.
The results of the four top-performing techniques with 10-fold cross-validation are shown in Table 2.
Table 2.
Four indicative metrics in the four top-performing AutoML models (raw dataset)
| Algorithm name | AUCW | APSW | F1W | ACC |
|---|---|---|---|---|
| StackEnsemble | 0.822 | 0.834 | 0.761 | 0.770 |
| VotingEnsemble | 0.821 | 0.834 | 0.755 | 0.767 |
| MaxAbsScaler, LightGBM | 0.819 | 0.831 | 0.756 | 0.766 |
| SparseNormalizer, XGBoostClassifier | 0.812 | 0.826 | 0.749 | 0.760 |
AutoML: automated machine learning, AUCW: area under the curve-weighted, APSW: average precision score-weighted, F1W: F1 score-weighted, ACC: accuracy.
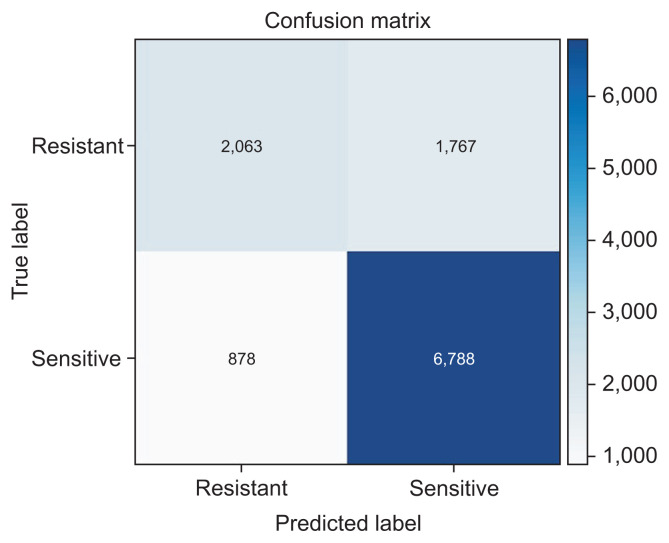
The best overall results were achieved by a stack ensemble technique with an AUCW metric of 0.822, an APSW of 0.834, an F1W of 0.761, and an accuracy of 0.770. The feature importance was 0.660 for the antibiotics, 0.348 for sex, 0.305 for age, 0.294 for the type of sample, and 0.112 for the Gram stain. Feature importance values vary between zero and one, with higher values indicating a stronger association with predictions. The sensitivity (recall) of the best model was 0.539, and the specificity (precision) was 0.896.
Ensemble learning [21,22] is an ML approach where multiple models are trained to solve the same problem. Stacking is an ensemble ML technique that combines multiple classifications via a meta-classifier. The default meta-classifier of the stack ensemble algorithm in Microsoft Azure AutoML for classification tasks is logistic regression. Decision-making based on the feedback of many experts is a common practice that serves as the foundation of human civilization. In recent decades, AI researchers have studied schemes that share a joint decision-making mechanism of this type. These schemes are commonly called ensemble learning and tend to reduce the variance between classifiers and improve the decision-making system’s robustness and accuracy. In practical terms, we also seek advice from various experts when we make a decision that has significant implications. For example, we consult with many physicians before agreeing to undergo a major medical procedure [22].
As shown in Figure 2, we can visualize the confusion matrix for our best-performing model (stack ensemble). A confusion matrix is a n × n matrix used to describe the performance of a classifier, where n is the number of classes. Each row in a confusion matrix represents the instances of the actual class in the dataset, and each column represents the instances of the class that was predicted by the classification model [18].
Figure 2.
Confusion matrix for the stack ensemble technique.
None of the metrics that are presented above can be a one-off solution for the model’s evaluation, but all of them can help healthcare professionals to get a better picture of the overall performance of the classification models through the targeted guidance from data scientists.
It is worth noting at this point that a portion of the same dataset was processed in previous research [23], where five AI algorithms were evaluated. Those results suggested that the multilayer perceptron classifier could serve as a suitable model, taking into account evaluations using both the F-measure (0.721) and the area under the ROC curve (0.746). The corresponding results of the best model (stack ensemble) in this research are an F1 score-weighted (F1W) of 0.761 and an area under the curve-weighted (AUCW) of 0.822. These results are consistent with previous findings, and the results improved because using the AutoML platform enabled us to test and compare several models with different configurations in a time-efficient manner.
The results presented above are based on raw data without considering the balance of the target binary class (i.e., in our case, antimicrobial susceptibility). In the examined dataset, the class attribute (antimicrobial susceptibility) contained 66.68% positive cases (sensitive to a specific antibiotic) and 33.32% negative cases (resistant to a particular antibiotic).
In previous articles [24–26], researchers investigated the task of monitoring and detecting hospital-acquired infections, a topic related to AMR. In these studies, a significant issue was class imbalance, a problem that can be noticed in many classification tasks in the medical domain. This means that, in general, the prevalence of one class in the examined dataset is much higher than that of another one. However, most ML techniques provided better generalization when the number of samples is similar for both classes. In the literature, several methods have been suggested [27] to solve the issue of imbalance. In this study, we utilized an oversampling approach that aims to increase the number of minority samples according to the majority class.
1. Synthetic Minority Oversampling Technique
SMOTE is a common oversampling method that was proposed to improve random oversampling, and its efficacy on high-dimensional data has been investigated in an earlier article [28]. After applying the aforementioned technique, the resulting dataset had 15,326 instances (an increase of about 33.3%), while the class attribute (antimicrobial susceptibility) contained 50.0% positive cases (sensitive to a specific antibiotic) and 50.0% negative cases (resistant to a specific antibiotic). The results of the four top-performing techniques in the transformed dataset after SMOTE processing are shown in Table 3.
Table 3.
Four indicative metrics of the four top-performing AutoML models (balanced dataset - SMOTE)
| Algorithm name | AUCW | APSW | F1W | ACC |
|---|---|---|---|---|
| StackEnsemble | 0.850 | 0.849 | 0.769 | 0.769 |
| VotingEnsemble | 0.850 | 0.849 | 0.768 | 0.768 |
| SparseNormalizer, XGBoostClassifier | 0.842 | 0.841 | 0.762 | 0.762 |
| SparseNormalizer, LightGBM | 0.837 | 0.835 | 0.756 | 0.756 |
AutoML: automated machine learning, SMOTE: Synthetic minority oversampling technique, AUCW: area under the curve-weighted, APSW: average precision score-weighted, F1W: F1 score-weighted, ACC: accuracy.
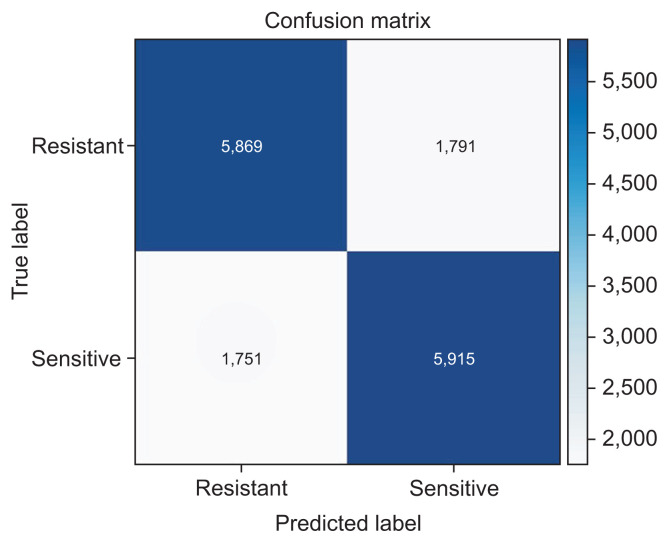
The stack ensemble technique also achieved the best results, with an AUCW metric of 0.850, an APSW of 0.849, an F1W of 0.769, and an accuracy of 0.769. We observed an improvement in all four metrics’ values compared to the best model of the corresponding raw dataset presented above in Table 3. The feature importance was 0.551 for the antibiotics, 0.334 for sex, 0.269 for age, 0.256 for the type of sample, and 0.100 for the Gram stain. The sensitivity (recall) of the best model (stack ensemble) was 0.766, which was greater than the corresponding value of the best model (0.539) that was deduced from the imbalanced dataset in the previous subsection. However, the specificity (precision) of 0.772 was lower than that of the previously presented model (0.896).
The confusion matrix of the best-performing technique is presented in Figure 3.
Figure 3.
Confusion matrix for the stack ensemble technique (balanced dataset).
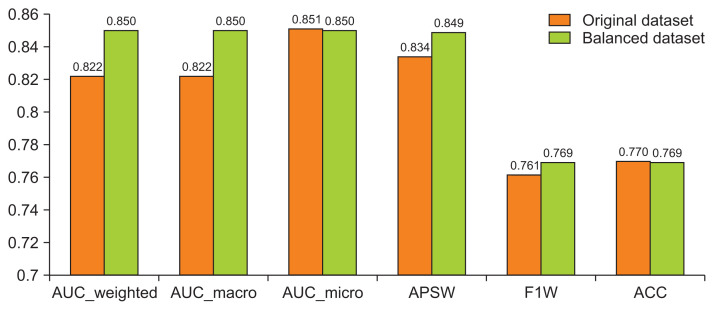
In Figure 4, apart from the aforementioned metrics, we also present the AUC_macro and AUC_micro for the two best performing models (stack ensemble) in both datasets, respectively. The AUC_macro is the arithmetic mean of the AUC for each class, and the AUC_micro is calculated by combining the true positives and false positives from each class.
Figure 4.
Performance metrics of the two stack ensemble models. AUC: area under the curve, APSW: average precision score-weighted, F1W: F1 score-weighted, ACC: accuracy.
IV. Discussion
The decision to start antibiotic therapy is faced in daily practice by clinicians and depends on complex clinical patient data and local epidemiology factors, as well as the information available at the time; the widespread dissemination of AMR further complicates empiric therapy decisions and raises the risk of therapy failure. Timely and appropriate initial therapy is of the utmost importance for sepsis outcomes. Molecular AMR diagnostics significantly reduce the time to achieve results compared to classical phenotypic tests, but aside from the high setup costs, the need for technical infrastructure, and staff training in bioinformatics, their major drawback is that they can detect only known resistance genes or mutations [29].
ML techniques, such as whole-genome sequencing, seem to be useful in AMR surveillance projects; however, the existing evidence does not support their use in guiding clinical decision-making for most bacterial species, according to the European Committee on Antimicrobial Susceptibility Testing [30].
The ML-based methodology proposed in this paper could empower physicians in decision-making while anticipating definitive results from the clinical microbiology laboratory on specific pathogen identification and antibiotic susceptibility testing, even in limited-resource settings. First, early recognition of patients at a high risk of being colonized or infected by strains resistant to one or more antibiotic classes leads to crucial patient and hospital ecosystem knowledge and subsequent improvement in healthcare resources management. More importantly, such systems may serve as a useful clinical decision support tool for physicians in selecting the appropriate empirical therapy. Thus, patient-tailored therapy can limit antibiotic misuse and, over time, reduce the prevalence of antibiotic-resistant bacteria.
Additionally, the proposed methodology is consistent with the practices of patient cohorting (placing patients who have been exposed to or infected with the same pathogen in the same inpatient room) or staff cohorting (assigning specific healthcare providers to care only for patients/residents known to be colonized or infected), which constitute an effective surveillance measure for multidrug-resistant infections that may prevent inadvertent patient-to-patient dissemination.
In this research, the clinical data of patients, such as the source of infection acquisition and the presence of active infection or colonization, were not included, as these were not readily accessible through the central hospital information system. Of course, if the antimicrobial susceptibility datasets also included the patient’s clinical details, the efficiency of the techniques that we present in this study could be considerably improved, even allowing for any final decision to be more explainable. Still, any inclusion of such information must incur the cost of retrieving the relevant data. This may involve a variety of hospital departments, thereby elevating the costs of communication and complicating the need to align protocols across different departments. After paying those costs, one would also need to review the extent to which the additional information gained (e.g., improved accuracy metrics) can be effectively incorporated into the practice of the hospital physicians, who may need to rethink the way they review their decisions in the light of confirming or opposing recommendations from a decision support system. This relates to the actual attitudes of physicians, who may need to consider modifying their practices, and the extent to which such considerations may also be triggered by a change in protocols or the extent to which such recommendations can be made within the prescribed time limits. All in all, we consider this study as a point in the spectrum of cost-effectiveness investigations that ML techniques are bound to trigger in the healthcare domain.
This study evaluated the results of applying AutoML of the Microsoft Azure platform to two internal medicine departments’ antimicrobial susceptibility datasets. In this article, we propose the use of AutoML as a decision tool for physicians since it can be more readily applied even by non-experts (e.g., a data scientist may be needed for a full-blown investigation, but a physician can gain some insight with a relatively smooth learning curve) and, as we showed, the deduced models have good performance.
AutoML platforms can be a beneficial tool for healthcare professionals with limited knowledge of the ML domain that can offer fast and reliable results. Ideally, data scientists’ participation could be important, especially in the stages of data pre-processing, by drawing actionable insights from the data, feature selection, and finally, evaluating the results.
Despite certain limitations of the study, our primary goal was to create an inexpensive ancillary tool to help clinicians rapidly identify patients carrying antibiotic-resistant strains and guide appropriate antibiotic treatment with greater confidence. In future work, dataset enhancement with clinical attributes will probably improve the AutoML algorithms’ performance.
Footnotes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
References
- 1.Gandra S, Barter DM, Laxminarayan R. Economic burden of antibiotic resistance: how much do we really know? Clin Microbiol Infect. 2014;20(10):973–80. doi: 10.1111/1469-0691.12798. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . No time to wait: securing the future from drug-resistant infections [Internet] Geneva, Switzerland: World Health Organization; 2019. [cited at 2021 Jul 27]. Available from: https://www.who.int/publications/i/item/no-time-to-wait-securing-the-future-from-drug-resistant-infections. [Google Scholar]
- 3.Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45(6):568–85. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Feretzakis G, Loupelis E, Sakagianni A, Skarmoutsou N, Michelidou S, Velentza A, et al. A 2-year single-centre audit on antibiotic resistance of Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae strains from an intensive care unit and other wards in a general public hospital in Greece. Antibiotics (Basel) 2019;8(2):62. doi: 10.3390/antibiotics8020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feretzakis G, Loupelis E, Petropoulou S, Christopoulos C, Lada M, Martsoukou M, et al. Using microbiological data analysis to tackle antibiotic resistance of Klebsiella pneumoniae. In: Mantas J, Hasman A, Gallos P, Kolokathi A, Househ MS, Liaskos J, editors. Health informatics vision: from data via information to knowledge. Amsterdam, The Netherlands: IOS Press; 2019. pp. 180–3. [DOI] [PubMed] [Google Scholar]
- 6.Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, et al. Diagnosis and treatment of adults with community-acquired pneumonia: an official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200(7):e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International evaluation of an AI system for breast cancer screening. Nature. 2020;577(7788):89–94. doi: 10.1038/s41586-019-1799-6. [DOI] [PubMed] [Google Scholar]
- 8.Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc J. 2019;6(2):94–8. doi: 10.7861/futurehosp.6-2-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waring J, Lindvall C, Umeton R. Automated machine learning: review of the state-of-the-art and opportunities for healthcare. Artif Intell Med. 2020;104:101822. doi: 10.1016/j.artmed.2020.101822. [DOI] [PubMed] [Google Scholar]
- 10.Feretzakis G, Loupelis E, Sakagianni A, Kalles D, Martsoukou M, Lada M, et al. Using machine learning techniques to aid empirical antibiotic therapy decisions in the intensive care unit of a general hospital in Greece. Antibiotics (Basel) 2020;9(2):50. doi: 10.3390/antibiotics9020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Aguero S, Mora-Jimenez I, Lerida-Garcia J, Alvarez-Rodriguez J, Soguero-Ruiz C. Machine learning techniques to identify antimicrobial resistance in the intensive care unit. Entropy (Basel) 2019;21(6):603. doi: 10.3390/e21060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oonsivilai M, Mo Y, Luangasanatip N, Lubell Y, Miliya T, Tan P, et al. Using machine learning to guide targeted and locally-tailored empiric antibiotic prescribing in a children’s hospital in Cambodia. Wellcome Open Res. 2018;3:131. doi: 10.12688/wellcomeopenres.14847.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacFadden DR, Coburn B, Shah N, Robicsek A, Savage R, Elligsen M, et al. Decision-support models for empiric antibiotic selection in Gram-negative bloodstream infections. Clin Microbiol Infect. 2019;25(1):108.e1–108.e7. doi: 10.1016/j.cmi.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 14.Kolozsvari LR, Konya J, Paget J, Schellevis FG, Sandor J, Szollosi GJ, et al. Patient-related factors, antibiotic prescribing and antimicrobial resistance of the commensal Staphylococcus aureus and Streptococcus pneumoniae in a healthy population: Hungarian results of the APRES study. BMC Infect Dis. 2019;19(1):253. doi: 10.1186/s12879-019-3889-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-Ami R, Rodriguez-Bano J, Arslan H, Pitout JD, Quentin C, Calbo ES, et al. A multinational survey of risk factors for infection with extended-spectrum beta-lactamase-producing enterobacteriaceae in nonhospitalized patients. Clin Infect Dis. 2009;49(5):682–90. doi: 10.1086/604713. [DOI] [PubMed] [Google Scholar]
- 16.Set up AutoML training with Python [Internet] Redmond (WA): Microsoft; 2021. [cited at 2021 Jul 28]. Available from: https://docs.microsoft.com/en-us/azure/machine-learning/how-to-configure-auto-train. [Google Scholar]
- 17.Bengio Y, Grandvalet Y. No unbiased estimator of the variance of k-fold cross-validation. J Mach Learn Res. 2004;5:1089–105. [Google Scholar]
- 18.SMOTE [Internet] Redmond (WA): Microsoft; 2019. [cited at 2021 Jul 28]. Available from: https://docs.microsoft.com/en-us/azure/machine-learning/algorithm-module-reference/smote. [Google Scholar]
- 19.Evaluate automated machine learning experiment results [Internet] Redmond (WA): Microsoft; 2020. [cited at 2021 Jul 28]. Available from: https://docs.microsoft.com/en-us/azure/machine-learning/how-to-understand-automated-ml. [Google Scholar]
- 20.Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27(8):861–74. [Google Scholar]
- 21.Sewell M. Ensemble learning. London, UK: University College London; 2011. [Google Scholar]
- 22.Zhang C, Ma Y. Ensemble machine learning: methods and applications. New York (NY): Springer Science & Business Media; 2012. [Google Scholar]
- 23.Feretzakis G, Loupelis E, Sakagianni A, Kalles D, Lada M, Christopoulos C, et al. Using machine learning algorithms to predict antimicrobial resistance and assist empirical treatment. Stud Health Technol Inform. 2020;272:75–8. doi: 10.3233/SHTI200497. [DOI] [PubMed] [Google Scholar]
- 24.Cohen G, Hilario M, Sax H, Hugonnet S. Asymmetrical margin approach to surveillance of nosocomial infections using support vector classification. Proceedings of the Intelligent Data Analysis in Medicine and Pharmacology; 2003 Oct 19ă22; Protaras, Cyprus. [Google Scholar]
- 25.Cohen G, Hilario M, Sax H, Hugonnet S, Pellegrini C, Geissbuhler A. An application of one-class support vector machine to nosocomial infection detection. Stud Health Technol Inform. 2004;107(Pt 1):716–20. [PubMed] [Google Scholar]
- 26.Cohen G, Hilario M, Sax H, Hugonnet S, Geissbuhler A. Learning from imbalanced data in surveillance of nosocomial infection. Artif Intell Med. 2006;37(1):7–18. doi: 10.1016/j.artmed.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 27.He H, Garcia EA. Learning from imbalanced data. IEEE Trans Knowl Data Eng. 2009;21(9):1263–84. [Google Scholar]
- 28.Blagus R, Lusa L. SMOTE for high-dimensional class-imbalanced data. BMC Bioinformatics. 2013;14:106. doi: 10.1186/1471-2105-14-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization . Molecular methods for antimicrobial resistance (AMR) diagnostics to enhance the Global Antimicrobial Resistance Surveillance System. Geneva, Switzerland: World Health Organization; 2019. [Google Scholar]
- 30.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect. 2017;23(1):2–22. doi: 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]