Abstract
Cancer immunotherapy has achieved positive clinical outcomes and is revolutionizing cancer treatment. However, cancer immunotherapy has thus far failed to improve outcomes for most “cold tumors”, which are characterized by low infiltration of immune cells and immunosuppressive tumor microenvironment. Enhancing the responsiveness of cold tumors to cancer immunotherapy by stimulating the components of the tumor microenvironment is a strategy pursued in the last decade. Currently, most of the agents used to modify the tumor microenvironment are small molecules or antibodies. Small molecules exhibit low affinity and specificity towards the target and antibodies have shortcomings such as poor tissue penetration and high production cost. Peptides may overcome these drawbacks and therefore are promising materials for immunomodulating agents. Here we systematically summarize the currently developed immunoactivating peptides and discuss the potential of peptide therapeutics in cancer immunology.
Keywords: Immunomodulation, Immuno-oncology, Immunotherapy, Tumor microenvironment, Therapeutic peptides
1. Introduction
Cancer immunotherapy has revolutionized the treatment of many different cancers and is now firmly established as a pillar of cancer treatment. Immune checkpoint inhibitors have been especially successful and have demonstrated remarkable responses in advanced-stage tumors that have been difficult to treat with conventional treatments such as cytotoxic chemotherapy [1]. However, based on current approvals, less than half of all patients with cancer are eligible to receive immune checkpoint inhibitor therapy, and it has been estimated that less than 15% of these patients respond to the treatment [2]. Even in tumor types for which immune checkpoint inhibitors are highly effective, complete responses are rare. Immune checkpoint inhibitors are particularly effective in tumors with high tumor mutational burdens and/or immune-inflamed tumors; by contrast, these agents have broadly failed to improve clinical outcomes for immune excluded-tumor or immune-desert tumors, such as colon cancer and pancreatic cancer [3]. The development of novel immunotherapies and therapy combinations that can extend the benefit of immunotherapy to these immune-resistant tumor types remains an important goal.
Paucity of tumor antigens, defects in the antigen-presenting process, impaired trafficking of immune cells to the tumor, and production of immunosuppressive cytokines in the tumor microenvironment (TME) are some of the proposed mechanisms leading to a cold tumor [4]. Various therapeutic strategies have been developed to overcome these immunosuppressive events. Induction of immunogenic cell death by chemotherapy, radiotherapy or the usage of oncolytic viruses, upregulation of molecules involved in the antigen presentation process by inhibitors of methyltransferases and histone deacetylases, administration of pro-inflammatory cytokines or neutralizing antibodies for immunosuppressive cytokines, and direct stimulation of the immune system by agonists of stimulator of interferon genes (STING) and pattern recognition receptors (PRRs) are promising approaches to turn cold tumors into hot tumors. Several clinical trials evaluating the efficacy of these therapeutics in combination with immune checkpoint inhibitors are being conducted [4-8].
Currently, all of the approved cancer immunotherapies and the vast majority of cancer immunotherapies in clinical development are small molecules or antibody therapies. Small molecules generally show low affinity and specificity against the targets and often cause side effects. Although therapeutic antibodies bind with their targets with high affinity and specificity, they too have several limitations. The most prominent disadvantage is poor tissue penetration. Since the interstitial hydrostatic pressure is high in tumors, convection of fluid from the microvasculature to the tissue is restricted in tumors. Therefore, extravasation mainly depends on the diffusion through the paracellular pores in the microvessels. This is problematic for antibodies since the diffusion rate of antibodies is low due to their large sizes. Antibodies undergo a large sieving effect while moving through the paracellular pores which further limits extravasation [9,10]. The distribution of antibodies within the tumor is heterogeneous, where antibodies are mainly found around the microvessels and not in avascular and hypoxic region [11]. The poor biodistribution of antibodies might partially explain the low overall response rate of immune checkpoint inhibitors in solid tumors.
The high cost of manufacturing antibodies is also a major drawback of antibody drugs. Since antibodies are large multimeric proteins with numerous post-translational modifications, massive culturing of mammalian cells and intricate purification are required, which makes the cost of manufacturing much higher than that of small molecular drugs [12]. Lastly, the long half-lives of antibodies might contribute to immune-related adverse events (irAEs). IrAEs are caused by the intervention of cancer immunotherapy with the system of immune tolerance which leads to excessive immunity against normal organs [13,14]. Agents with shorter half-lives are expected to be safer due to their ease of managing these side effects [15].
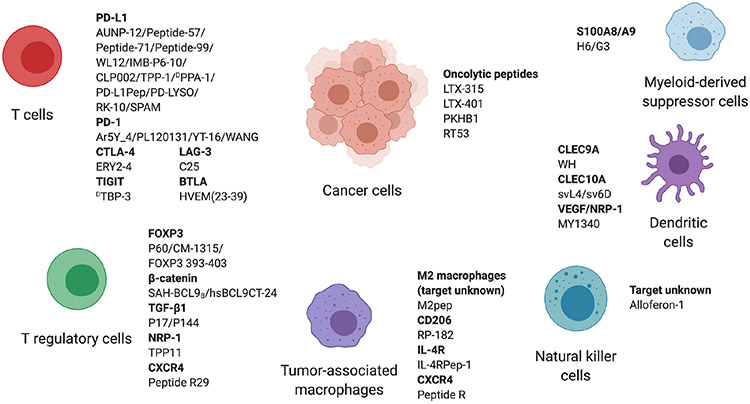
Therapeutic peptides have several advantages that can overcome the shortcomings of small molecular drugs and antibodies. Similar to antibodies, peptides show high affinity and specificity towards the target and are suitable for intervening with protein-protein interactions. Since the molecular size is smaller than antibodies, the penetration to tissues is generally better than antibodies [16]. Furthermore, some peptides can penetrate the cellular membrane, which is a feature that antibodies cannot achieve [17]. Unlike antibodies, peptides can be synthesized by chemical reactions such as solid-phase synthesis, which makes the production cost significantly lower than antibodies [18]. Peptides show few side effects and low toxicity because of their high specificity to the target, short half-life, and low immunogenicity. Another important characteristic is that accumulation in specific organs, which is the cause of severe organ failure in chemotherapies, is not seen in peptide treatment [16]. Due to all these advantages, intensive research has been conducted to develop peptides that modulate the TME and activate antitumoral immunity. We systematically review peptides that target different cells and receptors in the TME as illustrated in Fig. 1 and discuss the potential of peptides in the next generation of cancer immunotherapy. The usage of peptides as cancer vaccines, which is extensively reviewed elsewhere [19-21], is not discussed in this review.
Fig. 1.
Peptides targeting the tumor microenvironment.
2. Peptides targeting immune checkpoints
Immune checkpoints negatively regulate the immune system to maintain self-tolerance and prevent autoimmunity. Tumor cells often exploit this system by upregulating the expression of immune checkpoint molecules in the TME. Activation of immune checkpoint molecules leads to the suppression of antitumoral immunity, especially that of the adaptive T cell immunity. Therefore, dampening the immunosuppressive signaling pathway by blockade of the crucial ligand-receptor reactions is expected to restore antitumoral immunity and is considered as the central strategy in cancer immunology [22]. Inhibitors for cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), programmed cell death protein 1 (PD-1), and programmed death-ligand 1 (PD-L1) have been most successful and are currently used in the clinic. Although all the US Food and Drug Administration (FDA) approved drugs targeting these molecules are currently antibodies [23], intensive research has also been conducted to develop peptide inhibitors (Fig 2, Table 1).
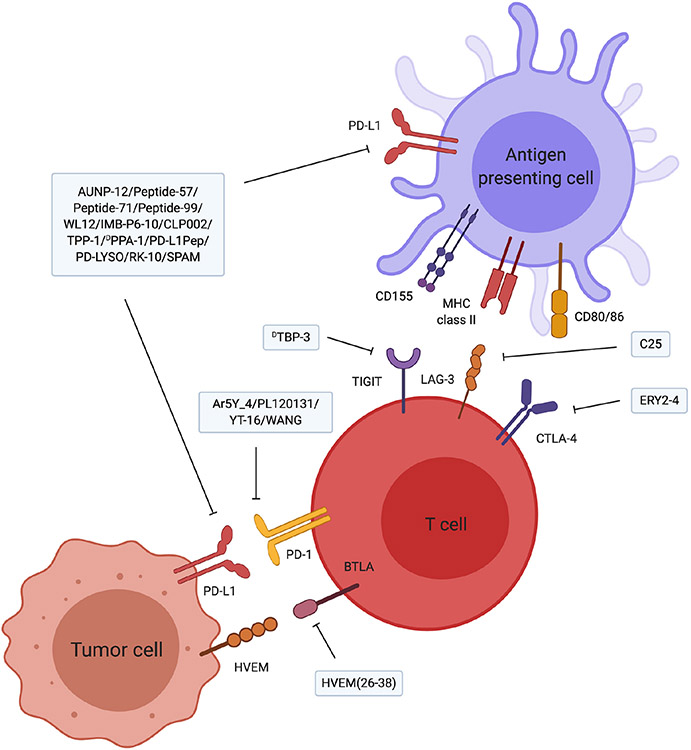
Fig. 2.
Peptides targeting immune checkpoint molecules. The majority of the peptides are inhibitors for the PD-1/PD-L1 pathway. Recently, peptides inhibiting newly identified immune checkpoints LAG-3 and TIGIT have been developed. Further development of peptides targeting immune checkpoints other than PD-1/PD-L1 is expected to improve the response rate of immune checkpoint inhibitors.
Table 1.
Peptides targeting immune checkpoint molecules.
| Mechanism of action |
Peptide name |
Sequence | Discovery method | Cancer model | Administration method |
Modifications for improving stability |
Ref |
|---|---|---|---|---|---|---|---|
| PD-L1 inhibitor | AUNP-12 | SNTSESFK(SNTSESF)FRVTQLAPKAQIKE | Crystal structure analysis | B16F10 melanoma, Renca renal cell carcinoma, CT26 colon cancer | Subcutaneous | [15,24] | |
| PD-L1 inhibitor | Peptide-57 | Cyclic[F-NMeA-NPHLSWSW-NMeNle-NMeNle-R-Scc]G | N/A | N/A | Cyclic structure, incorporation of unnatural amino acids | [25-27] | |
| PD-L1 inhibitor | Peptide-71 | Cyclic[F-NMeF-NMeNle-Sar-DV-NMeF-Y-Sar-WYL-Scc]G | N/A | N/A | Cyclic structure, incorporation of unnatural amino acids | [25-27] | |
| PD-L1 inhibitor | Peptide-99 | Cyclic[FLIVIRDRVFR-Scc]G | N/A | N/A | Cyclic structure, incorporation of unnatural amino acids | [25-27] | |
| PD-L1 inhibitor | IMB-P6-10 | LTCSLAPNIISAL | Derivative of PRDX5 | C16 colon cancer, CT26 colon cancer | Intraperitoneal | [34] | |
| PD-L1 inhibitor | CLP002 | WHRSYYTWNLNT | Screening by phage display | CT26 colon cancer | Intraperitoneal | [35] | |
| PD-L1 inhibitor | TPP-1 | SGQYASYHCWCWRDPGRSGGSK | Screening by bacterial surface display | H460-luc non-small cell lung cancer | Subcutaneous | [36,37] | |
| PD-L1 inhibitor | DPPA-1 | NYSKPTDRQYHF ※ All amino acids are D-enantiomer |
Screening by mirror image phage display | CT26 colon cancer | Subcutaneous, intraperitoneal | (D)-enantiomeric structure | [38] |
| PD-L1 inhibitor | PD-L1Pep-1 | CLQKTPKQC | Screening by phage display | CT26 colon cancer | Intravenous | [44] | |
| PD-L1 inhibitor | PD-L1Pep-2 | CVRARTR | Screening by phage display | CT26 colon cancer | Intravenous | [44] | |
| Binds to PD-L1 | WL12 | Cyclic[AcY-NMeA-NPHL-Hyp-WS-MeW-NMeNle-NMeNle-Orn-C]G | N/A | N/A | Cyclic structure, incorporation of unnatural amino acids | [28] | |
| Binds to PD-L1 | RK-10 | GSGSGSTYLCGAISLAPKAQIKESL | Crystal structure analysis | N/A | [50] | ||
| Binds to PD-L1 | SPAM | MPIFLDHILNKFWILHYA | Screening by mRNA display | N/A | [51] | ||
| PD-L1 degradation by lysosome | PD-LYSO | DKEMAATSAAIEDAVRRIEDMMNQYPYDVPDYAMDFSGLSLIKLKKQOH | Derivative of HIP1R | In vitro only | [45] | ||
| PD-1 inhibitor | Ar5Y_4 | GNWDYNSQRAQLYNQ | Crystal structure analysis | In vitro only | [29] | ||
| PD-1 inhibitor | PL120131 | Ac-GADYKRITVKVN | PD-L1 derivative | In vitro and 3D culture model only | [30] | ||
| PD-1 inhibitor | YT-16 | Cyclic[YRCMISYGGADYKCIT] | Virtual screening | In vitro only | Cyclic structure | [31] | |
| Binds to PD-1 | WANG | WANG-003: KRWWR WANG-004: FRWWR WANG-005: RRWQWR | Virtual screening | N/A | [49] | ||
| CTLA-4 inhibitor | ERY2-4 | CAWGQAILEGELAWLEGGGGGAGQLADLKRQLAWWKQAC | Screening by yeast surface display | In vitro only | [46] | ||
| TIGIT inhibitor | DTBP-3 | GGYTFHWHRLNP ※ All amino acids are D-enantiomer |
Screening by mirror image phage display | CT26 colon cancer, B16-OVA melanoma, 4 T1 breast cancer | Subcutaneous, intraperitoneal | (D)-enantiomeric structure | [42] |
| LAG-3 inhibitor | C25 | Cyclic[CVPMTYRAC] | Screening by phage display | CT26 colon cancer, B16 melanoma | Subcutaneous | Cyclic structure | [47] |
| Binds to BTLA | HVEM (23–39) | Ac-YRVKEACGELTGTVCEP | Derivative of HVEM | N/A | [52] |
NMeA: N-methylalanine, NMeNle: N-methylnorleucine, Scc: S-Carbamoylcysteine, NMeF: N-methylphenylalanine, Sar: Sarcosine, AcY: Acetyltyrosine, Hyp: Hydroxyproline, MeW: Methyltryptophan, Orn: Ornithine.
PD-1/PD-L1 signaling pathway has been the main target and the first peptide inhibitor was developed in 2014 [24]. The investigators identified the critical structure of PD-1 in the PD-1/PD-L1 interface based on the crystal structure of the PD-1/PD-L1 complex and combined several PD-1 derived loops and strands to develop a 29-amino acid branched peptide, AUNP-12 (NP-12) [15]. AUNP-12 inhibited the binding of PD-1/PD-L1 and rescued the proliferation capacity of lymphocytes in the presence of PD-L1. The immunomodulatory activity was confirmed in vivo and AUNP-12 showed significant antitumoral and anti-metastatic effects.
Miller et al. developed a library of macrocyclic peptides containing unnatural amino acids that bind to PD-L1 [25,26]. Among the peptides, the efficacy of peptide-57, peptide-71, and peptide-99 was investigated by Magiera-Mularz et al. [27]. Peptide-57 and peptide-71 had EC50 values of 566 nM and 293 nM respectively, whereas that of peptide-99 was 6.30 μM indicating a relatively lower affinity. The region on PD-L1 that the peptides bound overlapped with the PD-1 binding region. Chatterjee et al. applied WL12, another peptide in the library, to the imaging and detection of PD-L1 in tumors. They confirmed that WL12 binds specifically to hPD-L1 in vitro and in vivo, and developed imaging agents for positron emission tomography (PET) targeting PD-L1 [28]. So far, no study has demonstrated the immunomodulating activity of these macrocyclic peptides in vivo. Further research would be needed to fully reveal the potential of these peptides.
Several groups have taken in silico strategies to design inhibitory peptides for the PD-1/PD-L1 signaling pathway. By analyzing the crystal structure of the PD-1/PD-L1 complex, Li et al. identified 5 key anchor residues on PD-L1 which are critical for the binding with PD-1 [29]. Next, they selected pairs of peptides from a scaffold fragment library which can constitute the structure formed by the key anchor residues. The backbone of the peptide was constructed by connecting the pairs of peptides. In this way, they designed a peptide named Ar5Y_4 that mimics the structure of the PD-1 binding region of PD-L1. PL120131 is another peptide identified by structural analysis of the PD-1/PD-L1 complex [30]. PL120131 contains the sequence of hPD-L1 from glycine at position 120 to asparagine at 131, which was identified as the interaction interface of the PD-1/PD-L1 complex. Although the authors have not presented in vivo data demonstrating the immunomodulating effect of these peptides, both Ar5Y_4 and PL120131 were able to antagonize the suppressive effect of PD-1/PD-L1 signaling on T cells in vitro.
YT-16 is a PD-1 targeting cyclic peptide identified by virtual screening [31]. Abbas et al. determined the key residues on PD-1 that play a role in PD-1/PD-L1 interaction and designed a virtual peptide library based on the corresponding PD-L1 fragments interacting with these key residues. The affinity between the virtual peptides and PD-1 was predicted by simulating the docking of the virtual peptides and PD-1. YT-16 had the highest predicted affinity to PD-1 and the binding of YT-16 and PD-1 was confirmed experimentally. YT-16 enhanced the activity of T cells co-cultured with tumor cells in a dose-dependent manner.
IMB-P6-10 is a peptide derived from peroxiredoxin-5 (PRDX5). The main role of PRDX5 is to function as an antioxidant enzyme and prevent the accumulation of peroxide in cells [32]. Liu et al. found that PRDX5 exhibited antitumoral activity and predicted that hPRDX5 binds to hPD-L1 based on the result of a binding simulation [33]. Zou et al. experimentally demonstrated that PRDX5 inhibits the binding of PD-1/PD-L1, and proceeded to develop a peptide derived from PRDX5 [34]. They identified the α-helical structures in PRDX5 and determined the sequence that can interrupt the binding of PD-1 and PD-L1. Further modification led to the generation of IMB-P6-10, which restored the activity of T cells co-cultured with tumor cells and suppressed tumor growth in CT26 colon carcinoma model.
Apart from in silico strategies, biopanning approaches are also employed to discover inhibitory peptides of the PD-1/PD-L1 signaling. Liu et al. conducted a phage display screening to identify peptides that bound to the extracellular domain of PD-L1 [35]. Among the peptides they identified, CLP002 most efficiently blocked the interaction between PD-1 and PD-L1. TPP-1 is a PD-L1 binding peptide identified by a screening using bacterial surface display [36]. After running a screening using a random bacterial surface display library, they identified a consensus sequence CWCWR, which was enriched in the peptides that bound to PD-L1. To improve the affinity of the peptide, they further generated a focused library that consists of peptides containing the consensus sequence and additional random amino acids. As a result of the secondary screening, TPP-1 was identified as a peptide that binds to PD-L1 with high affinity. The immunoactivating and antitumoral effects of CLP002 and TPP-1 were demonstrated both in vitro and in vivo. Kuan et al. applied TPP-1 to PET imaging of PD-L1 and demonstrated that TPP-1 is also suitable for PD-L1 imaging [37].
Chang et al. conducted a screening using mirror-image phage display and developed PD-L1 binding (D)-enantiomeric peptides DPPA-1 and DPPA-2 [38]. Mirror-image phage display is a methodology developed by Schumacher et al. to identify (D)-peptide ligands [39]. The process of mirror-image phage display is as follows: first chemically synthesize a (D)-enantiomeric target protein. Next, conduct a phage display screening using a (L)-peptide library and identify (L)-peptides that bind to the target. Lastly, synthesize the (D)-enantiomeric form of the identified (L)-peptides. By symmetry, the generated (D)-peptides should bind to the natural (L)-enantiomeric target protein. Since living organisms almost exclusively consist of (L)-proteins, (D)-peptides are generally not recognized by the proteases and are more stable in the serum than their (L)-counterparts [40]. As expected, no degradation of DPPA-1 and DPPA-2 was observed after 24 h of incubation in 10% human serum. Although the authors did not assess the immunomodulatory activity of these peptides in detail, they demonstrated that DPPA-1 treatment suppressed tumor growth and improved survival in vivo. Sun et al. developed a paclitaxel containing nanoparticle decorated with DPPA-1 and CGKRK peptide, which is a peptide that binds to tumor neovascular endothelial cells [41]. This PD-1/PD-L1 pathway and angiogenesis dual targeting nanoparticle delivered paclitaxel to both PD-L1 expressing cells and endothelial cells and improved the survival of C6 glioma bearing mice.
Zhou et al. further applied the mirror-image phage display based screening technique to develop DTBP-3, which is a (D)-peptide inhibitor targeting T cell immunoglobulin and ITIM domain (TIGIT) [42]. TIGIT is a relatively recently discovered checkpoint molecule first reported in 2009. TIGIT is expressed on T cells and NK cells and inhibits the activity of these cells upon binding with its ligand [43]. DTBP-3 enhanced the activity and infiltration of immune cells in CT26 colon carcinoma model and demonstrated antitumoral and anti-metastatic effects.
The peptides mentioned so far have been selected by their affinity to purified target proteins. To identify peptides that bind to the natural conformation of PD-L1, Gurung et al. conducted a phage display screening to identify peptides that bound to PD-L1 expressed on the surface of live cells [44]. By negatively selecting the peptides that bound to non-transfected cells and positively selecting those that bound to PD-L1 overexpressing cells, they developed PD-L1Pep-1 and PD-L1Pep-2. Both peptides enhanced the activity of T cells in vitro and exerted antitumoral effect in vivo.
Wang et al. adopted a unique strategy utilizing the lysosomal protein degradation pathway to inhibit the PD-1/PD-L1 signaling pathway [45]. They analyzed the interactome of PD-L1 and identified Huntingtin-interactive protein 1-related protein (HIP1R) as a novel negative regulator of PD-L1. HIP1R contains a conserved domain that binds to PD-L1 and also a lysosomal sorting signal which targets the protein to the lysosome for proteolysis. PD-LYSO is a fusion peptide consisting of the PD-L1 binding domain and the lysosomal sorting signal of HIP1R. The transfection of PD-LYSO significantly induced the degradation of PD-L1 in tumor cells. No experimental data has demonstrated the efficacy of PD-LYSO treated from the outside of the cells and therefore the development of a formulation that could be administered as a drug would be necessary.
While the PD-1/PD-L1 pathway has been the main focus, peptides targeting other immune checkpoints have also been developed. Mudiyanselage et al. developed a yeast surface display library of helixloop-helix peptides and identified ERY2-4 as a CTLA-4 inhibitor by a screening using the library [46]. Zhai et al. identified C25, an antagonistic peptide of lymphocyte-activation gene 3 (LAG-3), by phage display screening [47]. LAG-3 has a similar structure to CD4 and binds to the MHC class II molecule. Upon binding with the ligand, LAG-3 transduces inhibitory signals in T cells [48]. C25 inhibited the activity of LAG-3 and showed immunoactivating and antitumoral effects both in vitro and in vivo.
PD-1 binding peptides WANG-003/WANG-004/WANG-005 [49], PD-L1 binding peptides RK-10 [50] and SPAM [51], and Herpesvirus entry mediator (HVEM) binding peptide HVEM(23–39) [52] are other peptides identified to bind immune checkpoint molecules. Although the binding capacity of these peptides has been demonstrated, no experimental data have shown immunoactivating capacities of these peptides.
3. Peptides targeting regulatory T cells
Regulatory T cells (Tregs) play a major role in forming an immunosuppressive environment in the tumor. In general, Tregs suppress immune reactions by several mechanisms: the release of inhibitory cytokines such as IL-10, IL-35, and TGF-β, granzyme dependent cytolysis of immune cells, disruption of effector cell metabolism by consuming scarce amino acids, generation of adenosine nucleosides which inhibits T cell activity, consumption of the survival-promoting cytokine IL-2, suppression of effector T cells via expression of immune checkpoint molecules, and inhibition of the maturation and the function of DCs [53-55]. Numerous clinical studies have shown that high frequency of tumor-infiltrating Tregs correlates with poor prognosis [56]. Tregs are critical targets for cancer immunotherapy and several peptides have been developed to downregulate the activity of Tregs (Fig. 3, Table 2).
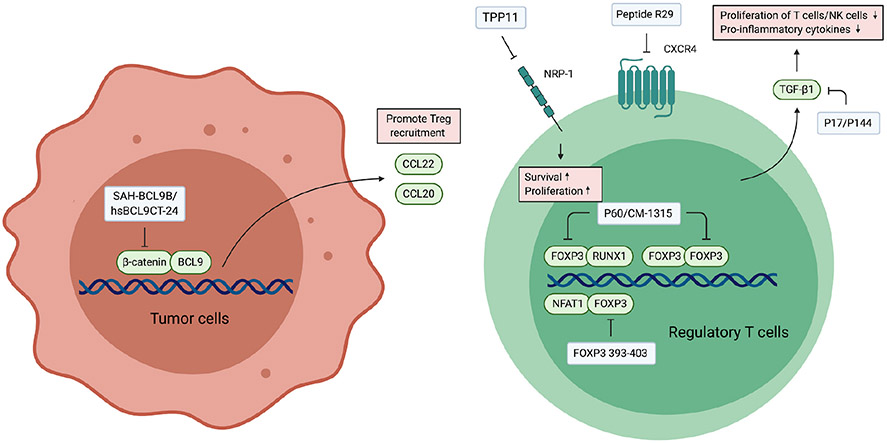
Fig. 3.
Peptides targeting Tregs. FOXP3 is a biomarker of Tregs which forms a complex with numerous transcription factors and chromatin modifying factors to regulate the expression of genes related to Treg differentiation and the maintenance of its immunosuppressive phenotype. Peptides targeting FOXP3 activate transcription factors such as NFAT1 and RUNX1 by releasing them from the inhibition by FOXP3. The activation of these transcription factors leads to the suppression of Treg activity. NRP-1 and CXCR4 are other targets highly expressed in intratumoral Tregs. TGF-β is a major immunosuppressive cytokine secreted by Tregs, and therefore inhibitors for TGF-β suppress the function of Tregs. Inhibition of tumor intrinsic β-catenin suppresses the secretion of Treg recruiting chemokines, which consequently reduce the infiltration of Tregs in the tumor.
Table 2.
Peptides targeting lymphocytes.
| Mechanism of action |
Peptide name |
Sequence | Discovery method |
Cancer model | Administration method |
Modifications for improving stability |
Ref |
|---|---|---|---|---|---|---|---|
| Peptides targeting Tregs | |||||||
| FOXP3 inhibitor | P60 | RDFQSFRKMWPFFAM | Screening by phage display | CT26 colon cancer | Intraperitoneal | [58] | |
| FOXP3 inhibitor | CM-1315 | Cyclic[RDAFQAFRKMWPFFAM] | Derivative of P60 | CT26 colon cancer | Intraperitoneal | [59] | |
| FOXP3 inhibitor | FOXP3 393–403 | KCFVRVESEKG | Derivative of FOXP3 | Hepa-129 hepatocellular carcinoma, TC-1 P3 (A15) lung cancer | Intratumoral | [61] | |
| β-catenin inhibitor | SAH-BCL9B | LSQEQLEHRERSLXTLRXIQRBLF ※ Amino acids at position X are stapled |
Crystal structure analysis | COLO320 colorectal adenocarcinoma, INA-6 multiple myeloma | Intraperitoneal | Peptide stapling | [73,74] |
| β-Catenin inhibitor | hsBCL9CT-24 | Ac-LQTLRXIQRXL-2-Nal ※ Amino acids at position X are stapled |
Derivative of SAH-BCL9B | COLO320 colorectal adenocarcinoma, CT26 colon cancer, LLC1 lung carcinoma, 4 T1 breast cancer | Intraperitoneal | Peptide stapling | [74] |
| TGF-β1 inhibitor | P17 | KRIWFIPRSSWYERA | Screening by phage display | E.G7-OVA T cell lymphoma, B16-OVA melanoma | Intraperitoneal | [76,79,80] | |
| TGF-β1 inhibitor | P144 | TSLDASIIWAMMQN | Derivative of TGFβR3 | E.G7-OVA T cell lymphoma, B16-OVA melanoma | Intratumoral | [75,80] | |
| NRP-1 inhibitor | Fc-TPP-11 | HTPGNSKPTRTPRR | Screening by yeast surface display | CT26 colon cancer, B16F10 melanoma | Intraperitoneal | Binding with the Fc region of antibodies | [82,83] |
| CXCR4 inhibitor | Peptide R29 | Ac-RA-cyclic[DCRFFC] | Derivative of peptide R | In vitro only | Cyclic structure, (D)-enantiomeric structure | [84,85] | |
| Peptides targeting natural killer cells | |||||||
| Induction of CD244 on NK cells | Alloferon-1 | HGVSGHGQHGVHG | Extraction from infected blow fly | P388 lymphoma, HCT116 colon cancer | Intraperitoneal | [87-89] | |
Forkhead box protein P3 (FOXP3) is a transcription factor that is constitutively expressed in Tregs and is involved in the development and maintenance of Tregs. FOXP3 forms a large complex with transcription factors and chromatin remodeling factors to modify the gene expression pattern [55]. Via proteomic analysis of the FOXP3 complex, Rudra et al. identified 361 proteins interacting with FOXP3 which suggests the heterogeneity of FOXP3 complex and its complicated mechanism of regulating gene expression [57]. Due to its role as a master regulator, FOXP3 has been the main target of therapeutics targeting Tregs.
Using phage display screening, Casares et al. developed a cell-penetrating FOXP3 binding peptide P60 [58]. P60 prevented the nuclear translocation of FOXP3 which resulted in enhanced activity of the two transcription factors essential for T cell activation, nuclear factor-κB (NFκB) and nuclear factor of activated T cells (NFAT). P60 restored the proliferation capacity of T cells cocultured with Tregs and increased the expression level of proinflammatory cytokines. Although monotherapy of P60 did not show a significant antitumoral activity, mice treated with P60 responded better to cancer vaccine. A follow-up study by Lozano et al. revealed that P60 suppresses Treg activity by inhibiting the homodimerization of FOXP3, which is required for its transcriptional regulating function, as well as its binding with Runt-related transcription factor 1 (Runx1), also known as acute myeloid leukemia 1 (AML1) [59]. Runx1 is a transcription factor that upregulates IL-2 and IFN-γ and its function is inhibited by FOXP3 [60]. P60 binds to the Runx1 interacting domain of FOXP3 and therefore competitively inhibits FOXP3/Runx1 interaction leading to the activation of Runx1. The authors further identified key residues on P60 for binding with FOXP3 and modified the sequence to augment the binding capacity of P60. CM-1315 is the final product, which possesses enhanced FOXP3 binding capacity and higher metabolic stability.
FOXP3 393-403 is another FOXP3 binding peptide [61]. FOXP3 393-403 is derived from the forkhead (FKH) domain of FOXP3 which is responsible for DNA binding. Crystal structure analysis revealed that the FKH domain is also involved in the interaction with NFAT1 [62]. In activated T cells, NFAT1 forms a complex with activator protein 1 (AP-1) and induces IL-2 production by binding to the promoter region of IL-2. In contrast, the NFAT1/FOXP3 complex represses the expression of IL-2 and promotes the expression of CD25 and CTLA4 [62]. FOXP3 393-403 disturbed the interaction between FOXP3 and NFAT1 which lead to the enhanced expression of proinflammatory cytokines and down-regulation of immunosuppressive molecules. They further demonstrated that FOXP3 393–403 inhibited the TGF-β dependent conversion from CD4+CD25− T cells to Tregs in vitro. In vivo experiments showed that FOXP3 393–403 treatment improved the efficacy of cancer vaccine and significantly suppressed tumor growth in combination with the tyrosine kinase inhibitor sorafenib. None of the FOXP3 targeting peptides has shown antitumoral activity on its own, and it appears that these peptides exert antitumoral activity by boosting the efficacy of other immunostimulatory treatments.
β-catenin, the key mediator of the canonical Wnt signaling pathway, is another therapeutic target for Treg downregulation. Under normal conditions, β-catenin is constantly degraded by proteasome so that β-catenin would only be activated when Wnt ligand binds its receptor [63]. However, loss of function mutations in the components of the degradation complex as well as gain of function mutations in β-catenin are frequently observed in tumor cells, which lead to unregulated activation of β-catenin and oncogenic transcription. The Cancer Genome Atlas program (TCGA) revealed that the Wnt signaling pathway was altered in 93% of colorectal tumors [64]. Tumor intrinsic Wnt/β-catenin signaling facilitates immune evasion of tumor cells and is associated with enhanced generation of Tregs, suppressed activation and infiltration of CD8+ T cells, conversion of DCs to an immunoregulatory phenotype, and decreased differentiation of CD8+ T cells into effector cells [65,66]. Of note, in contrast to tumor intrinsic signaling, Wnt/β-catenin in Tregs are shown to suppress the activity of Tregs. Upon activation of the Wnt signaling pathway, T cell factor 1 (TCF1), which is a transcription factor downstream of β-catenin, associates with FOXP3 and impairs its activity [67].
Kawamoto et al. analyzed the crystal structure of the β-catenin/B Cell Lymphoma 9 (BCL9) complex and developed a BCL9 derived peptide that disrupts the association of β-catenin and BCL9 [68]. BCL9 is a coactivator of β-catenin which is required for its full transcriptional activity [69]. From the observation that BCL9 increases the amount of nuclear β-catenin without altering the export and import rate of β-catenin, it is thought that BCL9 mainly regulates β-catenin activity by retaining β-catenin in the nucleus [70]. The BCL9 derived peptide was further modified by the peptide stapling technique, which is a strategy to constrain the mobility of α-helix by connecting 2 amino acids on the same side of the α-helix with a chemical linker. Constraining the peptide in the α-helical conformation leads to improved stability and higher affinity against the target [71]. Kawamoto et al. designed a triazole-stapled peptide [72], whereas Takada et al. applied hydrocarbon stapling and developed SAH-BCL9B [73] and its optimized version hsBCL9CT-24 [74]. When treated in vivo, the population of Tregs in tumors decreased while the population of CD8+ effector T cells and CD103+ DCs increased. hsBCL9CT-24 demonstrated significant antitumoral activity and also showed strong synergy with anti-PD-1 antibody. Mechanistically, hsBCL9CT-24 treatment resulted in less production of CCL20 and CCL22. Binding of these chemokines to the receptors expressed on Tregs, CCR6 and CCR4 respectively, induce migration [75,76]. On the other hand, the amount of CCL4 expressed by tumor cells, which is a chemokine that promotes dendritic cell infiltration, increased.
P17 and P144 are peptide inhibitors of the TGF-β signaling pathway. P144 is derived from the TGF-β1 binding region of human type III TGF-β1 receptor (TGFβRIII) [77], and P17 is a TGF-β1 binding peptide identified by phage display screening [78]. TGF-β is produced by Tregs at a high level and suppresses immune responses by downregulating the activities of T cells and NK cells [79]. Although the role of TGF-β on Treg development in vivo is controversial, at least in vitro, TGF-β upregulates FOXP3 expression in CD4+CD25− T cells and facilitates the development of Tregs [79,80]. Gil-Guerrero et al. demonstrated that the administration of P17 to stimulated splenic cells co-cultured with Tregs partially restored the proliferation capacity. Treatment of P17 and P144 did not show antitumoral effect by themselves in vivo, but enhanced the efficacy of tumor vaccine and adjuvant-based immunotherapy [81,82]. Interestingly, the benefits of these peptides were not seen in mice lacking CD25+ cells. Although TGF-β signaling is a pleiotropic signaling pathway affecting the phenotypes of both stromal and cancer cells, this result suggests that the antitumoral activities of P17 and P144 are mainly due to their effect on CD25+ cells.
Neuropilin-1 (NRP-1) is a co-receptor for semaphorins and vascular endothelial growth factor (VEGF). Upon binding of semaphorin-4a (Sema4a), NRP-1 alters the transcriptome of Tregs and promotes the immunosuppressive function and survival [83]. Fc-TPP-11 is a fusion of NRP-1 binding peptide and the immunoglobulin Fc region originally developed as an anti-angiogenic agent [84]. Jung et al. applied the peptide to Tregs and demonstrated that Fc-TPP-11 suppresses the proliferation and function of Tregs by antagonizing NRP-1 [85]. Importantly, Fc-TPP-11 selectively suppressed the function of intratumoral Tregs without impairing the function of peripheral Tregs. The authors demonstrated that the expression level of NRP-1 is significantly higher in intratumoral Tregs compared to Tregs in the peripheral blood, which could explain the difference of sensitivity between these cell types.
Santagata et al. demonstrated that intratumoral Tregs in renal cell carcinoma patients express a high level of CXCR4 and sought to target CXCR4 expressed on Tregs using peptide R29 [86]. Peptide R29 is a derivative of the CXCR4 antagonist peptide R with higher binding capacity and stability [87]. Peptide R has been shown to affect tumor-associated macrophages in the TME, which would be discussed later. Treatment of peptide R29 reduced the capability of patient-derived Tregs to suppress T cell proliferation and IFN-γ production in vitro, suggesting that CXCR4 is a potential target to inhibit the activity of Tregs. CXCR4 is ubiquitously expressed in tumor cells and other immune cells in the TME [88,89] and therefore peptide R29 could also affect the activity of other components of the TME.
4. Peptides targeting natural killer (NK) cells
NK cells could directly recognize and eliminate tumor cells by releasing cytolytic granules such as perforin and granzymes. However, the activity of NK cells is suppressed in tumors because of their limited recruitment to the tumor tissues, downregulation of activating receptors, and the production of anti-inflammatory cytokines by tumor cells and immunosuppressive cells in the TME [90]. Chernysh et al. discovered that alloferon-1, which is a peptide isolated from the larvae of blowfly infected with bacteria, enhances the cytotoxic activity of NK cells and possesses antitumoral and antiviral effects (Table 2) [91,92]. Further research revealed that alloferon-1 upregulates the activating receptor CD244 on NK cells and promotes the production of pro-inflammatory molecules [93]. The binding target and the detailed molecular mechanism of action remain unknown.
Fadda et al. proposed the concept of peptide antagonism as an approach to activate NK cells [94]. Killer-cell immunoglobulin-like receptor (KIR), which recognizes MHC class I molecules, is an important inhibitory receptor that regulates the activity of human NK cells. The characteristics of the peptides presented by the MHC class I molecules determine the affinity between KIR and the peptide/MHC class I complex. Peptides that induce strong binding stimulate the downstream signaling pathway of KIR and suppress the activity of NK cells whereas peptides that induce weak binding antagonize this inhibitory signaling. Therefore, peptides that induce weak interaction between KIR and the peptide/MHC class I complex may have the potential to activate NK cells. Although no antitumoral peptide has been developed based on the peptide antagonism concept so far, future studies may lead to the generation of NK cell-activating peptides with antitumoral features.
5. Peptides targeting tumor-associated macrophages (TAMs)
Macrophages are the major immune cells constituting the TME. Macrophages have been conventionally classified into classically activated macrophages (M1 macrophages) and alternatively activated macrophages (M2 macrophages) [95]. M1 macrophages are characterized by their capability to induce inflammation and eliminate tumor cells, whereas M2 macrophages exert anti-inflammatory activity and promote wound healing. Recent studies have demonstrated that macrophages are highly diverse and versatile and this dichotomic classification is getting outdated. The current view is that macrophages organize their gene expression patterns and cellular metabolism depending on the surrounding environment and acquire continuous phenotypes in which the M1 and M2 represent the extremes of the polarization spectrum [96,97]. Although the nomenclature and classification of macrophages are still controversial, in this review we would adhere to the nomenclature adopted by Mantovani et al., in which “M1” phenotype specifically indicates the phenotype of macrophages polarized by IFN-γ or bacterial products, and “M2” phenotype indicates the phenotype of macrophages polarized by IL-4 or IL-13. Regardless of the polarizing drivers, “M1-like” refers to the macrophages that induce antitumoral effect by its cytotoxic activity, and “M2-like” refers to the macrophages that suppress antitumoral immunity and promote tumor growth [98,99]. Clinical studies have revealed that the infiltration of macrophages in tumors correlated with poor prognosis in several cancer types [100]. In the TME, cytokines and metabolites produced by tumor cells, IL-4 and IL-13 released from TH2 helper cells, and other factors such as hypoxia shift the phenotype of TAMs towards M2-like macrophages [98]. TAMs promote tumor progression by several mechanisms including the stimulation of tumor cell proliferation, suppression of antitumor immunity, and upregulation of angiogenesis. The main strategies to target TAMs are inhibition of recruitment to the tumor, elimination of TAMs by specific delivery of cytotoxic agents to TAMs, and transforming TAMs from M1-like to M2-like macrophages (Fig. 4, Table 3) [101].
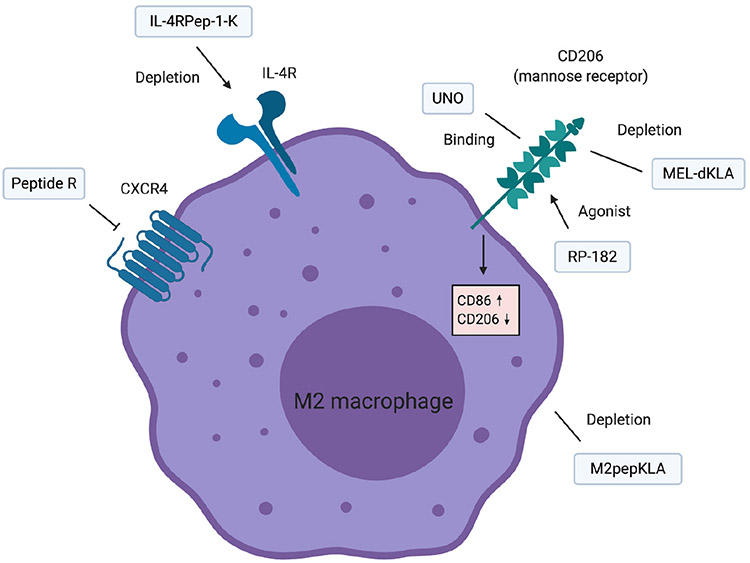
Fig. 4.
Peptides targeting TAMs. Several peptides binding CD206, which is a prominent biomarker of M2-like macrophages, have been developed. IL-4R is also highly expressed on M2-like macrophages. M2pepKLA is another peptide binding M2-like macrophages although the binding target is unknown. Peptide R is a CXCR4 inhibitory peptide which promotes the M1 feature of TAMs.
Table 3.
Peptides targeting myeloid cells.
| Mechanism of action |
Peptide Name |
Sequence | Discovery method | Cancer model | Administration method |
Modifications for improving stability |
Ref |
|---|---|---|---|---|---|---|---|
| Peptides targeting macrophages | |||||||
| Binds to M2 macrophages | M2pep | YEQDPWGVKWWY | Screening by phage display | CT26 colon cancer | Intravenous | [98-101] | |
| Binds to CD206+ cells | UNO | CSPGAKVRC | In vivo screening by phage display | 4 T1 breast cancer | [102] | ||
| Depletes CD206+ cells | MEL-dKLA | GIGAVLKVLTTGLPALISWIKRKRQQGGGGS-D[KLAKLAKKLAKLAK] | Naturally occurring peptide | Lewis lung carcinoma | Intraperitoneal | (D)-enantiomeric structure | [104,105] |
| Agonist of CD206 | RP-182 | KFRKAFKRFF | Shared motif of host defense peptides | KPC spontaneous pancreatic cancer model, K16 spontaneous pancreatic cancer model | Intraperitoneal | [106] | |
| Depletes IL-4R+ cells | IL-4RPep-1-K | CRKRLDRNC-GGG-KLAKLAKKLAKLAK | Screening by phage display | 4 T1 breast cancer | Intravenous | [108,109] | |
| Antagonist of CXCR4 | Peptide R | RA-cyclic[CRFFC] | NMR structure analysis | U87MG glioblastoma xenograft | Intraperitoneal | Cyclic structure | [111,112] |
| Peptides targeting dendritic cells | |||||||
| Binds to CLEC9A | WH | WPRFHSSVFHTH | Screening by phage display | B16-OVA melanoma | Subcutaneous | [115] | |
| Binds to CLEC10A | sv4D | [(VQATQSNQHTPRGGGS)2K]2K | Screening by phage display | ID8 ovarian cancer | Subcutaneous | [122] | |
| Binds to CLEC10A | sv6D | [(NQHTPRGGGS)2K]2K | Screening by phage display | ID8 ovarian cancer | Subcutaneous | [123] | |
| NRP-1 antagonist | MY1340 | [(TKPRKHG)2K]2KG | Derivative of the naturally occurring peptide tuftsin | H22 hepatocellular carcinoma | Subcutaneous | [124] | |
| Peptides targeting MDSCs | |||||||
| Depletion of MDSCs | Pep-H6 | MEWSLEKGYTIK | Screening by phage display | EL4 lymphoma | Intravenous | Fusion with the Fc region of antibodies | [133] |
| Depletion of MDSCs | Pep-G3 | WGWSLSHGYQVK | Screening by phage display | EL4 lymphoma | Intravenous | Fusion with the Fc region of antibodies | [133] |
Cieslewicz et al. conducted a phage display screening to identify peptides that bind to M2 macrophages [102]. By negatively selecting peptides that bound to M1 macrophages and positively selecting peptides that bound to M2 macrophages, they developed M2pep which preferentially binds to M2 macrophages over M1 macrophages. In a cell population harvested from tumors, the capability of M2pep to bind M2-like TAMs was significantly higher than that to bind M1-like TAMs. The administration of M2pepKLA, which is a fusion of M2pep and the pro-apoptotic KLA peptide, selectively eliminated M2-like TAMs in vivo. Further analysis showed that the binding efficacy of M2pep largely depends on the cancer cell type. M2pep strongly binds to TAMs harvested from 4 T1 breast tumors, whereas binding to TAMs harvested from HepG2 hepatocellular carcinoma was relatively moderate [103]. The identification of the currently unknown binding partner of M2pep would be necessary to fully understand the selectivity of M2pep.
Several attempts have been made to establish novel immunotherapies using M2pep. Pang et al. developed an M2pep coated nanoparticle to deliver PLX3397, an inhibitor of colony stimulating factor-1 receptor (CSF1R) which plays a crucial role in macrophage differentiation and survival [104]. The nanoparticles specifically internalized in M2-like TAMs in vivo and suppressed tumor growth. Li et al. developed an M2pep conjugated nanoparticle to deliver PI3K-γ inhibitor NVP-BEZ 235 and CSF1R siRNA [105]. The dual delivery of these agents resulted in a decrease of M2-like macrophages in pancreatic xenograft model. Remarkably, the infiltration of CD4+ T cells and CD8+ T cells was enhanced, whereas the infiltration of myeloid-derived suppressor cells (MDSCs) was diminished.
Mannose receptor (CD206), which is a molecule highly expressed on M2-like macrophages, has been the major target to develop peptides that bind to TAMs. Scodeller et al. performed an in vivo phage display screening to identify peptides that bind to peritoneal macrophages in mice bearing 4 T1 breast tumor and developed UNO [106]. In vivo imaging revealed that UNO internalized in CD206+ M2-like macrophages. Administration of UNO-conjugated nanoparticles loaded with paclitaxel lead to the accumulation of paclitaxel in CD206+ TAMs in vivo. Another peptide targeting CD206+ TAMs is melittin, which is an amphiphilic peptide found in honey bee venom [107]. Apart from the anti-tumoral effect caused by its nonspecific membrane disruptive attribute, Lee et al. discovered that melittin preferentially binds to and eliminates CD206+ M2-like macrophages which contribute to decreased angiogenesis and tumor suppression [108]. To boost the capability to eliminate M2 macrophages, they conjugated melittin with the proapoptotic peptide dKLA (MEL-dKLA) [109]. When intraperitoneally administered in Lewis lung carcinoma model, the number of M2-like macrophages decreased whereas the number of M1-like macrophages increased.
Jaynes et al. developed RP-182, an agonist of CD206 [110]. Under the hypothesis that a well-conserved sequence would play an important role in innate immunity, the authors screened a database of α-helical host defense peptides (HDPs) and detected structural homologies. As a result, they successfully identified a 10-mer structural motif well conserved across HDPs and further developed RP-182 which reconstitute the shared motif. In silico analysis predicted CD206 as the binding partner of RP-182 and visualization by electron microscopy confirmed that RP-182 acts as an agonist of CD206. RP-182 induced phagocytosis, autophagy, and apoptosis in CD206+ M2-like macrophages, but not in CD86+ M1-like macrophages. Interestingly, treatment with RP-182 induced the expression of CD86 and suppressed the expression of CD206 in M2 polarized bone marrow derived cells, indicating a reprogramming of macrophages from M2 to M1-like. RP-182 administration in spontaneous pancreatic tumor model suppressed the infiltration of M2-like macrophages, MDSCs, and Tregs in tumor whereas the infiltration of CD8+ T cells increased. Furthermore, cancer cell phagocytosis by TAMs was also enhanced by RP-182 treatment. These results suggest that RP-182 could enhance both macrophage-mediated innate immunity and T cell-mediated adaptive immunity to activate antitumoral immunity.
IL-4R is another molecule highly expressed on M2-like macrophages [111]. Hong et al. performed a phage display screening to identify a peptide that homes to atherosclerotic plaques and discovered IL-4RPep-1, which is a peptide that binds to IL-4R [112]. To apply the peptide to cancer immunotherapy, Vadevoo et al. developed IL-4RPep-1-K, which is a conjugate of IL-4RPep-1 and proapoptotic peptide KLA [113]. IL-4RPep-1-K treatment increased the population of M1-like macrophages, CD4+ T cells, and CD8+ T cells in vivo. Furthermore, the expression level of immunosuppressive molecules in the tumor tissue significantly decreased.
Peptide R is an antagonist of CXCR4 which mimics the structure of its ligand CXCL12. CXCR4 is overexpressed in more than 20 tumor types and is involved in many biological phenomena such as leukocyte trafficking, tumor proliferation, invasion, metastasis, and angiogenesis [88]. Peptide R was originally developed to target cancer cells and inhibit CXCL12 dependent migration and lung metastasis [114]. Mercurio et al. discovered that administration of peptide R reduces the number of CD11b+ myeloid cells and CD68+ activated microglia cells in glioma, indicating that peptide R has an immunomodulating function. Furthermore, the expression of iNOS on myeloid cells, which is a marker of M1-like macrophages, was enhanced indicating a polarization to M1-like [115]. Although the authors demonstrated that peptide R antagonize CXCR4 on tumor cells, they did not investigate its effect on macrophages and therefore whether the M1 polarizing effect is a result of a direct effect on macrophages or an indirect effect via the effect on tumor cells remains unclear. CXCR4 is also expressed in a wide range of immune cells including DCs, MDSCs, and neutrophils, and therefore the target of peptide R remains obscure.
6. Peptides targeting dendritic cells (DCs)
DCs are rare immune cells in the TME that initiate adaptive immunity against tumor cells by presenting tumor-associated antigens on MHC molecules, providing costimulatory signals to T cells, and releasing soluble factors that affect the differentiation of T cells. Tumor cells inhibit the activity of DCs by several means such as inhibiting the maturation of DCs, blocking the infiltration into TME, and interference with the DC activating signals. Administration of DC stimulating and mobilizing agents, blocking inhibitory signaling pathways, and delivering antigens using drug delivery systems targeting DCs are some of the approaches targeting DCs [116,117]. A few peptides have been developed to activate DCs (Table 3).
Yan et al. developed a peptide named WH which targets C-type lectin domain family 9 member A (CLEC9A) [118]. CLEC9A is selectively expressed on mouse CD8a+ DCs as well as on human BDCA3+ DCs which are functionally similar subsets [119-121]. These DCs are the major DC subsets that perform cross-presentation and play a major role in antiviral and anti-tumor immunity [122]. Therefore, delivering antigens to these specialized DCs is a promising strategy to boost antitumoral immunity. In vitro experiment demonstrated that the T cell stimulating potency of WH conjugated ovalbumin (WH-OVA) was higher than ovalbumin alone. When WH-OVA was administered in vivo, the expression levels of perforin, granzyme B, and IFN-γ in cytotoxic T cells were increased and less metastasis was observed in B16-OVA lung metastasis model. These results suggest that WH can be used to deliver tumor vaccines to DCs and improve their efficacy.
Asialoglycoprotein receptor-1 (ASGPR1) and the highly homologous C-type lectin domain family 10 member A (CLEC10A) are also C-type lectins expressed on DCs. These receptors specifically recognize N-acetylgalactosamine (GalNAc) residues attached to glycosylated proteins. Since terminal GalNAc occurs only when the regulatory pathways of protein glycosylation are disturbed, terminal GalNAc is generally restricted to cancer cells or pathogens, making ASGPR1 and CLEC10A specific detectors of these harmful species. Binding of the ligand and subsequent endocytosis of CLEC10A induces Ca2+ signal and activates transcription factors such as NFAT and NF-κB via activation of calcineurin and Ca2+/calmodulin-dependent kinase II (CaMKII). The activation of these transcription factors leads to the production of proinflammatory cytokines and maturation of DCs [123,124]. Eggink et al. conducted a phage display screening to identify peptides that bind to GalNAc-binding lectin [125]. After identifying a functional sequence that binds to the lectin, they developed a tetravalent peptide svL4 which contains 4 peptide chains of the binding sequence. sv6D, which has the same tetravalent structure but using a shorter binding sequence, was later developed as a peptide with higher affinity to lectins [126]. Both svL4 and sv6D bound to CLEC10A and ASGPR1 in vitro, and when administered in vivo, the number of mature DCs in the peritoneum cavity increased. sv6D extended survival of mice bearing ovarian cancer and were synergistic with paclitaxel and anti-PD-1 antibodies.
Mo et al. focused on the inhibitory role of VEGF on DC maturation and developed MY1340 which inhibits the interaction between VEGF and NRP-1 [127]. The role of VEGF on DC maturation was first demonstrated by Rotonda et al., which they discovered that treating CD34+ hematopoietic cells with recombinant VEGF inhibited their ability to induce T cell proliferation [128]. It was later confirmed in vivo that continuous infusion of VEGF suppresses maturation of DCs [129], whereas neutralizing VEGF by anti-VEGF antibodies enhanced the population of mature DCs in lymphoid tissues [130]. Oussa et al. demonstrated that NRP-1 deficient DCs were insensitive to the inhibitory effect of VEGF on LPS induced maturation, indicating that NRP-1 is a potential target to optimize DC maturation [131]. MY1340 inhibited the formation of the NRP-1/VEGF complex and canceled out the downregulating role of VEGF on DC maturation. When treated in vivo, MY1340 enhanced the maturation of DCs in the spleen and exerted antitumoral activity.
7. Peptides targeting myeloid derived suppressor cells (MDSCs)
Along with macrophages and DCs, MDSCs are another important population of myeloid cells constituting the TME. MDSCs are a heterogeneous population defined as non-macrophage immature myeloid cells with the ability to suppress immune activity. MDSCs suppress antitumoral immunity by several mechanisms, including consumption of key nutrients required for T cell activity, production of immunosuppressive cytokines, recruitment of other immunosuppressive cells, and expression of immune checkpoint molecules. MDSCs also promote tumor progression by supporting angiogenesis and metastasis [132]. Ample clinical evidence has demonstrated that the accumulation of MDSCs in the tumor correlates with poor prognosis [133-135].
Qin et al. isolated the Gr-1+CD11b+ splenic MDSCs from mice bearing EL4 thymomas and identified peptides that bound to these cells by phage display screening [136]. H6 and G3 are the 2 peptides identified in this process (Table 3). The authors further fused the peptides with the Fc region of antibody to develop peptibodies Pep-H6 and Pep-G3. In EL4 thymoma model, the peptibodies depleted MDSCs without affecting the population of DCs and lymphocytes and significantly inhibited tumor growth. Proteomic analysis and immunoprecipitation experiments revealed that both peptides bound to S100A8 and S100A9, which are highly expressed in MDSCs.
For the time being, H6 and G3 appear to be the only MDSC-targeting peptides developed. Several MDSC targeting small molecular drugs have been generated to either inhibit the recruitment of MDSCs in the tumor, deplete intratumoral MDSCs, or inhibit the immunosuppressive activity of MDSCs [137]. Identification of peptides targeting the molecules involved in these processes would be promising steps to develop peptide therapeutics targeting MDSCs. Lack of specific biomarkers of human MDSCs is a general problem hindering the development of MDSC-targeting therapies. Since many biomarkers are shared across myeloid cells, most MDSC-targeting agents exhibit multiple biological functions, making it difficult to evaluate the contribution of the MDSC-targeting effect of these agents. Identification of specific biomarkers of human MDSCs would be necessary to establish effective therapies targeting MDSCs.
8. Peptides inducing immunogenic cell death
Immunogenic cell death (ICD) is defined as a type of cell death that elicit immune responses. The immunogenicity of cell death depends on the display of neoepitopes and the release of damage associated molecular patterns (DAMPs) [138]. Immunostimulatory molecules such as high mobility group box 1 (HMGB1) and ATP are common DAMPs released upon ICD. Exposure of calreticulin is another phenomenon observed during ICD which promotes phagocytosis by DCs [138,139]. Several peptides have been developed to effectively cause ICD and boost antitumoral immunity (Table 4).
Table 4.
Peptides inducing immunogenic cell death.
| Mechanism of action |
Peptide name |
Sequence | Discovery method | Cancer model | Administration method |
Modifications for improving stability |
Ref |
|---|---|---|---|---|---|---|---|
| Rupture of the cellular membrane | LTX-315 | KKWWKKDipK | Based on structure-activity relationship analysis | B16F1 melanoma, MCA205 sarcoma, 4 T1 breast cancer | Intratumoral | [137-143] | |
| Rupture of the cellular membrane | LTX-401 | β(2,2)-amino acid derivative | Based on structure-activity relationship analysis | B16F1 melanoma, MCA205 sarcoma, JM1 hepatocellular carcinoma | Intratumoral | [144-147] | |
| Controversial | PKHB1 | DKRFYWMWKDK | Derivative of 4N1K | MEC-1 chronic lymphocytic leukemia, L5178Y-R lymphoma | Intraperitoneal | (D)-enantiomeric structure | [149-151] |
| Antagonist of AAC-11 | RT53 | RQIKIWFQNRRMKVVKKAKLNAEKLKDFKIRLQYFARGLQVYIRQLRLALQGKT | Derivative of AAC-11 | MCA205 sarcoma | Intratumoral | [152-155] |
Dip: Diphenylalanine.
LTX-315 is an oncolytic peptide that induces ICD [140]. The structure of LTX-315 was designed based on a structure-activity relationship study revealing that amphipathic structures with separated cationic residues and hydrophobic residues are essential for membrane-disturbing peptides [141]. In addition to the perturbation of the plasma membrane, Zhou et al. demonstrated that LTX-315 accumulates in the mitochondria and the Bax/Bak regulated permeabilization of mitochondrial membrane is also an essential step in the LTX-315 mediated cytotoxicity [142]. Although the cytotoxicity is not specific to cancer cells, Camilio et al. showed that cancer cells are more sensitive to LTX-315 compared to normal cells [143]. The selectivity of LTX-315 to cancer cells might be explained by the unique features of the cancer cell surface. Anionic molecules such as phosphatidylserine, sialic acid, and heparan sulfate are highly expressed on the cellular membrane of cancer cells. The upregulation of these molecules leads to a higher affinity to the cationic molecule LTX-315. The selectivity might also be explained by the greater membrane fluidity and cell surface area of cancer cells which lead to relatively unstable membrane structure and binding of more LTX-315 [143]. Intratumoral injection of LTX-315 improved the infiltration of T cells, increased the cytotoxic T cell/Treg ratio in tumors, and suppressed tumor growth in multiple tumor models [143-145]. In a phase 1/2 study using LTX-315 on solid tumor patients (NCT01986426), the number of intratumoral T cells was increased after treatment in 89% of evaluable biopsied patients receiving LTX-315 monotherapy [146]. Currently, a phase 2 study on sarcoma evaluating the safety and effectiveness to induce T cell infiltration is conducted (NCT03725605). LTX-401 is an amphipathic β(2,2)-amino acid derivative developed by the same research group which could also disrupt the cancer cell membrane and induce ICD [147]. Contrary to LTX-315, LTX-401 accumulates not in the mitochondria but the cytosol and the Golgi apparatus [148]. The efficacy in vivo was confirmed in multiple cancer models [147,149,150].
CD47 is an immunoglobulin frequently overexpressed on the surface of cancer cells. CD47 binds to signal-regulatory protein alpha (SIRPα) expressed on innate immune cells such as macrophages and inhibits phagocytosis [151]. Martinez-Torres et al. reported that PKHB1, a peptide that binds to CD47, induced programmed cell death of chronic lymphocytic leukemia B cells in a PLCα1 dependent manner [152]. Uscanga-Palomeque et al. further applied the peptide to T cell acute lymphoblastic leukemia cells and confirmed that leukemia cells released immunostimulatory DAMPs upon PKHB1 treatment [153]. In vivo experiments showed that the infiltration of leukocytes in lymphoid tissues increased in PKHB1 treated mice. Although the authors proposed that the PKHB1-induced cell death is caused by the activation of CD47, the underlying mechanism is somewhat controversial. Both studies have demonstrated that Ca2+ chelators abolish the cell-killing effect by PKHB1 whereas caspase inhibitors do not, suggesting a calcium-dependent, caspase-independent mechanism. However, they did not confirm the dependency of CD47 in their studies. Leclair et al. demonstrated that CD47 deficient T cells are equally susceptible to PKHB1 treatment as CD47 expressing T cells, indicating a CD47 independent mechanism [154].
RT53 is a peptide that was designed to antagonize anti-apoptotic clone 11 (AAC-11), which is a protein that inhibits E2F1 regulated apoptosis [155]. Rigou et al. demonstrated that AAC-11 binds to and inhibits Acinus, which is a protein that is activated upon cleavage by caspase and induces DNA fragmentation [156]. RT53 is derived from the protein-protein interaction module of AAC-11 and is capable of sensitizing cancer cells to apoptosis by disrupting the interaction of endogenous AAC-11 and Acinus. Jagot-Lacoussiere et al. demonstrated that RT53 also possesses membranolytic activity and induces rapid necrosis involving the release of DAMPs [157]. It was later confirmed in vivo that the number of T cells infiltrating the tumor was increased when RT53 was administered intratumorally [158].
9. Limitations and opportunities
Despite the beneficial properties for therapeutic applications in cancer, peptides have not been successful in the clinic so far. One of the major bottlenecks of peptides entering the clinic is the short half-lives in serum [159,160]. When injected in the body, peptides are rapidly removed by either enzymatic degradation by proteases, hepatobiliary excretion, or renal clearance. On the contrary, therapeutic antibodies are not susceptible to enzymatic degradation and are not filtered out at the glomeruli of the kidney because of their large sizes. The major degradation pathway of antibodies is the hydrolysis by lysosomes [12]. In the bloodstream, endothelial cells continuously take in antibodies by pinocytosis. The Fc regions of internalized IgG antibodies bind to the neonatal Fc receptors (FcRn) at the endosomes. The antibodies bound to FcRn would be recycled to the cellular surface and the molecules that did not bind to the FcRn would be degraded in the lysosome. This FcRn-mediated recycling significantly increases the half-lives of antibodies. The half-lives of peptides in vivo are typically a few minutes to a few hours, whereas half-lives of antibodies are typically around 3 weeks [12,161].
The difficulty of oral administration is another obstacle to clinical application. The bioavailability of peptides in the gastrointestinal tract is extremely low since peptides are degraded by the proteases and the acidic environment. Furthermore, the electrical charges of peptides disable penetration through the intestinal epithelium [159]. Both peptides and antibodies are not readily suited for oral administration and intravascular or subcutaneous injection would be the method for clinical administration. Since the half-lives of peptides are short, peptides would require frequent administration, which would impair the quality of life of the patient. Indeed, most in vivo experiments conducted using the peptides discussed so far administer peptides every day or every other day.
Generally, naked peptides have poor pharmacokinetic characteristics compared to antibodies. However, numerous techniques to improve the pharmacokinetics of peptides are being developed. Attaching peptides to larger molecules such as polyethylene glycol, albumin, immunoglobulin fragments, and nanoparticles reduces renal excretion and enzymatic degradation. Incorporation of unnatural amino acids, modification of the C-terminus and the N-terminus, adopting a cyclic structure, and usage of stapling technique are other strategies used to improve the stability of peptides [162]. Self-assembling peptides open possibilities for sustained delivery in multiple diseases including cancer [163,164]. Several methodologies for oral administration of peptides have also been developed. Usage of permeation enhancers, which interferes with the adhesion proteins of intestinal epithelial cells to promote the penetration of peptides through the intestinal epithelium, inhibitors of proteases, and nanoparticle carriers are some of the strategies used for oral administration of peptides [165].
10. Concluding remarks and future perspectives
In the current review, we introduced the peptides targeting the components of the TME and discussed the discovery methods, mechanisms of action, and their immunomodulatory effect in cancer models. Although various peptides demonstrating promising preclinical results have been developed, many targets remain unexplored for the development of therapeutic peptides. For example, peptides targeting immune checkpoint molecules are mostly aimed to disrupt the PD-1/PD-L1 complex and few peptides targeting other immune checkpoints have been developed. Other immune checkpoint molecules such as LAG-3, TIGIT, and T cell immunoglobulin and mucin-domain containing-3 (TIM-3), have shown promise as novel immune targets for cancer immunotherapy in preclinical and early clinical studies [166]. Peptides targeting these molecules could potentially be effective alone or in combination with inhibitors of the PD-1/PD-L1 pathway to further improve response rates and durability of response. Currently, there are limited biomarkers for TAMs, DCs, and MDSCs due to their high heterogeneity and ambiguous phenotypes. Detailed analysis of the cellular phenotypes of their subpopulation and identification of specific biomarkers are required for further development of peptide therapeutics. Recent advances in single-cell sequencing technologies and computational methods have contributed to the deconvolution of cellular heterogeneity and would continue to develop our knowledge. Combinations of multiple omics data beyond transcriptome such as proteomics and the profiling of epigenetic modifications would lead to the depiction of a high-resolution landscape of immunity and identification of therapeutic targets [167,168].
Several components of the TME have attracted relatively little interest for therapeutic development. Tumor associated neutrophils (TANs) are one of the untargeted immune cells in cancer immunotherapy. Analogous to macrophages, TANs are phenotypically classified into N1 and N2 neutrophils. N1 neutrophils exert cytotoxicity against tumor cells by production of reactive oxygen species, antibody-dependent cytotoxicity, and activation of other immune cells. On the other hand, N2 neutrophils suppress the activity of T cells by expression of PD-L1 and arginase-1. Specifically inhibiting the activity of N2 neutrophils or converting N2 neutrophils to N1 neutrophils would be promising strategies to target TANs [169]. Immunoactivating therapeutics targeting cancer associated fibroblasts (CAFs) are also scarce. CAFs suppress antitumoral immunity by secreting soluble factors that downregulate the activity of T cells and also disrupt the metabolism of T cells by consuming amino acids essential for T cell survival. Excessive production of the extracellular matrix by CAF inhibits the entry of immune cells in the tumor and contributes to the establishment of an immunosuppressive environment [170,171].
Peptides targeting angiogenesis could be reconsidered as agents with the potential to enhance the efficacy of immunotherapy. Recent preclinical and clinical studies have demonstrated the synergistic effect of immune checkpoint inhibitors and antiangiogenetic agents, which is explained by the concept of vascular normalization [172]. In tumors, overexpression of proangiogenic factors such as VEGF triggers the generation of abnormal blood vessels characterized by low pericyte coverage, loose cell junctions, and disrupted basement membrane. The hyperpermeable vasculature leads to poor perfusion of blood in the tumor, forming a hypoxic and acidic environment. Hypoxia modifies the TME into an immunosuppressive environment by several mechanisms such as expression of chemotactic factors recruiting Tregs and MDSCs, polarizing macrophages to an immunosuppressive phenotype, and further upregulation of VEGF which downregulates antitumoral immunity by inhibition of DC maturation. The inefficient vascular network hinders the delivery of drugs and immune cells to tumors which further contributes to the evasion of cancer cells from immunity [172-174]. Administrating low doses of antiangiogenic drugs would normalize the vasculature and improve blood perfusion in the tumor, which would alleviate hypoxia and its immunosuppressive effect [173]. Therefore, optimal dosing of antiangiogenic peptides may enhance antitumoral immunity. Indeed, Mirando et al. demonstrated that the administration of antiangiogenic integrin-binding peptide AXT201 in 4 T1 breast cancer leads to the decrease of Tregs and MDSCs and increase of activated T cells in the tumor [175]. Other antiangiogenic peptides, summarized by Rosca et al. [176] or Karagiannis and Popel [177], might also exert immunomodulating activities.
Although no peptide for cancer immunotherapy and only a few peptides for oncology in general have been approved for clinical usage so far [178], peptides possess valuable characteristics that small molecules and antibodies could not achieve. Poor pharmacokinetics have restricted their clinical applications, but several technologies have been developed to overcome this limitation. As we have seen above, these techniques are already being applied to immunomodulating peptides and further development would lead to success in the clinic. Recently, combination therapy with immune checkpoint inhibitors and other immunomodulating agents are attracting attention to treat cold tumors where monotherapy of immune checkpoint inhibitor is not effective [179]. Determining the optimal combinations of the immunomodulating peptides is another mission for the future.
Acknowledgements
This work was supported by NIH grants R01CA138264 and U01CA212007, and a graduate fellowship from Takenaka Scholarship Foundation (NF). The authors are grateful to Drs. Mark Yarchoan and Elisabeth M. Jaffee for critical comments on the manuscript.
Footnotes
Declaration of Competing Interest
The authors declare no competing interests.
References
- [1].Sun L, et al. , Clinical efficacy and safety of anti-PD-1/PD-L1 inhibitors for the treatment of advanced or metastatic cancer: a systematic review and meta-analysis, Sci. Rep 10 (2020) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Haslamn A, Gill J, Prasad V, Estimation of the percentage of US patients with cancer who are eligible for immune checkpoint inhibitor drugs, JAMA Netw. Open 3 (e200423) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yarchoan M, Hopkins A, Jaffee EM, Tumor mutational burden and response rate to PD-1 inhibition, N. Engl. J. Med 377 (2017) 2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bonaventura P, et al. , Cold tumors: a therapeutic challenge for immunotherapy, Front. Immunol 10 (2019) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Locy H, et al. , Immunomodulation of the tumor microenvironment: turn foe into friend, Front. Immunol 9 (2018) 2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Duan Q, Zhang H, Zheng J, Zhang L, Turning cold into hot: firing up the tumor microenvironment, Trends Cancer (2020) 1–14. [DOI] [PubMed] [Google Scholar]
- [7].Cao J, Yan Q, Cancer epigenetics, tumor immunity, and immunotherapy, Trends Cancer 6 (2020) 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Berraondo P, et al. , Cytokines in clinical cancer immunotherapy, Br. J. Cancer 120 (2019) 6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Thurber GM, Schmidt MM, Wittrup KD, Factors determining antibody distribution in tumors, Trends Pharmacol. Sci 29 (2008) 57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ryman JT, Meibohm B, Pharmacokinetics of monoclonal antibodies, CPT Pharmacometrics Syst. Pharmacol 6 (2017) 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Thurber GM, Schmidt MM, Wittrup KD, Antibody tumor penetration, Adv. Drug Deliv. Rev 60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chames P, Van Regenmortel M, Weiss E, Baty D, Therapeutic antibodies: successes, limitations and hopes for the future, Br. J. Pharmacol 157 (2009) 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Postow MA, Sidlow R, Hellmann MD, Immune-related adverse events associated with immune checkpoint blockade, N. Engl. J. Med 378 (2018) 158–168. [DOI] [PubMed] [Google Scholar]
- [14].Martins F, et al. , Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance, Nat. Rev. Clin. Oncol 16 (2019) 563–580. [DOI] [PubMed] [Google Scholar]
- [15].Sasikumar PG, et al. , A rationally designed peptide antagonist of the PD-1 signaling pathway as an immunomodulatory agent for cancer therapy, Mol. Cancer Ther 18 (2019) 1081–1091. [DOI] [PubMed] [Google Scholar]
- [16].Marqus S, Pirogova E, Piva TJ, Evaluation of the use of therapeutic peptides for cancer treatment, J. Biomed. Sci 24 (2017) 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guidotti G, Brambilla L, Rossi D, Cell-penetrating peptides: from basic research to clinics, Trends Pharmacol. Sci 38 (2017) 406–424. [DOI] [PubMed] [Google Scholar]
- [18].Jaradat DMM, Thirteen decades of peptide synthesis: key developments in solid phase peptide synthesis and amide bond formation utilized in peptide ligation, Amino Acids 50 (2018) 39–68. [DOI] [PubMed] [Google Scholar]
- [19].Hu Z, Ott PA, Wu CJ, Towards personalized, tumour-specific, therapeutic vaccines for cancer, Nat. Rev. Immunol 18 (2018) 168–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sahin U, Türeci Ö, Personalized vaccines for cancer immunotherapy, Science (80-. ) 359 (2018) 1355–1360. [DOI] [PubMed] [Google Scholar]
- [21].Peng M, et al. , Neoantigen vaccine: an emerging tumor immunotherapy, Mol. Cancer 18 (2019) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pardoll DM, The blockade of immune checkpoints in cancer immunotherapy, Nat. Rev. Cancer 12 (2012) 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vaddepally RK, Kharel P, Pandey R, Garje R, Chandra AB, Review of indications of FDA-approved immune checkpoint inhibitors per NCCN guidelines with the level of evidence, Cancers (Basel) 12 (1–19) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sasikumar PGN, Ramachandra M, Immunosuppression Modulating Compounds, 2014. [Google Scholar]
- [25].Miller MM, et al. , Macrocyclic Inhibitors of the PD-1/PD-L1 and CD80(B7-1)/ PD-L1 Protein/Protein Interactions, 2016. [Google Scholar]
- [26].Guzik K, et al. , Development of the inhibitors that target the PD-1/PD-L1 interaction—a brief look at progress on small molecules, peptides and macrocycles, Molecules 24 (2019) 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Magiera-Mularz K, et al. , Bioactive macrocyclic inhibitors of the PD-1/PD-L1 immune checkpoint, Angew. Chem. Int. Ed 56 (2017) 13732–13735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chatterjee S, et al. , Rapid PD-L1 detection in tumors with PET using a highly specific peptide, Biochem. Biophys. Res. Commun 483 (2017) 258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li Q, et al. , Discovery of peptide inhibitors targeting human programmed death 1 (PD-1) receptor, Oncotarget 7 (2016) 64967–64976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Boohaker RJ, et al. , Rational design and development of a peptide inhibitor for the PD-1/PD-L1 interaction, Cancer Lett. 434 (2018) 11–21. [DOI] [PubMed] [Google Scholar]
- [31].Abbas AB, et al. , Design and synthesis of A PD-1 binding peptide and evaluation of its anti-tumor activity, Int. J. Mol. Sci 20 (2019) 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Knoops B, Goemaere J, Van der Eecken V, Dedercq J-P, Peroxiredoxin 5: structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin, Antioxid. Redox Signal 15 (2011) 817–829. [DOI] [PubMed] [Google Scholar]
- [33].Liu J, et al. , The antitumor activity and preliminary modeling on the potential mechanism of action of human peroxiredoxin-5, Oncotarget 8 (2017) 27189–27198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zou S, et al. , Discovery of hPRDX5-based peptide inhibitors blocking PD-1/PD-L1 interaction through in silico proteolysis and rational design, Cancer Chemother. Pharmacol 85 (2020) 185–193. [DOI] [PubMed] [Google Scholar]
- [35].Liu H, et al. , Discovery of low-molecular weight anti-PD-L1 peptides for cancer immunotherapy, J. Immunother. Cancer 7 (2019) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li C, et al. , Peptide blocking of PD-1/PD-L1 interaction for cancer immunotherapy, Cancer Immunol. Res 6 (2018) 178–188. [DOI] [PubMed] [Google Scholar]
- [37].Kuan H, et al. , Developing native peptide-based radiotracers for PD-L1 PET imaging and improving imaging contrast by pegylation, Chem. Commun 55 (2019) 4162–4165. [DOI] [PubMed] [Google Scholar]
- [38].Chang HN, et al. , Blocking of the PD-1/PD-L1 interaction by a D -peptide antagonist for cancer immunotherapy, Angew. Chem. Int. Ed 54 (2015) 11760–11764. [DOI] [PubMed] [Google Scholar]
- [39].Schumacher TNM, et al. , Identification of D-Peptide Ligands Through Mirror-Image Phage Display, Science (80-. ) 271 (1996) 1854–1857. [DOI] [PubMed] [Google Scholar]
- [40].Garton M, et al. , Method to generate highly stable D-amino acid analogs of bioactive helical peptides using a mirror image of the entire PDB, Proc. Natl. Acad. Sci. U. S. A 115 (2018) 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sun Z, et al. , PD-1/PD-L1 pathway and angiogenesis dual recognizable nanoparticles for enhancing chemotherapy of malignant cancer, Drug Deliv. 25 (2018) 1746–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhou X, et al. , A novel d-peptide identified by mirror-image phage display blocks TIGIT/PVR for cancer immunotherapy, Angew. Chem. Int. Ed (2020) 1–6. [DOI] [PubMed] [Google Scholar]
- [43].Harjunpääa H, Guillerey C, TIGIT as an emerging immune checkpoint, Clin. Exp. Immunol 200 (2020) 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gurung S, et al. , Phage display-identified PD-L1-binding peptides reinvigorate T-cell activity and inhibit tumor progression, Biomaterials 247 (2020) 119984. [DOI] [PubMed] [Google Scholar]
- [45].Wang H, et al. , HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity, Nat. Chem. Biol 15 (2019) 42–50. [DOI] [PubMed] [Google Scholar]
- [46].Mudiyanselage TMRR, et al. , An immune-stimulatory helix-loop-helix peptide: selective inhibition of CTLA-4-B7 interaction, ACS Chem. Biol 15 (2020) 360–368. [DOI] [PubMed] [Google Scholar]
- [47].Zhai W, et al. , A novel cyclic peptide targeting LAG-3 for cancer immunotherapy by activating antigen-specific CD8+ T cell responses, Acta Pharm. Sin. B 10 (2020) 1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Andrews LP, Marciscano AE, Drake CG, Vignali DAA, LAG3 (CD223) as a cancer immunotherapy target, Immunol. Rev 276 (2017) 80–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang Y, et al. , PD-1-targeted discovery of peptide inhibitors by virtual screening, molecular dynamics simulation, and surface plasmon resonance, Molecules 24 (2019) 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Caldwell C, et al. , Identification and validation of a PD-L1 binding peptide for determination of PDL1 expression in tumors, Sci. Rep 7 (2017) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kamalinia G, et al. , mRNA display discovery of a novel programmed death ligand 1 (PD-L1) binding peptide (a peptide ligand for PD-L1), ACS Chem. Biol 1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Spodzieja M, et al. , Design of short peptides to block BTLA/HVEM interactions for promoting anticancer T-cell responses, PLoS One 12 (2017) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vignali DAA, Collison LW, Workman CJ, How regulatory T cells work, Nat. Rev. Immunol 8 (2008) 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ohue Y, Nishikawa H, Regulatory T (Treg) cells in cancer: can Treg cells be a new therapeutic target? Cancer Sci. 110 (2019) 2080–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lu L, Barbi J, Pan F, The regulation of immune tolerance by FOXP3, Nat. Rev. Immunol 17 (2017) 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tanaka A, Sakaguchi S, Targeting Treg cells in cancer immunotherapy, Eur. J. Immunol 49 (2019) 1140–1146. [DOI] [PubMed] [Google Scholar]
- [57].Rudra D, et al. , Transcription factor Foxp3 and its protein partners form a complex regulatory network, Nat. Immunol 13 (2012) 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Casares N, et al. , A peptide inhibitor of FOXP3 impairs regulatory T cell activity and improves vaccine efficacy in mice, J. Immunol 185 (2010) 5150–5159. [DOI] [PubMed] [Google Scholar]
- [59].Lozano T, et al. , Blockage of FOXP3 transcription factor dimerization and FOXP3/AML1 interaction inhibits T regulatory cell activity: sequence optimization of a peptide inhibitor, Oncotarget 8 (2017) 71709–71724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ono M, et al. , Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1, Nature 446 (2007) 685–689. [DOI] [PubMed] [Google Scholar]
- [61].Lozano T, et al. , Inhibition of FOXP3/NFAT interaction enhances T cell function after TCR stimulation, J. Immunol 195 (2015) 3180–3189. [DOI] [PubMed] [Google Scholar]
- [62].Wu Y, et al. , FOXP3 controls regulatory T cell function through cooperation with NFAT, Cell 126 (2006) 375–387. [DOI] [PubMed] [Google Scholar]
- [63].MacDonald BT, Tamai K, He X, Wnt/β-catenin signaling: components, mechanisms, and diseases, Dev. Cell 17 (2009) 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Muzny DM, et al. , Comprehensive molecular characterization of human colon and rectal cancer, Nature 487 (2012) 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Goldsberry WN, Londoño A, Randall TD, Norian LA, Arend RC, A review of the role of wnt in cancer immunomodulation, Cancers (Basel) 11 (1–19) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Li X, et al. , WNT/β-catenin signaling pathway regulating T cell-inflammation in the tumor microenvironment, Front. Immunol 10 (2019) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].van Loosdregt J, et al. , Canonical Wnt signaling negatively modulates regulatory T cell function, Immunity 39 (2013) 298–310. [DOI] [PubMed] [Google Scholar]
- [68].Kawamoto SA, et al. , Analysis of the interaction of BCL9 with β-catenin and development of fluorescence polarization and surface plasmon resonance binding assays for this interaction, Biochemistry 48 (2009) 9534–9541. [DOI] [PubMed] [Google Scholar]
- [69].Valenta T, Hausmann G, K Basler, The many faces and functions of Î 2-catenin, EMBO J. 31 (2012) 2714–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Krieghoff E, Behrens J, Mayr B, Nucleo-cytoplasmic distribution of β-catenin is regulated by retention, J. Cell Sci 119 (2006) 1453–1463. [DOI] [PubMed] [Google Scholar]
- [71].Ali AM, Atmaj J, Van Oosterwijk N, Groves MR, Dömling A, Stapled peptides inhibitors: a new window for target drug discovery, Comput. Struct. Biotechnol. J 17 (2019) 263–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kawamoto SA, et al. , Design of triazole-stapled BCL9 α-helical peptides to target the β-catenin/B-cell CLL/lymphoma 9 (BCL9) protein-protein interaction, J. Med. Chem 55 (2012) 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Takada K, et al. , Targeted disruption of the BCL9/ -catenin complex inhibits oncogenic Wnt signaling, Sci. Transl. Med 4 (2012), 148ra117–148ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Feng M, et al. , Pharmacological inhibition of β-catenin/BCL9 interaction overcomes resistance to immune checkpoint blockades by modulating Treg cells, Sci. Adv 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cook KW, et al. , CCL20/CCR6-mediated migration of regulatory T cells to the Helicobacter pylori-infected human gastric mucosa, Gut 63 (2014) 1550–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Curiel TJ, et al. , Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival, Nat. Med 10 (2004) 942–949. [DOI] [PubMed] [Google Scholar]
- [77].Ezquerro IJ, et al. , A synthetic peptide from transforming growth factor β type III receptor inhibits liver fibrogenesis in rats with carbon tetrachloride liver injury, Cytokine 22 (2003) 12–20. [DOI] [PubMed] [Google Scholar]
- [78].Dotor J, et al. , Identification of peptide inhibitors of transforming growth factor beta 1 using a phage-displayed peptide library, Cytokine 39 (2007) 106–115. [DOI] [PubMed] [Google Scholar]
- [79].Wan YY, Flavell RA, ‘Yin-Yang’ functions of transforming growth factor-β and T regulatory cells in immune regulation, Immunol. Rev 220 (2007) 199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Fantini MC, et al. , Cutting edge: TGF-β induces a regulatory phenotype in CD4 + CD25 – T cells through Foxp3 induction and down-regulation of Smad7, J. Immunol 172 (2004) 5149–5153. [DOI] [PubMed] [Google Scholar]
- [81].Gil-Guerrero L, et al. , In vitro and in vivo down-regulation of regulatory T cell activity with a peptide inhibitor of TGF-β1, J. Immunol 181 (2008) 126–135. [DOI] [PubMed] [Google Scholar]
- [82].Llopiz D, et al. , Peptide inhibitors of transforming growth factor-β enhance the efficacy of antitumor immunotherapy, Int. J. Cancer 125 (2009) 2614–2623. [DOI] [PubMed] [Google Scholar]
- [83].Delgoffe GM, et al. , Regulatory T cell stability is maintained by a neuropilin-1: semaphorin-4a axis, Nature 501 (2013) 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kim YJ, et al. , Immunoglobulin Fc-fused, neuropilin-1-specific peptide shows efficient tumor tissue penetration and inhibits tumor growth via anti-angiogenesis, J. Control. Release 216 (2015) 56–68. [DOI] [PubMed] [Google Scholar]
- [85].Jung K, et al. , A neuropilin-1 antagonist exerts antitumor immunity by inhibiting the suppressive function of intratumoral regulatory T cells, Cancer Immunol. Res 8 (2020) 46–56. [DOI] [PubMed] [Google Scholar]
- [86].Santagata S, et al. , Targeting CXCR4 reverts the suppressive activity of T-regulatory cells in renal cancer, Oncotarget 8 (2017) 77110–77120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Di Maro S, et al. , Exploring the N-terminal region of C-X-C motif chemokine 12 (CXCL12): identification of plasma-stable cyclic peptides as novel, potent C-X-C chemokine receptor type 4 (CXCR4) antagonists, J. Med. Chem 59 (2016) 8369–8380. [DOI] [PubMed] [Google Scholar]
- [88].Domanska UM, et al. , A review on CXCR4/CXCL12 axis in oncology: no place to hide, Eur. J. Cancer 49 (2013) 219–230. [DOI] [PubMed] [Google Scholar]
- [89].Susek KH, Karvouni M, Alici E, Lundqvist A, The role of CXC chemokine receptors 1-4 on immune cells in the tumor microenvironment, Front. Immunol 9 (2018) 2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Hu W, Wang G, Huang D, Sui M, Xu Y, Cancer immunotherapy based on natural killer cells: current progress and new opportunities, Front. Immunol 10 (2019) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Chernysh S, et al. , Antiviral and antitumor peptides from insects, Proc. Natl. Acad. Sci. U. S. A 99 (2002) 12628–12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Chernysh S, Irina K, Irina A, Anti-tumor activity of immunomodulatory peptide alloferon-1 in mouse tumor transplantation model, Int. Immunopharmacol 12 (2012) 312–314. [DOI] [PubMed] [Google Scholar]
- [93].Bae S, et al. , The effect of alloferon on the enhancement of NK cell cytotoxicity against cancer via the up-regulation of perforin/granzyme B secretion, Immunobiology 218 (2013) 1026–1033. [DOI] [PubMed] [Google Scholar]
- [94].Fadda L, et al. , Peptide antagonism as a mechanism for NK cell activation, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 10160–10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM, M-1/M-2 macrophages and the Th1/Th2 paradigm, J. Immunol 164 (2000) 6166–6173. [DOI] [PubMed] [Google Scholar]
- [96].Italiani P, Boraschi D, From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation, Front. Immunol 5 (2014) 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sica A, Mantovani A, Macrophage plasticity and polarization: in vivo veritas, J. Clin. Invest 122 (2012) 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P, Tumour-associated macrophages as treatment targets in oncology, Nat. Rev. Clin. Oncol 14 (2017) 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Murray PJ, et al. , Macrophage activation and polarization: nomenclature and experimental guidelines, Immunity 41 (2014) 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Qian BZ, Pollard JW, Macrophage diversity enhances tumor progression and metastasis, Cell 141 (2010) 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Sawa-Wejksza K, Kandefer-Szerszeń M, Tumor-associated macrophages as target for antitumor therapy, Arch. Immunol. Ther. Exp 66 (97–111) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Cieslewicz M, et al. , Targeted delivery of proapoptotic peptides to tumor-associated macrophages improves survival, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 15919–15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Kakoschky B, et al. , Selective targeting of tumor associated macrophages in different tumor models, PLoS One 13 (2018) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Pang L, et al. , Surface modification of polymeric nanoparticles with M2pep peptide for drug delivery to tumor-associated macrophages, Pharm. Res 36 (2019) 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Li M, et al. , Remodeling tumor immune microenvironment via targeted blockade of PI3K-γ and CSF-1/CSF-1R pathways in tumor associated macrophages for pancreatic cancer therapy, J. Control. Release 321 (2020) 23–35. [DOI] [PubMed] [Google Scholar]
- [106].Scodeller P, et al. , Precision targeting of tumor macrophages with a CD206 binding peptide, Sci. Rep 7 (2017) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Rady I, Siddiqui IA, Rady M, Mukhtar H, Melittin, a major peptide component of bee venom, and its conjugates in cancer therapy, Cancer Lett. 402 (2017) 16–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Lee C, Bae SJS, Joo H, Bae H, Melittin suppresses tumor progression by regulating tumorassociated macrophages in a Lewis lung carcinoma mouse model, Oncotarget 8 (2017) 54951–54965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lee C, et al. , Targeting of M2-like tumor-associated macrophages with a melittin-based pro-apoptotic peptide, J. Immunother. Cancer 7 (2019) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Jaynes JM, et al. , Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses, Sci. Transl. Med 12 (2020) 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Movahedi K, et al. , Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes, Cancer Res. 70 (2010) 5728–5739. [DOI] [PubMed] [Google Scholar]
- [112].Hong HY, et al. , Phage display selection of peptides that home to atherosclerotic plaques: IL-4 receptor as a candidate target in atherosclerosis, J. Cell. Mol. Med 12 (2008) 2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Vadevoo SMP, et al. , IL4 receptor-targeted proapoptotic peptide blocks tumor growth and metastasis by enhancing antitumor immunity, Mol. Cancer Ther 16 (2017) 2803–2816. [DOI] [PubMed] [Google Scholar]
- [114].Portella L, et al. , Preclinical development of a novel class of CXCR4 antagonist impairing solid tumors growth and metastases, PLoS One 8 (2013) 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Mercurio L, et al. , Targeting CXCR4 by a selective peptide antagonist modulates tumor microenvironment and microglia reactivity in a human glioblastoma model, J. Exp. Clin. Cancer Res 35 (2016) 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wculek SK, et al. , Dendritic cells in cancer immunology and immunotherapy, Nat. Rev. Immunol 20 (2020) 7–24. [DOI] [PubMed] [Google Scholar]
- [117].Gardner Á, de Mingo Pulido A, Ruffell B, Dendritic cells and their role in immunotherapy, Front. Immunol 11 (2020) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Yan Z, et al. , A novel peptide targeting Clec9a on dendritic cell for cancer immunotherapy, Oncotarget 7 (2016) 40437–40450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Caminschi I, et al. , The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement, Blood 112 (2008) 3264–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sancho D, et al. , Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin, J. Clin. Invest 118 (2008) 2098–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Huysamen C, Willment JA, Dennehy KM, Brown GD, CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes, J. Biol. Chem 283 (2008) 16693–16701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].van der Aa E, van Montfoort N, Woltman AM, BDCA3+CLEC9A+ human dendritic cell function and development, Semin. Cell Dev. Biol 41 (2015) 39–48. [DOI] [PubMed] [Google Scholar]
- [123].Cote R, Lynn Eggink L, Kenneth Hoober J, CLEC receptors, endocytosis and calcium signaling, AIMS Allergy Immunol. 1 (2017) 207–231. [Google Scholar]
- [124].Shumilina E, Huber SM, Lang F, Ca2+ signaling in the regulation of dendritic cell functions, Am. J. Physiol 300 (2011). [DOI] [PubMed] [Google Scholar]
- [125].Eggink LL, Hoober JK, A biologically active peptide mimetic of N-acetylgalactosamine/galactose, BMC Res. Notes 2 (2009) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Eggink LL, Roby KF, Cote R, Kenneth Hoober J, An innovative immunotherapeutic strategy for ovarian cancer: CLEC10A and glycomimetic peptides, J. Immunother. Cancer 6 (2018) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Mo Z, et al. , New peptide MY1340 revert the inhibition effect of VEGF on dendritic cells differentiation and maturation via blocking VEGF-NRP-1 axis and inhibit tumor growth in vivo, Int. Immunopharmacol 60 (2018) 132–140. [DOI] [PubMed] [Google Scholar]
- [128].Rotonda J, et al. , Production of vascular endothelial growth factor by human tumors inhibits the functional maturation of dendritic cells, Nat. Med 2 (1996) 619–625. [DOI] [PubMed] [Google Scholar]
- [129].Gabrilovich D, et al. , Vascular endothelial growth factor inhibits the development of dendritic cells and dramatically affects the differentiation of multiple hematopoietic lineages in vivo, Blood 92 (1998) 4150–4166. [PubMed] [Google Scholar]
- [130].Gabrilovich DI, Ishida T, Nadaf S, Ohm JE, Carbone DP, Antibodies to vascular endothelial growth factor enhance the efficacy of cancer immunotherapy by improving endogenous dendritic cell function, Clin. Cancer Res 5 (1999) 2963–2970. [PubMed] [Google Scholar]
- [131].Oussa NAE, et al. , VEGF requires the receptor NRP-1 to inhibit lipopolysaccharide-dependent dendritic cell maturation, J. Immunol 197 (2016) 3927–3935. [DOI] [PubMed] [Google Scholar]
- [132].Kumar V, Patel S, Tcyganov E, Gabrilovich DI, The nature of myeloid-derived suppressor cells in the tumor microenvironment, Trends Immunol. 37 (2016) 208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Hansen GL, Gaudernack G, Brunsvig PF, Cvancarova M, Kyte JA, Immunological factors influencing clinical outcome in lung cancer patients after telomerase peptide vaccination, Cancer Immunol. Immunother 64 (2015) 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Limagne E, et al. , Accumulation of MDSC and Th17 cells in patients with metastatic colorectal cancer predicts the efficacy of a FOLFOX-bevacizumab drug treatment regimen, Cancer Res. 76 (2016) 5241–5252. [DOI] [PubMed] [Google Scholar]
- [135].Gonda K, et al. , Myeloid-derived suppressor cells are increased and correlated with type 2 immune responses, malnutrition, inflammation, and poor prognosis in patients with breast cancer, Oncol. Lett 14 (2017) 1766–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Qin H, et al. , Generation of a new therapeutic peptide that depletes myeloid-derived suppressor cells in tumor-bearing mice, Nat. Med 20 (2014) 676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Fleming V, et al. , Targeting myeloid-derived suppressor cells to bypass tumor-induced immunosuppression, Front. Immunol 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G, Immunogenic cell death in cancer and infectious disease, Nat. Rev. Immunol 17 (2017) 97–111. [DOI] [PubMed] [Google Scholar]
- [139].Golden EB, Apetoh L, Radiotherapy and immunogenic cell death, Semin. Radiat. Oncol 25 (2015) 11–17. [DOI] [PubMed] [Google Scholar]
- [140].Zhou H, et al. , The oncolytic peptide LTX-315 triggers immunogenic cell death, Cell Death Dis. 7 (2016) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Rekdal Ø, et al. , Relative spatial positions of tryptophan and cationic residues in helical membrane-active peptides determine their cytotoxicity, J. Biol. Chem 287 (2012) 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Zhou H, et al. , The oncolytic peptide LTX-315 kills cancer cells through Bax/Bak-regulated mitochondrial membrane permeabilization, Oncotarget 6 (2015) 26599–26614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Camilio KA, Berge G, Ravuri CS, Rekdal Ø, Sveinbjørnsson B, Complete regression and systemic protective immune responses obtained in B16 melanomas after treatment with LTX-315, Cancer Immunol. Immunother 63 (2014) 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Yamazaki T, et al. , The oncolytic peptide LTX-315 overcomes resistance of cancers to immunotherapy with CTLA4 checkpoint blockade, Cell Death Differ. 23 (2016) 1004–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Camilio KA, et al. , Combining the oncolytic peptide LTX-315 with doxorubicin demonstrates therapeutic potential in a triple-negative breast cancer model, Breast Cancer Res. 21 (2019) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Spicer JF, et al. , A phase I/II study of the oncolytic peptide LTX-315 combined with checkpoint inhibition generates de novo T-cell responses and clinical benefit in patients with advanced solid tumors, J. Clin. Oncol 36 (2018) 3094. [Google Scholar]
- [147].Eike LM, Mauseth B, Camilio KA, Rekdal Ø, Sveinbjørnsson B, The cytolytic amphipathic β(2,2)-amino acid LTX-401 induces DAMP release in melanoma cells and causes complete regression of B16 melanoma, PLoS One 11 (2016) 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Zhou H, et al. , The oncolytic compound LTX-401 targets the Golgi apparatus, Cell Death Differ. 23 (2016) 2031–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Xie W, et al. , Tumor lysis with LTX-401 creates anticancer immunity, Oncoimmunology 8 (2019) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Mauseth B, et al. , The novel oncolytic compound LTX-401 induces antitumor immune responses in experimental hepatocellular carcinoma, Mol. Ther 14 (2019) 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Zhang W, et al. , Advances in anti-tumor treatments targeting the CD47/SIRPα Axis, Front. Immunol 11 (2020) 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Martinez-Torres AC, et al. , CD47 agonist peptides induce programmed cell death in refractory chronic lymphocytic leukemia B cells via PLCγ1 activation: evidence from mice and humans, PLoS Med. 12 (2015) 1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Uscanga-Palomeque AC, et al. , CD47 agonist peptide PKHB1 induces immunogenic cell death in T-cell acute lymphoblastic leukemia cells, Cancer Sci. 110 (2019) 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Leclair P, Kim MJ, Lim CJ, Peptide analogues PKHB1 and 4N1K induce cell death through CD47-independent mechanisms, Cancer Sci. 111 (2020) 1028–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Faye A, Poyet JL, Targeting AAC-11 in cancer therapy, Expert Opin. Ther. Targets 14 (2010) 57–65. [DOI] [PubMed] [Google Scholar]
- [156].Rigou P, et al. , The antiapoptotic protein AAC-11 interacts with and regulates Acinus-mediated DNA fragmentation, EMBO J. 28 (2009) 1576–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Jagot-Lacoussiere L, Kotula E, Villoutreix BO, Bruzzoni-Giovanelli H, Poyet JL, A cell-penetrating peptide targeting AAC-11 specifically induces cancer cells death, Cancer Res. 76 (2016) 5479–5490. [DOI] [PubMed] [Google Scholar]
- [158].Pasquereau-Kotula E, Habault J, Kroemer G, Poyet JL, The anticancer peptide RT53 induces immunogenic cell death, PLoS One 13 (2018) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Pernot M, Vanderesse R, Frochot C, Guillemin F, Barberi-Heyob M, Stability of peptides and therapeutic success in cancer, Expert Opin. Drug Metab. Toxicol 7 (2011) 793–802. [DOI] [PubMed] [Google Scholar]
- [160].Araste F, et al. , Peptide-based targeted therapeutics: focus on cancer treatment, J. Control. Release 292 (2018) 141–162. [DOI] [PubMed] [Google Scholar]
- [161].Werle M, Bernkop-Schnürch A, Strategies to improve plasma half life time of peptide and protein drugs, Amino Acids 30 (2006) 351–367. [DOI] [PubMed] [Google Scholar]
- [162].Di L, Strategic approaches to optimizing peptide ADME properties, AAPS J. 17 (2015) 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Lee S, et al. , Self-assembling peptides and their application in the treatment of diseases, Int. J. Mol. Sci 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Lima e Silva R, et al. , Tyrosine kinase blocking collagen IV–derived peptide suppresses ocular neovascularization and vascular leakage, Sci. Transl. Med 9 (2017) eaai8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Drucker DJ, Advances in oral peptide therapeutics, Nat. Rev. Drug Discov 19 (2020) 277–289. [DOI] [PubMed] [Google Scholar]
- [166].Qin S, et al. , Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4, Mol. Cancer 18 (2019) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Papalexi E, Satija R, Single-cell RNA sequencing to explore immune cell heterogeneity, Nat. Rev. Immunol 18 (2018) 35–45. [DOI] [PubMed] [Google Scholar]
- [168].Gomes T, Teichmann SA, Talavera-López C, Immunology driven by large-scale single-cell sequencing, Trends Immunol. 40 (2019) 1011–1021. [DOI] [PubMed] [Google Scholar]
- [169].Masucci MT, Minopoli M, Carriero MV, Tumor associated neutrophils. their role in tumorigenesis, metastasis, prognosis and therapy, Front. Oncol 9 (2019) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Liu T, et al. , Cancer-associated fibroblasts: an emerging target of anti-cancer immunotherapy, J. Hematol. Oncol 12 (2019) 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Sahai E, et al. , A framework for advancing our understanding of cancer-associated fibroblasts, Nat. Rev. Cancer 20 (2020) 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Yi M, et al. , Synergistic effect of immune checkpoint blockade and anti-angiogenesis in cancer treatment, Mol. Cancer 18 (2019) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Huang Y, Goel S, Duda DG, Fukumura D, Jain RK, Vascular normalization as an emerging strategy to enhance cancer immunotherapy, Cancer Res. 73 (2013) 2943–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Schaaf MB, Garg AD, Agostinis P, Defining the role of the tumor vasculature in antitumor immunity and immunotherapy, Cell Death Dis. 9 (2018) 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Mirando AC, et al. , Regulation of the tumor immune microenvironment and vascular normalization in TNBC murine models by a novel peptide, Oncoimmunology 9 (2020) 1760685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Rosca V, et al. , Anti-Angiogenic peptides for cancer therapeutics, Curr. Pharm. Biotechnol 12 (2011) 1101–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Karagiannis ED, Popel AS, A systematic methodology for proteome-wide identification of peptides inhibiting the proliferation and migration of endothelial cells, Proc. Natl. Acad. Sci. U. S. A 105 (2008) 13775–13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Lau JL, Dunn MK, Therapeutic peptides: historical perspectives, current development trends, and future directions, Bioorgan. Med. Chem 26 (2018) 2700–2707. [DOI] [PubMed] [Google Scholar]
- [179].Esteva FJ, Hubbard-Lucey VM, Tang J, Pusztai L, Immunotherapy and targeted therapy combinations in metastatic breast cancer, Lancet Oncol. 20 (2019) e175–e186. [DOI] [PubMed] [Google Scholar]