Abstract
Genome-wide association studies (GWAS) of schizophrenia have strongly implicated a risk locus in close proximity to the gene for miR-137. While there are candidate single-nucleotide polymorphisms (SNPs) with functional implications for the microRNA’s expression encompassed by the common haplotype tagged by rs1625579, there are likely to be others, such as the variable number tandem repeat (VNTR) variant rs58335419, that have no proxy on the SNP genotyping platforms used in GWAS to date. Using whole-genome sequencing data from schizophrenia patients (n = 299) and healthy controls (n = 131), we observed that the MIR137 4-repeats VNTR (VNTR4) variant was enriched in a cognitive deficit subtype of schizophrenia and associated with altered brain morphology, including thicker left inferior temporal gyrus and deeper right postcentral sulcus. These findings suggest that the MIR137 VNTR4 may impact neuroanatomical development that may, in turn, influence the expression of more severe cognitive symptoms in patients with schizophrenia.
Keywords: MIR137 VNTR, schizophrenia, genome sequencing, neuroanatomy, cognitive deficit
Introduction
Schizophrenia is a chronic psychiatric disorder characterized by a combination of positive, negative, and cognitive symptoms. These typically emerge during late adolescence or early adulthood and are attributed to neurodevelopmental changes that alter neural connectivity and transmission.1 Schizophrenia has a very significant genetic component with heritability estimated to be around 80%2 and a lifetime risk of 0.3%–0.7%.1 Large case-control genome-wide association studies (GWAS) have made significant progress in revealing the common variation broadly distributed across the genome (with over 150 significant hits) in association with schizophrenia.3–5 While this has implicated many genes and biological pathways, the functional variants and associated genes are often obscured by the genomic complexity. There is also an unknown heritable contribution from common structural variation, including genomic insertions and deletions (INDELs) and minisatellites, such as variable number tandem repeats (VNTR) that are not captured by the major genome-wide single-nucleotide polymorphism (SNP) genotyping platforms. In some cases, these may correspond to known association regions for common SNPs and even be causal variants in linkage disequilibrium (LD) with proxy SNPs used to map the loci, as is the case for the ASMT3 VNTR.6 In other circumstances, they may not be in LD with any SNPs represented in the large GWAS and may instead be an independent and unquantified component of missing heritability.
One candidate for this circumstance is the MIR137 VNTR. This 15-nucleotide repeat variant (rs58335419) is in close proximity to the miR-137 precursor sequence, including the wild-type variant (VNTRw) with 3 repeats, and 2 relatively common minor alleles with 4 repeats (VNTR4) and 5 repeats (VNTR5). While these variants do not appear to be in LD with the established genome-wide-associated SNPs,7,8 their close proximity to pre-miR-137 (6 bp upstream) suggests that it may directly influence the maturation of the microRNA (miRNA). This was initially found in a melanoma study, where the VNTR was observed to modulate the processing and function of the miRNA in melanocytes.9 The VNTR is also thought to be part of an internal promoter domain for MIR137 (Imir137), which can reportedly modulate expression levels of miR-137 in an allele-specific and stimulus-inducible manner by the synaptic plasticity-associated repressor element-1 silencing transcription factor (REST).7 In at least one previous study, miR-137 expression levels were significantly decreased in SH-SY5Y cells expressing VNTR4 and VNTR8 variants as compared to the VNTRw-containing cells.10
MIR137 generates miR-137, a miRNA that is enriched in the brain and regulates pathways involved in neurogenesis, neural development, and maturation, as well as synaptogenesis, all of which are critical in etiology of schizophrenia.11 miR-137 was shown to promote neuronal differentiation in various stem cells, including adult mouse and human neural stem cells,12 while inhibiting proliferation of embryonic neuronal stem cell through the regulation of target genes,13 suggesting a miR-137 role in the modulation of neuronal transition from pluripotentency to differentiated states. Overexpression of miR-137 has been shown to alter the expression of the genes implicated in schizophrenia pathways, including major histocompatibility complex, calcium channels and synapses.14 Evidence from postmortem transcriptomic studies has also shown that miR-137 expression could be dysregulated in brain. For example, miR-137 levels were significantly reduced in the schizophrenia-risk haplotype carriers in the human dorsolateral prefrontal cortex.15 Analysis of the miR-137 targets genes in the brain transcriptomes has shown that the majority of them have peak expression before the adulthood, suggesting the possible contribution of these genes to the disruption of cognitive function and gray matter density occurring over the lifespan of patients with schizophrenia.16
The implication of MIR137 in the pathology of schizophrenia is also supported by GWAS studies through its position in one of the more clearly demarcated risk alleles that are strongly associated with the disorder.3,17 Five miR-137 target genes, including TCF4, CSMD1, CACNA1C, C10orf26, and ZNF804A were also identified as schizophrenia-associated loci in GWAS18,19 and many other targets were enriched with variants that were nominally significant,17 collectively supporting a role for MIR137 in the etiology of schizophrenia. The risk haplotype has also been associated with several cognitive and brain endophenotypes, including performance in verbal episodic memory and attention, altered fronto-amygdala connectivity, and reduced white matter integrity.20–23 These findings suggest that the miRNA may regulate pathways associated with neural connectivity and its altered function could contribute to the manifestation of cognitive symptoms in carriers with schizophrenia. While MIR137 is a strong candidate for the causal variation associated with schizophrenia at this locus, its proximity to other genes, including DPYD, which has also been implicated in neurological disorders, suggests that there is some uncertainty about the fine mapping that remains to be resolved.
Given that the MIR137 VNTR has been shown to alter the expression of miR-1377,10 and that these effects appear to be independent of the genome-wide-associated SNPs, we hypothesized that this variant may have phenotypic implications for brain development and cognitive function in schizophrenia. In support of this, we observed that the VNTR4MIR137 variant was significantly enriched in individuals with a cognitive deficit (CD) subtype of schizophrenia compared to healthy controls (HC). In a subset with brain imaging data, we also show that the VNTR4 is associated with significantly thicker gray matter in the left inferior temporal gyrus and deeper right postcentral sulci compared to individuals with the wild-type variant.
Methods
Participants
Data from participants meeting International Classification of Disease (ICD)-10 criteria24 for schizophrenia or schizoaffective disorder (n = 299) and HC (n = 131) were obtained from the Australian Schizophrenia Research Bank (ASRB), an established register of clinical, cognitive, neuroimaging, and genetic data collected by scientific collaborators across 5 Australian states and territories.25 Diagnoses were confirmed using the OPCRIT algorithm26 applied to interviewer ratings on the Diagnostic Interview for Psychosis (DIP).27 All participants were fluent English speakers and had no history of an organic brain disorder, brain injury accompanied by greater than 24 h of amnesia, mental retardation defined as an IQ <70, movement disorder, current substance dependence, or electroconvulsive therapy in the preceding 6 months. For this study, all participants were of European ancestry. The healthy participants additionally had no personal or family history of psychotic disorder in their first-degree biological relatives. The majority of the cases were using typical or atypical antipsychotics, mood stabilizers, and/or antidepressants; however, no dosage-equivalent information was collected by the ASRB. Sociodemographic and clinical characteristics, as well as VNTR frequencies of the cognitive subgroups are summarized in table 1.
Table 1.
Genetic, sociodemographic, clinical, and cognitive characteristics of study participants
| HC (n = 131) | CS (n = 150) | CD (n = 149) | Statistics | Direction | |
|---|---|---|---|---|---|
| VNTR4 (n) | 12 | 21 | 27 | ||
| Age, mean (SD) | 39.08 (13.32) | 40.10 (11.19) | 39.08 (9.78) | F(2,281.41) = 0.414, P = .661 | — |
| Sex, n (M/F) | 68/68 | 96/55 | 110/46 | X 2 (2) = 13.246, P = .001 | |
| Years of education (SD) | 15.07 (3.16) | 13.91 (2.71) | 12.23 (2.57) | F(2,284.18) = 37.402, P < .001 | HC > CS**, HC > CD***, CS > CD*** |
| Handedness, n (L/R) | 19/117 | 12/139 | 15/141 | X 2(2) = 2.943, P = .230 | — |
| WTAR, mean (SD) | 105.82 (11.48) | 106.15 (8.70) | 89.82 (14.93) | F(2,276.92) = 73.859, P < .001 | HC > CD***, CS > CD*** |
| WASI, mean (SD) | 114.23 (12.79) | 112.15 (11.43) | 91.22 (11.59) | F(2,439) = 171.576, P < .001 | HC > CD***, CS > CD*** |
| Positive Symptoms Scores (DIP items) | |||||
| Lifetime, mean (SD) | — | 10.93 (4.30) | 9.02 (3.97) | t(246) = 3.654, P < .001 | CS > CD |
| Present state, mean (SD) | — | 2.70 (3.83) | 3.40 (3.57) | t(245) = 1.474, P = .142 | |
| Negative Symptoms Scores (SANS), mean (SD) | — | 19.70 (16.63) | 33.98 (20.03) | t(295) = 6.666, P < .001 | CD > CS |
| Illness duration, mean (SD) | — | 16.31 (10.54) | 16.02 (9.15) | t(296.23) = 0.259, P = .796 | — |
| Typical antipsychotics, n (%) | — | 7 | 17 | X 2(1) = 3.295, P = .114 | — |
| Atypical antipsychotics, n (%) | — | 127 | 128 | X 2(1) = 1.004, P = 1.000 | — |
| Antidepressants, n (%) | — | 47 | 44 | X 2(1) = 0.170, P = .738 | — |
| Mood stabilizers, n (%) | — | 24 | 19 | X 2(2) = 0.913, P = .440 | — |
Note: HC, healthy controls; CS, cognitively spared schizophrenia cases; CD, schizophrenia cases with cognitive deficits; VNTR4, 4-repeats variable number tandem repeat; M/F, males/females; L/R, left/right; DIP, Diagnostic Interview for Psychosis; SANS, Scale for the Assessment of Negative Symptoms; WTAR, Wechsler Test of Adult Reading; WASI, Wechsler Abbreviated Scale of Intelligence.
*P < .05; **P < .01; ***P < .001.
Clinical Assessments
Ratings for positive symptoms were indexed using present state and lifetime ratings of hallucination (items 49–53) and delusion items (items 58–64) from the DIP27 and negative symptoms were indexed using the Scale for Assessment of Negative Symptoms.28 Premorbid and current intelligence levels were assessed with the Wechsler Test of Adult Reading29 and the Wechsler Abbreviated Scale of Intelligence,30 respectively, and handedness was assessed with the Edinburgh Handedness Inventory.31
Cognitive Clustering
Grade of Membership analysis was performed with the DSIGoM software (beta version 1.01, Decision Systems, 1999; http://www.dsisoft.com) using cognitive performance data collected from schizophrenia patients only, as detailed in our previous work32 (see supplementary material). Use of this method applied previously to ASRB participants partitioned the clinical cases into groups presenting severe cognitive deficits (CD; n = 149) relative to those with relatively spared cognitive function (CS; n = 150) as described in Green et al.32 Sociodemographic and clinical characteristics, as well as VNTR frequencies, of the cognitive subgroups are summarized in table 1.
DNA Sequencing
Whole-genome sequencing was performed on the ASRB cohort for 299 schizophrenia cases (comprising 149 CD and 150 CS cases) and 131 HC, at the Garvan Institute of Medical Research: Kinghorn Centre for Clinical Genomics (Darlinghurst, New South Wales, Australia). A 20-ml blood sample collected at the time of the clinical interview was used to extract DNA. High-integrity genomic DNA from peripheral blood mononuclear cells (1 μg) was sequenced using the Illumina HiSeq X platform as described33 with 2 × 150 base pair paired-end reads. The sequencing coverage was a minimum mean yield of 30×. The Phred quality score of 75% of bases was greater than 30.
Variant Calling
Raw reads (FASTQ) were aligned to the hg19 reference using Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) before compression, sorting, and indexing with Samtools.34 Picard tools (http://broadinstitute.github.io/picard/) were then used to mark and remove PCR duplicates. Quality score recalibration and indel realignment were then carried out using the GATK3 BaseRecalibrator and IndelRealigner, respectively.35,36 Variants within the region defined by Ripke et al3 were called using the HaplotypeCaller, followed by joint genotyping with GenotypeGVCFs. Variant calling was conducted using at least 20 reads per sample covering the VNTR site with a minimum of 4 reads per allele. We used best practices as per GATK guidelines.37 VCF files were converted into a matrix with zygosity for each allele. Principal component analysis was performed to adjust for the population stratification and was used to remove non-European samples. VNTR4 carriers included 48 cases (CD = 27; CS = 21) and 12 controls and noncarriers included 251 cases (CD = 122; CS = 129) and 119 controls.
Magnetic Resonance Imaging Acquisition and Preprocessing
Participants with available genetic, cognitive, and magnetic resonance imaging (MRI) data included 174 clinical participants with a diagnosis of schizophrenia (n = 155) or schizoaffective disorder (n = 19; together referred to as the schizophrenia group) and 97 HC. Among the schizophrenia cases, 106 were classified as CS, and 68 classified as CD (see table 2). Details of MRI scans and processing are provided in the supplementary material.
Table 2.
Sociodemographic, clinical characteristics, genotype, and allelic frequencies for the cognitive subgroups with magnetic resonance imaging scans
| HC (n = 97) | CS (n = 106) | CD (n = 68) | Statistics | Direction | |
|---|---|---|---|---|---|
| Age, mean (SD) | 41.87 (13.41) | 39.97 (11.16) | 37.78 (8.16) | Welch = 3.104, df = 2,175.13, P = .047, η p 2 = 0.019 | ns |
| Sex, n (M/F) | 47/50 | 72/34 | 46/19 | X 2 = 12.054, df = 2, P = .002 | |
| Years of education (SD) | 15.11 (2.92) | 13.79 (2.81) | 12.32 (2.42) | F = 20.562, df = 2,268, P < .001, η p 2 = 0.127 | HC > CS**, CD***, CS > CD** |
| Handedness, n (L/R) | 16/81 | 9/97 | 8/60 | X 2 = 3.049, df = 2, P = .218 | |
| WTAR, mean (SD) | 105.54 (10.82) | 106.58 (9.03) | 92.40 (14.17) | Welch = 27.485, df = 2,147.88, P < .001, η p 2 = 0.223 | HC, CS > CD*** |
| WASI, mean (SD) | 115.81 (11.43) | 113.12 (9.76) | 94.32 (11.39) | F = 89.720, df = 2,268, P < .001, η p 2 = 0.395 | HC, CS > CD*** |
| Positive Symptoms Scores (DIP items) | |||||
| Lifetime, mean (SD) | — | 10.89 (4.31) | 8.93 (3.84) | t = 2.748, df = 136, P = .007, d = 0.48 | |
| Present state, mean (SD) | — | 2.48 (3.44) | 3.54 (3.37) | t = 1.803, df = 136, P = .074, d = 0.31 | |
| Negative Symptoms Scores (SANS), mean (SD) | — | 18.15 (14.81) | 29.80 (18.38) | t = 4.453, df = 161, P < .001, d = 0.70 | |
| Illness duration, mean (SD) | — | 15.92 (10.57) | 14.38 (8.76) | t = 1.044, df = 160.93, P = .298, d = 0.16 | |
| Typical antipsychotics, n (%) | — | 8 (8%) | 4 (6%) | X 2 = 0.778, df = 1, P = .418 | |
| Atypical antipsychotics, n (%) | — | 87 (82%) | 56 (82%) | X 2 = 0.641, df = 1, P = 1.000 | |
| Antidepressants, n (%) | — | 32 (30%) | 19 (28%) | X 2 = 1.161, df = 1, P = .531 | |
| Mood stabilizers, n (%) | — | 12 (11%) | 7 (10%) | X 2 = 0.126, df = 1, P = 1.000 | |
| Scan sites, n | X 2 = 29.707, df = 8, P < .001 | ||||
| Brisbane | 19 | 41 | 36 | ||
| Melbourne | 45 | 26 | 11 | ||
| Newcastle | 10 | 7 | 4 | ||
| Perth | 9 | 10 | 5 | ||
| Sydney | 14 | 22 | 12 | ||
| Genotypes, n (%) | X 2 = 10.710, df = 4, P = .030 | ||||
| GG | 10 (10%) | 1 (1%) | 2 (3%) | ||
| GT | 30 (31%) | 33 (31%) | 23 (34%) | ||
| TT | 57 (59%) | 72 (68%) | 43 (63%) | ||
| VNTR4, present/absent (% present) | 12/85 (14%) | 18/88 (20%) | 18/50 (36%) | X 2 = 5.516, df = 2, P = .063 | |
| Imaging | |||||
| Total GMV, mm3 (SD) | 686.82 (71.86) | 705.09 (77.86) | 682.56 (67.83) | ||
| Total WMV, mm3 (SD) | 535.48 (61.34) | 554.55 (63.91) | 544.90 (71.62) | ||
| Total CSF, mm3 (SD) | 325.25 (65.02) | 351.03 (64.98) | 349.29 (66.79) | ||
| TIV, mm3 (SD) | 1547.55 (144.41) | 1610.68 (153.61) | 1576.75 (153.17) |
Note: HC, healthy controls; SZ/SZA, clinical cases diagnosed with schizophrenia (SZ, n = 155) or schizoaffective disorder (SZA, n = 19); df, degrees of freedom; d, Cohen’s d; M/F, males/females; L/R, left/right; DIP, Diagnostic Interview for Psychosis; SANS, Scale for the Assessment of Negative Symptoms; WTAR, Wechsler Test of Adult Reading; WASI, Wechsler Abbreviated Scale of Intelligence; GMV, gray matter volume; WMV, white matter volume; CSF, cerebro-spinal fluid; TIV, total intracranial volume.
P < .05 are presented in bold typeface. *P < .05; **P < .01; ***P < .001.
Statistical Analysis
VNTR4 Enrichment Among Cognitive Subtypes of Schizophrenia
To test for the enrichment of VNTR4 among cognitive subtypes of schizophrenia as, well as in the diagnosis groups, a binomial logistic regression model was constructed and adjusted for sex, SNP rs1625579, and the first 3 principal components in 299 participants with schizophrenia (149 CD and 150 CS) compared to 131 HC. Wald test was used to calculate the P values for both models; analysis was performed using Statsmodels libraries from Python.38 The significance threshold was set at P < .05.
MIR137 VNTR Polymorphism in Association With Altered Brain Morphology
Descriptive statistics for the subsample of participants included in the neuroimaging analyses were performed using SPSS 24 (IBM, SPSS Inc.), with significance set at P < .05 (2 tailed). Indices of effect size (partial eta-squared, ηp2, and Cohen’s d) are reported. For neuroimaging analyses (CAT12), measures of total gray matter volume (GMV), white matter volume (WMV), and cerebro-spinal fluid (CSF) volumes were first extracted. To determine the main effects of the cognitive subgroups, of the presence of the VNTR4 and their potential interaction on these tissue-specific indices, a 3 (groups: HC vs CS vs CD) by 2 (VNTR4: present vs absent) MANCOVA was used (initial threshold P < .05). Where appropriate, this was followed by (within-group) univariate ANCOVAs with Bonferroni correction applied for the number of dependent variables (ie, GMV, WMV, and CSF, P < .017). Age, sex, SNP rs1625579 genotype (GG/GT/TT; to control its potential independent effect on brain morphology), and scanning location were entered as covariates for all imaging analysis. Total intracranial volume was also included as a covariate for whole-brain tissue-specific masks and voxel-based morphometry (VBM) but not for cortical thickness, gyrification, sulcal depth, analyses. A similar 2 (groups: HC vs SZ) by 2 (VNTR4: present vs absent) MANCOVA was conducted for reference, with results presented in supplementary material. A whole-brain 3 (HC vs CS vs CD) by 2 (VNTR4: present vs absent) ANCOVA was conducted on whole-brain volumes (VBM). To account for and exclude voxels related to potential artifacts or misclassified at the gray/white matter border, we applied an internal gray matter threshold of 0.2 for VBM analyses. Similar to VBM analyses, indices of cortical thickness, gyrification, and sulcal depth were used as dependent variables in a series of whole-brain ANCOVAs [P(FWEc) < .05]. For each index of brain morphology (whole-brain volume, cortical thickness, sulcal depth, and gyrification) the statistical inference was made separately using an initial voxel-level threshold of P < .001 uncorrected, to which a cluster-level family-wise error (FWE) correction was applied [P(FWEc) < .05]. Regions showing significant GMV differences were identified using the Neuromorphometrics atlas in DARTEL space, as implemented in CAT12 for VBM analyses, and using the Desikan–Killiany atlas39 for surface-based analyses. For reference, a series of similar 2 (groups: HC vs SZ) by 2 (VNTR4: present vs absent) ANCOVAs were performed for each index of brain morphology with results presented in supplementary material.
Results
The VNTR4 Is Associated With CD Subtypes of Schizophrenia
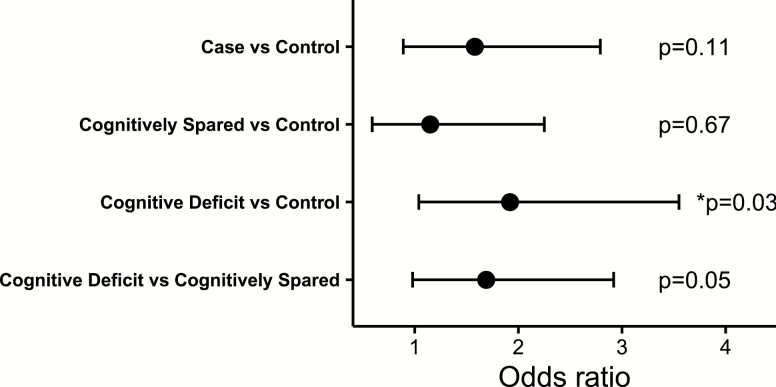
To investigate the potential impact of MIR137 VNTR length, we used DNA sequencing data obtained for individuals in the ASRB as this variant cannot be imputed from GWAS SNP data. Three alleles of the VNTR were observed, including VNTRw, VNTR4, and VNTR5 with a frequency of 0.91, 0.085, and 0.001, respectively. Participants carrying the rare VNTR5 variant were excluded from these analyses as we were underpowered to determine its association. The VNTR4 genotype was observed to be enriched within the CD subtype relative to the control group (P = .03, odds ratio [OR] = 1.92, 95% CI [1.04, 3.55]; figure 1). This was confirmed by the absence of difference between the CS group and controls (P = .67, OR = 1.15, 95% CI [0.59, 2.25]), CD and CS groups (P = .05, OR = 1.69, 95% CI [0.98, 2.92]), and combined case-control comparison (P = .11, OR = 1.58, 95% CI [0.89, 2.79]; figure 1).
Fig. 1.
Association of MIR137 variable number tandem repeat (VNTR) with cognitive deficit subtype. Logistic regression analyses showed that the 4-repeats VNTR (VNTR4) genotype was enriched in the cognitive deficit subtype (P = .03, odds ratio [OR] = 1.92) but not cognitively spared group (P = .67, OR = 1.15) as compared to the control group. No significant difference was observed between the cognitive deficit and cognitively spared groups (P = .05, OR = 1.69). Also there was no association between the VNTR4 and diagnostic groups (P = .11, OR= 1.58).
MIR137 VNTR Polymorphism Is Associated With Altered Cortex Morphology
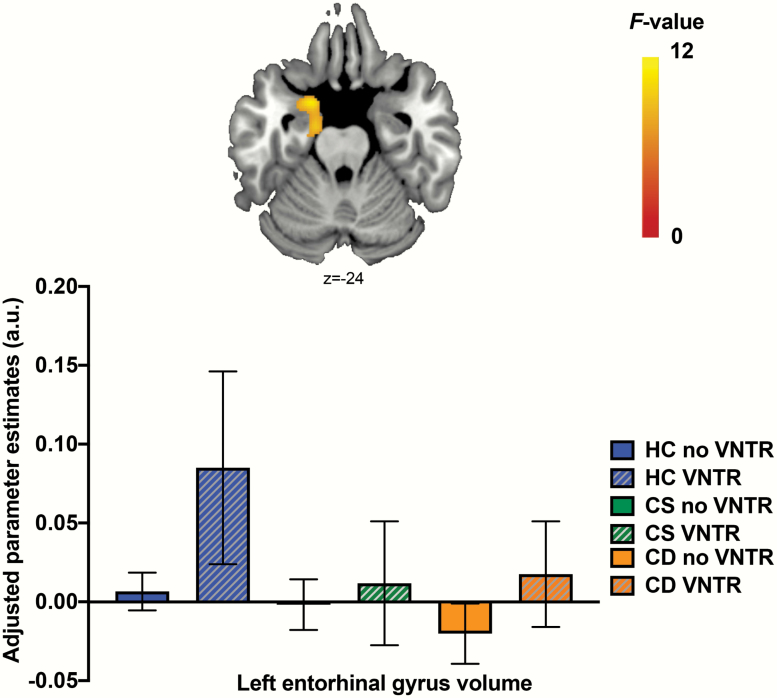
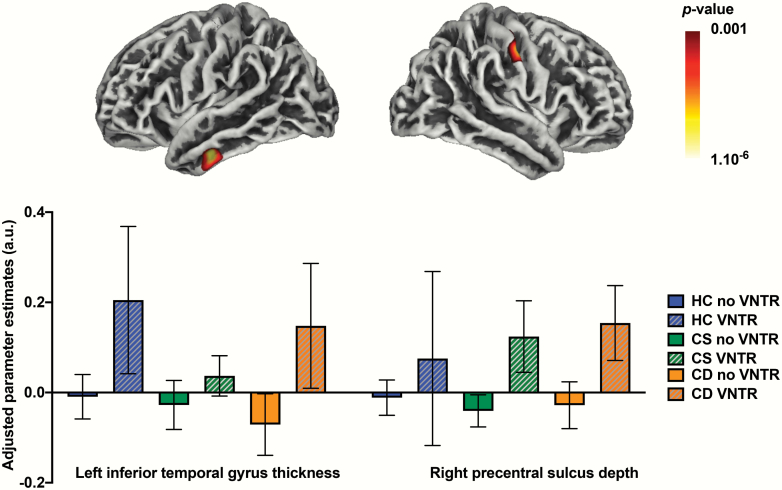
Sociodemographic and clinical characteristics of the imaging cohort, a subset of the main cohort, are summarized in table 2 and in supplementary material. The 3 × 2 ANCOVA on whole-brain tissue masks showed that there were no significant main effect of group (Wilk’s lambda = 0.955, F6,508 = 1.985, P = .066, ηp2 = 0.023), presence of the VNTR4 (Wilk’s lambda = 0.979, F3,254 = 1.846, P = .139, ηp2 = 0.021), or their interaction (Wilk’s lambda = 0.980, F6,508 = 0.841, P = .539, ηp2 = 0.010) on these measures. Whole-brain VBM analyses showed a main effect of cognitive subgroups on the volume of the left entorhinal gyrus (see table 3 and figure 2), with the HC group showing significantly larger volume than both the CS and CD group in this region (all post hoc P < .001). There were no significant effects of the presence of the VNTR4 or group-by-VNTR4 interaction on GMV. In addition, there were significant main effects of the presence of the VNTR4 on indices of cortical thickness and sulcal depth (see table 3 and figure 3), with VNTR4 carriers showing thicker left inferior/middle temporal gyrus (post hoc P < .001) and deeper right postcentral sulcus (post hoc P < .001) compared to noncarriers. There were no significant main effects of group or group-by-VNTR4 interaction on these indices, and there were no significant effects of group, presence of the VNTR4, or their interaction on indices of cortical gyrification.
Table 3.
Peaks of clusters showing significant main effect of groups (A. HC vs CS vs CD; B. HC vs SZ) or of presence of the VNTR4 on whole-brain indices of gray matter volume (VBM), cortical thickness, sulcal depth, and gyrification
| Hem | Cluster region | MNI coordinates | Cluster size | Peak F-statistic | Peak z-score | Cluster P(FWEc) | ||
|---|---|---|---|---|---|---|---|---|
| x | y | z | ||||||
| 3 (Groups: HC, CS, CD) × 2 (VNTR: present, absent) ANCOVA | ||||||||
| Main effect of cognitive subgroup—VBM | ||||||||
| L | Entorhinal gyrus | −21 | 4 | −27 | 796 | 12.27 | 4.31 | .032 |
| Main effect of VNTR—thickness | ||||||||
| L | Inferior/middle temporal gyrus | −50 | −15 | −29 | 453 | 19.24 | 4.15 | .032 |
| Main effect of VNTR—sulcal depth | ||||||||
| R | Postcentral gyrus | 52 | −15 | 42 | 994 | 21.09 | 4.35 | .005 |
| 48 | −16 | 53 | 14.32 | 3.55 | ||||
Note: Main peaks within the cluster of interest are in bold.
Hem, hemisphere; MNI, Montreal Neurologic Institute; FWEc, family-wise error correction for multiple comparisons at the cluster level; L, left; R, right; VBM: voxel-based morphometry; VNTR4, 4-repeats variable number tandem repeat; HC, healthy controls; CS, cognitively spared schizophrenia cases; CD, schizophrenia cases with cognitive deficits; SZ, clinical cases diagnosed with schizophrenia.
Fig. 2.
Whole-brain gray matter volume changes associated with cognitive subgroups of schizophrenia cases compared to healthy controls (HC). Volume of the left entorhinal gyrus was significantly larger in HC compared to both the cognitively spared (CS) and cognitive deficits (CD) groups (post hoc P < .001). a.u.: arbitrary units; color bar represents F-statistics; coordinates are reported in Montreal Neurologic Institute space; error bars represent 95% CI; VNTR, variable number tandem repeat.
Fig. 3.
Whole-brain surface-based gray matter changes associated with the variable number tandem repeat (VNTR) genotype. Left inferior/middle temporal gyrus (left) and right postcentral sulcus (right) were significantly thicker and deeper, respectively, in 4-repeats VNTR (VNTR4) carriers compared to noncarriers (post hoc P < .001). a.u.: arbitrary units; color bar represents P-value; error bars represent 95% CI.
Discussion
This study investigated the MIR137 VNTR rs58335419 heterogeneity in association with cognitive and neuroanatomical phenotypes in a relatively large cohort of schizophrenia cases and healthy individuals from the ASRB. Carriers of the VNTR4 were enriched in a CD subtype of schizophrenia, identified via latent class analysis of cognitive performance across multiple neuropsychological tests. While no significant difference was observed in direct comparison of cases and controls, we suspect this may be due to the disproportionate number of cases having relatively mild cognitive impairment. This was consistent with previous studies that also reported no association between the MIR137 variants (including VNTR) and SZ40 or SZ and/or BD.10 Interestingly, when we further investigated the neuroanatomical features associated with the VNTR4 genotype, we observed thicker left inferior/middle temporal gyrus and deeper right postcentral sulcus compared to the group with the VNTR wild type.
Our findings, therefore, suggest that the MIR137 variants, particularly the VNTR4, have a stronger association with cognitive and neuroanatomical endophenotypes of SZ than the disease diagnosis. This is consistent with previous reports suggesting that phenotypic changes associated with the MIR137 risk allele are independent of psychiatric diagnoses and may be more closely aligned with cognitive endophenotypes.21,41,42 For example, lower performance to a Stroop task, which has been consistently reported in schizophrenia,43,44 was found in healthy VNTR4 carriers compared to wild-type participants.45 Another study reported more cognitive deficits involving episodic memory and attentional control impairments in individuals carrying the schizophrenia GWAS-associated variant rs1625579 in a mixed cohort of schizophrenia and bipolar disorder.20 The same variant was associated with the P300 electrophysiological endophenotype in schizophrenia46 and impaired performance in verbal episodic memory and attention.20 Previous work from our group demonstrated that patients carrying this variant in combination with more severe negative symptoms predicted membership of a subtype of schizophrenia cases presenting severe cognitive deficits.32
In line with our findings concerning altered brain morphology among VNTR4 carriers, previous functional neuroimaging studies have implicated the inferior temporal gyrus in multiple cognitive processes, including language, visual perception, and multimodal sensory integration.47–49 The inferior temporal gyrus is also specifically associated with the recognition of objects/patterns using visual categories,48,49 which is often impaired in schizophrenia.50 The postcentral sulcus is at the border of the postcentral gyrus and parietal lobules; the former is somatosensory cortex whose abnormal processing contributes to deficits seen in neurological disorders,51 and the later play key roles in the control of action. Genetic variation in the MIR137 locus has also been associated with brain morphological and functional changes; eg, marker SNP rs1625579 was associated with small white matter density, reduced hippocampal volume, and larger lateral ventricles in schizophrenia patients compared to HC.23 Similar studies have also reported evidence of the association between the rs1625579 and decreased brain volume, increased brain functional activity, altered fronto-amygdala connectivity, and reduced white matter integrity.21,22,52
MIR137 encodes for miR-137, a brain-enriched miRNA that has been shown to regulate several processes thought to contribute to the pathogenesis of schizophrenia, including neurodevelopment, neural maturation and transmission, adult neurogenesis, and synaptogenesis.11 The schizophrenia-associated MIR137 variants are thought to exert functional consequence through the alteration of miR-137 expression. For example, neurons harboring the schizophrenia-risk haplotype have decreased levels of miR-137 compared to cells carrying the alternative allele.53 This finding is consistent with reduced levels of miR-137 associated with the risk allele in postmortem tissue from the human dorsolateral prefrontal cortex.15 Expression of miR-137 was shown to be directly altered by the VNTR, with reduced levels observed to emerge from the VNTR4-containing cells compared to their wild-type counterparts.10 MiR-137 target genes were also differentially expressed in transcriptomic analysis of the VNTR4-containing cells and enriched in pathways involved in synaptogenesis, synapse organization, and synaptic transmission.10 More recently it was reported that the longer variants including VNTR4 modulate alternative splicing of the pri-miR-137, resulting in reduced expression of miR-137 in fetal and adult human brains.54 However, previous reports, including ours, have shown that postmortem alteration in the miR-137 expression in the cerebral cortex was not significantly different in SZ.19,55–57 This is probably due to the fact that the samples were not stratified by genotype and were also subject to modulation by a variety of environmental exposures. It is also possible that the miR-137 expression is more critical and variable during the early development of the nervous system and, therefore, not detected in postmortem samples with the disorder.
This study has limitations associated with sample size that needs to be acknowledged. Future studies with larger sample size are, therefore, warranted to specifically test the association of this multiallelic variant between cognitive endophenotypes and in relation to diagnosis of the disorder. We were also underpowered to examine the effect of the rare VNTR5 variant, which had an allele frequency of 0.01 in our cohort. We suspect that the VNTR4 may also be associated with a diagnosis of schizophrenia and this signal may emerge in a much larger sequenced sample as we were also underpowered to detect an association with the GWAS risk SNP.
In summary, these findings suggest that the MIR137 VNTR4 genotype is enriched in individuals with a severe CD subtype of schizophrenia. The VNTR4 variant was also associated with altered brain morphology in the inferior temporal gyrus and postcentral sulci. We suspect that the functional significance of the VNTR4 is associated with dysregulation of miR-137, supporting the gene’s role in the cognitive aspects of the disorder more generally and the mounting evidence of its significance to brain development and function. It is also plausible that this miRNA is a candidate for therapeutic manipulation in individuals with schizophrenia, particularly those that have genomic variation in its gene and/or its biosynthesis and effector pathways. The significance of this cannot be underestimated given the unmet need for drugs that target the most intractable cognitive symptoms of schizophrenia that are associated with poor functional outcomes. More broadly, our observations suggest that some common structural variants in the genome that are not currently tagged by proxy SNP content on the major genotyping platforms may account for a significant burden of the missing heritability of complex traits and warrant further attention in future efforts to capture common genetic liability of disease. In some cases, such as the VNTR rs58335419 analyzed in the current study, these variants may provide additional support to the gene targets of causal variation in undifferentiated GWAS loci.
Funding
This study was supported by the NSW Health Collaborative Genomics grant program (M.C., M.G., and V.C.), a National Alliance for Research on Schizophrenia and Depression Independent Investigator Grant (M.C.), and National Health and Medical Research Council project grants (1067137, 1147644, and 1051672). Data and samples were also collected by the Australian Schizophrenia Research Bank (ASRB), supported by an Australian National Health and Medical Research Council Enabling Grant (386500), the Pratt Foundation, Ramsay Health Care, and the Viertel Charitable Foundation. The ASRB were also supported by the Schizophrenia Research Institute (Australia), utilizing infrastructure funding from NSW Health and the Macquarie Group Foundation. E.M. is supported by a University of Newcastle RHD scholarship. J.A. is supported by a University of Newcastle RHD scholarship and an Emlyn and Jennie Thomas Postgraduate Medical Research Scholarship. W.R. is supported by an Australian Postgraduate Research Award. M.C. is supported by a National Health and Medical Research Council Senior Research Fellowship (1121474).
Conflict of interest: The authors declare no conflict of interest.
Supplementary Material
References
- 1.van Os J, Kapur S. Schizophrenia. Lancet. 2009;374(9690):635–645. [DOI] [PubMed] [Google Scholar]
- 2.Gejman PV, Sanders AR, Duan J. The role of genetics in the etiology of schizophrenia. Psychiatr Clin North Am. 2010;33(1):35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z, Chen J, Yu H, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49(11):1576–1583. [DOI] [PubMed] [Google Scholar]
- 5.Pardiñas AF, Holmans P, Pocklington AJ, et al. ; GERAD1 Consortium; CRESTAR Consortium . Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M, Jaffe AE, Straub RE, et al. A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat Med. 2016;22(6):649–656. [DOI] [PubMed] [Google Scholar]
- 7.Warburton A, Breen G, Rujescu D, Bubb VJ, Quinn JP. Characterization of a REST-regulated internal promoter in the schizophrenia genome-wide associated gene MIR137. Schizophr Bull. 2015;41(3):698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warburton A, Breen G, Bubb VJ, Quinn JP. A GWAS SNP for schizophrenia is linked to the internal MIR137 promoter and supports differential allele-specific expression. Schizophr Bull. 2016;42(4):1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bemis LT, Chen R, Amato CM, et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68(5):1362–1368. [DOI] [PubMed] [Google Scholar]
- 10.Strazisar M, Cammaerts S, van der Ven K, et al. MIR137 variants identified in psychiatric patients affect synaptogenesis and neuronal transmission gene sets. Mol Psychiatry. 2015;20(4):472–481. [DOI] [PubMed] [Google Scholar]
- 11.Mahmoudi E, Cairns MJ. MiR-137: an important player in neural development and neoplastic transformation. Mol Psychiatry. 2017;22(1):44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun G, Ye P, Murai K, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins AL, Kim Y, Bloom RJ, Kelada SN, Sethupathy P, Sullivan PF. Transcriptional targets of the schizophrenia risk gene MIR137. Transl Psychiatry. 2014;4:e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guella I, Sequeira A, Rollins B, et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J Psychiatr Res. 2013;47(9):1215–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright C, Turner JA, Calhoun VD, Perrone-Bizzozero N. Potential impact of miR-137 and its targets in schizophrenia. Front Genet. 2013;4:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43(10):969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. 2013;18(1):11–12. [DOI] [PubMed] [Google Scholar]
- 19.Kim AH, Parker EK, Williamson V, McMichael GO, Fanous AH, Vladimirov VI. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophr Res. 2012;141(1):60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummings E, Donohoe G, Hargreaves A, et al. Mood congruent psychotic symptoms and specific cognitive deficits in carriers of the novel schizophrenia risk variant at MIR-137. Neurosci Lett. 2013;532:33–38. [DOI] [PubMed] [Google Scholar]
- 21.Mothersill O, Morris DW, Kelly S, et al. Effects of MIR137 on fronto-amygdala functional connectivity. Neuroimage. 2014;90:189–195. [DOI] [PubMed] [Google Scholar]
- 22.Kuswanto CN, Sum MY, Qiu A, Sitoh YY, Liu J, Sim K. The impact of genome wide supported microRNA-137 (MIR137) risk variants on frontal and striatal white matter integrity, neurocognitive functioning, and negative symptoms in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(5):317–326. [DOI] [PubMed] [Google Scholar]
- 23.Lett TA, Chakravarty MM, Chakavarty MM, et al. The genome-wide supported microRNA-137 variant predicts phenotypic heterogeneity within schizophrenia. Mol Psychiatry. 2013;18(4):443–450. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization. ICD-10: International Statistical Classification of Diseases and Related Health Problems. 10th rev. ed.New York, NY: World Health Organization; 2008. [Google Scholar]
- 25.Loughland C, Draganic D, McCabe K, et al. Australian Schizophrenia Research Bank: a database of comprehensive clinical, endophenotypic and genetic data for aetiological studies of schizophrenia. Aust N Z J Psychiatry. 2010;44(11):1029–1035. [DOI] [PubMed] [Google Scholar]
- 26.McGuffin P, Farmer A, Harvey I. A polydiagnostic application of operational criteria in studies of psychotic illness. Development and reliability of the OPCRIT system. Arch Gen Psychiatry. 1991;48(8):764–770. [DOI] [PubMed] [Google Scholar]
- 27.Castle DJ, Jablensky A, McGrath JJ, et al. The diagnostic interview for psychoses (DIP): development, reliability and applications. Psychol Med. 2006;36(1):69–80. [DOI] [PubMed] [Google Scholar]
- 28.Andreasen NC The Scale for the Assessment of Negative Symptoms (SANS). Iowa City, IA: The University of Iowa; 1983. [Google Scholar]
- 29.Wechsler D Wechsler Test of Adult Reading: WTAR. New York, NY: Psychological Corporation; 2001. [Google Scholar]
- 30.Wechsler D Wechsler Abbreviated Scale of Intelligence (WASI). New York, NY: The Psychological Corporation; 1999. [Google Scholar]
- 31.Oldfield RC. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9(1):97–113. [DOI] [PubMed] [Google Scholar]
- 32.Green MJ, Cairns MJ, Wu J, et al. ; Australian Schizophrenia Research Bank . Genome-wide supported variant MIR137 and severe negative symptoms predict membership of an impaired cognitive subtype of schizophrenia. Mol Psychiatry. 2013;18(7):774–780. [DOI] [PubMed] [Google Scholar]
- 33.Reay WR, Atkins JR, Quide Y, Carr VJ, Green MJ, Cairns MJ. Polygenic disruption of retinoid signalling in schizophrenia and a severe cognitive deficit subtype. Mol Psychiatry. 2020;25:719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poplin R, Ruano-Rubio V, DePristo MA, et al. (2018) ‘Scaling accurate genetic variant discovery to tens of thousands of samples’, bioRxiv, doi: 10.1101/201178, July 24, 2018, preprint: not peer reviewed.
- 38.Seabold S, Perktold J. Statsmodels: econometric and statistical modeling with python. Paper presented at: 9th Python in Science Conference; June 28–30, 2010; Austin, TX.
- 39.Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. [DOI] [PubMed] [Google Scholar]
- 40.Egawa J, Nunokawa A, Shibuya M, et al. Resequencing and association analysis of MIR137 with schizophrenia in a Japanese population. Psychiatry Clin Neurosci. 2013;67(4):277–279. [DOI] [PubMed] [Google Scholar]
- 41.van Erp TG, Guella I, Vawter MP, et al. Schizophrenia miR-137 locus risk genotype is associated with dorsolateral prefrontal cortex hyperactivation. Biol Psychiatry. 2014;75(5):398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B, Zhang X, Hou B, et al. The impact of MIR137 on dorsolateral prefrontal-hippocampal functional connectivity in healthy subjects. Neuropsychopharmacology. 2014;39(9):2153–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westerhausen R, Kompus K, Hugdahl K. Impaired cognitive inhibition in schizophrenia: a meta-analysis of the Stroop interference effect. Schizophr Res. 2011;133(1-3):172–181. [DOI] [PubMed] [Google Scholar]
- 44.Chen EY, Wong AW, Chen RY, Au JW. Stroop interference and facilitation effects in first-episode schizophrenic patients. Schizophr Res. 2001;48(1):29–44. [DOI] [PubMed] [Google Scholar]
- 45.González-Giraldo Y, González-Reyes RE, Forero DA. A functional variant in MIR137, a candidate gene for schizophrenia, affects Stroop test performance in young adults. Psychiatry Res. 2016;236:202–205. [DOI] [PubMed] [Google Scholar]
- 46.Decoster J, De Hert M, Viechtbauer W, et al. Genetic association study of the P300 endophenotype in schizophrenia. Schizophr Res. 2012;141(1):54–59. [DOI] [PubMed] [Google Scholar]
- 47.Cabeza R, Nyberg L. Imaging cognition II: an empirical review of 275 PET and fMRI studies. J Cogn Neurosci. 2000;12(1):1–47. [DOI] [PubMed] [Google Scholar]
- 48.Herath P, Kinomura S, Roland PE. Visual recognition: evidence for two distinctive mechanisms from a PET study. Hum Brain Mapp. 2001;12(2):110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mesulam MM. From sensation to cognition. Brain. 1998;121 (Pt 6):1013–1052. [DOI] [PubMed] [Google Scholar]
- 50.Butler PD, Javitt DC. Early-stage visual processing deficits in schizophrenia. Curr Opin Psychiatry. 2005;18(2):151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borich MR, Brodie SM, Gray WA, Ionta S, Boyd LA. Understanding the role of the primary somatosensory cortex: Opportunities for rehabilitation. Neuropsychologia. 2015;79(Pt B):246–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakamoto K, Crowley JJ. A comprehensive review of the genetic and biological evidence supports a role for MicroRNA-137 in the etiology of schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2018;177(2):242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siegert S, Seo J, Kwon EJ, et al. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat Neurosci. 2015;18(7):1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pacheco A, Berger R, Freedman R, Law AJ. A VNTR regulates miR-137 expression through novel alternative splicing and contributes to risk for schizophrenia. Sci Rep. 2019;9(1):11793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15(12):1176–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beveridge NJ, Tooney PA, Carroll AP, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17(8):1156–1168. [DOI] [PubMed] [Google Scholar]
- 57.Mellios N, Galdzicka M, Ginns E, et al. Gender-specific reduction of estrogen-sensitive small RNA, miR-30b, in subjects with schizophrenia. Schizophr Bull. 2012;38(3):433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.