Abstract
The receptor binding and proteolysis of Spike of SARS-CoV-2 release its S2 subunit to rearrange and catalyze viral-cell fusion. This deploys the fusion peptide for insertion into the cell membranes targeted. We show that this fusion peptide transforms from intrinsic disorder in solution into a wedge-shaped structure inserted in bilayered micelles, according to chemical shifts, 15N NMR relaxation, and NOEs. The globular fold of three helices contrasts the open, extended forms of this region observed in the electron density of compact prefusion states. In the hydrophobic, narrow end of the wedge, helices 1 and 2 contact the fatty acyl chains of phospholipids, according to NOEs and proximity to a nitroxide spin label deep in the membrane mimic. The polar end of the wedge may engage and displace lipid head groups and bind Ca2+ ions for membrane fusion. Polar helix 3 protrudes from the bilayer where it might be accessible to antibodies.
Keywords: viral-cell fusion, fusion peptide, membrane mimic, bicelle, phospholipid, NMR spectroscopy, solution structure, dynamics, paramagnetic relaxation enhancements, nitroxide spin label
Graphical Abstract
Introduction
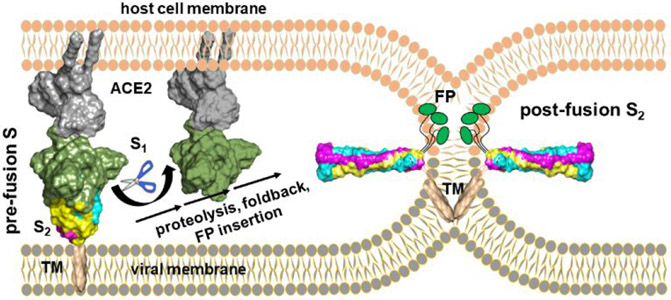
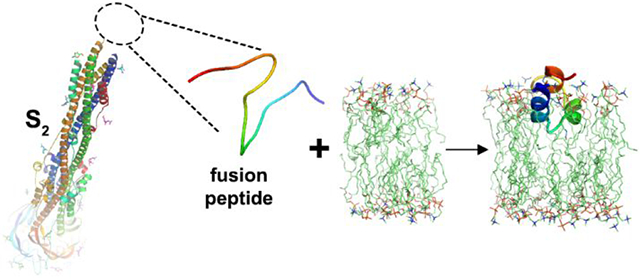
Glycoproteins in the envelopes of viruses, such as Spike (S) of coronaviruses, catalyze fusion of viral and host cell membranes in order for infection to proceed1-3. Coronaviral Spike comprises S1 and S2 subunits (Fig. 1). The prefusion state of the glycoprotein stores the fusion peptide on its surface and becomes primed to a fusion-competent metastable state ready to rearrange. In (β-coronaviruses, receptor binding and proteolysis of S 4-7 then jettisons the S1 subunit and triggers rearrangement of the fusogenic S2 subunit into an extended form that exposes on its tips the hydrophobic fusion peptide8,9 for the ensuing steps of fusion1-3 (Fig. 2). The extended conformation of the mature S2 subunit forms a bridge between host cell membrane and the viral envelope. The N-terminus deploys the fusion peptide for insertion into a host cell membrane while the C-terminus remains anchored in the viral envelope. Accompanying membrane insertion, the trimeric, fusogenic subunit (S2 in β-coronaviruses) folds back upon itself into a six-helix bundle to draw the bound cell membrane and viral envelope into apposition1-3,8-11 (Fig. 2). Clustering of hairpin-like fusion proteins in the virus-cell interface multiplies the favorable free energies from folding back into hairpins in order to pay the thermodynamic costs of fusion1-3,15. Joining of the outer leaflets of the bilayers in hemifusion and stalk intermediates are thought to precede opening of the fusion pore16.
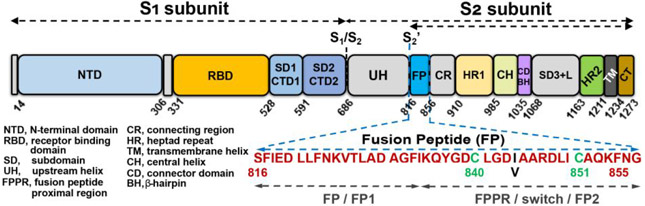
Figure 1. Subunits, domains, and fusion peptide sequence of SARS-CoV-2 Spike.
The sequence of the fusion peptide (FP) of this study is shown in red. Its nomenclature in other studies is labeled with gray text.
Figure 2. Transformation of prefusion S to mature S2 at a hemifusion stalk.
Maturation of S2 frees FP for membrane insertion and fusion. The structures of postfusion S2, ACE2, an ACE2-S complex, and the TM domain are from refs 11-14, respectively.
Compared to the rapidly evolving S1 subunit, the fusogenic S2 subunit is highly conserved (91% between SARS-CoV-2 and SARS-CoV). S2 was proposed for targeting by antibodies and peptides with potential for interfering in fusion and infection broadly among coronaviruses9,11,17-21, which is an attractive prospect as virulent variants of SARS-CoV-2 evolve and other (β-coronaviruses emerge in human populations. In 13 convalescent SARS-CoV patients, the antigen from S2 that distinguished their antibodies was a 26-residue portion of the proposed fusion peptide, suggesting that it could be an antigen for potentially neutralizing antibodies22. This epitope lies within the map of the fusion peptide (FP) of SARS-CoV and SARS-CoV-2 identified as genuine based on its ability to restrict the mobility of the head groups and nearby fatty acyl groups in the presence of calcium ions23,24. The S2’ site of proteolysis at Ser816 in SARS-CoV-2 6,9 forms the mature N-terminus. The 41-residue sequence reported for the functional FP begins at Ser816 23,24. We and others21 accept this region as the most authoritative location of FP. It includes a disulfide-bonded motif23 that others have referred to as FPPR9 or the “switch domain”10 (Fig. 1). This motif is missing from the structural coordinates of loosely packed prefusion states. In compact prefusion states of S in which all receptor binding domains (RBDs) are down, packing of the S1 subunit with the switch domain of S2 induces a few different structures in the switch 9,10,13. This switch of S2 is coupled to the up or down state of the RBDs via the intervening C-terminal domain 1, and was deemed central in conformational control and rearrangement of S 9,10,13. Asp614, site of the D614G mutation, can form hydrogen bonds and salt bridges with FP to stabilize it13. FP (and the switch domain within it; Fig. 1) has been absent from the electron density and structural coordinates of the rod-shaped, postfusion state of S2 where it is likely to be highly exposed at the tips of the rod 8,9,11.
The fusion peptides of HIV gp41, influenza HA2, and ebolavirus GP2 were characterized well in micelles by NMR spectroscopy25-31. Mimics of lipid bilayers known as bilayered micelles or bicelles, prepared at molar ratios that form a small bilayer with partial mixed micelle character32-35, have enabled NMR structural characterizations of proteins associated or inserted in a more bilayer-like environment14,36-43. To characterize exposed and bilayer-bound states of FP from SARS-CoV-2 (displayed in vivo from S2 in prehairpin intermediate and postfusion states21), NMR spectroscopy in solution has been exploited herein to gain understanding of FP in its fluid environments. FP undergoes a marked transformation from intrinsic disorder when exposed in aqueous solution to a compact, wedge-shaped structure with the hydrophobic, narrow end inserted in bicelles. The results expand our understanding of the SARS-2 FP in terms of its dynamic shifts in shape, expected bilayer interactions and distortion, and a conserved surface that might be accessible to agents such as antibodies.
Results
We prepared the N-terminal 42 residues of the mature S2 subunit of SARS-CoV-2 Spike, from Ser816 to Gly857 (Fig. 1), in a manner that added no foreign residues to the termini of this FP sequence once purified. We expressed an N-terminal His tag followed by protein GB1 and a TEV protease cleavage site ahead of the Ser816 - Gly857 region of interest (Fig. S1 in Supporting Information). The GB1–FP fusion appeared in the insoluble fraction from the E. coli host. This was dissolved in urea and purified by affinity for Ni2+-conjugated NTA resin. The denatured protein was folded with formation of the disulfide crosslink. The folding proceeded upon dilution and dialysis vs. denaturant-free solution at pH 8 with a 10- fold excess of thiol reductant over oxidant, followed by dialysis without redox reagents. FP was separated from this GB1-FP fusion by TEV proteolysis and chromatography on NTA resin followed by Q Sepharose. Correct mass and homogeneity were verified LC-MS. Verification of disulfide formation also used MS/MS of tryptic peptides, 13C chemical shifts of the cysteines, and loss of thiol reactivity. See Methods in SI for details.
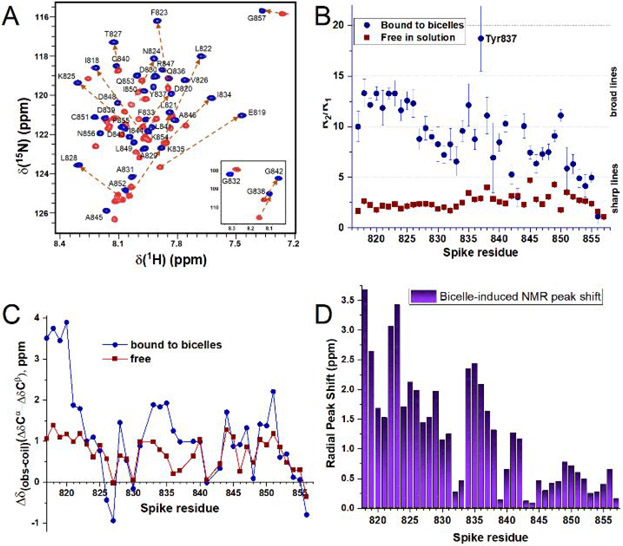
For relevance to viral entry via endocytosis, pH 5.0 was used in NMR studies. Short-lived samples of SARS-2 FP increased in solubility and longevity by at least two-fold with an I844V substitution. The NMR peaks were assigned by triple resonance NMR in solution as described in SI. The NMR spectra of the free state of FP (at 32°C) exhibit random coil chemical shifts, poor dispersion, and sharp lines with 15N R2 averaging 4.3/s and R2/R1 averaging 2.5 (Fig. 3A,B). These suggest intrinsic disorder. The secondary 13C chemical shifts are modestly positive from Phe817 – Lys825 and Val844 – Ala852 in the free state (Fig. 3C). This evidence, alongside the sharp NMR lines and lack of NOEs, suggests dynamic disorder that possibly samples helix transiently, i.e., with dynamic helix-coil transitions. 15N relaxation in the region spanning the Cys840 to Cys851 disulfide crosslink indicates slowed mobility in the free state: While the 15N R1·R2 product44 averages only 5.5/s2 from Phe817 – Ile834, it averages 9.9/s2 from Lys835 – Ala852 (Fig. S2A). The spectral density at low frequency, J(0) from 15N relaxation 45-47, follows a similar pattern with J(0) averaging 0.63 ns/rad from Phe817 – Ile834 and 1.1 ns/rad from Lys835 – Ala852 (Fig. S2B in SI). J(0) and 15N R1·R2 establish dynamic disorder in the N-terminal half of FP to Ile834, with some slowing of the dynamics in the C-terminal half spanning the disulfide. The extent of intrinsic disorder in free FP is consistent with its absence in large part from most electron density maps.
Figure 3. The fusion peptide of SARS-CoV-2 is disordered in aqueous solution but helical in bicelles.
The free state is represented by red squares and the state bound to bicelles by blue circles. The bicelles were composed of 120 mM DH7PC and 60 mM DMPC. NMR spectra were measured at pH 5.0, 32 °C, 800 MHz. (A) The 15N TROSY free in solution is superposed with that in bicelles. (B) 15N NMR relaxation rate constants R2 and R1 are plotted as a ratio. (C) The secondary 13Cα and 13Cβ chemical shifts are plotted as a difference. (D) The bicelle-induced disorder-to-order transition is accompanied by large shifts of the backbone amide NMR peaks, calculated as radial changes using eq. S1 in SI.
We prepared small isotropic bicelles using one long-chain phospholipid, i.e., DMPC, per two per two short-chain phospholipids, i.e., DH7PC. The heptanoyl fatty acyl chains of DH7PC decrease the critical micelle concentration (CMC) and presence of free detergent monomers to ~1 mM48. Addition of the small bicelles induces a disorder-to-order transition in FP that disperses the amide NMR peaks both at pH 5 (Fig. 3A) and pH 7 (Fig. S3). The bicelle-induced chemical shift perturbations are largest at the N-terminus and generally decrease toward the C-terminus (Fig. 3D). Elevated secondary 13C chemical shifts suggest formation of helices around Phe817 – Asn824, Ala831 – Tyr837, Val844 – Ala847, and Leu849 – Cys851 (Fig. 3C). 15N relaxation indicates that insertion into bicelles restricts the segmental motions, especially for the residues on the N-terminal side of the disulfide, which average 18.5 in R2/R1 and 2.8 ns/rad in J(0) (Figs. 3B, S2B). Tyr817 - Val827 experience the biggest rigidification, with 15N R2/R1 averaging 12.6, suggesting a rotational correlation time of about 16 ns using eq. S2. These observations imply folding of FP in bicelles. The evidence of rigidification correlates with the propensity for forming helix in the bicelles (Fig. 3C).
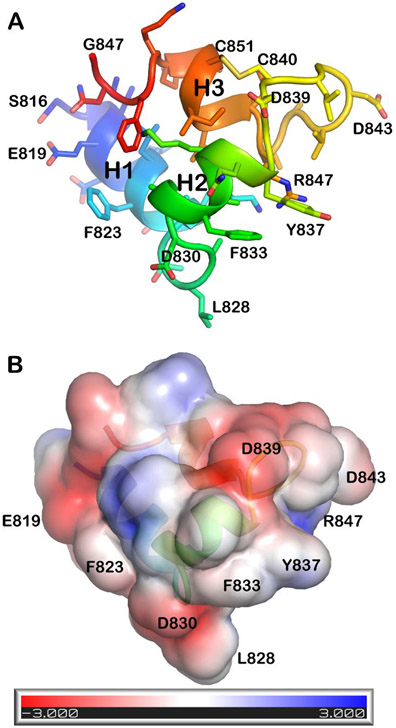
The induction of structure in FP by bicelles is supported by NOEs. NMR structural models were calculated from dozens of long- and medium-range NOEs (Fig. S4) assigned manually due to overlap of methyl NMR peaks. The pairwise RMSD values to the most representative structure (model 2) are 0.90 ± 0.40 Å for the backbone atoms and 1.39 ± 0.56 Å for all heavy atoms of residues Phe817 – Phe855 in the ensemble of 15 models (Fig. S5, Tables S1, S2). The first α-helix from Phe817 to Val826 abuts the second α-helix from Ala831 to Tyr837 (Fig. 4A). The medium-range NOEs suggest a less uniform and less stable helix from Ala845 through Gln853 (Figs. 4A, S5). The NMR structural models of FP associated with bicelles contrast hypotheses that its membrane-inserted structure could be a single helix, a very long helix, or a bipartite platform spread widely on the membrane. Instead, the three helices appear to be closed into a globular but narrow fold in which each helix forms a side (Fig. 4A,B). Helix 1 is positioned nearly perpendicular to helix 2. Helix 3 partially inserts between them and seems to brace helices 1 and 2 into an L-shape facing outward (Fig. 4A). The narrow dimension thins to the hydrophobic tip at Leu828 in the loop between helices 1 and 2 (Fig. 4). Helix 3 is formed within a disulfide-bridged hairpin where it adopts 3-10 helix from Ala845 through Leu849, bends at Ile850, and appears to fray on the C-terminal side of disulfide-linked Cys851 (Fig. S5). The fraying of the C-terminal end of helix 3 is suggested by its relative paucity of NOEs and its greater backbone dynamics (Figs. 3B, S2).
Figure 4. The fusion peptide of SARS-CoV-2 closes into a wedge-shaped structure in bicelles.
The lowest energy model from the ensemble of NMR structures (PDB: 7MY8, model 1) is plotted in a shared orientation with coloring of the chain from blue at the N-terminus to red at the C-terminus. A) Side chains and backbone ribbon are plotted. (B)The solvent-accessible surface is colored by electrostatic potential using APBS Tools 2.1 49-51.
The closed structure of FP associated with bicelles is superimposed in Fig. S6 on the open conformations of the corresponding Phe817 – Asn856 sequence in cryo-EM structural models of tightly packed prefusion forms of S having all RBDs down. The compact cryo-EM models vary in the orientation and presence of the second and third helices in this sequence. The closed conformation of the NMR structure of FP in bicelles aligns poorly with all of these more open conformations observed by cryo-EM (Fig. S6). Thus, FP appears to rearrange to a distinct fold when associated with a lipid bilayer-like environment. This provides a new snapshot of a step required for viral-cell fusion in infections by SARS-CoV-2.
We investigated features of the fusion peptide that might interact with membranes. Helices 1 and 2 are amphipathic while helix 3 is polar (Figs. 4B, S7). The evidence of two Ca2+-binding sites24 draws attention to the anionic side chains, which might bind divalent calcium ions together with phosphoryl moieties of phospholipids in order to promote membrane fusion. CaCl2 was titrated into bicelle-associated FP at pH 7.0 at low ionic strength to enhance the modest affinity for Ca2+. NMR detection of titrations can suggest binding sites on the basis of peaks that shift or broaden54. The amide NMR peaks of the backbone were shifted minimally. Some amide peaks were broadened by CaCl2 however (Fig. S8). Among the six carboxylic acid residues, addition of CaCl2 most weakened the amide peaks of Asp843 and Asp839 to 43% and 61% of the original peak heights (Fig. S8). Consequently, Asp843 and Asp839 are most likely to participate in binding of CaCl2. More modest effects of CaCl2 on the amide peaks of conserved Glu819, Asp820, and Asp848 (Fig. S8) are equivocal about any participation in calcium binding. The failure of CaCl2 to broaden the amide peak of Asp830 and the evidence of insertion of Asp830 among the fatty acyl chains (see below) diminish the possibility of Asp830 binding calcium. The negative charge of Asp830 might instead serve to neutralize the partial positive charge of the helix dipole of helix 2.
About 61% of the accessible surface area (ASA) of bicelle-bound FP is hydrophobic, i.e., 2070 ± 150 Å2 of the total ASA of 3400 ± 220 Å2. Helix 1 directs hydrophobic Phe817, Leu821, and Phe823 outward while helix 2 displays hydrophobic Phe833 and Tyr837 (Figs. 4, S7). Leu828 projects out from the 1-2 loop. These exposed, hydrophobic side chains, especially Leu828 and conserved Phe823 and Tyr837 (Figs. 4, S7, S9) are candidates for contacts with lipids. The especially large bicelle-induced shifts of the amide NMR peaks of residues 818, 823, and 837 (Fig. 3B) support the hypothesis of the exposed aromatic groups inserting into the bicelles. Consistent with this possibility, the amide groups at the C-terminal ends of helices 1 and 2 lack cross peaks to water in NOESY spectra. This implies the unlikelihood of either chemical exchange with water or proximity to water, as well as the likelihood of hydrogen bonds within the helices. This lack of water access could be correlated with the stability of helices 1 and 2, their insertion in the membrane, or both.
More direct insight into bicelle insertion was sought. Detection of NOEs to aromatic groups by 13C TROSY55 afforded an opportunity to reveal contacts between the aromatic side chains and fatty acyl groups. Methyl groups of the DH7PC acyl chains have NOEs to aromatic δ peaks of Phe817, Phe833, and Tyr837, as well as to the ε peak of Tyr837 (Fig. S10). This suggests that these groups might penetrate about half the acyl thickness of a leaflet of the bicelles (since heptanoyl chains are half as long as the myristoyl chains packed nearby in the best models of the bicelles34). Phe817, Phe823, Phe833, and Leu841 each appear to have NOEs to a distinct internal methylene group of a fatty acyl chain (Fig. S10). Phe823 and Leu841 each have an additional NOE to what could be the C2 position of DMPC just below its glycerol backbone.
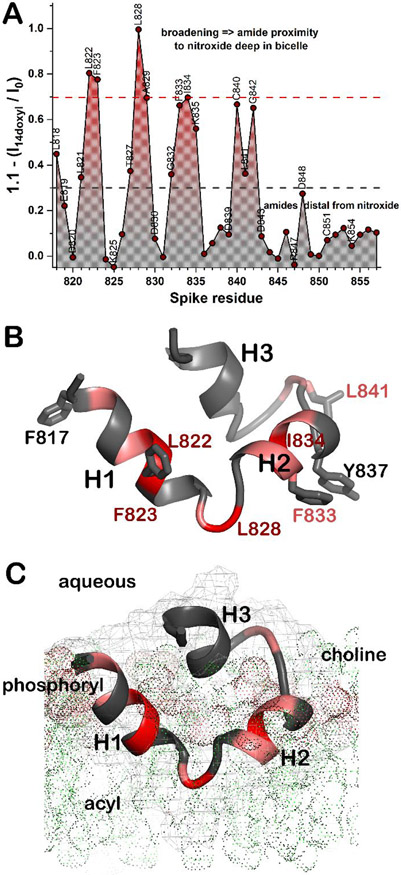
Proximity of the backbone of FP to the center of the bicelles was measured by doping the bicelles with a phospholipid harboring a nitroxide spin label near the methyl end of the acyl chain. The NMR peaks of the amide groups of the fusion peptide most broadened by addition of the paramagnetic probe of 16:0 – 14-doxyl PC belong to Leu822, Phe823, Leu828, Ala829, and Ile834 (Fig. 5A), Phe833, Cys840, and Gly842 were also very broadened. These backbone positions experiencing paramagnetic relaxation enhancements (PREs) lie near side chains with NOEs to fatty acyl chains (Fig. 5B). Thus, the PREs and NOEs to lipids corroborate one another and suggest that the hydrophobic and narrowed end of the wedge-shaped fold of FP is inserted in among acyl chains of the bicelles (Fig. 5). By contrast, the backbone amide groups of the carboxylate-containing amino acids and C-terminal end of FP have minimal PREs suggesting their relative distance from the interior of the bicelles (Fig. 5). These observations serve as the basis for these working hypotheses needing further testing and revision: (a) The narrowed tip of the wedge, i.e. the 1-2 loop around Leu828, points toward the interior of bilayers. (b) The broad, polar end with carboxylic acids and helix 3 points outward into the aqueous phase (Fig. 5C).
Figure 5. PRE and NOE evidence of FP insertion into bicelles.
About one nitroxide spin-labeled 14-doxyl PC probe per leaflet was added to the bicelles. Proximity to the nitroxide label deep in the bicelles is symbolized with red for strong paramagnetic broadening of the amide NMR line. Gray symbolizes minimal broadening. (A) The PREs are estimated from amide NMR peak heights before and after addition of 14-doxyl PC. The 15N HSQC used for detection was modified to include a 1H PROJECT CPMG train of 8 ms 39,52. (B) Residues with PREs to amide groups ≥ 0.7 in A are red while those between 0.7 and 0.3 are pink. Side chains that appear to have NOEs to phospholipid acyl chains (Fig. S10) are plotted. (C) A hypothesis regarding insertion of FP in a leaflet of DMPC is shown with lipids symbolized by dots. A faint mesh marks the surface of FP. The CHARMM-GUI Membrane Builder53 was used.
Discussion
Insertion of the wedge shape of the SARS-2 FP into a lipid bilayer can be hypothesized to displace lipid head groups outward more than the interior ends of their fatty acyl chains for two reasons. First, the polar end of the SARS-2 FP domain expected to contact head groups is broader than the hydrophobic tip that appears to be inserted among the acyl chains. Second, the depth of insertion of SARS-2 FP into bilayers is likely to be limited by the charged residues of its broad polar end. The pattern of NOEs between the fatty acyl chains and the FP (Figs. 5, S10) corroborates the limited depth of insertion. The enthalpically favorable binding of FP to the membrane mimic probably provides substantial free energy15,24 to pay the costs of forming the high-energy intermediate N of membrane fusion when the bilayers bulge toward one another16,56. The proposed investment in formation of intermediate N by insertion of a fusion peptide was forecasted to lower the cost of forming the subsequent stalk and modified-stalk intermediates of membrane fusion16.
The SARS-2 FP increases the rigidity of the head groups and glycerol backbone regions of liposomes more when Ca2+ is present at 1 to 2 mM 23,24. A potential mechanism for this ordering could be calcium ions bridging between the carboxylate group of Asp843, and possibly of Asp839 and Asp848, and phosphoryl linkages to the head groups. The charged ends of the basic side chains should be attracted to the phosphoryl moieties, potentially enhancing the restriction. Indirect ordering of the leaflet distal from the side of influenza HA fusion peptide insertion was also observed in molecular dynamics (MD) simulations56.
Concluding Remarks
During maturation of the S2 subunit of SARS-CoV-2 Spike when the S1 subunit is released, the fusion peptide appears likely to become intrinsically disordered when exposed to water. Upon membrane insertion, the FP adopts a fold of three helices that had not been observed or anticipated. Stable and largely hydrophobic helices 1 and 2, plus the loop between them, insert among the fatty acyl chains of bicelle mimics of lipid bilayers. A neighboring belt of charged residues may be positioned to interact favorably with the phosphoryl linkages in phospholipids and calcium ions. The polar, conserved, and more dynamic helix 3 is comparatively distant from the acyl phase of the bilayer mimic and may emerge into the aqueous phase. Any vulnerability of conserved helix 3 to hypothetical antibodies would become valuable if such antibodies could broadly neutralize beta-coronaviruses.
Supplementary Material
ACKNOWLEDGMENTS
National Science Foundation (U.S.) RAPID award 2030473 supported this work. NIH S10RR022341 contributed to the 800 MHz spectrometer. Fabio Gallazzi performed LC-MS. Brian P. Mooney verified the crosslink by MS/MS.
ABBREVIATIONS
- ASA
accessible surface area
- CPMG
Carry-Purcell-Meiboom-Gill
- DH7PC
1,2-diheptanoyl-sn-glycero-3-phosphocholine
- 16:0-14 doxyl PC
1-palmitoyl-2-stearoyl-(14-doxyl)-sn-glycero-3-phosphocholine
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- FP
fusion peptide
- HR1
heptad repeat 1
- HR2
heptad repeat 2
- MS
mass spectrometry
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- NTA
nitriloacetic acid
- PRE
paramagnetic relaxation enhancement
- RBD
receptor binding domain
- S
spike
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
Footnotes
Supporting Information.
Additional experimental details, materials, methods, references for methods, results in Supporting Figures S1-S10, and statistics of structural restraints and quality metrics in Supporting Tables S1 and S2
“This material is available free of charge via the Internet at http://pubs.acs.org.”
REFERENCES
- (1).White JM; Delos SE; Brecher M; Schornberg K Structures and Mechanisms of Viral Membrane Fusion Proteins: Multiple Variations on a Common Theme. Crit. Rev. Biochem. Mol. Biol 2008, 43 (3), 189–219. 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Harrison SC Viral Membrane Fusion. Virology 2015, 479–480, 498–507. https://doi.org/ 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kielian M. Mechanisms of Virus Membrane Fusion Proteins. Annu. Rev. Virol 2014, 1 (1), 171–189. 10.1146/annurev-virology-031413-085521. [DOI] [PubMed] [Google Scholar]
- (4).Shang J; Wan Y; Luo C; Ye G; Geng Q; Auerbach A; Li F Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci 2020, 117 (21), 11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Walls AC; Park Y-J; Tortorici MA; Wall A; McGuire AT; Veesler D Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181 (2), 281–292.e6. https://doi.org/ 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hoffmann M; Kleine-Weber H; Schroeder S; Krüger N; Herrler T; Erichsen S; Schiergens TS; Herrler G; Wu N-H; Nitsche A; Müller MA; Drosten C; Pöhlmann S SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181 (2), 271–280.e8. https://doi.org/ 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ou X; Liu Y; Lei X; Li P; Mi D; Ren L; Guo L; Guo R; Chen T; Hu J; Xiang Z; Mu Z; Chen X; Chen J; Hu K; Jin Q; Wang J; Qian Z Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and Its Immune Cross-Reactivity with SARS-CoV. Nat. Commun 2020, 11 (1), 1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Walls AC; Tortorici MA; Snijder J; Xiong X; Bosch B-J; Rey FA; Veesler D Tectonic Conformational Changes of a Coronavirus Spike Glycoprotein Promote Membrane Fusion. Proc. Natl. Acad. Sci 2017, 114 (42), 11157–11162. 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Cai Y; Zhang J; Xiao T; Peng H; Sterling SM; Walsh RM; Rawson S; Rits-Volloch S; Chen B Distinct Conformational States of SARS-CoV-2 Spike Protein. Science (80-. ). 2020, 369 (6511), 1586–1592. 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zhou T; Tsybovsky Y; Gorman J; Rapp M; Cerutti G; Chuang G-Y; Katsamba PS; Sampson JM; Schön A; Bimela J; Boyington JC; Nazzari A; Olia AS; Shi W; Sastry M; Stephens T; Stuckey J; Teng I-T; Wang P; Wang S; Zhang B; Friesner RA; Ho DD; Mascola JR; Shapiro L; Kwong PD Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a PH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains. Cell Host Microbe 2020, 28 (6), 867–879.e5. https://doi.org/ 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Fan X; Cao D; Kong L; Zhang X Cryo-EM Analysis of the Post-Fusion Structure of the SARS-CoV Spike Glycoprotein. Nat. Commun 2020, 11 (1), 3618. 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yan R; Zhang Y; Li Y; Xia L; Guo Y; Zhou Q Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science (80-. ). 2020, 367 (6485), 1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Xu C; Wang Y; Liu C; Zhang C; Han W; Hong X; Wang Y; Hong Q; Wang S; Zhao Q; Wang Y; Yang Y; Chen K; Zheng W; Kong L; Wang F; Zuo Q; Huang Z; Cong Y Conformational Dynamics of SARS-CoV-2 Trimeric Spike Glycoprotein in Complex with Receptor ACE2 Revealed by Cryo-EM. Sci. Adv 2021, 7 (1). 10.1126/sciadv.abe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Fu Q; Chou JJ A Trimeric Hydrophobic Zipper Mediates the Intramembrane Assembly of SARS-CoV-2 Spike. J. Am. Chem. Soc 2021, 143 (23), 8543–8546. doi: 10.1021/jacs.1c02394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Li Y; Han X; Tamm LK Thermodynamics of Fusion Peptide–Membrane Interactions. Biochemistry 2003, 42 (23), 7245–7251. 10.1021/bi0341760. [DOI] [PubMed] [Google Scholar]
- (16).Kuzmin PI; Zimmerberg J; Chizmadzhev YA; Cohen FS A Quantitative Model for Membrane Fusion Based on Low-Energy Intermediates. Proc. Natl. Acad. Sci 2001, 98 (13), 7235–7240. 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Elshabrawy HA; Coughlin MM; Baker SC; Prabhakar BS Human Monoclonal Antibodies against Highly Conserved HR1 and HR2 Domains of the SARS-CoV Spike Protein Are More Broadly Neutralizing. PLoS One 2012, 7 (11), e50366–e50366. 10.1371/journal.pone.0050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ng O-W; Keng C-T; Leung CS-W; Peiris JSM; Poon LLM; Tan Y-J Substitution at Aspartic Acid 1128 in the SARS Coronavirus Spike Glycoprotein Mediates Escape from a S2 Domain-Targeting Neutralizing Monoclonal Antibody. PLoS One 2014, 9 (7), 1–11. 10.1371/journal.pone.0102415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Xia S; Liu M; Wang C; Xu W; Lan Q; Feng S; Qi F; Bao L; Du L; Liu S; Qin C; Sun F; Shi Z; Zhu Y; Jiang S; Lu L Inhibition of SARS-CoV-2 (Previously 2019-NCoV) Infection by a Highly Potent Pan-Coronavirus Fusion Inhibitor Targeting Its Spike Protein That Harbors a High Capacity to Mediate Membrane Fusion. Cell Res. 2020, 30 (4), 343–355. 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Xia S; Yan L; Xu W; Agrawal AS; Algaissi A; Tseng C-TK; Wang Q; Du L; Tan W; Wilson IA; Jiang S; Yang B; Lu L A Pan-Coronavirus Fusion Inhibitor Targeting the HR1 Domain of Human Coronavirus Spike. Sci. Adv 2019, 5 (4). 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Tang T; Bidon M; Jaimes JA; Whittaker GR; Daniel S Coronavirus Membrane Fusion Mechanism Offers a Potential Target for Antiviral Development. Antiviral Res. 2020, 178, 104792. https://doi.org/ 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhang H; Wang G; Li J; Nie Y; Shi X; Lian G; Wang W; Yin X; Zhao Y; Qu X; Ding M; Deng H Identification of an Antigenic Determinant on the S2 Domain of the Severe Acute Respiratory Syndrome Coronavirus Spike Glycoprotein Capable of Inducing Neutralizing Antibodies. J. Virol 2004, 78 (13), 6938–6945. 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lai AL; Millet JK; Daniel S; Freed JH; Whittaker GR The SARS-CoV Fusion Peptide Forms an Extended Bipartite Fusion Platform That Perturbs Membrane Order in a Calcium-Dependent Manner. J. Mol. Biol 2017, 429 (24), 3875–3892. https://doi.org/ 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lai AL; Freed JH SARS-CoV-2 Fusion Peptide Has a Greater Membrane Perturbating Effect than SARS-CoV with Highly Specific Dependence on Ca2+. J. Mol. Biol 2021, 433 (10), 166946. https://doi.org/ 10.1016/j.jmb.2021.166946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Han X; Bushweller JH; Cafiso DS; Tamm LK Membrane Structure and Fusion-Triggering Conformational Change of the Fusion Domain from Influenza Hemagglutinin. Nat. Struct. Biol 2001, 8 (8), 715–720. 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- (26).Li Y; Tamm LK Structure and Plasticity of the Human Immunodeficiency Virus Gp41 Fusion Domain in Lipid Micelles and Bilayers. Biophys. J 2007, 93 (3), 876–885. https://doi.org/ 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Gregory SM; Harada E; Liang B; Delos SE; White JM; Tamm LK Structure and Function of the Complete Internal Fusion Loop from Ebolavirus Glycoprotein 2. Proc. Natl Acad. Sci 2011, 108 (27), 11211–11216. 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Jaroniec CP; Kaufman JD; Stahl SJ; Viard M; Blumenthal R; Wingfield PT; Bax A Structure and Dynamics of Micelle-Associated Human Immunodeficiency Virus Gp41 Fusion Domain,. Biochemistry 2005, 44 (49), 16167–16180. 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- (29).Lorieau JL; Louis JM; Bax A The Complete Influenza Hemagglutinin Fusion Domain Adopts a Tight Helical Hairpin Arrangement at the Lipid:Water Interface. Proc. Natl Acad. Sci 2010, 107 (25), 11341–11346. 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Holmstrom KO; Somersalo S; Mandal A; Palva TE; Welin B Improved Tolerance to Salinity and Low Temperature in Transgenic Tobacco Producing Glycine Betaine. J. Exp. Bot 2000, 51 (343), 177–185. [DOI] [PubMed] [Google Scholar]
- (31).Ghosh U; Xie L; Jia L; Liang S; Weliky DP Closed and Semiclosed Interhelical Structures in Membrane vs Closed and Open Structures in Detergent for the Influenza Virus Hemagglutinin Fusion Peptide and Correlation of Hydrophobic Surface Area with Fusion Catalysis. J. Am. Chem. Soc 2015, 137 (24), 7548–7551. 10.1021/jacs.5b04578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Glover KJ; Whiles JA; Wu G; Yu N; Deems R; Struppe JO; Stark RE; Komives EA; Vold RR Structural Evaluation of Phospholipid Bicelles for Solution-State Studies of Membrane-Associated Biomolecules. Biophys J 2001, 81 (4), 2163–2171. https://doi.org/ 10.1016/S0006-3495(01)75864-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Piai A; Fu Q; Dev J; Chou JJ Optimal Bicelle Size q for Solution NMR Studies of the Protein Transmembrane Partition. Chem. – A Eur. J 2017, 23 (6), 1361–1367. 10.1002/chem.201604206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Caldwell TA; Baoukina S; Brock AT; Oliver RC; Root KT; Krueger JK; Glover KJ; Tieleman DP; Columbus L Low-q Bicelles Are Mixed Micelles. J. Phys. Chem. Lett 2018, 9 (15), 4469–4473. 10.1021/acs.jpclett.8b02079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hutchison JM; Shih K-C; Scheldt HA; Fantin SM; Parson KF; Pantelopulos GA; Harrington HR; Mittendorf KF; Qian S; Stein RA; Collier SE; Chambers MG; Katsaras J; Voehler MW; Ruotolo BT; Huster D; McFeeters RL; Straub JE; Nieh M-P; Sanders CR Bicelles Rich in Both Sphingolipids and Cholesterol and Their Use in Studies of Membrane Proteins. J. Am. Chem. Soc 2020, 142 (29), 12715–12729. 10.1021/jacs.0c04669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lee D; Walter KFA; Brückner A-K; Hilty C; Becker S; Griesinger C Bilayer in Small Bicelles Revealed by Lipid–Protein Interactions Using NMR Spectroscopy. J. Am. Chem. Soc 2008, 130 (42), 13822–13823. 10.1021/ja803686p. [DOI] [PubMed] [Google Scholar]
- (37).Morrison EA; DeKoster GT; Dutta S; Vafabakhsh R; Clarkson MW; Bahl A; Kern D; Ha T; Henzler-Wildman KA Antiparallel EmrE Exports Drugs by Exchanging between Asymmetric Structures. Nature 2012, 481 (7379), 45–50. https://doi.org/http://www.nature.com/nature/journal/v481/n7379/abs/nature10703.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Liu Y; Kahn RA; Prestegard JH Dynamic Structure of Membrane-Anchored Arf*GTP. Nat Struct Mol Biol 2010, 17 (7), 876–881. https://doi.org/http://www.nature.com/nsmb/journal/v17/n7/abs/nsmb.1853.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Koppisetti RK; Fulcher YG; Jurkevich A; Prior SH; Xu J; Lenoir M; Overduin M; Van Doren SR Ambidextrous Binding of Cell and Membrane Bilayers by Soluble Matrix Metalloproteinase-12. Nat. Commun 2014, 5. 10.1038/ncomms6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Prior SH; Fulcher YG; Koppisetti RK; Jurkevich A; Van Doren SR Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors. Structure 2015, 23 (11). 10.1016/j.str.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Piai A; Fu Q; Cai Y; Ghantous F; Xiao T; Shaik MM; Peng H; Rits-Volloch S; Chen W; Seaman MS; Chen B; Chou JJ Structural Basis of Transmembrane Coupling of the HIV-1 Envelope Glycoprotein. Nat. Commun 2020, 11 (1), 2317. 10.1038/s41467-020-16165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Piai A; Fu Q; Sharp AK; Bighi B; Brown AM; Chou JJ NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology. J. Am. Chem. Soc 2021. 10.1021/jacs.1c01762. [DOI] [PubMed] [Google Scholar]
- (43).Smrt ST; Draney AW; Singaram I; Lorieau JL Structure and Dynamics of Membrane Proteins and Membrane Associated Proteins with Native Bicelles from Eukaryotic Tissues. Biochemistry 2017, 56 (40), 5318–5327. 10.1021/acs.biochem.7b00575. [DOI] [PubMed] [Google Scholar]
- (44).Kneller JM; Lu M; Bracken C An Effective Method for the Discrimination of Motional Anisotropy and Chemical Exchange. J. Am. Chem. Soc 2002, 124 (9), 1852–1853. 10.1021/ja017461k. [DOI] [PubMed] [Google Scholar]
- (45).Peng JW; Wagner G Mapping of Spectral Density Functions Using Heteronuclear NMR Relaxation Measurements. J. Magn. Reson 1992, 98, 308–332. [Google Scholar]
- (46).Peng JW; Wagner G Frequency Spectrum of NH Bonds in Eglin C from Spectral Density Mapping at Multiple Fields. Biochemistry 1995, 34, 16733–16752. [DOI] [PubMed] [Google Scholar]
- (47).Farrow NA; Zhang O; Szabo A; Torchia DA; Kay LE Spectral Density Function Mapping Using 15N Relaxation Data Exclusively. J. Biomol. NMR 1995, 6, 153–162. [DOI] [PubMed] [Google Scholar]
- (48).Lu Z; Van Horn WD; Chen J; Mathew S; Zent R; Sanders CR Bicelles at Low Concentrations. Mol Pharm 2012, 9 (4), 752–761. 10.1021/mp2004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Baker NA; Sept D; Joseph S; Holst MJ; McCammon JA Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc Natl Acad Sci U S A 2001, 98 (18), 10037–10041. 10.1073/pnas.181342398181342398 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Lerner MG PyMOL APBS Plugin. http://mglerner.com/software.php. (accessed 2021-7-16). [Google Scholar]
- (51).Jurrus E; Engel D; Star K; Monson K; Brandi J; Felberg LE; Brookes DH; Wilson L; Chen J; Liles K; Chun M; Li P; Gohara DW; Dolinsky T; Konecny R; Koes DR; Nielsen JE; Head-Gordon T; Geng W; Krasny R; Wei G-W; Holst MJ; McCammon JA; Baker NA Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 2018, 27 (1), 112–128. https://doi.org/ 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Aguilar JA; Nilsson M; Bodenhausen G; Morris GA Spin Echo NMR Spectra without J Modulation. Chem. Commun 2012, 48 (6), 811–813. [DOI] [PubMed] [Google Scholar]
- (53).Jo S; Lim JB; Klauda JB; Im W CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J 2009, 97 (1), 50–58. https://doi.org/ 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Matsuo H; Walters KJ; Teruya K; Tanaka T; Gassner GT; Lippard SJ; Kyogoku Y; Wagner G Identification by NMR Spectroscopy of Residues at Contact Surfaces in Large, Slowly Exchanging Macromolecular Complexes. J Am Chem Soc 1999, 121 (42), 9903–9904. [Google Scholar]
- (55).Pervushin K; Riek R; Wider G; Wüthrich K Transverse Relaxation-Optimized Spectroscopy (TROSY) for NMR Studies of Aromatic Spin Systems in 13C-Labeled Proteins. J. Am. Chem. Soc 1998, 120 (25), 6394–6400. 10.1021/ja980742g. [DOI] [Google Scholar]
- (56).Lagüe P; Roux B; Pastor RW Molecular Dynamics Simulations of the Influenza Hemagglutinin Fusion Peptide in Micelles and Bilayers: Conformational Analysis of Peptide and Lipids. J. Mol. Biol 2005, 354 (5), 1129–1141. https://doi.org/ 10.1016/j.jmb.2005.10.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.