Abstract
Luciferase-based biosensors have a wide range of applications and assay formats, including their relatively recent use in the study of viruses. Split luciferase, bioluminescence resonance energy transfer, circularly permuted luciferase, cyclic luciferase, and dual luciferase systems have all been used to interrogate the structure and function of prominent viruses infecting humans, animals, and plants. The utility of these assays is demonstrated by numerous studies which have not only successfully characterized interactions between viral and host cell proteins but that have also used these systems to identify viral inhibitors. In the present COVID-19 pandemic, luciferase-based biosensors are already playing a critical role in the study of the culprit virus SARS-CoV-2 as well as in the development of serological assays and drug development via high-throughput screening. In this review paper, we provide a summary of existing luciferase-based biosensors and their applications in virology.
Introduction
Biosensors are tools employed in molecular biology that allow for the measurement of biological phenomena. Historically, biosensors have been used to interrogate protein–protein interactions, but a wide breadth of applications has evolved beyond this over recent decades, including disease diagnosis and monitoring, as well as in vivo imaging.1,2 They have also proved useful in the generation of serological assays and the identification of inhibitory drugs via high-throughput screening. In the present COVID-19 pandemic, biosensors may prove to be invaluable, by providing timely information about interactions between the SARS-CoV-2 virus or its variants and host cells.
Traditionally, biosensors are composed of a bioreceptor that allows for the binding of two molecules of interest and the generation of a signal, as well as a transducer for signal detection.3 A common optical signal used in biosensors is bioluminescence, the light produced and emitted by a living organism as a result of chemical reactions occurring within itself.4 Bioluminescent biosensors have historically been used in macroscopic imaging due to their low brightness; however, modifications to bioreceptors and signal detectors are allowing for newer applications in microscopic imaging (e.g., the LV200 system).1 These systems offer increased sensitivity, facilitating the study of photosensitive cells, quantitative analyses, and single-cell resolution. Bioluminescent biosensors have further been adapted for high-throughput screening for chemical biology and drug discovery applications due to their ability to maintain sensitivity, signal strength, and biological fidelity in automated systems.4 Bioluminescent biosensors have several advantages compared to fluorescent biosensors, which make the former increasingly valuable for the development of new detection tools with higher sensitivity and specificity. These include having a higher dynamic range which allows for quantification with minimal background as well as their improved performance in in vivo models. Herein, we provide an overview of the various applications of biosensors to the field of virology. We specifically review the use of split luciferase, bioluminescence resonance energy transfer, circularly permuted luciferase, cyclic luciferase, and dual luciferase systems in the study of viruses and development of novel therapeutics. Finally, we highlight the recent use of biosensors in studying SARS-CoV-2 and emphasize opportunities for future work.
Luciferase Reporters
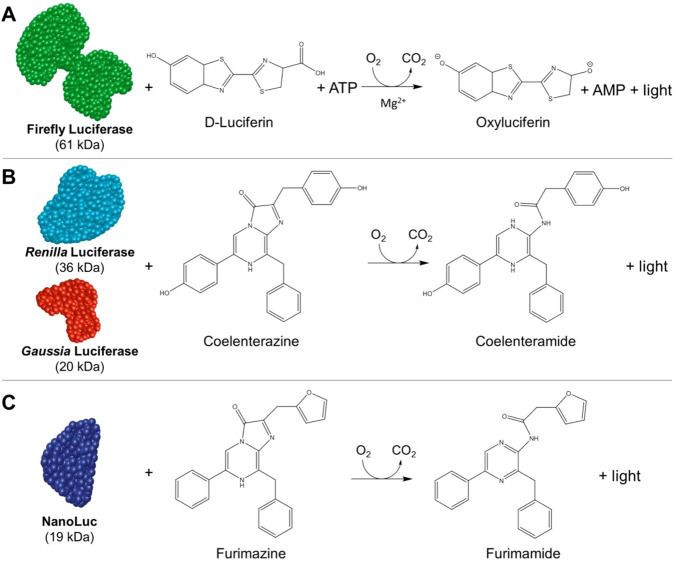
Firefly luciferase (FLuc) and Renilla luciferase (RLuc) are two well-characterized luciferase reporter systems. FLuc was first cloned from the North American firefly Photinus pyralis and catalyzes the oxidation of d-luciferin in the presence of ATP and magnesium ions, emitting a yellow-green light at 560 nm (Figure 1A).5 Luciferin–luciferase reactions have a high quantum yield, and consequently, a significant amount of light is emitted for each chemical reaction that occurs. The high quantum yield coupled with the relatively low toxicity of luciferin makes it an ideal system for a wide range of applications, such as the detection of target protein activity, both in vitro and in vivo.4,6−9 In contrast, light production from RLuc is ATP-independent and uses the substrate coelenterazine to emit a blue light in the presence of oxygen (Figure 1B). Light emission by RLuc additionally requires activation by calcium ions.6 As luminescence assays do not require an external light source to emit light, bioluminescent-based methods can have a high degree of sensitivity despite having a signal weaker than that of fluorescence-based methods and additionally offer a lower degree of interference from light scattering and background fluorescence.1,4 Furthermore, FLuc and RLuc have short intracellular half-lives compared to those of non-enzymatic fluorescent protein reporters and can consequently be used to measure the dynamic changes in reporter transcription levels in cell-based assays.4
Figure 1.
Bioluminescence is light produced as a result of luciferase enzymes reacting with their substrates. (A) FLuc catalyzes the oxidation of d-luciferin in the presence of ATP and Mg2+ to produce light and AMP byproducts. (B) RLuc and GLuc both catalyze a reaction with coelenterazine to emit light in the presence of molecular oxygen. (C) NLuc reacts with a derivative of coelenterazine called furimazine to produce light in the presence of oxygen. Adapted from ref (11). Copyright 2016 American Chemical Society.
More recent bioluminescent systems have employed Gaussia luciferase (GLuc) and nanoluciferase (NLuc) reporters. Like RLuc, GLuc is a luciferase that uses the substrate coelenterazine; however, it originates from the marine copepod Gaussia princeps (Figure 1B). A major advantage of GLuc is that it is naturally secreted from mammalian cells in an active form which avoids the need for cell lysis, making it particularly useful in studies of living cell growth or protein interaction kinetics in live cells.10 NanoLuc (NLuc) is a luciferase developed to overcome the size and stability limitations of FLuc and RLuc. It is derived from the luciferase of deep-sea shrimp, Oplophorus gracilirostris, and reacts with furimazine, an analogue of coelenterazine, in order to produce light (Figure 1C). It offers a 150-fold increase in luminescence compared to that with FLuc and RLuc, in addition to its smaller size and enhanced stability.11 While FLuc, RLuc, GLuc, and NLuc are the most common luciferases used in biosensors, approximately 30 naturally occurring luciferin systems have been discovered to date (Table 1).6 Indeed, bioluminescent systems of bacteria, dinoflagellates, and euphausiids are also used in various analytical techniques as well as those from the click beetle, glow worm, and various species of marine creatures.4
Table 1. Characteristics of the Most Common Naturally Occurring and Synthetic Luciferases and Their Current Use in Biosensors.
| luciferase | organism | substrate | cofactor(s) | size (kDa) | emission wavelength (nm) | used for biosensor (Y/N) |
|---|---|---|---|---|---|---|
| North American Firefly (FLuc) | Photinus pyralis | d-luciferin | ATP and Mg | 61 | 560 | Y |
| click beetle | Pyrophorus plagiophthalamus | d-luciferin | ATP and Mg | 64 | ∼600 | Y |
| Renilla (RLuc) | Renilla reniformis | coelenterazine | Ca2+ | 36 | 480 | Y |
| Renilla mutant (RLuc8) | Renilla reniformis | coelenterazine | Ca2+ | 36 | 535 | Y |
| Gaussia (GLuc) | Gaussia princeps | coelenterazine | N/A | 20 | 470 | Y |
| OLuc | Oplophorus gracilirostris | coelenterazine | N/A | 19 | 460 | N |
| NanoLuc | Oplophorus gracilirostris | furimazine | N/A | 19 | 460 | Y |
| bacterial luciferase (Lux) | bacteria (i.e., Photorhabdus luminescens, Vibrio harveyi) | tetradecanal | oxygen and reduced riboflavin phosphate | 77 | 480 | Y |
Applications of Luciferase Biosensors in Virology
There are many different assay formats in which luciferase biosensors are used, each with their own advantages. Split luciferase complementation assays are most commonly used to detect protein–protein interactions but also have been creatively applied to investigate other aspects of viral replication. Bioluminescence resonance energy transfer (BRET) offers an alternative approach for studying protein–protein interactions and may be useful for characterizing higher-order interactions. Circularly permuted luciferases and cyclic luciferases have been infrequently used to detect viral protease activity. Finally, dual luciferase reporter systems are often used to analyze protein structure and function. In the remainder of the review, we explore these five types of assays and their specific uses in virology.
Split Luciferase
The split luciferase complementation assay is a type of protein complementation assay (PCA) that can be used to monitor cellular events through the detection of protein–protein interactions. PCAs are based on the reconstitution of a bisected reporter gene, wherein fragments of the reporter gene are fused with two proteins of interest.12−17 The interaction of the proteins of interest causes the reporter fragments to be brought into close proximity to one another and regain their reporter function, generating a signal (Figure 2). In split luciferase systems, luciferase enzymes are divided into two fragments (often between the critical hydrophilic and hydrophobic amino acid region).12 The luciferase enzymatic activity is restored upon assembly of the N-terminal fragment with the C-terminal fragment.18 In general, the N-terminus of a luciferase enzyme determines its spectral characteristics, whereas the C-terminus contributes to its enzymatic function.19−21 In contrast to PCAs with green fluorescent protein, luciferase-based PCAs allow for a reversible approach for the detection of temporal dynamics of protein–protein interactions.22 In their application to virology, split luciferase assays have proven incredibly useful in studying protein–protein interactions between viral and host cell proteins. They have also been used to quantify viral protease activity and have further been applied successfully in examining viral cell entry by enveloped viruses. In this section, we provide a comprehensive overview of the use of split luciferase assays in virology, with specific examples from numerous phylogenetically distinct viruses.
Figure 2.
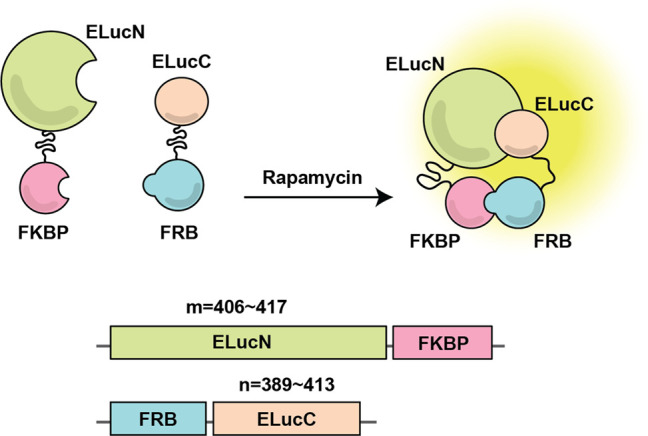
Construction of a split luciferase PCA. In this example, the N-terminus (ELucN) and C-terminus (ELucC) of enhanced green-emitting luciferase (ELuc) are fused to proteins of interest, FK506-binding protein (FKBP) and FKBP rapamycin binding (FRB) domain, respectively. These two proteins interact in the presence of rapamycin, which allows for the ELuc fragments to be brought into close proximity and restored bioluminescence.
Hepatitis B Virus
The split luciferase assay has been used to identify compounds which augment hepatitis B virus (HBV) assembly.23 In the HBV life cycle, two types of protein–protein interactions are required for capsid formation. Core protein dimerization constitutes one of these interactions and refers to the interaction between two core protein monomers to form dimers, which is subsequently followed by association of dimers into larger multimers.24 Nucleocapsid assembly presents an opportunity for therapeutic intervention as any disruption in core protein dimerization will disrupt viral replication. Split luciferase PCAs have been recently used to rapidly detect core protein dimer formation in a quantitative manner for drug discovery. In 2018, Wei et al. created multiple biosensors interrogating dimer formation using FLuc, RLuc, GLuc, and NLuc as well as negative control biosensors wherein the core protein interaction domains were disrupted.23 In generating these biosensors, the authors specifically sought out a construct that would not disturb the functioning of the core proteins while maintaining split luciferase enzyme activity. A RLuc-based biosensor was selected based on these characteristics and was used to screen for an inhibitor among a library of 672 drugs. Two were identified, Arbidol and 20-deoxyingenol. This study serves as a proof-of-concept for future investigations whereby the assay may be adapted and applied to study protein–protein interactions required for the assembly of other viruses.
Flaviviruses
Flavivirus is a genus of positive-strand RNA viruses in the family Flaviviridae. Various luciferase tools have been developed to study this genus. A luciferase-based approach has been used to investigate potential antivirals for the treatment of hepatitis C virus (HCV) infection. HCV replication relies on the maturation of a viral polyprotein precursor which is cleaved into 10 proteins by HCV viral proteases. HCV NS3/4A serine protease cleaves the polyprotein at four sites to liberate five proteins, including NS5A/B.25−27 In vivo methods to test HCV NS3/4A protease activity have been highly sought after as they would allow for the rapid identification of protease inhibitors while simultaneously providing insights into the efficacy, toxicity, and bioavailability of the candidate therapeutics. A split luciferase biosensor has been useful in the creation of an assay for measuring protease activity.28 Split FLuc termini NLuc- and CLuc- were fused to two interacting peptides (pepA and pepB) which were further fused head-to-tail with intervening cleavage sites for the NS3/4A protease. Cleavage by the NS3/4A protease allowed for the release of the NLuc and CLuc fragments, enabling their interaction and restoration of the luciferase activity to produce bioluminescence (Figure 3). Indeed, the level of luciferase activity was shown to be proportional to the amount of active protease in the presence of IFN-α, which has anti-HCV activity. The system was additionally tested and showed good inhibition using NS3/4A-specific shRNAs. Interestingly, this biosensor was further useful in vivo in mice and allowed for temporal monitoring of luminescent activity through repeated imaging.
Figure 3.
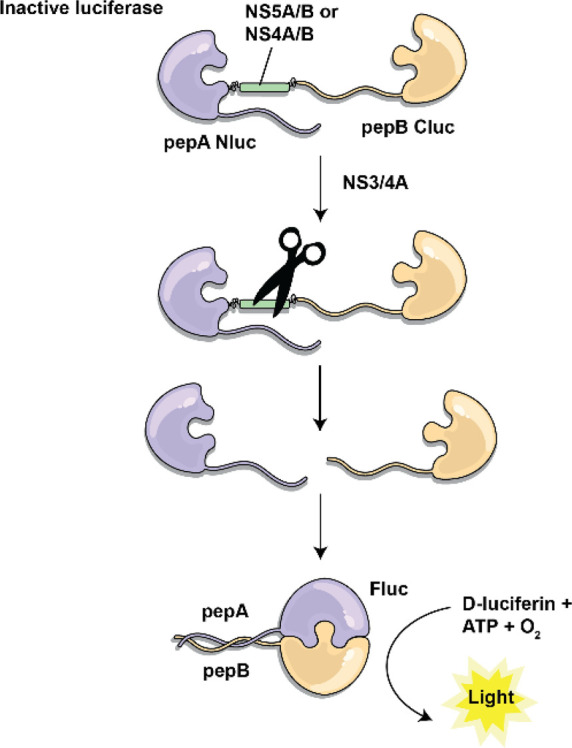
Schematic of a split luciferase biosensor for the detection of HCV NS3/4A protease activity. NLuc and CLuc were fused to interacting peptides (pepA and pepB) which were further linked via an intervening cleavage site, NS4A/B or NS5A/B. When NS3/4A cleaves the construct, pepA and pepB are able to interact and the FLuc fragments are brought into proximity, generating bioluminescence. NS: nonstructural protein. NS3/4A: protease. NS5A/B and NS4A/B: cleavage sites for NS3/4A.
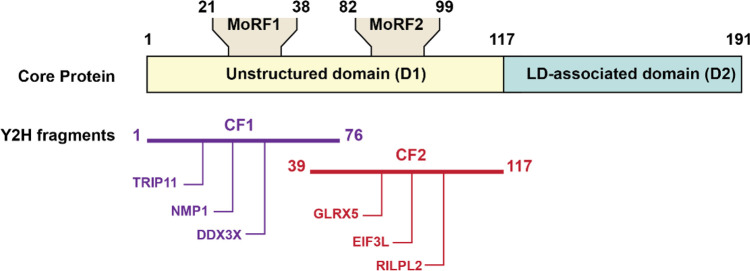
Outside of drug discovery, split luciferase assays have been used to address questions about protein–protein interactions between host cell surface molecules and HCV capsid proteins. These interactions often depend on molecular recognition features (MoRFs)—short sequences embedded in intrinsically disordered regions—within the participating proteins. In 2015, Dolan et al. utilized yeast two-hybrid and split luciferase complementation assays to identify HCV MoRFs.29 The split luciferase complementation assay was based on split FLuc normalized to GFP controls (Figure 4). Four of the six candidate binding partners (DDX3X, NPM1, TRIP11, and RILPL2) that were identified in the yeast two-hybrid assay were confirmed in the luciferase assay, which provided a useful means to identify false positive interactions.
Figure 4.
Schematic representation of potential cellular binding partners of HCV core protein MoRFs. The core fragments used in yeast two-hybrid screening (black bars) have MoRFs (gray rectangles). Each core fragment has three cellular binding partners (ovals). The split luciferase assay is normalized to GFP controls. DDX3X, NPM1, TRIP11, and RILPL2 were identified in the luciferase assay. MoRF: molecular recognition features. LD: lipid droplet. Y2H: yeast two-hybrid. CF: core fragment. Ovals: cellular binding partners.
The aforementioned NLuc protein has also been utilized in split luciferase assays. A commercially available system for this was developed by Promega in their “NanoLuc Binary Technology (NanoBiT)” luminescent protein-fragment complementation assay.30,31 A dissection point was created between the NLuc residues 156 and 157, which generates two peptide fragments: an 18 kDa large BiT (LgBiT) and a smaller 11 amino acid peptide (1.5 kDa) (Figure 5). The cleavage site for NLuc was optimized among 90 circularly permuted variants, wherein the N- and C-termini were relocated and then joined by a cleavable linker to identify fragments that require a proximity constraint to maintain luciferase activity.30 The fragments identified were then adapted to improve their analytical proficiency and stability, and mutations to the 11 amino acid fragment resulted in two popular alternative peptides for binding LgBiT: HiBiT and SmBiT. HiBiT has a high affinity for LgBiT, which makes it ideal for protein tagging and detection. Common experiments using this approach tag a target protein of interest with HiBiT and then reagents containing purified LgBiT and used for detection. In contrast, SmBiT has a lower affinity for LgBiT, which makes it ideal for studying protein–protein interactions. One key advantage of the NanoBiT system is the small size of HiBiT and SmBiT, which minimizes their interference with normal protein function when fusion constructs are created. Overall, NanoBiT provides high detection sensitivity with low steric bulk.
Figure 5.
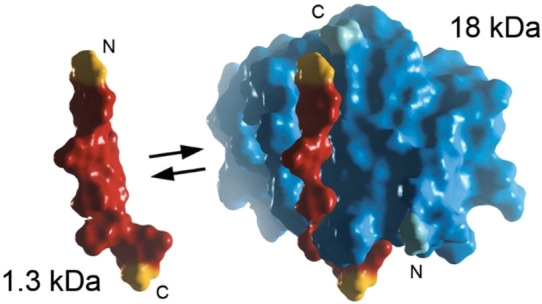
Illustration of the NLuc system for split luciferase assays. The NLuc enzyme can be divided into two subunits: a 1.3 kDa peptide (red) and a larger 18 kDa polypeptide called LgBiT (blue). Mutations to the small 1.3 kDa peptide yielded two well-known fragments: SmBiT and HiBiT (not shown). Adapted from ref (30). Copyright 2016 American Chemical Society.
A split luciferase assay using NanoBiT has been used to analyze viral entry of subviral particles (SVPs) and virus-like particles (VLPs) from flaviviruses. SVPs and VLPs are useful tools in the study of viral entry and release as they do not possess complete viral genomes and are therefore replication-incompetent. Despite this, SVPs and VLPs retain the ability to fuse and bud from target cells.32,33 Sasaki and colleagues generated SVPs (from E and prM proteins) and VLPs (from E, M, and C proteins) from West Nile virus, which were tagged with HiBiT (SVP-HiBiT and VLP-HiBiT, respectively).34 The authors showed that LgBiT and substrate-containing supernatant could be used to detect SVP-HiBiT budding from an infected cell. Similarly, VLP-HiBiT viral infection of cells could further be detected using LgBiT-expressing cells. These series of experiments demonstrate the utility of the NanoBiT system for interrogating factors affecting viral entry and release which could easily be scaled for use in high-throughput screening especially since SVPs and VLPs can be safely used and overcome biosafety restrictions.
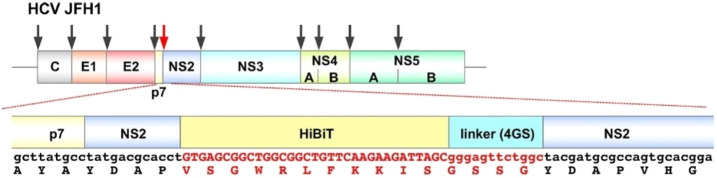
The NanoBiT system has also been useful for overcoming a major hurdle in the study of life cycles of many Flaviviridae. Typically, the flavivirus genome is not conducive to the insertion of large foreign genes, such as those usually used for luminescence or fluorescence. The HiBiT tag, at only 11 amino acids in length, overcomes this limitation. Accordingly, Tamura and colleagues engineered a HiBiT-tagged dengue virus, Japanese encephalitis virus (JEV), HCV, and bovine diarrhea virus and found that the propagation of each of these constructs was comparable to that of parental viruses.35 The optimal insertion sites for the HiBiT tag was determined based on their retention of infectivity and luciferase expression for each virus. For example, the location for insertion of HiBiT in HCV was determined by constructing 10 cDNA clones carrying the HiBiT gene along with a linker sequence in the N-terminus of each viral protein (Figure 6). Of these clones, the viruses carrying HiBiT in the N-terminus of E1, E2, or NS2 were successfully recovered. The recombinant HCV and JEV viruses expressing HiBiT were then used in a screen of a commercially available library of protease inhibitors which identified multiple compounds that significantly suppressed luciferase expression in infected cells.35−37 Thus, viruses encoding HiBiT can be useful tools for screening antiviral agents as well as for monitoring viral dynamics in vitro and in vivo.
Figure 6.
Schematic representation of the genome of a HiBiT-tagged HCV. Arrows indicate locations for insertion of HiBiT in the HCV genome to construct 10 different candidate tagged viruses. The clone with placement of the tag at NS2 (red arrow) provided the highest viral titer and stronger luciferase activity. HCV JFH1: hepatitis C virus (HCV) genotype 2a isolate. E1 and E2: two subunits of the envelope glycoprotein found in HCV. NS2, NS3, NS4, and NS5: nonstructural proteins in HCV. p7: membrane-associated ion channel protein. Linker (4GS): glycine–serine-rich linker. Adapted with permission from ref (35). Copyright 2018 American Society for Microbiology.
A third application of split luciferases to the study of flaviviruses was a screen for orthosteric inhibitors of the Zika virus protease complex, a high-priority drug target due to its essential role in viral replication. This complex is composed of the cofactor NS2B and the NS3 protease that cooperate with host proteases to cleave the viral polyprotein precursor.38,39 Various protein fusion constructs were created based on the split FLuc assay, with constructs NLuc-NS2B aa 49–66 (named NLuc-E66stop) and GST-CLuc-NS3 (named GCN) selected for their high signal when combined.40 A known orthosteric inhibitor of NS2B-NS3 interactions, SK-12, served as a positive control and caused a significant reduction in luciferase activity. The split luciferase assay was then adapted to allow for quantitative high-throughput screening of 2816 drugs from the National Institutes of Health Chemical Genomics Center Pharmaceutical Collection library. The screen identified three drugs (temoporfin, niclosamide, and nitazoxanide) as potential drug candidates inhibiting the protease complex. Collectively, these studies highlight how split luciferase assays have proved incredibly useful for the study of flavivirus replication and identification of antiviral agents.
Orthomyxoviridae
The influenza A virus (IAV) polymerase (PA-PB1-PB2) is a heterotrimer that plays an essential role in the virus life cycle by synthesizing viral mRNAs and genomic negative sense RNA via a complementary positive sense RNA intermediate.41 While IAV vaccines have been effective in curbing infections, the high mutation rate of IAV necessitates the development of annual vaccines as well as new antivirals for those individuals who develop severe disease. The viral polymerase is an attractive drug target composed of three viral subunits (PB1, PB2, and PA), and inhibitors impairing the protein–protein interactions between polymerase subunits are highly sought after. In a recent study, Zhang et al. generated a split luciferase reporter for the interaction between polymerase subunits PA and PB1 wherein N-terminal FLuc was fused to PB1 and C-terminal FLuc was fused to PA (Figure 7).18 Once again, the authors of this study tested three fusion partners to generate their biosensor, and ultimately, the GC-PAC/PB1N-NLuc pair was selected for drug screening as it produced a high luciferase signal. The reporter was then used to screen 10 000 compounds from the Myria Screen Diversity Collection, and two potential PA–PB1 interaction inhibitors, R160792 and R151785, were successfully identified. The drugs were then tested against a panel of human influenza A and B viruses, with both R160792 and R151785 showing antiviral activity.
Figure 7.
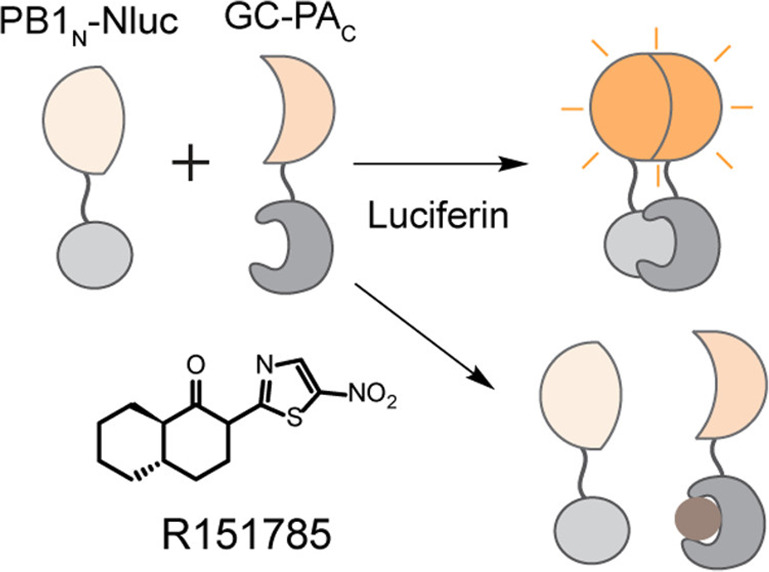
Schematic representation of split NLuc biosensor for the quantification of interactions between PA and PB1 subunits of influenza A virus and influenza B virus polymerase. The constructs GC-PAC/PB1N-NLuc produce a strong bioluminescence signal when the PA and PB1 constructs interact (top panel). PA–PB1 inhibitors (e.g., small molecule R151785) disrupt the interaction between PA and PB1 and therefore reduce biosensor signal. PB1 and PA: subunits in influenza virus polymerase. PB1N and PAC: N-terminus of PB1 and C-terminus of PA. GC: GST-CFLuc or glutathione S-transferase tagged C-terminal luciferase. R151785: potent PA–PB1 interaction inhibitor. Adapted from ref (18). Copyright 2020 American Chemical Society.
A split luciferase assay has been used to examine other aspects of influenza virus biology including the identification of barriers to influenza A virus (IAV) infection in poultry.42 An understanding of host–virus protein interactions underlying IAV replication is likely to form the basis for the development of antivirals to protect against zoonotic transmission and also may eventually be used to inform mechanisms to reduce the susceptibility of poultry to IAV infection. The ability of both avian- and mammalian-adapted IAV to replicate in chicken cells is reliant on an interaction between the IAV polymerase with the chicken host factor ANP32A but not ANP32B, whereas human homologues of ANP32A and ANP32B support the polymerase activity of human-adapted IAV. Long et al. used a split luciferase complementation assay to quantify interactions between avian-adapted influenza virus polymerase and chicken ANP32A/B proteins and to determine the effects of genetic mutations on these interactions. Specifically, the authors found that avian-adapted influenza polymerase relies solely on chicken ANP32A, as chicken ANP32B was inactive, to support viral replication. In order to demonstrate this, the PB1 polymerase subunit of an avian-adapted IAV was fused to one-half of GLuc and ANP32A or B was fused to the other half (Figure 8). ANP32A complementation produced a stronger luminescent signal when compared to those from ANP32B or a mutated ANP32A construct that was modified to contain amino acids 129I and 130N from ANP32B.
Figure 8.
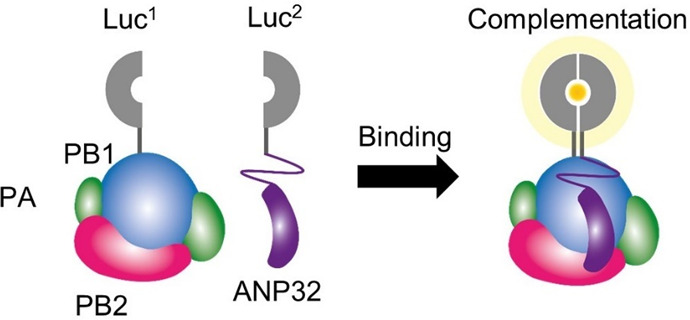
Schematic representation of the split GLuc assay to examine the interaction between avian IAV polymerase and chicken ANP32A/B proteins. When the polymerase-containing PB1 fused to luciferase fragment 1 (Luc1) and ANP32A or B fused to luciferase fragment 2 (Luc2) interact, the luciferase fragments produce a bioluminescence signal. PB1: polymerase basic 1 protein. PB2: polymerase basic 2 protein. PA: polymerase acidic protein. ANP32: acidic nuclear phosphoprotein 32 kDa. ANP32A: protein responsible for viral replication in avian IAV. ANP32B: variant of ANP32A. Adapted with permission from ref (42). Copyright 2019 Long et al.
Measles Morbillivirus
The role of the measles morbillivirus (MeV)C protein in viral replication is relatively poorly understood. In contrast, the phosphoprotein is a polymerase cofactor that has been implicated in viral transcription and replication.43,44 A split Gaussia luciferase assay has been used to study the role of the C protein in MeV replication and to determine whether it is a component of the replication complex. Split domains of GLuc were fused to the measles virus phosphoprotein and C protein, and the coexpression of the two viral proteins resulted in GLuc activity, reflecting the phosphoprotein–C protein interaction.44 A second assay in which both halves of GLuc were fused to C proteins was used to suggest that C proteins can also form homo-oligomers. Interestingly, further split luciferase assays were used in this study to support a lack of interaction between the C protein with the nucleocapsid protein and the RNA-dependent RNA polymerase. Ultimately, split luciferase assays formed a key methodology for the characterization of the MeV C protein as a ribonucleocapsid-associated protein within the viral replication complex, where it serves to increase replication accuracy and processivity of the polymerase.
Herpesviruses
The Epstein–Barr virus (EBV), also known as human herpesvirus 4, is the causative agent for infectious mononucleosis and has been studied using a split Renilla complementation assay. The EBV protein kinase BGLF4 is expressed during the early and late stages of the lytic cycle of EBV and has been identified in all human herpesviruses.45 BGLF4 phosphorylates a number of viral cellular proteins, such as the cell cycle regulator protein p27, and is also responsible for modulating nuclear pore structure and transportation, which facilitates the import of EBV lytic proteins.46 BGLF4 associates to Hsp90, a heat-shock protein which stabilizes BGLF4 expression in cells, and maintains the structure and function of various interacting proteins. A split RLuc complementation assay was developed by Wang et al. to detect interactions between the EBV protein kinase BGLF4 and the heat-shock protein Hsp90.47 Multiple configurations of the reporter were tested, with the combination of BGLF4-LN and LC-Hsp90 performing best in baseline assays. The relative contributions of BGLF4 functional domains to its interaction with Hsp90 were investigated using a series of deletion mutants. Through these experiments, the authors identified BGLF4 amino acids 250–295 as being essential to this interaction, with specific residues Phe-254, Leu-266, and Leu 267 being of particular importance. This finding provides key insights into the pathological mechanisms of EBV replication and may serve as a basis for further studies aimed at the development of inhibitors of this interaction.
Split luciferase assays also have proven fruitful in studying virus entry into host cells. A split Renilla luciferase assay for this purpose was designed by Kondo et al., who created a clever approach to studying the kinetics of HIV entry.48 In brief, in this approach, two populations of cells are created: the first which expresses the virus glycoprotein involved in attachment to the host cell receptor and half of the luciferase enzyme, and a second population expressing the viral receptor and the complementary half of luciferase. These populations are then cocultured, and ligand/receptor binding causes cell fusion, content mixing, and luciferase complementation. This creative assay illustrates the broad utility of split luciferase assays in characterizing the dynamics of complex cellular processes which are temporally and spatially regulated.
As an enveloped virus, Herpes simplex virus (HSV) host cell entry is dependent on the interactions of HSV glycoproteins gD, gH/gL, and gB, which mediate viral envelope/host cell membrane fusion while undergoing large conformational changes; however, the specific protein–protein interactions involved in this complex process remain to be fully understood.49,50 A series of studies using split luciferase assays and fluorescence complementation assays to investigate HSV-glycoprotein-mediated cell fusion have been conducted to study the role of glycoprotein gB in membrane fusion (Figure 9).51−55 In these studies, two populations of cells are created (one expressing the HSV receptor on the host cell, nectin-1, and half of RLuc; the second expressed viral glycoproteins gD, gH/gL, and gB and the complementary half of RLuc). Coculture of these two populations allows for nectin-1/glycoprotein binding and cell–cell fusion, as well as complementation of the luciferase enzyme activity. After characterizing the assay performance, the authors were further able to use this assay to interrogate many aspects of membrane fusion. As an example, in one set of experiments, they test the contributions of specific gB residues to the process of fusion using mutant gB glycoproteins. In doing so, they compared the fusion kinetics of wild-type gB, where initiation of cell fusion could be detected after 7 min and the rate remained constant for 8 h, with those of the mutant gB constructs. Conclusions could be drawn about conformational changes in gB during fusion by examining the dynamics observed in mutant phenotypes.
Figure 9.
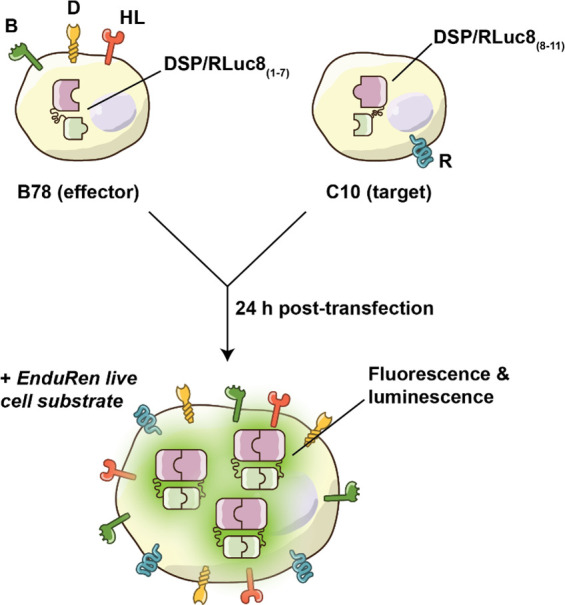
Schematic representation of split luciferase assay to measure cell–cell fusion mediated by the HSV glycoprotein. Effector cells are transfected with the HSV glycoprotein and one-half of RLuc and GFP. Target cells are transfected with the other half of RLuc and GFP. After 24 h, RLuc substrate coelenterazine (EnduRen) is added and the two cell types are mixed. If the cells fuse together, the RLuc halves can reunite to produce a bioluminescence signal. B78: mouse melanoma cells. C10: human colon cells. DSP: dual split protein. RLuc8: variant of RLuc. B, D, HL (on B78): gB, gD, and a heterodimer gH/gL which are four glycoproteins contained within the viral envelope. R (on C10): receptor.
Henipaviruses
The Henipavirus genus of viruses includes the nonpathogenic Cedar virus as well as several closely related highly pathologic viruses including Hendra virus (HeV) and Nipah virus (NiV).56 Virus entry of HeV and NiV is mediated by host cell ephrin-B2 and ephrin-B3 expression.57,58 Similar to the previously described studies on virus–host membrane fusion, a luciferase-based approach has been used to monitor membrane fusion kinetics of wild-type and recombinant Cedar viruses and to interrogate the role of ephrin-B2 and ephrin-B3 in this process. Laing et al. transfected HeLa-USU cells that are nonpermissive for fusion and virus infection with one-half of split RLuc and transfected another culture of HeLa-USU cells with the other half of the split luciferase and further infected this second group of cells with a recombinant Cedar virus (containing a GFP gene) (Figure 14).57 When the cell cultures were combined and luciferin substrate was added, content mixing between the cell populations occurred, and the levels of fusion could be assessed by luciferase assay. This assay showed that the HeLa cells overexpressing the ephrin-B2 receptor had the highest and fastest level of cell–cell fusion, whereas the standard cells only supported a small amount of fusion.
Figure 14.
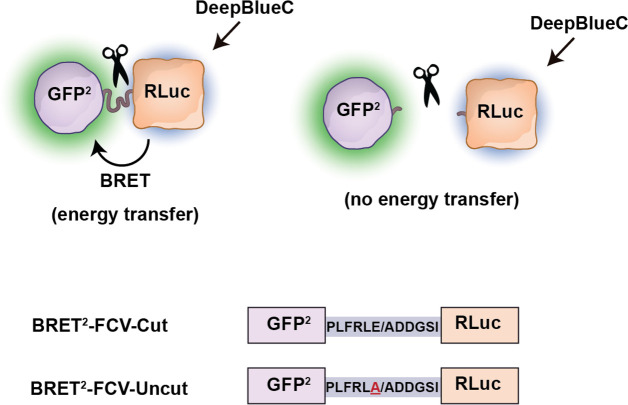
Schematic representation of wild-type and mutant BRET2-based probes for detection of FCV protease activity. Top: Energy transfer dissipates, and BRET2 signal disappears when GFP2 and RLuc are distanced upon cleavage by the FCV protease. Bottom: Amino acid sequences of the cut and uncut FCV protease cleavage motifs are shown. The mutation is underlined at the bottom uncut construct. FCV protease cleaves peptide bonds right after the glutamic acid (E), indicated by a slash. GFP2: green fluorescent protein 2. RLuc: Renilla luciferase. DeepBlueC: substrate of Rluc.
Human Papillomavirus
As a final example of the utility of split luciferase assays in virology, the split luciferase assay has been scaled to a high-throughput format for studies of “interactomics”. Neveu et al. used this approach to provide a comprehensive assessment of interactions between human papillomavirus (HPV) E6 and E7 proteins and host proteins.59 E6 and E7 proteins from 11 HPV genotypes were fused to split GLuc, and the other half of GLuc was fused to nearly 100 different cellular targets of interest. The normalized luciferase ratio from each of these interactions was then used to perform agglomerative hierarchical clustering, an analytical technique which groups the viral protein genes and sequences by their similarity. The split luciferase assay elucidated virus–host interactions by identifying E6 and E7 cellular targets, provided insight into the tropism of the viruses, and further demonstrated the potential for split luciferase assays to be scaled to answer questions about protein interaction networks.
Bioluminescence Resonance Energy Transfer
BRET is a relatively common and versatile technique for studying protein–protein interactions in living cells. Based on the principle of Förster resonance energy transfer, BRET occurs when there is a nonradiative energy transfer between a light-emitting enzyme (donor) and a fluorescent acceptor which have overlapping emission and absorption spectra (Figure 10).60 BRET occurs naturally in some marine species, such as the sea pansy, and involves RLuc and GFP, which act as the energy donor and acceptor, respectively.61 The components of this system were purified and served as the first nonradiative energy transfer system that was assembled in vitro.62 This system is often used to study protein–protein interactions by fusing interacting proteins of interest to respective BRET partners and measuring fluorophore activation.
Figure 10.
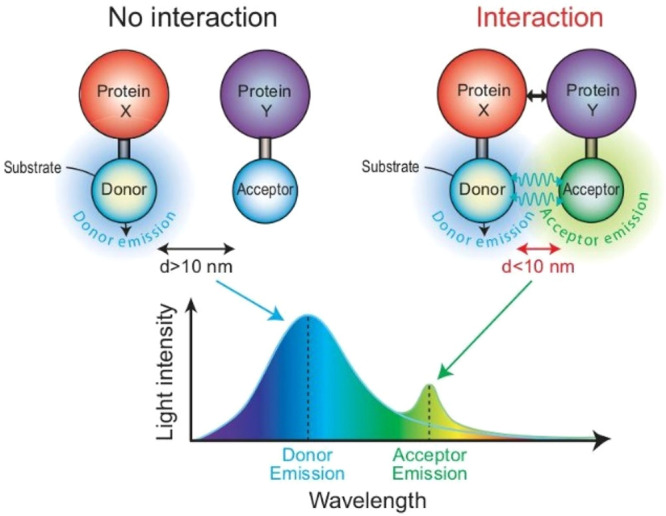
Schematic representation of a BRET biosensor. The donor and acceptor have overlapping emission and absorption spectra. Nonradiative energy is only transferred between a light-emitting donor and a fluorescent acceptor when the two are less than 10 nm apart. Proteins X and Y are interacting proteins fused to the donor and acceptor and allow for the two substrates to interact and emit light or BRET signal from the acceptor. Protein interactions can be determined by BRET by comparing donor and acceptor emission wavelengths, which differ in light intensity over wavelength. Adapted with permission from ref (60). Copyright 2008 Wiley-VCH Verlag GmbH & Co.
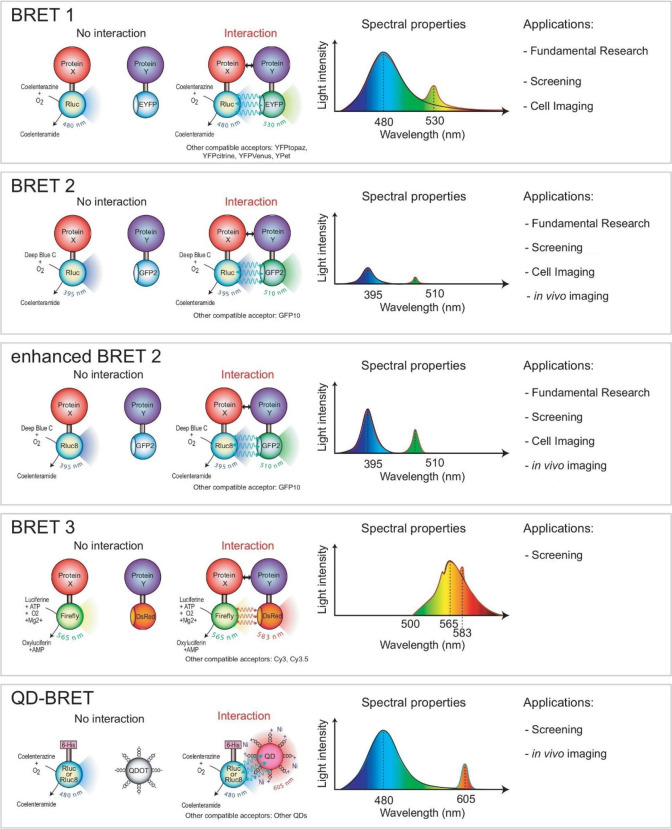
Like split luciferase techniques, BRET systems have undergone iterative modifications to generate partner pairings with varying applications and spectral properties. The original version, BRET1, is based on an interaction between RLuc and enhanced yellow fluorescent protein, and it is commonly used in fundamental research, drug candidate screening, and cell imaging.63 BRET2, in contrast, uses coelenterazine 400a as an alternative substrate for RLuc, shifting the maximal light emission of RLuc to facilitate excitation of GFP.64 This modification allows for greater separation between the donor and acceptor wavelengths and is better suited for assays that require a low signal-to-noise ratio, such as screening assays.65 BRET2 is also particularly useful for in vivo imaging. Further modifications to BRET1 and 2 have yielded enhanced BRET2 (with greater signal intensity compared to that with BRET2) as well as BRET3 and QD-BRET. In order to achieve lower cellular autofluorescence and a prolonged emission signal, BRET3 uses FLuc is used as a donor and DsRed fluorescent protein as the acceptor.60 Quantum dots are used in QD-BRET as acceptors (with a form of RLuc as the donor), which provides the advantages of distinct separation between donor and acceptor emission peaks as well as improved performance in screening assays and in vivo imaging. The most commonly used BRET systems are summarized in Figure 11.60
Figure 11.
Schematic representation of different BRET methods. Each method is characterized by different BRET donors and acceptors, spectral properties, and applications. BRET2 uses coelenterazine 400a, which shifts the emission of RLuc to that of GFP and has a larger separation of donor and acceptor emission peaks. Enhanced BRET2 has a light intensity stronger than that of BRET2. BRET3 uses FLuc and DsRed, which has a prolonged emission signal. QD-BRET uses quantum dots and has a distinct separation of donor and acceptor emission peaks. GFP2: green fluorescent protein 2. RLuc8: variant of Renilla luciferase. DsRed: red fluorescent protein. 6-His: polyhistidine tag. QD or QDot: quantum dots. Adapted with permission from ref (60). Copyright 2008 Wiley-VCH Verlag GmbH & Co.
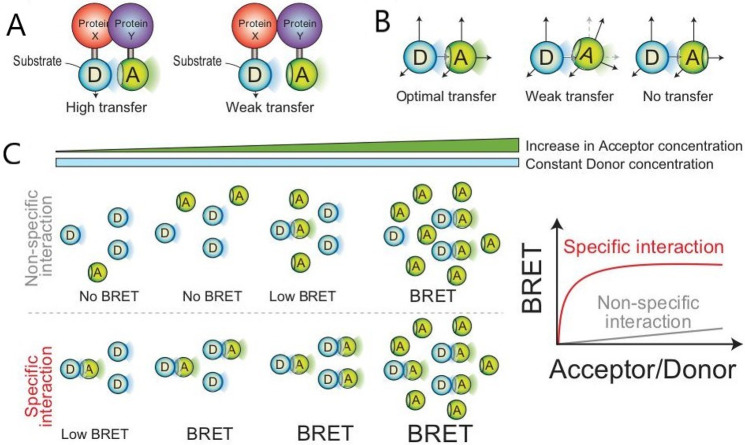
Regardless of the particular BRET system used, the intensity of the BRET signal largely depends on three factors: the distance between the donor and acceptor, the relative orientation of the BRET partners, and the donor to acceptor ratio (Figure 12). Distance is a particularly important factor, as for resonance energy transfer to occur the distance between the energy donor and acceptor must be less than 10 nm. The intensity of the signal is further proportional to the distance between the two molecules and increases as this nears 1 nm.66,67 In their application to studying protein–protein interactions, BRET partners are fused to two proteins of interest. If the two proteins interact, the BRET partners are brought into close proximity, resonance energy transfer occurs, and a signal which corresponds to the acceptor re-emission can be detected.63,68,69 Otherwise, if no interaction occurs, light only emits from the initial energy donor (luciferase). The orientation of BRET partners is also critical, due to the dipole–dipole nature of the transfer mechanism. A negative signal from a BRET protein–protein interaction assay must be carefully interpreted as the absence of a BRET might indicate that the protein partners do not interact but could also result from the BRET partners being in a suboptimal orientation to interact.67,70 Peptide linkers can be used to augment the energy donor and acceptor insertion site in an attempt to change in the orientation of the BRET partners and overcome this problem.65 Finally, the relative expression of donors and acceptors affects the ultimate signal produced. If BRET partners are concentrated within specific subcellular locations, such as within cell organelles or membranes, interaction is increased.60,61 These factors are essential to consider in designing and interpreting BRET assays.
Figure 12.
Schematic representation of parameters that affect the BRET signal strength. (A) First, the distance between partners, as the distance between partners decreases from 10 nm, the signal intensity increases. (B) Second, the orientation of the acceptor, as a strong energy transfer only occurs when the dipoles of the donor and acceptor are perfectly aligned, which means peptide linkers may be required to change the insertion sites and, ultimately, the orientation of the partners. (C) Finally, with the acceptor/donor ratio, as the concentration of acceptors increases in defined spaces of organelles, there are more interactions between the partners and a stronger signal. Adapted with permission from ref (60). Copyright 2008 Wiley-VCH Verlag GmbH & Co.
BRET protein–protein interaction assays have been applied to virology and are frequently used to determine viral protease activity and characterize protein–protein interactions/dimerization. In this section, we highlight key studies using this technique to investigate the biology of human immunodeficiency virus, calicivirus, respiratory syncytial virus, herpesvirus, as well as the host-mediated antiviral innate immune response.
Human Immunodeficiency Virus
Much like the aforementioned studies using split luciferase to assess viral protease activity, BRET2 has similarly been used to measure protease activity in human immunodeficiency virus type 1 (HIV-1). Hu et al. created a fusion construct with donor RLuc and acceptor-humanized GFP expressed on either sides of an HIV-1 protease cleavage site (Gag-p2/Gag-p7) (Figure 13).71 When expressed as a single fusion protein, the BRET partners exist in close proximity to each other, allowing for energy transfer and signal detection. However, coexpression with HIV-1 protease causes the construct to be cleaved and the BRET signal is lost. Protease inhibitors such as Saquinavir or Amprenavir, as well as a mutant form of the protease cleavage site, resulted in stable BRET signals, demonstrating the ability of this assay to accurately test protease inhibition.
Figure 13.
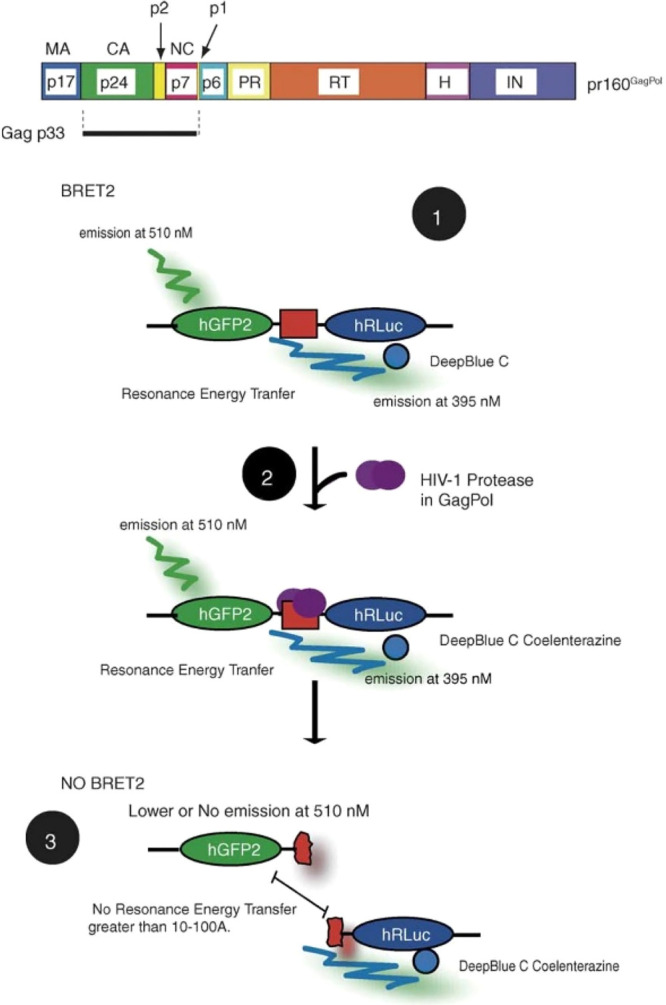
Schematic representation of BRET-based biosensor to detect HIV-1 protease activity. In the top panel, the proteins in pr160GagPol and protease cleavage sites are shown. In the bottom panel, the process of BRET2 assay is shown: (1) RLuc substrate DeepBlue C is added and interacts with hRLuc, and resonance energy is transferred to hGFP2; (2) HIV-1 protease cleaves its substrate between hGFP2 and hRLuc; (3) resonance energy transfer is low, and there is less light emission from the acceptor molecule. The inset legend on the right panel displays all of the PR cleavage sites and mutant sites tested. pr160GagPol: polyprotein encoded by HIV-1. Gag p33: Gag processing intermediate. p1 and p2: spacer peptides. MA: matrix protein. CA: capsid protein. NC: nucleocapsid protein. PR: protease. RT: reverse transcriptase. H: RNaseH. IN: integrase. hRLuc: humanized sea pansy Renilla reniformis luciferase; donor molecule. hGFP2: humanized green fluorescent protein; acceptor molecule. Adapted with permission from ref (71). Copyright 2005 Elsevier B.V.
In another study, BRET was used to test for a putative protein–protein interaction between the HIV-1 Nef protein, which is essential for efficient viral replication, and the host factor CD4.72 Cluet and colleagues hypothesized that the formation of a Nef/CD4 complex leads to Nef-induced downregulation of CD4. To test this, Nef was fused to RLuc and CD4 was fused to enhanced yellow fluorescent protein. Indeed, in the presence of the RLuc substrate coelenterazine, luminescence was transferred from RLuc to the enhanced yellow fluorescent protein and a signal corresponding to the enhanced yellow fluorescent protein emission spectrum was observed, supporting a close interaction between these two proteins.
Calicivirus
BRET2 has been used to detect calicivirus protease activity in cell-free systems and in vitro in infected cells.73 Feline calicivirus (FCV) traditionally causes upper respiratory disease in cats, although newer, more virulent strains of FCV have been reported to cause systemic hemorrhagic disease.74 The FCV protease plays an essential role in the formation of mature infectious virus particles and represents an attractive target for the development of FCV therapeutics.75,76 Oka and colleagues used BRET2 to identify inhibitors of the FCV protease (Figure 14).73,77 Their construct consisted of a FCV protease cleavage site bordered by RLuc and GFP. They further created a second control BRET2 system with a mutant cleavage motif, which they used to confirm the specificity of the system. This study highlights a convenient approach for the generation of experimental and control BRET biosensors for measuring enzyme activity that could presumably be scaled for screening.
Respiratory Syncytial Virus
Respiratory syncytial virus (RSV) infects epithelial cells in the respiratory tract and can cause serious complications such as pneumonia and bronchiolitis in children, immunosuppressed individuals, or the elderly.78,79 The structure of RSV is fairly complex. RSV is an enveloped virus with its genome contained within a nucleocapsid.80 A layer of matrix protein exists external to the nucleocapsid, between the nucleocapsid and the envelope, where they are thought to play a role in virus assembly and retention of the virus shape. Matrix proteins are thought to assemble into oligomers, although evidence supporting this has historically been limited.81,82 BRET, in conjunction with confocal laser scanning microscopy, was used to test for potential self-interaction between RSV matrix proteins in mammalian cells.83 To show this, a BRET donor and acceptor were created from matrix proteins. RLuc–matrix protein was expressed in the presence of the yellow fluorescent protein–matrix protein, and a signal corresponding to transferred emission was observed, supporting the existence of oligomerization between matrix proteins. Mutations affecting this dimerization led to a reduced BRET signal.
Herpesvirus
BRET has also proven to be useful in characterizing more complex protein–protein interactions. Through the formation of heteromeric complexes, G-protein-coupled receptors (GPCRs) are able to affect the functioning of other GPCRs. Nijmeijer et al. used bimolecular complementation as well as BRET (RLuc and mVenus) to reveal the formation of hetero-oligomeric complexes between the EBV-encoded GPCR BILF1 and human chemokine receptor CXCR4.84 They further found that BILF1 impairs chemokine CXCL12 binding to CXCR4, thereby weakening CXCR4 signaling. Split luciferase complementation, BRET, and fluorescence complementation techniques were then used in combination to study higher-order protein–protein interactions. Fusion of alphaBTX-binding site (BBS) to BILF1 allows detection of receptor proteins by high affinity binding of TMR-alphaBTX or alphaBTX. Coexpression of BBS-BILF1 and CXCR4 both fused to opposite halves of RLuc resulted in complementation and luciferase expression, and the split mVenus assay showed similar results (Figure 15). The reconstituted L1/L2 and V1/V2 heterodimers were mixed, and the BRET signal detected between the two complexes showed higher-order BILF1-CXCR4 protein complexes. All combinations of RLuc8 and mVenus complementation had an observable BRET signal, showing that BILF1 and CXCR4 form complexes consisting of a minimum of four protomers.
Figure 15.
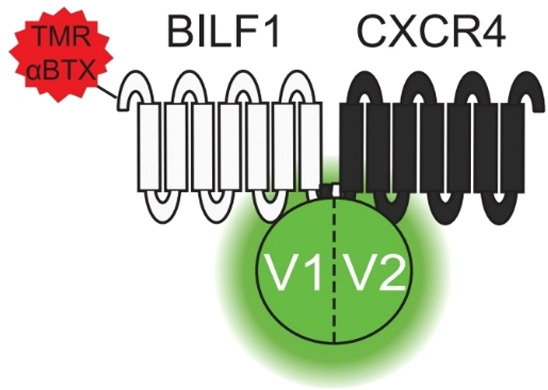
Schematic representation of BBS-BILF1 protomer in EBV labeled with TMR-alphaBTX, fused to venus luciferase. Split RLuc (not shown) and split venus luciferase were used in the assays. Fusion of BILF1 to TMR-alphaBTX allows detection of receptor proteins. Coexpression of BBS-BILF1 and CXCR4, fused to fragments of mVenus, resulted in complementation and signal production. The same was shown for RLuc, which indicates that BILF1 and CXCR4 form complexes of at least four protomers. V1, V2: two halves of mVenus. mVenus: Venus luciferase, a yellow fluorescent protein. BBS: αBTX-binding site. BILF1: Epstein–Barr virus-encoded F protein-coupled receptor. CXCR4: human chemokine receptor. TMR-alphaBTX: α-bungarotoxin-tetramethylrhodamine. Adapted with permission from ref (84). Copyright 2010 The American Society for Biochemistry and Molecular Biology, Inc.
Innate Antiviral Immune Response
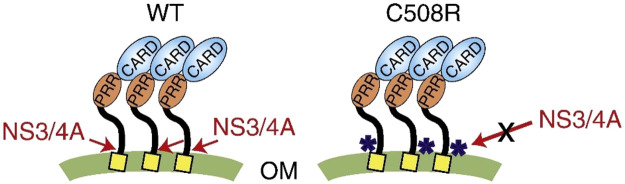
In mammals, mitochondrial factors act as one of many cellular mechanisms for antiviral innate immunity.85 The response to viral antigens depends in part on the mitochondrial antiviral signaling (MAVS) scaffold protein located on the outer mitochondrial membrane and activation of the cytoplasmic retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) signaling pathway leading to upregulation of NF-kB and IRF-3 through the recruitment of various downstream effectors. Upon infection with an RNA virus, RLRs translocate to the mitochondrial surface where they will interact with MAVS and trigger changes in MAVS concentration.86 This process has been implicated in the response to HCV, Dengue virus, Japanese encephalitis virus, Rabies lyssavirus, and influenza A viruses.87,88 A BRET system based on RLuc and Venus has been used to identify homodimer formation by MAVS (Figure 16). The formation of these homodimers leads to activation of MAVS and binding to downstream signaling. Furthermore, the BRET signal was disturbed by inhibitors of the RLR pathway, demonstrating that these inhibitors can differentially affect MAVS dimerization.86
Figure 16.
Schematic representation of wild-type MAVS and MAVS with cysteine-508 residue mutated to arginine in a BRET assay. NS3/4A cleaves MAVS at cysteine-508 and prevents MAVS–MAVS interactions. The wild-type MAVS can be cleaved, and the mutated RLuc-MAVS and Venus-MAVS cannot be cleaved. The BRET assay was set up to ultimately detect MAVS homo-oligomerization and used RLuc as the donor and Venus as the acceptor, both of which are fused to MAVS. MAVS: mitochondrial antiviral signaling. NS3/4A: hepatitis C virus protease. PRR: proline-rich region. CARD: caspase activation and recruitment domain. OM: mitochondrial outer membrane. Venus: yellow fluorescent protein. Adapted with permission from ref (86). Copyright 2013 Elsevier B.V.
Circularly Permutated and Cyclic Luciferases
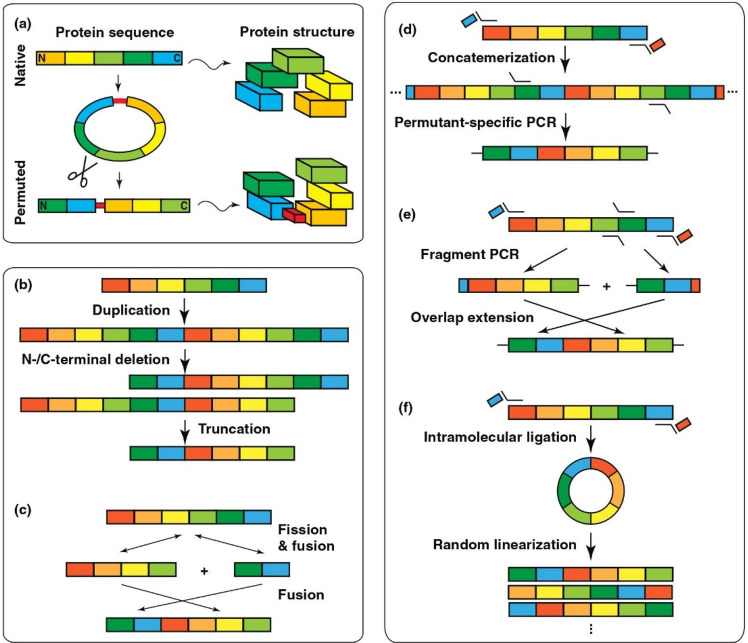
Circular permutation refers to a mechanism of reorganizing the sequence of a polypeptide chain to alter tertiary/quaternary structure, substrate binding affinity, catalytic activity, and dynamics and furthermore has been used to design biosensors.89 In this process, the N- and C-termini of a native protein are connected using a covalent linker, and the resulting circular structure is cleaved at a distinct existing peptide bond to create new ends (Figure 17a). Unlike most traditional methods of protein engineering, circular permutation does not result in amino acid substitutions, rather only changes the order of residues. Circular permutation also occurs naturally as a result of either duplications/deletions or fission/fusion of gene fragments (Figure 17b,c). In its application to luminescent biosensors, luciferase is inactivated by circular permutation, and the fragments are connected by a cleavage site corresponding to a protease under investigation (Figure 17d–f and Figure 18). In this way, circular permutation is reminiscent of the application of split luciferase assays to measuring protease assays. Indeed, if the protease of interest is present and functional, it will cleave the peptide at the protease site and the circularly permutated luciferase regains its active conformation.
Figure 17.
Schematic representation of circular permutation in nature and in the laboratory. (a) Protein sequence reorganization by circular permutation. The ends of the protein sequence are ligated, and a peptide bond within the protein is cleaved to create new ends. Natural circular permutation via (b) duplication (repetition of sequences)/deletion (omission of sequences) and (c) fission/fusion (sequences breaking apart and joining back together on different ends). (d,e) Individual gene permutations at the gene level could be created by permutant-specific and fragment PCR. (f) Generation of comprehensive permutation libraries by intramolecular ligation and random linearization. Adapted with permission from ref (89). Copyright 2011 Elsevier Ltd.
Figure 18.
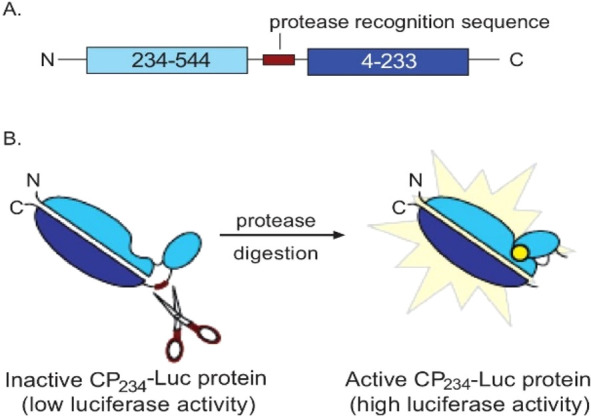
Schematic representation of an example assay with circularly permuted (CP) luciferase at Asp234 (CP234-Luc). CP-luciferase was created by joining the N- and C-termini of luciferase via inserting a protease recognition sequence. (A) New N- and C-termini were added, respectively, at amino acid positions 234 and 233. (B) Cyclization of the luciferase lowers its luciferase activity. Cognate protease digestion at the protease recognition sequence reactivates the CP234-Luc protein. Adapted with permission from ref (90). Copyright 2008 Wigdal et al.; Licensee Bentham Open.
The most notable application of circularly permutated luciferase to the study of virology was its use in the identification of potential inhibitors of Middle East respiratory syndrome coronavirus (MERS-CoV). MERS-CoV infection causes a respiratory syndrome characterized by severe respiratory distress and a mortality rate as high as 50%.92,93 No specific antiviral treatments exist for treatment of MERS-CoV infections; however, there is tremendous interest in the development of inhibitors of MERS-CoV proteases. Two main proteases have been found to be critical in the MERS-CoV replication cycle—papain-like proteases and 3-chymotrypsin-like proteases.94 In a search for inhibitors of these proteases, Killianski and colleagues created circularly permuted luciferase constructs with linkers corresponding to the papain-like protease and 3-chymotrypsin-like protease cleavage sites (Figure 19).95 Inhibitors were hypothesized to show minimal bioluminescence in the circular permutation assay as the activity of the protease is needed to restore the functioning of luciferase. Indeed, a known inhibitor (chloropyridine ester 5) for the related SARS-CoV protease caused a sharp decline in the luciferase activity of the circularly permutated MERS-CoV 3-chymotrypsin-like protease luciferase construct, suggestive protease inhibition. Thus, circularly permutated luciferase assays may provide an additional means for interrogating protease function in studying viral replication and identifying factors which augment these reactions.
Figure 19.
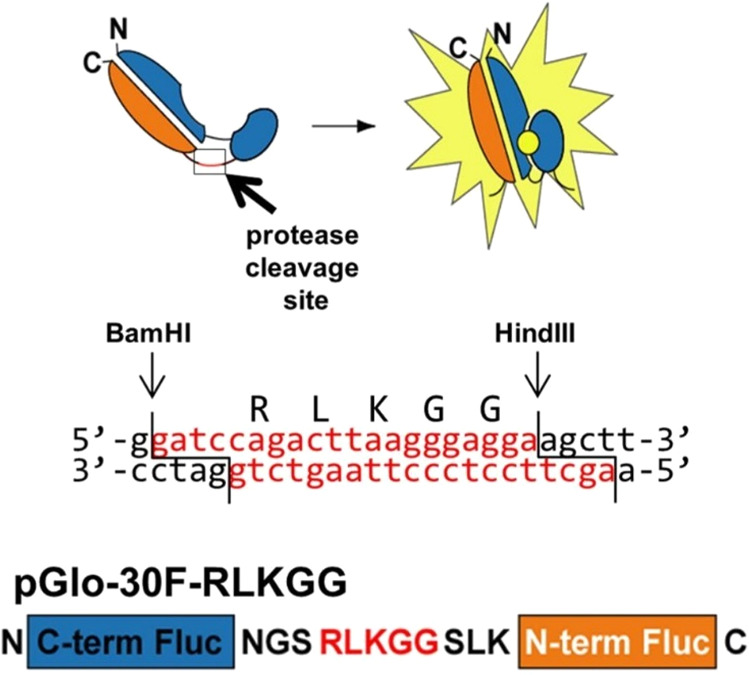
Schematic representation of the circularly permuted luciferase construct linked by the amino acid sequence RLKGG for assessing MERS-CoV papain-like protease (PLpro) activity in cells. For coronaviruses including MERS-CoV, PLpro processing of viral polyproteins is responsible for the formation of the MERS-CoV replicase complex following host cell entry and viral mRNA translation. Effective inhibitors of PLpro will lead to inhibition of MERS-CoV replication and will thus show little to no bioluminescence in the presence of the circularly permuted luciferase construct and the protease. This assay is effective for evaluating protease activity and to screen for potential inhibitors. pGlo-30F: vector backbone. RLKGG: coronavirus (CoV) papain-like protease (PLpro) cleavage site. BamHI, HindIII: restriction enzyme cleavage sites. While sharing high sequence identity, the PLpro of pathogenic coronaviruses such as SARS-CoV and SARS-CoV-2 exhibit distinct host substrate preferences.96 Nonetheless, it may be possible to identify a PLpro inhibitor that can offer broad suppression of coronavirus replication. Adapted with permission from ref (95). Copyright 2013 American Society for Microbiology.
Cyclic luciferase biosensors are structured in a nearly identical process with the N- and C-termini of luciferase linked with the sequence for a specific protease. However, in cyclic luciferase assays, the inactive biosensor exists in a cyclic conformation which restricts its movement and maintains it in a less-active form.90,91 When the activated protease digests its substrate sequence, the luciferase is opened, allowed to adopt its native confirmation, and recovers its activity. Cyclic luciferase is generally constructed with the terminal ends of FLuc flanked with DnaE intein, which is used to catalyze the protein trans-splicing reaction at a very high rate and can cyclize proteins without affecting their activities (Figure 20).91,97
Figure 20.
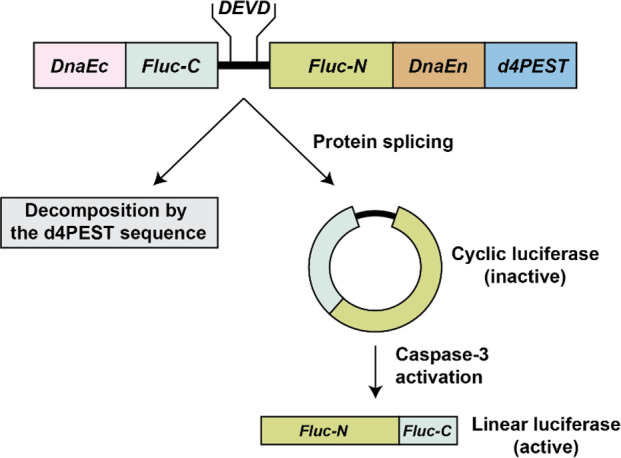
Schematic representation a cyclic luciferase assay for the detection of caspase-3 activity. DnaE fragments flank the FLuc fragments, which are connected via the amino acid sequence DEVD. N- and C-termini of FLuc ligate after translation and protein splicing, forming an inactive cyclic luciferase. Caspase-3 will cut the cyclic luciferase at DEVD, activating it and creating bioluminescence. A degradation sequence d4PEST is also included in this fusion protein design, such that only cyclic forms of the biosensor are allowed to exist. DEVD: substrate sequence of caspase-3; Asp-Glu-Val-Asp. DnaE: catalytic α subunit of DNA polymerase III. PEST: sequence which is known to accelerate degradation of a fused protein. d4PEST: mutated murine PEST sequence.
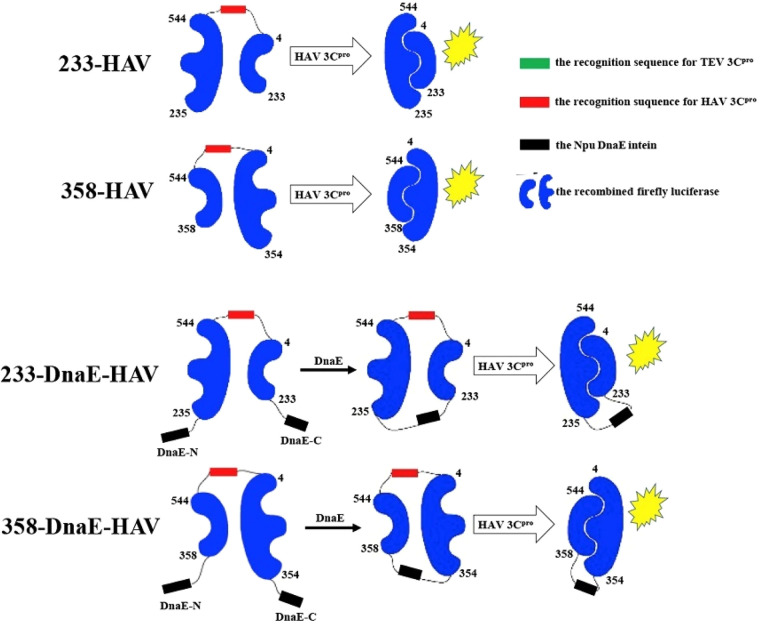
Cyclic luciferase-based biosensors have been used to study the activity of hepatitis A virus (HAV) and its relationship with interferon signaling in the human body.91 This virus contains a 3C cysteine protease which is critical for replication and proliferation of the virus and which also is responsible for cleavage of the human NF-kB essential modulator (NEMO), a kinase required for interferon signaling.98 In order to assess the activity of HAV 3C cysteine proteases, the HAV 3C cysteine protease cleavage recognition sequence of NEMO was fused to FLuc to create a cyclized luciferase-based biosensor. Four different conformations were tested—two constructs based on a split luciferase approach (233H, 358H) and two based on the cyclic luciferase approach (233DH and 358DH, where “D” indicates fusion to DnaE). When cells were transfected with the HAV 3C protease and the reporters, all four recombinant proteins were successfully cleaved (Figure 21). However, the 233DH biosensor proved to be the most sensitive and reliable. The specificity of this assay was tested using proteases from other viruses such as noroviruses, SARS-CoV, MERS-CoV, and porcine epidemic diarrhea virus, none of which showed a significant increase in luciferase activity. Overall, this study underscores the need for experiments testing luciferase biosensor design using varying conformations (including split luciferase, circularly permutated luciferase, and cyclic luciferase designs) in order to optimize biosensor signal intensity and specificity.
Figure 21.
Schematic representation of the generation of 233H, 358H, 233DH, and 358DH biosensors for detecting HAV 3C cysteine protease activity. 233-HAV has the recognition sequence for HAV 3Cpro (red rectangle) connecting the two halves of FLuc at amino acid positions 4 and 544, while 233-DnaE-HAV has additional DnaE fragments (black rectangles) attached to the N- and C-termini of FLuc. All four plasmids are cleaved and give a signal when they are cotransfected with HAV 3C protease plasmid. 233DH and 358DH only have complementation when both DnaE and HAV 3Cpro are present. H: hepatitis A virus. D: fusion to DnaE. HAV: hepatitis A virus. 3Cpro: HAV 3C cysteine protease. Npu: Nostoc punctiforme. DnaE: DNA polymerase III intein. Adapted with permission from ref (91). Copyright 2017 and Elsevier Inc.
Dual Luciferase Assay
Dual luciferase systems employ two different reporter genes and are most commonly used to quantify gene expression. In a typical assay, RLuc and FLuc are expressed under control or experimental promoters on a plasmid or within a genome. The luciferase expressed under a control promoter (usually RLuc) is often used to normalize the assay with assay results expressed as a ratio of FLuc to RLuc.99 It should be noted that FLuc and RLuc are the most common luciferases selected for these assays due to their different substrates and emission allowing their activities to be measured in a single reaction tube. Numerous kits and plasmids are commercially available for rapid and simple assembly of dual luciferase assays. In this section, we highlight the use of dual luciferase assays in studies of virology by considering examples from the Chilo iridescent virus, Ebolaviruses, cytomegalovirus, and HCV research.
Chilo Iridescent Virus
As a first example of dual luciferase assays in viral research, one such reporter system has been used to characterize the promoters of the Chilo iridescent virus (CIV) DNA polymerase and capsid protein genes.100 Iridoviruses have been studied as potential biological control agents due to the lethality of their infections in certain insect species.100−102 Deletion mutagenesis in combination with a luciferase reporter system was used by Nalçacioğlu and colleagues to determine essential sequences for promoter activity. Specifically, progressive 5′ deletions were constructed and fused to a FLuc reporter gene, whose activity was normalized against the control RLuc plasmid. This study illustrates the very common use of luciferase reporter vectors for the development of transcriptional reporters for promoters of interest and the relative ease with which essential elements can be identified for further study.
Ebolaviruses
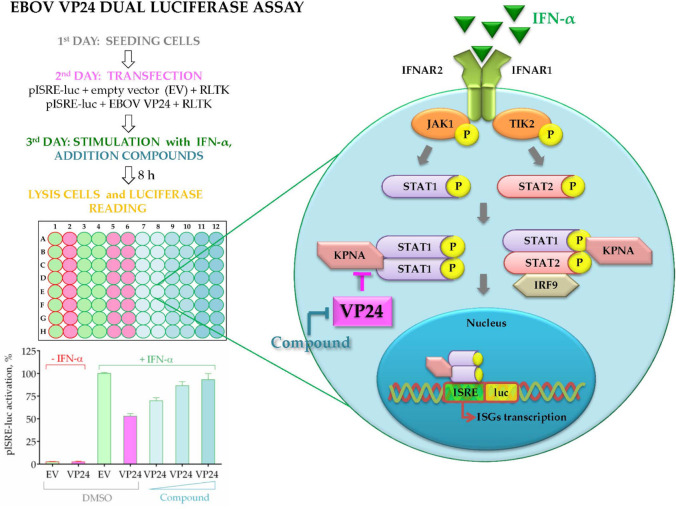
Beyond promoter characterization, other factors regulating the transcription of genes relevant to viral biology have been studied using dual luciferase assays. Fanunza et al. have used a dual luciferase assay to determine the effect of the Zaire Ebolavirus protein 24 (VP24) on interferon signaling.103 Four species of Ebolaviruses including the Zaire Ebolavirus are the causative agents for Ebolavirus disease (EVD) in humans and are responsible for recurrent outbreaks including the major health crisis in 2014 and ongoing outbreak in the Democratic Republic of the Congo in 2021. The Ebolavirus VP24 allows the virus to evade the interferon system, a key component of the antiviral immune response.104−107 In this study, a dual luciferase assay was used to identify compounds rescuing IFN-induced gene expression from the suppressive effect of VP24. HEK293T cells were cotransfected with a VP24 expression plasmid and pISRE-luc (interferon stimulated response element-luciferase). The authors of this study showed that their assay could easily be scaled for the screening of large numbers of molecules. Their approach to this is summarized in Figure 22.
Figure 22.
Schematic representation of the dual luciferase assay used to quantify inhibition of interferon signaling by Zaire Ebolavirus viral protein 24 (VP24) and to screen for inhibitors of this interaction. Cells were seeded and cotransfected with VP24 and pISRE-luc. The cells were stimulated and then lysed to measure luciferase activity. The expression of VP24 led to an inhibition of ISRE and a weaker luciferase signal. Effective inhibitors of VP24 could be added at the stimulation step, and the cells will thus produce a stronger signal. ISRE: interferon-stimulated response element. pISRE-luc: plasmid of interferon-stimulated response element attached to luciferase. RLTK: Renilla luciferase. IFN-α: interferon alpha. DMSO: dimethyl sulfoxide. Adapted with permission from ref (103). Copyright 2018 Fanunza et al.
Cytomegalovirus
Dual luciferase assays have also been used to characterize immune responses against viral infection, including the response to human cytomegalovirus (HCMV) by human IFN-inducible IFI16 protein. IFI16 acts an innate immune sensor of intracellular DNA and suppresses viral DNA synthesis, although the precise molecular mechanisms underlying these activities have been historically unclear.108 Gariano et al. created luciferase reporter constructs using wild-type and mutated versions of the HCMV DNA polymerase promoter UL54 and a LacZ plasmid as a control. Luciferase activity of the wild-type promoter was greatly increased upon infection by HCMV in comparison to HCMV-uninfected cells. In contrast, IFI16 overexpression prior to HCMV infection greatly suppressed luciferase activity. This suppression could be relieved in mutant copies of the UL54 promoter lacking a short sequence called the inverted repeat element 1 (IR-1), implicating this region in IFI16-induced HCMV DNA polymerase suppression.
Hepatitis C Virus
While it is evident that dual luciferase assays are useful for characterizing the transcriptional control of genes of interest, they have also been useful for studying post-transcriptional regulation. An example of this involves HCV replication which has been shown to depend on cellular microRNA-122 (miR-122).109 Binding of miR-122 to the HCV polyprotein transcript protects it from degradation and improves translation. There has been interest in the development of molecular sponges from circular RNAs as new HCV therapeutics that function by sequestering miR-122. Jost and colleagues have created a luciferase reporter system for the indirect assessment of miR-122-supported gene expression.109 In their assay, the HCV polyprotein open reading frame was replaced by the FLuc gene, allowing FLuc to be expressed in a miR-122-modulated manner (Figure 23). The authors further showed that they could generate circular RNAs which suppressed reporter activity. Thus, dual luciferase assays hold value not only for determining factors affecting the transcription of viral proteins of interest but also for interrogating post-transcriptional and translational regulation.
Figure 23.
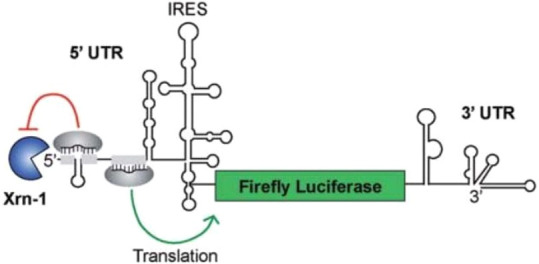
Schematic representation of the hepatitis C virus/luciferase reporter construct. The HCV polyprotein open reading frame was replaced by the FLuc gene. MicroRNA-122 binding sites 1 and 2 are depicted by gray rectangles, with Argonaute/microRNA (gray ovals) bound indicating its function in inhibiting Xrn-1 and stimulating translation of luciferase. Xrn-1: exoribonuclease. UTR: untranslated region. IRES: internal ribosome entry site. Adapted with permission from ref (109). Copyright 2018 Jost et al.
Roles for Luciferase-Based Biosensors in the Study of SARS-CoV-2
Evidently, luciferase-based biosensors of various forms have served as useful tools in addressing scientific questions in virology. In providing this overview of the literature, we have been motivated by the notion that broader dissemination of these techniques will facilitate future work in the field of virology and may simultaneously inspire creative solutions to present day problems including the SARS-CoV-2 pandemic.
A key tool in solving this and future pandemics will be surrogate markers for understanding various aspects of culprit viral function and detecting infections/immunity in patients. Challenges in the current testing methods for SARS-CoV-2 antigens and antibodies include temporal dynamics of viral antigens/antibodies affecting interpretation and low viral loads, as well as current methods being relatively time-consuming, resource intensive, costly, or insensitive.110−114
Bhalla and colleagues recently described the features of an ideal biosensor for research and clinical use in pandemics.115 Ideally, a biosensor would be highly sensitive and selective; have a quick response time, multiplexing capabilities, and sensing modes; be disposable with a long shelf life; as well as easy to use, cost-effective, mass manufacturable, and autonomous (Figure 24). Clearly, the development of a biosensor possessing all of these attributes represents an enormously challenging task; however, biosensors with even some of these characteristics would be exceedingly useful for drug discovery, vaccine development, early infection detection, and disease monitoring. Luciferase biosensors hold value in the detection of viral proteins or antibodies from SARS-CoV-2 and the aforementioned luciferase biosensors possess many of the features outlined by Bhalla et al. Proteins for use in these assays include the SARS-CoV-2 spike (S; full length or receptor binding subunit S1) or nucleocapsid (N) proteins (which are the main antigens currently used in serological assays) as well as the proteins/complexes with which they interact, including the SARS-CoV-2 ribonucleocapsid and host cell receptors involved in virus entry: angiotensin-converting enzyme 2 (ACE2) and the transmembrane protease serine 2 (TMPRSS2) (Figure 25).112,116−121
Figure 24.
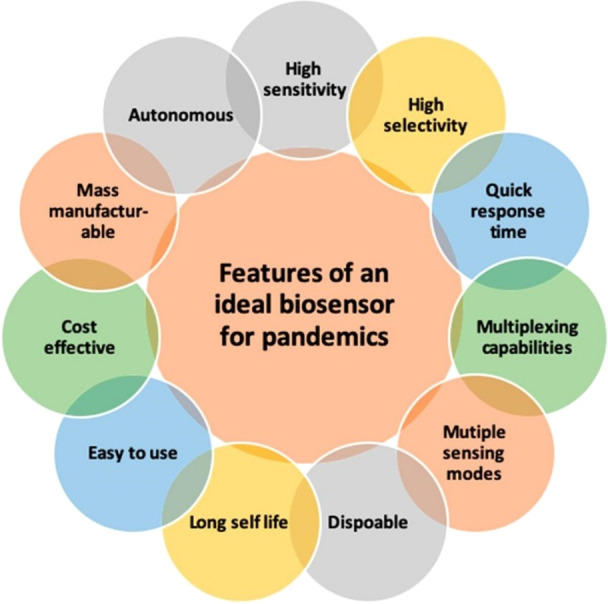
Features of an ideal biosensor for pandemics. These features include the following: high sensitivity, high selectivity, quick response time, multiplexing capabilities, multiple sensing modes, disposability, long shelf life, ease of use, cost effectiveness, mass manufacturing ability, and autonomy. Adapted from ref (115). Copyright 2020 American Chemical Society.
Figure 25.
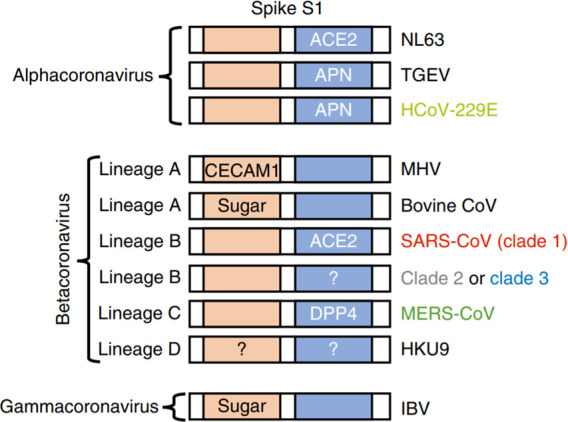
Schematic of coronavirus spike proteins and their receptors. Human variants of betacoronavirus receptors and a common receptor for alphacoronaviruses were tested for their ability to mediate cell entry of clade 2 and 3 spike chimeras. Orange rectangles represent N-terminal RBD found in spike S1 of lineage A betacoronaviruses and gammacoronaviruses. Blue rectangles represent C-terminal RBD found in spike S1 of alphacoronaviruses and lineage B betacoronavirus. RBD: Receptor-binding domain. S1: receptor-binding subunit. CECAM1: carcinoembryonic antigen-related cell adhesion molecule 1. HCoV: human coronavirus. IBV: infectious bronchitis virus. MHV: mouse hepatitis virus. TGEV: transmissible gastroenteritis coronavirus. Adapted with permission from ref (120). Copyright 2020 Letko et al. under exclusive license to Springer Nature Limited.
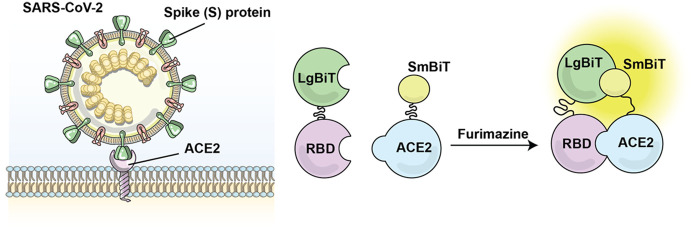
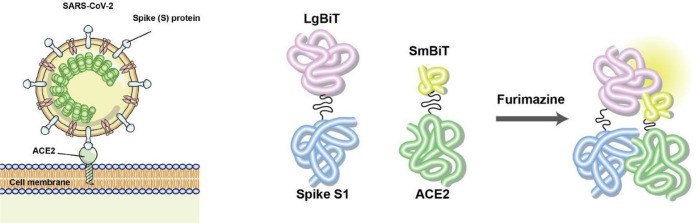
Work toward the development of luciferase-based biosensors for studying SARS-CoV-2 has already been undertaken by ourselves and others. Indeed, through the use of these tools, we have quickly gained insights into the basic biology underlying SARS-CoV-2 infection and have also generated foundational data supporting the use of luciferase-based biosensors in drug development and viral/serologic testing. We recently reported our construction of a nanoluciferase-based biosensor for use in the determination of critical host and viral factors mediating SARS-CoV-2 viral entry.122 Here, the receptor-binding domain (RBD) from the SARS-CoV-2 spike protein was fused to LgBiT, while the ACE2 ectodomain was fused to SmBiT. Secretory signals were added to the component biosensor parts to allow for both cell lysate and supernatant-based assays, and a flexible glycine linker was used to overcome potential steric limitations. We then studied the effect of known and novel mutations on the biosensor’s activity and ultimately discovered that N-glycosylation was essential for RBD–ACE2 binding. This work provided compelling preliminary data that agents modulating S protein glycosylation might hold therapeutic potential for disrupting viral/host cell binding during or prior to infection as well as for modulating S protein immunogenicity and antigenicity. We have subsequently employed this biosensor in characterizing key residues in ACE2 that mediate SARS-CoV-2 binding, which has provided a notable indication that ACE2 polymorphisms may augment RBD–ACE2 binding and susceptibility to infection across and within species.123
More recently, we shared our construction of an alternative NanoBiT split luciferase assay for the detection of SARS-CoV-2 virus/host cell binding (Figure 26). Rather than examining RBD–ACE2 binding, this biosensor specifically interrogates the spike S1 domain and ACE2 ectodomain interaction.124 We characterized the stability, dynamic range, and sensitivity of this biosensor and further showed that it could be used to identify inhibitors of the protein–protein interaction as recombinant RBD and ACE2 could be used as competitive inhibitors to block the interaction and decrease signal intensity. In developing a second biosensor for detecting SARS-CoV-2–host cell binding, we foresee a potential application for the RBD-ACE2 biosensor and S1 domain–ACE2 biosensors to be used in parallel as controls for each other and to investigate specific aspects of this protein–protein interaction.
Figure 26.
Construction of SARS-CoV-2 S1-NanoBiT split luciferase assay. Left: Schematic representation of SARS-CoV-2 viral entry via viral spike protein and cellular ACE2 interaction. Right: Outline of the NanoBiT split luciferase assay for Spike S1 and ACE2 binding upon addition of substrate furimazine. Adapted with permission from ref (124). Copyright 2021 Elsevier B.V.
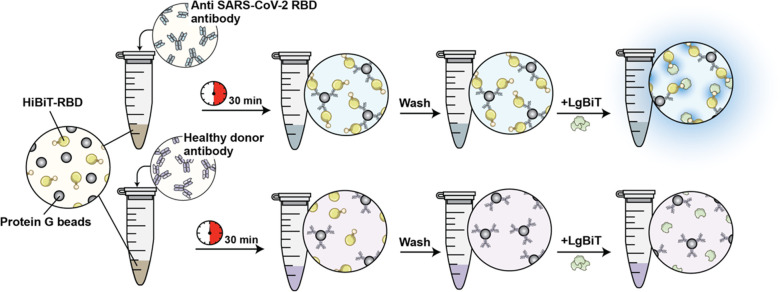
In a more translational application of this technology, we have further generated a NanoBiT-based biosensor which reliably detects SARS-CoV-2 seroconversion in patient samples.125 In this assay, recombinant RBD is fused with HiBiT and this fusion protein was incubated with patient serum and protein G beads. If RBD-binding antibodies are present in circulation, RBD-HiBiT/antibody/protein G complexes are precipitated and can be detected using LgBiT as a detection reagent (Figure 27). Indeed, patient samples from uninfected individuals showed extremely low luminescence signals (with no false positives), whereas those infected but nonhospitalized and infected and hospitalized individuals showed increasingly high luminescence. Notably, we were easily able to adapt this assay to a 384-well format where only 5 μL of serum or 10 ng of antibody were required per reaction.
Figure 27.
Schematic representation of the NanoBiT serological assay. RBD fused to HiBiT (HiBiT-RBD) is incubated in the patient sample along with protein G beads. LgBiT detects RBD-binding antibodies in the sample by binding to antibody-bound conjugates and fluorescing (top panel). Adapted with permission from ref (125). Copyright 2021 Azad et al.
Yao and colleagues recently published a paper demonstrating another approach on SARS-CoV-2 rapid serology using bioassays.126 Their novel assay, named SATiN, can be performed directly in liquid-phase patient sera to enable rapid detection of SARS-CoV-2 antibodies. SATiN uses two different tripart nanoluciferase (tNLuc) tags as antibody probes fused to both SARS-CoV-2 S and protein G. When proteins G and S interact in the presence of the third tNLuc component and substrate, the NLuc enzyme is reconstituted and readily detected using conventional methodologies.
Elledge et al.127 used nanoluciferase technology to develop several novel biosensors. They constructed anti-SARS-CoV-2 antibody biosensors by fusing split nanoluciferase (NanoLuc) fragments SmBiT and LgBiT to SARS-CoV-2 viral protein antigens. They demonstrated that their assay is rapid, low-cost, and solution-based and able to detect antibodies in serum, plasma, whole blood, and to a lesser extent saliva.
Xie and colleagues128 have revealed another interesting application of nanoluciferase for studying SARS-CoV-2. They developed a rapid neutralization assay, established a high-throughput assay for reliable antiviral screening, and screened exploratory and FDA-approved anti-infective drugs for potential COVID-19 repurposing. In their study, a stable SARS-CoV-2-NLuc construct with NLuc inserted into the ORF7 of the viral genome served as a platform for rapid serodiagnosis and high-throughput drug screening. Patient sera were used to validate their platform as a neutralization assay.
Collectively, these data demonstrate the utility of bioluminescent biosensors for interrogating various aspects of virology. Their relative ease of construction and validation makes bioluminescent biosensors a ready tool for application to urgent problems, including the present SARS-CoV-2 pandemic.
Conclusions and Future Perspectives
In this review, we have provided an overview of the broad utility of luciferase-based biosensors in the field of virology and have highlighted examples of the multitude of scientific questions that can be addressed using biosensor methodologies. We have further aimed to underscore key principles for the successful design and implementation of luciferase-based biosensors including the need for a combinatorial approach to biosensor design as well as the use of appropriate controls (i.e., function-negative mutant biosensors) in experimental design. Luciferase-based biosensors have provided invaluable and timely information to scientists about the interactions of different viruses and their variants with the host cell (Table 2).
Table 2. Applications of Luciferase-Based Biosensors and Their Assay Format.
| application of biosensor | type of biosensor | assay format | type of Luciferase | ref |
|---|---|---|---|---|
| detection of protein dimerization | split luciferase | in vitro | Renilla luciferase | (23) |
| split luciferase | in vitro | Gaussia luciferase | (44) | |
| BRET | in vitro | Renilla luciferase | (83,86) | |
| analysis of viral entry | split luciferase | in vitro | nanoluciferase | (34,122,123) |
| split luciferase | in vitro | Renilla luciferase | (48,49,54,55,57) | |
| creation of reporter viruses for screening of antiviral agents | split luciferase | in vitro/in vivo | nanoluciferase | (35) |
| screening for inhibitors of viral protease | split luciferase | in vitro | firefly luciferase | (40) |
| circularly permutated luciferase | in vitro | Renilla luciferase | (95) | |
| cyclic luciferase | in vitro | firefly luciferase | (91) | |
| BRET | in vitro | Renilla luciferase | (71,73) | |
| split luciferase | in vivo/in vitro | firefly luciferase | (28) | |
| screening for inhibitors of viral protein kinase | split luciferase | in vitro | Renilla luciferase | (47) |
| screening for inhibitors of viral polymerase | split luciferase | in vitro | firefly luciferase | (18) |
| studying immune system downregulation by viral infection | BRET | in vitro | Renilla luciferase | (72) |
| quantifying inhibition of interferon signaling | dual luciferase | in vitro | firefly luciferase/Renilla luciferase | (103) |
| studying interaction between virus and host proteins | split luciferase | in vitro | Gaussia luciferase | (42,59) |
| split luciferase | in vitro | firefly luciferase | (29) | |
| split luciferase | in vitro | nanoluciferase | (124) | |
| studying effects of sequence changes of genes | split luciferase | in vitro | Gaussia luciferase | (44,59) |
| dual luciferase | in vitro | firefly luciferase/Renilla luciferase | (100) | |
| studying viral protein structure | BRET | in vitro | Renilla luciferase | (84) |
| identifying microRNA binding sites | dual luciferase | in vitro | firefly luciferase/Renilla luciferase | (109) |
| screening for inhibitors for protein dimerization | split luciferase | in vitro | Renilla luciferase | (23) |
| detecting viral protease activity | split luciferase | in vitro/in vivo | firefly luciferase | (28) |
| detection of antibodies against SARS-CoV-2 | split luciferase | in vitro | nanoluciferase | (127) |
| split luciferase | in vitro | nanoluciferase | (128) | |
| characterizing immune response against viral infection | dual luciferase | in vitro | firefly/Renilla luciferase | (108) |
| detection of antibody seroconversion | split luciferase | in vitro | nanoluciferase | (125) |
| split luciferase | in vitro | nanoluciferase | (126) |
By comparison, many versions of fluorescent proteins with almost every possible excitation and emission wavelengths have been characterized. Despite the relatively limited number of bioluminescent proteins, luciferase-based biosensors have already made a great impact in the field of virology. Future studies aimed at identification of new luciferases with either a unique light emission spectrum or substrate may broaden this impact even further, allowing for multiplexed integration of biosensors and the interrogation of multiple interactions simultaneously.
Here, we have summarized various proof-of-concept biosensors for studying various aspects of virus biology. The SARS-CoV-2 virus is likely to be no exception to this trend, with our own and others’ data showing the potential for these biosensors to contribute to our fundamental knowledge about the virus and its interactions. The majority of the mentioned biosensors have been used to generate novel serological assays or to detect ACE2–spike interactions of SARS-CoV-2. Luciferase-based biosensors may still lead to the identification of inhibitory drugs via high-throughput screening. Lastly, other aspects of SARS-CoV-2 such as viral growth, replication, and release from cells can be explored to design other biosensors in future.
Acknowledgments
This work was possible by the generous support from the Ottawa Hospital Foundation, the Thistledown Foundation, the Terry Fox Research Institute, and the Canadian Cancer Society. This work was also funded by a FastGrant for COVID-19 Science to C.S.I., and a grant from the Canadian Institutes of Health Research to J.C.B. T.A. is funded by a CIHR Banting Fellowship. M.J.F.C. received funding support from MITACS in the form of a MITACS cluster Accelerate fellowship.
Author Contributions
¶ T.A., H.J.J.vR., J.M., R.R., and M.J.F.C. contributed equally to this work.
The authors declare no competing financial interest.
References
- Yeh H. W.; Ai H. W. Development and Applications of Bioluminescent and Chemiluminescent Reporters and Biosensors. Annu. Rev. Anal. Chem. 2019, 12 (1), 129–150. 10.1146/annurev-anchem-061318-115027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra P. Biosensors and their applications - A review. J. Oral Biol. Craniofac Res. 2016, 6 (2), 153–9. 10.1016/j.jobcr.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. K.; Govender P. P.; Tiwari A.. Advances in Biomembranes and Lipid Self-Assembly. Polymeric Micellar Structures for Biosensor Technology; Academic Press, 2016; Chapter 6, pp 143–161. [Google Scholar]
- Thorne N.; Inglese J.; Auld D. Illuminating Insights into Firefly Luciferase and Other Bioluminescent Reporters Used in Chemical Biology. Chem. Biol. 2010, 17 (6), 646–657. 10.1016/j.chembiol.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick A.; et al. Biotechnological Advances in Luciferase Enzymes. Bioluminescence - Analytical Applications and Basic Biology; IntechOpen, 2019; pp 1–23. [Google Scholar]
- Kaskova Z.; Tsarkova A.; Yampolsky I. 1001 lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev. 2016, 45 (21), 6048–6077. 10.1039/C6CS00296J. [DOI] [PubMed] [Google Scholar]
- Smirnova D. V.; Ugarova N. N. Firefly Luciferase-based Fusion Proteins and their Applications in Bioanalysis. Photochem. Photobiol. 2017, 93 (2), 436–447. 10.1111/php.12656. [DOI] [PubMed] [Google Scholar]
- Torkzadeh-Mahani M.; et al. Design and development of a whole-cell luminescent biosensor for detection of early-stage of apoptosis. Biosens. Bioelectron. 2012, 38 (1), 362–368. 10.1016/j.bios.2012.06.034. [DOI] [PubMed] [Google Scholar]
- Ataei F.; Torkzadeh-Mahani M.; Hosseinkhani S. A novel luminescent biosensor for rapid monitoring of IP3 by split-luciferase complementary assay. Biosens. Bioelectron. 2013, 41, 642–648. 10.1016/j.bios.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Tannous B. A. Gaussia luciferase reporter assay for monitoring biological processes in culture and in vivo. Nat. Protoc. 2009, 4 (4), 582–91. 10.1038/nprot.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England C. G.; Ehlerding E. B.; Cai W. NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjugate Chem. 2016, 27 (5), 1175–1187. 10.1021/acs.bioconjchem.6b00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M.; Ozawa T. Split luciferase complementation for analysis of intracellular signaling. Anal. Sci. 2014, 30 (5), 539–44. 10.2116/analsci.30.539. [DOI] [PubMed] [Google Scholar]
- Azad T.; et al. A gain-of-functional screen identifies the Hippo pathway as a central mediator of receptor tyrosine kinases during tumorigenesis. Oncogene 2020, 39 (2), 334–355. 10.1038/s41388-019-0988-y. [DOI] [PubMed] [Google Scholar]
- Azad T.; Tashakor A.; Hosseinkhani S. Split-luciferase complementary assay: applications, recent developments, and future perspectives. Anal. Bioanal. Chem. 2014, 406 (23), 5541–5560. 10.1007/s00216-014-7980-8. [DOI] [PubMed] [Google Scholar]
- Azad T.; Janse van Rensburg H. J.; Lightbody E. D.; Neveu B.; Champagne A.; Ghaffari A.; Kay V. R.; Hao Y.; Shen H.; Yeung B.; Croy B. A.; Guan K. L.; Pouliot F.; Zhang J.; Nicol C. J. B.; Yang X. A LATS biosensor screen identifies VEGFR as a regulator of the Hippo pathway in angiogenesis. Nat. Commun. 2018, 9, 1061. 10.1038/s41467-018-03278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri K.; et al. A kinome-wide screen using a NanoLuc LATS luminescent biosensor identifies ALK as a novel regulator of the Hippo pathway in tumorigenesis and immune evasion. FASEB J. 2019, 33 (11), 12487–12499. 10.1096/fj.201901343R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri K.; et al. Identification of Celastrol as a Novel YAP-TEAD Inhibitor for Cancer Therapy by High Throughput Screening with Ultrasensitive YAP/TAZ-TEAD Biosensors. Cancers 2019, 11 (10), 1596. 10.3390/cancers11101596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; et al. Discovery of Influenza Polymerase PA-PB1 Interaction Inhibitors Using an In Vitro Split-Luciferase Complementation-Based Assay. ACS Chem. Biol. 2020, 15 (1), 74–82. 10.1021/acschembio.9b00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef L. G.; et al. Multiplex detection of protein-protein interactions using a next generation luciferase reporter. Biochim. Biophys. Acta, Mol. Cell Res. 2016, 1863 (2), 284–92. 10.1016/j.bbamcr.2015.11.031. [DOI] [PubMed] [Google Scholar]
- Branchini B. R.; et al. Mutagenesis evidence that the partial reactions of firefly bioluminescence are catalyzed by different conformations of the luciferase C-terminal domain. Biochemistry 2005, 44 (5), 1385–93. 10.1021/bi047903f. [DOI] [PubMed] [Google Scholar]
- Nakatsu T.; et al. Structural basis for the spectral difference in luciferase bioluminescence. Nature 2006, 440 (7082), 372–6. 10.1038/nature04542. [DOI] [PubMed] [Google Scholar]
- Ohmuro-Matsuyama Y.; Ueda H.. Protein–Protein Interaction Assays Using Split-NanoLuc. Bioluminescence - Analytical Applications and Basic Biology; IntechOpen, 2019. [Google Scholar]
- Wei X.-F.; Gan C.-Y.; Cui J.; Luo Y.-Y.; Cai X.-F.; Yuan Y.; Shen J.; Li Z.-Y.; Zhang W.-L.; Long Q.-X.; Hu Y.; Chen J.; Tang N.; Guo H.; Huang A.-L.; Hu J.-L. Identification of Compounds Targeting Hepatitis B Virus Core Protein Dimerization through a Split Luciferase Complementation Assay. Antimicrob. Agents Chemother. 2018, 62, 12. 10.1128/AAC.01302-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A.; et al. Core protein: A pleiotropic keystone in the HBV lifecycle. Antiviral Res. 2015, 121, 82–93. 10.1016/j.antiviral.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penin F.; et al. Structural biology of hepatitis C virus. Hepatology 2004, 39, 5–19. 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- Tomei L.; et al. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J. Virol. 1993, 67 (7), 4017–26. 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D.; Penin F.; Rice C. M. Replication of hepatitis C virus. Nat. Rev. Microbiol. 2007, 5 (6), 453–63. 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Wang L.; et al. Bioluminescence imaging of Hepatitis C virus NS3/4A serine protease activity in cells and living animals. Antiviral Res. 2010, 87 (1), 50–6. 10.1016/j.antiviral.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Dolan P. T.; et al. Intrinsic disorder mediates hepatitis C virus core-host cell protein interactions. Protein Sci. 2015, 24 (2), 221–35. 10.1002/pro.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon A. S.; et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11 (2), 400–8. 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- Hall M. P.; et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012, 7 (11), 1848–57. 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T. C.; Diamond M. S.. Flaviviruses. Fields Virology, 6th ed.; Wolters Kluwer, 2013; pp 747–794. [Google Scholar]
- Schalich J.; et al. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 1996, 70 (7), 4549–57. 10.1128/jvi.70.7.4549-4557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M.; et al. Development of a rapid and quantitative method for the analysis of viral entry and release using a NanoLuc luciferase complementation assay. Virus Res. 2018, 243, 69–74. 10.1016/j.virusres.2017.10.015. [DOI] [PubMed] [Google Scholar]
- Tamura T.; Fukuhara T.; Uchida T.; Ono C.; Mori H.; Sato A.; Fauzyah Y.; Okamoto T.; Kurosu T.; Setoh Y. X.; Imamura M.; Tautz N.; Sakoda Y.; Khromykh A. A.; Chayama K.; Matsuura Y. Characterization of Recombinant Flaviviridae Viruses Possessing a Small Reporter Tag. J. Virol. 2018, 92, 2. 10.1128/JVI.01582-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.; et al. Hepatitis C virus core protein and cellular protein HAX-1 promote 5-fluorouracil-mediated hepatocyte growth inhibition. J. Virol. 2009, 83 (19), 9663–71. 10.1128/JVI.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P.; Whitten-Bauer C.; Chisari F. V. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (1), 291–6. 10.1073/pnas.0912966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J.; Grakoui A.; Rice C. M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J. Virol. 1991, 65 (11), 6042–50. 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B.; et al. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 1991, 65 (5), 2467–75. 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; et al. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res. 2017, 27 (8), 1046–1064. 10.1038/cr.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaert A.; Naesens L. The Influenza Virus Polymerase Complex: An Update on Its Structure, Functions, and Significance for Antiviral Drug Design. Med. Res. Rev. 2016, 36 (6), 1127–1173. 10.1002/med.21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J. S; Idoko-Akoh A.; Mistry B.; Goldhill D.; Staller E.; Schreyer J.; Ross C.; Goodbourn S.; Shelton H.; Skinner M. A; Sang H.; McGrew M. J; Barclay W. Species specific differences in use of ANP32 proteins by influenza A virus. eLife 2019, 8, e45066 10.7554/eLife.45066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel J.; et al. Sequence of events in measles virus replication: role of phosphoprotein-nucleocapsid interactions. J. Virol. 2014, 88 (18), 10851–63. 10.1128/JVI.00664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller C. K.; Bloyet L.-M.; Donohue R. C.; Huff A. L.; Bartemes W. P.; Yousaf I.; Urzua E.; Claviere M.; Zachary M.; de Masson d’Autume V.; Carson S.; Schieferecke A. J.; Meyer A. J.; Gerlier D.; Cattaneo R.; et al. The C Protein Is Recruited to Measles Virus Ribonucleocapsids by the Phosphoprotein. J. Virol. 2020, 94 (4), e01733-19 10.1128/JVI.01733-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee M. S.; Lawrence G. L.; Barrell B. G. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J. Gen. Virol. 1989, 70 (5), 1151–1160. 10.1099/0022-1317-70-5-1151. [DOI] [PubMed] [Google Scholar]
- Chang C. W.; et al. BGLF4 kinase modulates the structure and transport preference of the nuclear pore complex to facilitate nuclear import of Epstein-Barr virus lytic proteins. J. Virol. 2015, 89 (3), 1703–18. 10.1128/JVI.02880-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; et al. The split Renilla luciferase complementation assay is useful for identifying the interaction of Epstein-Barr virus protein kinase BGLF4 and a heat shock protein Hsp90. Acta Virol 2016, 60 (1), 62–70. 10.4149/av_2016_01_62. [DOI] [PubMed] [Google Scholar]
- Kondo N.; et al. Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain. J. Biol. Chem. 2010, 285 (19), 14681–8. 10.1074/jbc.M109.067090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher J. R.; et al. Functional fluorescent protein insertions in herpes simplex virus gB report on gB conformation before and after execution of membrane fusion. PLoS Pathog. 2014, 10 (9), e1004373 10.1371/journal.ppat.1004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertel P. E.; et al. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 2001, 279 (1), 313–24. 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- Atanasiu D.; et al. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 2010, 84 (23), 12292–9. 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D.; et al. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (47), 18718–23. 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D.; et al. Bimolecular complementation defines functional regions of Herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 2010, 84 (8), 3825–34. 10.1128/JVI.02687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D.; et al. Dual split protein-based fusion assay reveals that mutations to herpes simplex virus (HSV) glycoprotein gB alter the kinetics of cell-cell fusion induced by HSV entry glycoproteins. J. Virol 2013, 87 (21), 11332–45. 10.1128/JVI.01700-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saw W. T.; et al. Using a split luciferase assay (SLA) to measure the kinetics of cell-cell fusion mediated by herpes simplex virus glycoproteins. Methods 2015, 90, 68–75. 10.1016/j.ymeth.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton B. A. Nipah virus: transmission of a zoonotic paramyxovirus. Curr. Opin. Virol. 2017, 22, 97–104. 10.1016/j.coviro.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Laing E. D.; Amaya M.; Navaratnarajah C. K.; Feng Y.-R.; Cattaneo R.; Wang L.-F.; Broder C. C. Rescue and characterization of recombinant cedar virus, a non-pathogenic Henipavirus species. Virol. J. 2018, 15 (1), 56. 10.1186/s12985-018-0964-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnarajah C. K.; Rosemarie Q.; Cattaneo R. A Structurally Unresolved Head Segment of Defined Length Favors Proper Measles Virus Hemagglutinin Tetramerization and Efficient Membrane Fusion Triggering. J. Virol. 2016, 90 (1), 68–75. 10.1128/JVI.02253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu G.; et al. Comparative analysis of virus-host interactomes with a mammalian high-throughput protein complementation assay based on Gaussia princeps luciferase. Methods 2012, 58 (4), 349–59. 10.1016/j.ymeth.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacart J.; et al. The BRET technology and its application to screening assays. Biotechnol. J. 2008, 3 (3), 311–24. 10.1002/biot.200700222. [DOI] [PubMed] [Google Scholar]
- El Khamlichi C.; Reverchon-Assadi F.; Hervouet-Coste N.; Blot L.; Reiter E.; Morisset-Lopez S. Bioluminescence Resonance Energy Transfer as a Method to Study Protein-Protein Interactions: Application to G Protein Coupled Receptor Biology. Molecules 2019, 24 (3), 537. 10.3390/molecules24030537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward W. W.; Cormier M. J. An energy transfer protein in coelenterate bioluminescence. Characterization of the Renilla green-fluorescent protein. J. Biol. Chem. 1979, 254 (3), 781–8. 10.1016/S0021-9258(17)37873-0. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Piston D. W.; Johnson C. H. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (1), 151–6. 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand L.; et al. The BRET2/arrestin assay in stable recombinant cells: a platform to screen for compounds that interact with G protein-coupled receptors (GPCRS). J. Recept. Signal Transduction Res. 2002, 22 (1–4), 533–41. 10.1081/RRS-120014619. [DOI] [PubMed] [Google Scholar]
- Hamdan F. F.; Percherancier Y.; Breton B.; Bouvier M. Monitoring protein-protein interactions in living cells by bioluminescence resonance energy transfer (BRET). Curr. Protoc. Neurosci. 2006, 34, 5.23.1–5.23.20. 10.1002/0471142301.ns0523s34. [DOI] [PubMed] [Google Scholar]
- Wu P.; Brand L. Resonance energy transfer: methods and applications. Anal. Biochem. 1994, 218 (1), 1–13. 10.1006/abio.1994.1134. [DOI] [PubMed] [Google Scholar]
- Kasprzak A. A. The use of FRET in the analysis of motor protein structure. Methods Mol. Biol. 2007, 392, 183–97. 10.1007/978-1-59745-490-2_13. [DOI] [PubMed] [Google Scholar]
- Boute N.; Jockers R.; Issad T. The use of resonance energy transfer in high-throughput screening: BRET versus FRET. Trends Pharmacol. Sci. 2002, 23 (8), 351–4. 10.1016/S0165-6147(02)02062-X. [DOI] [PubMed] [Google Scholar]
- Pfleger K. D.; Eidne K. A. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET). Nat. Methods 2006, 3 (3), 165–74. 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- Clegg R. M.; et al. Observing the helical geometry of double-stranded DNA in solution by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U. S. A. 1993, 90 (7), 2994–8. 10.1073/pnas.90.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.; et al. A human immunodeficiency virus type 1 protease biosensor assay using bioluminescence resonance energy transfer. J. Virol. Methods 2005, 128 (1–2), 93–103. 10.1016/j.jviromet.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluet D.; et al. Detection of human immunodeficiency virus type 1 Nef and CD4 physical interaction in living human cells by using bioluminescence resonance energy transfer. J. Virol. 2005, 79 (13), 8629–36. 10.1128/JVI.79.13.8629-8636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T.; et al. Bioluminescence technologies to detect calicivirus protease activity in cell-free system and in infected cells. Antiviral Res. 2011, 90 (1), 9–16. 10.1016/j.antiviral.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J.; et al. Virulent systemic feline calicivirus infection: local cytokine modulation and contribution of viral mutants. J. Feline Med. Surg 2006, 8 (1), 55–61. 10.1016/j.jfms.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S. V.; Sosnovtseva S. A.; Green K. Y. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 1998, 72 (4), 3051–9. 10.1128/JVI.72.4.3051-3059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S. V.; Garfield M.; Green K. Y. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 2002, 76 (14), 7060–72. 10.1128/JVI.76.14.7060-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford A. D.; et al. Feline calicivirus. Vet. Res. 2007, 38 (2), 319–35. 10.1051/vetres:2006056. [DOI] [PubMed] [Google Scholar]
- Branche A. R.; Falsey A. R. Respiratory syncytial virus infection in older adults: an under-recognized problem. Drugs Aging 2015, 32 (4), 261–9. 10.1007/s40266-015-0258-9. [DOI] [PubMed] [Google Scholar]
- Stein R. T.; et al. Respiratory syncytial virus hospitalization and mortality: Systematic review and meta-analysis. Pediatr Pulmonol 2017, 52 (4), 556–569. 10.1002/ppul.23570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso C. L.; et al. Taxonomy of the order Mononegavirales: update 2016. Arch. Virol. 2016, 161 (8), 2351–60. 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster A.; et al. Dimerization of matrix protein is required for budding of respiratory syncytial virus. J. Virol. 2015, 89 (8), 4624–35. 10.1128/JVI.03500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M.; et al. The Thr205 phosphorylation site within respiratory syncytial virus matrix (M) protein modulates M oligomerization and virus production. J. Virol 2014, 88 (11), 6380–93. 10.1128/JVI.03856-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisan M.; Di Antonio V.; Radeghieri A.; Palu G.; Ghildyal R.; Alvisi G. Molecular Requirements for Self-Interaction of the Respiratory Syncytial Virus Matrix Protein in Living Mammalian Cells. Viruses 2018, 10 (3), 109. 10.3390/v10030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijmeijer S.; et al. The Epstein-Barr virus-encoded G protein-coupled receptor BILF1 hetero-oligomerizes with human CXCR4, scavenges Gαi proteins, and constitutively impairs CXCR4 functioning. J. Biol. Chem. 2010, 285 (38), 29632–41. 10.1074/jbc.M110.115618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A. P.; Shadel G. S.; Ghosh S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011, 11 (6), 389–402. 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki O.; et al. A structural perspective of the MAVS-regulatory mechanism on the mitochondrial outer membrane using bioluminescence resonance energy transfer. Biochim. Biophys. Acta, Mol. Cell Res. 2013, 1833 (5), 1017–27. 10.1016/j.bbamcr.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Kato H.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441 (7089), 101–5. 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Yoneyama M.; Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009, 227 (1), 54–65. 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- Yu Y.; Lutz S. Circular permutation: a different way to engineer enzyme structure and function. Trends Biotechnol. 2011, 29 (1), 18–25. 10.1016/j.tibtech.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Wigdal S. S.; et al. A novel bioluminescent protease assay using engineered firefly luciferase. Curr. Chem. Genomics 2008, 2, 16–28. 10.2174/1875397300802010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; et al. Assessing activity of Hepatitis A virus 3C protease using a cyclized luciferase-based biosensor. Biochem. Biophys. Res. Commun. 2017, 488 (4), 621–627. 10.1016/j.bbrc.2017.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z. A.; et al. Family cluster of Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 2013, 368 (26), 2487–94. 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- Zaki A. M.; et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367 (19), 1814–20. 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Perlman S.; Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7 (6), 439–50. 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilianski A.; et al. Assessing Activity and Inhibition of Middle East Respiratory Syndrome Coronavirus Papain-Like and 3C-Like Proteases Using Luciferase-Based Biosensors. Journal of Virology 2013, 87 (21), 11955–11962. 10.1128/JVI.02105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587 (7835), 657–662. 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A.; Umezawa Y.; Ozawa T. Detection of apoptosis using cyclic luciferase in living mammals. Methods Mol. Biol. 2009, 574, 105–14. 10.1007/978-1-60327-321-3_9. [DOI] [PubMed] [Google Scholar]
- Wang D.; et al. Hepatitis A virus 3C protease cleaves NEMO to impair induction of beta interferon. J. Virol. 2014, 88 (17), 10252–8. 10.1128/JVI.00869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb D. S.; Reed R.; Marciniak R. A. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryotic Cell 2005, 4 (9), 1539–49. 10.1128/EC.4.9.1539-1549.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalçacioğlu R.; et al. Promoter analysis of the Chilo iridescent virus DNA polymerase and major capsid protein genes. Virology 2003, 317 (2), 321–9. 10.1016/j.virol.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Kleespies R. G.; Tidona C. A.; Darai G. Characterization of a new iridovirus isolated from crickets and investigations on the host range. J. Invertebr. Pathol. 1999, 73 (1), 84–90. 10.1006/jipa.1998.4821. [DOI] [PubMed] [Google Scholar]
- Hernández O.; Maldonado G.; Williams T. An epizootic of patent iridescent virus disease in multiple species of blackflies in Chiapas, Mexico. Med. Vet Entomol 2000, 14 (4), 458–62. 10.1046/j.1365-2915.2000.00258.x. [DOI] [PubMed] [Google Scholar]
- Fanunza E.; Frau A.; Sgarbanti M.; Orsatti R.; Corona A.; Tramontano E. Development and Validation of a Novel Dual Luciferase Reporter Gene Assay to Quantify Ebola Virus VP24 Inhibition of IFN Signaling. Viruses 2018, 10 (2), 98. 10.3390/v10020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanty S.; Bray M. Pathogenesis of filoviral haemorrhagic fevers. Lancet Infect. Dis. 2004, 4 (8), 487–98. 10.1016/S1473-3099(04)01103-X. [DOI] [PubMed] [Google Scholar]
- Reid S. P.; et al. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol. 2006, 80 (11), 5156–67. 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S. P.; et al. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 2007, 81 (24), 13469–77. 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo M.; et al. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J. Virol. 2010, 84 (2), 1169–75. 10.1128/JVI.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariano G. R.; et al. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 2012, 8 (1), e1002498 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost I.; Shalamova L. A.; Gerresheim G. K.; Niepmann M.; Bindereif A.; Rossbach O. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018, 15 (8), 1032–1039. 10.1080/15476286.2018.1435248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q. X.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26 (6), 845–848. 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- To K. K.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020, 20 (5), 565–574. 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; et al. Comparison of immunoglobulin G responses to the spike and nucleocapsid proteins of severe acute respiratory syndrome (SARS) coronavirus in patients with SARS. Clin. Vaccine Immunol. 2007, 14 (7), 839–46. 10.1128/CVI.00432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26 (7), 1033–1036. 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A.; Pirofski L. A. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020, 130 (4), 1545–1548. 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N.; et al. Opportunities and Challenges for Biosensors and Nanoscale Analytical Tools for Pandemics: COVID-19. ACS Nano 2020, 14 (7), 7783–7807. 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J.; et al. Profile of Immunoglobulin G and IgM Antibodies Against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020, 71 (16), 2255–2258. 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F.; Stadlbauer D.; Strohmeier S.; Nguyen T. H. O.; Chromikova V.; McMahon M.; Jiang K.; Arunkumar G. A.; Jurczyszak D.; Polanco J.; Bermudez-Gonzalez M.; Kleiner G.; Aydillo T.; Miorin L.; Fierer D. S.; Lugo L. A.; Kojic E. M.; Stoever J.; Liu S. T. H.; Cunningham-Rundles C.; Felgner P. L.; Moran T.; Garcia-Sastre A.; Caplivski D.; Cheng A. C.; Kedzierska K.; Vapalahti O.; Hepojoki J. M.; Simon V.; Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036. 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. X.; Liu I.; Elledge S. K.; Glasgow J. E.; Lim S. A.; Loudermilk R.; Chiu C. Y.; Wilson M. R.; Leung K. K.; Wells J. A.. A SARS-CoV-2 serological assay to determine the presence of blocking antibodies that compete for human ACE2 binding. medRxiv, 2020; https://doi.org/10.1101/2020.05.27.20114652. [Google Scholar]
- Stadlbauer D.; Amanat F.; Chromikova V.; Jiang K.; Strohmeier S.; Arunkumar G. A.; Tan J.; Bhavsar D.; Capuano C.; Kirkpatrick E.; Meade P.; Brito R. N.; Teo C.; McMahon M.; Simon V.; Krammer F. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc Microbiol 2020, 57 (1), e100 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M.; Marzi A.; Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol 2020, 5 (4), 562–569. 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.; et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B 2020, 10 (7), 1228–1238. 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad T.; Singaravelu R.; Taha Z.; Jamieson T. R.; Boulton S.; Crupi M. J.F.; Martin N. T.; Brown E. E.F.; Poutou J.; Ghahremani M.; Pelin A.; Nouri K.; Rezaei R.; Marshall C. B.; Enomoto M.; Arulanandam R.; Alluqmani N.; Samson R.; Gingras A.-C.; Cameron D. W.; Greer P. A.; Ilkow C. S.; Diallo J.-S.; Bell J. C. Nanoluciferase complementation-based bioreporter reveals the importance of N-linked glycosylation of SARS-CoV-2 S for viral entry. Mol. Ther. 2021, 29, 1984. 10.1016/j.ymthe.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. E.; et al. Characterization of Critical Determinants of ACE2-SARS CoV-2 RBD Interaction. Int. J. Mol. Sci. 2021, 22 (5), 2268. 10.3390/ijms22052268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad T.; et al. SARS-CoV-2 S1 NanoBiT: a Nanoluciferase complementation-based biosensor to rapidly probe SARS-CoV-2 receptor recognition. Biosens. Bioelectron. 2021, 180, 113122. 10.1016/j.bios.2021.113122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad T.; et al. A High-Throughput NanoBiT-Based Serological Assay Detects SARS-CoV-2 Seroconversion. Nanomaterials 2021, 11 (3), 807. 10.3390/nano11030807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z.; Drecun L.; Aboualizadeh F.; Kim S. J.; Li Z.; Wood H.; Valcourt E. J.; Manguiat K.; Plenderleith S.; Yip L.; Li X.; Zhong Z.; Yue F. Y.; Closas T.; Snider J.; Tomic J.; Drews S. J.; Drebot M. A.; McGeer A.; Ostrowski M.; Mubareka S.; Rini J. M.; Owen S.; Stagljar I. A homogeneous split-luciferase assay for rapid and sensitive detection of anti-SARS CoV-2 antibodies. Nat. Commun. 2021, 12 (1), 1806. 10.1038/s41467-021-22102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. K.; Zhou X. X.; Byrnes J. R.; Martinko A. J.; Lui I.; Pance K.; Lim S. A.; Glasgow J. E.; Glasgow A. A.; Turcios K.; Iyer N. S.; Torres L.; Peluso M. J.; Henrich T. J.; Wang T. T.; Tato C. M.; Leung K. K.; Greenhouse B.; Wells J. A. Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. Nat. Biotechnol. 2021, 10.1038/s41587-021-00878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X.; Muruato A. E.; Zhang X.; Lokugamage K. G.; Fontes-Garfias C. R.; Zou J.; Liu J.; Ren P.; Balakrishnan M.; Cihlar T.; Tseng C.-T. K.; Makino S.; Menachery V. D.; Bilello J. P.; Shi P.-Y. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020, 11, 5214. 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]