ABSTRACT
Vibrio vulnificus is a pathogen that accounts for one of the highest mortality rates and is responsible for most reported seafood-related illnesses and deaths worldwide. Owing to the threats of pathogens with β-lactamase activity, it is important to identify and characterize β-lactamases with clinical significance. In this study, the protein sequence of the metallo-β-lactamase (MBL) fold metallohydrolase from V. vulnificus (designated Vmh) was analyzed, and its oligomeric state, β-lactamase activity, and metal binding ability were determined. BLASTp analysis indicated that the V. vulnificus Vmh protein showed no significant sequence identity with any experimentally identified Ambler class B MBLs or enzymes containing the MBL protein fold; it was also predicted to have a signal peptide of 19 amino acids at its N terminus and an MBL protein fold from amino acid residues 23 to 216. Recombinant V. vulnificus Vmh protein was overexpressed and purified. Analytical ultracentrifugation and electrospray ionization-mass spectrometry (MS) data demonstrated its monomeric state in an aqueous solution. Recombinant V. vulnificus Vmh protein showed broad degrading activities against β-lactam antibiotics, such as penicillins, cephalosporins, and imipenems, with kcat/Km values ranging from 6.23 × 102 to 1.02 × 104 M−1 s−1. The kinetic reactions of this enzyme exhibited sigmoidal behavior, suggesting the possibility of cooperativity. Zinc ions were required for the enzyme activity, which was abolished by adding the metal chelator EDTA. Inductively coupled plasma-MS indicated that this enzyme might bind two zinc ions per molecule as a cofactor.
KEYWORDS: metallo-β-lactamase (MBL) protein fold, Vibrio vulnificus, cooperativity, β-lactamases, β-lactam antibiotics, LC-MS, ICP-MS, analytical ultracentrifugation
INTRODUCTION
The human pathogen Vibrio vulnificus accounts for one of the highest mortality rates among known foodborne pathogens and is responsible for most reported seafood-related illnesses and deaths worldwide (1, 2). V. vulnificus is an autochthonous estuarine microorganism, and its ecology in seawater is markedly affected by temperature and salinity (3, 4). Higher numbers of V. vulnificus occur in seawater at a temperature above 20°C and a salinity range of 5 to 20 ppt; as a result, V. vulnificus is found in subtropical waters throughout the world (5). V. vulnificus is transmitted via handling or ingestion of contaminated seafood or exposure to seawater through an open wound (1, 6). V. vulnificus infection can be fatal, as it causes two distinct syndromes: primary septicemia and necrotizing wound infection (7, 8). Most patients develop sepsis and severe cellulitis with rapid development to ecchymoses and bullae, and mortality rates are greater than 50% for primary septicemia (9) and about 20% for wound infections (7). The period between the onset of symptoms and the subsequent clinical outcome is short, and immediate antibiotic therapy for suspected cases is considered critical (10, 11).
In clinical therapy, patients with septicemia or serious wound infections caused by V. vulnificus infections are recommended to be treated with a combination of doxycycline and ceftazidime or fluoroquinolone and minocycline (11, 12). Although most reports have shown that V. vulnificus has low levels of antibiotic resistance to fluoroquinolones (11, 13), third-generation cephalosporins (14, 15), tetracycline (16), imipenem (13), chloramphenicol (4), and erythromycin (17), the overuse or misuse of antibiotics in human health care and aquaculture might have accelerated antibiotic resistance among V. vulnificus isolates. Baker-Austin et al. (18) reported that a substantial proportion of V. vulnificus isolates from the U.S. coastal lines are resistant to eight or more antibiotics, such as cephalosporin, doxycycline, and tetracycline, all of which are frequently used in clinical treatment for V. vulnificus infections. In Korea, 31 V. vulnificus strains were isolated from water and seafood samples, and 24 isolates showed resistance against at least one of the eight tested classes of antibiotics, with 13 isolates showing multidrug resistance against cephazolin, cephalothin, ampicillin, and amikacin (19). Shaw et al. (13) screened 120 V. vulnificus isolates from the Chesapeake Bay and observed an intermediate resistance of about 78% of the V. vulnificus isolates against chloramphenicol. In Malaysia and Qatar, 60 V. vulnificus isolates from clams and cockles were resistant to two or more antibiotics, including penicillin, cephalothin, vancomycin, and erythromycin (20).
Chromosomal and plasmid-encoded β-lactamases produced by bacteria to hydrolyze β-lactam antibiotics, such as penicillins and cephalosporins, are the major causes of β-lactam resistance. Until 2010, more than 890 unique β-lactamases have been identified in bacterial isolates from nature sources (21). For vibrios, Lin et al. (22) reported a putative resistance gene in Vibrio cholerae encoding metallo-β-lactamase (MBL) VarG, which was found to confer resistance against penicillins, cephalosporins, and carbapenems. Chiou et al. (23) reported a novel class A β-lactamase CARB-17 gene in Vibrio parahaemolyticus, which was responsible for intrinsic penicillin resistance. A novel carbapenemase VCC-1 gene in the V. cholerae genome was characterized as hydrolyzing penicillins, first-generation cephalosporins, aztreonam, and carbapenems (24).
Owing to outbreaks and threats of pathogens with β-lactam drug resistance, it is important to identify and characterize β-lactamases with clinical significance. This study identified an MBL protein fold metallohydrolase from V. vulnificus exhibiting β-lactam antibiotic-degrading activities by presenting its kinetic parameter against β-lactam drugs and investigated its oligomeric state and metal binding stoichiometry.
RESULTS AND DISCUSSION
Analysis of the protein sequence of MBL fold metallohydrolase from V. vulnificus.
Vibrio vulnificus is a pathogen that accounts for one of the highest mortality rates and is responsible for most reported seafood-related illnesses and deaths worldwide; however, there has not been any MBL gene to be experimentally identified in its genome. We analyzed the whole genome sequence of V. vulnificus, and a gene predicted to encode MBL fold metallohydrolase (designated Vmh) in chromosome II of V. vulnificus was obtained from the National Center for Biotechnology Information (NCBI) database. The gene vmh comprises 846 nucleotides and encodes a protein of 281 amino acids. The Vmh sequence was found in all 28 V. vulnificus strains in the NCBI database; in addition, Vmh homologs (protein sequence identity, >30%) were also found in other vibrios, such as the human vibriosis-causing agents V. parahaemolyticus, V. alginolyticus, V. flavialis, V. mimicus, and V. metschnikovii (25).
BLASTp (26) analysis indicated that V. vulnificus MBL fold metallohydrolase Vmh showed no significant sequence identity with any experimentally identified Ambler class B MBLs or enzymes containing the MBL protein fold (27), such as glyoxalase II, arylsulfatase, and mRNA 3′ processing proteins (28). According to the SignalP server (29) and Pfam protein database (30), it is predicted that the MBL fold metallohydrolase Vmh from V. vulnificus has a signal peptide of 19 amino acids at its N terminus and an MBL protein fold from amino acid residues 23 to 216.
Class B β-lactamases, also known as MBLs, typically have an αβ/βα sandwich fold and require zinc ions to catalyze the hydrolysis of the β-lactam ring. The hydrolysis of the β-lactam ring function is important for pathogenic bacteria to show resistance to β-lactam antibiotics widely used in clinical treatment (27). Several structural and functional studies have indicated that the conserved amino acid residues of class B β-lactamases are involved in zinc binding and the hydrolysis activity of the β-lactam ring (27).
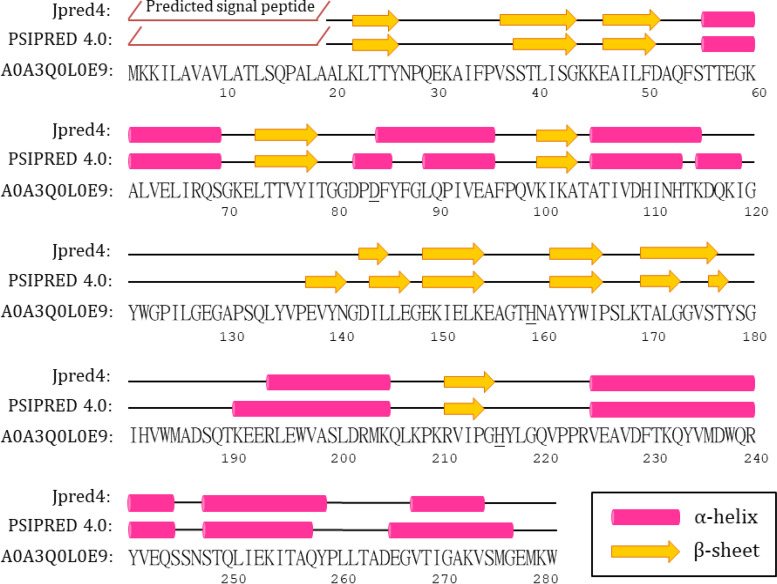
The secondary structure of V. vulnificus MBL fold metallohydrolase Vmh was predicted using the programs Jpred4 (31) and PSIPRED 4.0 (32, 33), as shown in Fig. 1. The H-X-H-X-D motif was suggested to be involved with zinc binding in MBLs, and the H116, H118, and H196 and D120, C221, and H263 amino acid residues have been reported to be involved in the binding of metal Zinc1 and Zinc2, respectively (27, 34). H116 could be replaced by an asparagine (Asn116) in the MBLs from subclass B2, and C221 could be replaced by a histidine (H121) in the MBLs from subclass B3 (35). However, the H-X-H-X-D motif could not be clearly identified in the Vmh protein from V. vulnificus. By using BLAST (26), we compared the protein sequence of the Vmh with those of other identified proteins in the protein database, and the results indicated that Vmh shared the highest sequence identity with VIM-2 from Pseudomonas aeruginosa and BcII from Bacillus cereus, both of which belong to subclass B1 MBLs. It was indicated that Vmh shares 31.75% and 21.76% identity with VIM-2 (E value of 5 × 10−6; coverage of 27%) and BcII (E value of 5.8 × 10−2; coverage of 64%), respectively. The multiple sequence alignment of Vmh, VIM-4 from P. aeruginosa, and BcII from Bacillus cereus was accomplished by using MAFFT (36). As shown in Fig. S1 in the supplemental material, H263 could be identified but the H-X-H-X-D motif could not be clearly identified in the Vmh protein.
FIG 1.
Predicted secondary structure of the V. vulnificus MBL fold metallohydrolases (Vmh) by using PSIPRED and Jpred. The putative H196 and H263 are underlined.
It has been reported that H196 in Zinc1 binding is located in the loop between the second and third β-strands of the C-terminal β-sheet structures in the B1 subgroup of MBLs, such as VIM-4 and BcII three-dimensional (3D) structures. Since H159 is in the loop between the second and third β-strands of the predicted C-terminal β-sheet in the Vmh protein from V. vulnificus, it might be involved in zinc binding as the Zinc1 ligand. The conserved residues VI/VPGH, which have been reported to contain H263 for Zinc2 binding in Pseudomonas aeruginosa VIM and Bacillus cereus BcII (34, 37), were found in the V. vulnificus Vmh protein (residues 212 to 216). According to the secondary structure prediction, H216 is located after the C-terminal β-strand (Fig. 1). This is similar to H263, which is located after the C-terminal β-strand as well as in the edge of the β-sheet, in VIM-4 and BcII 3D structures (34, 37).
Overexpression and identification of the V. vulnificus Vmh protein.
To determine the kinetic parameters of the V. vulnificus Vmh protein, the vmh gene was amplified from the genome of V. vulnificus, whose sequence information was obtained from the NCBI database (RefSeq accession no. NZ_CP012882.1). The first 19 amino acid residues of the N terminus of the V. vulnificus Vmh protein were predicted as the signal peptide using SignalP 5.0 (38). The vmh gene (with 57 bp deleted [Δ57 bp]) was cloned into the pET26b vector to target the periplasm space and transformed into Escherichia coli C43(DE3). The expression of Vmh was induced with 0.2 mM isopropyl β-d-1-thiogalactopyranoside and purified using a nickel affinity column. The purified Vmh protein was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and liquid chromatography-mass spectrometry (LC-MS) to determine the expression efficiency and confirm its identity. The expressed and purified Vmh protein has 262 amino acid residues (with 19 signal peptide amino acid residues deleted, Δ19SP) with a predicted molecular weight of about 30.76 kDa, and it showed a major band of about 32 kDa on SDS-PAGE (Fig. S2A). Further identification of the trypsin-digested Vmh protein using the LC-tandem MS (LC-MS/MS) method showed 92% sequence coverage, which provided solid evidence for identifying the Vmh protein (Fig. S2B). These data indicated that the Vmh protein was successfully overexpressed in E. coli C43(DE3) and purified for subsequent characterization studies.
Determination of the oligomeric state of Vmh from V. vulnificus.
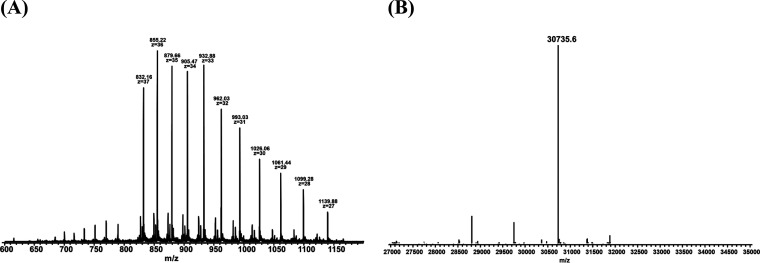
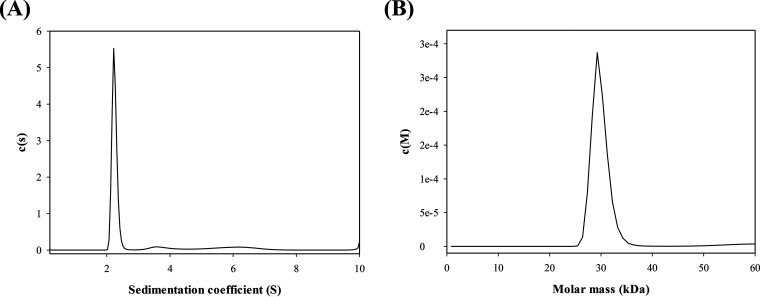
The functional and structural states of the purified V. vulnificus Vmh protein were characterized using size exclusion chromatography (SEC), high-resolution electrospray ionization-MS (ESI-MS), and analytical ultracentrifugation (AUC). Analytical SEC was used to determine the molecular weight of Vmh to investigate its oligomeric state. The Vmh protein was applied to the SEC column and showed a sharp peak at an elution volume of approximately 88.5 ml (Ve), giving a Kav (gel-phase distribution coefficient) value of 0.529, corresponding to an estimated Mr of 34.4 kDa for Vmh (Fig. S3). The SEC results suggested that the Vmh protein was in the monomeric state. To determine the accurate molecular weight of the Vmh protein, a high-resolution ESI-MS method was performed in this study. The mass spectrum in Fig. 2A shows the mass-to-charge signals of Vmh with multiple charges. The experimental monoisotopic mass of Vmh in Fig. 2B was determined by the deconvolution of signal clusters in Fig. 2A. The theoretical monoisotopic mass of Vmh was 30,736.8 Da (based on the formula of Vmh, C1397H2167N361O407S7), so the mass error of the experimental monoisotopic mass was about −39 ppm, indicating that purified Vmh formed mainly monomer in solution. Through AUC analysis, the sedimentation velocity (SV) was calculated to determine the oligomeric state of Vmh. The results showed that SV is shape dependent with a sedimentation coefficient of 2.2 S (Fig. 3A), corresponding to an estimated Mr of 29.8 kDa for the Vmh protein (Fig. 3B). All three methods confirmed that the purified V. vulnificus Vmh protein has high purity and is in the monomeric state.
FIG 2.
(A) Mass spectrum of recombinant Vmh by high-resolution ESI-MS; (B) monoisotopic molecular weight of Vmh determined by deconvolution of high-resolution MS spectrum.
FIG 3.
(A) Sedimentation coefficient and (B) predicted molecular weight of Vmh by sedimentation velocity analytical ultracentrifuge.
In previous studies, the oligomeric state of MBL protein could be in the form of either monomer or dimer. NDM-1, a subclass B1 MBL protein with a molecular weight of about 24 kDa, has been reported in a monomeric or dimeric form in solution based on the results of gel filtration chromatography (39). The loop-10, loop-8, and α-3 residues of NDM-1 proteins were suggested to be the key contacts for dimerization (40). It has also been demonstrated that both monomer and dimer forms of NDM-1 have full activity. In another study, based on gel filtration chromatography and matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) MS analyses, the recombinant MBL protein ImiS belonging to subclass B2 was determined to be a monomer form with a molecular mass of 25,247 Da (41). L1, another subclass B3 MBL protein, was reported to exist as a tetramer by gel filtration chromatography and X-ray crystallography (42–44). In addition, the MBL VarG from V. cholerae was confirmed to be in dimeric form with a molecular mass of 83,359 Da by SEC, AUC, and LC-MS/MS (22).
Kinetic parameters of β-lactam antibiotic hydrolysis by Vmh.
The kinetic parameters of recombinant Vmh for the hydrolysis of β-lactam antibiotics were determined against various β-lactam antibiotics, such as penicillins (ampicillin, carbenicillin, and piperacillin), cephalosporins (cephalothin, cefuroxime, ceftazidime, cefepime, and moxalactam), carbapenems (imipenem and meropenem), and monobactams (aztreonam). The classification of β-lactamases has been aligned based on their ability to hydrolyze specific types of β-lactams and the inhibitory properties of inhibitors. Most class A, C, and D β-lactamases have a substrate preference for hydrolyzing penicillins and cephalosporins. In contrast, class B β-lactamases have a relatively broader substrate spectrum for β-lactam antibiotics, except for relatively higher activity against carbapenems (21). According to the Ambler classification, metallo-β-lactamases were divided into three subclasses, B1, B2, and B3. It has been reported that the metallo-β-lactamases from various subclasses could exhibit different β-lactamase activities and catalytic properties for various classes of β-lactam antibiotics. For example, the metallo-β-lactamases from subclass B1 and B3 have a relatively broader substrate spectrum for β-lactam antibiotics, including carbapenems (45), whereas the metallo-β-lactamases from subclass B2 showed a high carbapenemase activity and weak activity toward penicillins and cephalosporins (46).
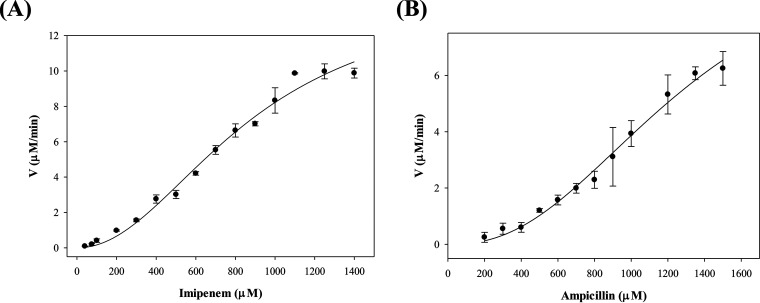
In preliminary experiments, we tested the β-lactamase activity of Vmh at various zinc concentrations. Not surprisingly, no activity was observed for the Vmh enzymes without the addition of zinc (data not shown). Our preliminary data showed a result similar to that of the study on MBL BcII by Jacquin et al. (47). BcII hydrolysis activity was determined in the presence of various zinc concentrations, the results showed a linear increase in activity with the addition of zinc, and a plateau was reached with saturation of zinc ions. Figure 4 shows the velocities of recombinant V. vulnificus Vmh degrading ampicillin and imipenem of various concentrations. Intriguingly, the kinetic curves for ampicillin and imipenem degradations (Fig. 4) and other β-lactam antibiotics (data not shown) were fitted to sigmoidal equations. The results showed that the kcat/Km ratios ranged from 6.23 × 102 to 1.02 × 104 M−1 s−1 for all tested β-lactams, and the highest kcat/Km ratios were observed with imipenem (1.02 × 104 M−1 s−1) with a Hill coefficient of 2.05 (Table 1), whereas no activity was observed against the monobactam aztreonam (data not shown). In addition, it is shown in Fig. 4 that Vmh exhibited sigmoidal behavior for the hydrolysis of ampicillin and imipenem. In general, the kinetic properties of other MBLs showed broad-spectrum hydrolysis of β-lactams, including penicillins, cephalosporins, and carbapenems, but not monobactams (21). These results showed that recombinant V. vulnificus Vmh also exhibited broad-spectrum activity against different β-lactam antibiotic classes, consistent with typical MBLs.
FIG 4.
Kinetic curve of antibiotic degradation by Vmh. (A) Imipenem; (B) ampicillin.
TABLE 1.
Steady-state kinetic parameters of Vmh protein of β-lactam antibiotics
| β-Lactam | Concn range (μM) | [Enzyme] (μM) | Vmax (μM/min) | Km (mM) | Kcat (s−1) | Kcat/Km (M−1 s−1) | n a |
|---|---|---|---|---|---|---|---|
| Ampicillin | 200–1,500 | 0.027 | 11.39 | 1.32 | 7.03 | 5.33 × 103 | 2.38 |
| Carbenicillin | 250–2,000 | 0.027 | 5.21 | 1.12 | 3.22 | 2.88 × 103 | 2.74 |
| Piperacillin | 300–1,000 | 0.027 | 6.38 | 1.63 | 3.94 | 2.42 × 103 | 3.25 |
| Cephalothin | 100–750 | 0.027 | 0.38 | 0.31 | 0.23 | 7.42 × 102 | 3.14 |
| Cefuroxime | 100–1,000 | 0.027 | 1.88 | 0.59 | 1.16 | 1.97 × 103 | 2.28 |
| Ceftazidime | 200–800 | 0.027 | 0.71 | 0.69 | 0.43 | 6.23 × 102 | 1.47 |
| Cefepime | 200–700 | 0.027 | 1.74 | 0.58 | 1.07 | 1.84 × 103 | 2.23 |
| Moxalactam | 100–1,200 | 0.027 | 1.15 | 0.54 | 0.71 | 1.31 × 103 | 3.88 |
| Imipenem | 40–1,400 | 0.027 | 14.65 | 0.89 | 9.04 | 1.02 × 104 | 2.05 |
| Meropenem | 50–900 | 0.027 | 3.98 | 0.55 | 2.46 | 4.47 × 103 | 1.38 |
an, Hill coefficient; n >1, positive cooperativity.
In a previous study, a novel subclass B2 MBL, PFM-1, showed limited hydrolysis capability on carbapenems and low catalytic efficiency in the hydrolysis of imipenem and meropenem, with kcat/Km values of 2.3 × 104 and 3.1 × 104 M−1 s−1, respectively (48). In addition, a subclass B3 MBL, BJP-1, has been reported to exhibit broad substrate catalytic activity against penicillins, cephalosporins, and imipenems, with kcat/Km values from 2 × 102 to 8.3 × 105 M−1 s−1 (49). Another study showed that a subclass B1 MBL, VIM-1, demonstrates high catalytic efficiency to carbenicillin, imipenem, and some cephalosporins, with a kcat/Km value of 1.0 × 106 M−1 s−1, but has low catalytic efficiency to penicillin G, ampicillin, and sulbactam, with a kcat/Km value of 2 × 104 M−1 s−1 (50). However, VIM-2, another MBL in subclass B1, showed high catalytic efficiency to penicillin G, ampicillin, and imipenem, with kcat/Km values ranging from 1.4 × 106 to 4.0 × 106 M−1 s−1, indicating that MBLs have distinct substrate preferences even in the same subclass (51).
Intriguingly, the sigmoidal curves showing rates of β-lactam antibiotic degradation might indicate the cooperative activity of monomeric V. vulnificus Vmh, although the possibility of substrate-induced dimerization of V. vulnificus Vmh could not be ruled out. This behavior of concentration dependence of the hydrolysis rate of Vmh is different from Michaelis-Menten kinetics and is worthy of further investigation. The best-studied kinetic cooperativity in monomeric enzymes is that of human glucokinase, which is the primary glucose sensor, plays a key role in glucose homeostasis regulation, and displays a sigmoidal kinetic response to increasing glucose concentrations (52). The kinetic cooperativity in monomeric enzymes generally displays a hysteretic response attributed to the structural transitions of enzymes. The hysteretic response refers to those enzymes that respond slowly to a rapid change in ligand or substrate concentrations or due to the relatively slow transitions of enzymes from one state to another (53, 54). Porter and Miller (55) presented an overview for monomeric enzymes, such as the mnemonic model (56), the ligand-induced slow transition (LIST) model (57), and the random-order model (58), to explain the cooperativity of monomeric enzymes. The mnemonic model indicates two enzyme states (low- and high-affinity states) in the catalytic cycle that depend on substrate concentrations. The LIST model proposes that the two conformations of monomeric enzymes with cooperativity possess different affinities for their substrates, and the equilibrium between these two conformations is controlled by the substrate concentration. The random-order model does not rely on enzyme conformational heterogeneity or low interconversion rates (58). The investigation of monomeric cooperativity would certainly require experimental methods that can probe the conformational changes associated with substrate in response to substrate concentration and association. High-resolution nuclear magnetic resonance (NMR) (59) and single-molecule fluorescence spectroscopy (60) might be adapted for elucidating the conformational heterogeneity of protein complexes and protein dynamics.
Effects of various metals and EDTA on Vmh activity.
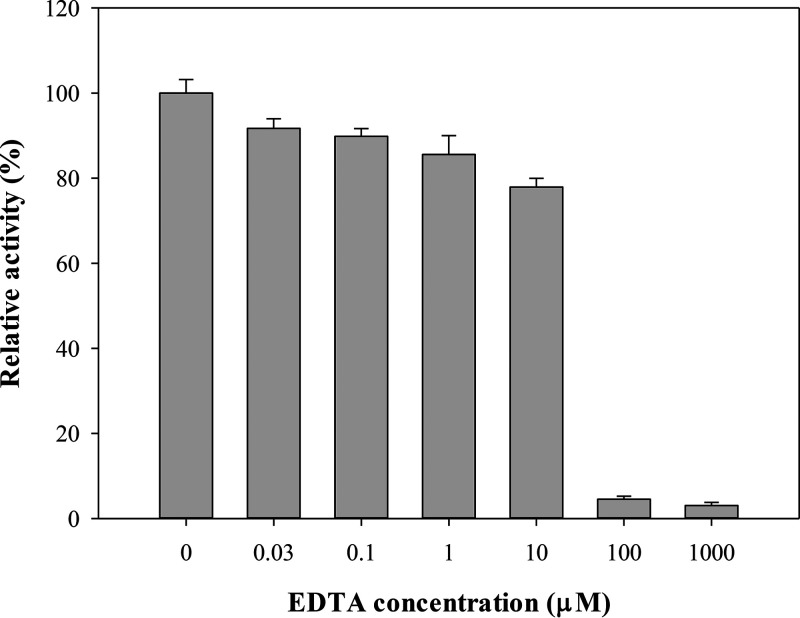
In general, subclass B1 and B3 MBLs bind two zinc ions at their active sites to exert their activity, whereas subclass B2 MBLs are monozinc enzymes with strong carbapenemase activity and are inhibited in binding of the second zinc ions (61). The common feature of all MBLs is the conserved motif of the two zinc ion binding sites, but different metal ions can also bind to these two binding sites and affect the catalytic activities of Vmh. To determine the effect of different metal ions on Vmh protein activity, the hydrolysis efficiency of β-lactam antibiotic imipenem in the presence of Vmh and different metal ions (Zn2+, Co2+, Mn2+, and Ca2+) was monitored. Because EDTA is known to be a metal binding inhibitor by stripping metal ions out of the binding sites in proteins (62), EDTA was used to examine whether metal ions are required for Vmh activity in this study. The recombinant Vmh protein was added to MOPS (morpholinepropanesulfonic acid) buffer containing ZnSO4, MnSO4, Cd(CH3COO)2, CoCl2, or CaCl2, and the hydrolysis efficiency of imipenem was measured individually. The results showed that Vmh proteins with Co2+ had slight activity (4%) compared with Vmh in the presence of Zn2+, and no activity was observed in Vmh with Mn2+ and Ca2+ (data not shown). Of all tested metals, only Cd2+ could substitute for Zn2+ with 96% relative activity, and it might be because they share a similar chemical property. The results showed that the hydrolysis activity decreased as the concentration of EDTA increased; moreover, the activity was abolished when the EDTA concentration reached 100 μM (Fig. 5). These data indicated that zinc ions are required for the recombinant Vmh protein to perform hydrolysis activity.
FIG 5.
Effect of EDTA on imipenem degradation. Vmh concentration, 0.027 μM; ZnSO4 concentration, 50 μM.
In previous studies, the hydrolysis activity of β-lactamase II in the presence of different metals, including Co2+, Cd2+, Mn2+, HgII+, Cu2+, Ni2+, Mg2+, and Ca2+, was measured. Compared with the control group in the presence of Zn2+, β-lactamase II showed low activity in the presence of Co2+, Cd2+, Mn2+, and HgII+ and no activity in a solution containing Cu2+, Ni2+, Mg2+, and Ca2+ (63). On the other hand, Yang and Bush (64) reported that β-lactamase AsbM1 from Aeromonas sobria AER 14M showed higher relative activity in the presence of Mn2+, Mg2+, and Ca2+, whereas Zn2+ and Cd2+ had an inhibitory effect on the hydrolysis of imipenem. It has been reported that BcII from B. cereus has different hydrolysis activities against β-lactam antibiotics in the presence of different metals, and the affinities of the metal ions decreased in the order Zn2+ > Cd2+ > Co2+ > Mn2+ (65).
Determination of zinc binding contents using ICP-MS.
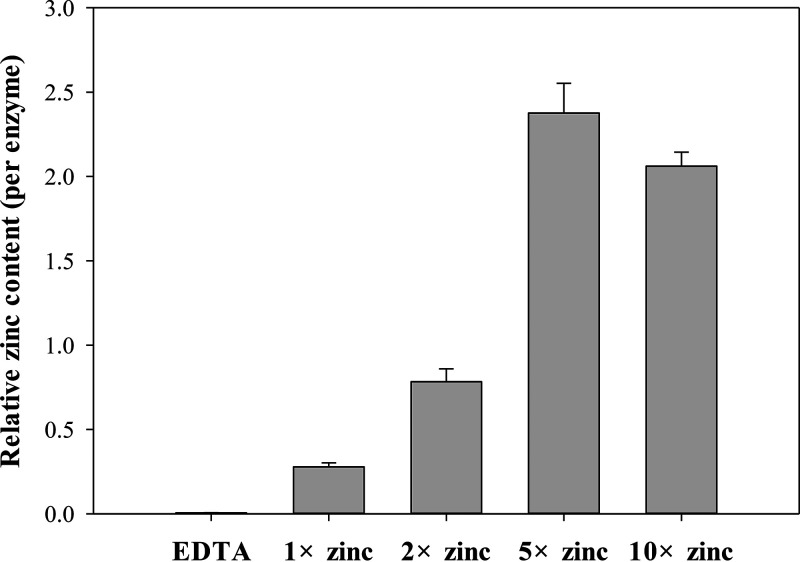
The zinc content of MBL was determined using inductively coupled plasma-MS (ICP-MS), which provides the total amount of zinc in the solution. The importance of zinc ions has been reported from the crystal structures of some zinc proteins, and they play an important role in substrate binding and catalysis (66). As shown in Fig. 6, after adding different amounts of zinc ions (1-, 2-, 5-, and 10-fold Vmh concentrations) to the Vmh solution (1-fold), the zinc content for each group was determined using ICP-MS, giving Zn/Vmh ratios of approximately 0.3 ± 0.02, 0.8 ± 0.08, 2.3 ± 0.18, and 2.1 ± 0.08, respectively. Vmh was found to be saturated with zinc ions by adding zinc at concentrations of 5 and 10 times that of Vmh, suggesting that the binding ratio of zinc to Vmh was 2 (Fig. 6).
FIG 6.
Determination of zinc content by ICP-MS of recombinant VmhΔSP.
In addition, the far-UV circular dichroism (CD) spectra of the Vmh in the presence and absence of zinc were determined. As shown in Fig. S4, the purified Vmh showed a CD spectrum similar to that of BcII and NDM, both of which belong to subclass B1 MBLs. The secondary-structure analyses for Vmh using the CDSSTR (67), CONTINLL (68), and SELCON3 (69) algorithms were performed with the CD data on the DichroWeb analysis server (70), and three algorithms showed similar predictions for Vmh secondary structures. Among all, the prediction result from the CDSSTR algorithm has the lowest normalized root mean square (0.033), and it indicated that Vmh has 30% α-helices, 23% β-strand, 19% β-turns, and 28% unordered secondary structures. Generally, the MBLs showed approximately 17% to 39% α-helices and 12% to 48% β-sheet content (41, 71, 72). In addition, only a slight difference between the far-UV CD spectra of the apo- and dizinc forms of Vmh was observed (Fig. S4), suggesting that the EDTA-treated Vmh did not go through a major transition for denaturation upon metal removal.
Palzkill (73) has reported that the subclass B1 and B3 MBLs bind two zinc ions at their active sites to exert their activities, whereas subclass B2 MBLs are monozinc enzymes with intense carbapenemase activity and are inhibited in the binding of the second zinc ions. Crowder et al. (42) determined the metal content of the purified subclass B3 MBL protein L1, and the zinc content ratio was two per enzyme molecule, which was similar to that of the subclass B1 MBL proteins IMP-1 (74) and BcII (75) and the subclass B3 MBL protein FEZ-1 (76). The subclass B3 MBL protein GOB-1 has a glutamine residue at position 116, a zinc binding residue in all known class B1 and B3 MBL structures. Horsfall et al. (77) replaced the Gln116 residue with Q116A, Q116N, and Q116H GOB-1 mutants to investigate the effect of the Gln116 residue, and the zinc contents for the wild type and mutants were determined. The results indicated that the wild-type GOB-1 contained two zinc ions per enzyme, whereas the Q116A and Q116N GOB-1 mutants had Zn/MBL ratios of 0.8 and 0.9, respectively, indicating the loss of zinc ions in the mutants.
Vmh showed better activity when saturated with two zinc ions and showed a broad substrate spectrum for β-lactam antibiotics. Based on the substrate spectrum and protein sequence analysis, it is suggested that the Vmh protein might be closer to the subclass B1 MBLs than the MBLs from subclasses B2 and B3.
MATERIALS AND METHODS
Strains and plasmids.
The E. coli strain NovaBlue(DE3) was used for genetic manipulation. E. coli strain C43(DE3) was used to overexpress VmhΔSP (with 19 signal peptide amino acid residues deleted) protein, and the purified VmhΔSP protein was analyzed to determine the oligomeric state, kinetic parameters, and mass spectrometry analysis. The pET26b vectors were used for vmh gene cloning and overexpression. V. vulnificus was purchased from the Bioresource Collection and Research Center (Hsinchu City, Taiwan). The signal peptide was predicted by using the SignalP 5.0 server.
Construction of pET26b-vmhΔSP.
The vmh gene was cloned from the V. vulnificus ATCC 27562 chromosome by using the PCR method. The truncated vmh gene was amplified by using primers 5′-AAAAACCCATGGGTGCACTGAAACTGACCACTT-3′ and 5′-AAAAAAGCGGCCGCCCACTTCATTTCA-3′ (57 bp deletion at the 5′ end), and then the amplified DNA was visualized on a 1% agarose gel containing SafeView DNA stain. The PCR product was digested with NcoI and NotI restriction enzymes and inserted into the pET26b vector. DNA purification was accomplished by using the Qiagen QIAquick gel extraction kit and Qiagen spin miniprep kit. Restriction digestion was performed by using NcoI and NotI restriction enzymes (Promega), and T4 DNA ligase (Promega) was used to catalyze the ligation of DNA fragments to the complementary ends of the linear vector. The plasmids carrying the vmh gene were transformed into E. coli C43(DE3) for protein overexpression.
Overexpression and purification of Vmh.
E. coli C43 harboring pET26b plasmid carrying the VmhΔSP gene was cultivated at 37°C, induced with 0.2 mM IPTG, and overexpressed at 24°C. The cells were collected by centrifugation (6,000 rpm, 4°C, 10 min), and cells were disrupted by osmotic shock. The cells were resuspended with ice-cold solution 1 (pH 8) containing 30 mM Tris, 1 mM EDTA, and 20% (vol/vol) sucrose, kept on ice for 20 min, and then harvested by centrifugation (5,000 rpm, 4°C, 10 min). As described above, the cell pellet was resuspended with ice-cold 5 mM MgSO4, kept on ice for 20 min, and then harvested by centrifugation (5,000 rpm, 4°C, 10 min), and the supernatant was collected (78). Buffer-solubilized Vmh proteins were purified by using a nickel affinity column (HiTrap chelating column; GE); the column was first washed with a 5- to 15-column volume of deionized water, and proteins were loaded onto a nickel affinity column by using a peristaltic pump at a rate of 1 ml/min. Vmh proteins were washed with A/B buffer in the presence of 50 to 300 mM imidazole (AKTA purifier). Recombinant Vmh protein was resolved by SDS-PAGE to determine the efficiency of overexpression and purification.
Vmh protein identification by using LC-MS/MS.
The excised band on the SDS gel was cut into cubes. Gel pieces were then transferred to a microtube and spun down on a bench-top microcentrifuge. The first step to destain the gel pieces excised from Coomassie blue-stained gels was to add 100 mM ammonium bicarbonate-acetonitrile (1:1, vol/vol) and to incubate them with vortexing for 30 min. Second, neat acetonitrile was added, followed by incubation at room temperature with vortexing until the gel pieces became white and shrank, and then acetonitrile was removed. A 20 mM dithiothreitol (DTT) solution was added to completely cover the gel pieces, which were incubated for 30 min at 55°C; after removal of the DTT solution, 55 mM iodoacetamide (IAA) solution was added, followed by incubation for 60 min at room temperature in the dark. The gel pieces were then shrunk by centrifugation, and all liquid was removed. Enough trypsin buffer (20 ng/μl) was then added to cover the dry gel pieces, followed by incubation for 15 min on ice, the liquid was removed, and enough buffer solution was added to cover the gel pieces again, followed by incubation for 12 h at room temperature. Finally, extraction solution (50% acetonitrile–5% formic acid) was added, followed by sonication for 15 min, and the buffer was moved to a new microtube, which was put into the vacuum centrifuge. Protein LC-MS analyses were performed on a Waters Acquity nano-UPLC in line with a Waters G2 quadrupole time of flight (Q-TOF) mass spectrometer. Protein samples (10 μg/ml) were directly infused onto a mass spectrometer through a syringe pump at a flow rate of 1 μl/min. The G2 Q-TOF mass spectrometer was run in positive-ion, high-resolution mode with detection in the range of 600 to 2,300 m/z. Source parameters were as follows: capillary voltage, 2.50 kV; source temperature, 90°C; desolvation temperature, 200°C; cone gas flow, 20 liters/h; desolvation gas flow, 500 liters/h. The protein peak was deconvoluted by the MassLynx MaxEnt1 function according to the following parameters: output resolution, 1.0 Da/channel; uniform Gaussian width at half height, 0.75 Da; minimum intensity ratios, 30% for left and right; iteration to convergence for completion.
SEC.
Size exclusion chromatography (SEC) analyses were performed on an AKTA purifier (Amersham) using a Superdex200 prep-grade HiLoad 16/60 column, equilibrated with 20 mM Tris-HCl (pH 8), 50 mM NaCl, and 10% (vol/vol) glycerol, and run at 1 ml/min. The protein Mr was calculated from a calibration curve constructed using CARB-17 (29.3 kDa), ovalbumin (44 kDa), conalbumin (75 kDa), aldolase (158 kDa), and dextran blue (void volume) standards. The Kav values were calculated using their elution volumes (Ve), the column volume (Vc), and the void volume (Vo) in the equation Kav = (Ve − Vo)/(Vc − Vo). A calibration curve was constructed by plotting the log of the Mr of the standards against their Kav values.
AUC.
Sedimentation velocity (SV) experiments were performed with a Beckman-Coulter (Fullerton, CA, USA) XL-A analytical ultracentrifuge. For SV analytical ultracentrifugation analysis (AUC), sample and buffer were loaded into 12-mm standard double-sector Epon charcoal-filled centerpieces and mounted in an An-60 Ti rotor. SV experiments were performed at a rotor speed of 50,000 rpm at 20°C. The sample signal was monitored at 280 nm, and the raw experimental data were analyzed by SEDFIT software.
LC-MS analysis.
High-resolution and high-mass-accuracy liquid chromatography-mass spectrometry (LC-MS) experiments were performed on an LTQ-FT Ultra (linear quadrupole ion trap Fourier transform ion cyclotron resonance) mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a standard ESI source, an Agilent 1100 series binary high-performance liquid chromatography pump (Agilent Technologies, Palo Alto, CA), and a Famos autosampler (LC Packings, San Francisco, CA). The Vmh protein sample was injected (5 μl) at a 50-μl/min flow rate on a BioResolve RP MAb polyphenyl column (1 mm [inside diameter] by 150 mm, 5 μm, 100 Å; Waters). The chromatographic separation was done using 0.1% formic acid in water as mobile phase A and 0.1% formic acid in 80% acetonitrile as mobile phase B at a 50-μl/min flow rate. The separation gradient was initially at 2% buffer B for 2 min, increasing to 98% buffer B at 110 min. Wide selected ion monitoring (SIM) MS conditions included a mass range of m/z 600 to 1,200 and resolution of 100,000 at m/z 400. The electrospray voltage was maintained at 4.0 kV, and the capillary temperature was maintained at 275°C.
Drug degradation assays and the effects of various metals.
The β-lactamase activity of purified Vmh was monitored as the decrease in β-lactam absorbance that results from the opening of the β-lactam ring during hydrolysis. The reactions were performed at 25°C in a mixture containing purified Vmh, 50 mM MOPS (pH 7.5), 50 μM ZnSO4, and β-lactam antibiotics, such as ampicillin and imipenem, and the decrease in absorbance was monitored. The effects of other metal ions on the imipenem degradation by Vmh was also monitored by measuring the activity in 50 mM MOPS (pH 7.5) containing 50 μM MnSO4, Cd(CH3COO)2, CoCl2, or CaCl2. The imipenem degradation activity of Vmh was also monitored in the presence of EDTA by measuring the activity in 50 mM MOPS (pH 7.5) containing 50 μM ZnSO4 in the presence of different concentrations of EDTA (0, 0.03, 0.1, 1, 10, 100, or 1,000 μM). The extinction coefficients and measured wavelengths for the drug degradation test, respectively, are 820 M−1 cm−1 and 235 nm for ampicillin and piperacillin, 400 M−1 cm−1 and 240 nm for carbenicillin, 6,500 M−1 cm−1 and 260 nm for cephalothin, 7,600 M−1 cm−1 and 260 nm for cefuroxime, 9,000 M−1 cm−1 and 260 nm for ceftazidime, 10,000 M−1 cm−1 and 260 nm for cefepime, 4,000 M−1 cm−1 and 265 nm for moxalactam, 10,940 M−1 cm−1 and 300 nm for meropenem, and 9,000 M−1 cm−1 and 300 nm for imipenem. The rate of β-lactam hydrolysis was measured as a function of the β-lactam concentration, and the data were fitted to either a hyperbolic or sigmoidal equation using SigmaPlot (Systat Software, Inc.).
Measurement of zinc concentration by ICP-MS.
The recombinant VmhΔHistag protein was overexpressed and purified as described in “Overexpression and purification of Vmh” with some modifications. The buffer containing recombinant VmhΔHistag obtained from osmotic shock was purified by using an ion-exchange column (HiTrap Q column; GE). A Q column was first equilibrated with 5 column volumes of the A buffer (20 mM Tris buffer, pH 7.5), and the VmhΔHistag protein was loaded onto the column using a peristaltic pump at a rate of 1 ml/min. The Vmh proteins were eluted with A buffer with a salt gradient of 1 to 1,000 mM NaCl (AKTA purifier). The purified VmhΔHistag protein was concentrated with a 5-kDa-cutoff Vivaspin (GE Health Care). The VmhΔHistag protein in the Vivaspin was treated with EDTA (20 mM in 50 mM TEAB [triethylammonium bicarbonate buffer]) for 1 h to remove the metals, and the EDTA was removed by buffer wash and concentration five times. The VmhΔHistag protein (20 μM) was then incubated with different amounts of zinc ions (1-, 2-, 5-, and 10-fold) for 1 h, and the sample was washed with 50 mM TEAB and concentrated for five times to remove unbound zinc ions. The Vivaspin containing the VmhΔHistag protein was spun for the proteins to precipitate onto the membrane. The membrane was collected and treated with concentrated nitric acid overnight to digest VmhΔHistag protein, and the metals were collected by centrifugation. The metals were analyzed on an inductively coupled plasma (ICP) mass spectrometer (Agilent 7700x; Agilent Technologies). The zinc concentration of Vmh protein was monitored and determined at an m/z of 64.
Far-UV circular dichroism.
Far-UV circular dichroism (CD) spectra for the metallated, apo, and remetallated forms of the Vmh proteins (0.1 mg/ml) were obtained using a JASCO J-815 spectropolarimeter at 25°C. The Vmh proteins were prepared by dialysis with 20 mM phosphate buffer (pH 7.4) at 4°C for 12 h. The metallated Vmh was obtained in the presence of 50 μM Zn2+. The apo-Vmh was obtained in the presence of 0.2 mM EDTA. The remetallated Vmh proteins were first treated with 0.2 mM EDTA and added with 0.5 mM Zn2+. The spectra were scanned at 25°C with 0.5-nm steps from 195 to 260 nm.
Statistical analysis.
Data were analyzed statistically using SPSS version 12.0 (SPSS, Inc., Chicago, IL, USA). One-way analysis of variance (ANOVA) was used to determine statistical differences between sample means, with the level of significance set at a P of <0.05. Multiple comparisons of means were done by Duncan’s test. All data are expressed as the mean ± standard deviation (SD).
ACKNOWLEDGMENTS
We thank Cheng-Kuan Su, National Chung Hsing University, for kindly providing his expertise on ICP-MS and the Mass Spectrometry Core Facility in Genomics Research Center of Academia Sinica for collecting mass spectrometry data. We thank Yu-Hou Chen, Institute of Biological Chemistry at Academia Sinica, for help on the analytical ultracentrifugation analysis. We thank Adrian R. Walmsley and Maria Ines Borges-Walmsley, Durham University, UK, for kindly providing the E. coli Kam3 strain and expression vectors.
This work was supported by the Center of Excellence for the Oceans, National Taiwan Ocean University, from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (NTOU-RD-AA-2019-1-02011-2 and NTOU-RD-AA-2021-1-02018).
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Jones MK, Oliver JD. 2009. Vibrio vulnificus: disease and pathogenesis. Infect Immun 77:1723–1733. 10.1128/IAI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canigral I, Moreno Y, Alonso JL, Gonzalez A, Ferrus MA. 2010. Detection of Vibrio vulnificus in seafood, seawater and wastewater samples from a Mediterranean coastal area. Microbiol Res 165:657–664. 10.1016/j.micres.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Blackwell KD, Oliver JD. 2008. The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina estuaries. J Microbiol 46:146–153. 10.1007/s12275-007-0216-2. [DOI] [PubMed] [Google Scholar]
- 4.Bier N, Schwartz K, Guerra B, Strauch E. 2015. Survey on antimicrobial resistance patterns in Vibrio vulnificus and Vibrio cholerae non-O1/non-O139 in Germany reveals carbapenemase-producing Vibrio cholerae in coastal waters. Front Microbiol 6:1179. 10.3389/fmicb.2015.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaysner CA, AbeytaC, Jr, Wekell MM, DePaolaA, Jr, Stott RF, Leitch JM. 1987. Virulent strains of Vibrio vulnificus isolated from estuaries of the United States West Coast. Appl Environ Microbiol 53:1349–1351. 10.1128/aem.53.6.1349-1351.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels NA. 2011. Vibrio vulnificus oysters: pearls and perils. Clin Infect Dis 52:788–792. 10.1093/cid/ciq251. [DOI] [PubMed] [Google Scholar]
- 7.Strom MS, Paranjpye RN. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microb Infect 2:177–188. 10.1016/S1286-4579(00)00270-7. [DOI] [PubMed] [Google Scholar]
- 8.Yun NR, Kim DM. 2018. Vibrio vulnificus infection: a persistent threat to public health. Korean J Intern Med 33:1070–1078. 10.3904/kjim.2018.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldhusen F. 2000. The role of seafood in bacterial foodborne diseases. Microbes Infect 2:1651–1660. 10.1016/s1286-4579(00)01321-6. [DOI] [PubMed] [Google Scholar]
- 10.MorrisJG, Jr, Tenney J. 1985. Antibiotic therapy for Vibrio vulnificus infection. JAMA 253:1121–1122. 10.1001/jama.1985.03350320041011. [DOI] [PubMed] [Google Scholar]
- 11.Tang HJ, Chang MC, Ko WC, Huang KY, Lee CL, Chuang YC. 2002. In vitro and in vivo activities of newer fluoroquinolones against Vibrio vulnificus. Antimicrob Agents Chemother 46:3580–3584. 10.1128/AAC.46.11.3580-3584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen SC, Lee YT, Tsai SJ, Chan KS, Chao WN, Wang PH, Lin DB, Chen CC, Lee MC. 2012. Antibiotic therapy for necrotizing fasciitis caused by Vibrio vulnificus: retrospective analysis of an 8 year period. J Antimicrob Chemother 67:488–493. 10.1093/jac/dkr476. [DOI] [PubMed] [Google Scholar]
- 13.Shaw KS, Rosenberg Goldstein RE, He X, Jacobs JM, Crump BC, Sapkota AR. 2014. Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland Coastal Bays. PLoS One 9:e89616. 10.1371/journal.pone.0089616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DM, Lym Y, Jang SJ, Han H, Kim YG, Chung CH, Hong SP. 2005. In vitro efficacy of the combination of ciprofloxacin and cefotaxime against Vibrio vulnificus. Antimicrob Agents Chemother 49:3489–3491. 10.1128/AAC.49.8.3489-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanetti S, Spanu T, Deriu A, Romano L, Sechi LA, Fadda G. 2001. In vitro susceptibility of Vibrio spp. isolated from the environment. Int J Antimicrob Agents 17:407–409. 10.1016/s0924-8579(01)00307-7. [DOI] [PubMed] [Google Scholar]
- 16.Chiang SR, Chuang YC. 2003. Vibrio vulnificus infection: clinical manifestations, pathogenesis, and antimicrobial therapy. J Microbiol Immunol Infect 36:81–88. [PubMed] [Google Scholar]
- 17.Thakur AB, Vaidya RB, Suryawanshi SA. 2003. Pathogenicity and antibiotic susceptibility of Vibrio species isolated from moribund shrimps. Indian J Mar Sci 32:71–75. [Google Scholar]
- 18.Baker-Austin C, McArthur JV, Lindell AH, Wright MS, Tuckfield RC, Gooch J, Warner L, Oliver J, Stepanauskas R. 2009. Multi-site analysis reveals widespread antibiotic resistance in the marine pathogen Vibrio vulnificus. Microb Ecol 57:151–159. 10.1007/s00248-008-9413-8. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Choresca CH, Shin SP, Han JE, Jun JW, Park SC. 2011. Occurrence and antibiotic resistance of Vibrio vulnificus in seafood and environmental waters in Korea. J Food Saf 31:518–524. 10.1111/j.1745-4565.2011.00329.x. [DOI] [Google Scholar]
- 20.Al-Dulaimi MMK, Abd Mutalib S, Abd Ghani M, Zaini NAM, Ariffin AA. 2019. Multiple antibiotic resistance (MAR), plasmid profiles, and DNA polymorphisms among Vibrio vulnificus isolates. Antibiotics (Basel) 8:68. 10.3390/antibiotics8020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54:969–976. 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin H-TV, Massam-Wu T, Lin C-P, Wang Y-JA, Shen Y-C, Lu W-J, Hsu P-H, Chen Y-H, Borges-Walmsley MI, Walmsley AR. 2017. The Vibrio cholerae var regulon encodes a metallo-beta-lactamase and an antibiotic efflux pump, which are regulated by VarR, a LysRtype transcription factor. PLoS One 12:e0184255. 10.1371/journal.pone.0184255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiou JC, Li RC, Chen S. 2015. CARB-17 family of beta-lactamases mediates intrinsic resistance to penicillins in Vibrio parahaemolyticus. Antimicrob Agents Chemother 59:3593–3595. 10.1128/AAC.00047-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangat CS, Boyd D, Janecko N, Martz S-L, Desruisseau A, Carpenter M, Reid-Smith RJ, Mulvey MR. 2016. Characterization of VCC-1, a novel Ambler class A carbapenemase from Vibrio cholerae isolated from imported retail shrimp sold in Canada. Antimicrob Agents Chemother 60:1819–1825. 10.1128/AAC.02812-15. (Erratum, 60:3263, 2016, .) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, Martinez-Urtaza J. 2018. Vibrio spp. infections. Nat Rev Dis Primers 4:8. 10.1038/s41572-018-0005-8. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phelan EK, Miraula M, Selleck C, Ollis DL, Schenk G, Mitić N. 2014. Metallo-β-lactamases: a major threat to human health. Am J Mol Biol 4:89–104. 10.4236/ajmb.2014.43011. [DOI] [Google Scholar]
- 28.Daiyasu H, Osaka K, Ishino Y, Toh H. 2001. Expansion of the zinc metallo-hydrolase family of the beta-lactamase fold. FEBS Lett 503:1–6. 10.1016/s0014-5793(01)02686-2. [DOI] [PubMed] [Google Scholar]
- 29.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 30.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43:W389–W394. 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 33.Buchan DWA, Jones DT. 2019. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res 47:W402–W407. 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lassaux P, Traore DA, Loisel E, Favier A, Docquier JD, Sohier JS, Laurent C, Bebrone C, Frere JM, Ferrer JL, Galleni M. 2011. Biochemical and structural characterization of the subclass B1 metallo-beta-lactamase VIM-4. Antimicrob Agents Chemother 55:1248–1255. 10.1128/AAC.01486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galleni M, Lamotte-Brasseur J, Rossolini GM, Spencer J, Dideberg O, Frere JM, Metallo-beta-lactamases Working Group. 2001. Standard numbering scheme for class B beta-lactamases. Antimicrob Agents Chemother 45:660–663. 10.1128/AAC.45.3.660-663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brem J, van Berkel SS, Zollman D, Lee SY, Gileadi O, McHugh PJ, Walsh TR, McDonough MA, Schofield CJ. 2016. Structural basis of metallo-beta-lactamase inhibition by captopril stereoisomers. Antimicrob Agents Chemother 60:142–150. 10.1128/AAC.01335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armenteros JJA, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 39.Thomas PW, Zheng M, Wu SS, Guo H, Liu DL, Xu DG, Fast W. 2011. Characterization of purified New Delhi metallo-beta-lactamase-1. Biochemistry 50:10102–10113. 10.1021/bi201449r. [DOI] [PubMed] [Google Scholar]
- 40.King D, Strynadka N. 2011. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci 20:1484–1491. 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crawford PA, Sharma N, Chandrasekar S, Sigdel T, Walsh TR, Spencer J, Crowder MW. 2004. Over-expression, purification, and characterization of metallo-beta-lactamase ImiS from Aeromonas veronii bv. sobria. Protein Expr Purif 36:272–279. 10.1016/j.pep.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 42.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. 1998. Overexpression, purification, and characterization of the cloned metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 42:921–926. 10.1128/AAC.42.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y, Maltseva N, Wilamowski M, Tesar C, Endres M, Joachimiak A. 2020. Structural and biochemical analysis of the metallo-beta-lactamase L1 from emerging pathogen Stenotrophomonas maltophilia revealed the subtle but distinct di-metal scaffold for catalytic activity. Protein Sci 29:723–743. 10.1002/pro.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ullah JH, Walsh TR, Taylor IA, Emery DC, Verma CS, Gamblin SJ, Spencer J. 1998. The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 angstrom resolution. J Mol Biol 284:125–136. 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 45.Felici A, Amicosante G, Oratore A, Strom R, Ledent P, Joris B, Fanuel L, Frere JM. 1993. An overview of the kinetic parameters of class B beta-lactamases. Biochem J 291:151–155. 10.1042/bj2910151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bebrone C. 2007. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol 74:1686–1701. 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 47.Jacquin O, Balbeur D, Damblon C, Marchot P, De Pauw E, Roberts GCK, Frere JM, Matagne A. 2009. Positively cooperative binding of zinc ions to Bacillus cereus 569/H/9 beta-lactamase II suggests that the binuclear enzyme is the only relevant form for catalysis. J Mol Biol 392:1278–1291. 10.1016/j.jmb.2009.07.092. [DOI] [PubMed] [Google Scholar]
- 48.Poirel L, Palmieri M, Brilhante M, Masseron A, Perreten V, Nordmann P. 2020. PFM-like enzymes are a novel family of subclass B2 metallo-beta-lactamases from Pseudomonas synxantha belonging to the Pseudomonas fluorescens complex. Antimicrob Agents Chemother 64. 10.1128/AAC.01700-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoczko M, Frere JM, Rossolini GM, Docquier JD. 2006. Postgenomic scan of metallo-beta-lactamase homologues in rhizobacteria: identification and characterization of BJP-1, a subclass B3 ortholog from Bradyrhizobium japonicum. Antimicrob Agents Chemother 50:1973–1981. 10.1128/AAC.01551-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franceschini N, Caravelli B, Docquier JD, Galleni M, Frere JM, Amicosante G, Rossolini GM. 2000. Purification and biochemical characterization of the VIM-1 metallo-beta-lactamase. Antimicrob Agents Chemother 44:3003–3007. 10.1128/AAC.44.11.3003-3007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Docquier JD, Lamotte-Brasseur J, Galleni M, Amicosante G, Frere JM, Rossolini GM. 2003. On functional and structural heterogeneity of VIM-type metallo-beta-lactamases. J Antimicrob Chemother 51:257–266. 10.1093/jac/dkg067. [DOI] [PubMed] [Google Scholar]
- 52.Cornish-Bowden A, Cárdenas ML. 2004. Glucokinase: a monomeric enzyme with positive cooperativity. Front Diabetes 16:125–134. 10.1159/000079011. [DOI] [Google Scholar]
- 53.Wu ZH, Xing JH. 2012. Functional roles of slow enzyme conformational changes in network dynamics. Biophys J 103:1052–1059. 10.1016/j.bpj.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Y, Li X, Morrow BR, Pothukuchy A, Gollihar J, Novak R, Reilly CB, Ellington AD, Walt DR. 2019. Single-molecule mechanistic study of enzyme hysteresis. ACS Cent Sci 5:1691–1698. 10.1021/acscentsci.9b00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Porter CM, Miller BG. 2012. Cooperativity in monomeric enzymes with single ligand-binding sites. Bioorg Chem 43:44–50. 10.1016/j.bioorg.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ricard J, Meunier JC, Buc J. 1974. Regulatory behavior of monomeric enzymes. 1. The mnemonical enzyme concept. Eur J Biochem 49:195–208. 10.1111/j.1432-1033.1974.tb03825.x. [DOI] [PubMed] [Google Scholar]
- 57.Ainslie GR, Shill JP, Neet KE. 1972. Transients and cooperativity: a slow transition model for relating transients and cooperative kinetics of enzymes. J Biol Chem 247:7088–7096. 10.1016/S0021-9258(19)44697-8. [DOI] [PubMed] [Google Scholar]
- 58.Pettersson G. 1986. Mechanistic origin of the sigmoidal rate behaviour of glucokinase. Biochem J 233:347–350. 10.1042/bj2330347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sugiki T, Kobayashi N, Fujiwara T. 2017. Modern technologies of solution nuclear magnetic resonance spectroscopy for three-dimensional structure determination of proteins open avenues for life scientists. Comput Struct Biotechnol J 15:328–339. 10.1016/j.csbj.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shashkova S, Leake MC. 2017. Single-molecule fluorescence microscopy review: shedding new light on old problems. Biosci Rep 37:BSR20170031. 10.1042/BSR20170031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez MM, Herman R, Ghiglione B, Kerff F, Gonzalez GD, Bouillenne F, Galleni M, Handelsman J, Charlier P, Gutkind G, Sauvage E, Power P. 2017. Crystal structure and kinetic analysis of the class B3 di-zinc metallo-beta-lactamase LRA-12 from an Alaskan soil metagenome. PLoS One 12:e0182043. 10.1371/journal.pone.0182043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flora SJS, Pachauri V. 2010. Chelation in metal intoxication. Int J Environ Res Public Health 7:2745–2788. 10.3390/ijerph7072745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davies RB, Abraham EP. 1974. Metal cofactor requirement of β-lactamase II. Biochem J 143:129–135. 10.1042/bj1430129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang YJ, Bush K. 1996. Biochemical characterization of the carbapenem-hydrolyzing beta-lactamase AsbM1 from Aeromonas sobria AER 14M: a member of a novel subgroup of metallo-beta-lactamases. FEMS Microbiol Lett 137:193–200. 10.1111/j.1574-6968.1996.tb08105.x. [DOI] [PubMed] [Google Scholar]
- 65.Badarau A, Page MI. 2006. The variation of catalytic efficiency of Bacillus cereus metallo-beta-lactamase with different active site metal ions. Biochemistry 45:10654–10666. 10.1021/bi060934l. [DOI] [PubMed] [Google Scholar]
- 66.Karsisiotis AI, Damblon CF, Roberts GCK. 2014. A variety of roles for versatile zinc in metallo-beta-lactamases. Metallomics 6:1181–1197. 10.1039/c4mt00066h. [DOI] [PubMed] [Google Scholar]
- 67.Sreerama N, Woody RW. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem 287:252–260. 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 68.van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC. 1990. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal Biochem 191:110–118. 10.1016/0003-2697(90)90396-q. [DOI] [PubMed] [Google Scholar]
- 69.Sreerama N, Venyaminov SY, Woody RW. 1999. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci 8:370–380. 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whitmore L, Wallace BA. 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89:392–400. 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 71.Cheng Z, Thomas PW, Ju L, Bergstrom A, Mason K, Clayton D, Miller C, Bethel CR, VanPelt J, Tierney DL, Page RC, Bonomo RA, Fast W, Crowder MW. 2018. Evolution of New Delhi metallo-beta-lactamase (NDM) in the clinic: effects of NDM mutations on stability, zinc affinity, and mono-zinc activity. J Biol Chem 293:12606–12618. 10.1074/jbc.RA118.003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toney JH, Wu JK, Overbye KM, Thompson CM, Pompliano DL. 1997. High-yield expression, purification, and characterization of active, soluble Bacteroides fragilis metallo-beta-lactamase, CcrA. Protein Expr Purif 9:355–362. 10.1006/prep.1996.0718. [DOI] [PubMed] [Google Scholar]
- 73.Palzkill T. 2013. Metallo-β-lactamase structure and function. Ann N Y Acad Sci 1277:91–104. 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horton LB, Shanker S, Mikulski R, Brown NG, Phillips KJ, Lykissa E, Prasad BVV, Palzkill T. 2012. Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site. Antimicrob Agents Chemother 56:5667–5677. 10.1128/AAC.01276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moali C, Anne C, Lamotte-Brasseur J, Groslambert S, Devreese B, Van Beeumen J, Galleni M, Frere JM. 2003. Analysis of the importance of the metallo-beta-lactamase active site loop in substrate binding and catalysis. Chem Biol 10:319–329. 10.1016/s1074-5521(03)00070-x. [DOI] [PubMed] [Google Scholar]
- 76.Mercuri PS, Bouillenne F, Boschi L, Lamotte-Brasseur J, Amicosante G, Devreese B, van Beeumen J, Frère JM, Rossolini GM, Galleni M. 2001. Biochemical characterization of the FEZ-1 metallo-beta-lactamase of Legionella gormanii ATCC 33297T produced in Escherichia coli. Antimicrob Agents Chemother 45:1254–1262. 10.1128/AAC.45.4.1254-1262.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horsfall LE, Izougarhane Y, Lassaux P, Selevsek N, Lienard BMR, Poirel L, Kupper MB, Hoffmann KM, Frere JM, Galleni M, Bebrone C. 2011. Broad antibiotic resistance profile of the subclass B3 metallo-beta-lactamase GOB-1, a di-zinc enzyme. FEBS J 278:1252–1263. 10.1111/j.1742-4658.2011.08046.x. [DOI] [PubMed] [Google Scholar]
- 78.Neu HC, Heppel LA. 1965. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem 240:3685–3692. 10.1016/S0021-9258(18)97200-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download AAC.00326-21-s0001.pdf, PDF file, 0.7 MB (761.1KB, pdf)