ABSTRACT
Klebsiella pneumoniae is an opportunistic Gram-negative pathogen that employs different strategies (resistance and persistence) to counteract antibiotic treatments. This study aimed to search for new means of combatting imipenem-resistant and persister strains of K. pneumoniae by repurposing the anticancer drug mitomycin C as an antimicrobial agent and by combining the drug and the conventional antibiotic imipenem with the lytic phage vB_KpnM-VAC13. Several clinical K. pneumoniae isolates were characterized, and an imipenem-resistant isolate (harboring OXA-245 β-lactamase) and a persister isolate were selected for study. The mitomycin C and imipenem MICs for both isolates were determined by the broth microdilution method. Time-kill curve data were obtained by optical density at 600 nm (OD600) measurement and CFU enumeration in the presence of each drug alone and with the phage. The frequency of occurrence of mutants resistant to each drug and the combinations was also calculated, and the efficacy of the combination treatments was evaluated using an in vivo infection model (Galleria mellonella). The lytic phage vB_KpnM-VAC13 and mitomycin C had synergistic effects on imipenem-resistant and persister isolates, both in vitro and in vivo. The phage-imipenem combination successfully killed the persisters but not the imipenem-resistant isolate harboring OXA-245 β-lactamase. Interestingly, the combinations decreased the emergence of in vitro resistant mutants of both isolates. Combinations of the lytic phage vB_KpnM-VAC13 with mitomycin C and imipenem were effective against the persister K. pneumoniae isolate. The lytic phage-mitomycin C combination was also effective against imipenem-resistant K. pneumoniae strains harboring OXA-245 β-lactamase.
KEYWORDS: resistance, persistence, bacteriophage therapy, drug repurposing, synergy, Klebsiella pneumoniae
INTRODUCTION
The emergence of multidrug-resistant (MDR) pathogens that are resistant to the vast majority of drugs used in clinical practice is a global health problem. Among these pathogens, Klebsiella pneumoniae is an opportunistic Gram-negative bacterium that causes severe difficult-to-treat infections, with many isolates displaying resistance to antimicrobials, especially to carbapenems such as imipenem (1). In recent years, reports of carbapenemase-producing strains of K. pneumoniae have increased worldwide; the increase has been attributed to long periods of hospitalization, the presence of comorbidities, inappropriate medical interventions, and overuse or misuse of antibiotics (2, 3).
However, resistance is not the only cause of the failure of antibiotic-based therapies, and persistence also has a strong impact (4). Persister cells are subpopulations of nongrowing, nonreplicative dormant bacteria that can reestablish infections once the antibiotic stress is removed (5). Bacterial persistence therefore plays an essential role in chronic, recurrent, and recalcitrant infections (6). Persister bacteria counteract antibiotic treatments without affecting the MIC of the drug (4). Unlike resistance, persistence is not inheritable; rather, it is thought to be caused by numerous bacterial pathways working together, e.g., accumulation of (p)ppGpp (7), activation of toxin-antitoxin modules (8), stress responses, quorum sensing (9), and ribosome dimerization (10), among others. Although the molecular mechanisms underlying this state have been extensively studied, there are currently very few effective treatments against this phenotype (11–13). Indeed, the clinical importance of persister cells is huge, as there is evidence that persisters contribute to the emergence of new resistant pathogens, constituting infectious reservoirs that are difficult to eradicate and possibly responsible for chronic infections (14–19).
Taking into consideration the clinical importance of resistant and persister isolates of K. pneumoniae, the development of new therapeutic strategies against these bacterial phenotypes is urgent. In this postantibiotic era, several strategies can potentially be used to fight infections, including the following: (i) drug repurposing, i.e., finding new indications for FDA-approved drugs, such as the use of the anticancerous agent mitomycin C as an antimicrobial molecule (20, 21), and (ii) combination therapies, i.e., the use of two or more drugs to kill pathogenic bacteria (22).
The repurposing or repositioning strategy is currently of great interest, as the discovery and development of new antibiotics are now limited and, in most cases, unsuccessful. The use of nonantibiotic compounds has been shown to reduce the emergence of cross-resistance, usually because the active molecule affects a target other than the antibiotic (12). Here, we used mitomycin C, an anticancer drug, as it has been reported on numerous occasions that this chemotherapeutic agent possesses antibacterial activity (23–25) and is effective in killing a broad range of bacterial cells, even those occurring in biofilms (24, 25).
Regarding the second strategy, numerous studies have reported potentially synergistic activity between bacteriophages and antibiotics in the eradication of MDR bacteria, even in biofilms with limited therapeutic options. Two cases have been reported of patients with prosthetic graft material colonized by MDR Pseudomonas aeruginosa in whom a combination of lytic phages and antibiotics resulted in a successful outcome (26, 27). Moreover, research in the last few decades has demonstrated that phages are safe to be administered as therapeutics and also have an enormous potential as antibiotic adjuvants (28, 29). Some phages may recognize bacterial efflux pumps as receptors, thus resensitizing the bacteria to antibiotics expelled by the pumps (30, 31). In addition, synergistic reactions between lytic phages and antibiotics can also occur (32–35).
In this study, mitomycin C and imipenem were combined with the lytic phage vB_KpnM-VAC13 and tested against two phenotypically and genotypically characterized clinical isolates of K. pneumoniae: an imipenem-resistant isolate (K2534) and a persister isolate (K3325). The study findings provide evidence of the therapeutic potential of a repurposed drug (mitomycin C) and a conventional antibiotic (imipenem) in combination with the lytic phage vB_KpnM-VAC13, both in vitro and in vivo.
RESULTS
Characterization of K2534 and K3325 isolates.
Two isolates (K2534 and K3325) were selected from the 16 clinical strains characterized by several phenotypic and genomic approaches (Table 1): the first was imipenem resistant (carrying OXA-245 derived from OXA-48, the most common carbapenemase in K. pneumoniae worldwide) and the second was a highly persistent to imipenem, as indicated by the tolerance disk test (TDtest) and time-kill assays with the antibiotic (36) (see Fig. S1 in the supplemental material).
TABLE 1.
Collection of clinical isolates of K. pneumoniae, their biological origin, phenotype against imipenem, presence of carbapenemases, and spot test of vB_KpnM-VAC13
| Bacterial strain | Biological origin | GenBank accession | STa | Capsular typeb | Phenotype to IMPc | Carbapenemase | Result of spot test of vB_KpnM-VAC13 |
|---|---|---|---|---|---|---|---|
| K. pneumoniae MGH78578 | ATCC | NC_009648 | 38 | KL52 | Persister | NA | + |
| K2521 | Rectal | WRXA00000000 | 340 | KL15 | NA | VIM-1 | + |
| K2522 | Axillary smear | WRWY00000000 | 512 | KL107 | Resistant | KPC-3 | + |
| K2523 | Urine | WRWW00000000 | 11 | KL24 | NA | OXA-48 | − |
| K2524 | Urine | WRWV00000000 | 258 | KL107 | NA | KPC-3 | + |
| K2530 | Wound | WRXH00000000 | 11 | KL24 | Resistant | OXA-245 | + |
| K2534 | Rectal sample | WRXG00000000 | 437 | KL36 | Resistant | OXA-245 | + |
| K2536 | Respiratory sample | WRXC00000000 | 11 | KL24 | Resistant | VIM-1 | + |
| K2691 | Blood | SAMEA3538911d | 11 | KL24 | NA | NA | − |
| K2707 | Blood | SAMEA3538945d | 11 | KL13 | NA | KPC-like | + |
| K2794 | Blood | NGe | Tolerant | NA | + | ||
| K2984 | Blood | SAMEA3649453d | 405 | KL151 | Persister | NA | − |
| K2986 | Blood | SAMEA3649454d | 307 | KL102 | Tolerant | NA | + |
| K2989 | Blood | SAMEA3649457d | 661 | KL24 | Persister | NA | + |
| K3325 | Blood | SAMEA3649525d | 42 | KL64 | Persister | NA | + |
| K3569 | Blood | NG | Sensitive | NA | − |
ST, sequence type (found in https://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
KL, K-loci for capsule type determination, found in Kaptive.
IMP, imipenem; NA, not applicable (absence of carbapenemase genes of KPC-2, KPC-3, OXA-48, OXA-245 or VIM-1, found in ResFinder).
BioSample code of complete genomes included in the European BioProject PRJEB10018.
NG, no genome available.
In addition, the mitomycin C and imipenem MICs were determined by the microdilution broth method, showing that K2534 is an imipenem-resistant strain, yielding an MICIMP of 8 mg/liter (according to the EUCAST guidelines, 2021). By using ResFinder, we found that this isolate harbors a gene encoding OXA-245 in its genome. However, for K3325, the imipenem MIC was 0.5 mg/liter; this isolate does not carry the carbapenemase gene but exhibits a clear persister phenotype in response to imipenem treatment, as observed by the time-kill curve (Fig. S1). Mitomycin C MICs of >25 mg/liter and 6.25 mg/liter were obtained for strains K2534 and K3325, respectively. However, in subsequent assays, the concentrations of mitomycin C used were 6 mg/liter for K2534 and 3 mg/liter for K3325 (one-fourth and one-half the MICs, respectively), as the antibacterial effect of this anticancer drug has been observed at concentrations of around 10 mg/liter (24) and we wanted to test doses close to the therapeutic doses (37).
Characterization of lytic bacteriophage vB_KpnM-VAC13.
The host range of vB_KpnM-VAC13 was determined by performing a spot test in 16 clinical isolates of K. pneumoniae (Table 1), showing that the lytic spectrum of this phage covered 75% of the strains. However, before studying the therapeutic potential, the phage was phenotypically characterized by using the K2534 isolate, which exhibited the highest efficiency of plating (EOP) (38). Infection, adsorption, and one-step growth curves were determined using K2534, and transmission electron microscopy (TEM) revealed the morphology of vB_KpnM-VAC13, showing a long contractile tail characteristic of the Myoviridae family (see Fig. S2).
Antimicrobial activity of the repurposed mitomycin C alone and in combination with the lytic phage vB_KpnM-VAC13.
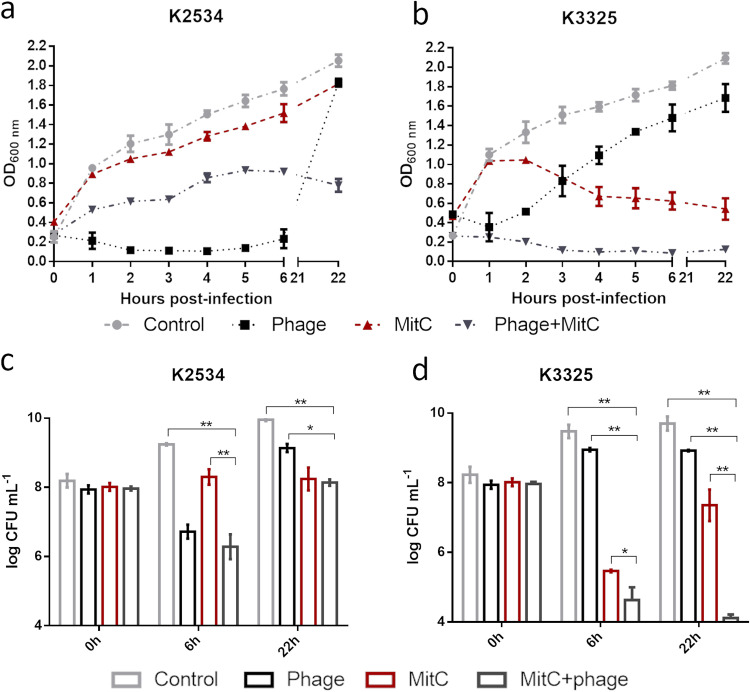
Assays of the antimicrobial activity of mitomycin C by time-kill curves with the K2534 strain revealed a small decrease in the optical density. In addition, no differences between this treatment and the control were observed when CFU were measured at 6 and 22 h. Nevertheless, this strain was very sensitive to the lytic phage vB_KpnM-VAC13, and when it was treated with the combination of the phage (at a multiplicity of infection [MOI] of 10) and mitomycin (one-fourth MIC), a reduction in growth was observed relative to that with mitomycin C alone and for the untreated control (Fig. 1a). Although there were no differences in the number of CFUs between the treatment with phage alone and the combination at 6 h, a significant reduction in the CFU was observed relative to those obtained with the mitomycin C (only) treatment (Fig. 1c). Moreover, at 22 h posttreatment, the emergence of phage-resistant mutants was reduced when the combination treatment was applied (Fig. 1a and c). Regarding K3325, the mitomycin treatment (one-half MIC) was effective at 6 and 22 h, as a significant reduction in growth was observed (Fig. 1b). Moreover, a significant reduction in CFU of almost 4-log was obtained at 6 h. However, the emergence of mitomycin resistance was observed at 22 h, although with significant differences relative to the control (Fig. 1d). Strain K3325 was poorly sensitive to phage vB_KpnM-VAC13, but when it was treated with the combination of mitomycin C and the phage, the reduction in the CFU caused by the combination was greater than the sum of the effects of both agents separately, suggesting a synergistic effect. Furthermore, the emergence of mitomycin C resistance was prevented (Fig. 1b and d).
FIG 1.
Optical density of K2534 (a) and K3325 (b) and number of CFU per milliliter of K2534 (c) and K3325 (d) in the presence of the lytic phage alone at an MOI of 10 (black), mitomycin C alone (dark red), and the combination of both (dark gray). Light gray symbols/lines represent the control in the absence of any drug. *, P < 0.05; **, P < 0.01. Absence of asterisk indicates no statistically significant difference.
Effect of the combination of imipenem and lytic phage vB_KpnM-VAC13.
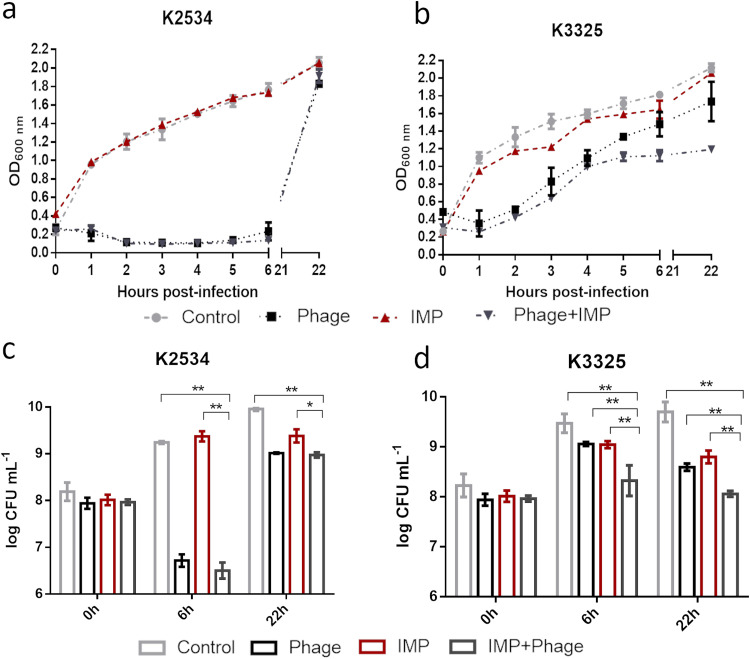
The antimicrobial effect of imipenem was assessed alone and in combination with phage vB_KpnM-VAC13 with both clinical isolates. For K2534, as an imipenem-resistant strain, no differences were observed between treatment with imipenem alone and the control. However, a significant reduction in growth and in the number of CFU was observed when K2534 was treated with the phage alone or in combination with the antibiotic, although no differences were obtained between these treatments at any of the time points (Fig. 2a and c). Regarding the persister isolate, K3325, a reduction in growth was observed with the combined treatment relative to that with imipenem or phage alone, especially from 6 h onwards, as verified by the number of CFU (Fig. 2b and d).
FIG 2.
Optical density of K2534 (a) and K3325 (b) and number of CFU per milliliter of K2534 (c) and K3325 (d) in the presence of the lytic phage alone at an MOI of 10 (black), imipenem alone (dark red), and the combination of both (dark gray). Light gray represents the control in the absence of any drug. *, P < 0.05; **, P < 0.01. Absence of asterisk indicates no statistically significant difference.
Effect of the combinations mitomycin C–vB_KpnM-VAC13 and imipenem–vB_KpnM-VAC13 on the emergence of resistant mutants.
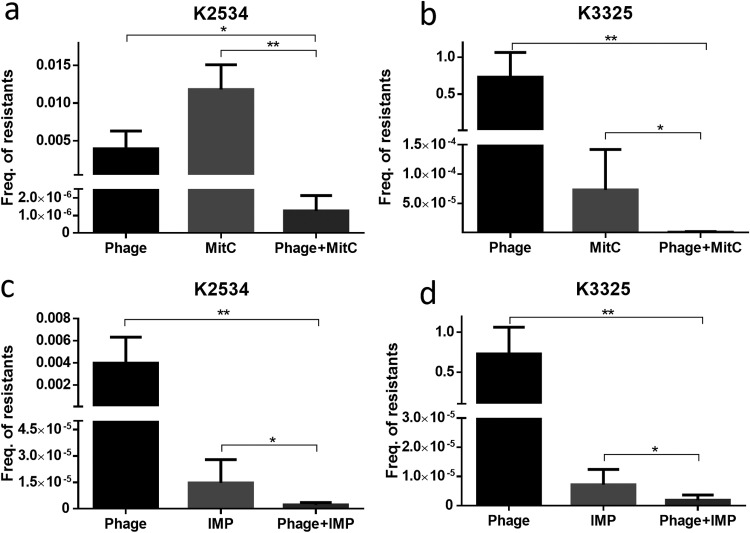
Regrowth of the K2534 strain in the presence of the imipenem-phage combination was observed at 22 h (Fig. 2). We therefore decided to determine the frequency of the emergence of resistant mutants in both strains for each individual treatment and for the combined treatments: vB_KpnM-VAC13 phage, mitomycin C and imipenem alone, and the combined drug-phage treatments. Statistically significant differences were obtained between the frequency of resistant mutants in the presence of the individual drugs and in the presence of the combination treatments for all strains tested (Fig. 3). The results showed a reduction in the emergence of mutants resistant to the phage and to the drugs alone. The frequency of resistance to the vB_KpnM-VAC13 phage was 3.96·10−3/24 h for K2534 and 7.29·10−1/24 h for K3325, while the frequencies of resistance to mitomycin C were 1.18·10−2 and 7.28·10−5/24 h, respectively, and the frequencies of resistance to imipenem were 1.48·10−5 and 7.15·10−6/24 h, respectively. Regarding mitomycin C-phage and imipenem-phage combinations, differences in the frequencies of resistant mutants were statistically significant for both isolates compared with those for the corresponding monotherapies (Fig. 3).
FIG 3.
Frequency of mutants of K2534 (a and c) and K3325 (b and d) resistant to the lytic phage vB_KpnM-VAC13 or the tested drugs (mitomycin C [MitC] and imipenem [IMP]), alone or in combination with the phage over 24 h. *, P < 0.05; **, P < 0.01. Absence of asterisk indicates the result was not statistically significant.
In vivo synergism between vB_KpnM-VAC13 and mitomycin C-imipenem in Galleria mellonella larvae.
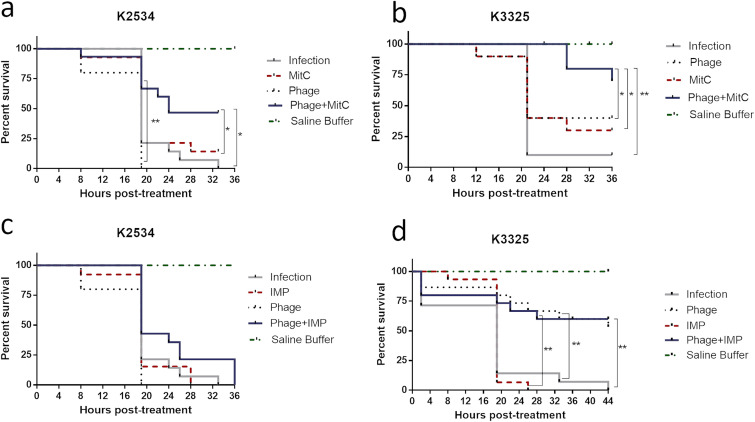
A survival assay was carried out with G. mellonella to assess the efficacy of the repurposed mitomycin C, the treatment with imipenem, and the combination of each with the lytic phage vB_KpnM-VAC13 in the complex in vivo system.
Regarding isolate K2534, an increase in the survival rate of larvae was promoted by the combination of mitomycin C and phage relative to the rates by each individual treatment. Moreover, this combination significantly protected the larvae relative to the untreated control (Fig. 4a). For isolate K3325, the in vitro results were confirmed in the in vivo G. mellonella model, in which the combination of mitomycin C and phage vB_KpnM-VAC13 promoted a significant reduction in the mortality rate of larvae relative to that of the infection group and the groups treated with mitomycin C only or with the phage only (Fig. 4b). Interestingly, the imipenem-lytic phage combination also led to a significant reduction in the mortality rate of larvae relative to that of the infection control (Fig. 4d), unlike the corresponding monotherapies, which did not significantly protect the larvae. Finally, some groups of uninfected larvae were included to test the strength of the antibacterial agents at the highest concentrations tested: 1010 PFU/ml of vB_KpnM-VAC13, 8 mg/liter of imipenem, and 6 mg/liter of mitomycin C. No differences in the mortality rates were observed relative to that for the group treated with saline (data not shown).
FIG 4.
Percentage of survival of G. mellonella larvae infected with K2534 or K3325. For each group, 15 larvae were included. *, P < 0.05; **, P < 0.01. Absence of asterisk indicates no statistically significant difference.
DISCUSSION
Antibiotic-resistant and persister bacteria constitute important threats in clinics, as these phenotypes place the success of antibiotic-based therapy at risk. We are currently facing an extraordinary crisis in which no new antibiotics are being discovered, and the most common bacteria that appear in clinics are already resistant to the majority of conventional antibiotics available. In this context, the repurposing of FDA-approved drugs with indications other than antimicrobial use is needed (39–41).
In this study, we combined the repurposed drug mitomycin C and also the conventional carbapenem imipenem with the lytic phage vB_KpnM-VAC13, with the aim of discovering synergistic relationships that might increase the chances of clearing infections caused by either imipenem-resistant isolates with β-lactamase OXA-245 (OXA 48 group) or persister isolates of K. pneumoniae in clinical settings. Interestingly, genetic analyses indicated that the gene encoding the β-lactamase OXA-48 group has the ability to spread among the enterobacterial species at a much higher rate than the KPC and NDM genes (42), which are currently present in 80% of K. pneumoniae isolates worldwide (43–45). The fact that combination therapies successfully decrease the viability of this strain opens the door to coadministration strategies to combat these problematic isolates.
To our knowledge, this is the first time that a repurposed drug such as mitomycin C has been combined with a lytic phage with the aim of enhancing the antibacterial effects of both compounds. Two antimicrobial agents used together are considered to display a synergistic relation when the effect of the combination is greater than the sum of the effects caused by both agents separately (46). Interestingly, for both K2534 (imipenem resistant) and K3325 (persister) strains, we observed synergistic effects of mitomycin C and the lytic phage, both in vitro and in vivo. Langeveld et al. reviewed the mechanisms leading to pharmacological synergy and reported three possible scenarios: a multitarget effect, in which combined compounds have different bacterial targets; pharmacokinetic or physicochemical effects, such as the improvement of solubility or bioavailability; and an effect whereby a specific bacterial resistance mechanism is targeted (47). Mitomycin C targets the guanosine bases adjacent in two DNA strands, leading to interstrand cross-linking reactions, whereas the phage vB_KpnM-VAC13 specifically lyses host cells after recognizing its receptor. Thus, the observed synergism between mitomycin C and the lytic phage appears to be due to a multitarget effect.
On the other hand, the combination of imipenem and the lytic phage in the K3325 (persister) strain reduced the optical density, and the number of viable CFU observed with the combination was equivalent to the sum of that for both effects separately (48). This was consistent with the result of the in vivo assay, in which the lytic phage both alone and in combination with imipenem significantly protected the larvae, probably due to the enhanced effect of the phage on the in vivo system; however, imipenem alone did not increase the survival rate. Although, in the in vitro assays, no drastic effect of vB_KpnM-VAC13 was achieved for K3325, we observed that combinations of the phage with mitomycin C and with imipenem prevented the emergence of resistant mutants to the corresponding monotherapies, thus explaining the success of these combinations in increasing the percentage of survival of the larvae in this isolate. However, we did not observe any synergy for the imipenem-resistant strain K2534 between imipenem and the lytic phage either in vitro or in vivo, because (i) the isolate harbors an OXA-245 β-lactamase able to hydrolyze imipenem and resist the actions of the antibiotic, and (ii) the difference in the antibacterial activity in vitro and the effect of the phage alone and in combination with imipenem was not sufficient. Nonetheless, we observed a statistically significant difference in the in vitro viability of the bacterial cells after exposure to imipenem alone and to the combination, both at 6 and 22 h. These results were consistent with those of the resistance assay, in which both combinations significantly decreased the frequency of resistance relative to monotherapies (42).
Overall, these results indicate that the repurposed drug mitomycin C can act as a potent enhancer of the lytic activity of the bacteriophage, in both imipenem-resistant and persister isolates of K. pneumoniae and also in an in vivo model. Although synergistic relationships between lytic phages and carbapenems have been observed in Acinetobacter baumannii, in which both agents compete for the same receptor (usually an efflux pump), this was not the case for the K2534 strain, in which the imipenem resistance was due to the enzymatic action of β-lactamase OXA-245 (31, 49). Therefore, for this isolate, the antibiotic was not resensitized by combining imipenem and the lytic phage, although the combined therapy drastically reduced the emergence of resistant mutants in vitro. It has very recently been reported that some bacterial strategies that confer phage resistance (mutation of phagic receptors or in the capsule biosynthesis clusters, production of extracellular polysaccharides, switch to a mucoid phenotype, etc.) involved the overproduction of β-lactamases, thus increasing the multidrug resistance of pathogens (50).
In conclusion, the findings of this study show, for the first time, that repurposing the anticancer drug mitomycin C may be useful for treating infections caused by imipenem-resistant and persister isolates of K. pneumoniae. Moreover, the combination of imipenem and lytic phage may be of interest for treating infections caused by persister isolates of this pathogen, although it was not effective against imipenem-resistant isolates in which the resistance mechanism was due to β-lactamase. The findings therefore indicate that combination therapies involving lytic phages and repurposed drugs could help to overcome some kinds of infections, suggesting a new way of potentiating the lytic activity of phages and preventing the development of phage-resistant mutants.
MATERIALS AND METHODS
Collection of K. pneumoniae clinical strains.
A total of 16 clinical strains of K. pneumoniae obtained from the Virgin Macarena University Hospital (Seville, Spain) and the National Centers for Microbiology (CNM; Carlos III Health Institute, Spain) were used (Table 1). The sensitivity of the isolates to imipenem was assessed by time-kill curve analysis and disk diffusion assay (TDtest), as previously described (36, 51). Strains K2534 (imipenem resistant with β-lactamase OXA-245) and K3325 (highly imipenem-persistent isolate) were selected for further treatment assays.
Determination of the MICs of mitomycin C and imipenem by broth microdilution assay.
Mitomycin C (from Streptomyces caespitosus) and imipenem were purchased from Sigma Aldrich.
Eleven serial double dilutions of each drug were prepared in Muller-Hinton broth (MHB) in 96-well microtiter plates. The wells were then inoculated with the corresponding K. pneumoniae strains to a final concentration of 5·105 CFU/ml. Inoculated MHB was included as a growth control, and one noninoculated row was included as a sterility control in each plate. All experiments were performed in triplicates, and the MIC was determined as the lowest concentration of the antimicrobial agent in which no visible bacterial growth was observed after incubation for 18 to 24 h at 37°C.
Characterization of lytic phage vB_KpnM-VAC13.
(i) Host range and EOP of vB_KpnM-VAC13. vB_KpnM-VAC13 is a lytic phage of K. pneumoniae isolated from sewage water. Its host range was established by performing the spot test with the clinical strains of K. pneumoniae (Table 1), according to the protocol described by Raya and H'bert (52). The semisolid top agar soft medium (TA-soft medium) was supplemented with 1 mM CaCl2 and then used to make plates by the top agar method. Each strain was tested in triplicates, and SM buffer (100 mM NaCl, 10 mM MgSO4, 20 mM Tris-HCl, pH 7.5) was included as a negative control.
The EOP was calculated as the ratio between the phage titer of the test strain and the titer of the host strain (K. pneumoniae subsp. pneumoniae ATCC 10031) (38). As the highest titer was obtained with K2534, further characterization of the lytic bacteriophage vB_KpnM-VAC13 was performed with this strain.
(ii) Transmission electron microscopy.
One milliliter of the lytic phage vB_KpnM-VAC13, diluted in SM buffer and carrying approximately 1011 PFU/ml, was negatively stained with 1% aqueous uranyl acetate before examination under a transmission electron microscope (TEM JEOL 1011).
(iii) Adsorption curve and one-step growth curve of vB_KpnM-VAC13 phage using K2534.
An overnight culture of clinical isolate K. pneumoniae K2534 was diluted 1:100 in LB broth supplemented with 1 mM CaCl2 and incubated at 37°C at 180 rpm until an early logarithmic phase (optical density at 600 nm [OD600] of 0.3). The lytic phage vB_KpnM-VAC13 was then added to the culture at a multiplicity of infection (MOI) of 0.01 (either for the adsorption curve or the one-step growth curve). For the first curve, which allowed us to determine the time of adsorption of vB_KpnM-VAC13 to the bacterial surface, the culture was maintained statically at room temperature (RT), and 1 ml of culture medium was removed for analysis every minute for 10 min. However, for the one-step growth curve, flasks were incubated at 37°C and 180 rpm, and aliquots were removed every 5 or 10 min. For both types of analysis, duplicate aliquots of culture medium were removed, centrifuged, and serially diluted, with some of these dilutions plated in TA supplemented with CaCl2 by the double-agar method.
Time-kill curves for mitomycin C and imipenem alone and in combination with the lytic phage vB_KpnM-VAC13.
Isolated colonies of K. pneumoniae strains K2534 and K3325 were inoculated in 5 ml of LB broth, incubated overnight at 37°C and 180 rpm, and then diluted 1:100 in LB plus 1 mM CaCl2 and incubated until an OD600 of 0.3. The phage vB_KpnM-VAC13 at an MOI of 10 (109 PFU/ml) was then added to the corresponding flasks, along with mitomycin C at one-fourth or one-half the MIC or imipenem at the respective MIC. OD600 was measured at 1-h intervals for 6 h, and bacterial concentrations (CFU/ml) were determined by counting the colonies present at 0, 6, and 22 h on LB agar plates. All experiments were performed in triplicates, and the data obtained were statistically analyzed with GraphPad v.6. The proportion of survivors was established in the control without antibacterial agent and in the presence of vB_KpnM-VAC13 and/or mitomycin C or imipenem.
Frequency of occurrence of resistance to vB_KpnM-VAC13 phage in K2534 and K3325 strains.
The frequency of resistant mutants was calculated as previously described (53). Briefly, overnight cultures of strains K2534 and K3325 were diluted 1:100 in LB and grown to an OD600 of 0.7. An aliquot of 100 μl of the culture containing 108 CFU/ml was serially diluted, and the corresponding dilutions were mixed with 100 μl of vB_KpnM-VAC13 at 109 PFU/ml and then plated by the top agar method in TA medium containing imipenem (8 mg/liter for K2534 and 0.5 mg/liter for K3325) or mitomycin C (6 mg/liter and 3 mg/liter, respectively). The plates were incubated at 37°C for 24 h, and the CFUs were then counted. The frequency of occurrence of resistant mutants was calculated by dividing the number of resistant bacteria (growing in the presence of the phage alone, mitomycin C or imipenem alone, and the drug-phage combinations) by the total number of bacteria plated in conventional LB agar.
Toxicity and efficacy of therapeutic combinations of mitomycin C and imipenem with the lytic phage vB_KpnM-VAC13 in the Galleria mellonella in vivo model.
The Galleria mellonella model was an adapted version of those developed by Göttig et al. (54) and Nath et al. (55), with the following modifications: healthy G. mellonella larvae acquired from a local pet store (Bichosa, Galicia, Spain) and lacking dark spots were selected according to their weights and, as previously performed by other research groups (56–58), allocated into groups ensuring equal representation of weights. Then, 10 larvae per group were injected in the last left proleg with 10 μl of a suspension containing 109 CFU/ml of K2534 and 107 CFU/ml of K3325, diluted in sterile saline buffer. For preparation of the inoculum, 5 ml of fresh LB broth was incubated overnight, grown at 37°C with shaking at 180 rpm, pelleted by centrifugation (4,000 × g, 15 min), and washed twice with saline buffer. The treatment groups were infected with K2534 or K3325. After 60 min, either 108 or 1010 PFU/ml of the phage (previously purified using an Amicon filter [59]), mitomycin C, or imipenem alone or the drugs in combination with the phage were injected via the last right proleg, at the same concentrations used in the in vitro assays. A control group of infected larvae injected with saline buffer alone (no treatment, control) was included. The treatment and control groups were placed in petri dishes and incubated in darkness at 37°C. Larval mortality was monitored regularly for 3 days, and larvae were considered dead when they did not move in response to touch (60). The survival curves were plotted using GraphPad Prism v.6, and data were analyzed using the Gehan-Breslow-Wilcoxon test calculating the P value.
ACKNOWLEDGMENTS
This study was funded by grants PI16/01163 and PI19/00878 awarded to M. Tomás within the State Plan for R+D+I 2013–2016 (National Plan for Scientific Research, Technological Development and Innovation 2008–2011) and cofinanced by the ISCIII-Deputy General Directorate for Evaluation and Promotion of Research–European Regional Development Fund and Instituto de Salud Carlos III FEDER, Spanish Network for the Research in Infectious Diseases (REIPI; RD16/0016/0001, RD16/0016/0006, and RD16/CIII/0004/0002) and by the Study Group on Mechanisms of Action and Resistance to Antimicrobials, GEMARA (SEIMC; http://www.seimc.org/). M. Tomás was financially supported by the Miguel Servet Research Program (SERGAS and ISCIII). I. Bleriot was financially supported by pFIS program (ISCIII, FI20/00302). O. Pacios and M. López were financially supported by grants IN606A-2020/035 and IN606B-2018/008, respectively (GAIN, Xunta de Galicia), and P. Domingo-Calap was financially supported by the ESCMID Research Grant 20200063.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Pitout JD, Nordmann P, Poirel L. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navon-Venezia S, Kondratyeva K, Carattoli A. 2017. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 41:252–275. 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 3.Reyes J, Aguilar AC, Caicedo A. 2019. Carbapenem-resistant Klebsiella pneumoniae: microbiology key points for clinical practice. Int J Gen Med 12:437–446. 10.2147/IJGM.S214305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 5.Wood TK, Knabel SJ, Kwan BW. 2013. Bacterial persister cell formation and dormancy. Appl Environ Microbiol 79:7116–7121. 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JS, Wood TK. 2017. Tolerant, growing cells from nutrient shifts are not persister cells. mBio 8:e00354-17. 10.1128/mBio.00354-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pacios O, Blasco L, Bleriot I, Fernandez-Garcia L, Ambroa A, López M, Bou G, Cantón R, Garcia-Contreras R, Wood TK, Tomás M. 2020. ppGpp and its role in bacterial persistence: new challenges. Antimicrob Agents Chemother 64:e01283-20. 10.1128/AAC.01283-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-García L, Blasco L, Lopez M, Bou G, García-Contreras R, Wood T, Tomas M. 2016. Toxin-antitoxin systems in clinical pathogens. Toxins (Basel) 8:227. 10.3390/toxins8070227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van den Bergh B, Fauvart M, Michiels J. 2017. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol Rev 41:219–251. 10.1093/femsre/fux001. [DOI] [PubMed] [Google Scholar]
- 10.Song S, Wood TK. 2020. ppGpp ribosome dimerization model for bacterial persister formation and resuscitation. Biochem Biophys Res Commun 523:281–286. 10.1016/j.bbrc.2020.01.102. [DOI] [PubMed] [Google Scholar]
- 11.Trastoy R, Manso T, Fernández-García L, Blasco L, Ambroa A, Pérez del Molino ML, Bou G, García-Contreras R, Wood TK, Tomás M. 2018. Mechanisms of bacterial tolerance and persistence in the gastrointestinal and respiratory environments. Clin Microbiol Rev 31:e00023-18. 10.1128/CMR.00023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pacios O, Blasco L, Bleriot I, Fernandez-Garcia L, González Bardanca M, Ambroa A, López M, Bou G, Tomás M. 2020. Strategies to combat multidrug-resistant and persistent infectious diseases. Antibiotics (Basel) 9:65. 10.3390/antibiotics9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furfaro LL, Payne MS, Chang BJ. 2018. Bacteriophage therapy: clinical trials and regulatory hurdles. Front Cell Infect Microbiol 8:376. 10.3389/fcimb.2018.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long Y, Fu W, Li S, Ren H, Li M, Liu C, Zhang B, Xia Y, Fan Z, Xu C, Liu J, Jin Y, Bai F, Cheng Z, Liu X, Jin S, Wu W. 2019. Identification of novel genes that promote persister formation by repressing transcription and cell division in Pseudomonas aeruginosa. J Antimicrob Chemother 74:2575–2587. 10.1093/jac/dkz214. [DOI] [PubMed] [Google Scholar]
- 15.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 16.Sebastian J, Swaminath S, Nair RR, Jakkala K, Pradhan A, Ajitkumar P. 2017. De novo emergence of genetically resistant mutants of Mycobacterium tuberculosis from the persistence phase cells formed against antituberculosis drugs in vitro. Antimicrob Agents Chemother 61:e01343-16. 10.1128/AAC.01343-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novak R, Henriques B, Charpentier E, Normark S, Tuomanen E. 1999. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature 399:590–593. 10.1038/21202. [DOI] [PubMed] [Google Scholar]
- 18.Windels EM, Michiels JE, Fauvart M, Wenseleers T, Van den Bergh B, Michiels J. 2019. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J 13:1239–1251. 10.1038/s41396-019-0344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilmaerts D, Windels EM, Verstraeten N, Michiels J. 2019. General mechanisms leading to persister formation and awakening. Trends Genet 35:401–411. 10.1016/j.tig.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Soo VWC, Kwan BW, Quezada H, Castillo-Juárez I, Pérez-Eretza B, García-Contreras SJ, Martínez-Vázquez M, Wood TK, García-Contreras R. 2017. Repurposing of anticancer drugs for the treatment of bacterial infections. Curr Top Med Chem 17:1157–1176. 10.2174/1568026616666160930131737. [DOI] [PubMed] [Google Scholar]
- 21.Domalaon R, Ammeter D, Brizuela M, Gorityala BK, Zhanel GG, Schweizer F. 2019. Repurposed antimicrobial combination therapy: tobramycin-ciprofloxacin hybrid augments activity of the anticancer drug mitomycin C against multidrug-resistant Gram-negative bacteria. Front Microbiol 10:1556. 10.3389/fmicb.2019.01556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viertel TM, Ritter K, Horz HP. 2014. Viruses versus bacteria-novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J Antimicrob Chemother 69:2326–2336. 10.1093/jac/dku173. [DOI] [PubMed] [Google Scholar]
- 23.Cruz-Muñiz MY, López-Jacome LE, Hernández-Durán M, Franco-Cendejas R, Licona-Limón P, Ramos-Balderas JL, Martinéz-Vázquez M, Belmont-Díaz JA, Wood TK, García-Contreras R. 2017. Repurposing the anticancer drug mitomycin C for the treatment of persistent Acinetobacter baumannii infections. Int J Antimicrob Agents 49:88–92. 10.1016/j.ijantimicag.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 24.Kwan BW, Chowdhury N, Wood TK. 2015. Combatting bacterial infections by killing persister cells with mitomycin C. Environ Microbiol 17:4406–4414. 10.1111/1462-2920.12873. [DOI] [PubMed] [Google Scholar]
- 25.Chowdhury N, Wood TL, Martínez-Vázquez M, García-Contreras R, Wood TK. 2016. DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol Bioeng 113:1984–1992. 10.1002/bit.25963. [DOI] [PubMed] [Google Scholar]
- 26.Tkhilaishvili T, Winkler T, Müller M, Perka C, Trampuz A. 2019. Bacteriophages as adjuvant to antibiotics for the treatment of periprosthetic joint infection caused by multidrug-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 64:e00924-19. 10.1128/AAC.00924-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D. 2018. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health 2018:60–66. 10.1093/emph/eoy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres-Barceló C, Hochberg ME. 2016. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol 24:249–256. 10.1016/j.tim.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Chan BK, Brown K, Kortright KE, Mao S, Turner PE. 2016. Extending the lifetime of antibiotics: how can phage therapy help? Future Microbiol 11:1105–1107. 10.2217/fmb-2016-0133. [DOI] [PubMed] [Google Scholar]
- 30.Chaturongakul S, Ounjai P. 2014. Phage-host interplay: examples from tailed phages and Gram-negative bacterial pathogens. Front Microbiol 5:442. 10.3389/fmicb.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blasco L, Ambroa A, Lopez M, Fernandez-Garcia L, Bleriot I, Trastoy R, Ramos-Vivas J, Coenye T, Fernandez-Cuenca F, Vila J, Martinez-Martinez L, Rodriguez-Baño J, Pascual A, Cisneros JM, Pachon J, Bou G, Tomas M. 2019. Combined use of the Ab105-2φΔCI lytic mutant phage and different antibiotics in clinical isolates of multi-resistant. Microorganisms 7:556. 10.3390/microorganisms7110556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaudhry WN, Concepcion-Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR. 2017. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One 12:e0168615. 10.1371/journal.pone.0168615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamal F, Dennis JJ. 2015. Burkholderia cepacia complex phage-antibiotic synergy (PAS): antibiotics stimulate lytic phage activity. Appl Environ Microbiol 81:1132–1138. 10.1128/AEM.02850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tagliaferri TL, Jansen M, Horz HP. 2019. Fighting pathogenic bacteria on two fronts: phages and antibiotics as combined strategy. Front Cell Infect Microbiol 9:22. 10.3389/fcimb.2019.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uchiyama J, Shigehisa R, Nasukawa T, Mizukami K, Takemura-Uchiyama I, Ujihara T, Murakami H, Imanishi I, Nishifuji K, Sakaguchi M, Matsuzaki S. 2018. Piperacillin and ceftazidime produce the strongest synergistic phage-antibiotic effect in Pseudomonas aeruginosa. Arch Virol 163:1941–1948. 10.1007/s00705-018-3811-0. [DOI] [PubMed] [Google Scholar]
- 36.Gefen O, Chekol B, Strahilevitz J, Balaban NQ. 2017. TDtest: easy detection of bacterial tolerance and persistence in clinical isolates by a modified disk-diffusion assay. Sci Rep 7:41284. 10.1038/srep41284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradner WT. 2001. Mitomycin C: a clinical update. Cancer Treat Rev 27:35–50. 10.1053/ctrv.2000.0202. [DOI] [PubMed] [Google Scholar]
- 38.Kutter E. 2009. Phage host range and efficiency of plating. Methods Mol Biol 501:141–149. 10.1007/978-1-60327-164-6_14. [DOI] [PubMed] [Google Scholar]
- 39.Peyclit L, Baron SA, Rolain JM. 2019. Drug repurposing to fight colistin and carbapenem-resistant bacteria. Front Cell Infect Microbiol 9:193. 10.3389/fcimb.2019.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Copp JN, Pletzer D, Brown AS, Van der Heijden J, Miton CM, Edgar RJ, Rich MH, Little RF, Williams EM, Hancock REW, Tokuriki N, Ackerley DF. 2020. Mechanistic understanding enables the rational design of salicylanilide combination therapies for Gram-negative infections. mBio 11:e02068-20. 10.1128/mBio.02068-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farha MA, Brown ED. 2019. Drug repurposing for antimicrobial discovery. Nat Microbiol 4:565–577. 10.1038/s41564-019-0357-1. [DOI] [PubMed] [Google Scholar]
- 42.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 43.Machuca J, López-Cerero L, Fernández-Cuenca F, Mora-Navas L, Mediavilla-Gradolph C, López-Rodríguez I, Pascual Á. 2019. OXA-48-like-producing Klebsiella pneumoniae in southern Spain in 2014–2015. Antimicrob Agents Chemother 63:e01396-18. 10.1128/AAC.01396-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitout JDD, Peirano G, Kock MM, Strydom KA, Matsumura Y. 2019. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev 33:e00102-19. 10.1128/CMR.00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mairi A, Pantel A, Sotto A, Lavigne JP, Touati A. 2018. OXA-48-like carbapenemases producing Enterobacteriaceae in different niches. Eur J Clin Microbiol Infect Dis 37:587–604. 10.1007/s10096-017-3112-7. [DOI] [PubMed] [Google Scholar]
- 46.Gómara M, Ramón-García S. 2019. The FICI paradigm: correcting flaws in antimicrobial in vitro synergy screens at their inception. Biochem Pharmacol 163:299–307. 10.1016/j.bcp.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Langeveld WT, Veldhuizen EJ, Burt SA. 2014. Synergy between essential oil components and antibiotics: a review. Crit Rev Microbiol 40:76–94. 10.3109/1040841X.2013.763219. [DOI] [PubMed] [Google Scholar]
- 48.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52:1. 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 49.Oechslin F. 2018. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 10:351. 10.3390/v10070351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Majkowska-Skrobek G, Markwitz P, Sosnowska E, Lood C, Lavigne R, Drulis-Kawa Z. 22March2021. The evolutionary trade-offs in phage-resistant Klebsiella pneumoniae entail cross-phage sensitization and loss of multidrug resistance. Environ Microbiol 10.1111/1462-2920.15476. [DOI] [PubMed] [Google Scholar]
- 51.Markowitz SM, Wells VD, Williams DS, Stuart CG, Coudron PE, Wong ES. 1991. Antimicrobial susceptibility and molecular epidemiology of beta-lactamase-producing, aminoglycoside-resistant isolates of Enterococcus faecalis. Antimicrob Agents Chemother 35:1075–1080. 10.1128/AAC.35.6.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raya RR, H'bert EM. 2009. Isolation of phage via induction of lysogens. Methods Mol Biol 501:23–32. 10.1007/978-1-60327-164-6_3. [DOI] [PubMed] [Google Scholar]
- 53.Lopes A, Pereira C, Almeida A. 2018. Sequential combined effect of phages and antibiotics on the inactivation of Escherichia coli. Microorganisms 6:125. 10.3390/microorganisms6040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Göttig S, Frank D, Mungo E, Nolte A, Hogardt M, Besier S, Wichelhaus TA. 2019. Emergence of ceftazidime/avibactam resistance in KPC-3-producing Klebsiella pneumoniae in vivo. J Antimicrob Chemother 74:3211–3216. 10.1093/jac/dkz330. [DOI] [PubMed] [Google Scholar]
- 55.Nath S, Moussavi F, Abraham D, Landman D, Quale J. 2018. In vitro and in vivo activity of single and dual antimicrobial agents against KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother 73:431–436. 10.1093/jac/dkx419. [DOI] [PubMed] [Google Scholar]
- 56.Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, Penadés JR, Ingmer H. 2016. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat Commun 7:13333. 10.1038/ncomms13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Astvad KMT, Meletiadis J, Whalley S, Arendrup MC. 2017. Fluconazole pharmacokinetics in Galleria mellonella larvae and performance evaluation of a bioassay compared to liquid chromatography-tandem mass spectrometry for hemolymph specimens. Antimicrob Agents Chemother 61:e00895-17. 10.1128/AAC.00895-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andrea A, Krogfelt KA, Jenssen H. 2019. Methods and challenges of using the greater wax moth (Galleria mellonella) as a model organism in antimicrobial compound discovery. Microorganisms 7:85. 10.3390/microorganisms7030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonilla N, Rojas MI, Netto Flores Cruz G, Hung SH, Rohwer F, Barr JJ. 2016. Phage on tap-a quick and efficient protocol for the preparation of bacteriophage laboratory stocks. PeerJ 4:e2261. 10.7717/peerj.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 53:2605–2609. 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download AAC.00900-21-s0001.pdf, PDF file, 7.4 MB (7.4MB, pdf)