ABSTRACT
Malaria parasites have three genomes: a nuclear genome, a mitochondrial genome, and an apicoplast genome. Since the apicoplast is a plastid organelle of prokaryotic origin and has no counterpart in the human host, it can be a source of novel targets for antimalarials. Plasmodium falciparum DNA gyrase (PfGyr) A and B subunits both have apicoplast-targeting signals. First, to test the predicted localization of this enzyme in the apicoplast and the breadth of its function at the subcellular level, nuclear-encoded PfGyrA was disrupted using CRISPR/Cas9 gene editing. Isopentenyl pyrophosphate (IPP) is known to rescue parasites from apicoplast inhibitors. Indeed, successful growth and characterization of PfΔGyrA was possible in the presence of IPP. PfGyrA disruption was accompanied by loss of plastid acyl-carrier protein (ACP) immunofluorescence and the plastid genome. Second, ciprofloxacin, an antibacterial gyrase inhibitor, has been used for malaria prophylaxis, but there is a need for a more detailed description of the mode of action of ciprofloxacin in malaria parasites. As predicted, PfΔGyrA clone supplemented with IPP was less sensitive to ciprofloxacin but not to the nuclear topoisomerase inhibitor etoposide. At high concentrations, however, ciprofloxacin continued to inhibit IPP-rescued PfΔGyrA, possibly suggesting that ciprofloxacin may have an additional nonapicoplast target in P. falciparum. Overall, we confirm that PfGyrA is an apicoplast enzyme in the malaria parasite, essential for blood-stage parasites, and a possible target of ciprofloxacin but perhaps not the only target.
KEYWORDS: apicoplast, CRISPR/Cas9, ciprofloxacin, DNA gyrase, Plasmodium falciparum
INTRODUCTION
Malaria is a global health problem, and continual emergence of resistance to existing antimalarial drugs underscores the importance of finding new drug targets and new antimalarials (1–6). There are three genomes in the malaria parasites: the nuclear genome, the mitochondrial genome, and the apicoplast genome (7–9). During cell division, DNA topoisomerases play a major role in DNA replication, DNA transcription, and DNA repair throughout the life cycle of parasites (10–13). There are two types of DNA topoisomerases (type I and type II) that are categorized based on their ability to break single-stranded or double-stranded DNA, respectively (10). Malaria parasites have two type I (topoisomerase I and III) and three type II (topoisomerase II and VI and DNA gyrase) enzymes (7). Biochemical and cellular functions of the majority of malarial topoisomerases remain uncertain, partly due to the difficulty in expressing active malaria enzymes. Previously, we successfully expressed full-length topoisomerase II and tested its sensitivity to some known enzyme and parasite proliferation inhibitors (14). Meanwhile, Chalapareddy et al. suggested that topoisomerase VI may be important for maintenance of the mitochondrial genome (15). Based on bioinformatics, DNA gyrase is known to carry an apicoplast-targeting signal peptide (16–18). Although heterologous expression of Plasmodium falciparum DNA gyrase A (PfGyrA) subunit was not successful, Dar et al. showed successful expression of PfGyrB subunit and its localization to the apicoplast (19). The functions of truncated forms of PfGyrA have been studied extensively in vitro in several independent studies (20–23). However, the essentiality, biological function, and sensitivity to inhibitors of PfGyrA have not been formally established for blood-stage malaria parasites.
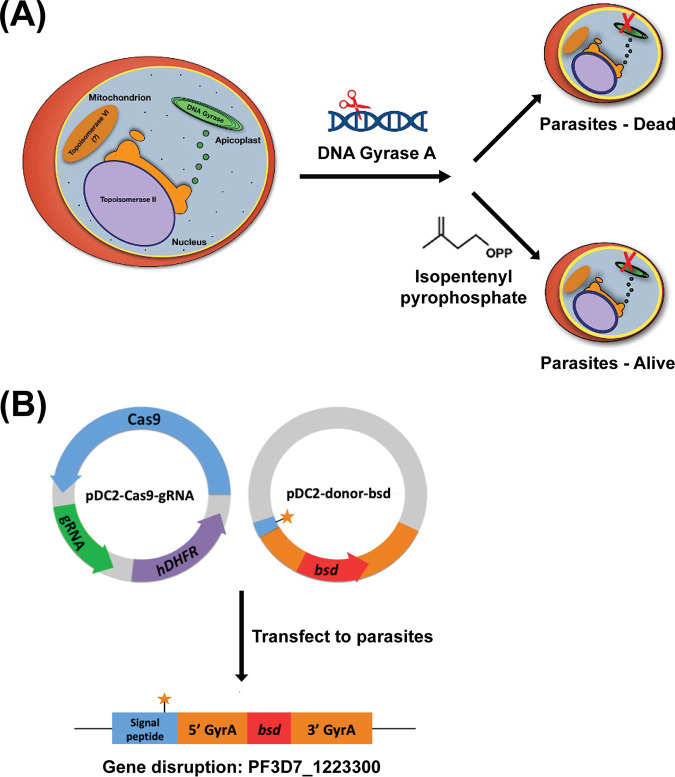
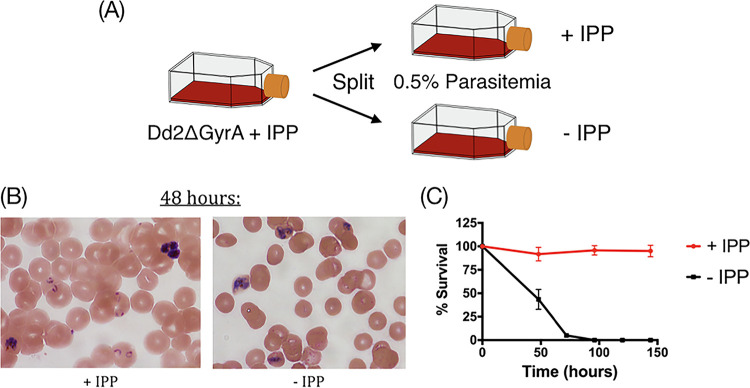
DNA gyrase is the only type II topoisomerase predicted to localize to the apicoplast and has no counterpart in the human host (17, 24, 25). Since Yeh and DeRisi showed that parasites lacking apicoplast functions can be chemically rescued with isopentenyl pyrophosphate (IPP) supplementation, we hypothesized that deletion of PfGyrA may be tolerated with the addition of IPP (Fig. 1A) (26). If successful, this would formally validate the importance of PfGyrA in malaria parasites and help us understand the mode of action of potential drugs that may target the apicoplast PfGyrA. CRISPR/Cas9 gene editing has been adapted for genome editing in Plasmodium falciparum (27–30). Here, we apply this gene-editing tool to demonstrate that PfGyrA is targeted to the apicoplast, that it is important for the function of the apicoplast, and that it is the primary, but perhaps not the sole, target of ciprofloxacin at high concentrations.
FIG 1.
Experimental model and design. (A) PfGyrA was hypothesized to be a type II topoisomerase located in the parasite apicoplast. Disruption of this enzyme was expected to kill parasites if its function is important to the maintenance of the apicoplast. However, genetically modified parasite without an apicoplast can be chemically rescued through supplementation with isopentenyl pyrophosphate. (B) Gene editing with CRISPR/Cas9. The Cas9 enzyme was encoded on a plasmid that also expresses gRNA and an hdfhr selectable marker. The donor plasmid carries the homology arms of the PfgyrA gene with a point mutation inserted on the 3′ end of the apicoplast-signal peptide. Transfected parasites would have the PfgyrA gene disrupted with the insertion of bsd.
RESULTS
CRISPR/Cas9-mediated gene knockout of PfGyrA.
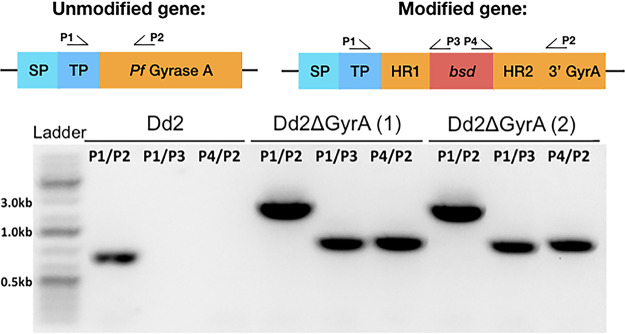
A plasmid, labeled pDC2-gRNA-Cas9, coexpressed Cas9 nuclease and human dihydrofolate reductase (encoded by hdhfr) and was selectable in malaria parasites with the antifolate WR99210 (Fig. 1A and B). The donor plasmid, labeled pDC2-donor-bsd, provided a repair template to leverage the homology-directed repair pathway of CRISPR/Cas9. The donor plasmid had homology regions from the targeted PfgyrA gene sequence, flanking the blasticidin-S-deaminase (BSD) selectable marker (Fig. 1B). The gRNA-Cas9 and donor plasmids were electroporated simultaneously into a parental P. falciparum Dd2 clone using the direct electroporation method (31, 32). At 48 h posttransfection, parasites were supplemented with IPP and maintained with WR99210 and blasticidin for 6 days, followed by culturing in IPP-supplemented medium. Parasites were visible microscopically on days 21 to 24 posttransfection in most flasks when IPP was supplemented in each culture. Genomic DNA was extracted from each positive transfected clone for downstream confirmation assays. PCR-based PfgyrA gene amplifications were performed on each transfected clone using different primer sets to confirm the disruption of PfgyrA gene (Fig. 2). Amplification with primers P1/P2 yielded an 898-bp fragment seen in the Dd2 clone, while amplification with the same primer set yielded a larger 2,490-bp fragment from the modified clones (Dd2ΔGyrA). Since the Dd2 clone did not include the bsd gene, no fragment was observed when the DNA was amplified with primers P1/P3 or P4/P2. On the other hand, amplification with primers P1/P3 or P4/P2 yielded a 1,110-bp or 1,102-bp fragment in the modified clones and hence confirmed the insertion of the selectable marker. Two out of 13 transfected clones carried the desired modification based on the PCR confirmation. DNA sequencing of the amplified PCR products further confirmed the disruption of the PfgyrA gene.
FIG 2.
Proof of PfgyrA disruption. PCR with different primers helped validate disruption of the PfgyrA gene. Genetically modified parasites yielded a larger PCR fragment when amplified with primers P1/P2. Amplification with primers P1/P3 and P4/P2 further confirmed the insertion of the bsd gene in the modified parasites. SP, signal peptide; TP, transit peptide; HR, homologous region.
Loss of apicoplast in Dd2ΔGyrA.
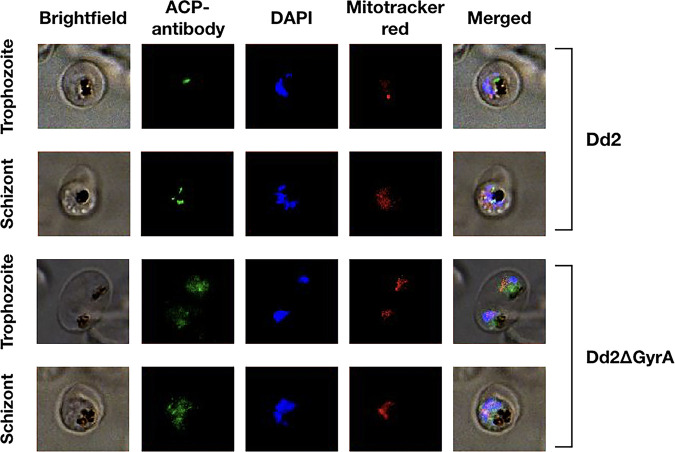
In order to determine whether the apicoplast remained intact after the disruption of the PfgyrA gene and growth in the presence of IPP, genetically pure populations of Dd2ΔGyrA were generated through limiting dilution. Apicoplast structure was visualized through staining of an apicoplast-targeted acyl-carrier protein (ACP) with an ACP antibody (33). As expected, the ACP localized to a discrete structure (the apicoplast) in the Dd2 clone, while ACP was diffused in the cytoplasm of the Dd2ΔGyrA clone (Fig. 3). As a control, the nucleus was stained with DAPI (4′,6-diamidino-2-phenylindole) and the mitochondrion was stained with Mitotracker CMXRos. Under these conditions, there was no difference in the staining pattern between Dd2 and Dd2ΔGyrA clones, suggesting that the nucleus and mitochondrion remained intact in these parasites.
FIG 3.
Loss of apicoplast immunofluorescence in Dd2ΔGyrA clone. The parasite apicoplast was stained with the ACP antibody, which was visualized with the anti-rabbit secondary antibody conjugated to Alexa Fluor 488. Nucleus was stained with DAPI, while mitochondrion was stained with the Mitotracker CMXRos. The apicoplast remained intact in the Dd2 clone, as ACPs were localized to a specific location. However, in the Dd2ΔGyrA clone, ACPs were dispersed throughout the cytoplasm of parasites, indicating loss of the apicoplast structure. Nuclear and mitochondrial genome remained intact in both Dd2 and Dd2ΔGyrA clones.
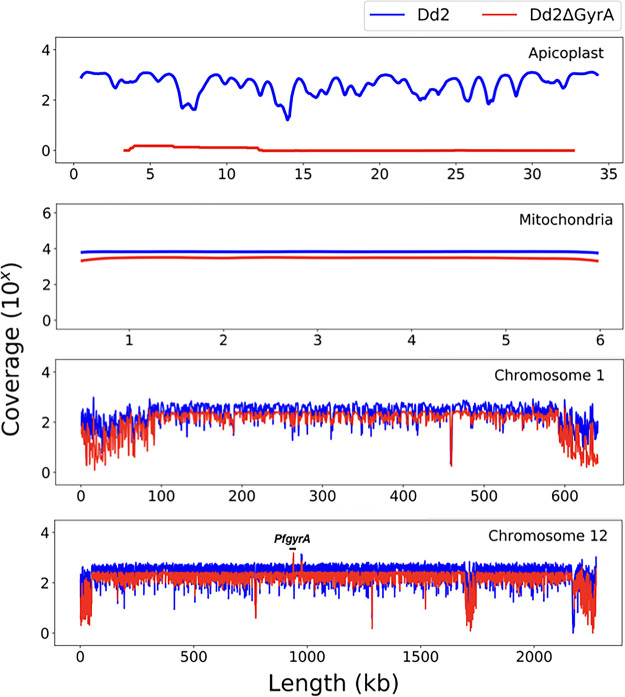
The loss of the apicoplast in Dd2ΔGyrA was further confirmed through whole-genome sequencing. The coverage of sequences from the whole genome of Dd2ΔGyrA was compared to that of a Dd2 clone. As shown in Fig. 4, Dd2ΔGyrA had lost its apicoplast DNA compared to the parental Dd2 clone. In contrast, as represented by chromosome 1 reads, both mitochondrial and nuclear genomes remained intact in both Dd2 and in Dd2ΔGyrA clones.
FIG 4.
Loss of the apicoplast genome in Dd2ΔGyrA clone. The genome sequence coverage plot (in red) confirmed loss of the apicoplast genome in Dd2ΔGyrA while still retaining the mitochondrial and nuclear genomes. The unmodified Dd2 clone has its apicoplast, mitochondrial, and nuclear genomes intact (in blue).
Dependence of Dd2ΔGyrA on IPP for growth.
In order to directly test whether Dd2ΔGyrA clone was dependent on IPP for growth, each flask was split into two. One flask was maintained in the presence of IPP, while the other flask had IPP removed. Parasites were observed microscopically (Fig. 5A). About 48 h after IPP removal, the parasite cytoplasm was less condensed and the morphology of parasites was distorted compared to that of parasites cultured in IPP-supplemented medium (Fig. 5B). While parasites proliferated normally in cultures supplemented with IPP, reduced parasite growth was observed in cultures at 48 h post IPP removal, and parasites were considered nonviable at 72 h post IPP removal (Fig. 5C).
FIG 5.
Dependence of Dd2ΔGyrA on IPP for growth. (A) Dd2ΔGyrA clone was split into two independent flasks: one received continuous supplementation of 200 μM IPP, while the other flask received no IPP. (B and C) Parasites with continuous IPP supplementation were able to proliferate and appeared healthy. Parasites in flask without IPP had no healthy rings and were not viable after 48 h.
IPP rescues parasites from delayed-death phenotype.
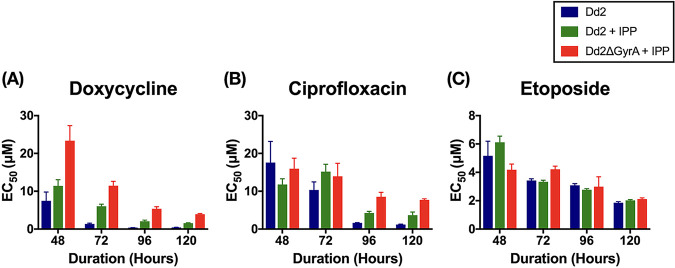
Previous studies have shown that parasites display a delayed-death phenotype when treated with antibacterials targeting apicoplast housekeeping processes (9, 34, 35). In those cases, parasites proliferated normally for one cycle but the daughter cells would lose their apicoplasts and could not complete another cycle. In order to determine the pharmacological susceptibilities of Dd2ΔGyrA grown with IPP, we applied doxycycline, ciprofloxacin, or etoposide for 48 h, 72 h, 96 h, or 120 h in drug-sensitivity assays. Doxycycline is an antibacterial known to inhibit bacterial protein synthesis, and ciprofloxacin is an antibacterial known to inhibit bacterial DNA gyrase. In both cases, inhibitory action on apicoplast was expected to cause a delayed-death phenotype in the parasite. On the other hand, etoposide is known to inhibit nuclear topoisomerase II and delayed-death phenotype was not expected. Indeed, Dd2 exhibited a delayed-death phenotype when treated with doxycycline or ciprofloxacin. When IPP was supplemented in the assay (Dd2+IPP), chemical rescue of apicoplast-specific inhibition was observed, consistent with previous findings involving other apicoplast targets (26, 36, 37). Dd2ΔGyrA supplemented with IPP displayed an inhibitor-response profile similar to that of a Dd2 clone (Fig. 6). This suggests that our CRISPR-generated Dd2ΔGyrA parasite line displays characteristics similar to those of the apicoplast-minus parasites generated through inhibition by small molecules. Inhibition by the control compound, etoposide, did not result in delayed death and did not allow chemical rescue with IPP. This indicated that our parasite line was not altered in regard to nuclear topoisomerase activity.
FIG 6.
Drug sensitivity of different antibacterial compounds. Delayed-death phenotype was observed when Dd2 clone was treated with doxycycline or with ciprofloxacin (panels A and B, blue bars). The delayed inhibition at 96 h or 120 h could be rescued with IPP in both Dd2 and Dd2ΔGyrA clones (panels A and B, green and red bars). Inhibition by etoposide (control) showed no delayed death and no chemical rescue with IPP (C). Error bars represent the standard deviation of the results.
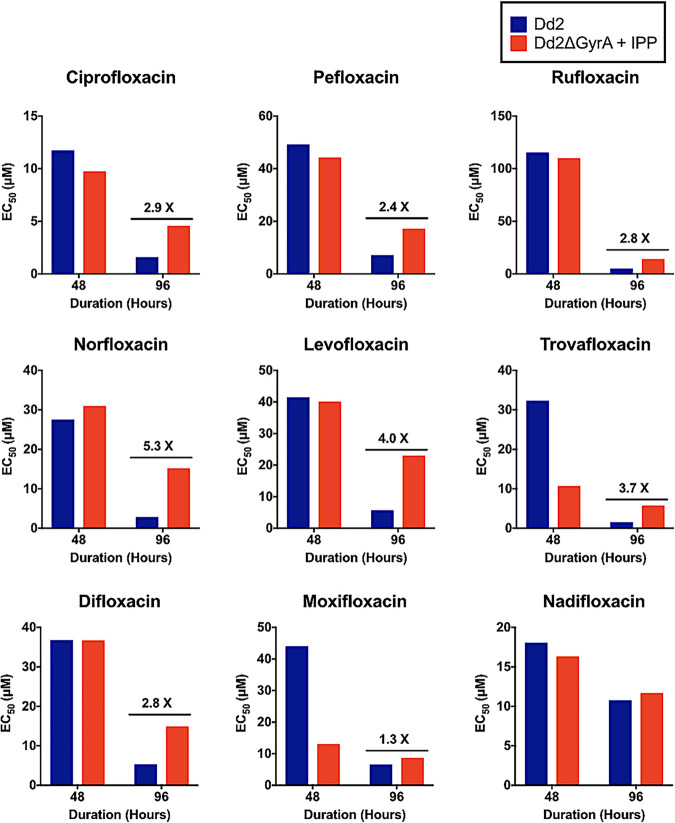
In follow-up studies to ciprofloxacin, various other commercially available fluoroquinolone compounds were tested to determine their effect on the Dd2ΔGyrA clone (Fig. S1). The majority of the fluoroquinolone compounds displayed inhibition and IPP-rescue profile similar to those of ciprofloxacin. Two compounds were different: moxifloxacin and nadifloxacin did not show delayed death or IPP-rescue effect when tested on the Dd2ΔGyrA clone (Fig. 7). Other compounds such as flumequine, cinoxacin, and nalidixic acid displayed 50% effective concentration (EC50) values of >125 μM, and their full inhibition profiles were not determined (results not shown).
FIG 7.
Drug sensitivity of fluoroquinolone derivatives. Dd2ΔGyrA clone exhibits similar inhibition and IPP-rescue profiles when tested with ciprofloxacin, pefloxacin, rufloxacin, norfloxacin, levofloxacin, trovafloxacin, and difloxacin. No significant delayed-death or IPP-rescue effects were seen in the Dd2ΔGyrA when tested with moxifloxacin or nadifloxacin. The bars were derived from EC50 curves at many different inhibitor concentrations.
DISCUSSION
In the present study, CRISPR/Cas9-mediated gene knockout of PfGyrA confirms its importance to the apicoplast in blood-stage forms of malaria parasites. Immunofluorescence and whole-genome sequencing showed that disruption of the PfgyrA nuclear gene led to the loss of the apicoplast. This supports previous suspicions based on bioinformatics that PfGyrA would be coded for in the nucleus and synthesized in the cytoplasm and that the protein product would be transported to the apicoplast, where it would have a major role in the maintenance of the integrity of the apicoplast (17).
The IPP chemical rescue strategy proved to be pivotal and necessary to allow for the potentially lethal disruption of the apicoplast-targeted DNA gyrase. Yeh and DeRisi first showed that IPP can be used to chemically rescue parasites that had been treated with apicoplast-targeting drugs (26). Similarly, Uddin et al. also demonstrated that a wide range of apicoplast-targeting drugs could be validated with the IPP chemical rescue method (37). For instance, for parasites that exhibit delayed-death phenotype when treated with doxycycline or ciprofloxacin, the inhibition could be chemically rescued with IPP supplementation (37). For the present work, it appears that the PfGyrA is only needed in the apicoplast and the IPP-rescue approach was sufficient to rescue the PfGyrA knockout parasites. Presumably, abrogation of an essential topoisomerase in the apicoplast (ΔPfGyrA) would lead to inefficient DNA replication and inefficient transcription in the apicoplast, followed by loss of translation and loss of multiple gene products encoded by the apicoplast. Ultimately, this would lead to a parasite without a functional apicoplast (experimentally, as long as IPP was provided exogenously to keep the apicoplast-less parasites alive).
Although the apicoplast can be lost through chemical inhibition by small molecules, there are several advantages of generating an apicoplast-minus parasite using a genomic approach directed at one specific gene with just one predicted function. First, the clean genetic knockout of PfGyrA offered an unambiguous comparison to characterize inhibitors like ciprofloxacin. Ciprofloxacin appears to have a single target at low concentrations (PfGyrA, which could be rescued with IPP). At higher ciprofloxacin concentrations, nonapicoplast targets in P. falciparum begin to be important as judged by inability to rescue parasites with IPP. Second, drug-sensitivity assays of various fluoroquinolone compounds also revealed that there may be more than one mode of action of this class of compounds on malaria parasites, one with an apicoplast target (DNA gyrase) sensitive to low micromolar drug concentrations and the other with a nonapicoplast target that required a higher concentration of ciprofloxacin (Fig. 7). On the surface, while the ciprofloxacin analogs appear similar to the parent compound, they showed variations in potency and mode of parasite inhibition. In particular, nadifloxacin and moxifloxacin (both fluoroquinolone derivatives) did not exhibit a delayed-death phenotype and were not rescued by IPP. The 50% inhibitory concentration (IC50) values for this class of compounds against 3D7 parasites range from 4.5 to 142.9 μg/ml. These results suggest that, unlike many ciprofloxacin analogs that appear to primarily be PfGyrA inhibitors and have delayed death, nadifloxacin and moxifloxacin are fast-acting antimalarials acting through a different mode of action. Overall, gene editing of PfGyrA allows for such formal dissection and comparisons of antimalarial activity directed at this high-value target. In the future, as we reach for novel potent inhibitors that specifically target PfGyrA from malaria parasites, the present validation model would allow us to stay on track.
MATERIALS AND METHODS
Plasmids construction.
Plasmids pDC2-gRNA-Cas9 and pDC2-donor-bsd were provided by Marcus Lee (Wellcome Sanger Institute, UK). Plasmid pDC2-gRNA-Cas9 was modified to include a 20-nucleotide gRNA sequence (ATAGGTAAATATCATCCACA) targeting PfgyrA. gRNA-oligonucleotide 1 (5′-ATTGATAGGTAAATATCATCCACA) and gRNA-oligonucleotide 2 (5′-AAACTGTGGATGATATTTACCTAT) were phosphorylated and annealed using T4 polynucleotide kinase in a thermocycler by incubating at 37°C for 30 min, increasing the heat to 94°C for 5 min, and then ramping down to 25°C at 5°C/min. Annealed gRNA-oligonucleotides were ligated with BbsI-digested plasmid. The sequencing primer (gRNA primer) was used for sequencing to confirm insertion of gRNA into the plasmid. To facilitate homology recombination, the plasmid pDC2-donor-bsd was modified to carry homology arms of PfgyrA (PlasmoDB ID PF3D7_1223300) flanking the selectable marker blasticidin-S deaminase (BSD). 5′ Homology arm of 307 bp starts from nucleotide +415 of the GyrA coding sequence, while 3′ homology arm of 306 bp starts from nucleotide +740. Nonsense mutations were inserted at nucleotides 437 to 438 (CT→GA) to introduce a stop codon at the beginning of the PfgyrA gene to stop further translation of the protein. 5′-GyrA-F and 5′-GyrA-R primers were used to amplify the 5′ homology arm from clone Dd2, GyrA-BSD-F and GyrA-BSD-R primers were used to amplify BSD selectable marker from the pDC2-donor-bsd plasmid, and 3′-GyrA-F and 3′-GyrA-R primers were used to amplify the 3′ homology arm from the Dd2 clone. These three PCR fragments were then assembled into a single piece that carried a BamHI site on the 5′ end and a SacI site on the 3′ end. The final assembled PCR construct was then digested with BamHI and SacI and ligated into the doubly digested pDC2-donor-bsd plasmid.
Parasite cultures and transfection.
Parasites were cultivated using established methods with some changes (38, 39). Cultures of Plasmodium falciparum clone Dd2 were grown in vitro at 37°C in solutions of 2% hematocrit (serotype A positive human erythrocyte) in RPMI 1640 medium (Invitrogen) containing l-glutamine and 25 mM HEPES and supplemented with 0.5% Albumax I and 0.1 g/liter hypoxanthine (ThermoFisher). Cultures were maintained in sterile, sealed flasks and flushed with a blood gas mixture (5% CO2, 5% O2, and 90% N2). Transfections were done with the direct electroporation method on sorbitol-synchronized ring-stage parasites as described previously (31, 32). Briefly, Dd2 clone was electroporated with 100 μg of each plasmid using Bio-Rad Gene Pulser II set at 0.31 kV and 960 μF. Selection pressure (2.5 nM WR99210 and 2 μg/μl blasticidin) and 200 μM IPP (Isoprenoids, LC and home supplies) were applied 48 h posttransfection. Media supplemented with IPP and selection pressure were renewed every day for 6 days. On day 8 posttransfection, selection pressure was removed and medium was supplemented with IPP and changed every other day. Parasite proliferation was monitored by Giemsa-stained thin-smeared blood samples taken at each medium change three times a week.
Synthesis of isopentenyl pyrophosphate.
Isopentenyl pyrophosphate was synthesized from 3-methyl-3-butene-1-ol according to the procedure of Davisson and coworkers depicted in Scheme 1 in the supplemental material (40). The synthesized IPP was first passed through cation exchange resin (ammonium form) for complete ion exchange. Subsequent purification on cellulose flash column chromatography resulted in a pure final product. Detailed synthesis procedures are outlined in the supplemental material.
Analysis of Dd2ΔGyrA clone.
Parasite pellets from Dd2ΔGyrA were lysed using 0.15% saponin solution, and genomic DNA was extracted using Qiagen DNA minikit, according to the manufacturer’s instructions (41). Targeted gene disruption and BSD-cassette integration were verified by PCR using BIO-X-ACT short mix (Bioline LLC) with different primer sets as described in Results (see Fig. 2). PCR products were then treated with EXO-SAP-IT PCR cleanup reagent (ThermoFisher) and sent for sequencing using the same primers.
Whole-genome sequencing analysis.
Genomic DNA of 400 ng from synchronized ring-stage Dd2ΔGyrA clone were sent to MedGenome, California for genomic libraries preparation and whole-genome sequencing. Paired-end reads were trimmed from bases with a quality score below 28 (Phred) using TrimGalore and then mapped to the 3D7 reference genome (Pf3D7v9) using BWA. Downstream processes were done following GATK (Genome Analysis Toolkit) best practices pipeline, where reads were sorted and duplicated reads were marked using Picard tools (42, 43). After Indel realignment and base recalibration steps using GATK software, final reads were compiled using SAMTools to generate variant calling files. Coverage of each chromosome including mitochondria and apicoplast was generated using SAMtools from the final pileup file, from which read depth at each nucleotide position was recorded. Results (see Fig. 4) were generated into coverage plots using Matplotlib.
Drug-sensitivity assay.
Drug-sensitivity assays were performed in 96-well plates containing serial dilution of test compounds in triplicate. Medium was supplemented with 200 μM IPP as indicated. To determine the EC50 of different compounds, plates of 0.5% parasitemia were incubated for 48 h or 72 h. To determine if parasites clones show delayed-death phenotype, cultures were initiated at 0.2% parasitemia and incubated for 96 h or 120 h, and 75% of the medium was exchanged every 48 h. For all assays, parasitized cells were stained using SYBR green I (Invitrogen) and counted by flow cytometry (BD Accuri C6). Parasite proliferation in each well was expressed as a percentage of the solvent control. EC50 was determined using GraphPad PRISM software.
Immunofluorescence microscopy.
Dd2 and Dd2ΔGyrA were incubated in 100 nM MitoTracker Red CMXRos (Invitrogen) stain for 30 min at 37°C. Cells were then prepared for microscopy as illustrated previously (44). Briefly, cells were washed and fixed in 4% paraformaldehyde (Electron Microscopy Sciences) with 0.0075% glutaraldehyde (Electron Microscopy Sciences) for 30 min at room temperature. Cells were then permeabilized with 0.1% Triton X-100 in PBS for 10 min and blocked with 3% bovine serum albumin/phosphate-buffered saline (BSA/PBS) for 2 h with end-over-end rotation. Cells were then incubated with 1:500 diluted ACP antibody for 1 h and then incubated with 1:500 diluted anti-rabbit secondary antibody conjugated with Alexa Fluor 488 for 30 min. Nuclear DNA was stained with 1 μg/ml DAPI for 10 min at room temperature. Cells were mounted onto slides with Hard Mount VectaShield (Vector Laboratories) and sealed. Fluorescence images were obtained on a Nikon Eclipse Ti-E equipped with a camera using a 100×/1.4 oil immersion objective. Images were analyzed using Nikon NIS-Elements software.
ACKNOWLEDGMENTS
We thank Marcus Lee (Wellcome Sanger Institute) for kindly providing us with the pDC2-gRNA-Cas9 and pDC2-donor-bsd plasmid constructs used to design our plasmids. We also thank Geoffrey McFadden (University of Melbourne) for providing us with the ACP antibody. Part of this work was included in the first author’s 2019 PhD dissertation at the University of Washington. This work was supported by a U.S. NIH grant from NIAID (R01 093380 and U19 AI089688) to P.K.R.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Burgess RW, Young MD. 1959. The development of pyrimethamine resistance by Plasmodium falciparum. Bull World Health Organ 20:37–46. [PMC free article] [PubMed] [Google Scholar]
- 2.Payne D. 1987. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol Today 3:241–246. 10.1016/0169-4758(87)90147-5. [DOI] [PubMed] [Google Scholar]
- 3.Hyde JE. 1990. The dihydrofolate reductase-thymidylate synthetase gene in the drug resistance of malaria parasites. Pharmacol Ther 48:45–59. 10.1016/0163-7258(90)90017-V. [DOI] [PubMed] [Google Scholar]
- 4.White NJ. 1992. Antimalarial drug resistance: the pace quickens. J Antimicrob Chemother 30:571–585. 10.1093/jac/30.5.571. [DOI] [PubMed] [Google Scholar]
- 5.Sibley CH, Hyde JE, Sims PF, Plowe CV, Kublin JG, Mberu EK, Cowman AF, Winstanley PA, Watkins WM, Nzila AM. 2001. Pyrimethamine–sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol 17:582–588. 10.1016/S1471-4922(01)02085-2. [DOI] [PubMed] [Google Scholar]
- 6.Taylor SM, Juliano JJ. 2014. Artemisinin combination therapies and malaria parasite drug resistance: the game is afoot. J Infect Dis 210:335–337. 10.1093/infdis/jiu142. [DOI] [PubMed] [Google Scholar]
- 7.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan M-S, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DMA, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511. 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mather MW, Vaidya AB. 2008. Mitochondria in malaria and related parasites: ancient, diverse and streamlined. J Bioenerg Biomembr 40:425–433. 10.1007/s10863-008-9176-4. [DOI] [PubMed] [Google Scholar]
- 9.Fichera ME, Roos DS. 1997. A plastid organelle as a drug target in apicomplexan parasites. Nature 390:407–409. 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 10.Wang JC. 1996. DNA topoisomerases. Annu Rev Biochem 65:635–692. 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 11.Mitscher LA. 2005. Bacterial topoisomerase inhibitors: quinolone and pyridone antibacterial agents. Chem Rev 105:559–592. 10.1021/cr030101q. [DOI] [PubMed] [Google Scholar]
- 12.Pommier Y, Leo E, Zhang H, Marchand C. 2010. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 17:421–433. 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Estrada C, Prada CF, Fernandez-Rubio C, Rojo-Vazquez F, Balana-Fouce R. 2010. DNA topoisomerases in apicomplexan parasites: promising targets for drug discovery. Proc Biol Sci 277:1777–1787. 10.1098/rspb.2009.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mudeppa DG, Kumar S, Kokkonda S, White J, Rathod PK. 2015. Topoisomerase II from human malaria parasites: EXPRESSION, PURIFICATION, AND SELECTIVE INHIBITION. J Biol Chem 290:20313–20324. 10.1074/jbc.M115.639039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalapareddy S, Chakrabarty S, Bhattacharyya MK, Bhattacharyya S. 2014. Radicicol-mediated inhibition of topoisomerase VIB-VIA activity of the. Mol Biol Physiol 1:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuegge J, Ralph S, Schmuker M, McFadden GI, Schneider G. 2001. Deciphering apicoplast targeting signals – feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene 280:19–26. 10.1016/S0378-1119(01)00776-4. [DOI] [PubMed] [Google Scholar]
- 17.Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Roos DS, Cowman AF, McFadden GI. 2003. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science 299:705–708. 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- 18.Weissig V, Vetro-Widenhouse TS, Rowe TC. 1997. Topoisomerase II inhibitors induce cleavage of nuclear and 35-kb plastid DNAs in the malarial parasite Plasmodium falciparum. DNA Cell Biol 16:1483–1492. 10.1089/dna.1997.16.1483. [DOI] [PubMed] [Google Scholar]
- 19.Dar MA, Sharma A, Mondal N, Dhar SK. 2007. Molecular cloning of apicoplast-targeted Plasmodium falciparum DNA gyrase genes: unique intrinsic ATPase activity and ATP-independent dimerization of PfGyrB subunit. Eukaryot Cell 6:398–412. 10.1128/EC.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghu Ram EVS, Kumar A, Biswas S, Kumar A, Chaubey S, Siddiqi MI, Habib S. 2007. Nuclear gyrB encodes a functional subunit of the Plasmodium falciparum gyrase that is involved in apicoplast DNA replication. Mol Biochem Parasitol 154:30–39. 10.1016/j.molbiopara.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Nagano S, Lin T-Y, Edula J, Heddle J. 2014. Unique features of apicoplast DNA gyrases from Toxoplasma gondii and Plasmodium falciparum. BMC Bioinformatics 15:416. 10.1186/s12859-014-0416-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagano S, Seki E, Lin T-Y, Shirouzu M, Yokoyama S, Heddle JG. 2015. Investigating the roles of the C-terminal domain of Plasmodium falciparum GyrA. PLoS One 10:e0142313. 10.1371/journal.pone.0142313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Girdwood SC, Nenortas E, Shapiro TA. 2015. Targeting the gyrase of Plasmodium falciparum with topoisomerase poisons. Biochem Pharmacol 95:227–237. 10.1016/j.bcp.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman CD, McFadden GI. 2013. Targeting apicoplasts in malaria parasites. Expert Opin Ther Targets 17:167–177. 10.1517/14728222.2013.739158. [DOI] [PubMed] [Google Scholar]
- 25.Kalanon M, McFadden GI. 2010. Malaria, Plasmodium falciparum; and its apicoplast. Biochem Soc Trans 38:775–782. 10.1042/BST0380775. [DOI] [PubMed] [Google Scholar]
- 26.Yeh E, DeRisi JL. 2011. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol 9:e1001138. 10.1371/journal.pbio.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio J-J. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 28.Lee MC, Fidock DA. 2014. CRISPR-mediated genome editing of Plasmodium falciparum malaria parasites. Genome Med 6:63. 10.1186/s13073-014-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner JC, Platt RJ, Goldfless SJ, Zhang F, Niles JC. 2014. Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nat Methods 11:915–918. 10.1038/nmeth.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng CL, Siciliano G, Lee MCS, de Almeida MJ, Corey VC, Bopp SE, Bertuccini L, Wittlin S, Kasdin RG, Le Bihan A, Clozel M, Winzeler EA, Alano P, Fidock DA. 2016. CRISPR-Cas9-modified pfmdr1 protects Plasmodium falciparum asexual blood stages and gametocytes against a class of piperazine-containing compounds but potentiates artemisinin-based combination therapy partner drugs. Mol Microbiol 101:381–393. 10.1111/mmi.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Sifri CD, Lei HH, Su XZ, Wellems TE. 1995. Transfection of Plasmodium falciparum within human red blood cells. Proc Natl Acad Sci U S A 92:973–977. 10.1073/pnas.92.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fidock DA, Nomura T, Wellems TE. 1998. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol Pharmacol 54:1140–1147. 10.1124/mol.54.6.1140. [DOI] [PubMed] [Google Scholar]
- 33.Mullin KA, Lim L, Ralph SA, Spurck TP, Handman E, McFadden GI. 2006. Membrane transporters in the relict plastid of malaria parasites. Proc Natl Acad Sci U S A 103:9572–9577. 10.1073/pnas.0602293103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahl EL, Rosenthal PJ. 2007. Multiple antibiotics exert delayed effects against the Plasmodium falciparum apicoplast. Antimicrob Agents Chemother 51:3485–3490. 10.1128/AAC.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodman CD, Su V, McFadden GI. 2007. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol 152. 10.1016/j.molbiopara.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Bowman JD, Merino EF, Brooks CF, Striepen B, Carlier PR, Cassera MB. 2014. Antiapicoplast and gametocytocidal screening to identify the mechanisms of action of compounds within the malaria box. Antimicrob Agents Chemother 58:811–819. 10.1128/AAC.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uddin T, McFadden GI, Goodman CD. 2018. Validation of putative apicoplast-targeting drugs using a chemical supplementation assay in cultured human malaria parasites. Antimicrob Agents Chemother 62:e01161-17. 10.1128/AAC.01161-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675. 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 39.Divo AA, Geary TG, Davis NL, Jensen JB. 1985. Nutritional requirements of Plasmodium falciparum in culture. I. Exogenously supplied dialyzable components necessary for continuous growth. J Protozool 32:59–64. 10.1111/j.1550-7408.1985.tb03013.x. [DOI] [PubMed] [Google Scholar]
- 40.Davisson VJ, Woodside AB, Neal TR, Stremler KE, Muehlbacher M, Poulter CD. 1986. Phosphorylation of isoprenoid alcohols. J Org Chem 51:4768–4779. 10.1021/jo00375a005. [DOI] [Google Scholar]
- 41.Bangham AD, Horne RW, Glauert AM, Dingle JT, Lucy JA. 1962. Action of saponin on biological cell membranes. Nature 196:952–953. 10.1038/196952a0. [DOI] [PubMed] [Google Scholar]
- 42.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van der Auwera GA. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–11.10.33. 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tonkin CJ, van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, Cowman AF, McFadden GI. 2004. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol Biochem Parasitol 137:13–21. 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and Table S1. Download AAC.00586-21-s0001.pdf, PDF file, 0.2 MB (159KB, pdf)