ABSTRACT
Aliarcobacter butzleri is an emergent enteropathogen for which resistance to several classes of antimicrobial agents has been described, although the underlying mechanisms have been poorly addressed. We aimed to evaluate the contribution of the resistance-nodulation-division-type (RND) efflux system, AreABC, to drug resistance in A. butzleri. A. butzleri strains were first tested against several antimicrobials with and without an efflux pump inhibitor. Then, erythromycin-resistant strains were screened for the presence of a premature stop codon in a putative transcriptional regulator of the AreABC system, areR. Lastly, antimicrobial susceptibility and ethidium bromide (EtBr) accumulation were evaluated using an areB knockout strain and a strain overexpressing the AreABC system through areR truncation. The presence of the efflux pump inhibitor resulted in increased susceptibility to most of the antimicrobials tested. A correlation between erythromycin resistance and the presence of premature stop codons in areR was observed. The truncation of areR resulted in increased expression of the AreABC system and decreased susceptibility to various antimicrobials. In contrast, areB inactivation resulted in increased susceptibility and a higher intracellular accumulation of EtBr. In conclusion, the AreABC efflux pump plays a role in the resistance of A. butzleri to multiple drugs and is regulated by a putative transcriptional repressor, areR. Our results support the importance of efflux pumps in this bacterium's resistance to major classes of antibiotics and other antimicrobials.
KEYWORDS: Aliarcobacter butzleri, antimicrobial resistance, efflux pump, resistance nodulation cell division, transcriptional repressor
INTRODUCTION
The genus Arcobacter was included in the Campylobacteraceae family in 1991, but in 2017, it was reclassified as a new family, Arcobacteraceae (1), followed by the proposal of a division of the genus Arcobacter into seven different genera (2). Considering the historical development of the genus Arcobacter, it is comprised of a large and heterogeneous group of bacteria, with 33 recognized species isolated from a wide range of habitats (3–6). Among these, Aliarcobacter butzleri is an emergent pathogen with a global distribution and is described as the fourth most prevalent pathogen found in diarrheal samples in humans among the Campylobacter-like organisms (7–9). A. butzleri has been associated with nondiarrheal gastrointestinal illness as well, causing abdominal pain, nausea, vomiting, or fever, and also has been associated with extraintestinal diseases such as bacteremia (3, 10–13). Besides its pathogenic potential, A. butzleri isolates from the environment, of both animal and human origin, have shown variable resistance to several classes of antibiotics, namely, to fluoroquinolones, macrolides, aminoglycosides, penicillins, and tetracyclines, among others, while being linked to high rates of multidrug resistance (14–18).
The A. butzleri genome is highly diverse, with this bacterium presenting a broad set of genes putatively involved in antibiotic resistance, including several efflux pump genes (19, 20). The analysis of 49 genomes showed the presence of 19 efflux pump systems, 8 belonging to the resistance-nodulation-cell-division (RND) family (20), a common family of efflux pumps known to be associated with antibiotic resistance in Gram-negative bacteria (21). Although A. butzleri resistance has been vastly described, only a few studies addressed the association between genomic background and resistance phenotype, with the underlying mechanisms being very poorly understood. Even so, a recent study by our team pointed to a correlation between resistance to erythromycin and the presence of premature stop codons in a putative transcriptional repressor found close to an RND-type system operon (20). In the present study, we aimed to evaluate the physiological contribution of RND efflux pumps to antimicrobial resistance in A. butzleri, focusing on the RND efflux pump system previously associated with erythromycin resistance, the AreABC system.
RESULTS
Effect of efflux systems in the resistance of A. butzleri to antimicrobials.
First, the effect of the efflux pump inhibitor (EPI), phenylalanine-arginine β-naphthylamide (PAβN), in the resistance to several antibiotics, a biocide, and a bile salt was evaluated using five A. butzleri strains previously shown to harbor several efflux pumps, including the AreABC system (20). The MIC determination in the presence and absence of PAβN showed that efflux pumps play a role in the resistance of A. butzleri to doxycycline, ciprofloxacin, ampicillin, cefotaxime, erythromycin, chloramphenicol, acriflavine, and sodium cholate (Table 1), although the effect was strain dependent. The presence of the EPI resulted in a 2- to 256-fold MIC reduction, the effect being more pronounced for erythromycin and for strains with higher levels of resistance.
TABLE 1.
MICs of antibiotics, biocides, and bile salts in the presence and absence of the efflux pump inhibitor PAβN in Aliarcobacter butzleri isolatesa
| Strain | MIC (µg/ml) of: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TET | DOX | CIP | AMP | CTX | ERY | GEN | CHL | ACR | SC | |
| Ab_2811 | 8 | 4 | 32 | 256 | 32 | 16 | 1 | 64 | 32 | 8,000 |
| +PAβN | 8 | 1 | 16 | 32 | 16 | 0.25 | 1 | 8 | 32 | 1,000 |
| CR641 | 4 | 4 | 0.25 | 16 | 16 | 16 | 2 | 16 | 32 | 4,000 |
| +PAβN | 8 | 1 | 0.125 | 8 | 2 | 0.125 | 2 | 4 | 32 | 1,000 |
| CR1143 | 4 | 1 | 16 | 256 | 16 | 4 | 1 | 8 | 32 | 2,000 |
| +PAβN | 8 | 1 | 16 | 128 | 16 | 0.125 | 1 | 4 | 32 | 2,000 |
| DQ40A1 | 1 | 0.25 | 0.03 | 8 | 4 | 1 | 1 | 4 | 16 | 4,000 |
| +PAβN | 2 | 0.125 | 0.015 | 4 | 1 | 0.25 | 1 | 2 | 8 | 2,000 |
| CR1132 | 2 | 0.25 | 0.03 | 16 | 8 | 2 | 1 | 8 | 16 | 4,000 |
| +PAβN | 4 | 0.25 | 0.03 | 16 | 2 | 0.5 | 1 | 4 | 8 | 1,000 |
+PAβN, MIC determined in the presence of 1/4× MIC, thus, 10 µg/ml for Ab_2811, 20 µg/ml for CR641, 5 µg/ml for CR1143 and CR1132, and 2.5 µg/ml for DQ40A1. Changes of at least twofold are indicated in bold type. Abbreviations: TET, tetracycline; DOX, doxycycline; CIP, ciprofloxacin; AMP, ampicillin; CTX, cefotaxime; ERY, erythromycin; GEN, gentamicin; CHL, chloramphenicol; ACR, acriflavine; SC, sodium cholate.
In silico analysis of the RND-type AreABC efflux system and its putative transcriptional regulator, AreR.
A. butzleri encodes several efflux pumps, namely, from the RND family, among which AreABC was found as a complete operon in 37 of the 49 whole genomes previously analyzed (20). At the genomic level, a contig of 5,700 bp containing three open reading frames (ORFs) was identified, with a putative transcriptional regulator ORF (tetR gene, here designated areR) located immediately upstream of the operon on the opposite strand. The length of areR was 540 bp, coding for a 179-amino-acid (aa) protein (20). The three ORFs composing the efflux pump operon were designated areA (1,155 bp), coding for a 384-aa membrane fusion protein; areB (3,168 bp), coding for a 1,055-aa inner membrane transporter; and areC (1,389 bp), coding for a 462-aa outer membrane lipoprotein, according to BLAST annotation. The three genes were found to be tandemly positioned with an overlap of four and eight nucleotides between areA and areB, and areB and areC, respectively, displaying a structural organization similar to the CmeABC efflux system of Campylobacter jejuni (22).
In addition, focusing on the conserved inner membrane component AreB, the homology with known and well-characterized systems was evaluated. Accordingly, a neighbor-joining tree analysis was constructed with representatives of RND inner membrane components of Escherichia coli (AcrB, AcrD, AcrF, MdtB, and MdtC), Pseudomonas aeruginosa (MexB, MexD, MexF, and MexY), and C. jejuni (CmeB) (Fig. S1 in the supplemental material). The analysis revealed that AreB clustered together with these RND family members, corroborating its role in the efflux system. Furthermore, AreB is predicted to possess 12 transmembrane domains (aa residues 11 to 28, 340 to 359, 366 to 385, 394 to 413, 438 to 461,474 to 497, 538 to 561, 875 to 894, 901 to 920, 931 to 950, 975 to 994, and 1,007 to 1,030), with two large extracellular loops between 1 and 2 and 7 to 8 domains, features from the inner membrane component of RND systems (23). Overall, the in silico analysis indicated that AreB is a putative transporter protein belonging to the RND family. By looking at the areR gene in 17 previously sequenced A. butzleri strains possessing a complete AreABC efflux system, we found that this gene codes for a 179-aa protein, although in 4 isolates, the predicted protein was found to be truncated due to premature stop codons (length range of 116 to 142 aa) or, in one case, due to the presence of an insertional sequence (Fig. S2) (20).
Correlation between areR premature stop codons and erythromycin resistance in A. butzleri.
In addition to the previous whole-genome sequencing of A. butzleri isolates (20), the areR sequences of an additional 50 isolates were analyzed by PCR and sequenced if positive. The complete predicted aa sequences of each isolate and the corresponding erythromycin MICs are presented in Table S1. For 14 erythromycin-susceptible strains, no areR amplification was obtained (Table S1 and Fig. S3), most likely due to the absence of the AreABC efflux system, as previously described (20). Among the 16 erythromycin-resistant strains with MICs ranging from 16 to 32 µg/ml, 13 had a truncated AreR, expanding the previously found AreR length variability (predicted length range, 12 to 163 aa) and explaining 81% of the erythromycin-resistant cases.
Effect of areB and areR impairment in AreABC efflux pump expression.
Two mutants from the A. butzleri DQ40A1 strain were generated by natural transformation. In one case, the inner membrane efflux transporter areB was inactivated by insertional mutation (mutant DQ40A1ΔareB), and in the other, the areR-encoding gene was replaced by its Ab_2811 counterpart (encoding a truncated protein) (mutant DQ40A1_trunc_areR). In the DQ40A1ΔareB mutant, the inactivation of the areB gene was confirmed (Fig. 1A). The real-time quantitative PCR (RT-qPCR) assay (Fig. 1B) showed that inactivation of areB had no effect on areA and areC expression (P > 0.05), while a truncated areR gene resulted in overexpression (19- to 28-fold increase) of all three efflux pump units (DQ40A1_trunc_areR mutant versus wild-type isogenic strain DQ40A1).
FIG 1.
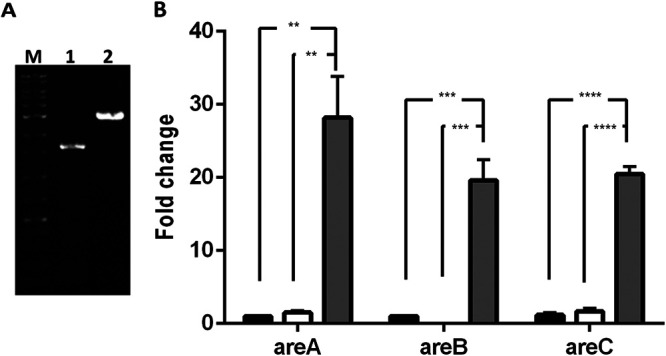
(A) PCR amplification of the areB gene. M, DNA marker, lane 1; DQ40A1ΔareB mutant, lane 2. DQ40A1 wild-type strain. (B) Influence of areB and areR impairment in the expression of efflux pump subunits areA, areB, and areC. Upregulation was normalized to 16S rRNA. The results correspond to at least three independent assays. Data are presented as mean ± SEM. Black column, DQ40A1 wild-type strain; white column, DQ40A1ΔareB mutant; gray column, DQ40A1_trunc_areR mutant; filled circles, DQ40A1; filled squares, DQ40A1ΔareB; filled triangles, DQ40A1_trunc_areR. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Contribution of AreABC to antimicrobial resistance.
To investigate the role of AreABC in antimicrobial resistance, the MICs of several antibiotics, acriflavine, sodium cholate, and ethidium bromide, were determined for A. butzleri DQ40A1 and for the mutants DQ40A1ΔareB and DQ40A1_trunc_areR (Table 2). No differences in the MIC values among wild-type and mutants were observed for ampicillin, cefotaxime, gentamicin, and sodium cholate, while DQ40A1ΔareB presented a 2-fold decrease in the MIC for erythromycin, acriflavine, and ethidium bromide. In contrast, the DQ40A1_trunc_areR mutant was less susceptible to tetracyclines, ciprofloxacin, erythromycin, chloramphenicol, acriflavine, and ethidium bromide, showing at least a 2-fold and up to a 16-fold increase in the MIC values compared to the wild-type strain. In particular, the mutant became resistant to erythromycin, with a 16-fold increase in MIC (1 and 16 µg/ml for DQ40A1 and DQ40A1_trunc_areR, respectively).
TABLE 2.
MICs of antibiotics and sodium cholate tested for Aliarcobacter butzleri DQ40A1 wild-type strain and its corresponding areB and areR mutantsa
| Strain | MIC (µg/ml) of: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TET | DOX | CIP | AMP | CTX | ERY | GEN | CHL | ACR | EtBr | SC | |
| DQ40A1 | 1 | 0.25 | 0.03 | 8 | 4 | 1 | 1 | 4 | 16 | 8 | 4,000 |
| DQ40A1ΔareB | 1 | 0.25 | 0.03 | 8 | 4 | 0.5 | 1 | 4 | 8 | 4 | 4,000 |
| DQ40A1_trunc_areR | 2 | 1 | 0.25 | 8 | 4 | 16 | 1 | 8 | 32 | 64 | 4,000 |
Changes of at least twofold are indicated in bold type. Abbreviations: TET, tetracycline; DOX, doxycycline; CIP, ciprofloxacin; AMP, ampicillin; CTX, cefotaxime; ERY, erythromycin; GEN, gentamicin; CHL, chloramphenicol; ACR, acriflavine; EtBr, ethidium bromide; SC, sodium cholate.
To confirm that the observed differences in MIC values were associated with functional changes in the efflux system, the intracellular accumulation of EtBr was evaluated for the wild-type and mutant strains. A time-dependent increase in fluorescence was observed for all of the strains, with the areB mutant showing a slightly higher increase in fluorescence, indicating a higher EtBr accumulation. A reduction in ethidium bromide accumulation was observed for the DQ40A1_trunc_areR mutant, confirming an overexpression of the AreABC efflux pump.
DISCUSSION
The antimicrobial resistance of A. butzleri has been widely reported, yet the underlying mechanisms are still unknown. In Gram-negative bacteria, one major mechanism of antimicrobial resistance is the active extrusion of compounds by efflux pumps. The sequence analysis of the A. butzleri genome demonstrated the presence of several efflux pump-encoding genes (19, 20, 24) which have never been characterized.
To further understand the role of efflux pumps in antimicrobial resistance, we started by evaluating the MIC of various compounds in the presence and absence of the EPI PAβN. The findings confirmed the role of efflux pumps in the resistance of A. butzleri to a broad range of antimicrobials, substantiated by the MIC decrease for several of the tested drugs in the presence of PAβN. It is noteworthy that the phenotype change from resistant to susceptible was observed for erythromycin and chloramphenicol. These results highlight the relevance of A. butzleri efflux pumps in the extrusion of antimicrobials, at least at low levels of resistance (Table 1). Our previous genomic analysis of A. butzleri isolates (20), together with the results obtained in this study, suggest that active efflux may be involved in resistance to macrolides since no other specific resistance mechanism was identified. Indeed, in A. butzleri, none of the well-described mechanisms in taxonomically close species, such as the occurrence of point mutations in domain V of the 23S rRNA gene or in the ribosomal proteins, or the presence of erythromycin resistance methylase-class genes, were observed (19, 20, 24). Thus, based on our results, we propose that macrolide resistance in A. butzleri is associated with truncated forms of the transcriptional repressor AreR. Since the proteins of the TetR transcriptional regulator family are involved in the repression of the transcription of efflux pump systems (25), this supports the role of AreABC in the macrolide resistance phenotype in A. butzleri. The truncation of transcriptional regulators has been previously associated with efflux pump overexpression. In C. jejuni, the inability of a truncated CmeR to bind to the promoter DNA explained the overexpression of the CmeABC efflux system (26, 27). Our data point to a regulation of the A. butzleri AreABC efflux pump similar to that occurring in Campylobacter. In addition, a low level of resistance of Campylobacter spp. to erythromycin has been associated with a mediation by efflux systems, mainly CmeABC, in the absence of a 23S rRNA target mutation (28–31). Furthermore, when testing over 100 A. butzleri isolates (15, 20; I. Venâncio, M. Oleastro, and S. Ferreira, unpublished data), high levels of resistance were not observed for erythromycin (MIC range,16 to 32 µg/ml), supporting the relevance of the role of the AreABC efflux system in erythromycin resistance.
To further clarify the role of the AreABC efflux pump system in antimicrobial resistance and the regulatory role of areR, mutants were generated by impairment of (i) the areB gene encoding the predicted inner membrane transporter, the component usually associated with substrate specificity; and (ii) the efflux pump transcriptional repressor areR gene. While the inactivation of areB had no impact on the expression of areA and areC genes compared to the native strain, the areR mutant showed a significant increase in the expression of the three efflux pump components, confirming that areR is a transcriptional repressor of areABC.
In addition, AreABC overexpression resulted in an MIC increase of various antimicrobials, in particular, erythromycin, where a 16-fold increase (from 1 to 16 µg/ml) was observed. Regarding ciprofloxacin, despite the 2-fold increase in MIC due to overexpression of AreABC, this effect was marginal compared to the level of resistance resulting from point mutations in the quinolone resistance-determining region of the A. butzleri gyrA gene (32–35). Overall, while the overexpression of AreABC (Fig. 1B) resulted in an MIC increase for several antibiotics, this was not observed when areB was interrupted instead, at least not by the same magnitude (Table 2), suggesting that areABC has a low basal level of expression in the wild-type strain. The EtBr accumulation assays further support the role of AreABC in active efflux in A. butzleri. Indeed, the upregulation of the AreABC efflux pump resulted in a reduction of ethidium bromide accumulation due to a greater rate of active export of this compound (Fig. 2).
FIG 2.
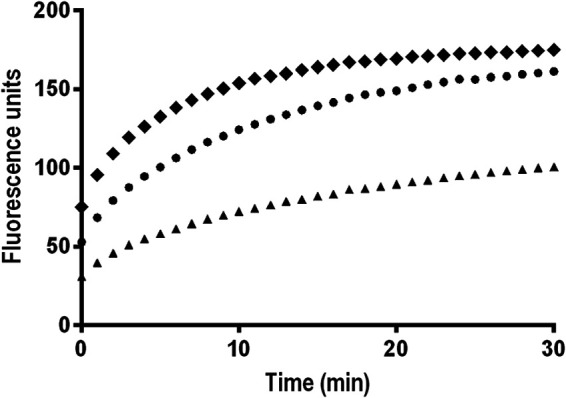
Ethidium bromide accumulation in Aliarcobacter butzleri DQ40A1 wild-type and corresponding areB and areR mutants. Data are presented as the mean of three independent assays. Filled circles, DQ40A1; filled squares, DQ40A1ΔareB; filled triangles, DQ40A1_trunc_areR.
Altogether, our results suggest that erythromycin is an important substrate of AreABC and that tetracyclines, ciprofloxacin, chloramphenicol, acriflavine, and ethidium bromide are also substrates of this transporter. In turn, ampicillin, cefotaxime, gentamicin, and sodium cholate do not seem to be a part of the spectrum of substrates of this efflux system (Table 2). In fact, the use of an EPI further indicated that the resistance mechanisms to gentamicin might not involve efflux pumps. Efflux pumps of the RND superfamily, such as CmeGHI of C. jejuni, have been associated with resistance to gentamicin, while this antibiotic is unlikely to be a substrate for other systems, like the CmeABC and CmeDEF (36, 37).
In summary, our findings show that the AreABC efflux pump presents a broad spectrum of substrates, the system is regulated by the transcriptional regulator AreR, and the occurrence of truncated forms of AreR is associated with macrolide resistance in A. butzleri. Overall, this work clearly identifies efflux pumps as relevant elements for antimicrobial resistance in A. butzleri and demonstrates their role in resistance to major antibiotic classes.
MATERIALS AND METHODS
Aliarcobacter butzleri strains and growth conditions.
A group of 72 nonrelated A. butzleri strains, isolated from clinical cases, environment, and food, were studied. The strains presented different enterobacterial repetitive intergenic consensus-PCR profiles, as previously described (15, 38). The A. butzleri DQ40A1 strain, harboring a complete areABC-encoded efflux pump system, was used for areB mutant construction. The A. butzleri Ab_2811 strain, also harboring a complete system but with a truncated areR gene, was used as a template for amplification of the truncated gene for the construction of an areABC-overexpressing mutant.
Antimicrobial susceptibility testing.
All strains were screened for erythromycin resistance using the agar dilution method; the resistance breakpoint was that of Campylobacter coli corresponding to an MIC of >8 µg/ml, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (39). The MIC determination for eight antibiotics (tetracycline, doxycycline, ciprofloxacin, ampicillin, cefotaxime, erythromycin, gentamicin, and chloramphenicol), as well as the biocide acriflavine and the bile salt sodium cholate, was performed by broth microdilution method (35) in the absence or presence of a subinhibitory concentration (1/4× of the previously determined MIC) of the EPI, PAβN.
Construction of the fragment used for insertional mutation of areB.
An isogenic areB mutant of A. butzleri DQ40A1 strain was constructed by insertional mutagenesis by interrupting an areB ORF with a kanamycin resistance gene. The kanamycin resistance marker was obtained by double digestion of pUC18-K2 plasmid with BamHI and KpnI endonucleases, followed by fragment purification. According to the published sequences of the areABC (EP16) operon (20), primers were designed to amplify the upstream and downstream regions of the areB gene (Table S2 in the supplemental material). Additional sequences were added to the 5′ region of areB_A2 and areB_B1 primers to allow hybridization with upstream and downstream regions of the kanamycin cassette, respectively. The final fragment (upstream region of areB, followed by the kanamycin cassette and the downstream region of areB) was constructed by overlap extension PCR. The purified product was then used for natural transformation of A. butzleri.
areR amplification and sequencing.
Template DNA from each strain was prepared by the boiling method and used in PCR targeting of the areR gene, using oligonucleotide primers tetR_F1, tetR_F2, and tetR_R1 (Table S2). The amplified fragments were both strand DNA sequenced by Sanger sequencing with the BigDye Terminator v1.1 Sequencing standard kit (PE Applied Biosystems Chemistry, Thermo Fisher Scientific, Germany) in the automated sequencer genetic analyzer ABI Prism3130xl (PE Applied Biosystems).
Mutagenesis by natural transformation.
The areB and areR mutants were obtained by natural transformation of the A. butzleri DQ40A1 strain as previously described (40), with the purified products corresponding to the fragment containing the inactivated areB and the fragment containing the areR truncated gene from Ab_2811 strain. The transformants were selected in blood agar, supplemented with 20 µg/ml of kanamycin (areB mutant) or with 4 µg/ml of erythromycin (areR mutant). The transformation with an areB-inactivated fragment was confirmed by PCR through analysis of the fragment size. Natural transformation with the PCR product of the truncated areR from A. butzleri Ab_2811 was confirmed by sequencing and analysis of the translated sequence.
RNA preparation and quantitative PCR assays.
A. butzleri strains were grown on tryptic soy agar (TSA) plates for 24 h at 30°C followed by overnight culture in tryptic soy broth (TSB), which was used to start a fresh culture allowed to grow until midexponential phase. The cultures were harvested by centrifugation and RNA isolated using the TripleXtractor reagent (GRiSP, Portugal), followed by a DNase I treatment. A total of 1 µg of RNA was reverse transcribed with the GRS cDNA Synthesis master mix (GRiSP, Portugal) according to the manufacturer’s instructions.
RT-qPCR was performed, and the constitutively expressed 16S rRNA gene was used for relative quantification. qPCR mixtures contained 5 µl of NZY qPCR green master mix (NZYTech Ltd., Portugal), 0.4-µM specific primers targeting areA, areB, and areC (Table S2 ), and 1 µl of cDNA to a final volume of 10 µl. The relative expression was determined using the comparative threshold cycle (2−ΔΔCT) method, and the ratios obtained after normalization were expressed as fold change.
Accumulation of ethidium bromide.
For the EtBr accumulation assay, midexponential-phase cells were harvested through centrifugation at 10,000 × g for 5 min, washed with phosphate-buffered saline (pH 7.2), and resuspended at an optical density at 620 nm (OD620) of 0.2. Each strain suspension was added in triplicate to a 96-well black plate with clear bottom for fluorescence and incubated at 30°C for 10 min. Ethidium bromide was added to a final concentration of 2 µg/ml, and fluorescence was measured for 30 min, with readings every minute, using excitation and emission wavelengths of 530 nm and 600 nm, respectively.
Bioinformatic analysis.
Homology searches, similarity, identity, and conserved domain analyses were performed through the NCBI internet server. Alignment and phylogenetic analyses of RND-type proteins were performed with MEGA software (version 7.0.25).
ACKNOWLEDGMENTS
Susana Ferreira acknowledges the Universidade da Beira Interior (UBI) and the Foundation for Science and Technology (FCT), both involved in the scientific employment contract according to DL57/2016. Cristiana Mateus acknowledges the fellowship reference BID/ICI-FCS/Santander Universidades-UBI/2019 within the scope of the protocol signed between the UBI and Santander Universidades. Sanger sequencing was performed at the Unidade de Tecnologia e Inovação (Departamento de Genética Humana, Instituto Nacional de Saúde Ricardo Jorge, Lisbon, Portugal).
This work was supported by the Foundation for Science and Technology (FCT) through funds from the state budget and by the European Regional Development Fund (ERDF) under the Portugal 2020 Program through the Regional Operational Program of the Center (Centro2020) and through the project with the reference UIDB/00709/2020.
Footnotes
Supplemental material is available online only.
Contributor Information
Susana Ferreira, Email: susana.ferreira@fcsaude.ubi.pt.
Mónica Oleastro, Email: monica.oleastro@insa.min-saude.pt.
REFERENCES
- 1.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K, Sievert SM, Simon J, Campbell BJ, Hanson TE, Woyke T, Klotz MG, Hugenholtz P. 2017. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol 8:682. doi: 10.3389/fmicb.2017.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pérez-Cataluña A, Salas-Massó N, Diéguez AL, Balboa S, Lema A, Romalde JL, Figueras MJ. 2018. Revisiting the taxonomy of the genus Arcobacter: getting order from the chaos. Front Microbiol 9:2077. doi: 10.3389/fmicb.2018.02077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira S, Oleastro M, Domingues F. 2019. Current insights on Arcobacter butzleri in food chain. Curr Opin Food Sci 26:9–17. doi: 10.1016/j.cofs.2019.02.013. [DOI] [Google Scholar]
- 4.Callbeck CM, Pelzer C, Lavik G, Ferdelman TG, Graf JS, Vekeman B, Schunck H, Littmann S, Fuchs BM, Hach PF, Kalvelage T, Schmitz RA, Kuypers MMM. 2019. Arcobacter peruensis sp.nov., a chemolithoheterotroph isolated from sulfide and organic rich coastal waters off Peru. Appl Environ Microbiol 85:e01344-19. doi: 10.1128/AEM.01344-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso R, Girbau C, Martinez-Malaxetxebarria I, Pérez-Cataluña A, Salas-Massó N, Romalde JL, Figueras MJ, Fernandez-Astorga A. 2020. Aliarcobacter vitoriensis sp. nov., isolated from carrot and urban wastewater. Syst Appl Microbiol 43:126091. doi: 10.1016/j.syapm.2020.126091. [DOI] [PubMed] [Google Scholar]
- 6.Collado L, Figueras MJ. 2011. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin Microbiol Rev 24:174–192. doi: 10.1128/CMR.00034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collado L, Gutiérrez M, González M, Férnandez H. 2013. Assessment of the prevalence and diversity of emergent campylobacteria in human stool samples using a combination of traditional and molecular methods. Diagn Microbiol Infect Dis 75:434–436. doi: 10.1016/j.diagmicrobio.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Ferreira S, Júlio C, Queiroz JA, Domingues F, Oleastro M. 2014. Molecular diagnosis of Arcobacter and Campylobacter in diarrhoeal samples among Portuguese patients. Diagn Microbiol Infect Dis 78:220–225. doi: 10.1016/j.diagmicrobio.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 9.Vandenberg O, Dediste A, Houf K, Ibekwem S, Souayah H, Cadranel S, Douat N, Zissis G, Butzler J-P, Vandamme P. 2004. Arcobacter species in humans. Emerg Infect Dis 10:1863–1867. doi: 10.3201/eid1010.040241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lappi V, Archer JRJR, Cebelinski E, Leano F, Besser JM, Klos RF, Medus C, Smith KE, Fitzgerald C, Davis JP. 2013. An outbreak of foodborne illness among attendees of a wedding reception in Wisconsin likely caused by Arcobacter butzleri. Foodborne Pathog Dis 10:250–255. doi: 10.1089/fpd.2012.1307. [DOI] [PubMed] [Google Scholar]
- 11.Van den Abeele AM, Vogelaers D, Van Hende J, Houf K. 2014. Prevalence of Arcobacter species among humans, Belgium, 2008–2013. Emerg Infect Dis 20:1746–1749. doi: 10.3201/eid2010.140433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandamme P, Pugina P, Benzi G, Van Etterijck R, Vlaes L, Kersters K, Butzler JP, Lior H, Lauwers S. 1992. Outbreak of recurrent abdominal cramps associated with Arcobacter butzleri in an Italian school. J Clin Microbiol 30:2335–2337. doi: 10.1128/jcm.30.9.2335-2337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb AL, Boras VF, Kruczkiewicz P, Selinger LB, Taboada EN, Inglis GD. 2016. Comparative detection and quantification of Arcobacter butzleri in stools from diarrheic and nondiarrheic people in Southwestern Alberta, Canada. J Clin Microbiol 54:1082–1088. doi: 10.1128/JCM.03202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreira S, Queiroz JA, Oleastro M, Domingues F. 2016. Insights in the pathogenesis and resistance of Arcobacter: a review. Crit Rev Microbiol 42:364–383. doi: 10.3109/1040841X.2014.954523. [DOI] [PubMed] [Google Scholar]
- 15.Vicente-Martins S, Oleastro M, Domingues FC, Ferreira S. 2018. Arcobacter spp. at retail food from Portugal: prevalence, genotyping and antibiotics resistance. Food Control 85:107–112. doi: 10.1016/j.foodcont.2017.09.024. [DOI] [Google Scholar]
- 16.Son I, Englen MD, Berrang ME, Fedorka-Cray PJ, Harrison MA. 2007. Antimicrobial resistance of Arcobacter and Campylobacter from broiler carcasses. Int J Antimicrob Agents 29:451–455. doi: 10.1016/j.ijantimicag.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Šilha D, Pejchalová M, Šilhová L. 2017. Susceptibility to 18 drugs and multidrug resistance of Arcobacter isolates from different sources within the Czech Republic. J Glob Antimicrob Resist 9:74–77. doi: 10.1016/j.jgar.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Rathlavath S, Kohli V, Singh AS, Lekshmi M, Tripathi G, Kumar S, Nayak BB. 2017. Virulence genotypes and antimicrobial susceptibility patterns of Arcobacter butzleri isolated from seafood and its environment. Int J Food Microbiol 263:32–37. doi: 10.1016/j.ijfoodmicro.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Fanelli F, Di Pinto A, Mottola A, Mule G, Chieffi D, Baruzzi F, Tantillo G, Fusco V. 2019. Genomic characterization of Arcobacter butzleri isolated from shellfish: novel insight into antibiotic resistance and virulence determinants. Front Microbiol 10:670. doi: 10.3389/fmicb.2019.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isidro J, Ferreira S, Pinto M, Domingues F, Oleastro M, Gomes JP, Borges V. 2020. Virulence and antibiotic resistance plasticity of Arcobacter butzleri: insights on the genomic diversity of an emerging human pathogen. Infect Genet Evol 80:104213. doi: 10.1016/j.meegid.2020.104213. [DOI] [PubMed] [Google Scholar]
- 21.Piddock LJV. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J, Michel LO, Zhang Q. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother 46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du D, van Veen HW, Murakami S, Pos KM, Luisi BF. 2015. Structure, mechanism and cooperation of bacterial multidrug transporters. Curr Opin Struct Biol 33:76–91. doi: 10.1016/j.sbi.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 24.Fanelli F, Chieffi D, Di Pinto A, Mottola A, Baruzzi F, Fusco V. 2020. Phenotype and genomic background of Arcobacter butzleri strains and taxogenomic assessment of the species. Food Microbiol 89:103416. doi: 10.1016/j.fm.2020.103416. [DOI] [PubMed] [Google Scholar]
- 25.Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grinnage-Pulley T, Zhang Q. 2015. Genetic basis and functional consequences of differential expression of the CmeABC efflux pump in Campylobacter jejuni isolates. PLoS One 10:e0131534. doi: 10.1371/journal.pone.0131534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao H, Yuan Z, Shen Z, Han J, Sahin O, Liu P, Zhang Q. 2013. Mutational and transcriptomic changes involved in the development of macrolide resistance in Campylobacter jejuni. Antimicrob Agents Chemother 57:1369–1378. doi: 10.1128/AAC.01927-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mavri A, Možina SS. 2012. Involvement of efflux mechanisms in biocide resistance of Campylobacter jejuni and Campylobacter coli. J Med Microbiol 61:800–808. doi: 10.1099/jmm.0.041467-0. [DOI] [PubMed] [Google Scholar]
- 29.Mamelli L, Prouzet-Mauléon V, Pagès J-M, Mégraud F, Bolla J-M. 2005. Molecular basis of macrolide resistance in Campylobacter: role of efflux pumps and target mutations. J Antimicrob Chemother 56:491–497. doi: 10.1093/jac/dki253. [DOI] [PubMed] [Google Scholar]
- 30.Payot S, Avrain L, Magras C, Praud K, Cloeckaert A, Chaslus-Dancla E. 2004. Relative contribution of target gene mutation and efflux to fluoroquinolone and erythromycin resistance, in French poultry and pig isolates of Campylobacter coli. Int J Antimicrob Agents 23:468–472. doi: 10.1016/j.ijantimicag.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Cagliero C, Mouline C, Payot S, Cloeckaert A. 2005. Involvement of the CmeABC efflux pump in the macrolide resistance of Campylobacter coli. J Antimicrob Chemother 56:948–950. doi: 10.1093/jac/dki292. [DOI] [PubMed] [Google Scholar]
- 32.Abdelbaqi K, Ménard A, Prouzet-Mauleon V, Bringaud F, Lehours P, Mégraud F. 2007. Nucleotide sequence of the gyrA gene of Arcobacter species and characterization of human ciprofloxacin-resistant clinical isolates. FEMS Immunol Med Microbiol 49:337–345. doi: 10.1111/j.1574-695X.2006.00208.x. [DOI] [PubMed] [Google Scholar]
- 33.Van den Abeele AM, Vogelaers D, Vanlaere E, Vanlaere E, Houf K. 2016. Antimicrobial susceptibility testing of Arcobacter butzleri and Arcobacter cryaerophilus strains isolated from Belgian patients. J Antimicrob Chemother 71:1241–1244. doi: 10.1093/jac/dkv483. [DOI] [PubMed] [Google Scholar]
- 34.Morejón IF, González A, Ferrús MA. 2017. Detection, identification, and antimicrobial susceptibility of Arcobacter spp. isolated from shellfish in Spain. Foodborne Pathog Dis 14:238–243. doi: 10.1089/fpd.2016.2202. [DOI] [PubMed] [Google Scholar]
- 35.Ferreira S, Correia DR, Oleastro M, Domingues F. 2018. Arcobacter butzleri ciprofloxacin resistance: point mutations in DNA gyrase A and role on fitness cost. Microb Drug Resist 24:915–922. doi: 10.1089/mdr.2017.0295. [DOI] [PubMed] [Google Scholar]
- 36.Akiba M, Lin J, Barton Y-W, Zhang Q. 2006. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother 57:52–60. doi: 10.1093/jac/dki419. [DOI] [PubMed] [Google Scholar]
- 37.Jeon B, Wang Y, Hao H, Barton Y-W, Zhang Q. 2011. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J Antimicrob Chemoth 66:79–85. doi: 10.1093/jac/dkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira S, Oleastro M, Domingues FC. 2017. Occurrence, genetic diversity and antibiotic resistance of Arcobacter sp. in a dairy plant. J Appl Microbiol 123:1019–1026. doi: 10.1111/jam.13538. [DOI] [PubMed] [Google Scholar]
- 39.European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2021. Breakpoint tables for interpretation of MICs and zone diameters, version 11.0. http://www.eucast.org.
- 40.Silva B, Nunes A, Vale FF, Rocha R, Gomes JP, Dias R, Oleastro M. 2017. The expression of Helicobacter pylori tfs plasticity zone cluster is regulated by pH and adherence, and its composition is associated with differential gastric IL-8 secretion. Helicobacter 22:e12390. doi: 10.1111/hel.12390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental tables and figures. Download AAC.00729-21-s0001.pdf, PDF file, 0.2 MB (189.6KB, pdf)


