Abstract
The biology, clinical phenotype and progression rate of chronic myelomonocytic leukemia (CMML) are highly variable due to diverse initiating and secondary clonal genetic events. To determine the effects of molecular features including clonal hierarchy in CMML, we studied whole-exome and targeted next-generation sequencing data from 150 patients with robust clinical and molecular annotation assessed cross-sectionally and at serial time points of disease evolution. To identify molecular lesions unique to CMML, we compared it to the related myeloid neoplasms (N = 586), including juvenile myelomonocytic leukemia, myelodysplastic syndromes (MDS) and primary monocytic acute myeloid leukemia and discerned distinct molecular profiles despite similar pathomorphological features. Within CMML, mutations in certain pathways correlated with clinical classification, for example, proliferative vs dysplastic features. While most CMML patients (59%) had ancestral (dominant/co-dominant) mutations involving TET2, SRSF2 or ASXL1 genes, secondary subclonal hierarchy correlated with clinical phenotypes or outcomes. For example, progression was associated with acquisition of new expanding clones carrying biallelic TET2 mutations or RAS family, or spliceosomal gene mutations. In contrast, dysplastic features correlated with mutations usually encountered in MDS (for example, SF3B1 and U2AF1). Classification of CMML based on hierarchies of ancestral and subclonal mutational events may correlate strongly with clinical features and prognosis.
INTRODUCTION
Chronic myelomonocytic leukemia (CMML) combines pathomorphological and clinical features of both myeloproliferative neoplasms (MPN) and myelodysplastic syndromes (MDS).1 Similar to MDS, CMML is characterized by an older age at presentation and progression to secondary acute myeloid leukemia (sAML). CMML has distinct pathogenesis and molecular features compared to morphologically similar juvenile myelomonocytic leukemia (JMML).1,2 The current diagnostic definition of CMML is predicated on persistent monocytosis, which frequently associates with reactive processes, resulting in a diagnostic challenge. Clinically, monocytosis in the context of neutrophilia may not distinguish CMML from atypical chronic myeloid leukemia nor chronic neutrophilic leukemia.3 It could also be that certain cases of monocytic AML actually represent late stages of a progressed aggressive CMML. Understanding the molecular makeup of CMML may improve its characterization and identify key prognostic variables involved in disease evolution.
Recent WHO diagnostic CMML criteria include recurrent somatic mutations to be interpreted in the right clinical context.1 High-throughput next-generation sequencing technologies (HT-NGS) enabled this.4–6 NGS helped define the clonal architecture of MDS, AML and other related diseases, paving way for the same principles to characterize the clonal architecture of CMML.7,8 While molecular lesions associated with CMML have been identified, these were derived largely from study cohorts analyzing a limited number of genes.9–13 Here we report a comprehensive clinical and NGS analysis of CMML patient samples taken longitudinally to define founder vs subclonal molecular events.
METHODS
Patients
We assessed 150 patients with CMML or sAML with a clear history of antecedent CMML (N = 27), 92 with JMML,2 and 64 patients with AML with monocytic differentiation7 from TCGA (Table 1). Clinical characteristics (2008 WHO classification3) of our study cohort were compared to lower-risk (N = 199) and higher-risk MDS (N = 231). The median follow up was 20 mo. (0–175). Patients were further risk stratified based on CMML-specific prognostic scoring system (CPSS) and clinical/molecular CPSS-Mol.14,15 Informed consents were collected in accordance to the declaration of Helsinki and institutional review board. Clinical data were collected at time of sample collection and survival analyses were calculated from the time of diagnosis to last follow up. Responses were assessed using International Working Group criteria and updated MDS/MPN response criteria.16,17 Cases with complete, partial response and hematological improvement were categorized as responders, while stable disease and progression during treatment were categorized as non-responders. Clinical course is depicted in Supplementary Figure 7 and Supplementary Table 7.
Table 1.
Clinical characteristics of patients with CMML
| Diagnosis a | N | Age median (years) | Gender M/F | Karyotype normal/abnormal (N) | Sub-classification | |
|---|---|---|---|---|---|---|
|
|
||||||
| MD-CMML (N) | MP-CMML (N) | |||||
|
| ||||||
| CMML-1 | 96 | 74 | 67/29 | 55/41 | 54 | 42 |
| CMML-2 | 27 | 70 | 22/5 | 13/14 | 18 | 9 |
| sAML | 27 | 69 | 15/12 | 15/12 | 14 | 13 |
| Total CMML | 150 | 104/46 | 83/67 | 86 | 64 | |
| M4/M5b | 64 | 59 | 34/30 | 41/16 | ||
| JMMLc | 92 | 19d | 61/31 | 77/15 | ||
| Sum | 156 | |||||
Abbreviations: CMML, chronic myelomonocytic leukemia; sAML, secondary acute myeloid leukemia; JMML, juvenile myelomonocytic leukemia; M, male; F, female; MD, myelodysplastic; MP, myeloproliferative; TDS, targeted deep sequencing; WES, whole-exome sequencing.
WHO 2008 CMML: 31 cases were analyzed by WES, 93 cases were analyzed by TDS and 26 cases were analyzed by Sanger sequencing.
TCGA cohort: 64 M4/M5 were analyzed by WES.
JMML: 13 out of 92 cases were sequenced by WES and all cases were analyzed by TDS.2
Median age in months. For cytogenetic information see Supplementary Table 3.
Whole-exome sequencing (WES)
Paired (tumor, germ line DNA, that is, CD3+ cells) was used to detect somatic mutations in 31 CMML patients to identify the most common mutations, which were confirmed by additional deep sequencing and/or Sanger sequencing. For WES, variants with a minimum of 10 reads deep with at least four positive reads were considered a mutant allele. Average coverage was 115 × and only variants with variant allelic frequency (VAF) >5% were used. Analytic calling algorithms were applied followed by confirmatory deep NGS to assert somatic nature of selected alterations and exclude possible artifacts (Supplementary Figure 1). Briefly, tumor DNA was extracted from patients’ marrow aspirates and blood. The data processing was divided into binary alignment map (BAM) file generation (http://samtools.sourceforge.net/) for paired normal/tumor samples and detection/identification of somatic point mutations and indels by comparing normal/tumor BAM files. Alignment of sequencing reads on hg19 genome was visualized using Integrative Genomics Viewer software (http://www.broadinstitute.org/igv)18 (Supplementary Figure 1).
Targeted NGS
A panel of 64 of the most commonly mutated genes in myeloid neoplasms was deep sequenced in an additional group of patient samples and further allowed categorization of the mutations by functional groups (Supplementary Tables 1 and 2). Analysis was split into three steps. First, raw, de-multiplexed and preprocessed reads (quality trimming, adapter removal; Trimmomatic V.032)19 were aligned to a human genome reference hg19 using Burrows Wheeler Aligner (BWA v0.7).20 Second, binary alignment map files were subjected to a variant extraction by GATK pipeline with Broad Institute Best Practices workflow.21,22 Finally, annotation of variants was performed with respect to their function by using a standard pipeline within Annovar with hg19 libraries.23 A summary of our sequencing analyses is provided in (Supplementary Tables 5 and 6).24 For targeted NGS, variants with a minimum of 20 reads deep with at least six positive reads were considered a mutant allele. Average coverage was 252 × and only variants with VAF >5% were used. VAFs of mutations were adjusted according to the zygosity/copy number confirmed by single-nucleotide polymorphism (SNP) -array. The adjusted VAFs of each mutation were analyzed by ranks or dichotomized for all identified mutations.
SNP-A analysis
SNP-array karyotyping for confirming metaphase cytogenetics and detecting copy number normal loss of heterozygosity was performed as previously described.11,12 Briefly, Affymetrix 250 K and 6.0 (Affymetrix, Santa Clara, CA, USA) SNP-arrays were used to evaluate copy number and loss of heterozygosity. Using our internal and publicly available databases (see URLs), the screening algorithm validated each lesion as somatic vs germ line.25,26 Non-somatic lesions were excluded from further analysis. Affected genomic positions in each lesion were visualized and extracted by CNAG (v3.0, Tokyo, Japan) or the Genotyping Console (Affymetrix) software.
Adjustment of variant allelic frequency
VAFs of mutations were adjusted according to the zygosity and copy number confirmed by SNP-array. VAF of homozygous mutations as well as mutations of the genes located on chromosome X in the male cases were reduced to the half value of raw data. Hemizygous mutation VAFs were adjusted based on the formula as ‘Adjusted VAF = a/(1+a), where a = raw VAF value.’ These adjustments of VAF were not required for heterozygous mutations. The adjusted VAF value of each mutation was categorized into large and small size dichotomized by mean VAF of all the identified mutations (Supplementary Materials and Methods).
Ancestral vs subclonal mutations
For distinction between ancestral/founder and secondary/subclonal mutations present in each case, we used the following criteria: (1) in cases with serial sample analyses, mutations appearing at progression to AML/CMML-2 but not present initially were deemed subclonal; (2) in each case, VAFs of significant mutations adjusted by copy number variations and zygosity were compared (Fisher’s exact test P<0.05) and the largest clone was deemed founder in that case, if significant; and (3) if criteria 1 and 2 were not applicable, bone marrow cells were cultured in semisolid media and subjected to single colony cell sequencing as previously described.11,27,28 Ambiguous results were further categorized into mosaic and inconclusive cases. Additional analyses included: (1) for each mutation the average VAF was calculated and mean values compared; (2) we also applied a ranking approach wherein, for each mutation, the proportion of cases in which that mutation was ancestral was calculated and the values compared to select the most likely ancestral events; and (3) cross-sectional analysis was performed and mutations with significantly higher VAF’s in advanced cases were deemed more likely to be subclonal events (Supplementary Materials and Methods).
Statistical analysis
Kaplan–Meier curves were used to display overall survival differences in patient groups assessed by Log-Rank Tests. Two-sided tests were used and P-value <0.05 were considered statistically significant. The statistical environment R was used for all computations. Forest plots were generated using the R package forest-plot.
RESULTS
Clinical features of CMML
Out of the 150 patients studied, 96 had CMML-1, 27 had CMML-2 and 27 had sAML with antecedent CMML. The median age at presentation in this cohort was 70 (35–89) years. Aberrant karyotypes were detected in 45% of the cohort, with trisomy 8 (10%), − 7/del7q (6%), del20q (5%), and –Y (5%) being the most common (Table 1 and Supplementary Table 3). Somatic segmental uniparental disomy (UPD) was common, found in 25% of patients, in particular UPD11q (CBL), 4q (TET2), 7q (EZH2) and 2p, 9p and 17p. The most common chromosomal microdeletions by SNP (that is, EZH2, CUX1 and LUC7L2) involved chr.7q (Supplementary Figure 2). On the basis of white blood cell count and clinical features at presentation the cohort was classified into CMML with proliferative (N = 86) or dysplastic features (N = 64; Supplementary Tables 3 and 4). Using the CPSS and CPSS-Mol, our findings validated the importance of the subtype of CMML and French-American-British (FAB) subtypes, CMML-specific cytogenetic risk groups, and transfusion dependency to risk stratify patients (P = 0.003; Supplementary Figure 6A). However, we were not able to validate the CPSS-Mol in our cohort (Supplementary Figure 6B).
Somatic mutations in CMML
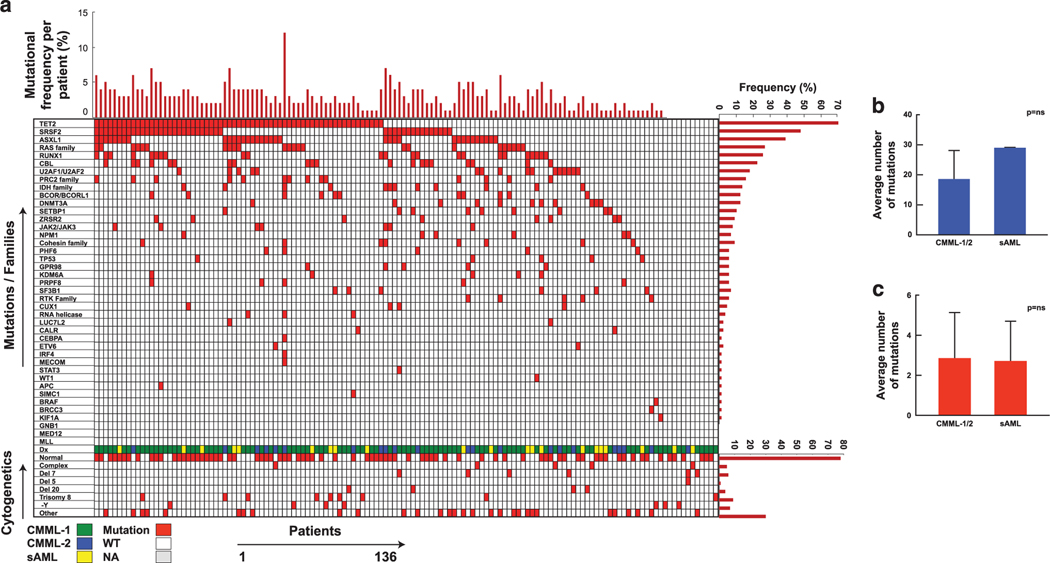
After removing polymorphisms and artifacts, WES identified 812 mutations in 672 genes; 91% of patients harbored ⩾ 1 somatic mutations (Figure 1a) with an average, 19 somatic mutations in CMML-1/2 and 29 mutations in sAML with antecedent CMML (Figure 1b), whereas the average mutational burden by targeted deep NGS was three per case in CMML-1/2 (Figure 1c). Somatic mutations were analyzed individually and by functional groups (Supplementary Tables 1 and 2). The most frequently mutated genes in CMML include TET2 (63/136; 46%), SRSF2 (43/113; 38%), ASXL1 (35/136; 26%), RUNX1 (23/129; 18%), CBL (20/136; 15%), U2AF1 (15/133; 11%), KRAS (13/136; 10%), DNMT3A (11/136; 8%), EZH2 (11/136; 8%) and NRAS (10/136; 7%) and the frequency of the remaining mutations ranged 0–7% (Figure 1). When the top 10, 20, 30 and 60 most frequently mutated genes were analyzed, they represent 80, 88, 90 and 91% of the patients with mutations commonly encountered in CMML, respectively. In addition, we detected novel somatic events, which have not been reported previously in CMML, including CRIPAK, FRG1, HYDIN and KDM6B, (Supplementary Table 6). By empirically clustering genes into their proximal functional families, we found that spliceosomal mutations were the most frequent family of genes mutated in our CMML cohort: 57% had ⩾ 1 mutation among SRSF2, U2AF1, PRPF8 and SF3B1 genes, followed by RAS, PRC2 and BCOR gene family mutations were found in 22%, 19% and 10% of the cohort respectively.
Figure 1.
Prevalence and distribution of somatic mutations in CMML. (a) Each column represents one patient and each row corresponds to one gene or family of genes. The color of each rectangle represents the status of the gene, an associated diagnosis and karyotype for each individual patient. The bar graphs represent the frequency of mutations for each individual patients, mutations and cytogenetics. For the purpose of this presentation mutations were grouped according to functional relationships (Supplementary Table 2). (b) Average number of somatic mutations detected by whole-exome sequencing and (c) average number of mutations detected by targeted deep sequencing.
When analyzing the mutational distributions based on descriptive features and clinical characteristics of the marrow morphology, we were able to categorize patients as having ‘proliferative’ or ‘dysplastic’ CMML; we identified that KRAS (18 vs 3%, P = 0.003), NRAS (13 vs 3%, P = 0.02), SRSF2 (49 vs 29%, P = 0.03) and TET2 (58 vs 37%, P = 0.02) mutations associated with proliferative more than dysplastic CMML. In 12/136 patients no mutations were found at the time of sampling. Of those, six patients had normal karyotypes. Serial analysis was not available for these 12 patients. Additional SNP analysis demonstrated homozygous mutations in UPD regions of CBL (N = 9), TET2 (N = 4), EZH2 (N = 3), TP53 (N = 2), DNMT3A (N = 2) and JAK2 (N = 2) (Supplementary Figure 2); four cases had a hemizygous mutation of either TET2, CUX1 or PHF6.
Comparative studies
Cross-sectional analyses between disease types demonstrated both distinct variations in the distribution of individual mutations and in some instances of concordance depending on the comparator disorder and specific genes (Figure 2 and Supplementary Figure 3). JMML had a distinct mutational spectrum, with the most common mutations in PTPN11, NF1, JAK3, KRAS and NRAS genes (Figure 2a). PTPN11, NF1 and JAK3 were more frequently mutated in JMML than in CMML (43 vs 2%, P<0.0001, 10 vs 5%, P<0.17; 11 vs 0%, P<0.0003). In contrast TET2, SRSF2, RUNX1 and ASXL1 mutations were more common in CMML. CBL and SETBP1 were mutated approximately equally in both cohorts. Comparing sAML-post CMML to non-core binding factor de novo monocytic AML, TET2 and ASXL1 mutations were more common in sAML with antecedent CMML (46 vs 6%, P<0.0001; 26 vs 0%, P<0.0001 respectively; Figure 2b). NPM1, FLT3 and DNMT3A mutations were more frequent in monocytic AML (39 vs 5%, P<0.0001; 33 vs 0.9%, P<0.0001; 39 vs 8%, P<0.0001, respectively). Comparing low- and high-risk MDS with early vs progressed CMML, we found advanced disease associated with higher mutational burdens in both MDS and CMML. CMML-1 was more enriched in TET2, ASXL1, SRSF2, RUNX1, CBL and RAS family mutations compared to lower-risk MDS, whereas SF3B1 was unique to lower-risk MDS (Figure 2c). When we compared CMML-2 and sAML with antecedent CMML to higher-risk MDS and sAML from MDS, we found that TET2, ASXL1, splicing factors, RUNX1, PHF6 and CBL mutations were more common in the CMML-2 and sAML from CMML (Figure 2d).
Figure 2.
CPSS The bar graphs represent the frequency of the mutations for each disease type. The stairway plots depict the concordance between two mutations. (a) CMML compared to JMML. (b) sAML-post CMML compared to non-core binding factor AML M4/M5. (c) CMML-1 compared to low-risk MDS. (d) CMML-2+sAML-post-CMML sAML compared to high-risk MDS+post-MDS sAML.
Clonal architecture and dynamics
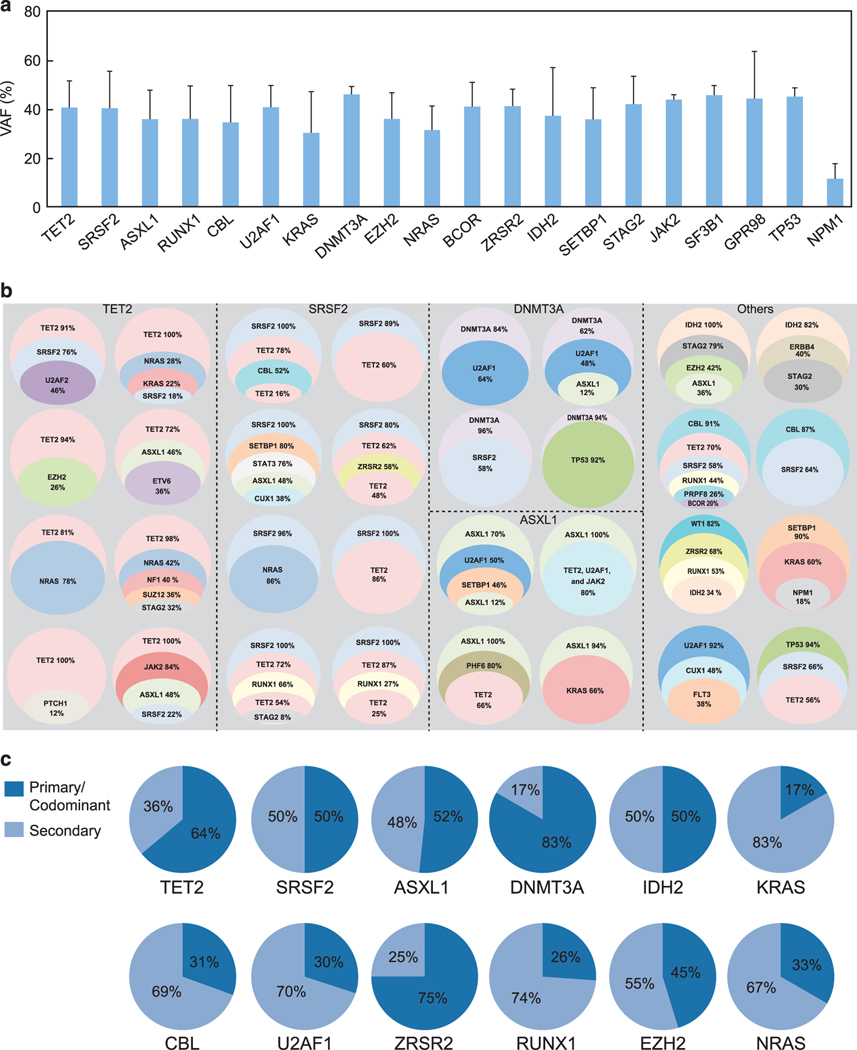
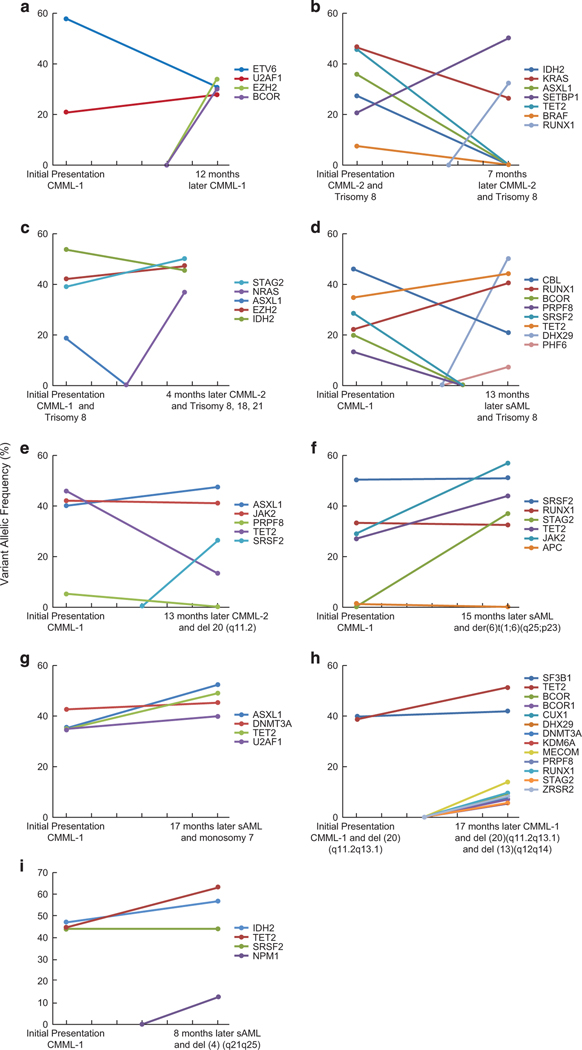
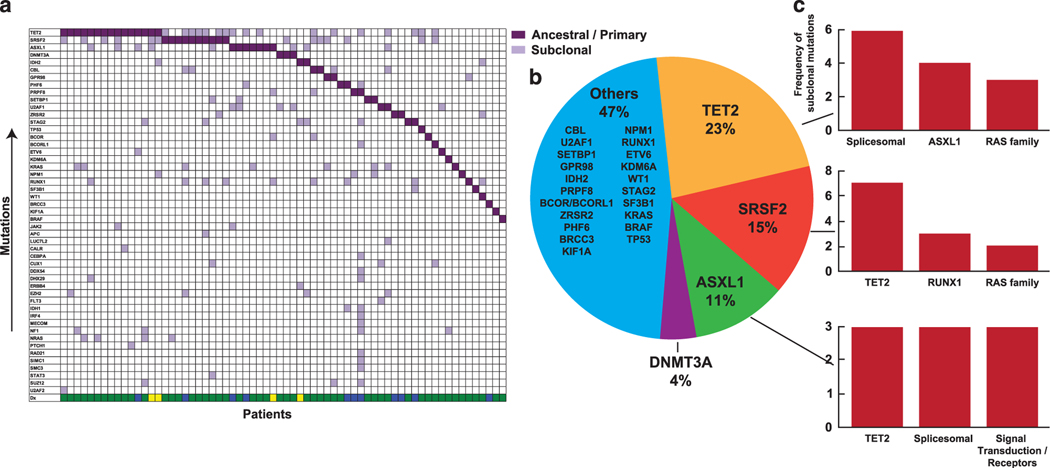
Clonal architecture can be recapitulated by analysis of mutational VAF for concurrently mutated genes as well as by serial sample analysis. Cross-sectional comparisons of the average VAF for the top 20 most frequently mutated genes allows for pathogenetic insights into potential ancestral mutations (Figure 3a); genes with the largest average VAF might indicate their founder role within clonal hierarchy. To further dissect the clonal architecture of CMML, we ranked mutations for each patient according to their clonal size, that is, the mutation with highest VAF was identified as the ancestral/dominant or co-dominant (when VAFs were similar). The CMML hallmark, TET2 mutation, was found to be an ancestral event in 15 cases (28%), co-dominant in 19 cases (36%) and a secondary event in 19 cases (36%). Similarly, SRSF2 was found to be an ancestral event in 10 cases (28%), a co-dominant event in eight cases (22%), and a secondary event in 18 cases (50%; Figure 3b). Similar estimates calculated for other genes show that CBL, U2AF1, RUNX1, EZH2 and KRAS may be dominant or secondary events, while some mutations are predominantly secondary (Figure 3b). To recapitulate clonal architecture, we quantified the clonal burden of the ancestral mutations and their sub-clones in CMML1/2 (Figure 3c). For instance, most TET2 mutations were dominant, while most RAS mutations were subclonal. Serially analyzed patients provided precise insights into the clonal hierarchy. For some patients (N = 14; Figure 3), such sequential analysis was available along with their treatments modalities (Figure 4 and Supplementary Table 8). In general, progression was associated with either expansion of specific sub-clones or with occurrence of new clones. Similar principles applied to chromosomal abnormalities. To that end, secondary clones could be subcategorized as emerging, increasing or vanishing as demonstrated for EZH2 and BCOR (Figure 4a), SETBP1 (Figure 4b) and ASXL1 (Figure 4c). In many cases (~60%) we identified a single unique ancestral/dominant event (Figure 5, left) for which a common dominant lesion frequency could be calculated (Figure 5, right). Moreover, for common ancestral mutations, we were able to assign most likely co-occurring subclonal events like TET2, RAS family and splicesomal mutations.
Figure 3.
Clonal architecture/hierarchy in CMML. (a) Mean VAF of the most frequently mutated genes in CMML. (b) Representative cases correspond to the circles grouped by first-hit/ancestral. Subclonal mutations are represented by individual colors circled areas proportional to subclonal burden. (c) For most commonly mutated genes proportion of cases with clonal vs subclonal mutations of any given gene is shown. Primary and subclonal status is determined by ranking.
Figure 4.
Serial analysis in CMML. (a–i) Serial analysis of nine individuals to illustrate the clonal dynamics. Patients were analyzed at presentation, along with the VAF of numerous mutations, followed by the acquisition of new mutations, cytogenetic abnormalities and progression.
Figure 5.
Ancestral and subclonal events in CMML. (a) Distribution of ancestral mutations (purple squares vs light purple squares correspond to subclonal events per patient in a vertical arrangement or for any given mutation in horizontal lines by prevalence. (b) Pie diagram shows the distribution of the most common ancestral events. (c) The bar graphs represent the most common subclonal events for the top three ancestral events in CMML.
Clinical impact of somatic events
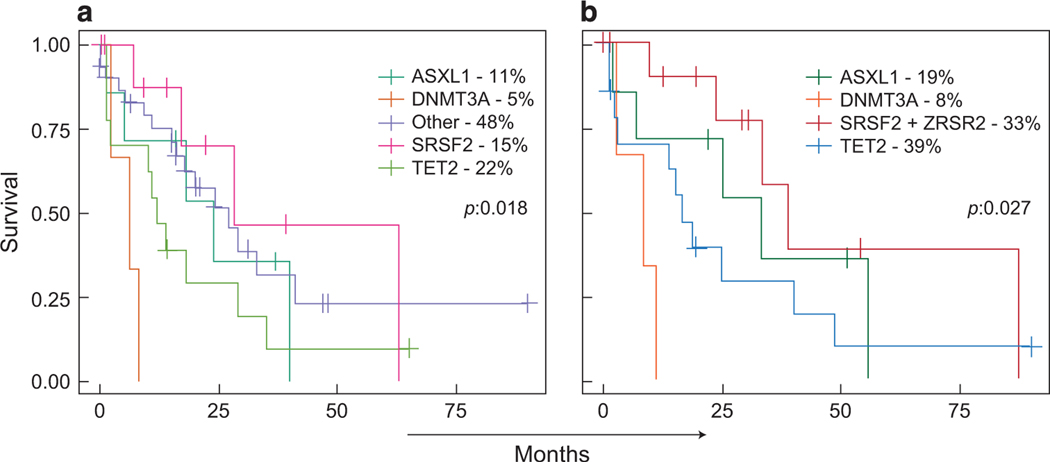
To understand the prognostic relevance of genetic events seen in CMML, we first performed a comprehensive survival analysis for the most frequent mutational events in functional groups, as well as individually (Supplementary Figures 4A and B). Any somatic lesions and/or mutations in EZH2 (P = 0.04), CUX1 (P = 0.003), DNMT3A (P = 0.005) and LUC7L2 (P = 0.006), (ancestral, co-dominant, secondary; Supplementary Figure 4A) had an adverse impact on overall survival for CMML patients. Functional grouping did not help to identify mutational groups predictive of poor prognosis except for the RNA helicase family (P = 0.006; Supplementary Table 2 and Supplementary Figure 4B). We also analyzed outcomes based on dysplastic or proliferative features within each individual group (Supplementary Figures 4Ca and b). In Supplementary Figures 4Ca and b for example, an adverse impact on survival was noted in the dysplastic group for LUC7L2 (P = 0.008), CUX1 (P = 0.007), EZH2 (P = 0.018), U2AF1 (P = 0.006) and DNMT3A (P = 0.045) mutations. In the proliferative group, DNMT3A (P = 0.048), IDH1 (P = 0.023) and SETBP1 (P = 0.006) mutations were associated with a poor outcome, in contrast to SF3B1 (P = 0.088) and TET2 (P = 0.097). Finally, given the hypothesis that ancestral mutations drive the pathogenesis of disease in myeloid neoplasms, evaluation of the dominant clone and its relationship to overall survival was investigated (Figure 6a). Patients with ancestral DNMT3A and ASXL1 had adverse outcomes, in comparison to those with TET2, SRSF2 and other ancestral mutations (22%, 15% and 48%; P = 0.018). When other less common ancestral mutations were omitted and mutations were grouped by functional groups, similar statistically significant findings were found (P = 0.027) (Figure 6b). Moreover, some ancestral events (for example, SRSF2) have a differential outcome in comparison to a secondary event in the same gene. However, for other genes, such a differential effect was not observed (Supplementary Figure 4D). Impact of somatic mutations on therapeutic outcomes of this cohort was evaluated for patients (56/136; azacitidine = 38; decitabine = 17; unknown = 1) receiving hypomethylating agent (HMA) therapy and according to International Working Group response criteria; overall response was 50% and a comparable results were obtained according to updated response criteria for MDS/MPN (Supplementary Table 7). While SRSF2, TET2, ASXL1 and IDH2 mutations seemed overrepresented among responders, they were not found to be predictive of response. Comparing these patients to non-responder male patients, age (>70 years), and primary proliferative phenotypes predicted HMA responses (odds ratio = 4.75, P = 0.03; Supplementary Figure 5). Other therapeutic modalities, not further analyzed due to limited sample size, included supportive care (that is, transfusions, growth factors), hematopoietic stem cell transplant, hydroxyurea, induction chemotherapy, splenectomy and clinical trials depending on individual clinical stage.
Figure 6.
The impact of ancestral events on survival in CMML. (a) Comparison of individual ancestral events. (b) Comparison between most common individual ancestral events and functional gene groups.
DISCUSSION
Molecular features are increasingly used as an essential diagnostic and prognostic component in the workup of myeloid neoplasms.4,7,29,30 In MPNs, certain molecular abnormalities, such as BCR-ABL and more recently mutations in CSF3R and SETBP1, identify subtypes. In MDS, we and others have described the molecular heterogeneity and complex hierarchy and dynamics of molecular evolution in these phenotypically diverse diagnoses. CMML, as an MDS/MPN overlap disorder, is expected to take this complexity one step further.
We demonstrated that, with rare exception and despite its classification as a distinct entity within the WHO classification for myeloid neoplasms, CMML cannot be typified by a unique molecular profile. Moreover, subclonal evolution from ‘typical’ molecular abnormalities such as TET2, SRSF2 and ASXL1 are more likely to define the disease and impact prognosis. We used first WES to identify complete mutational spectra. Top mutated genes were included into targeted sequencing panel applied the remainder of patients.
The order in which molecular abnormalities are acquired is important. Ancestral clonal lesions could potentially predetermine the prevailing phenotype and subsequent characteristics of the disease independent of secondary mutations. Alternatively, shared ancestral lesions (that is, CMML and MDS) could be followed by secondary phenotype-defining events in the form of secondary lesions. Finally, secondary or tertiary events may serve as a trigger of progression towards a more aggressive phenotype already fixed by earlier events. For example, mutations in the same gene may be either ancestral or secondary (that is, SRSF2) in different patients by chance, or they might be obligatory founder vs secondary lesions. Thus, it is possible that for some genes, the hierarchy/succession of the events is phenotype defining. For example, in AML core binding factor fusions are quintessential ancestral events that determine AML phenotype, and may be further modified by secondary events. This is also true for rare cases of CMML with balanced translocations.4 Our study shows that the majority of CMML cases are initiated by a few common and many uncommon ancestral events, but these are not unique to CMML—they can initiate other forms of myeloid neoplasms, such as MDS, and even AML.
Cross-sectional analyses of our CMML cohort confirmed common ancestral events (that is, TET2 and ASXL1) found in previous studies in which clonal hierarchy was elegantly demonstrated using single colony sequencing.31,32 Alluding to the importance of identifying ancestral mutations in CMML,31 acquisition of mutations was predominantly linear, with limited branching and early clonal dominance. We identified ancestral mutations by ranking mutations based on their VAF and their clonal size, which were further validated via serial sequencing. In about half of the patients, we were able to identify a single unique ancestral event (TET2, SRSF2 and ASXL1). In some cases, the distinction of a unique ancestral event was not possible due to the presence of co-dominant clones. For instance, we found TET2 mutations were frequently found to be co-dominant with SRSF2, ASXL1, ZRSR2, KRAS, NRAS and less commonly with EZH2.13 This pattern implies a downstream pathway convergence resulting from two distinct hits at the level of a hematopoietic stem cell, but this notion can be only confirmed by single colony sequencing. Instead we serially analyzed a subset of samples to allow for more precise analysis of clonal hierarchy and its dynamics.
The clonal size and serial analysis allowed us to further evaluate the role of sub-clones in the pathogenesis of CMML. We found that KRAS, NRAS, U2AF1, RUNX1 and CBL were commonly found to be secondary events. However, it was difficult to assess which particular clones played a role in disease progression because of co-occurrence of several mutations and concurrent acquisition of cytogenetic abnormalities.
In our cohort the most common molecular mutations in CMML included TET2, SRSF2, ASXL1, RUNX1 and RAS family a finding reported by others in smaller patients subsets.9,13,24,33–38 Similarly, we confirmed that proliferative CMML phenotypes showed a higher mutational frequency of KRAS and NRAS mutations than dysplastic CMML cases.38 However, unlike in previous studies of combined MDS and MDS/MPN patients treated with hypomethylating agent,39,40 analysis of treatment outcomes in our CMML cohort did not reveal differential responsiveness based on the presence of individual molecular lesions as shown, for example, for TET2.
When the mutational profiles of JMML and CMML were compared, we found that, despite their morphological similarity, distinct molecular patterns were evident. CMML harbored TET2, SRSF2, RUNX1 and ASXL1 mutations and JMML had RAS family mutations. Similar distinctions were found amongst non-core binding factor AML and sAML-post CMML, with TET2 and ASXL1 significantly more mutated in the latter and NPM1, FLT3 and DNMT3A more mutated in non-core binding factor AML cases. Despite these differences, due to a significant overlap in dominant and secondary events, we were unable to devise a predictive algorithm to precisely distinguish CMML from other related myeloid neoplasms on molecular grounds. Sub-classification of mutations into functional groups to consolidate the inherent molecular diversity did not help to discern pathognomonic features of CMML vs other related myeloid neoplasms.
It is likely that certain rare ancestral events, not covered by our sequencing panel, would be missed and that true founding events were not identified. Nevertheless, assignment of dominant events allows for formation of mutually exclusive broad categories and opens an avenue for outcome studies using subtype-defining clonal defects. To that end, we show that the results described in a recent the study can be further refined.15 In our study, we were able to identify a distinctive ancestral event in over half of our cohort, with some primary hits conferring a poor prognosis.
In conclusion, we showed for the first time that in CMML TET2, SRSF2 and ASXL1 are ancestral mutations. This population was reflective of those described in other studies,8,31 in demonstrating the clonal complexity of CMML and emphasizing the importance of identification of primary/ancestral events. Such lesions are always persistent including at progression from CMML to sAML or during relapse. They may offer new actionable targets for individuals with CMML subtypes with limited therapeutic options.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Edwards P Evans Foundation and Aplastic Anemia and MDS International Foundation, NIH-R01HL123904: NIH-R01HL118281, NIH-R01HL128425 for their contributions and support.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
REFERENCES
- 1.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016; 127: 2391–2405. [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi H, Okuno Y, Muramatsu H, Yoshida K, Shiraishi Y, Takahashi M et al. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet 2013; 45: 937–941. [DOI] [PubMed] [Google Scholar]
- 3.Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood 2011; 117: 5019–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014; 28: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malcovati L, Papaemmanuil E, Ambaglio I, Elena C, Gallì A, Della Porta MG et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood 2014; 124: 1513–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet 2017; 49: 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller CA, Wilson RK, Ley TJ. Genomic landscapes and clonality of de novo AML. N Engl J Med 2013; 369: 1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K et al. Clonal architecture of secondary acute myeloid leukemia. N Eng J Med 2012; 366: 1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood 2011; 118: 3932–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makishima H, Cazzolli H, Szpurka H, Dunbar A, Tiu R, Huh J et al. Mutations of e3 ubiquitin ligase cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J Clin Oncol 2009; 27: 6109–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makishima H, Jankowska AM, McDevitt MA, O’Keefe C, Dujardin S, Cazzolli H et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood 2011; 117: e198–e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 2012; 119: 3203–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meggendorfer M, Roller A, Haferlach T, Eder C, Dicker F, Grossmann V et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML). Blood 2012; 120: 3080–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Such E, Germing U, Malcovati L, Cervera J, Kuendgen A, Della Porta MG et al. Development and validation of a prognostic scoring system for patients with chronic myelomonocytic leukemia. Blood 2013; 121: 3005–3015. [DOI] [PubMed] [Google Scholar]
- 15.Elena C, Gallì A, Such E, Meggendorfer M, Germing U, Rizzo E et al. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood 2016; 128: 1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheson BD, Greenberg PL, Bennett JM, Lowenberg B, Wijermans PW, Nimer SD et al. Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood 2006; 108: 419–425. [DOI] [PubMed] [Google Scholar]
- 17.Savona MR, Malcovati L, Komrokji R, Tiu RV, Mughal TI, Orazi A et al. An international consortium proposal of uniform response criteria for myelodysplastic/myeloproliferative neoplasms (MDS/MPN) in adults. Blood 2015; 125: 1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G et al. Integrative genomics viewer. Nat Biotechnol 2011; 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo M, Banks E, Poplin R, Garimella K, Maguire J, Hartl C et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acid Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478: 64–69. [DOI] [PubMed] [Google Scholar]
- 25.Tiu RV, Gondek L, O’Keefe CL, Huh J, Sekeres MA, Elson P et al. New lesions detected by single nucleotide polymorphism array-based chromosomal analysis have important clinical impact in acute myeloid leukemia. J Clin Oncol 2009; 27: 5219–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huh J, Tiu RV, Gondek LP, O’Keefe CL, Jasek M, Makishima H et al. Characterization of chromosome arm 20q abnormalities in myeloid malignancies using genome-wide single nucleotide polymorphism array analysis. Genes Chromosomes Cancer 2010; 49: 390–399. [DOI] [PubMed] [Google Scholar]
- 27.Makishima H, Yoshida K, Nguyen N, Przychodzen B, Sanada M, Okuno Y et al. Somatic SETBP1 mutations in myeloid malignancies. Nat Genet 2013; 45: 942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014; 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A et al. Mutation in TET2 in myeloid cancers. N Eng J Med 2009; 360: 2289–2301. [DOI] [PubMed] [Google Scholar]
- 30.Kon A, Shin LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 2013; 45: 1232–1237. [DOI] [PubMed] [Google Scholar]
- 31.Itzykson R, Kosimder O, Renneville A, Morabito M, Preudhomme C, Berthon C et al. Clonal architecture of chronic myelomonocytic leukemias. Blood 2013; 121: 2186–2198. [DOI] [PubMed] [Google Scholar]
- 32.Itzykson R, Solary E. An evolutionary perspective on chronic myelomonocytic leukemia. Leukemia 2013; 27: 1441–1450. [DOI] [PubMed] [Google Scholar]
- 33.Dunbar AJ, Gondek LP, O’Keefe CL, Makishima H, Rataul MS, Szpurka H et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res 2008; 68: 10349–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kar SA, Jankowska A, Makishima H, Visconte V, Jerez A, Sugimoto Y et al. Spliceosomal gene mutations are frequent events in the diverse mutational spectrum of chronic myelomonocytic leukemia but largely absent in juvenile myelomonocytic leukemia. Haematologica 2013; 98: 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine RL, Loriaux M, Huntly BJ, Loh ML, Beran M, Stoffregen E et al. The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood 2005; 106: 3377–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyner JW, Erickson H, Deininger MW, Willis SG, Eide CA, Levine RL et al. High-throughput sequencing screen reveals novel, transforming RAS mutations in myeloid leukemia patients. Blood 2009; 113: 1749–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdel-Wahab O, Pardanani A, Patel J, Wadleigh M, Lasho T, Heguy A et al. Concomitant analysis of EZH2 and ASXL1 mutations in myelofibrosis, chronic myelomonocytic leukemia and blast-phase myeloproliferative neoplasms. Leukemia 2011; 25: 1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelsi-Boyer V, Trouplin V, Roquain J, Adélaïde J, Carbuccia N, Esterni B et al. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol 2010; 151: 365–375. [DOI] [PubMed] [Google Scholar]
- 39.Traina F, Visconte V, Elson P, Tabarroki A, Jankowska AM, Hasrouni E et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia 2014; 28: 78–87. [DOI] [PubMed] [Google Scholar]
- 40.Bejar R, Lord A, Stevenson K, Bar-Natan M, Pérez-Ladaga A, Zaneveld J et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood 2014; 124: 2705–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.