Abstract
Certain large DNA viruses, including those in the Marseilleviridae family, encode histones. Here we show that fused histone pairs Hβ-Hα and Hδ-Hγ from Marseillevirus are structurally analogous to the eukaryotic histone pairs H2B-H2A and H4-H3. These viral histones form “forced” heterodimers and a heterotetramer of four such heterodimers assembles DNA to form structures virtually identical to canonical eukaryotic nucleosomes.
Many living organisms, particularly in the domain Eukarya, assemble DNA into chromatin to regulate the structure and accessibility of their genomes to nuclear machinery critical for cellular functions1. The nucleosome is the primary, repeating unit of chromatin, and comprises an octamer of histone proteins wrapped by ~147 bp of DNA2,3. The four histone proteins, H2A, H2B, H3, and H4, harbor a conserved histone-fold (HF) dimerization motif. Canonical histones in Eukarya form obligate heterodimers in which H2A dimerizes exclusively with H2B and H3 with H4. Similarily, in Archaea, homodimers of HF-containing proteins can assemble higher-order structures with DNA4,5.
Histone genes are also present in the genomes of nucleocytoplasmic large DNA viruses (NCLDVs), a relatively recently discovered phylum of DNA viruses6. One member of this phylum, Marseilleviruses, have been found in human blood, although their primary host is an amoeba of the genus, Acanthamoeba7,8. Marseillevirus marseillevirus (Msv) is a giant virus with a massive 369-kbp genome that contains genes that encode two histone fusions, Hβ-Hα and Hδ-Hγ9. Phylogenomic analyses suggest a possible proto-eukaryotic origin of these histones predating the divergence of core histone variants, but they might also have been acquired through horizontal gene transfer, gene fusion, and sequence divergence10.
All Marseilleviridae genomes contain a pair of divergently transcribed genes encoding histone “doublets” featuring linked HF domains (Fig. 1A)10. Expanded phylogenetic analysis using the four separated histone domains aligned to eukaryotic core histones, including variants for H2A and H3, and several histone domains from Archaea, show that the four Msv histone domains (Hα Hβ, Hγ, Hδ) are viral counterparts to the four eukaryotic core histones (Extended Data Fig. 1A). While the precise origins of these histone fusions are still unclear, they are more closely related to eukaryotic core histones than to archaeal HFs. The structure of viral histones is currently unknown.
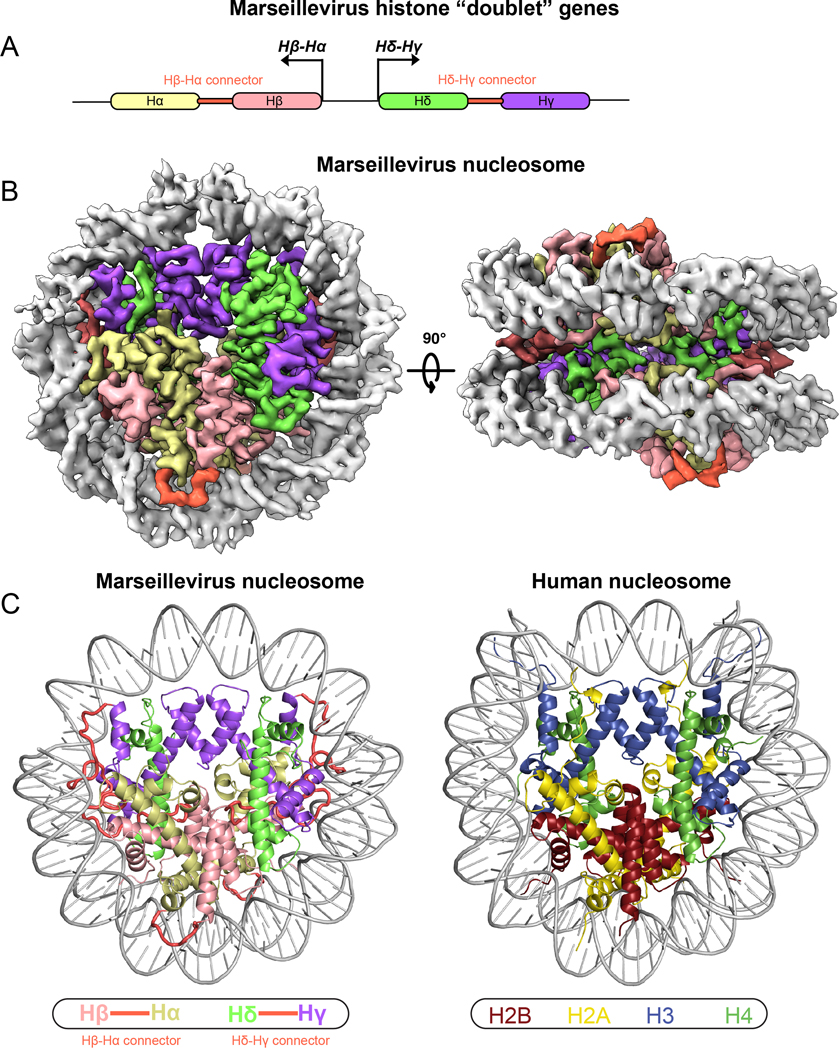
Fig. 1. Structure and conservation of Marseillevirus nucleosome.
a, Cartoon depiction of divergently transcribed histone “doublet” genes Hβ-Hα and Hδ-Hγ. b, Cryo-EM reconstruction of viral “nucleosomes” displayed in two separate views related by 90°. c, Comparison of the Marseillevirus nucleosome cryo-EM structure (left) and the human nucleosome (right, PDB ID 5AV9).
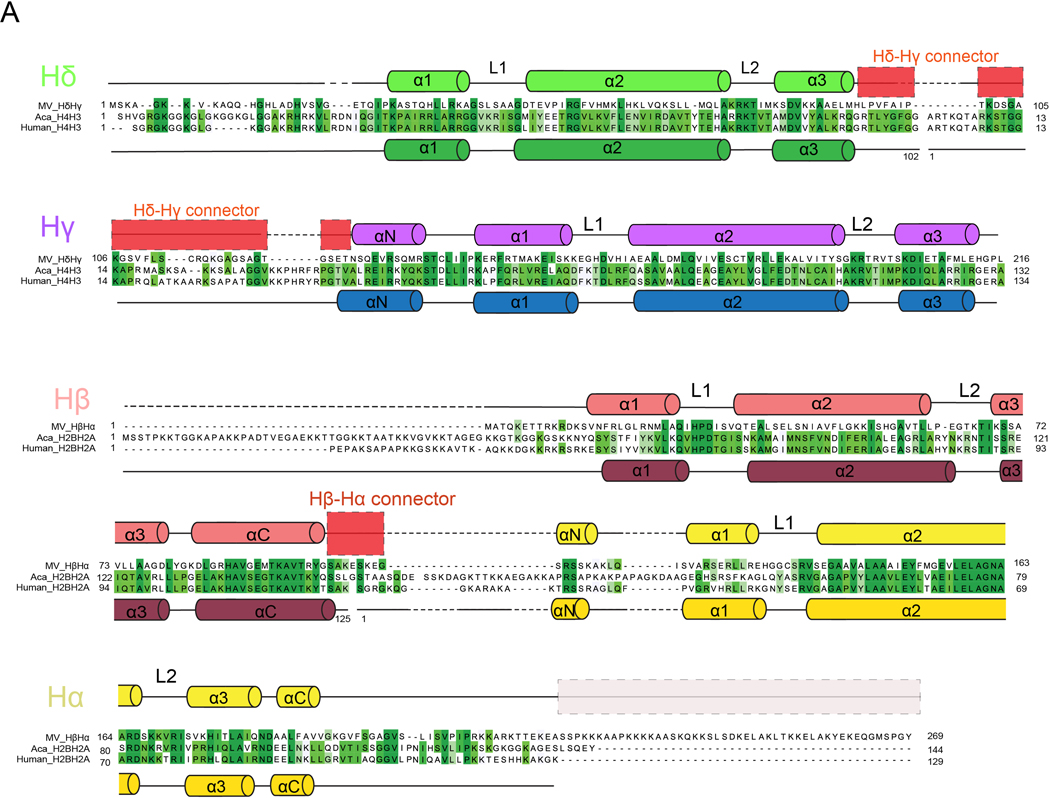
In order to understand the properties of these viral histones, we expressed Msv histone doublets in E. coli and assembled them with the Widom 601 DNA nucleosome positioning sequence (Extended Data Fig. 1B). Crosslinking mass spectrometry identified a substantial number of intermolecular contacts between Hβ-Hα and Hδ-Hγ doublets, suggesting that these proteins form higher-order assemblies in solution (Extended Data Fig. 1C and Supplementary Table 1). Analysis of these assemblies by negative stain microscopy showed shapes and sizes resembling eukaryotic nucleosomes (Extended Data Fig. 1D). Cryo-EM analysis of a subset of 146,506 particles resulted in an approximately 3.4-Å resolution map that revealed the structure of Marseillevirus nucleosomes (Fig. 1B, Extended Data Fig. 2 to 4, Supplementary Table 2). The general architecture of the viral nucleosomes closely resembles that of eukaryotic nucleosomes3,11 (Fig. 1C). Viral histone pairs heterodimerize, form a tetrameric structure similar to the eukaryotic octamer, but unlike the canonical eukaryotic histone octamer that organizes 145–147bp of DNA, the viral structure organizes only 121 bp (Fig. 1C and Extended Data Fig. 5A). Viral histone fusions fold into “forced dimer” structures that closely mimic those of obligate eukaryotic heterodimers (root mean square deviation, RMSD of 1.37 Å in H2B-H2A and 1.19 Å in H4-H3) (Fig. 2A and 2B). Interactions within a single polypeptide of Hβ- Hα and Hδ-Hγ result in a well-known handshake motif found in eukaryotic and archaeal histones3,4 (Fig. 2A and 2B). These interactions are mostly conserved, being fundamentally hydrophobic in the buried part between the helices, although some electrostatic contacts contribute to these interfaces as well3. The Hβ-Hα histone pair has 40.5 % identity and 57% similarity (over 180 residues) with human H2B and H2A, and Hδ-Hγ has 25.6% identity and 42% similarity (over 160 residues) with human H4 and H3 (Extended Data Fig. 6). The level of conservation of Msv histones with histone sequences of its host Acanthamoeba castellanii is similar (Extended Data Fig. 6). This sequence conservation explains the conservation of secondary structure, with a central HF consisting of three α-helices (α1, α2, and α3) connected by two loops (L1 and L2) (Fig. 2A, 2B and Extended Data Fig. 6). The unique feature of viral histones fusions are connectors between histones Hδ and Hγ and between Hβ and Hα (Fig. 1C and Extended Data Fig. 6). The N-termini of histones Hδ and Hβ, as well as C-terminus of Hα, contain extensions that resemble histone tails and the connector between Hδ-Hγ has some degree of conservation with the H3 tail (Extended Data Fig. 6).
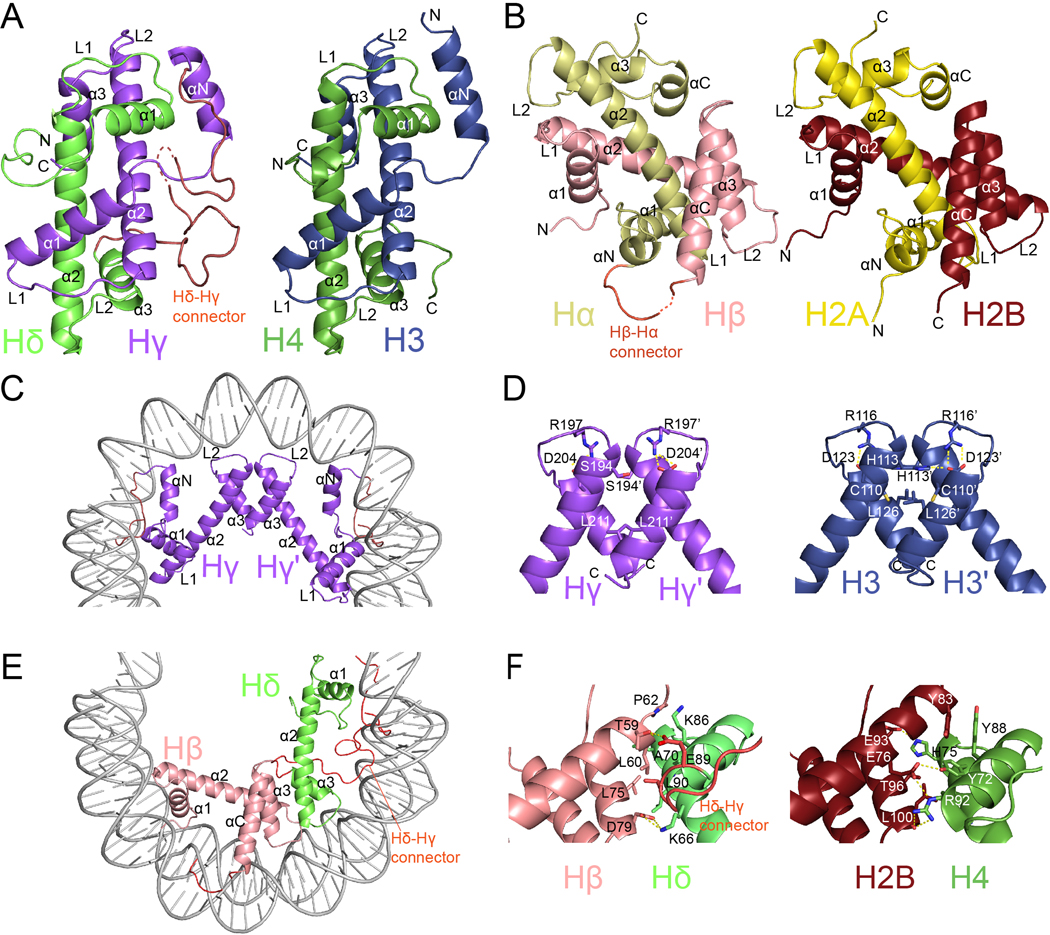
Fig. 2. |. General architecture of histones.
Comparison of Msv histone “forced dimers” (a and b left) with human histone heterodimers (a and b right). Overview of c, Hγ-Hγ’ and e, Hβ-Hδ four helix bundles. d, Detailed view of Hγ-Hγ’ (left) and H3-H3’ (right) interfaces f, Detailed view of Hβ-Hδ (left) and H2B-H4 (right) interfaces.
The viral heterotetramer, consisting of two Hβ-Hα and two Hδ-Hγ fusions, is equivalent to the eukaryotic core octamer. It is formed by specific interactions between Hγ-Hγ’ and Hβ-Hδ four-helix bundles with the geometry conserved with eukaryotic H3-H3’ and H2B-H4 four-helix bundles (Fig. 2C and 2E). In eukaryotes, H3-H3’ contacts are mediated by hydrophobic interactions and a buried intermolecular interaction between H113 of one monomer with D123 of the other monomer. D123 also establishes an intramolecular salt bridge with R116 (Fig. 2D). In Msv, the intramolecular salt bridge between Hγ R197 and D204 is conserved but the intermolecular electrostatic interaction is not, since the histidine is replaced with serine (with a shorter side chain). As in H3-H3’, the rest of the interactions that keep the Msv Hγ-Hγ’ together are mostly hydrophobic. Similarly, hydrophobic and electrostatic interactions between H2B and H4 at the interface between the tetramer and the dimers keep the eukaryotic octamer together. However, these interactions are not well conserved in Msv with substitutions of several critical residues at the interface between Hβ and Hδ (Fig. 2F). Specifically, residues Y72 and Y88 in H4 and Y83 in H2B, which form a hydrophobic cluster in eukaryotes are not conserved in Msv. Furthermore, the interactions between H4R92-H2BE76, and H4H75-H2BE93, are not present in Msv, instead a salt bridge is established between HδK66 and HβD79.
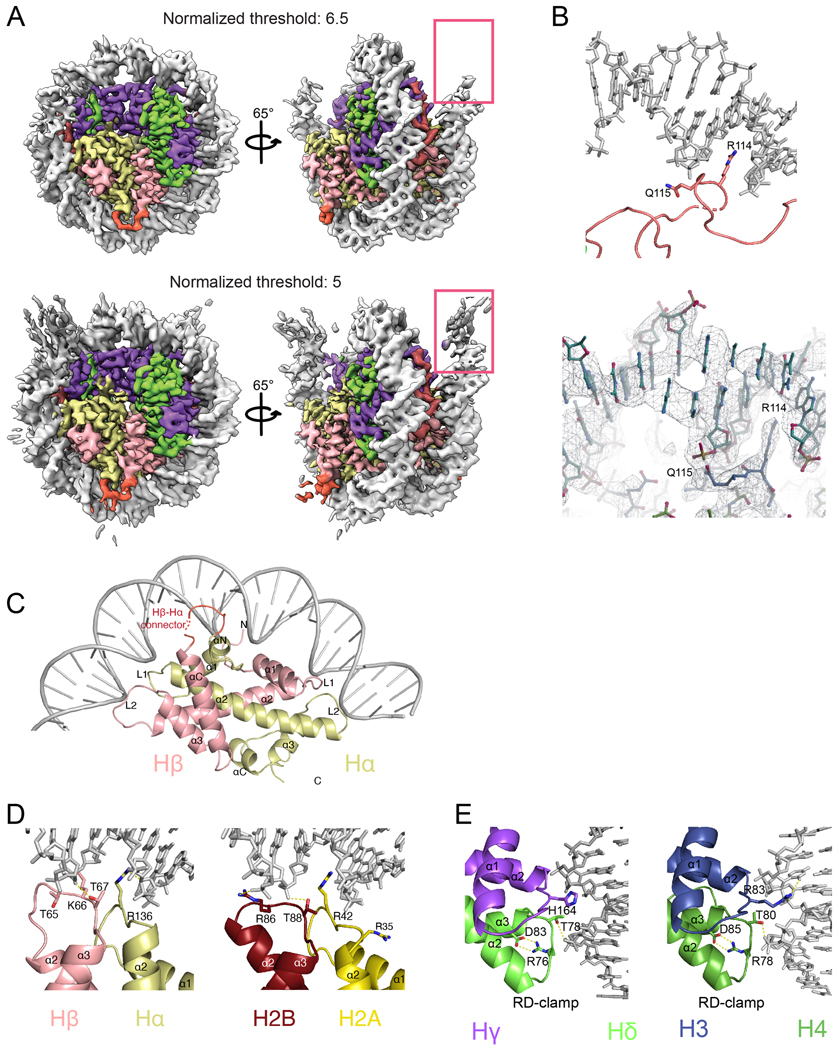
While 147 bp of DNA are visible in the reconstruction, only the central 121 bp are well ordered at high resolution and the 13 bp at the DNA entrance and exit of the viral nucleosome are less ordered (Fig. 1C and Extended Data Fig. 5A). Overall, the DNA structure and the main principles of histone–DNA interactions are conserved (Fig. 3A, 3B and Extended Data Fig. 5C–E). Interactions are mediated by histone residues and the DNA backbone in the minor groove. Each HF dimer organizes 27–28 bp via interactions made by α1 histone helices and L1–L2 loops (Fig. 3A and Extended Data Fig. 5C). In histone Hγ, an intramolecular RD “clamp” is conserved3 (Fig. 3B). However, intermolecular SR and HT pairs formed between Hδ and Hγ replace RT pairs between H4 and H3. Therefore, there is clear preservation of both the character of this interface and the interactions with the phosphodiester backbone, although the amino acid position and charge are swapped (Fig. 3B and Extended Data Fig. 5D and E). Similarly, the interface formed by Hβ-Hα with DNA is conserved with H2B-H2A dimers (Extended Data Fig. 5C).
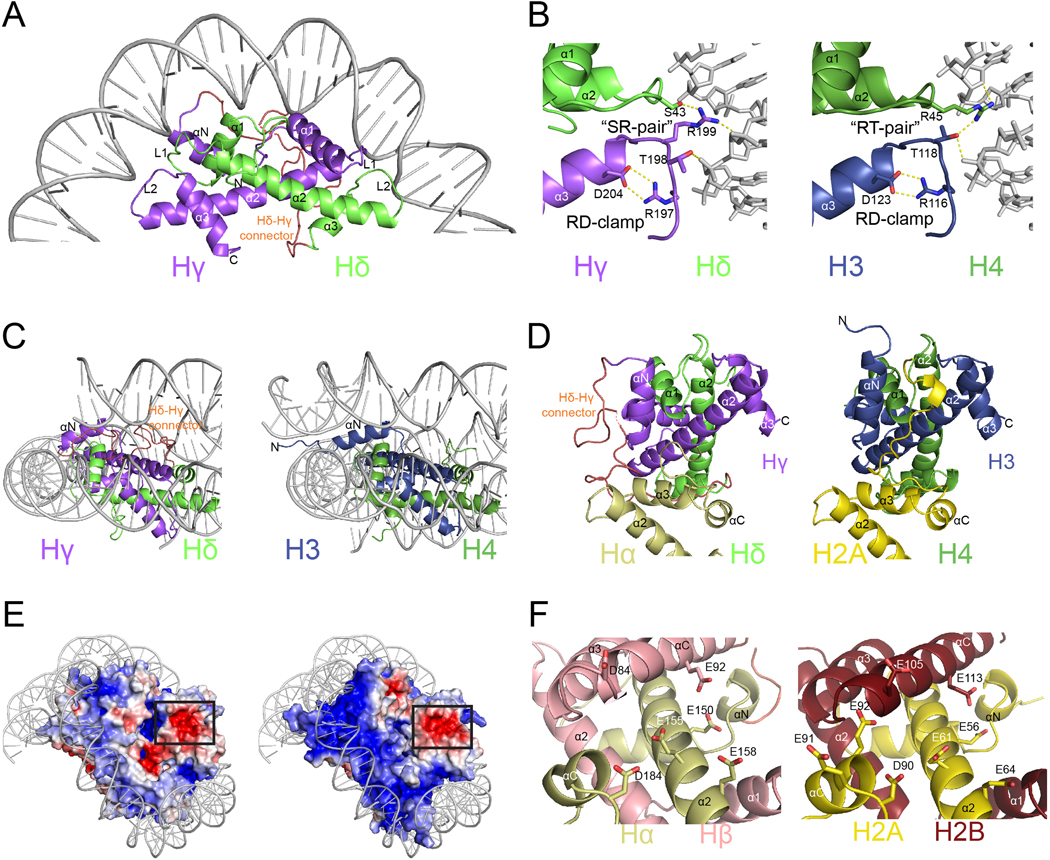
Fig. 3. |. DNA binding and acidic patch conservation.
a Hδ-Hγ “forced dimer”-DNA interface. b, Comparison of interactions between Hδ-Hγ with DNA (left) and human H4 and H3 with DNA (right). c, Close-up view of αN helices and the DNA ends of the Hγ (left) and H3 (right). d, Overview of the C-termini of Hα (left) and H2A (right). e, Representation of electrostatic surface potential of Msv (left) and human (right) nucleosomes. f, Close-up of the acidic patch in Marseillevirus Hβ-Hα (left) and H2B and H2A (right).
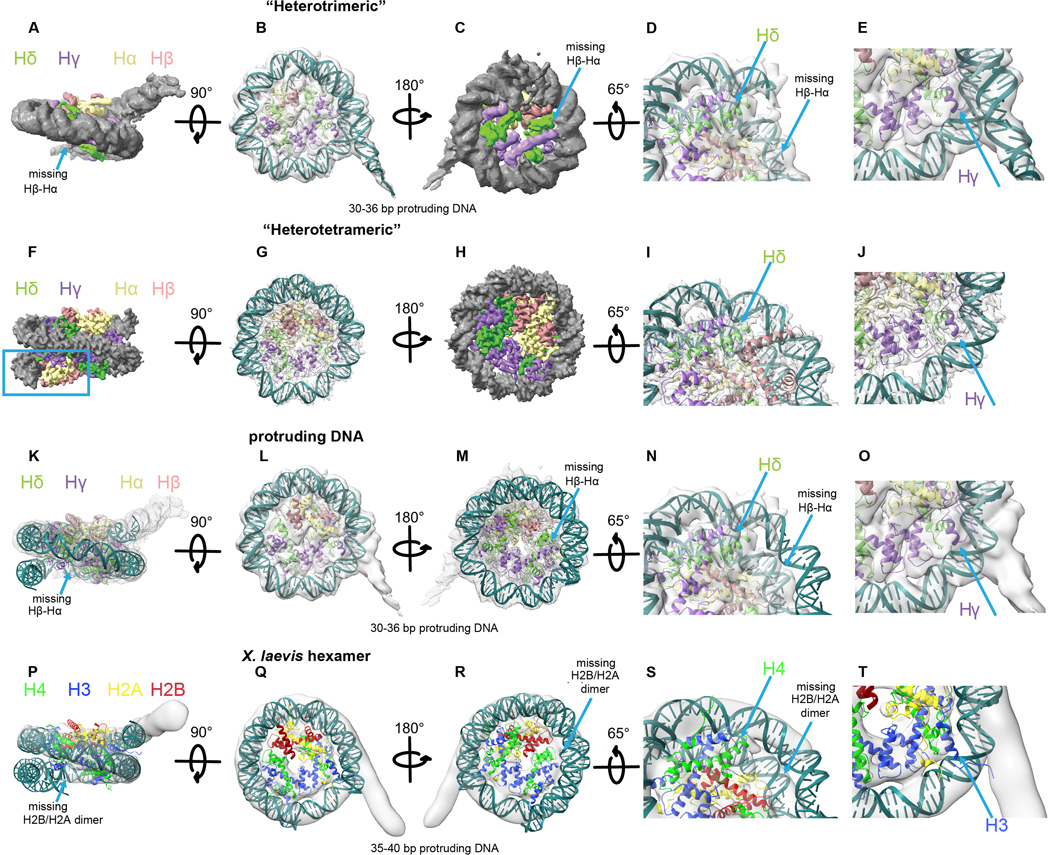
During cryo-EM data processing, we also obtained a class of particles that lack one copy of the Hβ-Hα (Extended Data Fig. 3). This viral heterotrimeric structure organizes 85–91 bp of DNA and resembles the structure of Xenopus laevis hexameric nucleosomes12 (Extended Data Fig. 7). The existence of this structure suggests instability of the Hβ-Hδ four-helix bundle interface and indicates a possible diversity of the structures that can be formed by viral histones.
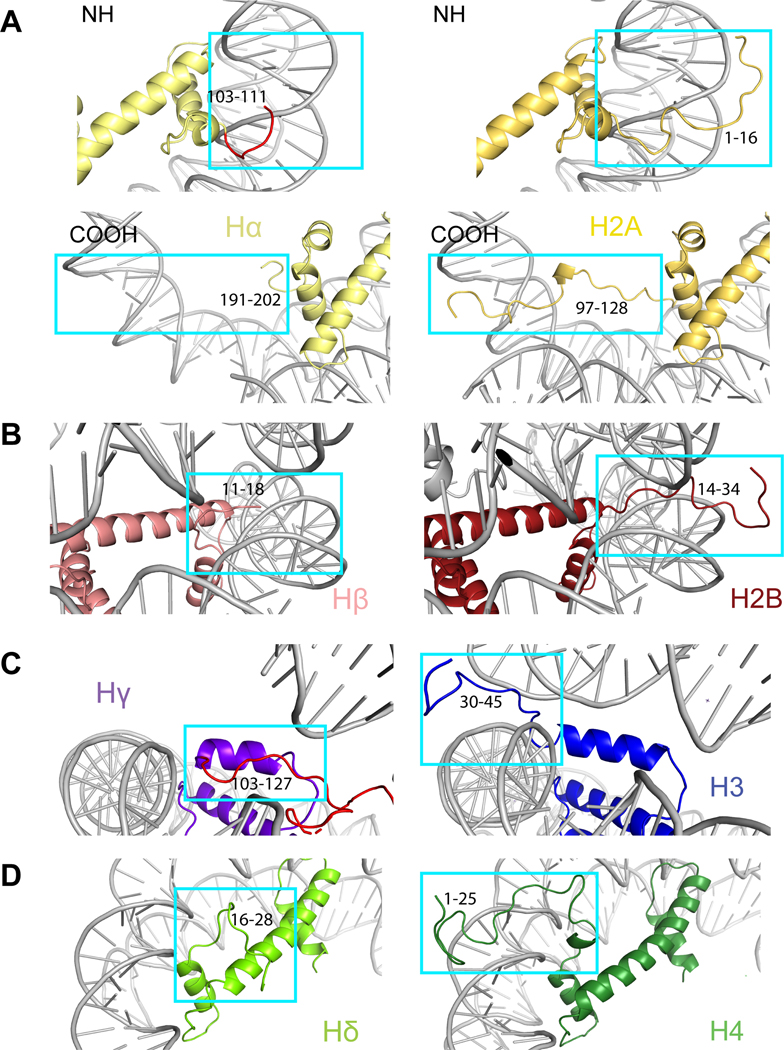
Viral histones contain extensions that resemble histone tails. In eukaryotes, histone tails exit nucleosomes between the gyres of the DNA (Extended Data Fig. 8). In the heterotetrametric Msv nucleosome, the first 10 residues of the Hβ N-terminal tail are disordered in our structure exactly at the exits between the DNA gyres, and the histone tail itself seems to be around 20 residues shorter than the tail of histone H2B (Extended Data Figs. 6 and 8B). Hα lacks the N-terminal tail and the first 14 residues of this histone are replaced with a short connector that turns and binds the C-terminus of Hβ (Fig. 2B and Extended Data Figs. 6 and 8A). The N-terminal tail of Hδ has a length similar to that of the eukaryotic histone H4; in our structure this tail turns and comes close to the C-terminus of Hγ (Fig. 2A and Extended Data Fig. 6). The N-terminus of Hδ is disordered prior to residue 16 and lacks the basic patch 16-KRHRK-20 (Extended Data Fig. 6 and 8D). The N-terminus of Hγ forms the connector with the C-terminus of Hδ (Fig. 2A and Extended Data Fig. 6). The portion of Hγ in this connector contains two lysines with possible homology to the H3 tail (positions 9 and 14 in eukaryotes, Extended Data Fig. 6), but does not extend and stabilize the terminal part of the DNA (Fig. 3C). Histone tails in eukaryotes are heavily modified, which adds a layer of regulation on nuclear processes13. We note that the tails in viral histones, especially Hδ, are charged and harbor residues that could potentially be modified post-translationally.
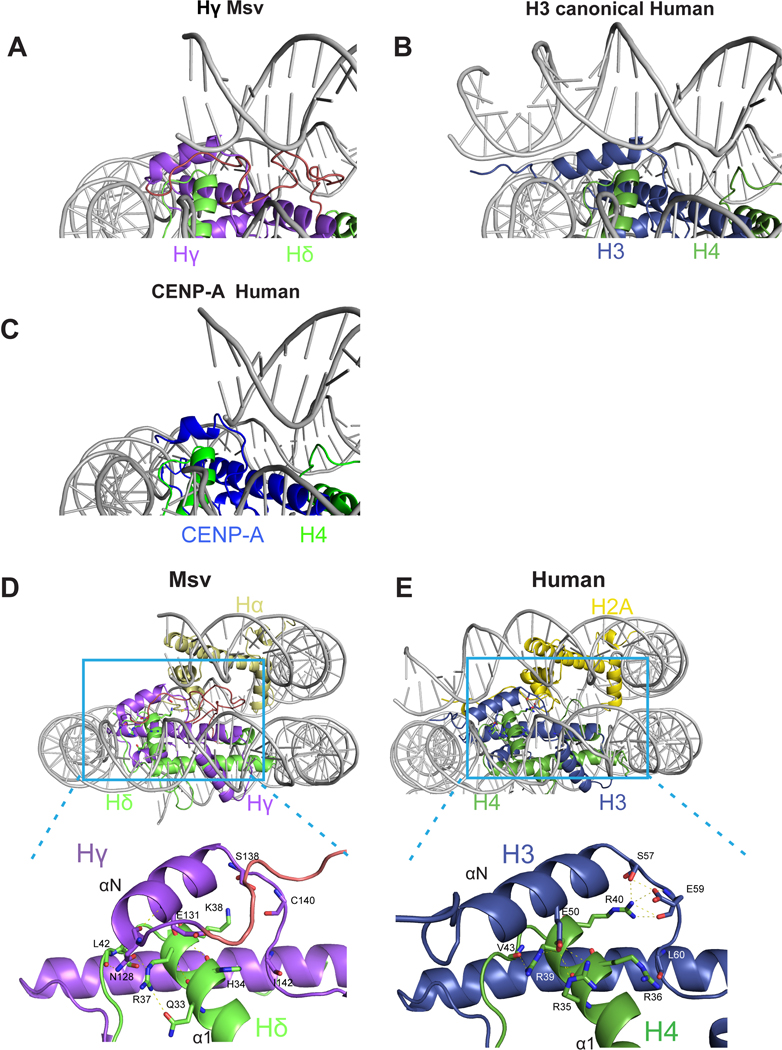
Specific differences in viral histones that might contribute to a different length of DNA interactions can be attributed to the differences in the N-tail of Hγ, the αN helix of Hγ, and the docking domain of Hα. First, the αN helix in Hγ is shorter than that of H3 (Fig. 3C and Extended Data Fig. 8C). In the eukaryotic nucleosome, the length of this helix is essential for maintaining DNA ends14. The observed destabilization of the DNA terminus due the weak Hγ contacts is reminiscent of the structure of the centromeric nucleosome containing the histone H3 variant CENP-A14 (Extended Data Fig. 9A–C). In eukaryotes, the H3 αN helix and adjacent loop make contacts with four arginines in α1 helix of H4. The interactions at this interface are not conserved in Msv, potentially contributing to destabilization of the Hγ αN helix (Extended Data Fig. 9D and E). The connector between the C terminus of Hδ and N-terminus of Hγ forms an arc that is sandwiched between two turns of the double helix of DNA (Fig. 3C and Extended Data Fig. 9A). Because Hγ lacks the canonical interaction with the DNA terminus, the connector interaction with DNA happens in a more central part of the nucleosome (Extended Data Fig. 8C and 9A). In this region, the connector Hδ-Hγ presents an R114/Q115 motif for interaction with the DNA (Extended Data Fig. 5B). In addition, the docking domain of H2A in eukaryotes interacts with the H3-H4 tetramer guiding the αN helix of H3 to interact with the last turn of DNA (Fig. 3D and Extended Data Fig. 9D and E). In our structure of the viral nucleosome, the last 67 amino acids of the docking domain in Hα are disordered (Fig. 3D).
The surface of the viral nucleosome has a similar charge to the eukaryotic nucleosome (Fig. 3E) and we found partial conservation of the acidic patch, one of the most critical region mediating interactions with arginine anchor proteins in eukaryotic nucleosomes15–17. In eukaryotes, the acidic patch includes six negatively charged residues in histone H2A (E56, E61, E64, D90, E91, and E92) and two in H2B (E105 and E113). In the viral nucleosome, four out of six residues in Hα (E150, E155, E158, and D184) and two Hβ residues (D84 and E92) are conserved (Fig. 3F).
The structural conservation of key residues responsible for higher-order assembly of viral histones highlights the conserved architecture of nucleosomes assembled from viral, archaeal and eukaryotic histones (Extended Data Fig.10). The function of viral histones is currently unknown, but it is tempting to speculate that viral nucleosome-like particles might have a role in condensing DNA during viral DNA packaging. The H4 tail in eukaryotes interacts via charge complementarity with an acidic patch on neighboring nucleosomes, which has profound consequences in chromatin fiber formation18,19. The conservation of charge in the Hδ tail and acidic patch in Marseillevirus suggests it might be used for the same purpose. Another, not mutually exclusive possibility is that viral nucleosome-like particles are incorporated into the genome of the host amoeba to impact gene expression. Both possibilities require further investigation.
Methods
Phylogenetic inference
Protein sequences were subjected to microhomology alignment-based trimming to reduce the number of tandem repeats after initial multiple sequence alignments using ClustalW and MUSCLE algorithms from the MEGA X software (v10.1.8). This trimming was typically done on the N-terminal ends flanking the histone domain. For example, the amoeba H4 peptide sequence (GGKGL)x2 was reduced to one occurrence of (GGKGL)x1. Gaps were then adjusted so that they mostly did not occur in the alpha-helices determined by the structures reported here. The final alignment consisted of 193 columns. The alignment was then formatted as a nexus file and subjected to 1.8 million generations of paired metropolis-coupled MCMC Bayesian inference using MrBayes v3.2.7a (temperature setting of 0.08 and burn-in setting of 20%). This tree completed with an average standard deviation of split frequencies of <1% (0.01674), and had swap frequencies of 0.57, and 0.58. FigTree v1.4.4 was used to graph and annotate the resulting tree from the Bayesian phylogenetic inference (Extended Data Fig.1A).
Expression and Purification
Marseillevirus histones, Hδ-Hγ with a C terminal 6XHis tag and Hβ-Hα, were ordered from GeneScript and cloned into a single pet-24B vector using Gibson Assembly (NEB). The plasmid was transformed into Escherichia coli BL2-codon plus(DE3)-RIL competent cells (Agilent technologies). Growth occurred in LB media with 1x Kanamycin antibiotic. After reaching OD600 between 0.4–0.6, cell cultures were induced with 0.5 mM IPTG at 37 °C. After 3 hours, cells were harvested (Sorvall LYNX6000) and lysed (AvestinEmulsiflexC3) using lysis Buffer containing 20 mM Tris, pH 7.5, 2 M NaCl, 5 mM Imidazole, 1 mM BME. Purification using Ni-NTA agarose beads (Qiagen) was then performed (Elution Buffer: 20 mM Tris, pH 7.5, 2 M NaCl, 300 mM Imidazole, 1 mM BME). The protein sample was further purified using a Superdex200 26/600 size-exclusion chromatography column (GE Healthcare), and fractions were collected and concentrated to 6 mg/ml for further analysis (Buffer: 10 mM Tris, pH 7.5, 2M NaCl, 1 mM EDTA, 5 mM BME).
Purification of Widom 601 DNA
Widom 601 nucleosome positioning sequence20 was generated from a plasmid containing eight copies of the positioning sequence, each flanked by the EcoRV cutting site. This plasmid was transformed into DH5αTM (ThermoFisher) competent cells and grown in 2ÝT-Amp media overnight. The Widom 601 DNA fragments were excised using EcoRV and purified using previously published protocols21.
Nucleosome Assembly
Marseillevirus viral nucleosome was generated by combining purified Widom 601 DNA and purified Marseillevirus heterotetramer at a ratio of 1:1.8, respectively. After overnight salt gradient dialysis in buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM DTT, 2M to 0.25M KCl,) using Gilson Rapid Pump, the sample was run on a Superdex200 10/300 size-exclusion chromatography column (Buffer: 20 mM Tris, pH 7.5, 1 mM EDTA, 1 mM BME). Fractions containing the peak were collected and concentrated for further analysis.
Negative staining
Negative staining was performed based on the following protocol22. 3 μL of the sample was applied to a freshly glow-discharged carbon-coated grid. The grid was washed with two drops of deionized water and stained with two drops of 0.75% uranyl formate. After blotting off the excess liquid, the grid was vacuum aspirated and left to air dry for 20 min. The samples were viewed on an FEI Talos L120C TEM with Gatan 4k x 4k OneView camera at a magnification of 73,000x with a calibrated pixel size of 0.2 nm (Extended Data Fig.1D). The most promising sample judged by integrity and distribution of nucleosome particles was chosen for further cryo-EM experiments.
Cryo-EM sample preparation, data acquisition, and processing
Msv nucleosome sample at 15 μM in Buffer A (20 mM HEPES, pH 7.5; 1 mM EDTA; 2 mM DTT) was incubated with an equal volume of Buffer A containing 0.05% of glutaraldehyde. The crosslinking reaction was incubated for 1 hour at 4°C and then quenched by adding 100 mM Tris pH 7.5. The sample was dialyzed for 3 hours to Buffer A and concentrated to 3.3 mg/mL. Cryo-EM grids of the Msv nucleosome particle were prepared following established protocol23. 3 μL of the Msv nucleosome particle was applied to 30 s glow-discharged Quantifoil gold grids (400 mesh, 1.2 μm hole size), blotted for 3 s using Vitrobot Mark IV (FEI Company) at 4 °C and 100% humidity. Grids were plunge-frozen in liquid ethane. Images were collected on FEI Titan Krios 300kV (Extended Data Fig. 2A) equipped with Gatan K3 Summit camera at a nominal magnification of 64,000x and a calibrated pixel size of 1.09 Å using Leginon24. Each image was collected as a movie stack for 2.5s, fractionated into 50 subframes; every frame was exposed for 0.05 s and to 1.3 e/Å 2 electron dose, accruing the total accumulated exposure of 65 e/Å 2 (Supplementary Table 2). A total of 4,503 images were collected at a nominal defocus range of −1 μm to −2.4 μm. Movie stacks acquired in counting mode were corrected for global and local motion (in 5×5 patches) using UCSF MotionCor2 v1.2.125, resulting in dose-weighted and un-weighted sums and binned 2x using Fourier binning.
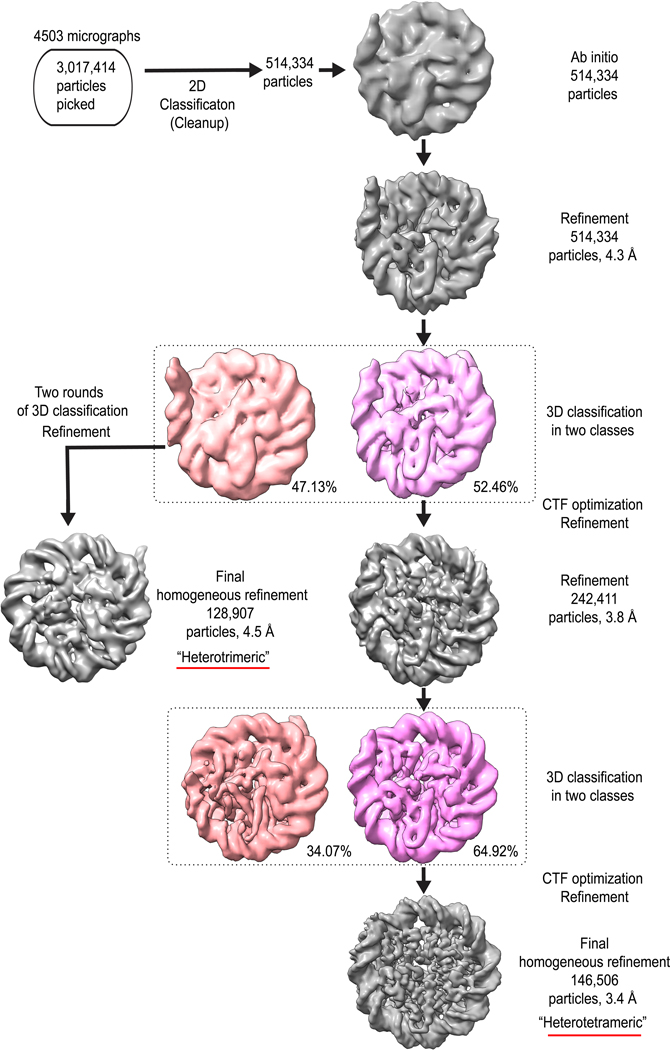
The resulting micrographs were processed in cryoSPARC26. The CTF parameters were estimated from the average of aligned frames with CTFFIND4 as implemented in cryoSPARC27. A small set of particles were picked manually (~1,000), then classified into 8 reference-free 2D classes. These 2D-classes were used for an automated template picker. Particles were then extracted and subjected to reference-free 2D classification into 50 classes. The “Ab initio” function was used to generate an initial 3D model. This resulted in a reconstruction with a clearly defined nucleosome density. Particles were then classified using heterogenous refinement. One of the classes contained the “octameric-like” viral heterotetramer structure, and another clearly was missing one of the “Hβ-Hα forced dimers” yielding “hexameric-like” viral heterotrimer structure (Extended Data Fig.3). For the viral heterotetramer class, a single round of heterogenous 3D classification, CTF optimization, and refinement produced the final 3.4 Å reconstruction; for the viral heterotrimer class, two rounds of 3D classification and refinement resulted in the final 4.5 Å reconstruction. The resolutions are reported at 0.143 Fourier Shell Correlation cutoff following gold-standard refinement28. The processing details and summaries are shown in Extended Data Fig. 2, 3, and Supplementary Table 2.
Model building and refinement.
The Msv nucleosome model was built using the reconstruction at 3.4 Å. Each of the Msv histones was split from the forced dimer and modeled based on homology alignment and 3D target-template (from coordinates PDB ID 1KX5), using the servers XtalPred29 and Swiss-model30. In addition, building was guided by the fitting of side chains to the density and the secondary structure elements determined by PSIPRED31. For DNA, we used the available X-ray crystal structure PDB ID 3LZ0 for initial rigid body fit into our cryo-EM map. DNA in 3LZ0 was then mutated to accommodate our 601 sequence (3TU4). The models of Msv histones were first manually fit into the map and then locally optimized using UCSF Chimera’s “Fit in map” function32. We then used Coot for local adjustments of secondary structure elements and side-chains into densities33. Linkers were built de novo manually using Coot. Model refinement was done using PHENIX (phenix.real_space_refine)34, beginning with only rigid body and then secondary structure, ADPs, rotamer and Ramachandran restraints in 100 iterations. The model was then visually inspected, and Ramachandran outliers and problematic regions were fixed manually in Coot (final refinement statistics are summarized in Supplementary Table 2). For the heterotrimeric Msv nucleosome, we used the heterotetrameric model docking into the cryo-EM reconstruction at 4.5 Å using Chimera, removing the missing chains of the Hβ-Hα and the part of the DNA that was missing in the reconstruction in Coot. The DNA that was protruding on one side was modeled using the tetranucleosome structure (PDB ID: 1ZBB). Chimera, Coot, and PyMOL were used in preparing figures of the model and cryo-EM densities32,33,35. Validation details are shown in Supplementary Table 2.
Nucleosome crosslinking for mass spectrometry.
Amine-specific crosslinking of histones within the nucleosome was done with bis(sulfosuccinimidyl)suberate (BS3, ThermoFisher Scientific). 20-μl reactions containing 0.75 mg/ml nucleosomes in crosslinking buffer (20 mM Hepes pH 7.5, 1 mM EDTA and 1 mM DTT) and 0.5–2 mM BS3 were incubated on ice for 2 hours before they were quenched with Tris-HCl pH 7.5 added to 50 mM from 0.5 M stock. After 10-min incubation with Tris, proteins were precipitated with acetone: 4 vol of acetone (cooled at –20 °C) were added, followed by 1-hour incubation at –20 °C. Protein precipitates were collected by 10-min centrifugation at 16,000g at room temperature. Pellets were rinsed with 80% acetone, air dried, and dissolved in 9 μl of buffer containing 50 mM ammonium bicarbonate, 10 mM DTT, and 8 M urea. Following a 10-min incubation at room temperature, samples were mixed with 1 μl 0.5 M iodoacetamide and left for 30 min in the dark. For digestion, 10-μl samples were diluted with 100 μl 50 mM ammonium bicarbonate, 5 mM DTT containing 10 ng/μl trypsin/LysC mixture (Promega) and incubated overnight at 25 °C. Digestion reactions were stopped by mixing with 10 μl 20% heptafluorobutyric acid and clarified by 10-min centrifugation at 16,000g. Finally, peptides were desalted using C18 OMIX tips (Agilent) according to the manufacturer’s protocol, dried under vacuum, and dissolved in 15 μl 0.1% formic acid.
Mass spectrometry
For LC-MS analysis of peptides, Dionex UltiMate 3000 chromatography system together with Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific) were used. Peptides were resolved on 50-cm long EASY-Spray reverse phase C18 column (Thermo Scientific) at a flow rate of 150 nl/min using 120-min linear gradient from 96% buffer A (0.1% formic acid in water) to 40% buffer B (0.1% formic acid in acetonitrile) followed by step elution with 98% buffer B over 5 min. Each cycle of data-dependent acquisition included recording full MS scan (orbitrap mass analyzer, resolution 60,000) followed by MS/MS scans (orbitrap mass analyzer, resolution 15,000) for 20 topmost abundant precursors of charge 4 to 6 fragmented by HCD with normalized energy 35. Monoisotopic precursor selection was enabled, quadrupole isolation window was set to 2 m/z, and dynamic exclusion window was set to 30 sec.
Mass spectrometry data processing and analysis
Raw data was processed with pLink236 using database of concatenated sequences of proteins constituting nucleosome and common contaminants. For peptides identification, up to 3 missed trypsin cleavages were allowed, variable modification with oxidation at methionine residue and carbamidomethyl at cysteine residue, cross-linker was set to BS3, FDR level was set to 1%, precursor and fragment tolerance were set to 10 ppm, and 50 ppm, respectively. Other parameters were left unchanged. A list of identified crosslinked peptides was imported into R environment for statistical computing and filtered to include only those with reported Q-value below 0.01. Crosslinking sites confirmed by at least two peptide-spectrum matches were considered for further analysis.
Reporting Summary statement
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability:
Crosslinking data are available from PRIDE via ProteomeXchange with identifier PXD024220. Cryo-EM density maps are deposited in the Electron Microscopy Data Bank, under accession numbers EMD-23529 and EMD-23530. The atomic model is deposited in the PDB, under accession numbers PDB 7LV8 and PDB 7LV9. All other data are available in the main manuscript or extended data section.
Extended Data
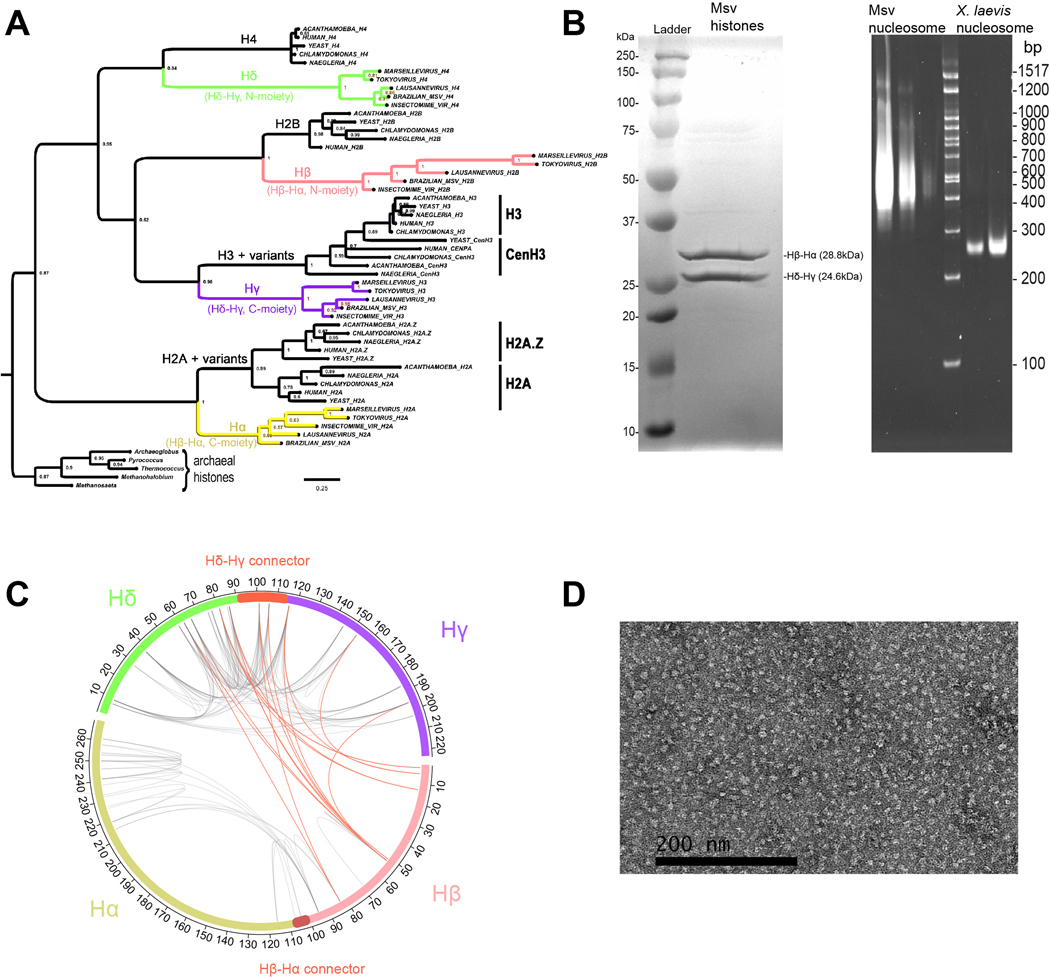
Extended Data Fig. 1. Phylogenetic and structural characterization of Marseillevirus histones and their higher-order assemblies.
a, A protein sequence alignment based on the Marseillevirus histone heterotetrameric structure was used to infer the phylogenetic relationships of the separate histone moieties (N- and C- terminal halves) from each the two histone doublet genes from three different Marseilleviridae genomes. This tree shows that each of the four viral histone domains (Hβ and Hα from the Hβ-Hα gene, and Hδ and Hγ from the Hδ-Hγ gene) is related to one of the four canonical eukaryotic core histone families. Furthermore, the C-terminal halves of each viral histone doublet are distantly related to eukaryotic core histone sub-families that come in distinctive functional variants (Hα → H2A + variants, and Hγ → H3 + variants). This phylogeny was computed using Bayesian inference on 193 alignment columns (Materials & Methods). Histone domains encoded by various archaeal genomes were used as an outgroup clade to determine root placement. b, SDS-PAGE gel of Msv histones stained with Coomassie blue (left) and PAGE native gel with different dilutions of Msv and Xenopus nucleosome assemblies stained with ethidium bromide (right). Each sample was run independently twice (n=2). c, Crosslinking mass spectrometry revealed intramolecular (gray lines) and intermolecular (red lines) contacts between histones. d, Negative stain electron microscopy image of glutaraldehyde crosslinked viral histone assemblies. This image shows presence of round, nucleosome shaped particles.
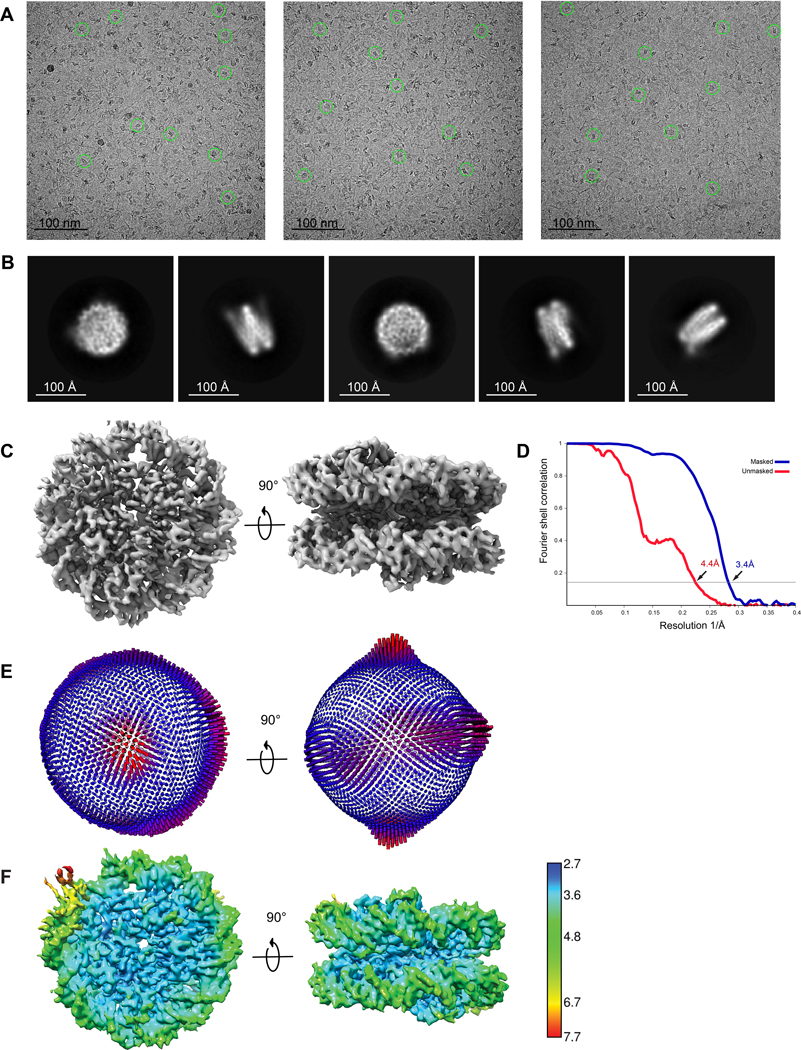
Extended Data Fig. 2. Cryo-EM analysis of sample of Marseillevirus nucleosome.
Data were collected on Titan Krios 300kV microscope. a, Raw cryo-EM images of Msv nucleosome were collected as described in Materials and Methods. Individual particles are highlighted in green circles. b, Representative 2D class averages selected from the dataset. c, Two representative views of the 3.4 Å 3D reconstruction Msv nucleosome. d, Fourier Shell Correlation plot of the 3.4 Å Msv nucleosome between two independently refined half maps (measured at FSC=0.143). e, Euler angle distribution of assignment of particles used to generate the final 3.4 Å reconstruction. The length of every cylinder is proportional to the number of particles assigned to the specific orientation. f, Two different views of the heat map of the Msv nucleosome cryo-EM density colored by local resolution.
Extended Data Fig. 3. 3D Classification scheme of samples of Marseillevirus nucleosome.
Detailed summary of classification of Msv dataset collected on Titan Krios operated at 300 kV. The two classes from the first 3D classification were further processed to leading to the “heterotetrameric” and “heterotrimeric” Msv nucleosomes.
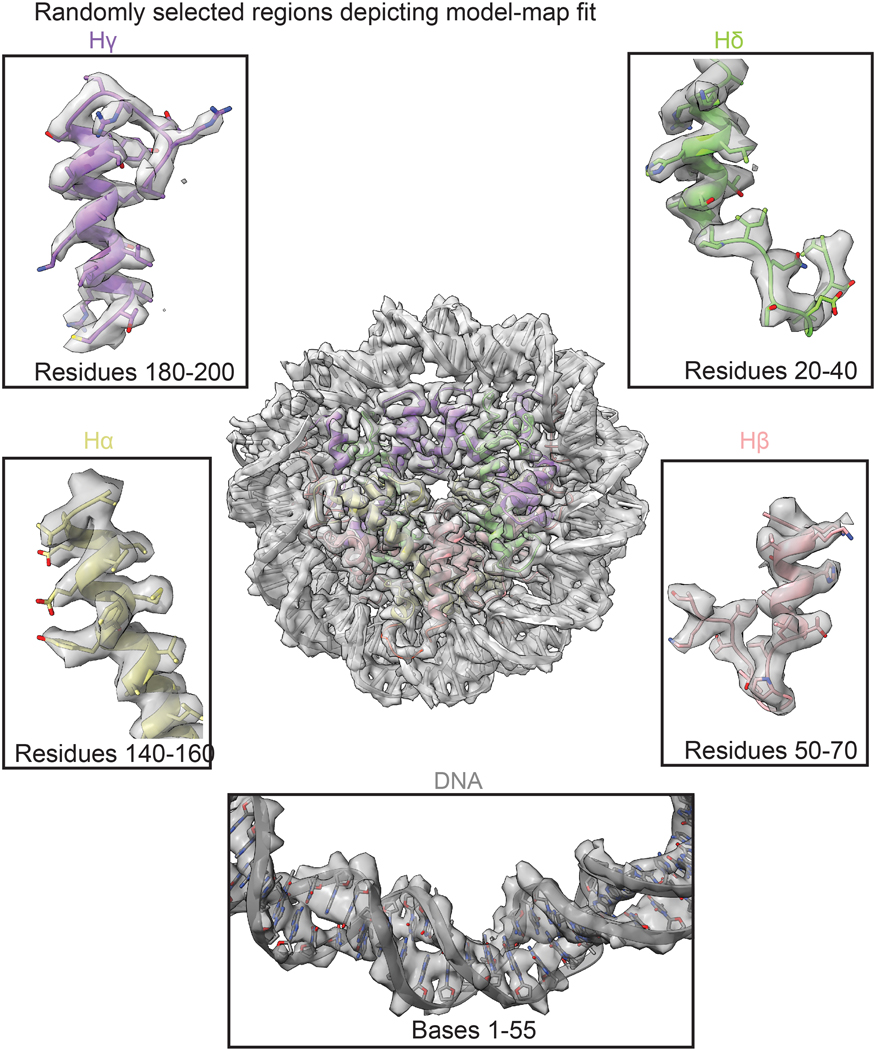
Extended Data Fig. 4. Gallery of the Coulomb map showing the fitting of the model to the density in some regions of the reconstruction.
Fitting of the model to the Cryo-EM density of different regions of the Msv histones Hα, Hβ, Hγ, Hδ. The model is color coded as in Fig. 1.
Extended Data Fig. 5. Specific contacts of Msv histone forced dimers with the DNA.
a, Cryo-EM density of Msv nucleosome at two different thresholds showing the flexibility of the DNA ends in the Msv nucleosome. Two different views of the Coulomb map at contouring level of 6.5 sigma (top) and 5 sigma (bottom). At a lower contouring, the full length of the DNA (147 bp) is still visible but only 121 bp are stabilized and coordinates can be assigned. The map is color coded as in Fig. 1. b, Close up of the model and cryo-EM map fitting of specific contacts between the Hδ-Hγ connector and the DNA. c, Overview of the Hβ-Hα DNA interface. d, Comparison of interactions between Hβ-Hα with DNA (left) and H2B and H2A with DNA (right). e, Interactions of Hδ-Hγ with the DNA (left) and H3 and H4 with the DNA (right). Human nucleosome (PDB ID 5AV9).
Extended Data Fig. 6. Sequence conservation between Msv and eukaryotic histones.
Sequence alignment depicting secondary structures of Msv, human and A. castellanii histones. The secondary structure is shown for Msv and Human histones only. No structural data is available for A. castellanii. Shades of green indicate identical (dark green) and similar (light green) residues. Sequence alignments were performed with Clustal Omega from UniPort. A summary of structural and sequence alignments is shown in Extended Data Fig. 10.
Extended Data Fig. 7. Msv “heterotrimeric” nucleosome.
a-e, Model of the “heterotrimeric” Msv nucleosome with modeled DNA fitting inside the “heterotrimeric” Msv cryo-EM map. a-c, Three different views, d, close up of the region where DNA is missing and e, close-up of the region where DNA is protruding. f-j, Model of the “heterotetrameric” Msv nucleosome fitting inside the “heterotetrameric” Msv cryo-EM map. f-h, Three different views, i and j, close ups of the regions shown in d and e. k-o, Model of the “heterotrimeric” Msv nucleosome without the modeled DNA fitting inside the “heterotrimeric” Msv cryo-EM map. k-m, Three different views, n and o, close ups of the regions shown in d and e. p-t, Docking of a model of a “hexameric” eukaryotic nucleosome (PDB ID 5AV9) produced taking off one of the dimers H2B and H2A and fitted inside the “hexameric” Xenopus cryo-EM map EMD-3929. Please note that we did not rebuilt the DNA in this model. p-r, Three different views, s and t,. close ups of the regions shown in d and e. Maps and models are color coded as Fig. 1. The blue square shows the missing DNA region and the arrow the missing Hβ-Hα (or H2B and H2A) histones.
Extended Data Fig. 8. Differences between Msv and eukaryotic histone tails.
Comparison of the N-terminal tails of the Msv nucleosome with the Xenopus nucleosome (PDB ID 1KX5) for: a, N-terminal Hα (top left) vs H2A (top right), C-terminal Hα (bottom left) vs H2A (bottom right). b, N-terminal Hβ vs H2B, c, N-terminal Hγ vs H3 and d, N-terminal Hδ vs H4. The models are color coded as in Fig. 1. The histone tail residues are indicated.
Extended Data Fig. 9. The contacts between Msv Hγ and DNA ends.
Comparison of interactions between DNA terminus with a, Hγ in Msv, b, canonical H3 in human (PDB ID 5AV9) and c, CENP-A in centromeric human nucleosomes (PDB ID 3AN2). d-e, Overview (top) and close-up (bottom) of the contacts established between Hδ α1 helix and Hγ αN helix of the d, Msv and e, human nucleosome (PDB ID 5AV9). Differences in the interface between Msv Hδ α1 helix with Hγ αN helix when compared to H4 α1 helix with H3 αN helix could contribute to increased flexibility of the DNA ends in the Msv nucleosome. The nucleosome is color coded as in Fig. 1.
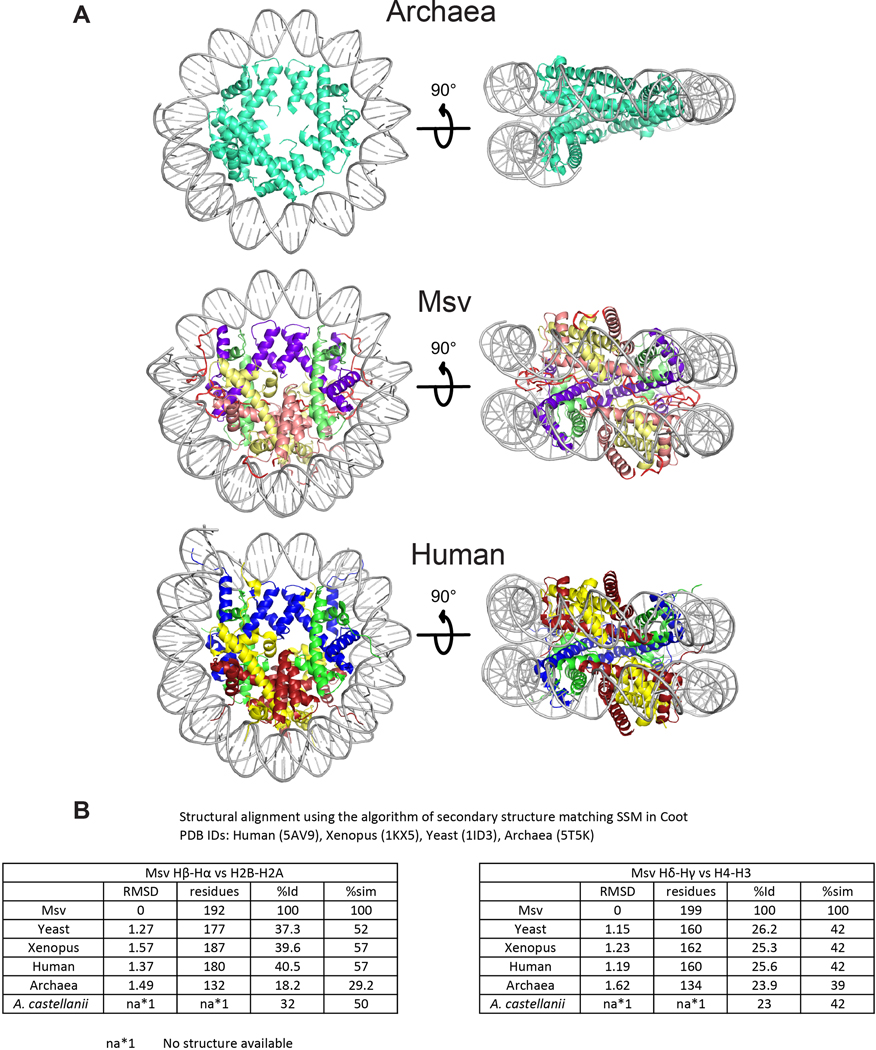
Extended Data Fig. 10. Comparison between Msv, eukaryotic and archaeal nucleosomes.
a, Two different views of (top) archaea (PDB ID 5T5K), (middle) Msv and (bottom) human (PDB ID 5AV9) nucleosomes. b, RMSD and sequence conservation between histones in different organisms. Structural alignment and identity were obtained using the SSM algorithm in Coot. All the values of similarity and the sequence alignment with A. castellanii were obtained with BLASTp from NCBI. Only one homodimer from Archaea was possible to be aligned to the heterodimers for other organisms.
Supplementary Material
Acknowledgments
We thank Dr. William Rice and Dr. Bing Wang for helping with data collection at NYU cryo-EM Shared Resource. We thank NYU Microscopy Laboratory for helping with negative stain microscopy. We thank the HPC Core at NYU Langone Health for computer access and support. We thank Dr. Jean-Paul Armache for the feedback. This work in the Armache laboratory was supported by grant from the David and Lucile Packard Foundation. S.A-A is supported by Molecular Biophysics T32 grant (5T32GM088118). E.N. and N.V. are supported by the NIH grant R01 GM127267, Blavatnik Family Foundation, and by the Howard Hughes Medical Institute. P.T. and S.H. are supported by the Howard Hughes Medical Institute. Some of this work was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310) with additional support from Agouron Institute (F00316), NIH (OD019994) and NIH (RR029300).
Footnotes
Competing interests: The authors declare no competing interests.
REFERENCES
- 1.Talbert PB, Meers MP & Henikoff S. Old cogs, new tricks: the evolution of gene expression in a chromatin context. Nat Rev Genet 20, 283–297 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–71 (1974). [DOI] [PubMed] [Google Scholar]
- 3.Luger K, Mader AW, Richmond RK, Sargent DF & Richmond TJ Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–60 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Mattiroli F. et al. Structure of histone-based chromatin in Archaea. Science 357, 609–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowerman S, Wereszczynski J. & Luger K. Archaeal chromatin ‘slinkies’ are inherently dynamic complexes with deflected DNA wrapping pathways. Elife 10(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yutin N, Wolf YI, Raoult D. & Koonin EV Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6, 223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aherfi S, Colson P. & Raoult D. Marseillevirus in the Pharynx of a Patient with Neurologic Disorders. Emerg Infect Dis 22, 2008–2010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arantes TS et al. The Large Marseillevirus Explores Different Entry Pathways by Forming Giant Infectious Vesicles. J Virol 90, 5246–55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer M. et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A 106, 21848–53 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erives AJ Phylogenetic analysis of the core histone doublet and DNA topo II genes of Marseilleviridae: evidence of proto-eukaryotic provenance. Epigenetics Chromatin 10, 55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakamori M. et al. Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation. Sci Rep 5, 17204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilokapic S, Strauss M. & Halic M. Histone octamer rearranges to adapt to DNA unwrapping. Nat Struct Mol Biol 25, 101–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taverna SD, Li H, Ruthenburg AJ, Allis CD & Patel DJ How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14, 1025–1040 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tachiwana H. et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476, 232–5 (2011). [DOI] [PubMed] [Google Scholar]
- 15.McGinty RK & Tan S. Recognition of the nucleosome by chromatin factors and enzymes. Curr Opin Struct Biol 37, 54–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valencia-Sanchez MI et al. Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination. Mol Cell 74, 1010–1019 e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armache KJ, Garlick JD, Canzio D, Narlikar GJ & Kingston RE Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science 334, 977–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorigo B, Schalch T, Bystricky K. & Richmond TJ Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol 327, 85–96 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Shogren-Knaak M. et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–7 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Lowary PT & Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Dyer PN et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol 375, 23–44 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Ohi M, Li Y, Cheng Y. & Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online 6, 23–34 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10, 584–90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suloway C. et al. Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Rohou A. & Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenthal PB & Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–45 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Slabinski L. et al. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics 23, 3403–5 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Schwede T, Kopp J, Guex N. & Peitsch MC SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 31, 3381–5 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGuffin LJ, Bryson K. & Jones DT The PSIPRED protein structure prediction server. Bioinformatics 16, 404–5 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–12 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–32 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8. (2015). [Google Scholar]
- 36.Chen ZL et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat Commun 10, 3404 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crosslinking data are available from PRIDE via ProteomeXchange with identifier PXD024220. Cryo-EM density maps are deposited in the Electron Microscopy Data Bank, under accession numbers EMD-23529 and EMD-23530. The atomic model is deposited in the PDB, under accession numbers PDB 7LV8 and PDB 7LV9. All other data are available in the main manuscript or extended data section.