Abstract
The research on human gut microbiome, regarded as the black box of the human body, is still at the stage of infancy as the functional properties of the complex gut microbiome have not yet been understood. Ongoing metagenomic studies have deciphered that the predominant microbial communities belong to eubacterial phyla Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, Cyanobacteria, Verrucomicrobia and archaebacterial phylum Euryarchaeota. The indigenous commensal microbial flora prevents opportunistic pathogenic infection and play undeniable roles in digestion, metabolite and signaling molecule production and controlling host’s cellular health, immunity and neuropsychiatric behavior. Besides maintaining intestinal health via short-chain fatty acid (SCFA) production, gut microbes also aid in neuro-immuno-endocrine modulatory molecule production, immune cell differentiation and glucose and lipid metabolism. Interdependence of diet and intestinal microbial diversity suggests the effectiveness of pre- and pro-biotics in maintenance of gut and systemic health. Several companies worldwide have started potentially exploiting the microbial contribution to human health and have translated their use in disease management and therapeutic applications. The present review discusses the vast diversity of microorganisms playing intricate roles in human metabolism. The contribution of the intestinal microbiota to regulate systemic activities including gut–brain–immunity crosstalk has been focused. To the best of our knowledge, this review is the first of its kind to collate and discuss the companies worldwide translating the multi-therapeutic potential of human intestinal microbiota, based on the multi-omics studies, i.e. metagenomics and metabolomics, as ready solutions for several metabolic and systemic disorders.
Graphic abstract
Keywords: Gut microbiome, Short chain fatty acids, Xenobiotic degradation, Immune system and gut–CNS axis, Therapeutic applications
Introduction
Human gut microbiome refers to the total microbial population in the human gastrointestinal tract (GIT) that includes bacteria and other microorganisms. The human body is the host to approximately 500–1000 species of gut microbes that encode 100-fold more unique genes than the host human genome (Ley et al. 2006). The huge diversity of gut microbiota remained unknown until culture independent high-throughput sequencing technology and powerful analytical and bioinformatics tools were developed. With the development and advancement of next generation sequencing technology, the long unexplored black box of the human body, i.e. the gut microbiome came into light and a deeper understanding of the same was designated as the second human genome (Mosca et al. 2016). Although the first one is inherited from the parents of an individual and is highly stable, the second genome is adapted from the environment after birth and remains continuously dynamic (D’Argenio and Salvatore 2015). The second genome, i.e. the acquired microbiome is therefore, highly dependent and subject to change upon several environmental factors, such as i) host genotype and ethnicity, ii) age and sex, iii) diet, iv) hormonal cycles, v) illness and therapies, and vi) travel history. Establishment of a microbial community is therefore, consequent to varied range of ecological interactions classified under five different heads: i) mutualism—beneficial interaction for both the partners), ii) amensalism—one impacts the other negatively, iii) commensalism—one impacts the other positively, iv) competition—both the competitors harm each other, v) predation and parasitism—one gets benefitted out of the other (Faust and Raes 2012; Mosca et al. 2016).
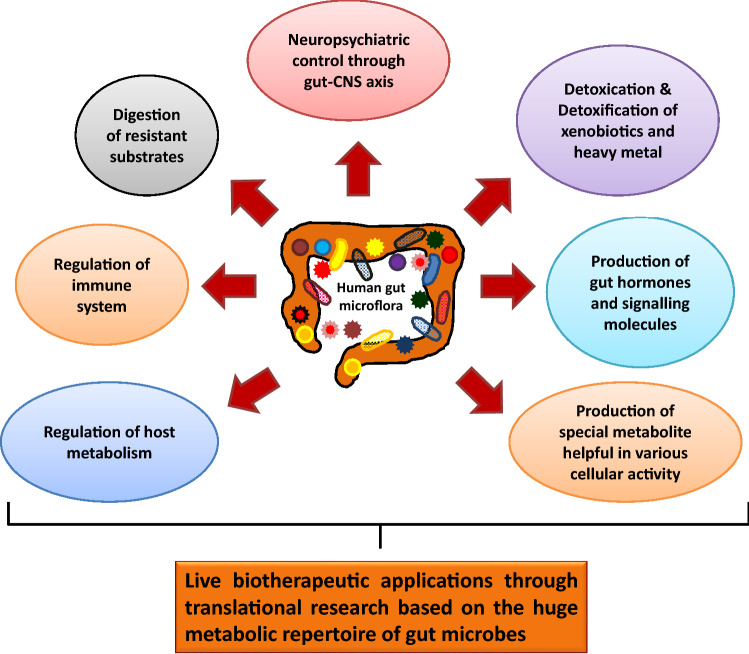
Mounting evidences indicate that the trillions of bacteria and archaea residing in the human gut are extensively related to host health regulating various metabolisms, such as fermentation and digestion of various biomolecules, xenobiotic degradation and heavy metal biotransformation, production of various immune regulators and special metabolites, energy production, epithelial homeostasis, short-chain fatty acid (SCFA) production and so on (Bäckhed et al. 2015; Li et al. 2019). The gut microbiome comprising thousands of bacterial species interact and crosstalk with the host cells in various ways and comprehensively with the host genome to determine human health. Disturbed ecology of the gut microbiome has been frequently reported to be associated with various human diseases, such as obesity and metabolic syndromes, non-alcoholic fatty lever disease, coronary heart disease, irritable bowel syndrome, inflammatory bowel disorders, allergy, asthma, etc. (Holmes et al. 2011; Mosca et al. 2016). Extensive research on gut microbiome through various alterations have shown that the secondary metabolites produced by the bacterial flora can signal the activity of distal tissues, such as liver, brain, muscle and adipose tissues. A healthy and normal microbiota not only maintains the gastrointestinal activities, but also controls various central nervous systemic activities, perturbation of which might trigger neuropsychiatric effects, such as anxiety, depression, schizophrenia and autism (Yarandi et al. 2016). The nature of microbial composition of human gut is increasingly being documented through various microbiome projects, such as the International Human Microbiome Consortium, the European Commission’s Metagenomics of the Human Intestinal Tract Project, the US National Institutes of Health’s Human Microbiome Project, the Human Microbiome Project (HMP) the Human Gastrointestinal Bacteria Genome Collection (HGG) and the Canadian Microbiome Initiative which finally converge into a single global network (Bäckhed et al. 2015; Gevers et al. 2012; Huttenhower et al. 2012). Several recent research findings that are intriguing yet very promising could pave the way for filling the knowledge gaps in microbiome–host interactions and their role in disease pathogenesis as well as potential therapeutic applications.
In view of the existing knowledge on gut microbiome the present review discusses an integrated approach to understand the diversity of the complex black box of our body and the specific roles of particular bacterial taxa related to human metabolism under varying environmental or physiological conditions. It further discusses as to how the extraordinarily diverse microbial community participates in the functioning of innate and adaptive immune responses and is also involved in the crosstalk with central nervous system where brain commands several gut functions like mucin production, peristalsis and gut immune functions. The potential use of gut microbiota by several start-up companies for therapeutic applications and commercialization is also discussed.
Diversity in the gut microbiome
Microbial colonization on the human body starts immediately following birth and the community composition is shaped by various environmental factors. Various molecular techniques have been employed to analyze the microbial diversity and abundances (Table 1). The infant gut microbiota is majorly predominated by members of Actinobacteria, Proteobacteria, Firmicutes and Bacteroidetes and the abundance of the phylum detected are according to the order of their appearance. However, in adults, the abundance shifts to Firmicutes followed by the members of Bacteroidetes and Actinobacteria. Although members of Proteobacteria, Fusobacteria, Cyanobacteria and Verrucomicrobia are present in adults, but are less represented (Eckburg 2005; Ley et al. 2006; Winston and Theriot 2020; Zhernakova et al. 2016). Trillions of bacteria belonging to thousands of different species majorly fall under the above mentioned phyla (D’Argenio and Salvatore 2015; Li et al. 2008). Human metabolic phenotypes are found to be modulated by the symbiotic intestinal gut microbiome. Moreover, their ratio of abundance might act as biomarkers indicating sex, age or diseased conditions. Li et al. (2008) in their study showed that a low ratio of Bacteroidetes to Firmicutes correlate with obesity and could be elevated by restricting the dietary calorific intake.
Table 1.
List of microbiological, biochemical and molecular techniques applied to analyze the microbiome components of an environment
| Sl. No | Technique | Principle | References |
|---|---|---|---|
| Microbiological and Biochemical Techniques | |||
| 1 | Plate counts | Direct cultivable bacterial count upon growth on media plates based on morphological differences | Kirk et al. 2004 |
| 2 | Community Level Physiological Profiling (CLPP) | Physiological profiling based on sole carbon source utilization properties of microbial communities helpful in detecting copiotrophic organisms | Lladó and Baldrian 2017 |
| 3 | Fatty Acid Methyl Ester analysis (FAME) | Gas chromatographic analysis of cellular fatty acids through | Ghosh et al. 2020 |
| 4 | Guanine plus Cytosine (GC) | GC content of the genomic DNA, a taxon level characteristic feature, is analyzed through melting temperature curve of DNA renaturation studies | Kirk et al. 2004 |
| Molecular Techniques | |||
| 1 | 16S rDNA sequencing approach |
a. 16S rRNA gene amplification, preparation of clone libraries (for metagenomic samples), forming operational taxonomic units (OTUs) upon Amplified Ribosomal DNA Restriction Analysis (ARDRA), selection of representative members and sequencing b. 16S rRNA gene amplicon library preparation, next generation high-throughput sequencing, OTU formation and bioinformatic analysis for phylogenetic classification |
Ghosh and Sar 2013 Dutta et al. 2018 |
| 2 | Polymorphism based techniques |
a. Denaturing Gradient Gel Electrophoresis (DGGE): 16S rRNA gene amplicon from bacterial genomic DNA differing in sequence composition is resolved electrophoretically based on their difference in denaturation at increasing concentrations of the denaturant in the gel b. Temperature Gradient Gel Electrophoresis (TGGE): Electrophoretic separation of 16S rRNA gene amplicon based on the varying melting temperature on a temperature gradient gel c. Amplified ribosomal DNA restriction analysis (ARDRA) or restriction fragment length polymorphism (RFLP) Terminal restriction fragment length polymorphism (T- RFLP): Polymorphism of restriction sites of 16S rRNA gene for a particular restriction enzyme is utilized to differentiate the microbial communities d. Single strand conformation polymorphism (SSCP): Separation based on the electrophoretic mobility of secondary structures formed out of DNA single strands under non-denaturing conditions |
Kirk et al. 2004 Theron and Cloete 2000 Schwieger et al., 1998 |
| 3 | Nucleic acid reassociation and hybridization techniques |
a. DNA-DNA hybridization: Genomic DNA hybridization with known microbial taxa to analyze distinctness of the taxon to species level with 70% as cornerstone b. DNA microarray: Microscopic DNA slides with spots of known DNA sequences to which the unknown DNA sequences are hybridised and fluorescence measured c. DNA Reassociation: Lower the DNA reassociation kinetics of a microbial community, higher is the diversity d. Reciprocal Hybridization of Community DNA: Reciprocal hybridization of total community DNA indicates the presence of same kinds of organisms in two samples based on the idea that only identical or very closely related species would show significant cross-hybridization of pure culture DNA |
Theron and Cloete 2000 Cho and Tiedje 2001 |
| 4 | Ribosomal Intergenic Spacer Analysis (RISA)/Automated Ribosomal Itergenic Spacer Analysis (ARISA) | DNA fingerprinting technique based on the amplification of the intergenic region between 16 and 23S rRNA genes in the rRNA operon. Different microbial taxon has characteristic and significant variability in the length and nucleotide sequence of this region. ARISA is an updated and more efficient technique to get high-resolution data involving a fluorescence-tagged oligonucleotide primer for PCR amplification and subsequent electrophoresis in an automated system | Ciesielski et al. 2013 |
| 5 | Flow cytometry | Flow cytometry conjugated with fluorescence-activated cell sorting (FACS) relying on fluorescent dyes for detection helps quantify and fractionate complex bacterial communities | Park et al. 2005 |
| 6 | Fluorescence In Situ Hybridization (FISH) | Fluorescently labelled DNA probes are used to target rRNA of defined taxonomic or phylogenetic groups for microbial identification. Recently, an updated technique named live-FISH combined with FACS has been developed to sort specific taxonomic groups of bacteria and culture them for their further taxon level identification | Batani, et al. 2019 |
DGGE fingerprint study to identify the key OTUs of specific and dominant intestinal bacterial groups provided a deeper level understanding (Li et al. 2008). The major OTUs were found to be predominated by Bacteroides coprocola, B. thetaiotaomicron and B. uniformis. These were shown to be important metabotypes which displayed correlation with various urinary metabotypes (Li et al. 2008). Studies on gut microbiome in an European population through fluorescence in situ hybridization (FISH) and flow cytometry is also found to be sex dependent as gender specific differences in ratio of Bacteroides to Prevotella were found to be higher for males than females (Mueller et al. 2006). Among these, B. thetaiotaemicron was found to be more abundant in males than in females which may serve as a potential marker for sex-discrimination (Li et al. 2008). Another study documented the prominent differences in the microbial community composition in obese and non-obese individuals through metagenomic high-throughput sequencing (Chatelier et al. 2013). Individuals with low gene count (LGC), i.e. < 480,000 genes were found to be associated with inflammatory bowel disorder (IBD), inflammation and obesity and were detected with low richness of gut microbiota. Others were considered as high-gene count (HGC) group, i.e. with > 480,000 genes who were non-obese and with little reported gut disorder. They could detect that 46 bacterial genera differed significantly in abundance. The phylogenetic shift showed an abundance of phyla Proteobacteria and Bacteroidetes encompassing genera Bacteroides, Parabacteroides, Ruminococcus (R. torques, R. gnavus), Campylobacter, Dialister, Porphyromonas, Staphylococcus and Anaerostipes in LGC individuals. Concurrently, HGC individuals were found to be highly associated with members of phyla, Verrucomicrobia, Actinobacteria and Euryarchaeota with genera Faecalibacterium, Bifidobacterium, Lactobacillus, Butyrivibrio, Alistipes, Akkermansia, Coprococcus and Methanobrevibacter. Evidently, high richness of the gut microbiome diversity is associated with healthy host metabolism.
Another factor that plays the most crucial role in maintaining the diversity of the bacterial taxa found in gut microbiome is the presence of predator bacterial communities. Predators are the key species that limits the population of the dominant species, preventing it from overgrowing and building up a high biomass which will directly hamper the species diversity. Mathematical simulation based on Lotka Voltera model suggested that the predators can drastically reduce the population of their preys helping in maintenance of the species diversity (Mosca et al. 2016). One of the best-known predatory bacteria are the members of Bdellovibrio and Bdellovibrio—like organisms (BALO). These have shown the best sensitivity towards gram-negative bacteria like Salmonella enteritidis and Pseudomonas fluorescens and have also been found to impact the growth of gram-positive Staphylococcus aureus. Besides, predation upon dominant species of the gut microbiome has also been reported through protists and bacteriophages.
A mini microbiome was constructed considering reference genomes in level of phyla obtained from Integrated Microbial Genomes–Human Microbiome Project (IMG/HMP) online platform was built to represent a healthy adult gut microbiome (Kaoutari et al. 2013). A list of 177 genomes was predominated by the members of Firmicutes (n = 104), Bacteroidetes (mostly Bacteroides spp. n = 29), Proteobacteria (n = 22) and Actinobacteria (n = 12). The mini microbiome also contained members of Fusobacteria (n = 2) and Cyanobacteria, Elusimicrobia, Lentisphaerae, Spirochaetes, Synergistetes, Tenericutes, Thermotogae and Verrucomicrobia, (n = 1 of each phylum).
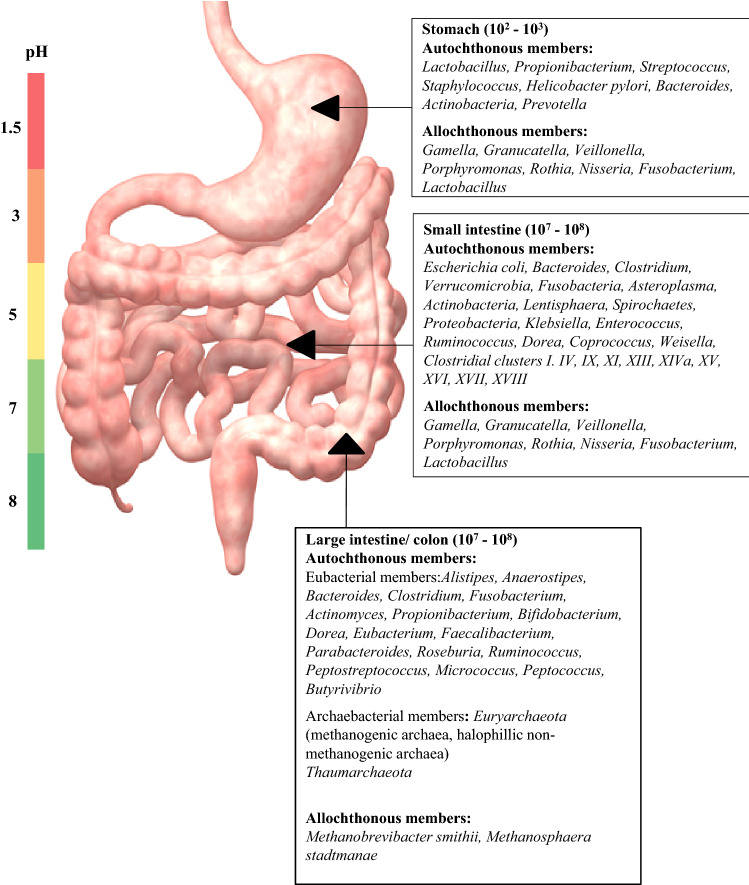
The human gut is segregated and partitioned so that the first shot of digestive activity on the food ingested is the host’s own. The gut is partitioned into stomach, duodenum, jejunum, ilium and colon, the environment of which is in the ascending order of pH and anaerobic from stomach to colon. The stomach is highly acidic (pH ~ 1.5) and was considered as sterile until Helicobacter pylori was discovered to be able to survive this hostile environment (Marshall and Warren 1984). Later investigations on possibility of existence of other microbial life in this acidic environment revealed that the gastric fluid was predominated by the members of Firmicutes, Bacteroidetes and Actinobacteria (Minalyan et al. 2017). The gastric mucosa was rather found to be richer in diversity with bacterial members belonging to Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria and Actinobacteria. In most individuals Streptococcus and Prevotella were found to be predominating apart from H. pylori. Other allochthonous species were found to belong to Gamella, Granulicatella, Veillonella, Porphyromonas, Rothia, Neisseria, Fusobacterium and Lactobacillus (Walter and Ley 2011). The microbial biomass in the gastric environment remains up to 102–3 cells/ml. This increases up to 108 cells/ml in the small intestine (SI). The SI mucosa is associated with members of phyla, Bacteroidetes and Firmicutes. The SI is partitioned into three sections—the duodenum with pH 5–7 and bacterial load of 103–4 cells/ml where Gram positive aerobes predominate, followed by jejunum and ileum with pH 7–9 and cell density of 104–8 cells/ml comprising strict to facultative anaerobic Gram-positive and Gram-negative bacteria. The microbiome composition of the intestinal lumen, known as mucosal and epithelial spaces of the gut (Swidsinski et al. 2005), is highly diverse and comprises members of Verrucomicrobia, Fusobacteria, Asteroplasma, Cyanobacteria, Actinobacteria, Lentisphaera, Spirochaetes, Bacteroidetes, Proteobacteria, Bacilli, Clostridial clusters I, IV, IX, XI, XIII, XIVa, XV, XVI, XVII, XVIII and uncultured Clostridiales and Mollicutes (Zoetendal et al. 2012). The predominating genera are E. coli, Klebsiella, Enterococcus, Bacteroides, Ruminococcus, Dorea, Clostridium, Coprococcus, Weisella and Lactobacillus. Allochthonous populations include Granulicatella, Streptococcus, Veilonella and Lactobacillus. The large intestine (LI) or the colon is strictly anaerobic with pH varying from 5 to 7. This area of the gut is the fermentation hub where various amino acids and butyrate–an SCFA, are fermented and production of various phenolic and indolic compounds take place (Smith and MacFarlane 1996, 1998). This compartment of the gut is home to the most complex bacterial diversity where the cell density reaches 1011 cells/ml. The high-bacterial diversity and abundance in the LI is due to several factors, such as i) larger volume, ii) moderate or less acidic pH, iii) low concentration of biliary salts and iv) longer retention time due to relatively slower peristalsis. Five major phyla- Firmicutes, Bacteroidetes, Actinobacteria, Verrucomicrobia and Proteobacteria covering a wide range of bacterial genera Clostridium, Fusobacterium, Bacteroidetes, Actinomyces, Propionibacterium are associated with the LI. Other Gram-positive cocci- micrococci, peptococci, peptostreptococci and ruminococci have been also reported to play crucial roles in the LI (Ramakrishna 2013; Walter and Ley 2011). These are majorly responsible for the SCFA production, i.e. acetate, butyrate and propionate. The specific bacteria responsible for particular SCFA synthesis have been discussed later in this review.
Apart from eubacterial members, archaebacterial members also constitute the LI microbiota with Methanobrevibacter smithii and Methanosphaera stadtmanae as the predominating species. M. smithii alone makes 10% of the colonic anaerobic bacterial population (Walter and Ley 2011; Jhangi et al. 2014) (Fig. 1). Archaeal growth is syntrophic with eubacterial H2, C, acetate, formate or methanol production in mammalian gut where these are incorporated as precursor materials by methanogens in their methanogenesis and energy production process playing a crucial role in energy balance (Hoffmann et al. 2013; Matarazzo et al. 2012; Ishaq et al. 2015). In contrast to these species, Methanomassiliicoccus luminyensis was found to significantly increase with age having remarkable metabolic properties, such as trimethylamine degradation with low immunogenic properties (Bang et al. 2017). Besides methanogens, members of halophilic archaea, Crenarchaeota and Thaumarchaeota have also been detected to colonize human intestinal tract through metagenomic investigations (Gaci et al. 2014). Archaeal diversity has been found to be maximum in the age group 25–60 years where apart from the predominant spp., abundance of M. oralis, M. arboriphilus, M. millerae, M. ruminantium, Methanosalsum zhilanaea, Methanomassiliicoccus luminyensis, Methanoculleus chikugoensis have been found (Nkamga et al. 2017; Guindo et al. 2021). Occurrence of Nitrososphaera, a member of Thaumarchaeota, although at a low abundance, have been found to be antagonistic with Methanobrevibacter (Hoffmann et al. 2013). However, the diversity is less in the age group below 15 years and above 70 years old. Predominant taxa in the former age group are Methanobreviibacteriales and Methanomassiliicocales and that in the later age group are M. smitthii, M. stadtmanae, M. luminyensis and Candidatus M. intestinalis. Representative taxa of halophillic archaea identified in human fecal samples of populations with higher intake of salty and sea foods have been found to be Halorubrum koreense, H. alimentarium, H. saccharovorum, Halococcus morrhuae and Halopherax massiliense (Nkamga et al. 2017). A recent study on the Korean gut archaeom revealed abundance of archaeal genera, Halolamina, Haloplanus, Halorubrum, Halobacterium, Haloterrigena, Natronomonas, Halarchaeum, Haloarcula, Halonotius and Halorussus (Kim et al. 2020).
Fig. 1.
Autochthonous and allochthonous members of bacterial taxa distributed in different compartments of GIT. The colour bar indicates pH range corresponding to the pH of the GIT compartment lying beside, respectively
A neighbor-joining phylogenetic tree based on the 16S rRNA sequences of all the microbial species detected in human gut microbiome has been constructed which reflects that the predominant phyla are Firmicutes, Proteobacteria, Bacteroides and Actinobacteria in eubacterial group and Euryarchaeota in the archaeal group (Fig. 2). Other phyla are moderately to sparsely represented.
Fig. 2.
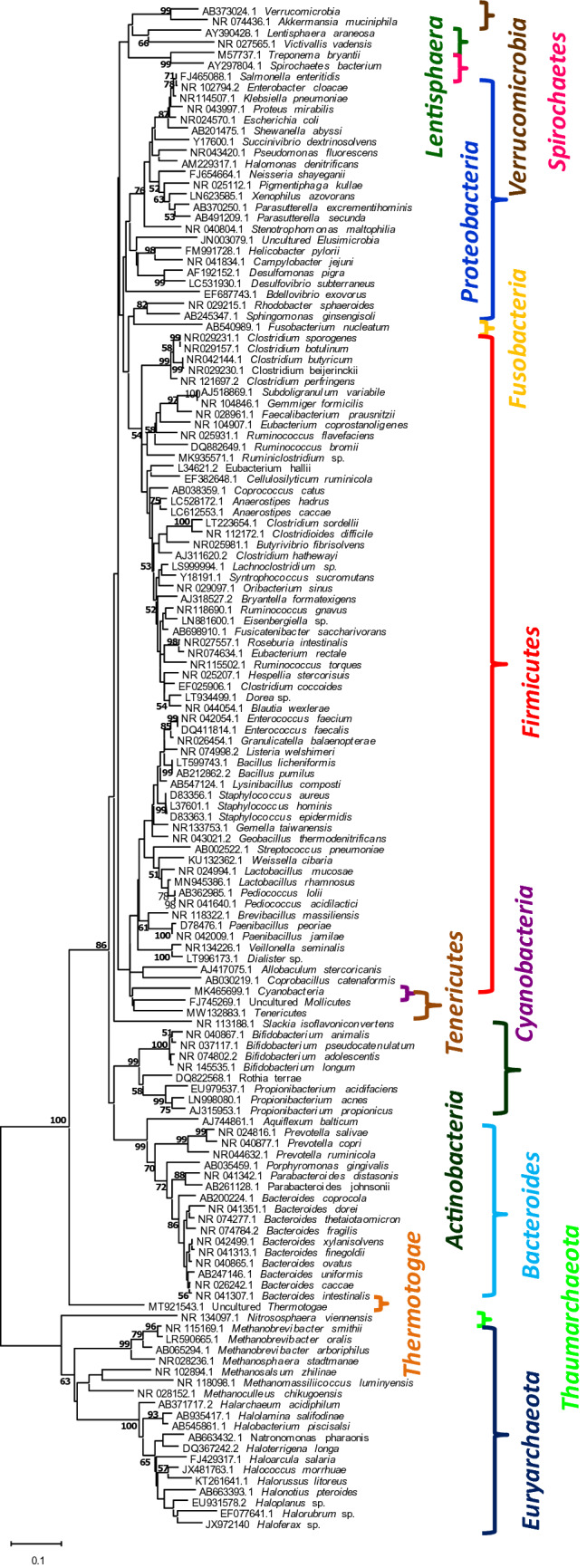
Neighbour joining tree showing phylogenetic relatedness among human gut bacterial and archaeal 16S rRNA genes based on Jukes Cantor model. Nucleotide sequences were retrieved from NCBI database and the values at the branches denote bootstrap values obtained upon 1000 iterations. Coloured brackets indicate various phyla Evolutionary analyses were conducted in MEGAX (Felsenstein 1985; Jukes and Cantor 1969; Kumar et al. 2016; Saitou and Nei 1987)
Role of gut microbiome in human metabolic activities
-
Digestion
Gut microbes and microbial enzymes play pivotal roles in digestion of human diet; however, the specific contributions of different species are not well understood. Many substrates in human diet are resilient to host enzymes and their digestion solely depends on gut microbial enzymes. Dietary resistant starch, a complex carbohydrate composed of amylase and amylopectin, is one such example which is essentially broken down to small chain fatty acids like butyrate, propionate, valerate and isovalerate, etc. Amylopectin, a polymer of glucose, is readily hydrolysed by amylase at 1–6 glycosidic bonds. However, amylose, a more linear glucose polymer with 1–4 glycosidic bonds, is the one that show resistance to degradation. The varying proportions of amylose to amylopectin that a starch molecule contains increases the degree of resistance to degradation. Besides, digestibility of starch molecule in the diet vastly depends on the structure, crystallinity, particle size and cooking. Depending on digestibility, Resistant Starch (RS) has been classified into 5 different types: RS type 1 (physically inaccessible), RS type 2 (native granular starch consisting of ungelatinized granules), RS type 3 (retrograded amylose) and RS type 4 (indigestible i.e. chemically modified) and RS type 5 (amylase- lipid complexes- often considered as slowly digested starch and not a true RS) (DeMartino and Cockburn 2020). Digestion of resistant starch has been found to be mainly associated with the members of Clostridium cluster IV- Faecalibacterium prausnitzii, Clostridium leptum and Ruminococcus bromii and XIV- Eubacterium rectal, Clostridium coccoides, Butyrivibrio fibrisolvens and Roseburia sp. whereas members of Bacteroidetes were found to be downregulated upon administration of high-resistant starch (HRS) diet (Maier et al. 2017; Ramakrishna 2013). F. prausnitzii, Ruminococcus, Prevotellaceae, Eubacterium rectale, Roseburia faecis and Akkermansia muciniphila are the specific taxa that were found to increase upon HRS diet (Maier et al. 2017). R. bromii, has been regarded as the keystone species in RS degradation. They have been found to possess special structures called amylosomes in which the amylolytic enzymes are uniquely arranged bound through cohesin and dockerin modules (DeMartino and Cockburn 2020). These multi-enzyme complexes remain attached to the cell surface via scaffolding proteins as found in cellulosomes (DeMartino and Cockburn 2020; Ze et al. 2012, 2015). Metaproteomic study has also reflected that most of the carbohydrate metabolizing enzymes and transportational molecular systems were affiliated to F. prausnitzii and Coprococcus comes, following an HRS diet. An HRS diet has shown upregulation in human enzymes related to lipid metabolism, such as colipase, pancreatic triglyceride lipase and bile salt-stimulated lipase while downregulating human α-amylase. Thus, an HRS diet would promote the growth and abundance of microbial taxa involved in resistant starch digestion and SCFA production in colon and is a coherent phenomenon with human lipid metabolism (Maier et al. 2017). On the contrary, a diet rich in starch and other easily digestible carbohydrates has been found to be associated with methanogenic archaeal members, M. smithii (Carberry et al. 2014; Hoffmann et al. 2013) which has shown to improve polysaccharide digestion and promote the production of acetate or formate for its own use. M. stadtmanae, found in omnivores, helps in pectin fermentation to produce methanol required to carry out its methanogenesis pathway (Dridi et al. 2009; Ishaq et al. 2016; Moses et al. 2015). Among various dietary lipids, cholesterol (majorly in Western diets) is one of the major components that poses serious health threats related to cardiovascular diseases. Upon ingestion followed by enterohepatic absorption, biliary excretion and circulation, cholesterol is subjected to gut microbial reduction to produce co-prostanol- a major (50%) steroid found in human feces. Eubacterium coprostanoligenes have been documented to be a cholesterol reducing coprostanol synthesizing gut bacteria (Gérard et al. 2004; Kenny et al. 2020; Koppel et al. 2017).
Another recalcitrant substrate is gluten which resists complete digestion via human enzymes. Gluten, rich in amino acids like proline and glutamine, is a crucial source of dietary protein derived from wheat and wheat-based diets (Fernandez-Feo et al. 2013; Helmerhorst et al. 2010; Zamakhchari et al. 2011). Human digestive proteases release incompletely digested metastable Pro/Gln-rich gliadin peptides, 30–40 residues long, into the gut lumen. Sometimes these induce abnormal immune responses in some individuals with genetic predisposition eliciting intestinal symptoms and severe mucosal damage, a condition known as coeliac disease (CD). However, upon arrival to large intestine after escaping digestion in the stomach and the small intestine, gluten proteins and peptides act as dietary compounds from which gut microbiota derive energy. The specific taxa found to show glutenasic activity are Bacillus licheniformis, B. subtilis, B. pumilus, Bacteroides fragilis, Bifidobacterium longum, Clostridium sordellii, C. perfringens, C. botulinum/sporogenes, C. butyricum/beijerinckii, Enterococcus faecalis, E. faecium, Propionibacterium acnes, Pediococcus acidilactici, Paenibacillus jamilae, Staphylococcus epidermidis, S. hominis and Stenotrophomonas maltophilia (Caminero et al. 2014; Herrán et al. 2017). The hydrolysis of gliadin peptide is again found to be a specific peptidase activity for a group of microbial taxa. The 33-mer peptidase activity was found to be higher in Lactobacillus mucosae, L. rhamnosus and Clostridium botulinum/sporogenes than other bacteria belonging to Enterococcus faecalis or Bacillus licheniformis (Caminero et al. 2014).
Other highly complex and variable plant cell wall polysaccharides, such as xylans, xyloglucans and pectins require the concerted action of different glycosidases to produce fermentable monosaccharides. For example, the breakdown of type I and type II rhamnogalacturonan that are two different components of pectin, requires at least a dozen enzymes or more for complete breakdown. Role of gut microbial enzymes have also been found in degradation of animal glycans, such as glycosaminoglycan substrates, such as hyaluronan, heparin and chondroitin (Cantarel et al. 2012; Stam et al. 2006). Other glycosylated proteins such as mucins (produced by intestinal epithelial cells) and peptidoglycans (bacterial cell wall component) also act as alternative energy source for the distal gut microbiota. Degradation of the total dietary carbohydrates is not solely possible through human enzymes as human produces only 17 active enzymes for digestion of food glycans. Hence, the huge and complex repertoire of our dietary polysaccharides are digested into fermentable compounds through the microbial carbohydrate active enzymes (CAZymes) produced in our gut. The CAZyme family of the gut microbiome encodes 15,882 enzymes and is composed of glycoside hydrolases (GH = 57%), polysaccharide lyases (PL = 2%), glycosyl transferases (GT = 35%) and carbohydrate esterases (CE = 6%) (Kaoutari et al. 2013). Majority of the CAZymes constituting GHs and PLs are produced by the members of Bacteroides caccae, B. dorei, B. finegoldii, B. fragilis, B. intestinalis, B. ovatus, B. thetaiotaomicron, B. uniformis, B. xylanisolvens, Bryantella formatexigens, Butyrivibrio fibrisolvens, Clostridium hathewayi, Enterobacter cloacea, Escherichia coli, Faecalibacterium prausnitzii, Klebsiella pneumoniae, Parabacteroides distasonis, Prevotella copri, P. salivae, Roseburia intestinalis, Ruminococcus flavefaciens, R. gnavus, Subdoligranulum variabile, Victivallis vadensis (Oliphant and Allen-Vercoe 2019). Archaeal members are also associated with a healthy digestion process and the relationship among the archaeal and eubacterial members are metabolically interdependent. Methanobrevibacter ruminantium is associated with diet high in fiber and structural carbohydrates, such as cellulose, hemicelluloses, lignin that majorly remains recalcitrant to animal and human digestive enzymes (Zhou et al. 2010). On the contrary, a diet rich in starch and readily digestable carbohydrates is associated with M. smithii which has also been shown to improve polysaccharide digestion and influence the production of acetate or formate for its own use (Hoffmen et al. 2013; Carberry et al. 2014; Ishaq et al. 2015). The archaeal diversity of an omnivore is comprised of Msp. stadtmanae as it requires methanol, a by-product of pectin fermentation, for its methanogenesis pathway (Ishaq et al. 2015).
-
Production of special metabolites
Degradation of dietary carbohydrates, lipids and proteins by the gut microbiome give rise to multitude of biochemical metabolites of both local and systemic action. These metabolites can be either potentially beneficial or harmful to the host depending on the concentration and site of action. Generally, a wide variety of metabolites are produced, such as SCFAs and alcohols from monosaccharides, ammonia, branched chain fatty acids, amines, sulfur compounds, phenols and indoles from amino acids, glycerol and choline derivatives from lipids and tertiary cycling of CO2 and hydrogen. Production of SCFAs like butyrate, acetate and propionate upon degradation of pyruvate are the most abundant faecal metabolites. These are extremely important for host health as butyrate apart from serving as the primary energy source for colonocytes, also improves the integrity of intestinal epithelial cells (IECs) by promoting tight junctions and cell proliferation, increases mucin production by Goblet cells, production of cytokines, TGF-β, IL-10 and IL-8 and induces differentiation of naïve T-cells. Both acetate and propionate also aid in anti-inflammatory process and cytokine production. Excess SCFAs can be incorporated in gluconeogenic and lipogenic process. (Oliphant and Allen-Vercoe 2019). F. prausnitzii, Eubacterium rectale, Eubacterium hallii and R. bromii are well known n-butyrate producers (Li et al. 2008; Louis et al. 2010). Bacteroides, Veillonella, Dialister and Salmonella (gram-negative bacteria) as well as gram-positive bacteria Coprococcus, Roseburia and Ruminococcus are key propionate producers (Covasa et al. 2019; Morrisson 2016) whereas, Akkermansia municiphilla, Bacteroides, Clostridium and Bifidobacterium longum and Bifidobacterium adolescentis are profound acetate producing bacteria (Derrien et al. 2004; Frost et al. 2014; Fukuda et al. 2011; Russell et al. 2013).
Eubacterium halii—an SCFA producer and Clostridium sporogenes is known to produce indole-3-propionic acid and Clostridium difficile has been known to produce 4-cresol, cholesterol and coprostanol (Wang et al. 2017). Pyruvate fermentation can also result in production of small amount of alcohols, such as ethanol, propanol, 2,3- butane-di-ol. Gut microbiota can also produce toxic methanol by processes other than fermentation. Members of Proteobacteria are known to be alcohol producers. Besides, members of genera Bifidobacterium, Clostridium, Eubacterium, Blautia, Coprococcus, Dorea, Lachnoclostridium, Roseburia, Lactobacillus, Faecalibacterium, Ruminiclostridium, Ruminococcus, Streptococcus, Veilonella and Escherichia are well-known alcohol producers including ethanol, propanol, 1,2- propanediol.
2,3-butanediol production. These species also produce various other SCFAs like formate, acetate, lactate, propionate, succinate, valerate, etc. (Oliphant and Allen-Vercoe 2019).
Dietary protein after digestion by pancreatic proteases and other proteolytic enzymes move to the small intestine where the peptides and amino acids that are generated during proteolysis, are further subjected to microbial fermentation. Microbes have lesser diverse enzymatic apparatus to degrade amino acids due to the obvious fact that the complex steps involved consume more energy than actually generated. Incorporation of the available amino acids in their anabolic processes rather than utilizing them as their energy substrates, however, is more preferred by the gut microbes. However, series of Stickland reactions take place by members of Clostridium sp. produces variety of SCFAs and branched chain fatty acids (BCFAs) like isovalerate, isobutyrate, 2- methyl butyrate. Members of Bacilli are capable of producing other SCFAs and BCFAs (Oliphant and Allen-Vercoe 2019). These processes may give rise to indolic and phenolic metabolites, which are otherwise not produced by human cells and may exert deleterious effects in the host. Degradation products of α-amino acids like tyrosine and phenylalanine include 4-hydroxyphenylpyruvate, 4-hydroxyphenylacetate, 4-hydroxyphenylpropionate and 4-hydroxyphenylacetate, phenol, p-cresol and 4-ethylphenol as well as phenylpyruvate, phenyllactate, phenylacetate and phenylpropionate, respectively. Degradation of tryptophan generates indole, 3-methyl indole (skatole), indole acetate and indole propionate (Windey et al. 2012). These metabolites have been found to be produced by bacterial members belonging to F. prausnitzii, Subdoligranulum variabile and Bifidobacterium pseudocatenulatum (Li et al. 2008). Production of toxic metabolites, such as indoxyl sulfate, p-cresyl sulfate, amines and ammonia have been shown to be related to nephrological issues, cardiovascular diseases in chronic kidney disease (CKD) in human host (Mafra et al. 2014; Oliphant and Allen-Vercoe 2019; Windey et al. 2012). The predominant proteolytic bacteria with strong peptidase activity in human faeces are the members of Bacteroides spp. and Clostridium spp., whereas members of Desulfomonas spp. and Desulfovibrio spp. are capable of oxidizing sulfur containing amino acids like cystine, cysteine, taurine and methionine to producing H2S. Depending on the concentration of H2S produced, it may act as “friend or foe” to the host (Blachier et al. 2019). Polyamines, such as putrescine, spermidine, agmatine, cadaverin, tyramine and histamine are small polycationic molecules with multitude of functions including gene regulation, stress resistance, cell growth and proliferation and differentiation (Mafra et al. 2014; Tofalo et al. 2019). Members of Bacteroides, Fusobacterium, Escherichia coli, Enterococcus faecalis, Bifidobacterium animalis sub sp. lactis and Lactobacillus rhamnosus have been shown to play profound roles in production of various polyamines (Tofalo et al. 2019).
-
Production of signaling molecules
The gut microbiome is capable of playing a multitude of functions related to host metabolism through its ability to produce extremely diverse repertoire of metabolites and gene products (D’Argenio and Salvatore 2015; Olivares et al. 2018; Yano et al. 2015). Depending on the dietary exposure, it also produces signaling molecules which contribute in controlling neuro-immuno-endocrine activities as well as peripheral metabolism (Turnbaugh et al. 2007). The inaccessible carbohydrates like resistant starch and plant polysaccharides in the host diet are utilized by the intestinal microbiota, the products of which in turn aid in release of several gut hormones by the enteroendocrine (EE) cells of the GI tract which are important peripheral host metabolism regulators. The microbial structural components, such as flagella and membrane-bound lipopolysaccharide (LPS), can also act as signaling molecules (Gordon 2002). The cell wall LPS of Gram-negative bacteria, e.g. Bacteroidetes phylum, is a potent ligand for toll like receptor 4 (TLR4). The activation of TLRs induce strong immunity- and inflammation effects (Lancaster et al. 2018; Takeuchi and Akira 2010), along with secretion of multitude of metabolically active hormones, such as glucagon-like peptide-1 (GLP-1) (Lebrun et al. 2017), 5-hydroxytryptamine (5-HT) (Kidd et al. 2008) and peptide tyrosine tyrosine (PYY) (Larraufie et al. 2017).
The proportion of various SCFAs, such as acetate, propionate and butyrate, produced by the intestinal microbes upon degradation of dietary fibers and resistant starch although depends on the diet, microbial composition and intestinal transit time, yet maintains a ratio of 3:1:1 in the intestinal lumen (Cummings et al. 1987; Mowat & Agace 2014; Topping & Clifton 2001). These SCFAs are found to be important neuro-immuno-endocrine regulators and play significant roles in microbiota-gut-brain crosstalk like motility, secretion and blood flow. SCFAs also controls peripheral activities like brown adipose tissue activation, regulation of liver mitochondrial function, body energy homeostasis and control over appetite and sleep (De Vadder et al. 2014; Éva et al. 2019; Li et al. 2008, 2019; Mollica et al. 2017; Silva et al. 2020; Bhattacharya et al. 2002). Following their production, colonocytes absorb SCFAs mainly via H+-dependent or sodium-dependent monocarboxylate transporters (MCTs and SMCTs, respectively) (Vijay and Morris 2014). While acetate is readily absorbed by the circulatory tissue for peripheral distribution, propionate is metabolized by hepatocytes and butyrate acts as fuel for colonocytes (Koh et al. 2016; Martin et al. 2019). Not much has yet been elucidated regarding the specific role of different taxa in metabolite production, however, members of Bacteroidetes are found to be enriched with carbohydrate metabolism genes and Firmicutes have been reported to harbor bile acid metabolism genes (David et al. 2014; Martin et al. 2019). These SCFAs, thus produced, act as important signaling molecules for enteroendochrine cells by i) inhibiting nuclear histone deacetylase (HD) and ii) stimulating G-protein coupled free fatty acid receptors 2 & 3 (FFAR2 and FFAR3) (Fellows et al. 2018; Larraufie et al. 2017; Offermans et al. 2014; Rooks and Garrett 2016; Waldecker et al. 2008).
Secondary bile acids, such as lithocholate (LCA) and deoxycholate (DCA) are important signaling molecules that have profound roles in peripheral metabolism through their action on two bile acid receptors expressed on EE cells, the G- protein coupled receptor TGR5 and the nuclear farnesoid receptor FXR (Ramírez-Pérez et al. 2018; Winston & Theriot 2020). These are more hydrophobic than the bile acids and are produced by deconjugation and dehydroxylation of bile salts by microbial bile salt hydrolases (BSH). Metagenomic studies have revealed that BSHs are expressed majorly within three phyla, i.e. Firmicutes (30%), Bacteroidetes (14.4%) and Actinobacteria (8.9%) (Jones et al. 2008; Winston and Theriot 2020). Members of genera Clostridium, Bacteroides, Lactobacillus, Bifidobacterium and Enterococcus and their roles in BSH expression have been vividly studied (Begley et al. 2005; Jones et al. 2008). The BSH of Firmicutes and Actinobacteria metabolizes all conjugated bile salts, whereas those of Bacteroidetes are specific to tauro-conjugated bile acids (Jones et al. 2008). The importance of secondary bile acids lies in the fact that being more hydrophobic help in better reabsorption of the bile salts through passive diffusion, thus limiting faecal loss (Martin et al. 2019; Winston and Theriot 2020).
The gut microbiota also exerts its control over host metabolism by regulating the release of an array of gut hormones and peptides like 5-HT, GLP-1, PYY, glucose-dependent insulinotropic peptide (GIP), cholecystokinin (CCK), ghrelin, leptin, pancreatic polypeptide (PP), oxyntomodulin and neurotensin (Bliss and Whiteside 2018; Covasa et al. 2019; Dockray 2014; Fukui et al. 2018) (Table 2). Serotonin or 5-HT is one among several other important regulatory factors which not only acts as a brain neurotransmitter, but also regulates diverse functions like platelet aggregation, bone development, immune responses, cardiac functions, promote homeostasis and control enteric motor and secretory reflexes. Serotonin is known to be secreted by the enterochromaffin cells upon sensing the nutrient condition in the intestinal lumen (Yano et al. 2015). Cultivable bacterial isolates have been shown to produce 5-HT under laboratory conditions. Therefore, it is still ambiguous if microbial de-novo synthesis of 5-HT by indigenous members of gut microbiota contribute to host 5-HT levels. However, the gut microbiota comprising spore forming microbes have been specifically found to promote 5-HT production in adult mice. The 5-HT concentrations in serum, colon and fecal samples were low for germ-free mice as compared to conventionally colonized specific pathogen-free controls (Wikoff et al. 2009). Glucagon like peptide, an incretin hormone and the neuroendocrine hormone PYY are the two regulatory hormones secreted by L-cells of ileum and colon. GLP-1 is released in response to glucose to augment insulin and impede the secretion of insulin. On the other hand, PYY is secreted postprandially from illeal and colonic endocrine cells inducing feeling of satiety and reducing food intake. This is therefore, directly related to obesity (Muller et al. 2007). The enzyme dipeptidyl peptidase IV (DPP-IV) can cleave GLP-1 as well as PYY leading to potentially anti-diabetic effects. The dynamic relation among the L-cells and microbial DPP like activity, SCFA signaling, bacterial lipopolysaccharide and indole production by gut microbes can exert potential influence on levels of host GLP-1 and PYY levels, thereby controlling glucose metabolism. This, in turn may also influence the microbial composition and microbial metabolite compositions. Indole, another major bacterial metabolite, derived from tryptophan metabolism stimulates GLP-1 (Chimerel et al. 2014). Another gastric inhibitory peptide GIP secreted from K-cells significantly contributes to insulin secretion postprandially. While DPP-IV and microbial DPP-IV like activity, such as in Prevotella or Lactobacillus (Olivares et al. 2018) attenuates the biological activity of GIP and GLP-1, it stimulates that of PYY by breaking down it to PYY3-36. Cholecystokinin, secreted by I—cells, is released in response to fat and protein content of the host diet. It acts by activation of CCK1 and CCK2 receptors located all over the tissues of GIT and CNS. Although, abundance of certain gut bacteria like commensal Bifidobacterium and Lactobacillus strains have been negatively correlated to the decreased levels of cholecystokinin and ghrelin in germ-free mice, yet no strong association has yet been found among the intestinal bacteria and levels of these hormones (Covasa et al. 2019; Martin et al. 2019; Pen & Welling 1983; Perry et al. 2016; Queipo-Ortuño et al. 2013), etc.
-
Xenobiotic degradation and heavy metal transformation
Xenobiotics are chemicals or synthetic substances metabolically extrinsic to host system which might accumulate within host extraneously or may be produced as a defense mechanism by certain host microbes (Atashgahi et al. 2018; Koppel et al. 2017). These include environmental pollutants produced in large volumes for industrial, agricultural and domestic use (Atashgahi et al. 2018) which might enter the environment at macro (μg/L to mg/L range) or micro levels (ng/L to μg/L range) (Meckenstock et al. 2015; Schwarzenbach et al. 2006). Pharmaceutical compounds like metformin, methotrexate, proton-pump inhibitors (PPIs), opioids, non-steroidal anti-inflammatory drugs (NSAIDs) and antibiotics, etc. immensely affect the gut microbiome structure. Other xenobiotic compounds, such as persistent organic compounds (POPs), pesticides, pharmaceuticals, e.g. psychotropic agents like olanzapine, personal care products (PPCPs), food additives, disinfection by-products (DBPs) (Atashgahi et al. 2018) can also potentially alter the gut microbiome diversity. Dietary compounds including polyphenolic phytochemicals like resveratrol and flavonoids, polyunsaturated fatty acids, artificial sweeteners and plant sterol esters have also been associated with changes in structural diversity of gut microbiome. Variety of other environmental and industrial chemicals, such as endocrine disrupting chemicals, heavy metals, pesticides and pollutants majorly impact the structural composition as well as functionality of the gut microbiome (Clarke et al. 2019).
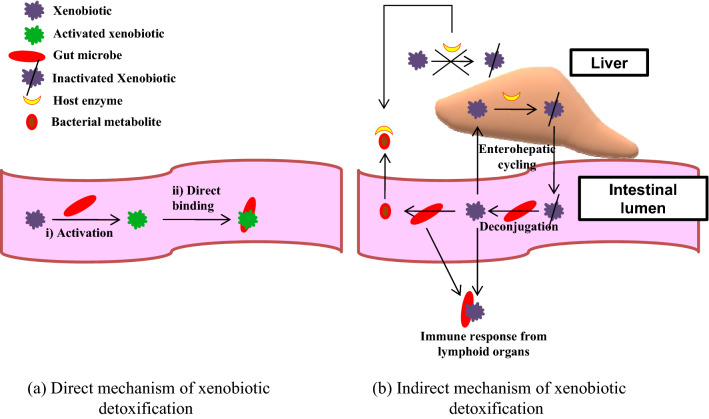
The human gut microbiota homes genetic information for multiple xenobiotic detoxification and sequestration and is capable of encoding a broad diversity of enzymes (Haiser and Turnbaugh 2013; Spanogiannopoulos et al. 2016). Factors, such as host genetics, age, geographical location of the host, diet, administration of drugs, hormonal status, gender, stress and circadian rhythm immensely control microbial metabolism of xenobiotic compounds (Clarke et al. 2019; Das et al. 2016). A cyclic metabolic interaction takes place between microbes and host cells. The anoxic environment of the gut facilitates a reductive and hydrolytic metabolism generating non-polar, low-molecular weight by-products that are readily absorbed by the host cells. These by-products are transported to the liver where the hepatic cells metabolize them to generate hydrophilic, polar metabolites via a diverse repertoire of oxidative and conjugative enzymes. These high-molecular weight polar metabolites are secreted through the bile and again reach the gut where they are further subjected to reductive and hydrolytic metabolism (Claus et al. 2017; Koppel et al. 2017; Sousa et al. 2008). Two mechanisms are involved in xenobiotic metabolism, i.e. direct mechanism which includes production of active compounds which are microbially detoxified in the gut lumen. In indirect mechanism the xenobiotic undergoes enterohepatic cycling where the host physiology is manipulated by the gut bacteria. Here, xenobiotic compounds are inactivated by conjugation in the liver; the conjugated compound then enters the intestinal lumen through bile where, microbial enzymes release the conjugate group, reactivating the compound. This active compound may now re-enter the circulation or may undergo microbial transformation to form microbial metabolite which now competes with the active xenobiotic compound for binding sites on the enzyme. These microbial metabolites may in turn stimulate immune responses through translocation or inflammation (Fig. 3) (Carmody and Turnbaugh 2014). The host may face beneficial, detrimental or even lethal outcomes of xenobiotic metabolism (Okuda et al. 1998; Satoh-Takayama et al. 2008; Shin et al. 2013). Exposure to xenobiotics may lead to significant alteration in gut microbiota composition and metabolic activity (Maurice et al. 2013) and, may increase predisposition to various diseases (Lee & Hase 2014; Lu et al. 2015; Šrut et al. 2018; Wang et al. 2011). Metabolism of xenobiotic in human occurs in two phases, i.e. Phase I—wherein, polar functional groups are exposed and Phase II—wherein, phase I groups are conjugated to more-polar metabolites. While the oxidative, reductive or hydrolytic reactions are performed by Phase I enzymes to generate hydroxyl groups, epoxides, thiols and amines; in Phase II, the xenobiotic molecule or the phase I metabolites are appended with glucuronyl, methyl, acetyl, sulfonyl and glutathionyl groups (Koppel et al. 2017; Pellock and Redinbo 2017). Enzymes used and reactions catalysed in xenobiotic degradation have been listed in Table 3.
Gut microbiome has been found to play profound roles in increasing or decreasing the activity of various pharmaceutical drugs when incubated with respective cell lines. Structural modification of anti-cancerous drugs by tumor-associated bacteria, such as E. coli or Listeria welshimeri have been experimentally found to be responsible for inter-individual differences in response to anti-cancerous chemotherapy. Likewise, many anti-inflammatory prodrugs rely on gut microbial activity to be transformed into active drugs, such as sulfasalazine containing azo linkages. Various gut associated azoreductases have been characterized till date, which are mainly involved in detoxification of various azo dyes, nitro-aromatic and azoic drugs. Diverse group of azoreductase enzymes with varying co-factor dependence, structure and functional mechanism, catalytic activity and aerobic or anaerobic microbial source have been isolated and characterized with a common potential to reduce azo compounds (Bryant and DeLuca 1991; Bürger and Stolz 2010; Matsumoto et al. 2010; Misal and Gawai 2018; Morrison et al. 2012; Vijay and Morris 2014) Aerobic bacterial strains belonging to genera Pseudomonas, Bacillus, Escherichia, Xenophillus, Pigmentiphaga, Rhodobacter, Enterococcus, Staphylococcus, Geobacillus, Brevibacillus, Lysinibacillus, Aquiflexum, Shewanella, Rhodococcus and Halomonas along with anaerobic members of Clostridium, Eubacterium, Butyrivibrio, Sphingomonas produce a large array of azoreductases varying in their temperature and pH optima, flavin/nicotinamide requirement, with aerobic/anaerobic natures (Misal and Gawai 2018).
Exposure to heavy metals have also been reported to perturb the gut microbiome significantly. A study by Li et al. (2019) on the effect of heavy metal exposure on gut metabolic health suggested that several metabolically important genera like Blautia, Eisenbergiella, Clostridium showed a decline in metabolic interactions. A significant decline in butyrate producing organism, such as Fusicatenibacter, Eisenbergiella, Syntrophococcus, Blautia, Clostridium XIVb, Cellulosilyticum, Oribacterium, Coprococcus, Anaerostipes, Hespellia and Lachnospiracea incertae sedis was found upon exposure to both As and Cd. Exposure was also found to affect bile acids, amino acids and taxa associated with metabolic health. Butyrate besides being the major energy source for the colonic epithelium, is also capable of improving insulin sensitivity and increase energy expenditure (Gao et al. 2018; Roediger 1980). Consequently, these downregulated genera along with Parasutterella and Gemmiger, are also associated with alleviation of T2DM (Helmerhorst et al. 2010; Li et al. 2018; Xu et al. 2015). Exposure to lead has been found to be associated with Succinivibrionaceae and Gammaproteobacteria in children (Bisanz et al. 2014) and Gammaproteobacterial members in adults (Eggers et al. 2019).
The specific bacterial groups in the human intestinal lumen act for detoxication rather than detoxification of the xenobiotic or heavy metals to which human body is exposed. Detoxication is the mechanism by which the drugs, mutagens and other harmful agents like heavy metals are removed from the body (Monachese et al. 2012). This helps to prevent the harmful agents to enter and impair the important organs of human body. Heavy metals are largely removed from the intestinal lumen by gram-positive bacteria by binding of metals to their cell walls. Ion exchange reactions of the exposed metal with peptidoglycan and teichoic acid, precipitation through nucleation reactions and complexation with nitrogen and oxygen ligands are main mechanisms through which these gram-positive bacteria, particularly Bacillus spp., having high peptidoglycan and teichoic acid content in their cell walls absorb the heavy metals. However, gram-negative bacteria show poorer metal absorbing capacity due to presence of a thin cell wall with lower peptidoglycan content.
The abundance of xenobiotic metabolizing enzyme repertoire in specific groups of bacteria vary across different nationalities (Das et al. 2016). Depending on the abundance pattern of various bacterial genera, the microbial groups have been categorized accordingly. The first category includes genera like Prevotella, Faecalibacterium, Dorea, Roseburia, Eubacterium, Ruminococcus, Bacteroides, etc. which showed a high-specific abundance across all regions. The next category included genera like Neisseria, Bacillus, Slackia, Coprobacillus, Treponema, etc. which were highly abundant across the European and American populations, but depleted relatively in the gut microbiomes of individuals of Asian nationalities. Genera like, Rhizobium, Rhodospirillum, Bradyrhizobium, Rhodopseudomonas, Methylobacterium, etc. comprised the third group which could be sparsely detected in the gut microbiome of European and American populations and was absent in that of Asians. Asian gut microbiome rather showed the presence of E. coli as one of the genera harboring xenobiotic metabolizing gene repertoire.
Table 2.
List of signaling molecules, secretory cells and bacterial genera aiding in secretion of specific signaling molecule
| Gut hormones | Secretory cells | Aiding microbes | References |
|---|---|---|---|
| Serotonin | Enterochromaffin cells | Clostridium spp., Escherichia, Enterococcus, Truchuris, Candida, Streptococcus | Covasa et al. 2019; Yano et al. 2015 |
| Glucagon like peptide 1 | Colonic L-cells | Bifidobacteria, Lactobacillus, Akkermansia muciniphila | Everard and Cani 2014; Greiner and Bäckhed 2016 |
| Peptide YY | Colonic L- cells | Bifidobacteria, Lactobacillus, Akkermansia muciniphila, Escherichia, Enterococcus and Truchuris | Covasa et al. 2019; Everard and Cani 2014; Greiner and Bäckhed 2016; Xu et al. 2015 |
| DPP 4 | Enterocytes, epithelial cells and immune cells | Prevotella and Lactobacillus | Klemann et al. 2016; Olivares et al. 2018; Zhong et al. 2015 |
| Glucose dependent insulinotrophic peptide | K- cells | NK | Fukui et al. 2018 |
| Cholecystokinin | I- cells | NK | Dockray 2014; Fukui et al. 2018 |
| Leptin | adipocyte | NK | Queipo-Ortuño et al. 2013 |
| Pancreatic polypeptide | F or PP cells | NK | Bliss and Whiteside 2018 |
| Ghrelin | cardiomyocytes | NK | Iglesias et al. 2004 |
| Oxyntomodulin | Pancreatic cells | NK | Bliss and Whiteside 2018 |
| Neurotensin | Gastrointestinal endocrine N cells | NK | Bliss and Whiteside 2018 |
| Motilin | Endocrine M- cells | NK | Chapman et al. 2016 |
| Insulin | B- cells | Lactobacillus, Bifidobacterium, Blautia coccoides, Eubacterium rectale, Prevotella | Zhang et al. 2015 |
| Glucagon | A cells | NK | Kelly et al. 2010 |
| Somatostatin | D cells | NK | Giloteaux et al. 2012 |
| Dopamine or noradrenaline | Nerve cells | Escherichia, Bacillus and Saccharomyces | Covasa et al. 2019 |
| Acetylcholine | Nerve cells | Lactobacillus | Covasa et al. 2019 |
| GABA | Β- cells | Lactobacillus, Bifidobacterium | Covasa et al. 2019 |
| Indole | NK | Escherichia, Bacteroides, Clostridium | Covasa et al. 2019 |
| Bile acids | Hepatocytes | Acetatifactor and Bacterroides | Covasa et al. 2019 |
NK not known
Fig. 3.
Direct (a) and indirect (b) mechanisms of xenobiotic detoxification
Table 3.
List of microbial enzymes involved in specific group of xenobiotic metabolism and the organisms producing the enzymes
| Enzymes used | Xenobiotic detoxified | Organism | References |
|---|---|---|---|
| Cytochrome P450 | fatty acids, 4-n-nonylphenol, pharmaceutical agents, such as S-mephenytoin, phenytoin, S-warfarin, tolbutamide, arachidonic acid, steroids and non-steroidal anti-inflammatory substances | Sphingomonas sp. strain KSM1, Bacillus megaterium, | Bracco et al. 2013; Di Nardo and Gilardi 2012; Elefterios Venizelos Bezirtzoglou 2012 |
| Nocardia farcinica IFM 10152 | |||
| Actinoplanes sp. | |||
| Monoamine oxidase | biogenic amines like histamine, benzyl amines, tyramine, dopamine, octopamine or norepinephrine, etc | Lactic acid bacteria, Klebsiella, Enterobacter, Escherichia, Salmonella, Serratia and Proteus, Pseudomonas aeruginosa IFO 3901, Sarcina lutea, Micrococcus luteus IFO 12708 and Brevibacterium ammoniagenes IAM 1641 | Murooka et al. 1996; Murooka et al. 1979 |
| Epoxide hydrolase | Epichlorohydrin, epibromohydrin, epoxyoctane, styrene epoxide and, lipids | Rhodococcus, Nocardia spp., Mycoplana rubra and Methylobacterium spp., Alternaria alternata | Krenn et al. 1999; Morisseau 2013; Theisen and Berger 2005 |
| Alcohol dehydrogenase | 2- chloroethanol | Acetohalobium, Acholeplasma, Achromobacter, Acidovorax, Anaerococcus, Coprococcus, Coprothermobacter, Coraliomargarita, Desulfarculus, Desulfatibacillum, Desulfitobacterium, Desulfobacca, Desulfobacterium, Gemmatimonas Geobacillus, Geobacter, Geodermatophilus, Geopsychrobacter, Haemophilus, Hafnia, Marinobacter, Marinomonas, Streptococcus gordonii | Das et al. 2016; Pavlova et al. 2013 |
| Aldehyde dehydrogenase | Acetaldehyde, folate, aromatic aldehydes, Fatty aldehydes, Glutamate γ-semialdehyde, Succinic semialdehyde, Methylmalonate semialdehyde and, Aminoaldehydes | Pseudomonas aeruginosa, Klebsiella pneumoniae, Escherichia coli | Nosova et al. 1998; Vasiliou et al. 2000 |
| Thiopurine methyltransferase | thiopurine drugs, such as 6-mercaptopurine | Acetobacter, Acetohalobium, Acholeplasma, Achromobacter, Acidaminococcus, Acidimicrobium, Acidiphilium, Acidithiobacillus, Acidobacterium, | Das et al. 2016 |
| N-acetyl transferase | 5-Aminosalicylate, sulfapyridine, mesalamine and sulfasalazine, etc | Helicobacter pylori, Klebsiella pneumoniae, Pseudomonas aeruginosa, | Chen et al. 2012; Chung et al. 2001; Ricart et al. 2002; Westwood et al. 2005 |
| Glutathione S-transferase | organophosphorus pesticides and cytotoxic aldehydes produced during lipid peroxidation | Acidiphilium, Acidithiobacillus, Enterobacter, Enterococcus, Pseudoanabaena, Pseudoalteromonas, Thiomicospira, Thiomonas, etc | Booth and O’Halloran 2001 |
| Azo reductase | dimethoxybenzidine-based dye- Direct Blue 15, acid yellow, amaranth, azodisalicylate, chicago sky blue, congo red, direct black 38, direct blue 6, direct brown 95, fast yellow, lithol red, methyl orange, methyl red, methyl yellow, naphthalene fast orange 2G, neoprontosil, new coccine, orange ii, phenylazo-2-naphtho1, ponceau 3R, ponceau SX, red 2G, red 10b, salicylazosulphapyridine, sunset yellow, tartrazine, trypan blue, brilliant black | Acidaminococcus, Aerococcus, Bacillus, Butyrivibrio sp., Bacteroides sp., Brevibacillus, Clostridium clostridiiforme., C.paraputrificum, C. nexile, Escherichia, Enterococcus faecalis, E. faecium, E. avium, E. durans, E. gallinarum, E. hirae Eubacterium hadrum, Fusobacterium, Halomonas, Lactobacillus, Lysinibacillus, Peptostreptococcus, Pigmentiphaga, Pneumococcus, Pseudomonas, Rhodobacter, Rhodococcus, Ruminococcus, Salmonella, Shigella, Shewanella, Sphingomonas, Staphylococcus, Streptococcus, Veillonella, Xenophyllus | Chung et al. 1992; Rafii et al. 1990; Zahran et al. 2019 |
| Alkane hydroxylase | Norcarane, n-Hexane, Iso-hexane, Cyclopentane, etc | Pseudomonas oleovorans, Pseudomonas aerofaciens, P. putida. Acinetobacter, Rhodococcus and Alcanivorax borkumensis, Hydrocarboniphaga effusa | Janssen et al. 2005 |
| Haloalkane dehalogenase | 1,2‐dibromoethane, 1,3‐dichloropropene, 1‐chlorobutane‐, 1‐chlorohexane‐ and, 1,6‐dichlorohexane, | Xanthobacter autotrophicus, Rhodococcus erythropolis, Mycobacterium, Pseudomonas pavonaceae | Janssen et al. 2005 |
| Oxalate | an oxalate:formate antiporter; formyl-CoA transferase and oxalyl-CoA decarboxylase | Oxalobacter formigenes | Carmody and Turnbaugh 2014 |
Control over immune system
The extraordinarily diverse and complex microbial community of gut microbiome has been found to participate in maturation and functioning of innate as well as adaptive immunity (Cénit et al. 2014; Cheng et al. 2019). The diverse microbial flora also plays important role in host defense against pathogens by repairing intestinal mucosal damage, production of various anti-microbial peptides and induction of secretion of interleukins IL-22, IL-17 and, IL-10 by host immune cells.
The first line of defense provided by intestinal bacteria is by competing for attachment sites and nutrient in the gut lining, thus preventing attachment of non-commensals or pathogenic strains- a mechanism known as competitive-exclusion effect. Further, these bacteria are also equipped with ability to produce several bacteriocins and elicit production of various anti-microbial peptides, such as α-defensins, β- defensins, angiogenins, C-lectins and RegIIIγ and Reg IIIβ by intestinal epithelial cells (IECs) (Bull and Plummer 2014; Cheng et al. 2019; Guarner and Malagelada 2003; Lazar et al. 2018). An example of bacteriocin production has been found in E.coli competing for amino acids and producing bacteriocin against enterohemorrhagic strain of E.coli (Belkaid 2015; Momose et al. 2008). Induction of other anti-microbial peptides by B. thetaiotaomicron, targeting other intestinal microbe have also been reported (Lazar et al. 2018). Intestinal microbes can even exert immunomodulatory effects so that the environment becomes hostile for the pathogenic species, e.g. Lactobacillus creating an acidic environment unfit for pathogenic invasion. The α-defensins are produced by intestinal Paneth cells upon stimulation by both Gram-negative and Gram- positive bacteria and bacterial metabolites, such as lipopolysaccharides, lipoteichoic acids, lipid A and muramyl dipeptide. Lactobacillus induces production of α-defensins upon infection with Helicobacter hepaticus. β-defensins are involved in direct killing or attenuation of the pathogenic microorganisms by penetrating into their cell membrane and chemoattraction of immune cells. Lactobacillus and E.coli induce β-defensin production upon infection with S. aureus, S. pyogenes, P. aeruginosa, E. coli and C. albicans (Cheng et al. 2020; Islam et al. 2004; Schlee et al. 2007; Seo et al. 2012; Steubesand et al. 2009; Wehkamp et al. 2004). C-type lectins, i.e. RegIIIγ and Reg IIIβ also play key roles in innate immunity by providing protection against specific pathogens like Enterococcus faecalis, Yersinia pseudotuberculosis and Listeria monocytogenes. Several interleukins have been documented to be produced upon infection with particular pathogens by different innate immune response cells. Enterobacteriaceae, pathobiont Proteus mirabilis, gut microbe derived ATP and SCFAs have been reported to induce production of IL-1β and IL-18. IL-22 is found to play central role in maintenance of mucosal barrier integrity produced in response to infection by diverse pathogens, such as Klebsiella pneumonia, Citrobacter rodentium, vancomycin resistant Enterococcus, Plasmodium chabaudi. Gut microbes like Lactobacillus, Allobaculum spp. E.coli, Clostridium and Bacteroides spp. utilize tryptophan and produce indole-3-aldehyde which in turn induce IL-22 production by innate lymphoid cells (ILCs) (Abt et al. 2016; Kinnebrew et al. 2010; Ota et al. 2011; Satoh-Takayama et al. 2008; Sellau et al. 2016; Xu et al. 2015; Zheng et al. 2008). IL-17 is another important cytokine produced by intraepithelial lymphocytes (IELs) modulated by commensal bacteria in response to pathogens like Salmonella typhimurium. IL-17 acts by both neutrophil recruitment and anti-microbial peptide production, thereby limiting invasion and dissemination of pathogenic microbes. Production of IL-17 is induced by Bacteroides in response to infection with S. typhimurium. IL-10 is an anti-inflammatory cytokine produced by macrophages in response to host damage by pathogens and play neutralizing roles to maintain intestinal homeostasis. Clostridium butyricum has been shown to induce IL-10 production in IL-10 deficient mice to prevent acute colitis. Probiotic strains of Lactobacilli and Bifidobacterium have been shown to modulate IL-10 production (Niers et al. 2005).
However, innate immune homeostasis is acquired by the intestinal macrophages through a mechanism called inflammation anergy (Smythies et al. 2005, 2010). This mechanism involves phenotypic modulation of intestinal macrophages with no or low innate response in contrast to the blood monocytes. This is due to the lack of or low expression of a key receptor CD14 protein involved in recognition of bacterial LPS or related antigens known as microbe-associated molecular proteins (MAMPs) (Smith et al. 1997, 2011). Other innate response receptors, such as CD 89, CD 64, CD 32, CD16, CD 11, CD 18, CD 25, CD 123 and proinflammatory cytokines, such as IL-1, IL-6, IL-12, RANTES, TNFβ, TNFα were found to be downregulated upon inflammatory stimulations (Cénit et al. 2014).
The adaptive immune response required for local and systemic homeostasis is highly regulated by the diverse composition of gut microbiota. Gut harbors an environment where continuous presence of microbes imposes a selective pressure on the gut-associated lymphoid tissue (GALT) to undergo dynamic remodeling. Intestinal CD4 + T cells present on the lamina propria of intestine are the key component of adaptive immunity which can differentiate into four major subtypes, i.e. T helper 1 cells (Th1), Th2, Th17 and regulatory T (Treg) cells, The characteristics and roles of different subtypes of T cells have been summarized in Table 4 (Wu and Wu 2012). Gut microbiota plays a major role in T cell differentiation, e.g. the polysaccharide A molecule of Bacteroides fragilis induces systemic Th1 response. Segmented Filamentous Bacteria (SFBs) influence TH17 cell differentiation through pro-inflammatory response and Th1 cells to a lesser extent. Overexpression of Th17 response may lead to auto-immune diseases. Clostridia spp. belonging to clusters XIVa and IV were found to be associated with IL-10 producing Treg cells (Atarashi et al. 2011).
Table 4.
Subtypes of T-cells, role played, health impact due to overexpression of the T cells and gut microbial taxa influencing T cell differentiation
| Subtype of T cell | Role played | Overexpression of T cells | Gut microbial taxa influencing T cell differentiation |
|---|---|---|---|
| Th1 | Host defence against intracellular microbial infection | Autoimmune disease | Bacillus fragilis |
| Th2 | Elimination of infection by parasites | Allergen specific IgE response | Roseburia intestinalis |
| Th 17 | Produces IL-17; a crucial cytokine involved in limiting invasion and dissemination of pathogens, such as Salmonella typhimurium | Autoimmune disease | Segmented filamentous bacteria (SFB) |
| T reg | Immune tolerance; promotes class switching to IgA in presence of specific antigen | Autoimmune disorder | Clostridial clusters IV. XIVa |
Microbiota-gut-central nervous system axis
The gut-brain axis is an integrated communication system that involves afferent and efferent neural, endocrine/hormonal, nutrient and immunological signals for the crosstalk of the gut microbiota and its metabolites with the central nervous system. It is a bidirectional communication system where in response to the gut microbial environment and its metabolite signaling, brain commands several gut functions, such as mucin production, peristalsis and gut immune functions. Factors, such as stress, variations in diet (Buffington et al. 2016), immune activation (Estes and McAllister 2016; Foley et al. 2014) and alterations in maternal microbiome during pregnancy due to use of antibiotics or probiotics (Russell et al. 2013; Tochitani et al. 2016), can modulate the microbiome, neurodevelopment and behavior of an individual (Baumgart and Carding 2007; Borre et al. 2014; Bull and Plummer 2014; Silva et al. 2020). Gut pathologies increase permeability of the intestinal barrier which leads to increase in translocation of bacterial products that can in turn, enhance the production of cytokines and impact the blood brain barrier (BBB). This leads to even more serious ill effects. Moreover, it is well documented that levels of several neurotransmitters are regulated by the gut microbes. These microbes have even been found to directly synthesize or modulate the synthesis of various other neurotransmitters like g-aminobutyric acid (GABA), serotonin (5-HT), dopamine (DA) and noradrenaline (NA) (Calvani et al. 2018; Fung et al. 2017; Sherwin et al. 2018) which can potentially influence microglial activation and several cerebral functions. Signal transducers, such as enterochromaffin cells can bind several microbial metabolites, secrete serotonin increasing its concentration in both blood and colon. Among other transducers are vagus nerve signaling involved in mediating satiety, stress and mood and microbial metabolites, such as SCFAs, the variation of which have shown various neuropathologies (Silva et al. 2020). There are speculations that SCFAs might have roles in the production of GLP-1, PYY and other gut hormones, such as 5-HT, GIP, ghrelin and CKK.
The microbiome-gut-brain axis involves multidirectional communication which includes metabolic, endocrine, neural and immune pathways (Joscelyn and Kasper 2014; Wang and Kasper 2014). The intrinsic nervous system of the GI tract is known as the enteric nervous system (ENS) and is a part of the autonomic nervous system. Due to its huge extent, degree of autonomy and capability to control gastric functions, such as absorption, secretions, integrity, proliferation, barrier function and defense alarm system in concert with CNS, it is also known as the ‘second brain’. Enteric neurons have been known to be activated by various bacterial toxins and metabolites, e.g. Lactobacillus rhamnosus- (strain JB-1) and B. fragilis-specific polysaccharide A (PSA) activates intestinal primary afferent neurons of the ENS in mice (Ochoa-Repáraz and Kasper 2016). Gut commensals including E. coli have also been documented to show symbiotic host-microbiota relationships via production of inositol 1,4,5 triphosphate through phytate metabolism. Besides, inducing growth of human tissue derived intestinal organoids, inositol 1, 4, 5 triphosphate has been shown to stimulate histone deacetylase 3 (HDAC3)—dependent proliferation and counteract the inhibitory effect of abundantly present butyrate upon colonic growth. Thus, the gut microbiota-derived metabolite inositol 1,4,5 triphosphate has been shown to activate a mammalian HDAC to promote intestinal epithelial repair (Wu et al. 2020).
Translation and commercialization of gut microbiome
Several investigators have proposed that the human microbiome should be used as an integral part of precision medicine approach as not only it could contribute to inter-individual variability in diseases but could also be a modifiable factor in terms of development of future therapeutics. Various researchers observed that personalized diets could be created through blood glucose response by integrating parameters, such as dietary habit, physical activity and gut microbiota for lowering blood glucose post-meal (Suez and Elinav 2017; Zeevi et al. 2015). Several promising research findings have documented the link between the gut microbiome, their therapeutic effects and various systemic diseases in recent times. Numerous start-up companies have initiated the translation of the research findings of several investigations on gut microbiome for fruitful therapeutic applications and subsequent commercialization. Companies developing microbiome therapy pipelines use various microbial approaches which include small molecule therapy (31%), e.g. prebiotics supporting growth of a particular group of bacteria of therapeutic importance; development and administration of single strain whole bacteria (26%). Few other approaches, such as application of microbial consortia and genetically modified single strain bacteria (12%) are also adopted. However, with increase in the number of bacterial populations number of factors playing role in cause and effect also increases. The least used approaches are phage cocktail (4%) and microbial ecosystems (4%) which call for several challenges including immune response, microbial succession and change in gut flora composition of an individual.
The microbiome therapeutic pipeline includes five stages of trials, i.e. preclinical, research, phase I, phase II and, phase III before commercialization of the product for public use. Several companies, such as SERES therapeutics have reported to meet the Phase 3 primary endpoint in developing oral microbiome therapeutic SER-109 to show statistically significant reduction in the rate of Clostridium difficile infection (CDI). Another company Rebiotix have also started Phase III trial for reduction of recurrent CDI. Faecal microbiota transplantation (FMT) has been found to be rigorously successful, with cure rates over 95%, in treating various infectious disease, such as recurrent CDI, ulcerative colitis, irritable bowel syndrome, etc. Cure for other diseases like obesity, autism, Alzheimer’s and Parkinson’s diseases are also being investigated with good prospects and highly expected to be successful through microbiome therapies.
Today around 200 companies are working across the globe to utilize the therapeutic capacity of the healthy gut microbiome to translate it for public use. Biomica is an emerging company working on microbiome-based therapeutics against immune mediated infectious diseases, immuno-oncology and, gastro-intestinal tract disorders. Finch therapeutics group is another concern that harnesses the gut microbiome. Companies adopting other approaches, i.e. on single strain cell development are A-mansia, BioGaia, Bio-Me, Ginkgo Bioworks, Next Biotix, etc. Others that focus on development of gut microbiome modulating drugs are Vedanta BioSciences, Snipr Biome, Ritter Pharmaceuticals, etc. The use of prebiotics/probiotics has been immensely adopted for the treatment of various gut related diseases affecting metabolism, immune system and CNS. Genetically modified Escherichia coli Nissle 1917 (EcN) is a well-known probiotic or Live Biotherapeutic Products (LBP) which is believed to impede the growth of opportunistic pathogens, including Salmonella spp. and other coliform enteropathogens, through the production of microcin proteins or production of iron-scavenging siderophores. It has been used to treat various gastrointestinal conditions, including inflammatory bowel disease and irritable bowel syndrome in its unengineered form. Besides, probiotics belonging to diverse genera Lactobacillus, Lactococcus, Lysteria monocytogenes, Bifidobacterium, Staphylococcus, Salmonella typhii, Clostridium and Bacteroidetes have also been engineered to be applied for biotherapeutic purpose to treat various therapeutic indications (Charbonneau et al. 2020). Companies like Acto Bio Therapeutics, Synlogic, Oragenics, Novome Biotechnologies and, CHAIN biotech, etc. are actively involved in bioengineering and production of various lantibiotics, immunomodulators and probiotics to treat systemic and metabolic disorders (Table 5). Several other companies have also worked on novel therapeutics based on small molecules, such as 4D Pharma, VAXIMM, Scioto Biosciences, etc. specially targeted to act as immunomodulators or live biotherapeutics to treat grave diseases as various cancers, neurodegenerative disorders and recurrent C. difficile prevention.
Table 5.
List of companies working on translational research on gut microbiome, location and their research focus
| Company | Location | Research focus |
|---|---|---|
| Adapsyn Bioscience | Canada | Production of novel bacterial metabolites |
| AOBiome | Boston | Restoration of ammonia oxidizing bacteria |
| A-Mansia | Louvain-la-Neuve | microbial products based on unique properties of the Akkermansia muciniphila bacterium |
| ActoBio Therapeutics | Ghent | Targeted and microbe-based therapeutic agents for locally delivering potential disease modifying therapeutics |
| 4D Pharma | UK | Novel therapeutics |
| Artugen Therapeutics | Concord | Novel Live Biotherapeutic Products to help patients living with infectious, inflammatory and oncologic diseases |
| Biohm | Cleveland | Probiotic comprising bacteria, fungi and enzymes to remove digestive plaques |
| BioGaia | Stockholm | Developing probiotics for gut and immune system |
| BioMe | Oslo | High-throughput microbiome analysis and probiotic development |
| Biomica | Park Rehovot | Microbiome-based therapeutics for the treatment of immune-mediated and infectious diseases, with specific focus on Immuno-Oncology and GI-related disorders |
| BiomX | Ness Ziona | Phage cocktails containing natural and/or engineered phage developed through algorithm and experimental validation for targeted killing of specific pathogenic bacteria |
| Biosortia Pharmaceuticals | San Diego | Development of drugs/therapeutics based on the cell to cell communication chemistry from mining of microbiome in the field of immuno-oncology and immunology |
| Boehringer Ingelheim | Fremont | Small Molecules, Biologics, Microbiome |
| Carverr | Brooklyn | Developing traceable probiotics and custom microbiomes |
| CHAIN Biotechnology | Nottingham, Marlow | Develop live biotherapeutics—these are novel drugs based on living bacteria found in the gut but engineered to deliver specific therapeutic molecules |
| ClostraBio | Chicago | Developing new therapeutics to treat food allergies and provide protective immunity |
| Consortia Therapeutics | La Jolla CA | Develops microbial therapies to prevent and treat human disease and allergies |
| Da Volterra | Paris | Development of microbiota protective therapy during antibiotic treatments to help prevent and cure human diseases |
| Diagnostic Solutions Laboratory | Alpharetta | Provides diagnostic solutions for identifying pathogenic organisms in stool through PCR and comprehensive stool testing for assessing GI health through DNA-based studies |
| EnteroBiotix | UK | Full therapeutic potential of fecal microbiota transplantation (FMT) through a field-leading GMP-compliant minimally manipulated microbiome platform and an ex vivo microbiome engineering platform |
| Enterome | Cambridge and Paris | Discovery and development of novel therapeutics upon understanding the gut microbe and immune system interaction; production of small molecules and peptides with a focus on cancer, autoimmune, inflammatory and metabolic diseases |
| Evolve Biosystems | Davis | Developing the next generation of products to establish, restore and maintain a healthy newborn gut microbiome |
| Federation Bio | South San Francisco | Therapeutic approaches of using beneficial microbes for curing human diseases like secondary hyperoxaluria, metabolic disorders, cancer, immune diseases |
| Finch Therapeutics | Somerville MA | Developing oral microbiome drugs for recurrent C. difficile, chronic hepatitis B, inflammatory bowel disease and children with autism and significant GI symptoms |
| Gusto Global | Morrisville NC | Development of novel live biotherapeutic products through advanced computational and microbiological tools for targeted immune modulation and optimization of metabolic pathways |
| Kaleido Biosciences | Bedford MA | Develop microbial metabolic therapies (MMT) to understand the disease pathways, such as the use of synthetic glycans, harnessing target enzymes across the microbial taxa, formulation of therapeutic products to mitigate serious complications of Covid-19, treat IBD, urea cycle disorders and hepatic encephalopathy |
| Locus Biosciences | Morrisville NC | Works on CRISPR-Phage (crPhage) platform combines the antibacterial power of CRISPR-Cas3 with the efficient, safe delivery of bacterial viruses called bacteriophage. The lead targets are E. coli, Clostridium difficile and organisms causing respiratory infections |
| MaaT | France | Patient specific biotherapeutics for improving the survival outcome of blood cancer through proprietary data collection and analysis on CGMP platform |
| Metabiomics | Aurora CO, Manassas VA, Chevy Chase MD; US | Developing a non-invasive microbiome test for the earlier and more accurate detection of colon polyps and colorectal cancer |
| Microbiotica Limited | Cambridge, UK | Identifies gut bacteria linked to phenotype with unprecedented precision in order to discover and develop live bacterial therapeutics and biomarkers |
| Novome Biotechnologies | South San Francisco CA; United States | Synthetic biology and microbial therapies; selecting potential bacteria from the human gut microbiome, engineering them to express therapeutic products to treat chronic disease |
| Oragenics | Florida, US | Production of lantibiotics helpful to recover microbial antibiotic resistance and prebiotics active in weight management in obese people |
| TargEDys | Rouen; France | Appetite regulation via molecular mimicry of pharmacological targets controlling gut brain axis involving bacterial, hormonal mimetic, proteins naturally occurring in the gut microbiome. They target to regulate/ moderate appetite in obese and/or old aged people |
| Sun Genomics | San Diego CA, USA | Customises probiotics constituting gut microbes, such as Lactobacillus acidophillus, L. rhamnosus, Bifidobacterium lactis |
| Synlogic | Cambridge, MA | Develop biotic medicines to treat various metabolic diseases like phenylketonuria, hyperoxaluria as well as immunomodulation therapies to treat diseases like cancer, IBS, etc |
| Sugarlogix | Berkeley CA, USA | Produces complex sugars with prebiotic functions which selectively feed the beneficial human gut bacteria strengthening the immune and nervous functions in turn |
| Seres Therapeutics | Cambridge, MA | Formulates and develops microbiome therapeutics to cure recurrent C. difficile, ulcerative colitis, metastatic melanoma and antibiotic resistant bacterial disease and graft versus host disease, |
| Vedanta BioSciences | Cambridge MA, United States | Leveraging live therapeutics made up of gut bacterial consortia which can stimulate immunoregulatory responses controlling allergic diseases as well as holds roles in cancer and vaccination |
| Xeno Biosciences | Los Angeles CA, United States | Engineer gut bacterial consortia addressing obesity and metabolism-related diseases; oral formulation of probiotic composition to cause weight loss by mimicking the microbiome changes induced by gastric bypass surgery |
Concluding remarks
The complex and dynamic population of microorganisms in the human gut and its relationship with human health and disease has been the subject of extensive research in the last two decades. Several research studies have reported that the human gut microbiome contribute immensely in regulating human health including metabolic diseases, neuropsychological disorders and immunological response. Exciting discoveries on targeted roles of microbial communities on host metabolism, increasing knowledge on keystone species relating to particular disease states through high-throughput sequencing data, understanding links between the bacterial activation of host cells through expression of special metabolites and production of host signaling molecules in turn could open up new avenues to treat infectious and chronic diseases in a much healthy and chemical-free manner. Deeper understanding of the roles of gut microbial species in host immune activation and regulation of central nervous system would be highly rewarding in treating several autoimmune diseases and neuropsychiatric disorders which either remain untreated or suffer from various adverse side-effects upon chemical therapy. Researchers worldwide have started utilizing the knowledge excavated from microbiome mining studies into the translational pipeline to treat and prevent various human diseases. However, even deeper insights would be needed to completely unravel the complexity of human gut microbiome or the second genome and realize its enormous potential to serve mankind.
Acknowledgements
The authors acknowledge KRC-NEERI for checking the manuscript through anti-plagiarism software iTHENTICATE (KRC No. CSIR-NEERI/KRC/2021/JAN/KZC/1). The authors duly acknowledge Department of Biotechnology, Government of India for funding the project BT/PR31826BIC/101/1211/2019 to Dr. Soma Ghosh.
Author contributions
Conceptualization and original draft preparation [SG], Review and editing [SP, SG].
Funding
The work was funded by Department of Biotechnology, Government of India (BT/PR31826BIC/101/1211/2019).
Availability of data and material
Not Applicable.
Code availability
Not Applicable.
Declarations
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors agree to publish.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abt MC, McKenney PT, Pamer EG. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol. 2016;14(10):609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atashgahi S, Shetty SA, Smidt H, de Vos WM. Flux, impact, and fate of halogenated xenobiotic compounds in the gut. Front Physiol. 2018;9(JUL):1–18. doi: 10.3389/fphys.2018.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Jun W. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Bang C, Vierbuchen T, Gutsmann T, Heine H, Schmitz RA. Immunogenic properties of the human gut-associated archaeon Methanomassiliicoccus luminyensis and its susceptibility to antimicrobial peptides. PLoS One. 2017;12(10):e0185919. doi: 10.1371/journal.pone.0185919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batani G, Bayer K, Böge J, et al. Fluorescence in situ hybridization (FISH) and cell sorting of living bacteria. Sci Rep. 2019;9:18618. doi: 10.1038/s41598-019-55049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369(9573):1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation yasmine. Cell. 2015;157(1):121–141. doi: 10.1016/j.cell.2014.03.011.Role. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya P, Jacks G, Ahmed KM, Routh J, Khan AA. Arsenic in groundwater of the Bengal Delta Plain aquifers in Bangladesh. B Environ Contam Tox. 2002;69(4):538–545. doi: 10.1007/s00128-002-0095-5. [DOI] [PubMed] [Google Scholar]
- Blachier F, Beaumont M, Portune KJ, Steuer N, Lan A, Audebert M, Khodorova N, Andriamihaja M, Airinei G, Benamouzig R. High-protein diets for weight management: Interactions with the intestinal microbiota and consequences for gut health. A position paper by the my new gut study group. Clin Nutr. 2019;38(3):1012–1022. doi: 10.1016/j.clnu.2018.09.016. [DOI] [PubMed] [Google Scholar]
- Bliss ES, Whiteside E. The gut-brain axis, the human gut microbiota and their integration in the development of obesity. Front Physiol. 2018 doi: 10.3389/fphys.2018.00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth LH, O’Halloran K. A comparison of biomarker responses in the earthworm Aporrectodea caliginosa to the organophosphorus insecticides diazinon and chlorpyrifos. Environ Toxicol Chem. 2001;20(11):2494–2502. doi: 10.1002/etc.5620201115. [DOI] [PubMed] [Google Scholar]
- Borre YE, O’Keeffe GW, Clarke G, Stanton C, Dinan TG, Cryan JF. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol Med. 2014;20(9):509–518. doi: 10.1016/j.molmed.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Bracco P, Janssen DB, Schallmey A. Selective steroid oxyfunctionalisation by CYP154C5, a bacterial cytochrome P450. Microb Cell Fact. 2013;12(1):1–11. doi: 10.1186/1475-2859-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant C, DeLuca M. Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacter cloacae. J Biol Chem. 1991;266(7):4119–4125. doi: 10.1016/S0021-9258(20)64294-6. [DOI] [PubMed] [Google Scholar]
- Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165(7):1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull MJ, Plummer NT. Part 1: The human gut microbiome in health and disease. Integr Med (encinitas) 2014;13(6):17. [PMC free article] [PubMed] [Google Scholar]
- Bürger S, Stolz A. Characterisation of the flavin-free oxygen-tolerant azoreductase from Xenophilus azovorans KF46F in comparison to flavin-containing azoreductases. Appl Microbiol Biotechnol. 2010;87(6):2067–2076. doi: 10.1007/s00253-010-2669-1. [DOI] [PubMed] [Google Scholar]
- Calvani R, Picca A, Lo Monaco MR, Landi F, Bernabei R, Marzetti E. Of microbes and minds: a narrative review on the second brain aging. Front Med. 2018;5:53. doi: 10.3389/fmed.2018.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminero A, Herrán AR, Nistal E, Pérez-Andrés J, Vaquero L, Vivas S, Ruiz de Morales JMG, Albillos SM, Casqueiro J. Diversity of the cultivable human gut microbiome involved in gluten metabolism: Isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol Ecol. 2014;88(2):309–319. doi: 10.1111/1574-6941.12295. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS One. 2012;7(6):e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carberry CA, Waters SM, Kenny DA, Creevey CJ. Rumen methanogenic genotypes differ in abundance according to host residual feed intake phenotype and diet type. Appl Environ Microbiol. 2014;80(2):586–594. doi: 10.1128/AEM.03131-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Turnbaugh PJ. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin Investig. 2014;124(10):4173–4181. doi: 10.1172/JCI72335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cénit MC, Matzaraki V, Tigchelaar EF, Zhernakova A. Rapidly expanding knowledge on the role of the gut microbiome in health and disease. Biochim Biophys Acta Mol Basis Dis. 2014;1842(10):1981–1992. doi: 10.1016/j.bbadis.2014.05.023. [DOI] [PubMed] [Google Scholar]
- Chapman MJ, Deane AM, O’Connor SL, Nguyen NQ, Fraser RJL, Richards DB, Hacquoil KE, Johnson LSV, Barton ME, Dukes GE. The effect of camicinal (GSK962040), a motilin agonist, on gastric emptying and glucose absorption in feed-intolerant critically ill patients: a randomized, blinded, placebo-controlled, clinical trial. Crit Care. 2016;20(1):232. doi: 10.1186/s13054-016-1420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau MR, Isabella VM, Li N, Kurtz CB. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat Commun. 2020;11(1):1–11. doi: 10.1038/s41467-020-15508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Shih K, Hu M, Li F, Liu C, Wu W, Tong H. Biostimulation of indigenous microbial communities for anaerobic transformation of pentachlorophenol in paddy soils of Southern China. J Agric Food Chem. 2012;60(12):2967–2975. doi: 10.1021/jf204134w. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Ning MX, Chen DK, Ma WT. Interactions between the gut microbiota and the host innate immune response against pathogens. Front Immunol. 2019;10:1–11. doi: 10.3389/fimmu.2019.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Sun F, Li S, Gao M, Wang L, Sarhan M, Abdel-Rahman MA, Li W, Kwok HF, Wu Y. Inhibitory activity of a scorpion defensin BmKDfsin3 against Hepatitis C virus. Antibiotics. 2020;9(1):33. doi: 10.3390/antibiotics9010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimerel C, Emery E, Summers DK, Keyser U, Gribble FM, Reimann F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep. 2014;9(4):1202–1208. doi: 10.1016/j.celrep.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JC, Tiedje JM. Bacterial species determination from DNA-DNA hybridization by using genome fragments and DNA microarrays. Appl Environ Microbiol. 2001;67(8):3677–3682. doi: 10.1128/AEM.67.8.3677-3682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K-T, Stevens SE, Cerniglia CE. The reduction of azo dyes by the intestinal microflora. Crit Rev Microbiol. 1992;18(3):175–190. doi: 10.3109/10408419209114557. [DOI] [PubMed] [Google Scholar]
- Chung J-G, Hsia T-C, Kuo HM, Li YC, Lee YM, Lin SS, Hung CF. Inhibitory actions of luteolin on the growth and arylamine N-acetyltransferase activity in strains of Helicobacter pylori from ulcer patients. Toxicol in Vitro. 2001;15(3):191–198. doi: 10.1016/S0887-2333(01)00015-7. [DOI] [PubMed] [Google Scholar]
- Ciesielski S, Bułkowska K, Dabrowska D, Kaczmarczyk D, Kowal P, Możejko J. Ribosomal intergenic spacer analysis as a tool for monitoring methanogenic archaea changes in an anaerobic digester. Curr Microbiol. 2013;67(2):240–248. doi: 10.1007/s00284-013-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G, Sandhu KV, Griffin BT, Dinan TG, Cryan JF, Hyland NP. Gut reactions: Breaking down xenobiotic–microbiome interactions. Pharmacol Rev. 2019;71(2):198–224. doi: 10.1124/pr.118.015768. [DOI] [PubMed] [Google Scholar]
- Claus SP, Guillou H, Ellero-Simatos S. Erratum: the gut microbiota: a major player in the toxicity of environmental pollutants? Npj Biofilms Microbiomes. 2017 doi: 10.1038/npjbiofilms.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covasa M, Stephens RW, Toderean R, Cobuz C. Intestinal sensing by gut microbiota: targeting gut peptides. Front Endocrinol. 2019 doi: 10.3389/fendo.2019.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings J, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Argenio V, Salvatore F. The role of the gut microbiome in the healthy adult status. Clin Chim Acta. 2015;451:97–102. doi: 10.1016/j.cca.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Das A, Srinivasan M, Ghosh TS, Mande SS. Xenobiotic metabolism and gut microbiomes. PLoS One. 2016;11(10):1–26. doi: 10.1371/journal.pone.0163099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1–2):84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- DeMartino P, Cockburn DW. Resistant starch: impact on the gut microbiome and health. Curr Opin Biotechnol. 2020;61:66–71. doi: 10.1016/j.copbio.2019.10.008. [DOI] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(5):1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- Di Nardo G, Gilardi G. Optimization of the bacterial cytochrome P450 BM3 system for the production of human drug metabolites. Int J Mol Sci. 2012;13(12):15901–15924. doi: 10.3390/ijms131215901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockray GJ. Gastrointestinal hormones and the dialogue between gut and brain. J Physiol Paris. 2014;592(14):2927–2941. doi: 10.1113/jphysiol.2014.270850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B, Henry M, El Khechine A, Raoult D, Drancourt M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS One. 2009;4(9):e7063. doi: 10.1371/journal.pone.0007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Gupta SD, Gupta A, Sarkar J, Roy S, Mukherjee A, Sar P. Exploration of deep terrestrial subsurface microbiome in Late Cretaceous Deccan traps and underlying Archean basement. India Sci Rep. 2018;8(1):1–16. doi: 10.1038/s41598-018-35940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers S, Safdar N, Sethi AK, Suen G, Peppard PE, Kates AE, Skarlupka JH, Kanarek M, Malecki KMC. Urinary lead concentration and composition of the adult gut microbiota in a cross-sectional population-based sample. Environ Int. 2019;133:105122. doi: 10.1016/j.envint.2019.105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefterios Venizelos Bezirtzoglou E. Intestinal cytochromes P450 regulating the intestinal microbiota and its probiotic profile. Microb Ecol Health Dis. 2012;23(1):18370. doi: 10.3402/mehd.v23i0.18370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes ML, McAllister AK. Maternal immune activation: implications for neuropsychiatric disorders. Science. 2016;353(6301):772–777. doi: 10.1126/science.aag3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Éva S, Massie AR, Levente K. Butyrate, a metabolite of intestinal bacteria, enhances sleep. Sci Rep. 2019;9(1):7035. doi: 10.1038/s41598-019-43502-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Cani PD. Gut microbiota and GLP-1. Rev Endocr Metab Disord. 2014;15(3):189–196. doi: 10.1007/s11154-014-9288-6. [DOI] [PubMed] [Google Scholar]
- Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10(8):538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- Fellows R, Denizot J, Stellato C, Cuomo A, Jain P, Stoyanova E, Balázsi S, Hajnády Z, Liebert A, Kazakevych J. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun. 2018;9(1):1–15. doi: 10.1038/s41467-017-02651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791 [DOI] [PubMed]
- Fernandez-Feo M, Wei G, Blumenkranz G, Dewhirst FE, Schuppan D, Oppenheim FG, Helmerhorst EJ. The cultivable human oral gluten-degrading microbiome and its potential implications in coeliac disease and gluten sensitivity. Clin Microbiol Infec. 2013;19(9):E386–E394. doi: 10.1111/1469-0691.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley KA, Ossenkopp K-P, Kavaliers M, MacFabe DF. Pre-and neonatal exposure to lipopolysaccharide or the enteric metabolite, propionic acid, alters development and behavior in adolescent rats in a sexually dimorphic manner. PLoS One. 2014;9(1):e87072. doi: 10.1371/journal.pone.0087072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost G, Sleeth ML, Sahuri-Arisoylu M, Lizarbe B, Cerdan S, Brody L, Anastasovska J, Ghourab S, Hankir M, Zhang S. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun. 2014;5(1):1–11. doi: 10.1038/ncomms4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469(7331):543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- Fukui H, Xu X, Miwa H. Role of gut microbiota-gut hormone axis in the pathophysiology of functional gastrointestinal disorders. J Neurogastroenterol Motil. 2018;24(3):367–386. doi: 10.5056/jnm18071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20(2):145. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaci N, Borrel G, Tottey W, O’Toole PW, Brugère JF. Archaea and the human gut: new beginning of an old story. World J Gastroenterol. 2014;20(43):16062. doi: 10.3748/wjg.v20.i43.16062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, Li T, Yin Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Mi. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard P, Béguet F, Lepercq P, Rigottier-Gois L, Rochet V, Andrieux C, Juste C. Gnotobiotic rats harboring human intestinal microbiota as a model for studying cholesterol-to-coprostanol conversion. FEMS Microbiol Ecol. 2004;47(3):337–343. doi: 10.1016/S0168-6496(03)00285-X. [DOI] [PubMed] [Google Scholar]
- Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, Nelson KE, White O, Methé BA, Huttenhower C. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10(8):e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Sar P. Identification and characterization of metabolic properties of bacterial populations recovered from arsenic contaminated ground water of North East India (Assam) Wat Res. 2013;47(19):6992–7005. doi: 10.1016/j.watres.2013.08.044. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Mohapatra B, Satyanarayana T, Sar P. Molecular and taxonomic characterization of arsenic (As) transforming Bacillus sp. strain IIIJ3–1 isolated from As-contaminated groundwater of Brahmaputra river basin, India. BMC Microbiol. 2020;20(1):1–20. doi: 10.1186/s12866-020-01893-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giloteaux L, Holmes DE, Williams KH, Wrighton KC, Wilkins MJ, Montgomery AP, Smith JA, Orellana R, Thompson CA, Roper TJ, Long PE, Lovley DR. Characterization and transcription of arsenic respiration and resistance genes during in situ uranium bioremediation. ISME J. 2012;7(2):370–383. doi: 10.1038/ismej.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. Pattern recognition receptors: doubling up for the innate immune response. Cell. 2002;111(7):927–930. doi: 10.1016/S0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- Greiner TU, Bäckhed F. Microbial regulation of GLP-1 and L-cell biology. Mol Metab. 2016;5(9):753–758. doi: 10.1016/j.molmet.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet. 2003;361(9356):512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- Guindo CO, Davoust B, Drancourt M, Grine G. Diversity of methanogens in animals’ gut. Microorganisms. 2021;9:13. doi: 10.3390/microorganisms9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, Turnbaugh PJ. Developing a metagenomic view of xenobiotic metabolism. Pharmacol Res. 2013;69(1):21–31. doi: 10.1016/j.phrs.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmerhorst EJ, Zamakhchari M, Schuppan D, Oppenheim FG. Discovery of a novel and rich source of gluten-degrading microbial enzymes in the oral cavity. PLoS One. 2010;5(10):e13264. doi: 10.1371/journal.pone.0013264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrán AR, Pérez-Andrés J, Caminero A, Nistal E, Vivas S, de Morales JMR, Casqueiro J. Gluten-degrading bacteria are present in the human small intestine of healthy volunteers and celiac patients. Res Microbiol. 2017;168(7):673–684. doi: 10.1016/j.resmic.2017.04.008. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8(6):e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19(7):349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias MJ, Piñeiro R, Blanco M, Gallego R, Diéguez C, Gualillo O, González-Juanatey JR, Lago F. Growth hormone releasing peptide (ghrelin) is synthesized and secreted by cardiomyocytes. Cardiovasc Res. 2004;62(3):481–488. doi: 10.1016/j.cardiores.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Ishaq SL, Moses PL, Wright A-DG. The pathology of Methanogenic archaea in human gastrointestinal tract disease. Gut Microb. 2016 doi: 10.5772/64637. [DOI] [Google Scholar]
- Islam FS, Gault AG, Boothman C, Polya DA, Charnock JM, Chatterjee D, Lloyd JR. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature. 2004;430(6995):68–71. doi: 10.1038/nature02638. [DOI] [PubMed] [Google Scholar]
- Janssen DB, Dinkla IJT, Poelarends GJ, Terpstra P. Bacterial degradation of xenobiotic compounds: evolution and distribution of novel enzyme activities. Environ Microbiol. 2005;7(12):1868–1882. doi: 10.1111/j.1462-2920.2005.00966.x. [DOI] [PubMed] [Google Scholar]
- Jhangi S, Gandhi R, Glanz B, Cook S, Nejad P, Ward D, Weiner H (2014) Increased Archaea species and changes with therapy in gut microbiome of multiple sclerosis subjects (S24. 001). Neurol 82(10 Supplement)
- Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA. 2008;105(36):13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joscelyn J, Kasper LH. Digesting the emerging role for the gut microbiome in central nervous system demyelination. Mult Scler J. 2014;20(12):1553–1559. doi: 10.1177/1352458514541579. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR (1969) Evolution of protein molecules. Mammalian Protein Metabolis 3:21–132
- Kaoutari AE, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11(7):497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Kelly C, Flatt PR, McClenaghan NH. Cell-to-cell communication and cellular environment alter the somatostatin status of delta cells. Biochem Bioph Res Commun. 2010;399(2):162–166. doi: 10.1016/j.bbrc.2010.07.036. [DOI] [PubMed] [Google Scholar]
- Kenny DJ, Plichta DR, Shungin D, Koppel N, Hall AB, Fu B, Vasan RS, Shaw SY, Vlamakis H, Balskus EP. Cholesterol metabolism by uncultured human gut bacteria influences host cholesterol level. Cell Host Microbe. 2020;28(2):245–257. doi: 10.1016/j.chom.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd M, Modlin IM, Gustafsson BI, Drozdov I, Hauso O, Pfragner R. Luminal regulation of normal and neoplastic human EC cell serotonin release is mediated by bile salts, amines, tastants, and olfactants. Am J Physiol-Gastr L. 2008;295(2):G260–G272. doi: 10.1152/ajpgi.00056.2008. [DOI] [PubMed] [Google Scholar]
- Kim JY, Whon TW, Lim MY, Kim YB, Kim N, Kwon M-S, Kim J, Lee SH, Choi H-J, Nam I-H. The human gut archaeome: identification of diverse haloarchaea in Korean subjects. Microbiome. 2020;8(1):1–17. doi: 10.1186/s40168-020-00894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates toll-like receptor 5—dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201(4):534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk JL, Beaudette LA, Hart M, Moutoglis P, Klironomos JN, Lee H, Trevors JT. Methods of studying soil microbial diversity. J Microbiol Methods. 2004;58(2):169–188. doi: 10.1016/j.mimet.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Klemann C, Wagner L, Stephan M, von Hörsten S. Cut to the chase: a review of CD26/dipeptidyl peptidase-4’s (DPP4) entanglement in the immune system. Clin Exp Immunol. 2016;185(1):1–21. doi: 10.1111/cei.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Koppel N, Rekdal VM, Balskus EP. Chemical transformation of xenobiotics by the human gut microbiota. Science. 2017;356(6344):1246–1257. doi: 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn W, Osprian I, Kroutil W, Braunegg G, Faber K. Bacterial epoxide hydrolases of opposite enantiopreference. Biotechnol Lett. 1999;21(8):687–690. doi: 10.1023/A:1005565108510. [DOI] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874 [DOI] [PMC free article] [PubMed]
- Lancaster GI, Langley KG, Berglund NA, Kammoun HL, Reibe S, Estevez E, Weir J, Mellett NA, Pernes G, Conway JRW. Evidence that TLR4 is not a receptor for saturated fatty acids but mediates lipid-induced inflammation by reprogramming macrophage metabolism. Cell Metab. 2018;27(5):1096–1110. doi: 10.1016/j.cmet.2018.03.014. [DOI] [PubMed] [Google Scholar]
- Larraufie P, Doré J, Lapaque N, Blottière HM. TLR ligands and butyrate increase Pyy expression through two distinct but inter-regulated pathways. Cell Microbiol. 2017;19(2):e12648. doi: 10.1111/cmi.12648. [DOI] [PubMed] [Google Scholar]
- Lazar V, Ditu LM, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, Picu A, Petcu L, Chifiriuc MC. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front Immunol. 2018;9(AUG):1–18. doi: 10.3389/fimmu.2018.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Yamada T. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Lebrun LJ, Lenaerts K, Kiers D, de Barros J-PP, Le Guern N, Plesnik J, Thomas C, Bourgeois T, Dejong CHC, Kox M. Enteroendocrine L cells sense LPS after gut barrier injury to enhance GLP-1 secretion. Cell Rep. 2017;21(5):1160–1168. doi: 10.1016/j.celrep.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Lee W-J, Hase K. Gut microbiota–generated metabolites in animal health and disease. Nat Chem Biol. 2014;10(6):416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M, Zhao L. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci. 2008;105(6):2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yi C-X, Katiraei S, Kooijman S, Zhou E, Chung CK, Gao Y, van den Heuvel JK, Meijer OC, Berbée JFP. Butyrate reduces appetite and activates brown adipose tissue via the gut-brain neural circuit. Gut. 2018;67(7):1269–1279. doi: 10.1136/gutjnl-2017-314050. [DOI] [PubMed] [Google Scholar]
- Li X, Brejnrod AD, Ernst M, Rykær M, Herschend J, Olsen NMC, Dorrestein PC, Rensing C, Sørensen SJ. Heavy metal exposure causes changes in the metabolic health-associated gut microbiome and metabolites. Environ Int. 2019;126(March):454–467. doi: 10.1016/j.envint.2019.02.048. [DOI] [PubMed] [Google Scholar]
- Lladó S, Baldrian P. Community-level physiological profiling analyses show potential to identify the copiotrophic bacteria present in soil environments. PLoS One. 2017;12(2):e0171638. doi: 10.1371/journal.pone.0171638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA: acetate CoA-transferase gene. Environ Microbiol. 2010;12(2):304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- Lu K, Mahbub R, Fox JG. Xenobiotics: interaction with the intestinal microflora. ILAR J. 2015;56(2):218–227. doi: 10.1093/ilar/ilv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafra D, Lobo JC, Barros AF, Koppe L, Vaziri ND, Fouque D. Role of altered intestinal microbiota in systemic inflammation and cardiovascular disease in chronic kidney disease. Future Microbiol. 2014;9(3):399–410. doi: 10.2217/fmb.13.165. [DOI] [PubMed] [Google Scholar]
- Maier TV, Lucio M, Lee LH, Verberkmoes NC, Brislawn CJ, Bernhardt J, Lamendella R, McDermott JE, Bergeron N, Heinzmann SS, Morton JT, González A, Ackermann G, Knight R, Riedel K, Krauss RM, Schmitt-Kopplin P, Jansson JK. Impact of dietary resistant starch on the human gut Microbiome, Metaproteome, and Metabolome. Mbio. 2017;8(5):1–16. doi: 10.1128/mBio.01343-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;323(8390):1311–1315. doi: 10.1016/S0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- Martin AM, Sun EW, Rogers GB, Keating DJ. The influence of the gut microbiome on host metabolism through the regulation of gut hormone release. Front Physiol. 2019;10:428. doi: 10.3389/fphys.2019.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo F, Ribeiro AC, Faveri M, Taddei C, Martinez MB, Mayer MPA. The domain Archaea in human mucosal surfaces. Clin Microbiol Infect. 2012;18(9):834–840. doi: 10.1111/j.1469-0691.2012.03958.x. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Mukai Y, Ogata D, Shozui F, Nduko JM, Taguchi S, Ooi T. Characterization of thermostable FMN-dependent NADH azoreductase from the moderate thermophile Geobacillus stearothermophilus. Appl Microbiol Biotechnol. 2010;86(5):1431–1438. doi: 10.1007/s00253-009-2351-7. [DOI] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152(1–2):39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckenstock RU, Elsner M, Griebler C, Lueders T, Stumpp C, Aamand J, van Breukelen BM. Biodegradation: updating the concepts of control for microbial cleanup in contaminated aquifers. Environ Sci Technol. 2015;49(12):7073–7081. doi: 10.1021/acs.est.5b00715. [DOI] [PubMed] [Google Scholar]
- Minalyan A, Gabrielyan L, Scott D, Jacobs J, Pisegna JR. The gastric and intestinal microbiome: role of proton pump inhibitors. Curr Gastroenterol Rep. 2017 doi: 10.1007/s11894-017-0577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misal SA, Gawai KR. Azoreductase: a key player of xenobiotic metabolism. Bioresour Bioprocess. 2018 doi: 10.1186/s40643-018-0206-8. [DOI] [Google Scholar]
- Mollica MP, Raso GM, Cavaliere G, Trinchese G, De Filippo C, Aceto S, Prisco M, Pirozzi C, Di Guida F, Lama A. Butyrate regulates liver mitochondrial function, efficiency, and dynamics in insulin-resistant obese mice. Diabetes. 2017;66(5):1405–1418. doi: 10.2337/db16-0924. [DOI] [PubMed] [Google Scholar]
- Momose Y, Hirayama K, Itoh K. Competition for proline between indigenous Escherichia coli and E. coli O157: H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157: H7. Antonie Leeuwenhoek. 2008;94(2):165–171. doi: 10.1007/s10482-008-9222-6. [DOI] [PubMed] [Google Scholar]
- Monachese M, Burton JP, Reid G. Bioremediation and tolerance of humans to heavy metals through microbial processes: a potential role for probiotics? Appl Environ Microbiol. 2012;78(18):6397–6404. doi: 10.1128/AEM.01665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisseau C. Role of epoxide hydrolases in lipid metabolism. Biochimie. 2013;95(1):91–95. doi: 10.1016/j.biochi.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JM, Wright CM, John GH. Identification, Isolation and characterization of a novel azoreductase from Clostridium perfringens. Anaerobe. 2012;18(2):229–234. doi: 10.1016/j.anaerobe.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Morrisson MS. Modernism, science, and technology. Bloomsbury Publishing; 2016. [Google Scholar]
- Mosca A, Leclerc M, Hugot JP. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front Microbiol. 2016;7:1–12. doi: 10.3389/fmicb.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses PL, Ishaq S, Gupta K, Maurer SM, Wright A-DG. Biodiversity of human gut methanogens varies with concentration of exhaled breath methane: 1256. Am J Gastroenterol Suppl. 2015;110:S552–S553. doi: 10.14309/00000434-201510001-01256. [DOI] [Google Scholar]
- Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14(10):667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, Cresci A, Silvi S, Orpianesi C, Verdenelli MC. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72(2):1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Médigue C, Koechler S, Barbe V, Barakat M, Talla E, Bertin PN. A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genet. 2007;3(4):e53. doi: 10.1371/journal.pgen.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murooka Y, Doi N, Harada T. Distribution of membrane-bound monoamine oxidase in bacteria. Appl Environ Microbiol. 1979;38(4):565–569. doi: 10.1128/aem.38.4.565-569.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murooka Y, Azakami H, Yamashita M. The monoamine regulon including syntheses of arylsulfatase and monoamine oxidase in bacteria. Biosci Biotechnol Biochem. 1996;60(6):935–941. doi: 10.1271/bbb.60.935. [DOI] [PubMed] [Google Scholar]
- Niers LEM, Timmerman HM, Rijkers GT, van Bleek GM, van Uden NOP, Knol EF, Kapsenberg ML, Kimpen JLL, Hoekstra MO. Identification of strong interleukin-10 inducing lactic acid bacteria which down-regulate T helper type 2 cytokines. Clin Exp Allergy. 2005;35(11):1481–1489. doi: 10.1111/j.1365-2222.2005.02375.x. [DOI] [PubMed] [Google Scholar]
- Nkamga VD, Henrissat B, Drancourt M. Archaea: essential inhabitants of the human digestive microbiota. Hum Microbiome J. 2017;3:1–8. doi: 10.1016/j.humic.2016.11.005. [DOI] [Google Scholar]
- Nosova T, Jokelainen K, Kaihovaara P, Heine R, Jousimies-Somer H, Salaspuro M. Characteristics of aldehyde dehydrogenases of certain aerobic bacteria representing human colonic flora. Alcohol Alcohol. 1998;33(3):273–280. doi: 10.1093/oxfordjournals.alcalc.a008391. [DOI] [PubMed] [Google Scholar]
- Offermans NSM, Vermeulen R, Burdorf A, Goldbohm RA, Kauppinen T, Kromhout H, van den Brandt PA. Occupational asbestos exposure and risk of pleural mesothelioma, lung cancer, and laryngeal cancer in the prospective Netherlands cohort study. J Occup Environ Med. 2014;56(1):6–19. doi: 10.1097/JOM.0000000000000060. [DOI] [PubMed] [Google Scholar]
- Okuda H, Ogura K, Kato A, Takubo H, Watabe T. A possible mechanism of eighteen patient deaths caused by interactions of sorivudine, a new antiviral drug, with oral 5-fluorouracil prodrugs. J Pharmacol Exp Ther. 1998;287(2):791–799. [PubMed] [Google Scholar]
- Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome. 2019;7(1):1–15. doi: 10.1186/s40168-019-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares M, Schüppel V, Hassan AM, Beaumont M, Neyrinck AM, Bindels LB, Benítez-Páez A, Sanz Y, Haller D, Holzer P, Delzenne NM. The potential role of the dipeptidyl peptidase-4-like activity from the gut microbiota on the Host Health. Front Microbiol. 2018;9:1–10. doi: 10.3389/fmicb.2018.01900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota N, Wong K, Valdez PA, Zheng Y, Crellin NK, Diehl L, Ouyang W. IL-22 bridges the lymphotoxin pathway with the maintenance of colonic lymphoid structures during infection with Citrobacter rodentium. Nat Immunol. 2011;12(10):941–948. doi: 10.1038/ni.2089. [DOI] [PubMed] [Google Scholar]
- Park HS, Schumacher R, Kilbane JJ. New method to characterize microbial diversity using flow cytometry. J Ind Microbiol Biotechnol. 2005;32(3):94–102. doi: 10.1007/s10295-005-0208-3. [DOI] [PubMed] [Google Scholar]
- Pavlova SI, Jin L, Gasparovich SR, Tao L. Multiple alcohol dehydrogenases but no functional acetaldehyde dehydrogenase causing excessive acetaldehyde production from ethanol by oral Streptococci. Microbiology. 2013;159:1437. doi: 10.1099/mic.0.066258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellock SJ, Redinbo MR. Glucuronides in the gut: Sugar-driven symbioses between microbe and host. J Biol Chem. 2017;292(21):8569–8576. doi: 10.1074/jbc.R116.767434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pen J, Welling GW. Influence of the microbial flora on the amount of CCK8-and secretin21–27-like immunoreactivity in the intestinal tract of mice. Comp Biochem Physiol B. 1983;76(3):585. doi: 10.1016/0305-0491(83)90298-5. [DOI] [PubMed] [Google Scholar]
- Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. Acetate mediates a microbiome–brain–β-cell axis to promote metabolic syndrome. Nature. 2016;534(7606):213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queipo-Ortuño MI, Seoane LM, Murri M, Pardo M, Gomez-Zumaquero JM, Cardona F, Casanueva F, Tinahones FJ. Gut microbiota composition in male rat models under different nutritional status and physical activity and its association with serum leptin and ghrelin levels. PloS One. 2013;8(5):e65465. doi: 10.1371/journal.pone.0065465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii F, Franklin W, Cerniglia CE. Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl Environ Microbiol. 1990;56(7):2146–2151. doi: 10.1128/aem.56.7.2146-2151.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna BS. Role of the gut microbiota in human nutrition and metabolism. J Gastroenterol Hepatol. 2013;28:9–17. doi: 10.1111/jgh.12294. [DOI] [PubMed] [Google Scholar]
- Ramírez-Pérez O, Cruz-Ramón V, Chinchilla-López P, Méndez-Sánchez N. The role of the gut microbiota in bile acid metabolism. Ann Hepatol. 2018;16(1):21–26. doi: 10.5604/01.3001.0010.5494. [DOI] [PubMed] [Google Scholar]
- Ricart E, Taylor WR, Loftus EV, O’Kane D, Weinshilboum RM, Tremaine WJ, Harmsen WS, Zinsmeister AR, Sandborn WJ. N-acetyltransferase 1 and 2 genotypes do not predict response or toxicity to treatment with mesalamine and sulfasalazine in patients with ulcerative colitis. Am J Gastroenterol Suppl. 2002;97(7):1763–1768. doi: 10.1111/j.1572-0241.2002.05838.x. [DOI] [PubMed] [Google Scholar]
- Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21(9):793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16(6):341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4(2):158–164. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425 [DOI] [PubMed]
- Satoh-Takayama N, Vosshenrich CAJ, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention J-J, Thiam K, Cerf-Bensussan N, Mandelboim O. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29(6):958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Schlee M, Wehkamp J, Altenhoefer A, Oelschlaeger TA, Stange EF, Fellermann K. Induction of human β-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infect Immun. 2007;75(5):2399–2407. doi: 10.1128/IAI.01563-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach RP, Escher BI, Fenner K, Hofstetter TB, Johnson CA, Von Gunten U, Wehrli B. The challenge of micropollutants in aquatic systems. Science. 2006;313(5790):1072–1077. doi: 10.1126/science.1127291. [DOI] [PubMed] [Google Scholar]
- Schwieger F, Tebbe CC. A new approach to utilize PCR–single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl Environ Microbiol. 1998;64(12):4870–4876. doi: 10.1128/AEM.64.12.4870-4876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellau J, Alvarado CF, Hoenow S, Mackroth MS, Kleinschmidt D, Huber S, Jacobs T. IL-22 dampens the T cell response in experimental malaria. Sci Rep. 2016;6(1):1–11. doi: 10.1038/srep28058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E, Weibel S, Wehkamp J, Oelschlaeger TA. Construction of recombinant E. coli Nissle 1917 (EcN) strains for the expression and secretion of defensins. Int J Med Microbiol Suppl. 2012;302(6):276–287. doi: 10.1016/j.ijmm.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Sherwin E, Dinan TG, Cryan JF. Recent developments in understanding the role of the gut microbiota in brain health and disease. Ann N Y Acad Sci. 2018;1420(1):5–25. doi: 10.1111/nyas.13416. [DOI] [PubMed] [Google Scholar]
- Shin SM, Yang JH, Ki SH. Role of the Nrf2-ARE pathway in liver diseases. Oxidative Med Cell Longev. 2013 doi: 10.1155/2013/763257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol. 2020;11(January):1–14. doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Microbiol. 1996;81(3):288–302. doi: 10.1111/j.1365-2672.1996.tb04331.x. [DOI] [PubMed] [Google Scholar]
- Smith EA, MacFarlane GT. Enumeration of amino acid fermenting bacteria in the human large intestine: Effects of pH and starch on peptide metabolism and dissimilation of amino acids. FEMS Microbiol Ecol. 1998;25(4):355–368. doi: 10.1016/S0168-6496(98)00004-X. [DOI] [Google Scholar]
- Smith PD, Janoff EN, Mosteller-Barnum M, Merger M, Orenstein JM, Kearney JF, Graham MF. Isolation and purification of CD14-negative mucosal macrophages from normal human small intestine. J Immunol Methods. 1997;202(1):1–11. doi: 10.1016/S0022-1759(96)00204-9. [DOI] [PubMed] [Google Scholar]
- Smith PD, Smythies LE, Shen R, Greenwell-Wild T, Gliozzi M, Wahl SM. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 2011;4(1):31–42. doi: 10.1038/mi.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, Orenstein JM, Smith PD. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Investig. 2005;115(1):66–75. doi: 10.1172/JCI200519229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies LE, Shen R, Bimczok D, Novak L, Clements RH, Eckhoff DE, Bouchard P, George MD, Hu WK, Dandekar S. Inflammation anergy in human intestinal macrophages is due to Smad-induced IκBα expression and NF-κB inactivation. J Biol Chem. 2010;285(25):19593–19604. doi: 10.1074/jbc.M109.069955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M, Pozniak A, Boffito M. Pharmacokinetics and pharmacodynamics of drug interactions involving rifampicin, rifabutin and antimalarial drugs. J Antimicrob Chemother. 2008;62(5):872–878. doi: 10.1093/jac/dkn330. [DOI] [PubMed] [Google Scholar]
- Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol. 2016;14(5):273. doi: 10.1038/nrmicro.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šrut M, Menke S, Höckner M, Sommer S. Earthworms and Cadmium—Heavy metal resistant gut bacteria as indicators for heavy metal pollution in soils? BioRxiv. 2018;10:295444. doi: 10.1101/295444. [DOI] [PubMed] [Google Scholar]
- Stam MR, Danchin EGJ, Rancurel C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel. 2006;19(12):555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- Steubesand N, Kiehne K, Brunke G, Pahl R, Reiss K, Herzig K-H, Schubert S, Schreiber S, Fölsch UR, Rosenstiel P. The expression of the β-defensins hBD-2 and hBD-3 is differentially regulated by NF-κB and MAPK/AP-1 pathways in an in vitro model of Candida esophagitis. BMC Immunol. 2009;10(1):1–16. doi: 10.1186/1471-2172-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suez J, Elinav E. The path towards microbiome-based metabolite treatment. Nat Microbiol. 2017;2(6):1–5. doi: 10.1038/nmicrobiol.2017.75. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005;11(8):1131. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Theisen S, Berger S. Screening of epoxide hydrolase producing microorganisms for biotransformation of deoxynivalenol. Mycotoxin Res. 2005;21(1):71–73. doi: 10.1007/BF02954823. [DOI] [PubMed] [Google Scholar]
- Theron J, Cloete TE. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit Rev Microbiol. 2000;26(1):37–57. doi: 10.1080/10408410091154174. [DOI] [PubMed] [Google Scholar]
- Tochitani S, Ikeno T, Ito T, Sakurai A, Yamauchi T, Matsuzaki H. Administration of non-absorbable antibiotics to pregnant mice to perturb the maternal gut microbiota is associated with alterations in offspring behavior. PLoS One. 2016;11(1):e0138293. doi: 10.1371/journal.pone.0138293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofalo R, Cocchi S, Suzzi G. Polyamines and gut microbiota. Front Nutr. 2019;6:1–5. doi: 10.3389/fnut.2019.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:3. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-liggett C, Knight R, Gordon JI. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliou V, Pappa A, Petersen DR. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem Biol Interact. 2000;129(1–2):1–19. doi: 10.1016/S0009-2797(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Vijay N, Morris ME. Role of monocarboxylate transporters in drug delivery to the brain. Curr Pharm Des. 2014;20(10):1487–1498. doi: 10.2174/13816128113199990462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008;19(9):587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain Behav Immun. 2014;38:1–12. doi: 10.1016/j.bbi.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung Y-M. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang B, Wu J, Jiang X, Tang H, Nielsen OH. Modulation of gut microbiota in pathological states. Engineering. 2017;3(1):83–89. doi: 10.1016/J.ENG.2017.01.013. [DOI] [Google Scholar]
- Wehkamp J, Harder J, Wehkamp K, Wehkamp-von Meissner B, Schlee M, Enders C, Sonnenborn U, Nuding S, Bengmark S, Fellermann K. NF-κB-and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium. Infect Immun. 2004;10:5750–5758. doi: 10.1128/IAI.72.10.5750-5758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood IM, Holton SJ, Rodrigues-Lima F, Dupret J-M, Bhakta S, Noble MEM, Sim E. Expression, purification, characterization and structure of Pseudomonas aeruginosa arylamine N-acetyltransferase. Biochem J. 2005;385(2):605–612. doi: 10.1042/BJ20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci. 2009;106(10):3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windey K, de Preter V, Verbeke K. Relevance of protein fermentation to gut health. Mol Nutr Food Res. 2012;1:184–196. doi: 10.1002/mnfr.201100542. [DOI] [PubMed] [Google Scholar]
- Winston JA, Theriot CM. Diversification of host bile acids by members of the gut microbiota. Gut Microbes. 2020;11(2):158–171. doi: 10.1080/19490976.2019.1674124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3(1):4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SE, Hashimoto-Hill S, Woo V, Eshleman EM, Whitt J, Engleman L, Alenghat T. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature. 2020;586(7827):108–112. doi: 10.1038/s41586-020-2604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lian F, Zhao L, Zhao Y, Chen X, Zhang X, Guo Y, Zhang C, Zhou Q, Xue Z. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 2015;9(3):552–562. doi: 10.1038/ismej.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarandi SS, Peterson DA, Treisman GJ, Moran TH, Pasricha PJ. Modulatory effects of gut microbiota on the central nervous system: How gut could play a role in neuropsychiatric health and diseases. J Neurogastroenterol Motil. 2016;22(2):201–212. doi: 10.5056/jnm15146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahran SA, Ali-Tammam M, Hashem AM, Aziz RK, Ali AE. Azoreductase activity of dye-decolorizing bacteria isolated from the human gut microbiota. Sci Rep. 2019;9(1):1–14. doi: 10.1038/s41598-019-41894-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamakhchari M, Wei G, Dewhirst F, Lee J, Schuppan D, Oppenheim FG, Helmerhorst EJ. Identification of Rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PloS One. 2011;6(9):e24455. doi: 10.1371/journal.pone.0024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6(8):1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X, David YB, Laverde-Gomez JA, Dassa B, Sheridan PO, Duncan SH, Louis P, Henrissat B, Juge N, Koropatkin NM. Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcus bromii. Mbio. 2015;6(5):e01058–e1115. doi: 10.1128/mBio.01058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, Suez J, Mahdi JA, Matot E, Malka G, Kosower N, Rein M, Zilberman-Schapira G, Dohnalová L, Pevsner-Fischer M, Segal E. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Li S, Gan RY, Zhou T, Xu DP, Li HB (2015) Impacts of gut bacteria on human health and diseases. Int J Mol Sci 16(4):7493–7519 [DOI] [PMC free article] [PubMed]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, De Sauvage FJ. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14(3):282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Maiseyeu A, Davis SN, Rajagopalan S. DPP4 in cardiometabolic disease: recent insights from the laboratory and clinical trials of DPP4 inhibition. Circ Res. 2015;116(8):1491–1504. doi: 10.1161/CIRCRESAHA.116.305665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Hernandez-Sanabria E, Guan LL. Characterization of variation in rumen methanogenic communities under different dietary and host feed efficiency conditions, as determined by PCR-denaturing gradient gel electrophoresis analysis. Appl Environ Microbiol. 2010;76(12):3776–3786. doi: 10.1128/AEM.00010-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Raes J, Van Den Bogert B, Arumugam M, Booijink CCGM, Troost FJ, Bork P, Wels M, De Vos WM, Kleerebezem M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6(7):1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not Applicable.
Not Applicable.