ABSTRACT
Along with respiratory tract disease per se, viral respiratory infections can also cause extrapulmonary complications with a potentially critical impact on health. In the present study, we used an experimental model of influenza A virus (IAV) infection to investigate the nature and outcome of the associated gut disorders. In IAV-infected mice, the signs of intestinal injury and inflammation, altered gene expression, and compromised intestinal barrier functions peaked on day 7 postinfection. As a likely result of bacterial component translocation, gene expression of inflammatory markers was upregulated in the liver. These changes occurred concomitantly with an alteration of the composition of the gut microbiota and with a decreased production of the fermentative, gut microbiota-derived products short-chain fatty acids (SCFAs). Gut inflammation and barrier dysfunction during influenza were not attributed to reduced food consumption, which caused in part gut dysbiosis. Treatment of IAV-infected mice with SCFAs was associated with an enhancement of intestinal barrier properties, as assessed by a reduction in the translocation of dextran and a decrease in inflammatory gene expression in the liver. Lastly, SCFA supplementation during influenza tended to reduce the translocation of the enteric pathogen Salmonella enterica serovar Typhimurium and to enhance the survival of doubly infected animals. Collectively, influenza virus infection can remotely impair the gut’s barrier properties and trigger secondary enteric infections. The latter phenomenon can be partially countered by SCFA supplementation.
KEYWORDS: bacterial translocation, gut microbial dysbiosis, short-chain fatty acids, enteric infection, influenza
INTRODUCTION
Even though vaccines and antiviral drugs are available, influenza still constitutes a serious health problem and a key economic issue. Although influenza virus infection generally causes mild-to-moderate disease, it can sometimes, depending on the strain’s virulence and the host’s health status, trigger severe disease. Seasonal influenza leads to ∼300,000 deaths a year worldwide (1). Most of these influenza-related deaths are due to acute respiratory distress symptoms, compromised pulmonary functions, multiorgan dysfunction, and secondary bacterial infections (2, 3). Along with lung disease per se, influenza is frequently associated with extrapulmonary complications, including intestinal disorders (4). Indeed, the results of preclinical and clinical studies indicate that influenza can lead to nausea, vomiting, and/or diarrhea (4–8). The causes, nature, and consequences of these associated intestinal disorders have yet to be fully characterized.
We and others have shown that influenza alters the composition and function of the gut microbiota (6–10). The consequences of this dysbiosis on the disease’s outcome remain to be fully defined (11). Wang and colleagues (6) were the first to report a link between gut dysbiosis and intestinal inflammation. An impact of gut dysbiosis on secondary bacterial infection in the intestine has also been suggested (7, 8). Lastly, our recent findings indicate that by reducing the production of short-chain fatty acids (SCFAs; the main metabolites of the gut microbiota), influenza favors secondary bacterial infection of the lungs (10). With regard to the role of SCFAs in gut homeostasis (12–17), we hypothesize herein that the influenza virus infection’s impairment of SCFA production weakens the gut’s barrier function and thus favors secondary enteric infections.
Short-chain fatty acids represent the end products of dietary fiber fermentation (for reviews, see references 18 and 19). The SCFAs supply energy to colonocytes and are critical for intestinal homeostasis, gut functions, and gut metabolism. Although the SCFAs’ role in the control of gut inflammation appears to be a function of the disease state, these compounds tend to reinforce the gut barrier, a critical property for controlling the dissemination of gut commensals, opportunistic pathogens, and microbial components. Moreover, SCFAs display antimicrobial activity by favoring the synthesis of antimicrobial components (including antimicrobial peptides) in the intestine (17–20). In the present study, we sought to specify the nature of gut disorders (including disruption of barrier functions) during an experimental influenza A virus (IAV) infection. We found that (i) decreased SCFA production is important for these effects and (ii) SCFA supplementation during IAV infection partially protects animals from secondary infection with the enteric pathogen Salmonella enterica serovar Typhimurium. Taken as a whole, our data emphasize that viral respiratory infections can remotely impact gut homeostasis and intestinal barrier functions and thereby favor secondary bacterial infections. Reduced production of gut microbiota-derived fermentative products (SCFAs) might have a critical role in these alterations.
RESULTS
Influenza virus infection is associated with intestinal inflammation and disorders.
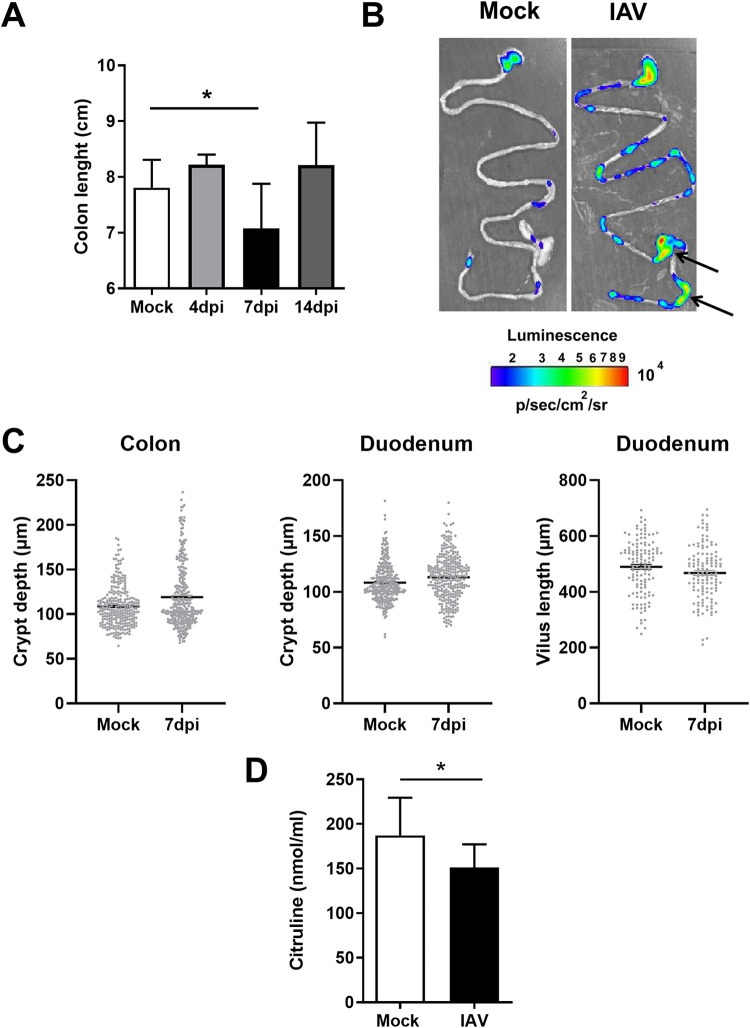
Influenza virus infection can be accompanied by intestinal symptoms, leading to intestinal inflammation and immune injury (5, 6). We first characterized the nature of these intestinal disorders in a mouse model of sublethal influenza virus infection (see Fig. S1A in the supplemental material). In line with a previous study (6), no IAV genomic RNA was detected in the intestine over the course of the infection (Fig. S1B). Relative to that in mock-infected mice, significant but transient colon shortening (a marker of inflammation) was observed at 7 days postinfection (dpi) in IAV-infected animals (Fig. 1A). The use of NF-κB–luciferase reporter mice also indicated an increase in intestinal NF-κB activity, a marker of stress and inflammation, at 7 dpi (Fig. 1B and Fig. S1C). The expression of NF-κB was most obvious in the cecum and colon but was also observed in the small intestine (Fig. 1B and Fig. S1D). In contrast (but in line with other published studies [7, 9]), the intestine had a normal histological appearance at 7 dpi, with no difference (i.e., no remodeling) in crypt depth in the colon and the crypt/villus length in the duodenum (Fig. 1C and Fig. S1E). Influenza virus infection was also associated with a low blood concentration of citrulline (a marker of the functional enterocyte mass and metabolic activity [21]) at 7 dpi (Fig. 1D). Overall, a sublethal influenza virus infection was associated with colon shortening, NF-κB activation, and a relative decrease in intestinal metabolic function but not major structural remodeling of the epithelium.
FIG 1.
Intestinal inflammation and disorders during IAV infection. Mice were infected or not (mock) with IAV (H3N2). (A) The colon length was measured at 4, 7, and 14 dpi (n = 6 to 13). For the mock control, colons were collected at day 7. (B) Ex vivo bioluminescence imaging was performed on guts collected from naive and IAV-infected (7 dpi) NF-κB–luciferase transgenic mice. Arrows indicate higher NK-κB expression. The scale indicates the average radiance. One representative image of at least 10 mice is shown. (C) Histological analysis of intestinal (colon and duodenum) sections in mock-treated mice and IAV-infected mice (7 dpi). Crypt depths and villus lengths were determined after hematoxylin and eosin coloration (n = 9 to 10/group). (D) Citrulline concentrations in the blood collected from mock-treated mice and IAV-infected mice (7 dpi) (n = 14). Results represent two pooled experiments. Significant differences were determined using the Mann-Whitney U test (C and D) and the Kruskal-Wallis ANOVA test (A). (*, P < 0.05).
Influenza leads to alterations in intestinal gene expression and impairment of the gut barrier.
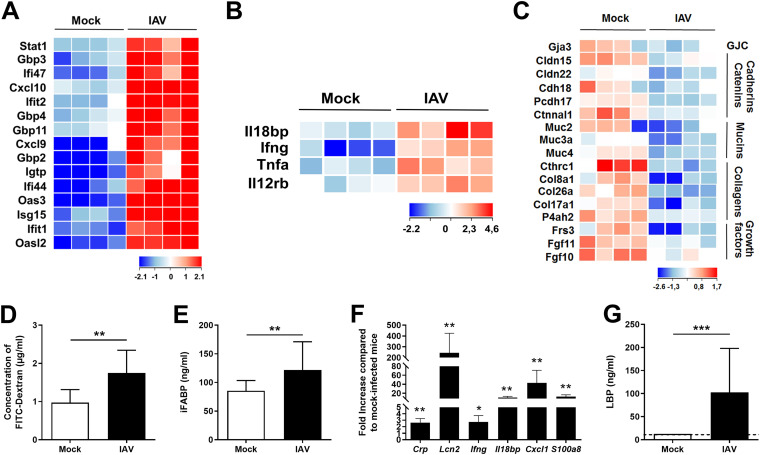
We next analyzed gene expression in the colons of the IAV-infected mice. The expression of a large number of genes (1,433 genes with a fold change of >2; adjusted P value, <0.05) was modulated at 7 dpi (Fig. S2A). An ontological analysis highlighted the upregulation of several families of genes with immune and inflammatory functions (Fig. S2B). Large numbers of interferon-stimulated genes (ISGs) and, to a lesser extent, NF-κB-dependent inflammatory genes were upregulated during influenza (Fig. 2A and B). A reverse transcription-quantitative PCR (RT-qPCR) analysis confirmed the transient alteration in gene expression in the colon (Fig. S2C). Many of the downregulated genes had cell functions (e.g., transport) and metabolic functions (e.g., O-linked glycosylation) (Fig. S2D). IAV infection was also associated with the altered expression of genes involved in mucin type O glycan biosynthesis (Fig. S2E). Transcripts of genes involved in barrier functions were also downregulated, albeit more moderately; these included the gap junction component Gja3, cadherin/catenin family members, collagens, and fibroblast growth factors (Fig. 2C and Fig. S2C).
FIG 2.
Altered colonic gene expression and increased gut permeability during IAV infection. (A to C) Transcriptomic analysis of colon samples from mock-treated and IAV-infected mice (7 dpi). Shown are heat maps representing ISGs (A), NF-κB-dependent inflammatory genes (B), and transcriptional expression of components involved in barrier functions and epithelial integrity (GJC, gap junction components) (C) (n = 4/group). (D) Fluorescence intensity quantified in the blood of mice 4 h after FITC-dextran oral administration (n = 8). (E) Intestinal fatty acid-binding protein (iFABP) concentration in the blood (n = 21 to 28). (F) Analysis of gene expression in liver by RT-qPCR (n = 5). (G) LPS-binding protein (LBP) concentration in the blood (n = 11 to 15). (D to G) Results are from two to three pooled experiments (7 dpi). Significant differences were determined using the Mann-Whitney U test (D, E, and G) and the Kruskal-Wallis ANOVA test (F) (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The barrier function is one of the gut’s most important features. Under normal conditions, an intact gut barrier prevents the excessive spreading of microbial components into the blood and peripheral tissues. Disease conditions can disrupt the barrier function and thus lead to bacterial translocation, systemic inflammation, and septic shock. IAV’s impact on the gut’s barrier property has not been characterized in detail and so warranted investigation. To this end, we orally administered fluorescein isothiocyanate (FITC)-dextran to IAV-infected mice and then assayed its concentration in plasma. Relative to the translocation of FITC-dextran from the gut lumen to the plasma in the control, enhanced translocation was observed at 7 dpi in infected mice (Fig. 2D). Accordingly, the blood concentration of intestinal fatty acid-binding protein (a systemic marker associated with altered intestinal permeability) was significantly elevated at 7 dpi (Fig. 2E). Disturbances in the intestinal barrier can result in the increased portal influx of bacteria or their products into the liver, where they can cause inflammation and acute injury. A quantitative RT-PCR analysis highlighted the upregulated expression of inflammatory genes in the liver, including acute-phase proteins (i.e., C-reactive protein, lipocalin-2), cytokines, chemokines, and antimicrobial peptides (Fig. 2F and data not shown). Lastly, the serum concentration of lipopolysaccharide (LPS)-binding protein (which is produced primarily as an acute-phase protein in the liver) was elevated at 7 dpi (Fig. 2G). Overall, influenza virus infection strongly modified gene expression in the gut and altered the intestinal epithelium’s barrier properties.
A restricted food intake mimicking influenza disease does not recapitulate intestinal inflammation and barrier dysfunction.
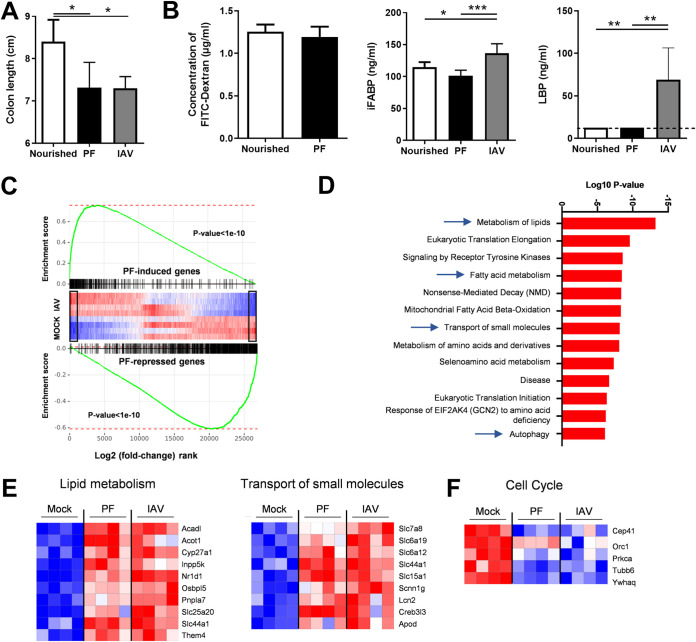
We and others have shown that reduced food intake and associated weight loss due to acute viral respiratory infection trigger an alteration of the composition of the gut microbiota (10, 22). As dysbiotic microbiotas can lead to gut inflammation and altered barrier properties, we investigated whether rapid reduction of food consumption, as observed during IAV infection, may lead to intestinal inflammation and gut disorders. To this end, we designed a pair-feeding experiment exactly as described previously (10). Pair-fed mice were sacrificed when weight loss attained 15% of the initial weight. As depicted in Fig. 3A, the colon length of pair-fed mice was significantly reduced, as is the case during an IAV infection. However, the intestinal barrier, as measured by FITC-dextran, intestinal fatty acid-binding protein, and LPS-binding protein in blood, was unchanged (Fig. 3B). Transcriptomic analysis of colons collected from pair-fed mice and IAV-infected mice revealed a global resemblance, as assessed by gene set enrichment analysis (GSEA) (Fig. 3C). However, among the major differences between the two signatures was the absence of ISG expression in pair-fed mice (Fig. S3A). Expression of genes belonging to the NF-κB pathway was also specifically altered in IAV-infected mice. In line with Fig. 3B, the expression of transcripts of genes involved in barrier functions were not decreased in the colons of pair-fed mice, unlike with IAV-infected mice (Fig. S3B). An ontological analysis highlighted the upregulation of several families of genes involved in metabolism, signaling, and transport in the colons of pair-fed mice (Fig. 3D). In particular, pathways involved in lipid (fatty acid) metabolism and the transport of small molecules were induced in pair-fed mice and IAV-infected mice (Fig. 3E). Families of genes found to be downregulated in pair-fed mice and in part in IAV-infected mice included genes involved in cell cycle and mitosis (Fig. 3F and Fig. S3C). Collectively, inappetence and rapid weight loss do not appear to play major roles in gut inflammation and an altered intestinal barrier during influenza virus infection.
FIG 3.
Analysis of gut disorders in pair-fed mice. The food access was restricted in order to mimic the weight loss of IAV-infected mice (7 dpi, ∼15% of body mass). (A) The colon lengths of pair-fed mice and normally nourished mice were measured. (B) FITC-dextran, intestinal fatty acid-binding protein (iFABP), and LPS-binding protein (LBP) concentrations in the blood. (C and D) Analysis of gene expression in the colons of pair-fed mice and IAV-infected mice. (C) GSEA plot showing enrichment of pair-feeding (PF)-induced (top) and pair-feeding-repressed (bottom) genes in mock-treated and IAV-infected mice. Black rectangles indicate statistically significant differences in the mock-versus-IAV comparison. Global IAV and pair-feeding signatures appear statistically correlated. (D) Gene set enrichment analysis was run on pair-feeding (PF)-induced genes in a comparison with those in the mock condition (fold change, >2; adjusted P value < 0.05) using the Reactome database. (E, left) Heatmap showing significantly modulated genes (fold change, >2; adjusted P value < 0.05) related to lipid metabolism from the Reactome database’s “Metabolism of lipids,” “Fatty acid metabolism,” and “Mitochondrial Fatty Acid Beta-Oxidation” pathways. (E, right) Heatmap representing the expression of genes from the Reactome database’s “Transport of small molecules” pathway. (F) Heatmap representing the expression of genes from the Reactome database’s “Cell Cycle” pathway. (A and B) Significant differences were determined using the Kruskal-Wallis ANOVA test and the Mann-Whitney U test (FITC dextran) (n = 6 to 12) (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
SCFA supplementation partially reverses intestinal barrier disruption.
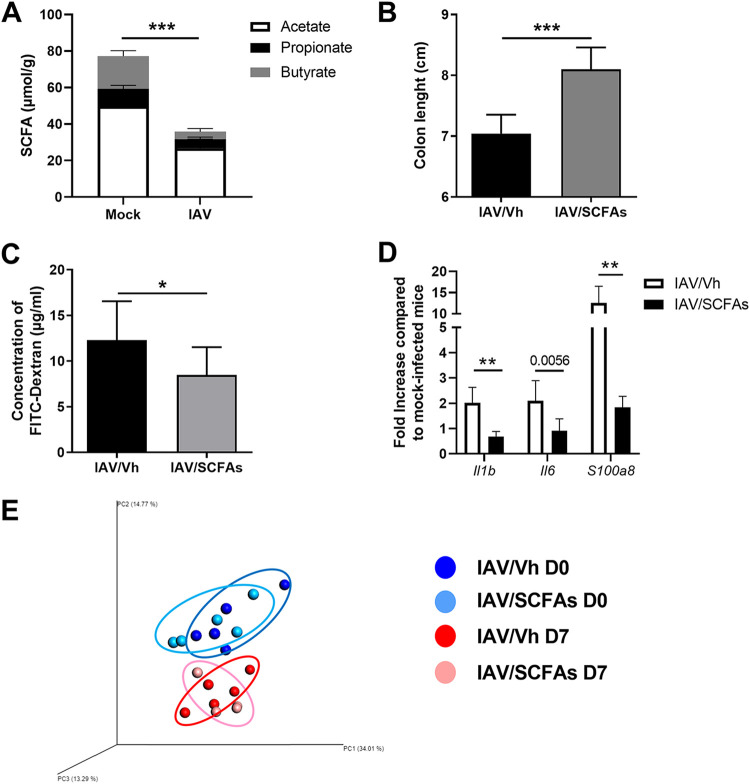
We have previously reported that influenza virus infection leads to gut dysbiosis and strongly reduces the production of the fermentative products SCFAs (10). In the IAV-infected mouse model studied here, we confirmed that cecal concentrations of acetate (the most abundant SCFA), propionate, and butyrate were abnormally low at 7 dpi (Fig. 4A). Although SCFAs are important for intestinal homeostasis and function, their role in gut inflammation is more context dependent (12, 15–17). We hypothesized that the drop in SCFA production during influenza virus infection might influence intestinal disorders. To this end, the mice’s drinking water was supplemented with acetate, propionate, and butyrate starting 2 days after IAV infection. Treatment with SCFAs moderately but significantly rescued the decrease in colon length in these animals and thus indicated that the inflammation was less intense (Fig. 4B). The translocation of FITC-dextran from the gut lumen to the plasma was reduced in SCFA-treated animals relative to that in controls (Fig. 4C). This finding indicates that SCFA treatment partially restored the intestine’s physical barrier. We then looked at whether the restoration of barrier function translated into less inflammation in the liver. As shown in Fig. 4D, the expression of inflammatory genes in the liver was significantly lower in mice fed SCFA-supplemented drinking water than in nonsupplemented mice. SCFA treatment may indirectly exert these effects by modulating the composition of the gut microbiota. To investigate this possibility, the feces from SCFA-treated and untreated mice, previously infected or not infected with IAV, were collected, and 16S rRNA sequencing was performed. An analysis of the beta diversity by principal-coordinate analysis (PCoA) clearly showed that, whatever the condition (mock or IAV infection), the bacterial populations from SCFA-treated mice and untreated mice did not differ (Fig. 4E). On the other hand, in line with the results in references 7, 8, and 10 and relative to mock-infected animals, PCoA clearly indicated an intergroup difference in the fecal microbiota at 7 dpi. A taxonomic analysis did not reveal any major changes at the phylum and lower taxonomic levels between SCFA-treated and untreated mice (Fig. S4A and S4B and data not shown). This indicates that SCFA treatment does not reverse intestinal barrier disruption by modulating the composition of the gut microbiota. Taken as a whole, the reduced production of SCFAs by the gut microbiota influences barrier leakage during influenza virus infection.
FIG 4.
Effect of SCFA supplementation during IAV infection on gut disorders. (A) Cecal concentrations of total and individual SCFAs in mock-treated and IAV-infected mice (7 dpi) (n = 7, one experiment out of at least six performed). (B to D) Mice were infected with IAV. Two days after infection, mice were treated with acetate, propionate, and butyrate (150 mM, 50 mM, 20 mM, respectively, in drinking water) or with vehicle (Vh). (B) The colon length was measured at 7 dpi (n = 6 to 9; one representative experiment out of two). (C) Fluorescence intensity quantified in the blood of mice 4 h after FITC-dextran oral administration (n = 14; two pooled experiments). (D) Gene expression in total liver was analyzed by RT-qPCR (n = 6; one representative experiment out of two). (E) Analysis of the gut microbiota’s composition was performed on feces collected from uninfected mice treated (bright blue) or not (dark blue) with SCFAs for 5 days and from mice infected 7 days earlier with IAV and treated (pink) or not (red) with SCFAs at 2 dpi (n = 4 to 5). Bacterial communities were clustered using PCoA of weighted UniFrac distance matrices (beta diversity). The first three principal coordinates (PC1, PC2, and PC3) are plotted for each sample, and the percentage variation in the plotted principal coordinates is indicated on the axes. Each spot represents one sample, and each group of mice is denoted by a different color. The distance between dots represents the extent of compositional difference. D0 and D7, days 0 and 7. Significant differences were determined using the Mann-Whitney U test (A to C) and the Kruskal-Wallis ANOVA test (D) (*, P < 0.05; **, P < 0.01, ***, P < 0.001).
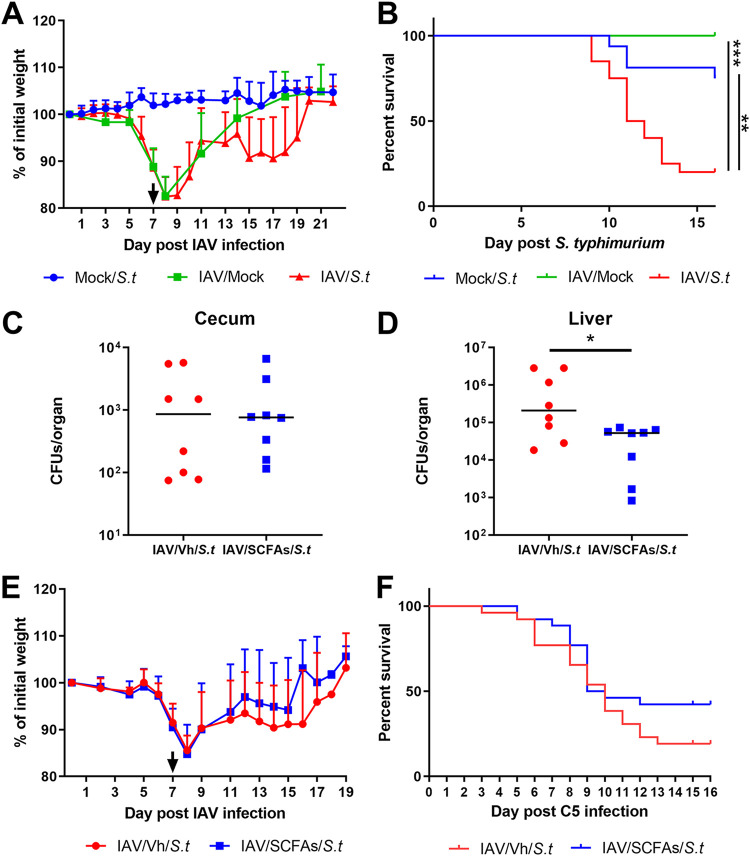
SCFA supplementation during influenza attenuates bacterial enteric infections.
Recent research indicates that gut alterations during an influenza virus infection may favor the local colonization and systemic dissemination of the foodborne pathogen Salmonella enterica serovar Typhimurium (7, 8). The underlying mechanism has yet to be defined. We postulated that the influenza-associated drop in intestinal SCFA production and epithelial permeability are causally related to these secondary enteric infections. To test this hypothesis, we developed a model of secondary enteric bacterial infection (Fig. S5A). Mice previously infected with IAV were orally exposed to S. Typhimurium. Unlike in other studies (7, 8), we deliberately did not treat the mice with antibiotics prior to S. Typhimurium infection. After preliminary dose-response experiments, a sublethal dose was inoculated into IAV-infected mice by gavage (7 dpi). We measured weight loss and the mortality index in doubly infected mice. In contrast to naive mice infected with S. Typhimurium or IAV alone, IAV-infected mice lost weight 6 to 7 days after S. Typhimurium infection (Fig. 5A and Fig. S5B). Lastly, IAV-infected mice succumbed to secondary enteric infection, while mice infected with S. Typhimurium or IAV alone did not (Fig. 5B). Of note, in contrast to IAV-infected mice and the control mice, pair-fed mice were not susceptible to secondary Salmonella infection (Fig. S5C).
FIG 5.
Effect of SCFA supplementation during IAV infection on secondary Salmonella Typhimurium (S.t) infection. (A and B) Mice were infected or not (mock) with IAV (H3N2). At 7 dpi, mice were (super)infected or not with S. Typhimurium. (A) Body weight evolution (percentages of initial body weights). (B) The survival of singly infected and doubly infected animals was monitored. (A and B) Results from two pooled experiments are shown (n = 16). (C to E) The same operation was repeated for IAV-infected mice treated or not with acetate, propionate, and butyrate (150 mM, 50 mM, and 20 mM, respectively, in drinking water) at 2 dpi. (C and D) The numbers of bacteria in the cecum (C) and liver (D) were determined 7 days after the bacterial challenge (n = 8; a representative experiment out of two). The body weight (E) and survival rate (F) of superinfected animals were monitored (n = 24; results from three pooled experiments are shown). (C and D) Significant differences were determined using the Mann-Whitney U test. (B, F) Rates of mouse survival were compared using Kaplan-Meier analysis and the log rank test (*, P < 0.05).
To investigate whether the local drop in SCFA levels during IAV infection was involved in secondary S. Typhimurium infection, the mice’s drinking water was supplemented with SCFAs from day 2 after influenza virus infection onwards (Fig. S5A). In oral infection models in mice, S. Typhimurium colonizes sites in the distal intestine, such as the cecum, before spreading to systemic tissues and shedding via the feces into the environment (23). SCFA treatment failed to lower the bacterial load in the cecum, indicating that SCFAs did not have a local impact on bacterial growth (Fig. 5C). Supplementation of SCFAs also failed to reduce the number of Salmonella organisms in the feces (not shown). This suggested that SCFA supplementation has no role in the local control of Salmonella infection. In line with this, SCFA supplementation failed to enhance the intestinal transcription of factors known to participate in the host defense against Salmonella (7), including cytokines and antimicrobial peptides (Fig. S5D). In contrast, SCFA supplementation reduced the translocation of bacteria, as revealed by the lower bacterial count in the liver (Fig. 5D). Lastly, the body weight loss due to secondary enteric infection and the mortality rate were lower in SCFA-supplemented mice, although the difference from controls was not statistically significant (Fig. 5E and F). We conclude that SCFA treatment during the course of influenza tended to reduce secondary enteric infection and mitigate the latter’s systemic consequences.
DISCUSSION
Our data support the hypothesis that viral respiratory infection can remotely trigger intestinal disorders (namely, mild inflammation and altered barrier functions) and thus have a major impact on secondary enteric infections. Furthermore, our data show that SCFA supplementation during an influenza virus infection ameliorated gut disorders and reduced secondary Salmonella infection. Given that gut dysfunction has been described in many critical illnesses, these relationships might have clinical relevance.
Several studies have highlighted the impact of influenza virus infection on gut homeostasis, with dysbiosis and inflammation (6–10, 22). In line with other researchers (7, 9), we did not observe major intestinal architecture remodeling at 7 dpi (i.e., when gut disorders peak). However, a shorter colon and altered intestinal metabolic functions (as revealed by systemic citrulline levels) indicated the presence of intestinal dysfunction in our mouse model. Furthermore, the present study is the first to have shown that influenza virus infection is associated with disruption of the intestine’s barrier functions. In line with the results of Deriu and colleagues (7), our overall transcript analysis indicated that the expression of a large panel of inflammatory (NF-κB-dependent) genes and immune genes (in particular, ISGs) was modulated in the intestine during an IAV infection. ISG expression probably reflects the increased systemic interferon levels during infection, since a quantitative PCR did not detect any genomic IAV RNA in the intestine (6, 10). Although speculative at this stage, a change in the enteric virome during influenza (exposure to virus-associated signals) might also influence local ISG expression (24). Influenza virus infection is also associated with a low expression of genes involved in the maintenance of barrier functions and with elevated paracellular permeability, as assessed by enhanced passage of FITC-dextran into the blood. This alteration in barrier function was combined with enhanced inflammatory gene expression in the liver, perhaps as a result of the portal translocation of bacterial components from the gut. Pair-feeding experiments indicated that reduced food consumption during influenza is not involved in gut inflammation and barrier disruption (and secondary Salmonella infection). There are probably many causes of intestinal barrier leakage. One can reasonably hypothesize that microbiota dysbiosis, altered microbiota-host interactions, and inflammation during influenza are causally linked to the alterations in barrier properties (25, 26). Hence, the loss of beneficial members of the microbial community (Lachnospiraceae and Lactobacillus), the overgrowth of pathosymbionts (Alphaproteobacteria, Gammaproteobacteria, and the Escherichia genus) and/or a shift in metabolism (such as altered fermentation) may accentuate the gut’s leakiness (10). Enhanced production of toxic metabolites may also be involved in barrier disruption. For instance, the excessive production of phenolic and sulfur-containing compounds by dysbiotic microbiota can alter tight junctions between cells and disrupt barrier functions (27, 28). Counts of mucin-degrading bacteria like Ruminococcus increase during influenza (10). These bacteria erode the colonic mucosa and favor interactions between luminal bacteria and the intestinal epithelium, which in turn may lead to inflammation and the impairment of barrier function (29).
The question of the impact of gut dysbiosis and intestinal disorders on influenza outcomes then arises. Increased leakiness of the gut barrier and dissemination of commensal bacteria and/or their components might lead to extraintestinal complications, such as acute liver injury, acute respiratory distress syndrome, bacterial respiratory tract coinfections, and systemic symptoms (e.g., a cytokine storm, circulatory collapse, sepsis, and multiorgan dysfunction) (11). These complications are observed in critically ill patients and constitute key causes of mortality (30–32). However, the extent to which gut perturbations cause extraintestinal disorders remains to be determined. Gut barrier dysfunction is a well-known feature of aging and chronic metabolic diseases, like obesity (25, 33–36). It would be interesting to look at whether these alterations amplify secondary extraintestinal complications in obese and older individuals, who are particularly at risk of developing critical illnesses during influenza (31). The dissemination of microbial components through the gut wall might affect not only inflammation and organ functions but also host metabolism (such as glucose homeostasis) and other outcomes (37–39). Our present data highlighted the altered expression of many inflammatory genes and metabolic genes (data not shown) in the liver. We also recently showed that the homeostasis of adipose tissue is dramatically altered during experimental influenza, with inflammation and metabolic perturbations (40). The impact of compromised gut barrier function on hepatic and adipose inflammation and metabolism warrants investigation.
Influenza virus infection is associated with a drop in the production of SCFAs; this phenomenon is due, at least in part, to reduced food (fiber) consumption by the diseased host (10). This drop may limit the energy supply to colonocytes, accentuate mucosal inflammation, and alter the intestine’s barrier functions. SCFAs are currently considered to be a promising adjunct treatment for active inflammatory bowel disease and diversion colitis (for a review, see reference 20). Various approaches (including enemas of butyrate and mixtures of acetate, propionate, and butyrate) have given differing clinical outcomes (41–43). In the present study, we observed a beneficial effect of SCFA supplementation on gut barrier functions, as measured by greater gut impermeability and diminished inflammatory gene expression in the liver. This effect was not associated with major changes in the composition of the gut microbiota. In contrast, we did not find a marked effect on local inflammatory gene expression (data not shown), even though SCFA supplementation rescued the colon shortening typically observed in a setting of inflammation. This is in line with suggestions that the SCFAs’ functional activities depend on the disease context and severity. Recent research indicates that gut disorders during influenza virus infection may favor the local colonization and systemic dissemination of the intestinal pathogenic bacterium Salmonella (7, 8), a leading cause of acute gastroenteritis and inflammatory diarrhea. Our data confirm these findings, although the underlying mechanism has yet to be defined. However, we postulated that the influenza-associated drop in intestinal SCFA production might cause, at least in part, secondary enteric infections. Acetate can protect against E. coli (13), while propionate and butyrate can protect against S. Typhimurium (44) and Citrobacter rodentium (45, 46). In these settings, SCFAs inhibited pathogen growth directly or (through host signaling) indirectly, leading to the production of cytokines and antimicrobial compounds (13, 45, 47, 48) and to increased bactericidal activity by macrophages (46). SCFAs can also prevent the abnormal expansion of antibiotic-resistant strains of Enterobacteriaceae, in part by increasing intracellular acidification (49). SCFAs (including propionate) have also been shown to regulate (favor) the growth and virulence of enteric pathogens, such as enterohemorrhagic and adherent-invasive E. coli (50, 51). In our experimental model, mice fed SCFAs before an S. Typhimurium infection did not show a lower degree of local bacterial colonization, indicating a lack of a direct effect on bacterial growth. In contrast, SCFA supplementation limited systemic bacterial dissemination, probably by reinforcing intestinal barrier properties, as our data show. Interestingly, SCFA supplementation tended to lower the degree of morbidity (i.e., improved weight loss recovery) and to increase the survival rate of doubly infected animals. Mechanisms through which SCFAs reduced Salmonella dissemination in our setting are still elusive. They might include a direct role in virulence factors involved in bacterial invasion and translocation (52) and/or an indirect role in host signaling pathways. Among them, activation of the G protein-coupled receptors free fatty acid receptor 3 and/or 2 (receptors for acetate and/or propionate) and/or inhibition of histone deacetylase (butyrate and, to a lower extent, propionate) are probable. Together with the fact that acetate protected against postinfluenza pneumococcal infection in the lungs (10), our finding highlights the potential benefit of using SCFAs to lower secondary influenza outcomes. In this context, the restoration of mucosa homeostasis might be achieved by stimulating SCFA production via prebiotics (e.g., high-fiber diets) or probiotics (i.e., SCFA producers themselves). The clinical efficacy of this adjunct treatment remains to be determined.
MATERIALS AND METHODS
Mice and ethics statement.
Specific-pathogen-free C57BL/6J mice (7 weeks old, male) were purchased from Janvier (Le Genest-St.-Isle, France). Mice were maintained in a biosafety level 2 facility in the Animal Resource Center at the Lille Pasteur Institute for at least 2 weeks prior to usage to allow appropriate acclimation. Unless specified, mice were fed a standard rodent chow (SAFE A04; SAFE, Augy, France) and had access to water ad libitum. This diet contains ∼11.8% fiber, including ∼10% water-insoluble fiber (3.6% cellulose) and 1.8% water-soluble fiber. All experiments complied with current national and institutional regulations and ethical guidelines (Institut Pasteur de Lille/B59-350009). The protocols were approved by the institutional ethical committee (Comité d’Ethique en Experimentation Animale [CEEA] 75), Nord Pas-de-Calais. All experiments were approved by the Education, Research and Innovation Ministry, France, under registration number APAFIS 13743-2018022211144403. Transgenic mice expressing the firefly luciferase gene under the control of a nuclear factor κB (NF-κB) promoter (NF-κB–luciferase reporter) have been previously described (53). NF-κB–luciferase transgenic BALB/c mice express the luciferase gene driven by two NF-κB sites from the κ light chain enhancer in front of a minimal fos promoter. They were obtained by backcrossing NF-κB–luciferase transgenic B10.A mice (a kind gift of Richard Flavell, Howard Hughes Medical Institute) with BALB/c mice to ensure the production of transgenic mice with white fur to avoid absorption of the light by the dark skin and fur of the B10.A mice.
Viruses and bacteria.
The mouse-adapted H3N2 IAV strain Scotland/20/1974 and the Salmonella enterica serovar Typhimurium strain C5 are described in references 54 and 55, respectively.
Infections and assessment of bacterial loads.
For infection with IAV alone, mice were anesthetized by intramuscular injection of 1.25 mg of ketamine plus 0.25 mg of xylazine in 100 μl of phosphate-buffered saline (PBS) and then intranasally (i.n.) infected with 50 μl of PBS containing (or not, in a mock sample) 30 PFU of IAV (54, 56). For infection with S. Typhimurium alone, gradual doses were used (oral gavage, 200 μl). For secondary infection, IAV-infected mice were intragastrically challenged at 7 dpi with S. Typhimurium (1 × 104 CFU, 200 μl). This dose is sufficient to allow bacterial outgrowth, dissemination, and death in mice previously infected with IAV. In doubly infected mice, bacteria in the cecum and liver were counted 7 days after the bacterial challenge by plating serial 10-fold dilutions of cecum or liver homogenates onto Hektoen agar plates. The plates were incubated at 37°C with 5% CO2 overnight, and viable bacteria were counted 24 h later. Survival and body weight were monitored daily after IAV infection, and mice were euthanized when they lost in excess of 20% of their initial body weight.
Measurement of NF-κB activity.
Bioluminescence measurements of the NF-κB-luciferase transgenic mice were measured using the IVIS 200 imaging system (PerkinElmer, Waltham, MA). Mice were anaesthetized, and luminescence was measured 7 days after IAV infection as described previously (53). Briefly, after anesthesia, luciferin was i.n. instilled (0.75 mg · kg of body weight–1). Living Image software (version 4.0; PerkinElmer) was used to measure the luciferase activities. Bioluminescence images were acquired for 1 min with the f-stop equal to 1 and binning equal to 8. A digital false-color photon emission image of the mouse was generated, and photons were counted within the whole-body area. Photon emission was measured as radiance, which corresponds to the sum of the photons per second from each pixel inside the region of interest (ROI) per number of pixels (photons per second per square centimeter per steradian).
Transcriptomic analysis.
Colon transcriptional profiling was performed using Agilent’s SurePrint G3 mouse GE 8×60K kit (G4852A). Minimum information about the microarray experiment (MIAME) was deposited in ArrayExpress at EMBL (http://www.ebi.ac.uk/microarray-as/ae; accession numbers E-MTAB-6707 [IAV] and E-MTAB-10036 [pair-feeding]). Arrays were hybridized according to the manufacturer's instructions and as previously described (57). Differentially expressed genes were identified using a moderated t test. A Benjamini-Hochberg false-discovery rate (FDR) was then used as a multiple-testing correction method, and a corrected P value cutoff of 5% was applied. A fold change cutoff of >2 was then added to select the differentially expressed genes between the mock and infected conditions. For functional analysis, data files were uploaded into the Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, Redwood City, CA). Pathway enrichment analysis was performed using the GSEA software (https://www.gsea-msigdb.org/gsea/index.jsp).
Measurement of SCFA concentrations and treatment with SCFAs.
Concentrations of SCFAs in the cecal content were determined after extraction with diethyl ether using GC-2014 gas chromatography with a model AOC-20i auto-injector (Shimadzu, Hertogenbosch, The Netherlands) as described previously (58). Results are expressed as micromoles per gram of cecal content. To assess the effects of SCFAs on intestinal permeability and defense against secondary enteric bacterial infection, mice infected with IAV were treated (drinking water) with a combination of acetate, propionate, and butyrate (200 mM, 50 mM, and 5 mM, respectively) after 2 days of IAV infection. Treatment was continued after secondary bacterial infection until animal sacrifice.
Assessment of gene expression and quantification of viral loads by quantitative RT-PCR.
Total RNA from colon tissues was extracted with the NucleoSpin RNA kit (Macherey-Nagel, Hoerdt, Germany). RNA was reverse transcribed with the high-capacity cDNA Archive kit (Life Technologies, USA). The resulting cDNA was amplified using SYBR green-based real-time PCR and the QuantStudio 12K Flex real-time PCR systems (Applied Biosystems, USA) according to the manufacturer’s protocol. Relative quantification was performed using the gene coding for glyceraldehyde 3-phosphate dehydrogenase (Gapdh). Specific primers were designed using Primer Express software (Applied Biosystems, Villebon sur Yvette, France). Relative mRNA levels (2–ΔCt, where Ct is the cycle threshold) were determined by comparing (a) the PCR Ct for the gene of interest and the housekeeping gene Gadph (ΔCt) and (b) ΔCt values for treated and control groups (ΔCt). Data are expressed as a fold change over the mean gene expression level in mock-treated mice. Quantification of viral RNA was performed as described in reference 10. Viral load is expressed as viral RNA normalized to the Gapdh expression level. Data were normalized to expression of the Gapdh gene and were expressed as Cts. The sequences of the oligonucleotides used in this study are shown in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| Gapdh | Forward, 5′-GCAAAGTGGAGATTGTTGCCA-3′ |
| Reverse, 5′-GCCTTGACTGTGCCGTTGA-3′ | |
| Iav m1 | Forward, 5′-AAGAACAATCCTGTCACCTCTGA-3′ |
| Reverse, 5′-CAAAGCGTCTACGCTGCAGTCC-3′ | |
| Crp | Forward, 5′-GTGCGCAGCTTCAGTGTCTTC-3′ |
| Reverse, 5′-AGCACCACCCACTCCAAAAGCA-3′ | |
| Lcn2 | Forward, 5′-GCCCAGGACTCAACTCAGAACTT-3′ |
| Reverse, 5′-GCTCATAGATGGTGCTGTACATCG-3′ | |
| Ifng | Forward, 5′-CAACAGCAAGGCGAAAAAG-3′ |
| Reverse, 5′-GTGGACCACTCGGATGAGCT-3′ | |
| Il18bp | Forward, 5′-ACTGAGCCCCACCCTACGAA-3′ |
| Reverse, 5′-GGAGCTGTCTTCAACCCATCC-3′ | |
| Cxcl1 | Forward, 5′-GCGCCTATCGCCAATGAGC-3′ |
| Reverse, 5′-GCAAGCCTCGCGACCATTC-3′ | |
| S100a8 | Forward, 5′-CACCATGCCCTCTACAAGAATG-3′ |
| Reverse, 5′-CACCATCGCAAGGAACTCC-3′ | |
| Cxcl10 | Forward, 5′-ACCCAAGTGCTGCCGTCAT-3′ |
| Reverse, 5′-CATTCTCACTGGCCCGTCAT-3′ | |
| Ifi44 | Forward, 5′-GGCCATGAGAACTCGTTTGACA-3′ |
| Reverse, 5′-ACTTCTGCACACTCGCCTTGTA-3′ | |
| Gbp4 | Forward, 5′-CACAAATGGCTCCCATTTGTC-3′ |
| Reverse, 5′-GGACCATCCAACAATAGCCACT-3′ | |
| Foxj1 | Forward, 5′-CCACCAAGATCACTCTGTCGG-3′ |
| Reverse, 5′-AGGACAGGTTGTGGCGGAT-3′ | |
| Muc2 | Forward, 5′-TGCTGACGAGTGGTTGGTGA-3′ |
| Reverse, 5′-TTAAGCGAAAGCCCTGGTGT-3′ | |
| Il1b | Forward, 5′-TCGTGCTGTCGGACCCATA-3′ |
| Reverse, 5′-GTCGTTGCTTGGTTCTCCTTGT-3′ | |
| Il6 | Forward, 5′-CAACCACGGCCTTCCCTACT-3′ |
| Reverse, 5′-CCACGATTTCCCAGAGAACATG-3′ | |
| S100a9 | Forward, 5′-GACACCCTGACACCCTGA-3′ |
| Reverse, 5′-GCCATCAGCATCATACACTCCTC-3′ |
Measurement of intestinal permeability.
To measure the perturbation of intestinal permeability following influenza virus infection or pair-feeding, fluorescein isothiocyanate (FITC)-dextran (dextran size, 4,000 kDa) dissolved in sterile saline was administered by oral gavage (200 μl, 60 mg/ml). After 4 h, mock-infected, IAV-infected mice and pair-fed mice were euthanized and their blood was collected by cardiac puncture. FITC-dextran in plasma was quantified using a Tecan (Männedorf, Switzerland) plate reader (excitation, 485 nm; emission, 528 nm). Serum citrulline, intestinal fatty acid-binding protein, and lipopolysaccharide (LPS)-binding protein were quantified by enzyme-linked immunosorbent assay (ELISA) (MyBioSource, CA).
Histological examination of gut sections.
Histological examination of the duodenum and proximal colon was performed on 4-μm-thick sections stained with hematoxylin, eosin, and saffron. Thirty crypts per mouse were analyzed to determine the average crypt depth and villus length. Slides were scanned using Panoramic Scan (3DHistech), and CaseViewer software (3DHistech) was used to measure crypt depth and villus length.
Sample collection, genomic DNA extraction and sequencing, and gut microbiota analysis.
To study the potential impact of SCFA treatment on the gut microbiota’s composition, feces were collected from uninfected mice treated or not treated with SCFAs for 5 days and from mice infected 7 days earlier with IAV and treated or not with SCFAs at 2 days postinfection. Extraction of microbial DNA and determination of microbial diversity and composition by 16S rRNA gene pyrosequencing (V3 and V4 hypervariable regions) were determined exactly as described previously (10). Taxonomic and diversity analyses were performed with the Metabiote Online v2.0 pipeline (GenoScreen, Lille, France), which is partially based on the software QIIME v1.9.1. Beta diversity (between samples) was used to examine changes in microbial community structure between mouse fecal group samples. Principal-component analyses (PCoA) of the Bray-Curtis distance index were performed to assess beta diversity. Differences in the relative abundances of individual taxa, between mouse cecal group samples, were assessed for significance using the Mann-Whitney U test, controlling for the false-discovery rate implemented within the software package QIIME.
Pair-feeding experiments.
To provide the pair-fed group with only as much food daily as is consumed by IAV-infected mice, intake was restricted during the last 3 days by 15% (day 4), 35% (day 5), and 85% (day 6) of mice (sacrifice at day 7) (10). Mice were anesthetized at day 0. Food was supplied twice a day to pair-fed animals, and water was available at all times. The ad libitum (normally nourished) group mice were allowed unrestricted access to food and water. At the sacrifice, pair-fed mice lost ∼15% of their body mass.
Statistical analysis.
Results are expressed as means ± standard deviations (SD) unless otherwise stated. All statistical analysis was performed using GraphPad Prism v6 software. A Mann-Whitney U test was used to compare two groups unless otherwise stated. Comparisons of more than two groups with each other were analyzed with the one-way analysis of variance (ANOVA) Kruskal-Wallis test (nonparametric), followed by Dunn’s posttest. The rates of survival of mice were compared using Kaplan-Meier analysis and the log rank test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Data availability.
MIAME was deposited in ArrayExpress at EMBL under accession numbers E-MTAB-6707 (IAV) and E-MTAB-10036 (pair-feeding). For the gut microbiota analysis, raw sequence data are accessible in the National Center for Biotechnology Information (project number PRJNA675027), biosample accession numbers SAMN16686871 to SAMN16686906.
ACKNOWLEDGMENTS
We acknowledge Richard Flavell (Howard Hughes Medical Institute, Princeton University, Princeton, NJ) for the gift of the NF-κB–luciferase transgenic B10.A mice and Hugues Lelouard (CIML, Marseille, France) for scientific discussions. We thank the animal facility (PLETHA) of the Pasteur Institute, Lille, France, for animal maintenance and the PICT platform (INRAE, Jouy-en-Josas, France) for microarray technical assistance.
This work was supported in part by the INSERM, CNRS, University of Lille, Pasteur Institute of Lille, Région des Hauts-de-France (FLUMICROBIOTE), and the Agence Nationale de la Recherche (AAP Générique 2017, ANR-17-CE15-0020-01, ACROBAT) (F.T.). V.S., M.G.M., and A.B. received salary support (Ph.D. fellowship) from Lille University and from the Fondation pour la Recherche Médicale (V.S.).
Footnotes
Supplemental material is available online only.
Contributor Information
François Trottein, Email: francois.trottein@pasteur-lille.fr.
Andreas J. Bäumler, University of California, Davis
REFERENCES
- 1.Coates BM, Staricha KL, Wiese KM, Ridge KM. 2015. Influenza A virus infection, innate immunity, and childhood. JAMA Pediatr 169:956–963. 10.1001/jamapediatrics.2015.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rynda-Apple A, Robinson KM, Alcorn JF. 2015. Influenza and bacterial superinfection: illuminating the immunologic mechanisms of disease. Infect Immun 83:3764–3770. 10.1128/IAI.00298-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCullers JA. 2014. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol 12:252–262. 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- 4.Sellers SA, Hagan RS, Hayden FG, Fischer WA. 2017. The hidden burden of influenza: a review of the extra-pulmonary complications of influenza infection. Influenza Other Respir Viruses 11:372–393. 10.1111/irv.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dilantika C, Sedyaningsih ER, Kasper MR, Agtini M, Listiyaningsih E, Uyeki TM, Burgess TH, Blair PJ, Putnam SD. 2010. Influenza virus infection among pediatric patients reporting diarrhea and influenza-like illness. BMC Infect Dis 10:3–7. 10.1186/1471-2334-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Li F, Wei H, Lian Z-X, Sun R, Tian Z. 2014. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J Exp Med 211:2397–2410. 10.1084/jem.20140625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deriu E, Boxx GM, He X, Pan C, Benavidez SD, Cen L, Rozengurt N, Shi W, Cheng G. 2016. Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type I interferons. PLoS Pathog 12:e1005572. 10.1371/journal.ppat.1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yildiz S, Mazel-Sanchez B, Kandasamy M, Manicassamy B, Schmolke M. 2018. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome 6:9. 10.1186/s40168-017-0386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groves HT, Cuthbertson L, James P, Moffatt MF, Cox MJ, Tregoning JS. 2018. Respiratory disease following viral lung infection alters the murine gut microbiota. Front Immunol 9:182. 10.3389/fimmu.2018.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sencio V, Barthelemy A, Tavares LP, Machado MG, Soulard D, Cuinat C, Queiroz-Junior CM, Noordine M-L, Salomé-Desnoulez S, Deryuter L, Foligné B, Wahl C, Frisch B, Vieira AT, Paget C, Milligan G, Ulven T, Wolowczuk I, Faveeuw C, Le Goffic R, Thomas M, Ferreira S, Teixeira MM, Trottein F. 2020. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep 30:2934–2947.e6. 10.1016/j.celrep.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Sencio V, Machado MG, Trottein F. 2021. The lung-gut axis during viral respiratory infections: the impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol 14:296–304. 10.1038/s41385-020-00361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461:1282–1286. 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469:543–547. 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 14.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. 2014. Activation of the receptor (Gpr109a) for niacin and the commensal metabolite butyrate suppresses colonic inflammation and carcinogenesis. Immunity 40:128–139. 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, Binge L, Thorburn AN, Chevalier N, Ang C, Marino E, Robert R, Offermanns S, Teixeira MM, Moore RJ, Flavell RA, Fagarasan S, Mackay CR. 2015. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun 6:6734–6748. 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 16.Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. 2016. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165:1332–1345. 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 17.Milligan G, Shimpukade B, Ulven T, Hudson BD. 2017. Complex pharmacology of free fatty acid receptors. Chem Rev 117:67–110. 10.1021/acs.chemrev.6b00056. [DOI] [PubMed] [Google Scholar]
- 18.Thorburn AN, Macia L, Mackay CR. 2014. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 40:833–842. 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Sun M, Wu W, Liu Z, Cong Y. 2017. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol 52:1–8. 10.1007/s00535-016-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN, Hermoso MA. 2019. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol 10:277. 10.3389/fimmu.2019.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jianfeng G, Weiming Z, Ning L, Fangnan L, Li T, Nan L, Jieshou L. 2005. Serum citrulline is a simple quantitative marker for small intestinal enterocytes mass and absorption function in short bowel patients. J Surg Res 127:177–182. 10.1016/j.jss.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Groves HT, Higham SL, Moffatt MF, Cox MJ, Tregoning JS. 2020. Respiratory viral infection alters the gut microbiota by inducing inappetence. mBio 11:e03236-19. 10.1128/mBio.03236-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam LH, Monack DM. 2014. Intraspecies competition for niches in the distal gut dictate transmission during persistent Salmonella infection. PLoS Pathog 10:e1004527. 10.1371/journal.ppat.1004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DPB, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW. 2015. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160:447–460. 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, Loukov D, Schenck LP, Jury J, Foley KP, Schertzer JD, Larché MJ, Davidson DJ, Verdú EF, Surette MG, Bowdish DME. 2017. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe 21:455–466.e4. 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner JR. 2009. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 9:799–809. 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 27.Hughes R, Kurth MJ, McGilligan V, McGlynn H, Rowland I. 2008. Effect of colonic bacterial metabolites on Caco-2 cell paracellular permeability in vitro. Nutr Cancer 60:259–266. 10.1080/01635580701649644. [DOI] [PubMed] [Google Scholar]
- 28.McCall IC, Betanzos A, Weber DA, Nava P, Miller GW, Parkos CA. 2009. Effects of phenol on barrier function of a human intestinal epithelial cell line correlate with altered tight junction protein localization. Toxicol Appl Pharmacol 241:61–70. 10.1016/j.taap.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derrien M, Belzer C, de Vos WM. 2017. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog 106:171–181. 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Florescu DF, Kalil AC. 2014. The complex link between influenza and severe sepsis. Virulence 5:137–142. 10.4161/viru.27103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalil AC, Thomas PG. 2019. Influenza virus-related critical illness: pathophysiology and epidemiology. Crit Care 23:258–264. 10.1186/s13054-019-2539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng VCC, To KKW, Tse H, Hung IFN, Yuen K-Y. 2012. Two years after pandemic influenza A/2009/H1N1: what have we learned? Clin Microbiol Rev 25:223–263. 10.1128/CMR.05012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bischoff SC, Barbara G, Buurman W, Ockhuizen T, Schulzke J-D, Serino M, Tilg H, Watson A, Wells JM. 2014. Intestinal permeability—a new target for disease prevention and therapy. BMC Gastroenterol 14:189. 10.1186/s12876-014-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti J-F, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. 2007. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56:1761–1772. 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 35.Scott KA, Ida M, Peterson VL, Prenderville JA, Moloney GM, Izumo T, Murphy K, Murphy A, Ross RP, Stanton C, Dinan TG, Cryan JF. 2017. Revisiting Metchnikoff: age-related alterations in microbiota-gut-brain axis in the mouse. Brain Behav Immun 65:20–32. 10.1016/j.bbi.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Boutagy NE, McMillan RP, Frisard MI, Hulver MW. 2016. Metabolic endotoxemia with obesity: is it real and is it relevant? Biochimie 124:11–20. 10.1016/j.biochi.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermúdez-Humarán LG, Smirnova N, Bergé M, Sulpice T, Lahtinen S, Ouwehand A, Langella P, Rautonen N, Sansonetti PJ, Burcelin R. 2011. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med 3:559–572. 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lelouvier B, Servant F, Païssé S, Brunet A-C, Benyahya S, Serino M, Valle C, Ortiz MR, Puig J, Courtney M, Federici M, Fernández-Real J-M, Burcelin R, Amar J. 2016. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: a pilot analysis. Hepatology 64:2015–2027. 10.1002/hep.28829. [DOI] [PubMed] [Google Scholar]
- 39.Alvarez-Silva C, Schierwagen R, Pohlmann A, Magdaleno F, Uschner FE, Ryan P, Vehreschild MJGT, Claria J, Latz E, Lelouvier B, Arumugam M, Trebicka J. 2019. Compartmentalization of immune response and microbial translocation in decompensated cirrhosis. Front Immunol 10:69. 10.3389/fimmu.2019.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ayari A, Rosa-Calatrava M, Lancel S, Barthelemy J, Pizzorno A, Mayeuf-Louchart A, Baron M, Hot D, Deruyter L, Soulard D, Julien T, Faveeuw C, Molendi-Coste O, Dombrowicz D, Sedano L, Sencio V, Le Goffic R, Trottein F, Wolowczuk I. 2020. Influenza infection rewires energy metabolism and induces browning features in adipose cells and tissues. Commun Biol 3:237. 10.1038/s42003-020-0965-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer R-J. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119. 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 42.Gill PA, van Zelm MC, Muir JG, Gibson PR. 2018. Review article: short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment Pharmacol Ther 48:15–34. 10.1111/apt.14689. [DOI] [PubMed] [Google Scholar]
- 43.Tominaga K, Kamimura K, Takahashi K, Yokoyama J, Yamagiwa S, Terai S. 2018. Diversion colitis and pouchitis: a mini-review. World J Gastroenterol 24:1734–1747. 10.3748/wjg.v24.i16.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K, Bouley DM, Vilches-Moure JG, Smith M, Sonnenburg JL, Bhatt AS, Huang KC, Monack D. 2018. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe 24:296–307.e7. 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiminez JA, Uwiera TC, Abbott DW, Uwiera RRE, Inglis GD. 2017. Butyrate supplementation at high concentrations alters enteric bacterial communities and reduces intestinal inflammation in mice infected with Citrobacter rodentium. mSphere 2:e00243-17. 10.1128/mSphere.00243-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, Chomka A, Ilott NE, Johnston DGW, Pires E, McCullagh J, Sansom SN, Arancibia-Cárcamo CV, Uhlig HH, Powrie F. 2019. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50:432–445.e7. 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarker P, Ahmed S, Tiash S, Rekha RS, Stromberg R, Andersson J, Bergman P, Gudmundsson GH, Agerberth B, Raqib R. 2011. Phenylbutyrate counteracts Shigella mediated downregulation of cathelicidin in rabbit lung and intestinal epithelia: a potential therapeutic strategy. PLoS One 6:e20637. 10.1371/journal.pone.0020637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jellbauer S, Perez Lopez A, Behnsen J, Gao N, Nguyen T, Murphy C, Edwards RA, Raffatellu M. 2016. Beneficial effects of sodium phenylbutyrate administration during infection with Salmonella enterica serovar Typhimurium. Infect Immun 84:2639–2652. 10.1128/IAI.00132-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorbara MT, Dubin K, Littmann ER, Moody TU, Fontana E, Seok R, Leiner IM, Taur Y, Peled JU, van den Brink MRM, Litvak Y, Bäumler AJ, Chaubard J-L, Pickard AJ, Cross JR, Pamer EG. 2019. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med 216:84–98. 10.1084/jem.20181639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ormsby MJ, Johnson SA, Carpena N, Meikle LM, Goldstone RJ, McIntosh A, Wessel HM, Hulme HE, McConnachie CC, Connolly JPR, Roe AJ, Hasson C, Boyd J, Fitzgerald E, Gerasimidis K, Morrison D, Hold GL, Hansen R, Walker D, Smith DGE, Wall DM. 2020. Propionic acid promotes the virulent phenotype of Crohn’s disease-associated adherent-invasive Escherichia coli. Cell Rep 30:2297–2305.e5. 10.1016/j.celrep.2020.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pobeguts OV, Ladygina VG, Evsyutina DV, Eremeev AV, Zubov AI, Matyushkina DS, Scherbakov PL, Rakitina DV, Fisunov GY. 2020. Propionate induces virulent properties of Crohn’s disease-associated Escherichia coli. Front Microbiol 11:1460. 10.3389/fmicb.2020.01460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawhon SD, Maurer R, Suyemoto M, Altier C. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA: short-chain fatty acids and Salmonella invasion. Mol Microbiol 46:1451–1464. 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- 53.Vidy A, Maisonnasse P, Da Costa B, Delmas B, Chevalier C, Le Goffic R. 2016. The influenza virus protein PB1-F2 increases viral pathogenesis through neutrophil recruitment and NK cells inhibition. PLoS One 11:e0165361. 10.1371/journal.pone.0165361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barthelemy A, Sencio V, Soulard D, Deruyter L, Faveeuw C, Le Goffic R, Trottein F. 2018. Interleukin-22 immunotherapy during severe influenza enhances lung tissue integrity and reduces secondary bacterial systemic invasion. Infect Immun 86:e00706-17. 10.1128/IAI.00706-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zoumpopoulou G, Foligne B, Christodoulou K, Grangette C, Pot B, Tsakalidou E. 2008. Lactobacillus fermentum ACA-DC 179 displays probiotic potential in vitro and protects against trinitrobenzene sulfonic acid (TNBS)-induced colitis and Salmonella infection in murine models. Int J Food Microbiol 121:18–26. 10.1016/j.ijfoodmicro.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 56.Barthelemy A, Ivanov S, Fontaine J, Soulard D, Bouabe H, Paget C, Faveeuw C, Trottein F. 2017. Influenza A virus-induced release of interleukin-10 inhibits the anti-microbial activities of invariant natural killer T cells during invasive pneumococcal superinfection. Mucosal Immunol 10:460–469. 10.1038/mi.2016.49. [DOI] [PubMed] [Google Scholar]
- 57.Le Goffic R, Leymarie O, Chevalier C, Rebours E, Da Costa B, Vidic J, Descamps D, Sallenave J-M, Rauch M, Samson M, Delmas B. 2011. Transcriptomic analysis of host immune and cell death responses associated with the influenza A virus PB1-F2 protein. PLoS Pathog 7:e1002202. 10.1371/journal.ppat.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Weirdt R, Possemiers S, Vermeulen G, Moerdijk-Poortvliet TCW, Boschker HTS, Verstraete W, Van de Wiele T. 2010. Human faecal microbiota display variable patterns of glycerol metabolism. FEMS Microbiol Ecol 74:601–611. 10.1111/j.1574-6941.2010.00974.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5. Download IAI.00734-20-s0001.pdf, PDF file, 1.26 MB (1.3MB, pdf)
Data Availability Statement
MIAME was deposited in ArrayExpress at EMBL under accession numbers E-MTAB-6707 (IAV) and E-MTAB-10036 (pair-feeding). For the gut microbiota analysis, raw sequence data are accessible in the National Center for Biotechnology Information (project number PRJNA675027), biosample accession numbers SAMN16686871 to SAMN16686906.