Abstract
Background
Carbapenem-resistant Klebsiella pneumoniae (CRKP) infection has attracted worldwide concern and became a serious challenge for clinical treatment. The aims of this study were to evaluate the molecular characteristics and risk factors for CRKP infection.
Methods
All the CRKP strains were screened for antimicrobial resistance genes, virulence genes, and integron by polymerase chain reaction (PCR). Plasmid typing was performed by plasmid conjugation assay and PCR-based replicon typing (PBRT). The genetic environments of blaKPC-2 and blaNDM-1 were analyzed by using overlapping PCR and molecular typing was performed by multi-locus sequence typing (MLST). Risk factors for CRKP infection were analyzed by logistic regression model.
Results
All the 66 CRKP isolates were multidrug-resistant, but all of them were susceptible to tigecycline and polymyxin B. Among the CRKP isolates, 42 blaKPC-2-positive strains were identified carrying IncFII plasmids. Meanwhile, 24 blaNDM-positive strains were found on lncX3 plasmids, including 20 blaNDM-1 isolates and 4 blaNDM-5 isolates. Most of CRKP isolates contained several virulence genes and the class I integron (intl1). The genetic environments of blaKPC-2 and blaNDM-1 revealed that the conserved regions (tnpA-tnpR-ISkpn8-blaKPC-2) and (blaNDM-1-bleMBL-trpF-tat) were associated with the dissemination of KPC-2 and NDM-1. ST11 was the most common type in this work. Hematological disease, tracheal cannula, and use of β-lactams and β-lactamase inhibitor combination were identified as independent risk factors for CRKP infection.
Conclusion
This study established the resistance pattern, molecular characteristics, clonal relatedness, and risk factors of CRKP infection. The findings of the novel strain that co-harboring blaNDM-5 and blaIMP-4, and the novel ST4495 indicated that the brand-new types have spread in Southwest China, emphasizing the prevent and control the further dissemination of CRKP isolates are highly needed.
Keywords: carbapenem-resistant Klebsiella pneumoniae, molecular characteristics, genetic environments, plasmid, risk factors
Introduction
Klebsiella pneumoniae, belonging to the family Enterobacteriaceae, is an important pathogen that causes opportunistic infections in hospitalized patients, and plays a primary role in pneumonia and neonatal sepsis.1 With the increase of prevalence of multidrug-resistant (MDR) strains that producing extended-spectrum β-lactamase (ESBL) and/or carbapenemase, K. pneumoniae has emerged as a major threat in clinical and public health. Carbapenem-resistant K. pneumoniae (CRKP) has become one of the particularly important problems worldwide due to the gradually higher morbidity and subsequently worrying mortality.2,3 According to the 2020 CHINET Resistance Monitoring Network, K. pneumoniae resistance to imipenem has risen rapidly from 16.1% in 2016 to 23.3% in 2020.
The resistance mechanism of Enterobacteriaceae to carbapenem antibiotics is usually caused by two major mechanisms: (i) Nonenzymatic resistance mechanisms and (ii) Hydrolysis of carbapenems by carbapenemases production.4 Nonenzymatic resistance mechanisms mainly include overexpression of efflux pump-encoding genes and the mutation of porins.5,6 As for later mechanism, carbapenemases are classified into three groups (ie, A, B, D) based on their molecular structure. Class A and D carbapenemases require an acyl enzyme that forms from an active serine site to hydrolyze their substrates, while class B metalloenzymes utilize the active zinc ion site to induce hydrolysis of carbapenem.7 Klebsiella pneumoniae carbapenemase (KPC) is the most common class A enzyme in Enterobacteriaceae and have widely disseminated due to the encoding genes located on plasmid, especially for KPC-2.8 In addition, New Delhi metallo-β-lactamase (NDM) is one of the important member in class B carbapenemases, and the NDM-1 is the most prevalent type globally. NDM-1-producing clinical strains have caused several outbreaks in China since it was first reported in 2012.9–11 In other regions, such as Colombia, Italy, and Brazil, these strains also lead to health crisis.12–14 Furthermore, a more pathogenic and virulent phenotype, also known as hypervirulent K. pneumoniae (HVKP), has been well-separated quickly. Importantly, the emergence of multidrug-resistant HVKP (MDR-HVKP) has been gradually reported due to the horizontal transfer of mobile genetic elements, bringing a remarkable challenge to healthcare system.15–17
The objectives of this study were to systematically analyze the clinical characteristics, molecular epidemiology, and risk factors for CRKP infection in a tertiary hospital. These findings will provide valuable information for further monitoring and controlling the dissemination of KPC and NDM K. pneumoniae strains in Southwest China.
Materials and Methods
Data Collection
The isolates were defined as CRKP if they were resistant to at least one of the carbapenem agents, including imipenem, ertapenem, and meropenem (www.cdc.gov/HAI/organisms/cre). A total of 66 non-repetitive clinical CRKP isolates were collected from September 2016 to August 2019 in the Affiliated Hospital of Southwest Medical University (Luzhou, China), and all the CRKP isolates were confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker Daltonics, Bremen, Germany).
In order to investigate the risk factors for CRKP infection, a stepwise matching method at a ratio of 1:1 has been used to identify appropriate control cases from patients who are infected with carbapenem-susceptible K. pneumoniae (CSKP). The same site of infection, the same gender, age ± 2 years, and the same year of hospital admission were considered as the matching criteria. The relative clinical data were retrospectively collected from medical records of each patient, including basic demographic information, underlying diseases and comorbidities, invasive procedures, antibiotic treatment, and clinical outcomes.
Antimicrobial Susceptibility Testing (AST)
The antimicrobial susceptibilities of all the 66 clinical CRKP isolates to 16 antimicrobials were tested by VITEK 2 Compact system (bioMérieux, Marcy l’Etoile, Lyon, France), including cefepime, cefotaxime, cefazolin, cefuroxime, ceftazidime, cefoxitin, piperacillin/tazobactam, ampicillin/sulbactam, amikacin, tobramycin, gentamicin, levofloxacin, ciprofloxacin, sulperazone, compound sulfamethoxazole, and aztreonam. The minimum inhibitory concentrations (MICs) of meropenem, imipenem, ertapenem, tigecycline, and polymyxin B were determined by broth microdilution method, and the results were interpreted according to the standards of the Clinical and Laboratory Standards Institute (CLSI) 2020-M100.18 Pseudomonas aeruginosa ATCC27853 and Escherichia coli ATCC25922 were used as quality control strains (purchased from China National Health Inspection Center).
Screening of CRKP and Phenotypic Detection of Carbapenemase
The metallo-β-lactamase-producing isolates were differentiated from all the 66 CRKP by using the imipenem-EDTA double-disk synergy method and the carbapenem inactivation method (CIM). Briefly, the bacterial suspension (0.5 McFarland standard) was diluted from the overnight culture, and then smeared on Mueller-Hinton (MH) agar plate (Haibo, Qingdao, China). A disk containing 10 μg imipenem (OXOID, ThermoFisher Scientific, Massachusetts, USA) and a blank disk with 1.5mg/mL EDTA were placed 10 mm between each disk on the plate. After 18 h incubation, an enlarged zone of inhibition was considered as EDTA-synergy test positive.18,19
The CIM was performed as described previously.18,20 A single colony of CRKP isolates was inoculated into a tube containing 2 mL trypticase soy broth (TSB) (Haibo, Qingdao, China) and a tube containing 2 mL TSB with 5mM EDTA, respectively. A 10 μg meropenem disk was placed in each tube. After 4 h incubation, these disks were took out and placed on a MH agar plate that was inoculated with a lawn of the meropenem-susceptible Escherichia coli ATCC25922 (0.5 McFarland standard). The results were interpreted according to CLSI.18
The Detection of Resistance Genes, Virulence Genes, and Integrase-Associated Genes
All of template DNAs were extracted by bacterial DNA Kit (Tiangen, Beijing,China) and used to detect carbapenemase genes, ESBL genes, AmpC β-lactamase genes, virulence genes, and integrase-associated genes by polymerase chain reaction (PCR). The primers were shown in Tables S1 and S2. The PCR conditions were described previously.21–23 Positive products were sequenced by Shanghai Jieli Biotechnology, and the variable region of the class I integron (intl1) and the insertion sequence common region I (ISCR I) were analyzed by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Plasmid Conjugation and Analysis
In order to evaluate whether the carbapenemase genes are located on the plasmid and whether these genes can be horizontally transferred, a plasmid conjugation transfer experiment was performed. Four hundred μL donor strain (CRKP strains) and 200 μL receptor strain (sodium azide-resistant E. coli strain J53) at logarithmic phase were added into a glass tube containing 800 μL Luria-Bertani (LB) broth and cultured at 37°C for 18 h. Transconjugants were selected by using LB plates that contains sodium azide (180 μg/mL) and imipenem (0.5 μg/mL).21 The transconjugants were verified by MALDI-TOF MS, and the conjugative carbapenemase genes were confirmed by PCR. In addition, the successful conjugative plasmids were analyzed by PCR-based replicon typing according to previous study.22
Genetic Environments of KPC-2-Carrying Plasmids and NDM-1-Carrying Plasmids
The overlapping PCR was applied to investigate the genetic environments of the blaKPC-2 and blaNDM-1 (Tables S3 and S4).23,24 The PCR products were sequenced by Shanghai Jieli Biotechnology, and then analyzed by NCBI GenBank database.25
Molecular Epidemiological Study
Multi-locus sequence typing (MLST) was performed to explore the genetic correlation of all the clinical CRKP isolates.26 The positive products were sequenced by Shanghai Jieli Biotechnology, and the results were submitted to K. pneumoniae MLST database (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef) for comparison.
Statistical Analysis
All analyses and graphs were performed using SPSS v.24.0 software (SPSS Inc., Chicago, USA). The chi-square test or Fisher’s exact test was used to analyze categorical variables. Continuous variables were presented as means ± standard deviation (SD), and were evaluated by Student’s t-tests or Mann–Whitney U-test. Multivariable logistic regression analysis was operated to identify independent risk factors for CRKP infection. All biologically plausible variables with a value of P < 0.1 within univariate analysis were included in the following multiple logistic regression model. P < 0.05 was considered as statistically significant, and all probability values were two-tailed distribution.
Results
Distribution of Clinical CRKP Isolates
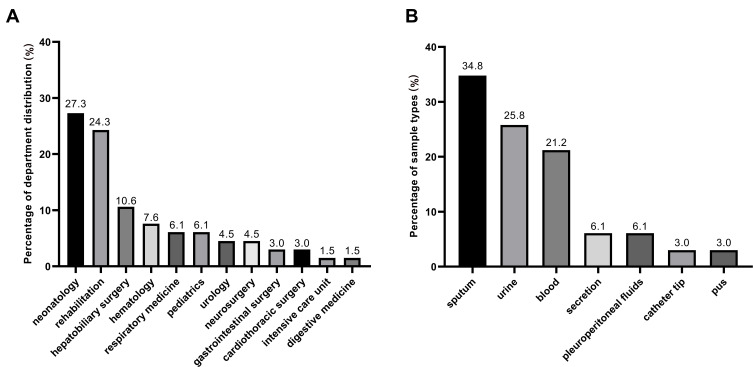
A total of 66 non-repetitive clinical CRKP isolates were collected from September 2016 to August 2019 in Southwest China. The clinical CRKP isolates were obtained from patients admitted to the 12 different departments, including the majority of neonatology department (27.3%, n = 18) and rehabilitation department (24.3%, n = 16) (Figure 1A). These isolates were mainly originated from sputum (34.8%, n = 23), followed by urine (25.8%, n = 17), blood (21.2%, n=14), secretion (6.1%, n= 4), pleuroperitoneal fluids (6.1%, n = 4), catheter tip (3.0%, n = 2), and pus (3.0%, n = 2) (Figure 1B).
Figure 1.
Distribution of 66 clinical CRKP strains. (A) Department distribution of 66 CRKP strains; (B) sample types of 66 CRKP strains.
Antimicrobial Susceptibility Profiles
All of 66 clinical CRKP isolates were multidrug-resistant and resistance rates to 12 antimicrobials reached 100%, which were cefepime, imipenem, piperacillin/tazobactam, ceftriaxone, ampicillin/sulbactam, cefotaxime, cefazolin, cefuroxime, ertapenem, ceftazidime, meropenem, and cefoxitin. In addition, the resistance rates to sulperazon, aztreonam, tobramycin, gentamicin, levofloxacin, ciprofloxacin, cotrimoxazole, and amikacin were 95.5%, 87.9%, 83.3%, 83.3%, 81.9%, 80.3%, 65.2%, and 54.5%, respectively. Remarkably, all the CRKP strains in this study were sensitive to tigecycline and polymyxin B (Table 1).
Table 1.
Antimicrobial Susceptibility Profiles of 66 Clinical CKRP Strains
| Antimicrobial Agent | No. | %R | No. | %I | No. | %S |
|---|---|---|---|---|---|---|
| Amikacin | 36 | 54.5 | 0 | 0 | 30 | 45.5 |
| Cefepime | 66 | 100 | 0 | 0 | 0 | 0 |
| Imipenem | 66 | 100 | 0 | 0 | 0 | 0 |
| Piperacillin/tazobactam | 66 | 100 | 0 | 0 | 0 | 0 |
| Ceftriaxone | 66 | 100 | 0 | 0 | 0 | 0 |
| Ampicillin/sulbactam | 66 | 100 | 0 | 0 | 0 | 0 |
| Cefotaxime | 66 | 100 | 0 | 0 | 0 | 0 |
| Cefazolin | 66 | 100 | 0 | 0 | 0 | 0 |
| Cefuroxime | 66 | 100 | 0 | 0 | 0 | 0 |
| Ertapenem | 66 | 100 | 0 | 0 | 0 | 0 |
| Tobramycin | 55 | 83.3 | 4 | 6.1 | 7 | 10.6 |
| Sulperazon | 63 | 95.5 | 2 | 3.0 | 1 | 1.5 |
| Ceftazidime | 66 | 100 | 0 | 0 | 0 | 0 |
| Meropenem | 66 | 100 | 0 | 0 | 0 | 0 |
| Levofloxacin | 54 | 81.9 | 0 | 0 | 12 | 18.1 |
| Cefoxitin | 66 | 100 | 0 | 0 | 0 | 0 |
| Aztreonam | 58 | 87.9 | 0 | 0 | 8 | 12.1 |
| Ciprofloxacin | 53 | 80.3 | 0 | 0 | 13 | 19.7 |
| Gentamicin | 55 | 83.3 | 0 | 0 | 11 | 16.7 |
| Tigecycline | 0 | 0 | 0 | 0 | 66 | 100 |
| Compound sulfamethoxazole | 43 | 65.2 | 0 | 0 | 23 | 34.8 |
| Polymyxin B | 0 | 0 | 0 | 0 | 66 | 100 |
Abbreviations: S, susceptible; I, intermediate; R, resistant.
Carbapenem Resistance Phenotype and Molecular Characteristics
Both EDTA-synergy test and CIM test were shown that 24 (36.4%) clinical CRKP strains produced class B carbapenemases. The sequencing results confirmed that all of which isolates contained blaNDM, in which 20 isolates carried blaNDM-1 and 4 isolates carried blaNDM-5, and 3 isolates possessed blaNDM and blaIMP simultaneously. The rest of 42 (63.6%) isolates contained blaKPC-2. Notably, in addition to the production of carbapenemase, 100% and 95.5% of the clinical CRKP isolates were positive for ESBL and AmpC genes, respectively. Furthermore, virulence genes were detected in all isolates, 54 (81.8%) isolates detected three different virulence genes. Especially for strain Kpn497 and Kpn131, they carried 12 virulence genes and 11 virulence genes, respectively (Table 2).
Table 2.
Genotypes of 66 Clinical CRKP Strains
| N | Carbapenemase | ESBLs | AmpC | Int | Virulence Gene | Gene Cassette | ISCR | MLST | Replicon Type |
|---|---|---|---|---|---|---|---|---|---|
| 16 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ISCR I | ST11 | IncFII |
| 26 | NDM-1 | SHV+CTX-M | DHA | entB+mrkD | ISCR I | ST4495 | lncX3 | ||
| 27 | NDM-1 | SHV+CTX-M | DHA | entB+fimH+mrkD | ISCR I | ST4495 | lncX3 | ||
| 30 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ISCR I | ST11 | IncFII |
| 32 | NDM-1 | SHV+CTX-M | DHA | entB+fimH+mrkD | ISCR I | ST4495 | lncX3 | ||
| 34 | NDM-1 | SHV+CTX-M | DHA | entB+fimH+mrkD | ISCR I | ST4495 | lncX3 | ||
| 36 | NDM-1 | SHV+CTX-M | DHA | entB+fimH+mrkD | ISCR I | ST4495 | lncX3 | ||
| 37 | NDM-1 | SHV+CTX-M | DHA | entB+fimH+mrkD | ISCR I | ST4495 | lncX3 | ||
| 39 | NDM-1 | SHV | DHA | entB+mrkD | ISCR I | ST4495 | lncX3 | ||
| 40 | NDM-1 | SHV | DHA+ACC | entB+fimH | ISCR I | ST37 | lncX3 | ||
| 45 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | ||
| 46 | NDM-1 | SHV+CTX-M | DHA+ACC | entB+fimH+mrkD | ISCR I | ST4495 | lncX3 | ||
| 49 | NDM-1 | SHV+CTX-M | DHA+ACC | entB+fimH+mrkD | ISCR I | ST4495 | lncX3 | ||
| 51 | KPC-2 | SHV+CTX-M | ACC | entB+fimH+mrkD | ST11 | ||||
| 56 | NDM-1 | SHV+CTX-M | DHA+ACC | entB+fimH+mrkD | ISCR I | ST4495 | lncX3 | ||
| 57 | NDM-1 | SHV+CTX-M | DHA+ACC | entB+fimH+mrkD | ISCR I | ST4495 | lncX3 | ||
| 58 | NDM-1+IMP-4 | SHV+CTX-M | DHA+ACC | entB+fimH | ST4495 | lncX3 | |||
| 101 | NDM-1 | SHV | ACC | entB+fimH+mrkD | ST37 | lncX3 | |||
| 131 | KPC-2 | SHV+TEM | ACC | IntI | entB+fimH+mrkD+rmpA/rmpA2+iucA+terB+aerobactin+HI1B+iroN+iutA | addA2 | ISCR I | ST11 | IncFII |
| 210 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 211 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 214 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 215 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 221 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ISCR I | ST11 | IncFII |
| 223 | KPC-2 | SHV+TEM+CTX-M | ACC | entB+fimH+mrkD | ST147 | ||||
| 227 | KPC-2 | SHV+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 228 | KPC-2 | SHV+CTX-M | ACC | entB+fimH+mrkD | ST11 | IncFII | |||
| 230 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 232 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 233 | KPC-2 | SHV | ACC | IntI | entB+fimH+mrkD | dfrA12, addA2 | ST147 | IncFII | |
| 234 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | InFIB | |
| 241 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 244 | KPC-2 | SHV | ACC | IntI | entB+fimH+mrkD | dfrA12 | ST23 | IncFII | |
| 245 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 251 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 252 | KPC-2 | SHV+TEM+CTX-M | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | ||
| 257 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | ||
| 258 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 267 | NDM-1 | SHV | ACC | IntI | entB+fimH+mrkD | addA2 | ISCR I | ST11 | lncX3 |
| 271 | KPC-2 | SHV+TEM+CTX-M | ACC | entB+fimH+mrkD | ST11 | IncFII | |||
| 285 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 354 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 406 | KPC-2 | SHV+TEM | DHA | IntI | fimH+mrkD | OXA-10, addA1, aacA4 | ISCR I | ST318 | IncFII |
| 436 | KPC-2 | SHV+TEM | DHA | IntI | entB+fimH+mrkD | addA2 | ST17 | IncFII | |
| 440 | NDM-1 | SHV+TEM | ACC | IntI | mrkD | addA2 | ST307 | lncX3 | |
| 445 | NDM-1 | SHV+TEM | ACC | mrkD | ST307 | lncX3 | |||
| 473 | NDM-1+IMP-4 | SHV+TEM | ACC | IntI | entB+fimH+mrkD | addA2 | ISCR I | ST152 | lncX3 |
| 475 | NDM-1 | SHV+TEM | ACC | IntI | fimH+mrkD | addA2 | ISCR I | ST15 | lncX3 |
| 478 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ISCR I | ST11 | IncFII |
| 479 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH | addA2 | ST11 | IncFII | |
| 480 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 483 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 489 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | dfrA12, addA2 | ST11 | ||
| 490 | KPC-2 | SHV | ACC | IntI | entB+fimH+mrkD | dfrA27, arr3 | ST467 | IncFII | |
| 494 | NDM-5 | SHV+CTX-M | DHA | IntI | entB+fimH+mrkD | addA2 | ISCR I | ST2407 | lncX3 |
| 495 | KPC-2 | SHV+TEM+CTX-M | IntI | entB+fimH+mrkD | dfrA12 | ISCR I | ST11 | IncFII | |
| 496 | KPC-2 | SHV+TEM | ACC | IntI | entB+fimH+mrkD | gcuF, dfrA12 | ST405 | ||
| 497 | KPC-2 | SHV+TEM | IntI | entB+fimH+mrkD+rmpA/rmpA2+iucA+ terB+wcaG+ aerobactin+HI1B+iroN+iutA | addA2 | ST11 | IncFII | ||
| 498 | KPC-2 | SHV+CTX-M | ACC | IntI | entB+mrkD | dfrA12 | ST11 | IncFII | |
| 499 | NDM-5 | SHV+CTX-M | DHA | IntI | entB+fimH+mrkD | addA2 | ISCR I | ST2407 | lncX3 |
| 500 | KPC-2 | SHV+TEM+CTX-M | ACC | entB+fimH+mrkD | ST11 | ||||
| 502 | NDM-5 | SHV+CTX-M | DHA | IntI | entB+fimH+mrkD | addA2 | ST2407 | lncX3 | |
| 504 | NDM-5+IMP-4 | SHV+TEM+CTX-M | DHA | IntI | entB+fimH+mrkD | addA2 | ST11 | lncX3 | |
| 505 | KPC-2 | SHV+TEM+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII | |
| 506 | NDM-1 | SHV+CTX-M | DHA | IntI | entB+fimH+mrkD | addA2 | ST4495 | lncX3 | |
| 507 | KPC-2 | SHV+CTX-M | ACC | IntI | entB+fimH+mrkD | addA2 | ST11 | IncFII |
Abbreviations: ESBL, extended-spectrum β-lactamase; Int, integron; ISCR, insertion sequence common region.
Plasmid and Integron-Associated Analysis
KPC-encoding plasmids were positive in 35 transconjugants and all of these plasmids belonged to IncFII. Meanwhile, NDM-encoding plasmids that belong to IncX3 were detected in 24 transconjugants (Table 2).
Nearly 85% (n = 56) strains were intl1-positive, of which 46 strains contained variable region. A total of 6 cassette arrays were found among them. The cassette arrays were revealed in this study included: addA2 (82.6%), ddfrA12 (6.5%), dfrA12-addA2 (4.3%), dfrA27-arr3 (2.2%), gcuF-dfrA12 (2.2%), and blaOXA-10-addA1-aacA4 (2.2%) (Table 2) In addition, ISCRI was detected in 25 strains.
Genetic Environments of blaKPC-2 and blaNDM-1
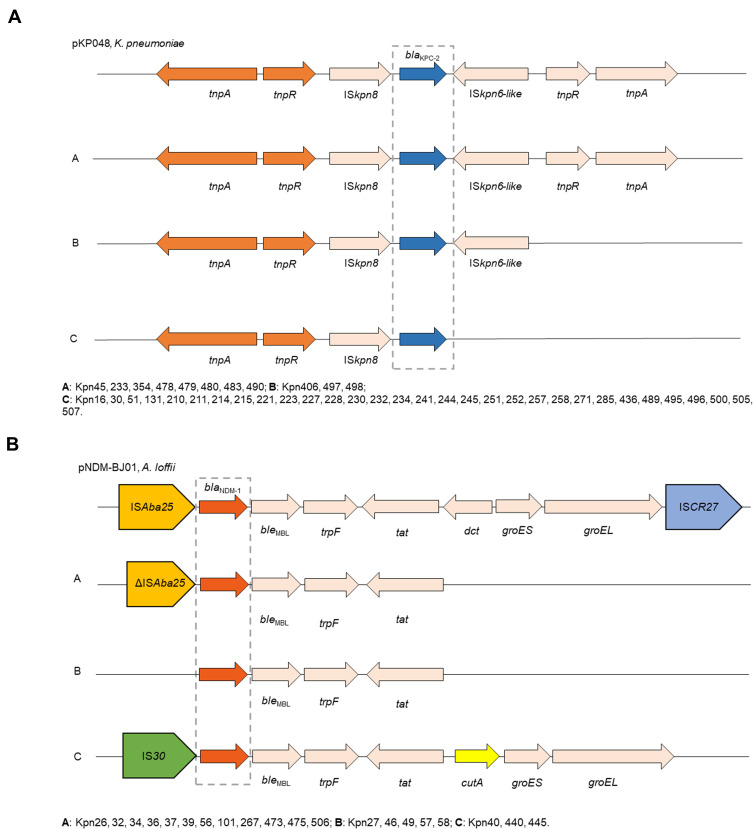
All of 42 KPC-2-producing strains could be divided into three different types (A, B, and C) based on the genetic structures that compared with pKP048 (GenBank Accession No.FJ628167). Type C was the most prevalent (n = 31), followed by type A (n = 8) and type B (n = 3). Type A exhibited the same structure as pKP048. The downstream of ISKpn6-like was deletion in type B, and three genetic elements downstream of blaKPC-2 were deletion in type C, including ISKpn6-like, tnpR, and tnpA (Figure 2A).
Figure 2.
Comparison of the genetic elements surrounding the blaKPC-2 and blaNDM-1 genes identified in this study. (A) Comparison of the genetic environments of blaKPC-2, reference sequences: K. pneumoniae (pKP048, GenBank Accession No.FJ628167); (B) comparison of the genetic environments of blaNDM-1, reference sequences: A. lwoffii (pNDM-BJ01, GenBank accession NO. JQ001791).
Furthermore, the genetic structure of pNDM-BJ01 (GenBank accession No. JQ001791) was used to compare with each NDM-1-producing strain, revealing A, B, and C types among these strains. As for type A (n = 12), the ISAba125 upstream of the blaNDM−1 was truncated, all downstream genes of tat were complete deletion. ISAba125 upstream of the blaNDM−1 was deletion in type B compared with type A. Type C presented as the IS30 upstream of blaNDM-1 and followed by bleMBL, trpF, tat, cutA, groES, groEL (Figure 2B).
MLST of Clinical CRKP Isolates
A total of 13 MLSTs were identified among all the clinical CRKP isolates, and ST11 (56.2%, n = 37) was the most frequent ST type, followed by ST4495 (19.7%, n = 13), ST2407 (4.6%, n = 3), ST147 (3.0%, n = 2), ST307 (3.0%, n = 2), ST37 (3.0%, n = 2), ST15 (1.5%, n = 1), ST17 (1.5%, n = 1), ST152 (1.5%, n = 1), ST23 (1.5%, n = 1), ST405 (1.5%, n = 1), ST318 (1.5%, n = 1), and ST467 (1.5%, n = 1).
Risk Factors and Multivariate Analysis of CRKP Infection
Statistically significant differences were observed for respiratory disease (P = 0.034), renal disease (P = 0.039), hematological disease (P = 0.02), tracheal cannula (P < 0.001), and use of β-lactams/β-lactamase inhibitor combination (P = 0.009) between CRKP and CSKP groups (Table 3). Multivariate analysis revealed that hematological disease (odds ratio [OR], 2.568; 95% confidence interval [95% CI], 1.106 to 5.964; P = 0.028), tracheal cannula (OR, 4.883; 95% CI, 1.797 to 13.265; P = 0.002), and use of β-lactams/β-lactamase inhibitor combination (OR, 4.271; 95% CI, 1.760 to 10.365; P = 0.001) were independent risk factors for CRKP infection.
Table 3.
Clinical Characteristics of CRKP and CSKP Strains
| Variable | CRKP (n=66) | CSKP (n=66) | P-value |
|---|---|---|---|
| Demographic, n (%) or IQR | |||
| Age (years) | 44.5 (0,60) | 46.5 (0.61) | 0.754 |
| Sex (male) | 47 (71.2%) | 46 (69.7%) | 0.894 |
| Length of hospital stays | 15 (7, 27) | 25 (10, 38) | 0.1 |
| Admission to ICU | 14 (21.1%) | 7 (10.6%) | 0.096 |
| Co-morbidity, n (%) | |||
| Malignant disease | 7 (10.6%) | 15 (22.7%) | 0.062 |
| Diabetes mellitus | 11 (16.7%) | 12 (18.2%) | 0.819 |
| Hypertension | 16 (24.2%) | 16 (24.2%) | 1 |
| Heart disease | 9 (13.6%) | 11 (16.7%) | 0.627 |
| Hepatobiliary disease | 12 (18.2%) | 15 (22.7%) | 0.517 |
| Respiratory disease | 45 (68.2%) | 33 (50%) | 0.034 |
| Renal disease | 7 (10.6%) | 16 (24.2%) | 0.039 |
| Urinary tract infection | 10 (15.2%) | 9 (13.6%) | 0.804 |
| Craniocerebral disease | 15 (22.7%) | 9 (13.6%) | 0.176 |
| Hematological disease | 32 (48.5%) | 19 (28.8%) | 0.02 |
| Gastrointestinal disease | 8 (12.1%) | 5 (7.6%) | 0.381 |
| Invasive procedures and devices | |||
| Tracheal cannula | 24 (36.4%) | 7 (10.6%) | <0.001 |
| Central venous catheter | 5 (7.6%) | 12 (18.2%) | 0.069 |
| Foreign material in the body | 14 (21.2%) | 7 (10.6%) | 0.096 |
| Surgical operations after admission | 9 (13.6%) | 16 (24.2%) | 0.12 |
| Colostomy | 3 (4.5%) | 1 (1.5%) | 0.612 |
| Gastrostomy | 2 (3%) | 0 | 0.476 |
| Antibiotic treatment, n (%) | |||
| Penicillins | 2 (3%) | 5 (7.6%) | 0.437 |
| First, second-generation cephalosporins | 7 (10.6%) | 15 (22.7%) | 0.21 |
| Third, fourth-generation cephalosporins | 17 (25.8%) | 19 (28.8%) | 0.696 |
| Aminoglycosides | 4 (6.1%) | 1 (1.5%) | 0.362 |
| Quinolones | 20 (30.3%) | 21 (31.8%) | 0.851 |
| Metronidazole | 0 | 1 (1.5%) | 1 |
| Carbapenems | 30 (45.5%) | 32 (48.5%) | 0.727 |
| β-lactams and β-lactamase inhibitor combination | 40 (60.6%) | 25 (37.9%) | 0.009 |
| Clinical outcomes, n (%) | |||
| Patient outcome: mortality | 5 (7.6%) | 0 | 0.068 |
Note: Bold indicates P < 0.05.
Abbreviations: IQR, interquartile range; CR, carbapenem-resistant; CS, carbapenem-susceptible; ICU, intensive care unit.
Discussion
The clinical CRKP strains were first reported in 1997,27,28 and this type of resistance did not become common within that decade. However, with the increasing use of carbapenems in recent years, CRKP strains have been distributed around the world at a boosting rate, and it was listed as one of the critical-priority bacteria by World Health Organization (WHO).29 In this work, the antimicrobial susceptibility profiles, molecular characteristics, plasmid and integron-associated analysis, genetic environments of blaKPC-2 and blaNDM-1, and MLST were evaluated among 66 clinical CRKP isolates. Meanwhile, the risk factors for CRKP infection were also investigated. All the CRKP strains were MDR or even extensively drug-resistant (XDR), in this case, the tigecycline and polymyxin B were the last resort options for CRKP infection. However, the treatment was hardly successful because of the low blood concentrations when tigecycline was used in monotherapy.30 Adverse events, such as hypofibrinogenemia, also resulted in treatment termination associated with tigecycline therapy.31 In addition, nephrotoxicity and unclear dosing also limited the widespread clinical usage of polymyxin B.32,33 Importantly, tigecycline- and polymyxin B-resistant CRKP strains have been reported in many regions,34,35 therefore, the optimal therapeutic strategy for CRKP infection still needs to be explored.
Focusing on the phenotype and molecular analysis of CRKP isolates, the results revealed that the primary resistance mechanism of CRKP was carbapenemase production, which was consistent with other researches.36,37 The blaKPC-2 and blaNDM-1 were two major carbapenemase genes in our study, accounting for 63.6% and 30.3%, respectively. KPC-2 is the most commonly identified enzyme among over 20 reported KPC variants so far,38 and the majority of NDM-1-producers also harbored various other resistance genes but mostly were still susceptible to polymyxin B and tigecycline,39 which were highly correlated with our results. Moreover, IMP is another important metallo-β-lactamase, and blaIMP-1 emerged in Japan in the late 1980s.40 The blaIMP-4 was one of the most prevalent blaIMP variants since it was first identified in Acinetobacter spp. in the mid-1990s,41 particularly in China and Australia.42,43 It is worth noting that blaIMP often co-exists with other resistance genes,44 which is consistent with our results. In our work, 3 strains were identified co-carrying blaNDM and blaIMP genes, including two isolates with blaNDM-1 and blaIMP-4 and one with blaNDM-5 and blaIMP-4. Coexistence of blaNDM-1 and blaIMP-4 was still rarely distributed in China since first reported in 2012, but the strain co-harboring blaNDM-5 and blaIMP-4 has never been published before.45
In this work, blaKPC-2 was only found on IncFII plasmids, and blaNDM was only detected on IncX3 plasmids. The IncFII plasmid family is commonly low copy number, not only carries multiple antimicrobial resistance and virulence genes, but also can replicate and disseminate in many different species of Enterobacteriaceae.46 Furthermore, several studies have revealed that IncX3 plasmids were the predominant plasmid type that harboring a variety of blaNDM genes.47–49 Of note, IncX3 plasmids can frequently disseminate in humans, animals, and environment, and more commonly spread in Asia, including China,49,50 Myanmar,51 South Korea,52 and India.53 In addition, 46 CRKP strains carried variable regions of intl1, and 7 resistance gene cassettes were detected among them. The addA2 (40/46) and addA1 (1/46) were the genes responsible for aminoglycoside resistance. The dfrA12 (6/46), dfrA27 (1/46) contributed to trimethoprim resistance. Furthermore, a β-lactams-resistant gene blaOXA-10, a gentamicin-resistant gene arr3, and a rifampicin-resistant gene aacA4 were also detected.
A previous study has proved that the genetic environment of blaKPC-2 in pKP048 from China was considered as a combination of the Tn3-based transposon and the partial Tn4401 structure.54 In our study, KPC-2 producers presented a similar structure (tnpA-tnpR-ISkpn8-blaKPC-2), but the environment surrounding blaKPC-2 was partly different from other reports in China due to the specific insertions and deletions.55,56 Additionally, all the NDM-1 producers carried the highly conserved region (blaNDM−1-bleMBL-trpF-tat) surrounding the blaNDM−1 gene, which was identified not only in K. pneumoniae, but also in Acinetobacter lwoffii and Enterobacter Cloacae.25,57,58
Furthermore, the results showed that more than 80% clinical CRKP strains simultaneously carried virulence genes entB, fimH, and mrkD. The enterobactin that encoded by entB is one of the ubiquitous siderophores in K. pneumoniae, which is an important factor involving in iron acquisition.59 The fimH and mrkD mediate bacterial adhesion through encoding type I and type III fimbriae of K. pneumoniae implied that these two genes may contribute to biofilm formation and development. Zhang et al have also observed that 95% tested K. pneumoniae isolates co-carrying fimH and mrkD genes in their study.60 Additionally, the genes peg-344, iroB, iucA, rmpA, and rmpA2 locate on the HVKP virulence plasmid and are regarded as regulators of hypervirulent phenotype.61 Notably, two carbapenem-resistant HVKP (CR-HVKP) strains were found in this work, which contained blaKPC-2 gene. There are two hypotheses to explain the evolution of this novel phenotype. First, CRKP strains acquire HVKP-specific virulence plasmid; second, HVKP strains obtain the carbapenem-resistant genes by acquisition of resistance plasmid or by the insertion of resistance determinants into HVKP-specific virulence plasmid.59,62 Compared with the second hypothesis, the first one was more likely to occur.63–65
According to the results of MLST, the 42 KPC-2-producing CRKP strains were grouped into 7 ST types, of which 35 strains belonged to ST11. It has been reported that most of KPC-2-producing strains belong to clone group (CG) 258; meanwhile, ST11 and ST258 are the dominant ST types.66 ST258 has spread around the world since it emerged in the early 21st century, particularly in North America, Latin America, and several European countries.66,67 However, ST11 is more prevalent in Asia, and usually accounts for more than half of all KPC-2-producing CRKP strains in China.47,68,69 In addition, the 24 NDM-producing CRKP strains were classified into 8 ST types, and ST4495 was the most common ST type. Remarkably, ST4495 (4-1-99-1-9-5-5) has never been detected before, indicating a novel clone carrying blaNDM-1 has spread in Southwest China.
Hematological disease, tracheal cannula, and exposure to β-lactams and β-lactamase inhibitor combination were independent risk factors for CRKP infection. Many studies have demonstrated that the history of tracheal cannula was independent risk factor for CRKP infection.70–73 Meanwhile, exposure to antibiotic was also associated with CRKP infection, such as exposure to quinolones,74 β-lactams and β-lactamase inhibitor combination, and carbapenems,75 which was consistent with our study. Notably, hematological disease was rarely found to be a risk factor for CRKP infection in other studies, but it is reasonable due to patients with hematological disease were accompanied with severe immune function deficiency and were more prone to be infected. There are some limitations in our study, first, the number of patients included in this study is relatively small, although this is a common problem in studies assessing risk factors for multidrug-resistant microbial infections.74 Secondly, the study was performed in a single-center setting, so that some important risk factors may be missed.
Conclusion
The current study revealed the high prevalence of CRKP infection caused by NDM-1 and KPC-2 producers, and ST11 was the most common lineage. To the best of our knowledge, the novel ST4495 and co-exist of blaNDM-5 and blaIMP-4 were the first reported in this work. Moreover, plasmid, integron, ISCR, and other mobile genetic elements endowed bacteria a rapid adaptation ability in changing environments; meanwhile, the genetic environments of blaKPC-2 and blaNDM-1 showed polymorphism. Logistic regression model indicated that hematological disease, tracheal cannula, and use of β-lactams and β-lactamase inhibitor combination were independent risk factors for CRKP infection, suggesting that appropriate clinical management and antimicrobials treatment were necessary for retarding the selection of CRKP strains. Effective measures to prevent and control the further dissemination of CRKP strains are highly needed.
Acknowledgments
This work was supported by the grants from Sichuan Science and Technology Program (2021YFH001, 2021YFS0329,), Luxian Government and Southwest Medical University Cooperation Program (2020LXXNYKD-04).
Data Sharing Statement
The data used and/or analyzed in this study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
This study was approved by the Institutional Review Board of affiliated hospital of southwest medical university (KY2020043). As for neonate, a parent or legal guardian provided informed consent for these patients, and that this study was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflict of interest.
References
- 1.Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–359. doi: 10.1038/s41579-019-0315-1 [DOI] [PubMed] [Google Scholar]
- 2.Li M, Wang X, Wang J, et al. Infection-prevention and control interventions to reduce colonisation and infection of intensive care unit-acquired carbapenem-resistant Klebsiella pneumoniae: a 4-year quasi-experimental before-and-after study. Antimicrob Resist Infect Control. 2019;8(1):8. doi: 10.1186/s13756-018-0453-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoxha A, Kärki T, Giambi C, et al. Attributable mortality of carbapenem-resistant Klebsiella pneumoniae infections in a prospective matched cohort study in Italy, 2012–2013. J Hosp Infect. 2016;92(1):61–66. doi: 10.1016/j.jhin.2015.06.018 [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin Infect Dis. 2019;69(Suppl 7):S521–s528. doi: 10.1093/cid/ciz824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22(1):161–182. Table of Contents. doi: 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54(3):969–976. doi: 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4:48. doi: 10.3389/fmicb.2013.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Sun L, Ding B, et al. Outbreak of NDM-1-producing Klebsiella pneumoniae ST76 and ST37 isolates in neonates. Eur J Clin Microbiol Infect Dis. 2016;35(4):611–618. doi: 10.1007/s10096-016-2578-z [DOI] [PubMed] [Google Scholar]
- 10.Zheng R, Zhang Q, Guo Y, et al. Outbreak of plasmid-mediated NDM-1-producing Klebsiella pneumoniae ST105 among neonatal patients in Yunnan, China. Ann Clin Microbiol Antimicrob. 2016;15(1):10. doi: 10.1186/s12941-016-0124-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Xu X, Li Z, et al. An outbreak of a nosocomial NDM-1-producing Klebsiella pneumoniae ST147 at a teaching hospital in mainland China. Microb Drug Resist. 2014;20(2):144–149. doi: 10.1089/mdr.2013.0100 [DOI] [PubMed] [Google Scholar]
- 12.Escobar Pérez JA, Olarte Escobar NM, Castro-Cardozo B, et al. Outbreak of NDM-1-producing Klebsiella pneumoniae in a neonatal unit in Colombia. Antimicrob Agents Chemother. 2013;57(4):1957–1960. doi: 10.1128/AAC.01447-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monteiro J, Inoue FM, Lobo APT, Ibanes AS, Tufik S, Kiffer CRV. A major monoclonal hospital outbreak of NDM-1-producing Klebsiella pneumoniae ST340 and the first report of ST2570 in Brazil. Infect Control Hosp Epidemiol. 2019;40(4):492–494. doi: 10.1017/ice.2018.333 [DOI] [PubMed] [Google Scholar]
- 14.Ripabelli G, Sammarco ML, Salzo A, Scutellà M, Felice V, Tamburro M. New Delhi metallo-β-lactamase (NDM-1)-producing Klebsiella pneumoniae of sequence type ST11: first identification in a hospital of central Italy. Lett Appl Microbiol. 2020;71(6):652–659. doi: 10.1111/lam.13384 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Zhao C, Wang Q, et al. High prevalence of hypervirulent klebsiella pneumoniae infection in china: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60(10):6115–6120. doi: 10.1128/AAC.01127-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327–336. doi: 10.1093/jac/dkz446 [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, Wu Q, He L, et al. Clinical and molecular characteristics of carbapenem-resistant hypervirulent klebsiella pneumoniae isolates in a tertiary hospital in Shanghai, China. Infect Drug Resist. 2021;14:2697–2706. doi: 10.2147/IDR.S321704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI M100. Wayne PA: CLSI; 2020. [Google Scholar]
- 19.Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001;7(2):88–91. doi: 10.1046/j.1469-0691.2001.00204.x [DOI] [PubMed] [Google Scholar]
- 20.Sfeir MM, Hayden JA, Fauntleroy KA, et al. EDTA-modified carbapenem inactivation method: a phenotypic method for detecting metallo-β-lactamase-producing enterobacteriaceae. J Clin Microbiol. 2019;57(5):e01757–18. doi: 10.1128/JCM.01757-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang T, Chen C, Wen J, et al. Resistance of klebsiella pneumoniae strains carrying bla (NDM-1) gene and the genetic environment of bla (NDM-1). Front Microbiol. 2020;11:700. doi: 10.3389/fmicb.2020.00700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi: 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 23.Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic features of blaNDM-1-positive enterobacteriaceae. Antimicrob Agents Chemother. 2011;55(11):5403–5407. doi: 10.1128/AAC.00585-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra S, Upadhyay S, Sen MR, Maurya AP, Choudhury D, Bhattacharjee A. Genetic acquisition of NDM gene offers sustainability among clinical isolates of Pseudomonas aeruginosa in clinical settings. PLoS One. 2015;10(1):e0116611. doi: 10.1371/journal.pone.0116611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia X, Dai W, Ma W, et al. Carbapenem-resistant E. cloacae in Southwest China: molecular analysis of resistance and risk factors for infections caused by NDM-1-producers. Front Microbiol. 2018;9:658. doi: 10.3389/fmicb.2018.00658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradford PA, Urban C, Mariano N, Projan SJ, Rahal JJ, Bush K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC beta-lactamase, and the foss of an outer membrane protein. Antimicrob Agents Chemother. 1997;41(3):563–569. doi: 10.1128/AAC.41.3.563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacKenzie FM, Forbes KJ, Dorai-John T, Amyes SG, Gould IM. Emergence of a carbapenem-resistant Klebsiella pneumoniae. Lancet. 1997;350(9080):783. doi: 10.1016/S0140-6736(05)62567-6 [DOI] [PubMed] [Google Scholar]
- 29.Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 30.Ruiz J, Ramirez P, Villarreal E, et al. Effect of pharmacokinetic/pharmacodynamic ratio on tigecycline clinical response and toxicity in critically ill patients with multidrug-resistant Gram-negative infections. SAGE Open Med. 2020;8:2050312120958897. doi: 10.1177/2050312120958897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, Wang J, Liu H, Ma W, Zhou S, Zhou J. Risk factors for tigecycline-induced hypofibrinogenaemia. J Clin Pharm Ther. 2020;45(6):1434–1441. doi: 10.1111/jcpt.13250 [DOI] [PubMed] [Google Scholar]
- 32.Avedissian SN, Liu J, Rhodes NJ, et al. A review of the clinical pharmacokinetics of polymyxin B. Antibiotics. 2019;8(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zavascki AP, Nation RL. Nephrotoxicity of polymyxins: is there any difference between colistimethate and polymyxin B? Antimicrob Agents Chemother. 2017;61(3):e02319–16. doi: 10.1128/AAC.02319-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao L, Lv Y, Li Y. Analysis of the drug resistance of carbapenem-resistant Klebsiella pneumoniae in the China antimicrobial resistance surveillance trial program, 2007–2018. Microb Drug Resist. 2020;26(8):944–950. doi: 10.1089/mdr.2019.0299 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem-resistant enterobacteriaceae infections: report from the China CRE network. Antimicrob Agents Chemother. 2018;62(2):e01882–17. doi: 10.1128/AAC.01882-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng B, Dai Y, Liu Y, et al. Molecular epidemiology and risk factors of carbapenem-resistant Klebsiella pneumoniae infections in Eastern China. Front Microbiol. 2017;8:1061. doi: 10.3389/fmicb.2017.01061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumudunie WGM, Wijesooriya LI, Namalie KD, Sunil-Chandra NP, Wijayasinghe YS. Epidemiology of multidrug-resistant enterobacteriaceae in Sri Lanka: first evidence of bla(KPC) harboring Klebsiella pneumoniae. J Infect Public Health. 2020;13(9):1330–1335. doi: 10.1016/j.jiph.2020.04.010 [DOI] [PubMed] [Google Scholar]
- 38.Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi: 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35(1):147–151. doi: 10.1128/AAC.35.1.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu YW, Afzal-Shah M, Houang ET, et al. IMP-4, a novel metallo-beta-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob Agents Chemother. 2001;45(3):710–714. doi: 10.1128/AAC.45.3.710-714.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung GH, Gray TJ, Cheong EY, Haertsch P, Gottlieb T. Persistence of related bla-IMP-4 metallo-beta-lactamase producing Enterobacteriaceae from clinical and environmental specimens within a burns unit in Australia - a six-year retrospective study. Antimicrob Resist Infect Control. 2013;2(1):35. doi: 10.1186/2047-2994-2-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng W, Zhou D, Wang Q, et al. Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci Rep. 2016;6(1):33419. doi: 10.1038/srep33419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kizny Gordon A, Phan HTT, Lipworth SI, et al. Genomic dynamics of species and mobile genetic elements in a prolonged blaIMP-4-associated carbapenemase outbreak in an Australian hospital. J Antimicrob Chemother. 2020;75(4):873–882. doi: 10.1093/jac/dkz526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Z, Wang Y, Tian L, et al. First report in China of Enterobacteriaceae clinical isolates coharboring blaNDM-1 and blaIMP-4 drug resistance genes. Microb Drug Resist. 2015;21(2):167–170. doi: 10.1089/mdr.2014.0087 [DOI] [PubMed] [Google Scholar]
- 46.Villa L, García-Fernández A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65(12):2518–2529. doi: 10.1093/jac/dkq347 [DOI] [PubMed] [Google Scholar]
- 47.Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon EJ, Kang DY, Yang JW, et al. New Delhi Metallo-beta-lactamase-producing enterobacteriaceae in South Korea between 2010 and 2015. Front Microbiol. 2018;9:571. doi: 10.3389/fmicb.2018.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Tong MK, Chow KH, et al. Occurrence of highly conjugative incx3 epidemic plasmid carrying bla (NDM) in enterobacteriaceae isolates in geographically widespread areas. Front Microbiol. 2018;9:2272. doi: 10.3389/fmicb.2018.02272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho PL, Li Z, Lo WU, et al. Identification and characterization of a novel incompatibility group X3 plasmid carrying bla NDM-1 in enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microb Infect. 2012;1(11):e39. doi: 10.1038/emi.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugawara Y, Akeda Y, Sakamoto N, et al. Genetic characterization of blaNDM-harboring plasmids in carbapenem-resistant Escherichia coli from Myanmar. PLoS One. 2017;12(9):e0184720. doi: 10.1371/journal.pone.0184720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JS, Jin YH, Park SH, et al. Horizontal transfer of bla(NDM-1)-carrying IncX3 plasmid between carbapenem-resistant enterobacteriaceae in a single patient. J Infect. 2020;81(5):816–846. doi: 10.1016/j.jinf.2020.07.013 [DOI] [PubMed] [Google Scholar]
- 53.Choudhury NA, Paul D, Chakravarty A, Bhattacharjee A, Dhar Chanda D. IncX3 plasmid mediated occurrence of bla(NDM-4) within Escherichia coli ST448 from India. J Infect Public Health. 2018;11(1):111–114. doi: 10.1016/j.jiph.2017.06.008 [DOI] [PubMed] [Google Scholar]
- 54.Shen P, Wei Z, Jiang Y, et al. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among enterobacteriaceae in China. Antimicrob Agents Chemother. 2009;53(10):4333–4338. doi: 10.1128/AAC.00260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo Y, Yang J, Ye L, et al. Characterization of KPC-2-producing Escherichia coli, Citrobacter freundii, Enterobacter cloacae, Enterobacter aerogenes, and Klebsiella oxytoca isolates from a Chinese hospital. Microb Drug Resist. 2014;20(4):264–269. doi: 10.1089/mdr.2013.0150 [DOI] [PubMed] [Google Scholar]
- 56.Cai JC, Zhang R, Hu YY, Zhou HW, Chen GX. Emergence of Escherichia coli sequence type 131 isolates producing KPC-2 carbapenemase in China. Antimicrob Agents Chemother. 2014;58(2):1146–1152. doi: 10.1128/AAC.00912-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wailan AM, Paterson DL, Kennedy K, Ingram PR, Bursle E, Sidjabat HE. Genomic characteristics of NDM-producing enterobacteriaceae isolates in Australia and their blaNDM genetic contexts. Antimicrob Agents Chemother. 2016;60(1):136–141. doi: 10.1128/AAC.01243-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wailan AM, Sidjabat HE, Yam WK, et al. Mechanisms involved in acquisition of blaNDM genes by IncA/C2 and IncFIIY plasmids. Antimicrob Agents Chemother. 2016;60(7):4082–4088. doi: 10.1128/AAC.00368-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):e00001–19. doi: 10.1128/CMR.00001-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, Zhang X, Wu Q, et al. Clinical, microbiological, and molecular epidemiological characteristics of Klebsiella pneumoniae-induced pyogenic liver abscess in southeastern China. Antimicrob Resist Infect Control. 2019;8(1):166. doi: 10.1186/s13756-019-0615-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9):e00776–18. doi: 10.1128/JCM.00776-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei DD, Wan LG, Deng Q, Liu Y. Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in Mainland China. Diagn Microbiol Infect Dis. 2016;85(2):192–194. doi: 10.1016/j.diagmicrobio.2015.03.012 [DOI] [PubMed] [Google Scholar]
- 63.Feng Y, Lu Y, Yao Z, Zong Z. Carbapenem-resistant hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob Agents Chemother. 2018;62(7):e02644–17. doi: 10.1128/AAC.02644-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60(1):709–711. doi: 10.1128/AAC.02173-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu L, Tang L, Wang S, et al. Co-location of the bla(KPC-2), bla(CTX-M-65), rmtB and virulence relevant factors in an IncFII plasmid from a hypermucoviscous Klebsiella pneumoniae isolate. Microb Pathog. 2018;124:301–304. doi: 10.1016/j.micpath.2018.08.055 [DOI] [PubMed] [Google Scholar]
- 66.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–696. doi: 10.1016/j.tim.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Tella D, Tamburro M, Guerrizio G, Fanelli I, Sammarco ML, Ripabelli G. Molecular epidemiological insights into colistin-resistant and carbapenemases-producing clinical Klebsiella pneumoniae isolates. Infect Drug Resist. 2019;12:3783–3795. doi: 10.2147/IDR.S226416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu J, Yu J, Chen F, et al. Emergence and establishment of KPC-2-producing ST11 Klebsiella pneumoniae in a general hospital in Shanghai, China. Eur J Clin Microbiol Infect Dis. 2018;37(2):293–299. doi: 10.1007/s10096-017-3131-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao H, Liu Y, Wang R, Wang Q, Jin L, Wang H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine. 2020;51:102599. doi: 10.1016/j.ebiom.2019.102599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akgul F, Bozkurt I, Sunbul M, Esen S, Leblebicioglu H. Risk factors and mortality in the Carbapenem-resistant Klebsiella pneumoniae infection: case control study. Pathog Glob Health. 2016;110(7–8):321–325. doi: 10.1080/20477724.2016.1254976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiao Y, Qin Y, Liu J, et al. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection/colonization and predictors of mortality: a retrospective study. Pathog Glob Health. 2015;109(2):68–74. doi: 10.1179/2047773215Y.0000000004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Guo LY, Song WQ, Wang Y, Dong F, Liu G. Risk factors for carbapenem-resistant K. pneumoniae bloodstream infection and predictors of mortality in Chinese paediatric patients. BMC Infect Dis. 2018;18(1):248. doi: 10.1186/s12879-018-3160-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng X, Wang J-F, Xu W-L, Xu J, Hu J. Clinical and molecular characteristics, risk factors and outcomes of carbapenem-resistant Klebsiella pneumoniae bloodstream infections in the intensive care unit. Antimicrob Resist Infect Control. 2017;6(1):102. doi: 10.1186/s13756-017-0256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Falagas ME, Rafailidis PI, Kofteridis D, et al. Risk factors of carbapenem-resistant Klebsiella pneumoniae infections: a matched case control study. J Antimicrob Chemother. 2007;60(5):1124–1130. doi: 10.1093/jac/dkm356 [DOI] [PubMed] [Google Scholar]
- 75.Hu Y, Ping Y, Li L, Xu H, Yan X, Dai H. A retrospective study of risk factors for carbapenem-resistant Klebsiella pneumoniae acquisition among ICU patients. J Infect Dev Ctries. 2016;10(3):208–213. doi: 10.3855/jidc.6697 [DOI] [PubMed] [Google Scholar]