Abstract
Brain regions communicate with each other via tracts of myelinated axons, commonly referred to as white matter. Here we identified common genetic variants influencing white matter microstructure using diffusion magnetic resonance imaging of 43,802 individuals. Genome-wide association analysis identified 109 associated loci, 30 of which were detected by tract-specific functional principal component analysis. A number of loci colocalized with brain diseases, such as glioma and stroke. Genetic correlations were observed between white matter microstructure and 57 complex traits and diseases. Common variants associated with white matter microstructure altered the function of regulatory elements in glial cells, particularly oligodendrocytes. In summary, this large-scale tract-specific study advances the understanding of the genetic architecture of white matter and its genetic links to a wide spectrum of clinical outcomes.
One Sentence Summary:
Common variants in multiple genes alter brain structural connectivity and correlate with a variety of clinical endpoints.
Brain functions depend on effective communications across brain regions (1). White matter comprises roughly half of the human brain and contains most of the brain’s long-range communication pathways (2). White matter tracts build a complex network of structural connections (3, 4). Cerebral white matter tracts form connections among cerebral cortical areas and between cortical and subcortical regions (5, 6). Aspects of this connectivity can be estimated via measures of cellular microstructure (7). Evidence from neuroscience has further suggested that cerebral white matter microstructure may underpin brain function and dysfunction, and connectivity differences are relevant to a wide variety of neurological and psychiatric disorders, such as attention-deficit/hyperactivity disorder (ADHD) (8), major depressive disorder (MDD) (9), schizophrenia (10), and Alzheimer’s disease (11). White matter microstructural differences and abnormalities can be captured in vivo by diffusion magnetic resonance imaging (dMRI). Using dMRI data, microstructural connectivity can be quantified in diffusion tensor imaging (DTI) models (12) and measured by several DTI-derived metrics, including fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (AD), radial diffusivity (RD), and mode of anisotropy (MO) (Fig. 1A). Among them, FA serves as the primary metric of interest in many studies (13). It is a robust measure of overall integrity/directionality and is highly sensitive to general connectivity changes. On the other hand, MD, AD, and RD directly quantify the absolute magnitude of directionalities. They are more sensitive to specific types of microstructural changes and have also been used in a variety of clinical studies (14). In addition, MO can characterize the anisotropy type, describing whether the shape of the diffusion tensor is more linear or planar (15).
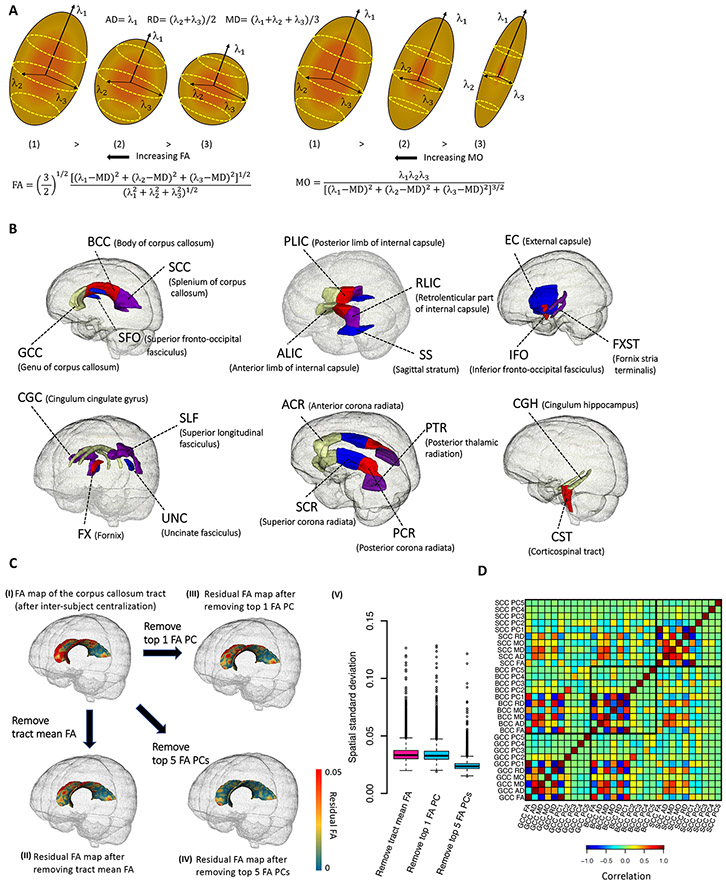
Fig. 1. Illustration of white matter traits and DTI parameters.
(A) Schematic representation of the five diffusion tensor images (DTI)-derived metrics. Axial diffusivity (AD) is the diffusion along the long axis (λ1), radial diffusivity (RD) is the diffusion of two small axes (average of λ2 and λ3), and mean diffusivity (MD) is the average diffusion regardless of direction (mean of λ1 , λ2 , and λ3). Fractional anisotropy (FA) and mode of anisotropy (MO) are two more complicated measures of general directionality. (B) Annotation of the 21 white matter tracts in human brain. (C) Comparison between tract mean FA and tract-specific FA principal components (PCs) on the corpus callosum tract (CC, including GCC, BCC, and SCC). (I) illustrates an example skeletonized FA map within the corpus callosum tract after inter-subject centralization; (II) displays the residual FA map after removing the within-subject tract mean FA; In (III) and (IV), instead of removing the within-subject mean as in (II), we removed the top one and five FA PCs, respectively. (V) illustrates the standard deviation across the voxels in residual FA map for each subject in the UKB (n = 36,624). The standard deviations are similar between the residual FA maps after removing tract mean FA (in (II)) and the first FA PC (in (III)), suggesting that this PC mainly accounts for the mean FA. Comparing (II) with (IV), other four FA PCs can capture more spatial variations that are ignored by the tract mean FA, and thus reduce the standard deviations of residuals in (V). (D) Correlation between the DTI parameters among GCC, BCC, and SCC tracts. Ten parameters are generated in each tract, including the mean FA, mean MD, mean AD, mean RD, mean MO, as well as the top five FA PCs. The five FA PCs are orthogonal to each other and the first FA PC can be highly correlated with mean FA.
White matter differences in general population cohorts are under strong genetic control. Both family and population-based studies have reported that DTI measurements of white matter microstructure have generally high heritability with estimates varying across different age groups (16) and tracts (17). For example, heritability estimates of tract-averaged FA ranged from 53% to 90% in twin study of the Human Connectome Project (HCP) (18). Recent genome-wide association studies (GWAS) of UK Biobank (UKB) reported an average SNP-based heritability of 48.7% across different tracts (19). Several GWAS (17, 19-23) have been performed to identify loci associated with inter-individual variation in white matter microstructure but shared at least two major limitations: (i) sample size and (ii) spatial specificity. First, the current largest published GWAS of dMRI phenotypes had a sample size of 17,706 (19). Similar to other brain-related traits, white matter has a complex and extremely polygenic genetic architecture (19). Large sample size is essential to boost GWAS power in order to identify many common risk variants with small effect sizes and uncover the shared genetic influences with other complex traits. Second, previous GWAS mainly focused on global dMRI measures of the whole brain (20, 21) or tract-averaged (mean) values (17, 19). Global and tract-averaged measures can capture the largest variations in white matter, while reducing the burden to test multiple neuroimaging traits, particularly suitable for GWAS with limited sample size; however, these measures may lose information, because microstructural differences and changes may not have a uniformly consistent pattern across the whole tract. Heterogeneous variation patterns typically exist within voxel-wise DTI maps of the 3D tract curve, which may be more relevant to specific underlying biological processes. For example, a previous study found that the association between bipolar disorder and FA is specific to one given segment of the anterior limb of internal capsule tract connecting prefrontal cortex with the thalamus and brain stem (24). Due to these limitations, a large number of genetic factors influencing white matter may still be undiscovered. Consequently, with few exceptions (for example, stroke (20) and cognitive traits (19)), the shared genetic influences between white matter and other complex traits are unknown. Uncovering these potential genetic links may identify important brain regions that are involved in clinical outcomes, especially for brain disorders.
To overcome these limitations, here we collected individual-level dMRI from five data resources: the UKB (25), Adolescent Brain Cognitive Development (ABCD (26)), HCP (27), Pediatric Imaging, Neurocognition, and Genetics (PING (28)), and Philadelphia Neurodevelopmental Cohort (PNC (29)). We harmonized image processing by using the ENIGMA-DTI pipeline (30, 31) and obtained voxel-wise DTI maps for 43,802 subjects (after quality controls), including 36,624 in UKB. We mainly focused on 21 predefined cerebral white matter tracts (Fig. 1B) and generated two groups of phenotypes (19). The first group contains 110 tract-averaged parameters for FA, AD, MD, MO, and RD in 21 tracts and across all white matter tracts (5 × 22). For the second group, we applied functional principal component analysis (FPCA (32)) to generate 105 tract-specific principal components (PCs) for FA by taking the top five PCs of the voxel-wise map within each tract. FPCA is a well established approach in neuroimaging and functional data analysis (33). In DTI models, FPCA can characterize the strongest variation components of FA within each tract, which are expected to provide more microstructural details about axonal organization and myelination omitted by tract-averaged values, while relieving multiple testing burdens. More importantly, these PCs may represent FA changes that are more relevant to specific clinical outcomes. We used the UKB imaging data released up through 2018 (referred to as UKB phases 1 and 2 hereafter, n = 17,706) to generate the loadings of FPCA and applied them to compute the FA PCs in other independent cohorts. Figure 1C illustrates the difference between tract-averaged FA and tract-specific FA PCs for the corpus callosum, which is the largest white matter tract in the human brain. The first FA PC performed similarly to (but slightly better than) the tract-averaged FA, and the other top ranked FA PCs (which are orthogonal to the first FA PC, Fig. 1D) further captured FA variations in subfields of the corpus callosum. We then performed a GWAS for these 215 phenotypes to discover the genetic architecture of white matter and explore the genetic links to a plethora of clinical endpoints in different trait domains. Our GWAS results have been made publicly available and can be browsed through our Brain Imaging Genetics Knowledge Portal (BIG-KP) https://bigkp.org/.
GWAS discovery and validation for 215 DTI parameters
Our heritability analysis utilized data from UKB subjects of British ancestry (n = 34,024). All of the 110 DTI mean parameters had significant SNP heritability (34) (h2) after Bonferroni adjustment (P < 4.1 × 10−25, Fig. 2A and Table S1). The h2 estimates varied from 22.4% to 66.5% (mean h2 = 47.6%), which were comparable with previous results (17, 19). For the 105 tract-specific FA PC parameters, we found that 102 had significant h2 (h2 range = (8.9%, 67.3%), mean = 34.9%, P < 6.5 × 10−6). For example, all of the top five PCs in the external capsule, superior longitudinal fasciculus, and splenium of corpus callosum had moderate to high heritability (h2 range = (30.6%, 67.3%), P < 2.3 × 10−47). The 3rd PC of the corticospinal tract (5.5%), 5th PC of the cingulum hippocampus (4.3%), and 4th PC of the superior fronto-occipital fasciculus (3.6%) had nominally significant h2 estimates (P < 0.03), which became insignificant after Bonferroni adjustment. To examine the robustness of heritability estimates of the 105 FA PCs, we reran the heritability analysis separately using the UKB phases 1 and 2 data (n = 17,706) and the newly released independent data in 2020 (named UKB phase 3, n = 15,918, removing the relatives of subjects in early released dataset). The correlation between the two sets of heritability estimates was 0.92 (mean h2 = 34.6% vs. 33.6%, two-sample t test P = 0.64, Table S2), suggesting that the FA PCs are heritable and have consistent heritability in independent cohorts. Overall, heritability analysis showed that the major microstructural variations of voxel-wise FA maps captured by unconventional tract-specific FA PCs are under genetic control. Those heritable FA variation patterns may have higher power than mean FA to identify the shared genetic influences with other complex traits and clinical outcomes.
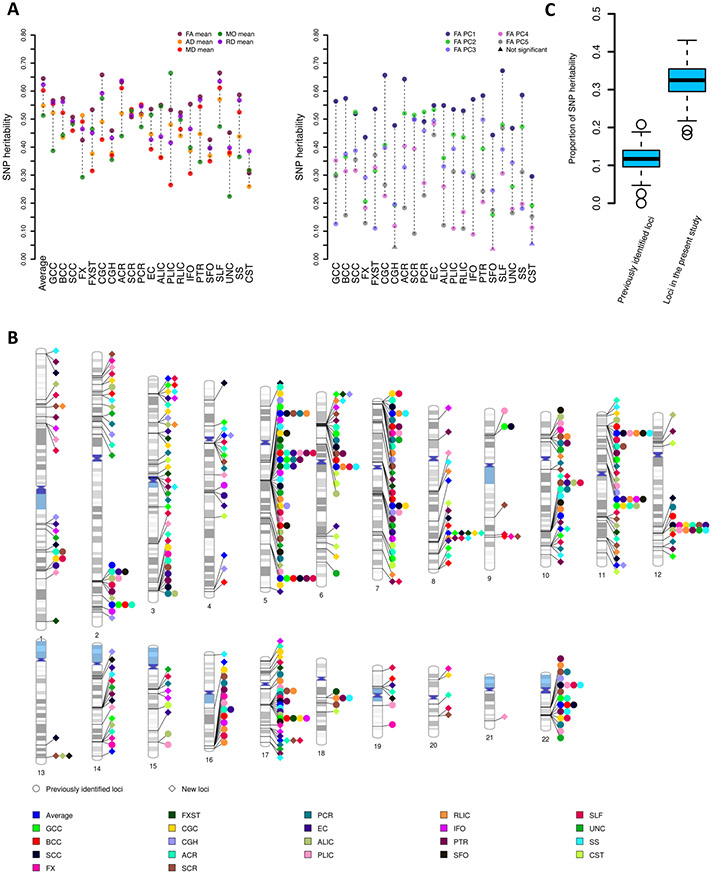
Fig. 2. SNP heritability and the associated genomic regions of DTI parameters.
(A) SNP heritability of 215 DTI parameters, including 110 mean parameters (left panel, mean values of axial diffusivity (AD), radial diffusivity (RD), mean diffusivity (MD), fractional anisotropy (FA), and mode of anisotropy (MO) in 21 tracts and the whole brain, 5 × 22) and 105 FA principal components (PCs) (right panel, top 5 FA PCs of 21 tracts, 5 × 21). Average, global average across 21 tracts; Full names of the 21 white matter tracts can be found in Fig. 1(B). (B) Ideogram of genomic regions influencing DTI parameters (P < 2.3 × 10−10), including 42 previously identified regions and 109 additional regions identified in the present study. The colors represent the 21 white matter tracts (and the global average). Each signal point indicates that at least one of the 10 DTI parameters (5 mean parameters and 5 FA PCs) of this tract is associated with the genomic region. (C) Proportion of SNP heritability of the 215 DTI parameters that can be explained by the 44 genomic regions identified in previous study (11.7%) and 151 regions identified in the current study (32.3%).
We used a multiple-stage design in our GWAS of the 215 DTI parameters (Fig. S1). First, we checked the robustness of GWAS results within the UKB British cohort. Specifically, GWAS were separately performed in two independent datasets: the UKB phases 1 and 2 data (n = 17,706) and the independent UKB phase 3 data (n = 15,918). We calculated the replication slope between the two set of GWAS results, which was the correlation of the standardized effect size of variants estimated from two independent GWAS (35). This analysis was restricted to top (P < 1 × 10−6) independent lead variants after LD (linkage disequilibrium)-based clumping (window size 250, LD r2 = 0.01). The replication slope was 0.8 (standard error = 0.01, P < 2 × 10−16), indicating strong similarity between these top variant effect size estimates. In addition, for each of the 215 DTI parameters, we used LD score regression (LDSC) (36) to calculate genetic correlation (gc) between DTI measurements from the two GWAS. The mean gc estimate was 1.01 (standard error = 0.17, Table S3) across these parameters, 209 of which were significant after adjusting for multiple comparisons by using the Benjamini-Hochberg procedure to control false discovery rate (FDR) at 5% level. Genetic correlation estimates near 1 indicate a consistent genetic basis for these phenotypes measured in different cohorts. In conclusion, these findings indicate that GWAS results of DTI parameters are highly consistent within the UKB British cohort.
Second, we performed GWAS for these 215 DTI parameters using the combined UKB data of British ancestry (n = 34,024). All Manhattan and quantile-quantile (QQ) plots can be browsed through our BIG-KP server (https://bigkp.org/). The LDSC intercepts (37) were close to one, suggesting no genomic inflation of test statistics due to confounding factors (mean intercept = 1.008, standard error = 0.008; Table S4). At a stringent significance level 2.3 × 10−10 (5 × 10−8/215, additionally adjusted for the 215 phenotypes studied), FUMA (38) identified 539 independent significant lead variants (LD r2 < 0.1) involved in 1,312 significant associations with 173 DTI parameters (106 mean and 67 PC parameters, Table S5). These variant-level associations were further summarized into 996 significant locus-trait associations (Table S6). Genetic variants had broad effects across all of the 21 white matter tracts. We replicated 42 of the 44 genomic regions discovered in the largest previous GWAS (19) and identified 109 additional regions associated with DTI parameters (Fig. 2B). On average, the 151 (42+109) significant genomic regions can explain 32.3% of SNP heritability for the 215 DTI parameters, while the 44 previously identified genomic regions can only explain 11.7% of SNP heritability (Fig. 2C and Table S7). Of the 109 genomic regions we identified, 30 were only detected by FA PC parameters. For example, the third FA PC of posterior thalamic radiation detected 3 additional regions (Fig. S2). In summary, our results illuminate the broad genetic control of white matter microstructural differences and the contribution of tract-specific FA PCs in identifying genetic variants associated with white matter tracts. The genetic effects are spread across a large number of genomic regions, consistent with the observed polygenic genetic architecture of many brain-related traits (39).
Next, to replicate our results in UKB British GWAS, we performed additional analysis in nine independent validation datasets. GWAS was separately performed on five datasets consisting of individuals of European ancestry, including the UKB White but Non-British (UKBW, n = 1,938), ABCD European (ABCDE, n = 3,821), HCP (n = 334), PING (n = 461), and PNC (n = 537); and on four non-European validation datasets: the UKB Asian (UKBA, n = 435), UKB Black (UKBBL, n = 227), ABCD Hispanic (ABCDH, n = 768), and ABCD African American (ABCDA, n = 1,257). For each DTI parameter, the genetic correlation between discovery GWAS and the meta-analyzed European validation GWAS (total n = 7,091) was estimated by LDSC (36). The mean genetic correlation estimate was 0.93 (standard error = 0.36) across the 215 DTI parameters, 136 of which were significant at FDR 5% level (Table S8). We then checked whether the locus-trait associations detected in UKB British GWAS can be replicated in these validation datasets. For the 996 significant associations, 203 (20.4%, in 50 genomic regions) passed the 5 × 10−5 (0.05/996) Bonferroni significance level in the meta-analyzed European validation GWAS, and 532 (53.4%, in 92 regions) were significant at 1 × 10−3 level. We also meta-analyzed the four non-European GWAS (total n = 2,687) and found that 64 associations (6.4%, in 19 regions) passed the Bonferroni significance level and 367 (36.8%, in 75 regions) were significant at 1 × 10−3 level. Moreover, we performed a third meta-analysis to combine all of the nine validation datasets, after which the number of replicated associations moved up to 258 (25.9%, in 52 regions) and 678 (68%, in 115 regions) at Bonferroni and the 1 × 10−3 significance levels, respectively (Table S9). Overall, our results suggest that the associated genetic loci discovered in UKB British GWAS can be strongly validated in independent studies, despite the fact that these studies may use different cohorts and/or MRI scanners. Therefore, we performed a final meta-analysis to combine all of these datasets (n = 43,802). At the 2.3 × 10−10 significance level, the meta-analyzed GWAS detected 193 significant genomic regions, including 144 (95.4%) of the 151 regions identified in UKB British GWAS (Table S10).
Additionally, we tested for replication by using polygenic risk scores (PRS) (40) derived from UKB British GWAS. At FDR 5% level (215 × 5 tests), the mean number of significant PRS in the five European validation GWAS datasets was 201 (range = (193, 210), Table S11). Almost all (213/215) DTI parameters had significant PRS in at least one dataset and 173 had significant PRS in all of them, showing the high generalizability of our discovery GWAS results. Across the five validation datasets, the mean additional variance that can be explained by PRS (incremental R-squared, adjusting for the effects of sex, age, and top ten genetic PCs) was 1.8% (range = (0.6%, 4%)) for the 173 consistently significant DTI parameters. The largest R-squared was on the 4th PC of the external capsule (range = (1.8%, 5.3%), P range = (1.2 × 10−19, 5.6 × 10−12)). Finally, we constructed PRS on four non-European validation datasets. At FDR 5% level, the number of significant PRS decreased to 172, 46, 144, 147 in UKBA, UKBBL, ABCDH and ABCDA, respectively (Table S12). These findings show that UKB British GWAS results have high generalizability in European cohorts, but the generalizability is reduced in cross-population applications, highlighting the importance of recruiting sufficient samples from global diverse populations in future genetic discovery of white matter.
Concordance with previous GWAS
We carried out association lookups for independent significant variants (38) (P < 2.3 × 10−10, and variants within LD, r2 ≥ 0.6) detected in our UKB British discovery GWAS. In the NHGRI-EBI GWAS catalog (41), white matter microstructure had shared genetic influences with a wide range of other complex traits in different trait domains, including brain structures, glioma (glioblastoma (GBM) and non-GBM tumors), stroke, vascular risk factors (such as diabetes, high blood pressure, obesity, and smoking), cognitive traits, neurological disorders (such as Alzheimer's disease and Parkinson's disease), psychiatric disorders (such as schizophrenia and autism spectrum disorder (ASD)), psychological traits (such as neuroticism and well-being spectrum), sleep, anthropometric measurements, bone mineral density, and educational attainment. These results are summarized in Table S13.
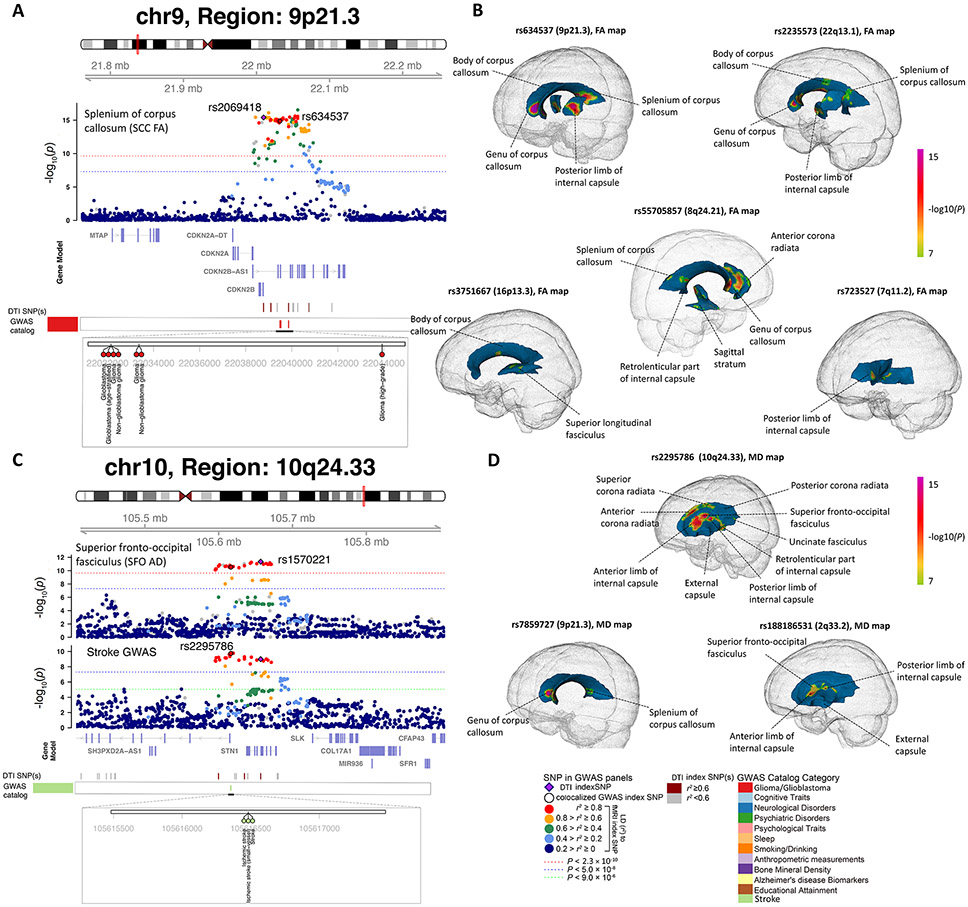
Our results tagged 17 genomic regions that had been associated with brain structures, such as 3q28 and 17q21.31 with intracranial volume (42); 22q13.1, 10p12.31, 16q24.2, 3q28, and 12q23.3 with lateral ventricular volume (43); 5q14.3, 6q25.1, 13q34, 2p16.1, 17q25.1, 10q24.33, and 2q33.2 with white matter hyperintensity (20, 44); 10q26.13, 14q23.1, and 5q12.3 with hippocampal subfield volumes (45); and 15q14 with brain imaging measurements (17). Moreover, we observed colocalizations with glioma in 9p21.3, 22q13.1, 16p13.3, 7p11.2, and 8q24.21 (46) (Fig. 3A and Figs. S3-S6). These colocalizations were detected by multiple mean parameters and FA PCs. To illustrate the genetic effect pattern in spatial brain maps, we took the lead index variants from the five glioma-associated regions and performed target-variant analysis on voxel-wise FA and MD data. After Bonferroni adjustment (P < 8.5 × 10−8), glioma risk variants were associated with multiple white matter tracts, including the corpus callosum, internal capsule, superior longitudinal fasciculus, anterior corona radiata, and sagittal stratum (Fig. 3B and Fig. S7A). Reduced microstructure integrity and altered brain structural connectivity have been observed in glioma (47). Early invasive glioma growth along white matter tracts is one of the main challenges in treating glioma (48). The corpus callosum serves as a hub between hemispheres, and glial cells may migrate along the corpus callosum during glioma cell infiltration (49). In addition, three regions were colocalized with stroke, including 10q24.33, 9p21.3, and 2q33.2 (50) (Fig. 3C, Fig. 3D, Fig. S7B, and Fig. S8). Stroke risk variants from these regions had genetic effects in the corpus callosum, superior fronto-occipital fasciculus, corona radiata, internal capsule, and external capsule. Ischemic white matter disorganization and the associated neuropsychiatric disorders (such as depression) have been frequently reported after stroke (51). White matter integrity is closely related to the pathologic stages of stroke and can predict the outcome of stroke recovery. For example, the MD of ischemic regions stays reduced during the acute phase of stroke, then increases over time to become pseudonormalized in the later-subacute phase, and continues rising in the chronic phase (52). Moreover, these three colocalized regions were also associated with cardiovascular disease, type 2 diabetes (T2D), obesity, and high blood pressure, suggesting that the genetic relationship between white matter microstructure and stroke may be partially mediated through vascular risk factors.
Fig. 3. Selected genetic loci that were associated with both DTI parameters and brain diseases.
(A) In 9p21.3, we observed colocalization (LD r2≥ 0.6) between the mean fractional anisotropy (FA) of splenium of corpus callosum (SCC FA, index variant rs2069418) and glioma (both glioblastoma and non-glioblastoma tumors, index variant rs634537). (B) We illustrated the voxel-wise genetic effects of 5 colocalized glioma GWAS index variants (rs634537, rs2235573, rs55705857, rs3751667, and rs723527) on FA. The genetic effects were obtained by performing voxel-wise target-variant analysis for the 5 colocalized glioma significant variants. We displayed the voxels passing the Bonferroni significant level (P < 8.5 × 10−8) in the voxel-wise target-variant analysis. White matter tracts that had significant voxels were labelled in each map. (C) In 10q24.33, we observed colocalization between the mean axial diffusivity (AD) of superior fronto-occipital fasciculus (SFO AD, index variant rs1570221) and stroke (index variant rs2295786). (D) We illustrated the voxel-wise genetic effects of 3 colocalized stroke GWAS index variants (rs2295786, rs7859727, and rs18818651) on mean diffusivity (MD).
The 17q21.31 inversion region and the 6p22.1 and 6p22.2 in the extended major histocompatibility complex (MHC) region have complicated LD patterns. In these regions, white matter associated variants colocalized with risk variants of many brain disorders and cognition, such as schizophrenia (53), Alzheimer's disease (54), corticobasal degeneration (55), Parkinson's disease (56), progressive supranuclear palsy (55), ASD (57), alcohol use disorder (58), depression (59), educational attainment (60), and cognitive ability (61) (Figs. S9-S11). Colocalizations were also found in 3q28 with Alzheimer's disease biomarkers (Fig. S12), in 3p21.31 with intelligence (Fig. S13), in 3p22.1 with amyotrophic lateral sclerosis (ALS), and in 1q23.2 with multiple sclerosis. Many colocalized loci of cognitive traits and educational attainment can only be detected by FA PCs. In addition, we also observed colocalizations with psychological traits (such as neuroticism (62), well-being spectrum (63), and general risk tolerance (64)), anthropometric traits (such as body mass index (BMI) (35)), smoking/drinking (65), and sleep (including sleep duration (66) and chronotype (67)). Overall, genetic links were found between white matter microstructure and a wide spectrum of clinical outcomes and complex traits. Integrating the GWAS of white matter with these clinical outcomes may help understand the underlying mechanisms leading to the changes in brain structural connectivity and risk of brain disorders.
An atlas of genetic correlations with other complex traits
Because of the shared loci associated with both white matter microstructure and other complex traits, we systematically examined their pairwise genetic correlations by using publicly available summary-level data of 70 other complex traits via LDSC (Table S14). We mainly focused on brain-related complex traits with large GWAS sample size, such as neurological and psychiatric disorders, cognitive traits, and cardiovascular risk factors. There were 727 significant pairs between 57 complex traits and 171 DTI parameters at FDR 5% level (70 × 215 tests, P range = (3 × 10−13, 2.4 × 10−3), Table S15), 41% (298/727) of which were detected by PC parameters. We found that DTI parameters were widely correlated with brain disorders (such as stroke, MDD, ADHD, schizophrenia, ALS, Alzheimer's disease, and ASD), cognition, educational attainment, chronotype, insomnia, neuroticism, risk tolerance, automobile speeding, high blood pressure, T2D, coronary artery disease, BMI, white matter hyperintensities, drinking, smoking, manual occupation, and lung function (Figs. S14-S16).
We replicated previously reported genetic correlations with cognitive/educational traits (19), drinking behavior (19), stroke (17, 20), and MDD (19, 20), and more tract-specific details were revealed. For example, stroke (any subtypes) and ischemic stroke subtypes (50) (large artery stroke, cardioembolic stroke, and small vessel stroke) showed broad genetic correlations with the corpus callosum, corona radiata, internal capsule, external capsule, superior longitudinal fasciculus, superior fronto-occipital fasciculus, and uncinate fasciculus (∣gc∣ range = (0.3, 0.42), P range = (3.2 × 10−9, 2.4 × 10−3), Figs. 4A-4B), matching previous findings in the literature (68, 69). We further observed that small vessel stroke subtype had specific, but stronger genetic correlations with the anterior limb of internal capsule and superior fronto-occipital fasciculus (∣gc∣ range = (0.56, 0.69), P range = (3.7 × 10−4, 1.2 × 10−3)). In contrast, there were no significant genetic correlations detected for large artery and cardioembolic stroke, demonstrating the potentially much stronger genetic links between white matter tracts and small vessel stroke subtype. This finding is consistent with previous observation that white matter changes are associated with small vessel stroke, but not with non-small vessel events (70).
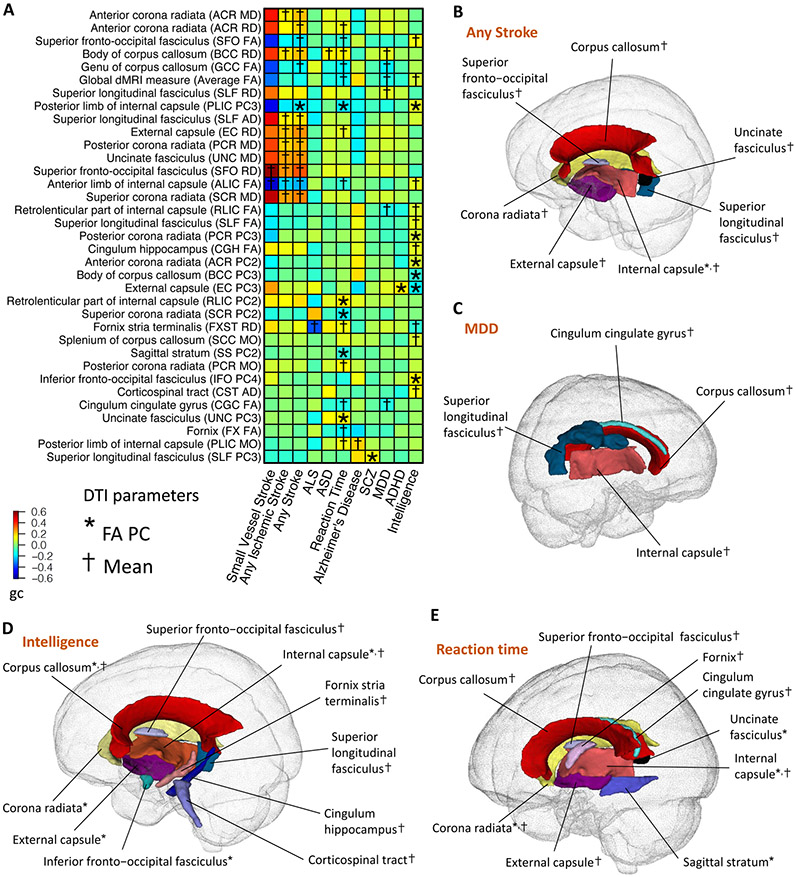
Fig. 4. Selected pairwise genetic correlations between DTI parameters of white matter tracts and brain disorders and cognitive functions.
(A) The asterisks (for FA PCs) and daggers (for mean parameters) highlight significant genetic correlations after controlling the FDR at 5% level. The y-axis lists the DTI parameters of white matter tracts and the x-axis provides the names of brain-related traits/disorders. The colors represent genetic correlations (gc). FA, fractional anisotropy; AD, axial diffusivity; MD, mean diffusivity; MO, mode of anisotropy; RD, radial diffusivity; PC, principal component of FA; ALS, amyotrophic lateral sclerosis; ASD, autism spectrum disorder; MDD, major depressive disorder; SCZ, schizophrenia; ADHD, attention-deficit/hyperactivity disorder. (B-E) Location of the white matter tracts whose DTI parameters were genetically correlated with (B) stroke (any subtype); (C) MDD; (D) intelligence; and (E) reaction time. The colors describe different white matter tracts.
In addition, we observed genetic correlations between MDD and the corpus callosum, superior longitudinal fasciculus, internal capsule, and cingulum cingulate gyrus (∣gc∣ range = (0.16, 0.19), P range = (1.8 × 10−7, 2.1 × 10−3), Fig. 4C). Anatomical deficits in the corpus callosum are related to inter-hemispheric disconnections involved in the pathology of MDD (71). Lower FA in the superior longitudinal fasciculus, internal capsule, and cingulum has also been reported in patients with MDD (14, 72). For intelligence and reaction time, we found genetic correlations with the FA of multiple white matter tracts, such as the internal capsule, corpus callosum, superior fronto-occipital fasciculus, corona radiata, external capsule, fornix, and superior longitudinal fasciculus (∣gc∣ range = (0.15, 0.18), P range = (3 × 10−9, 2.2 × 10−3), Figs. 4D-4E). Higher FA can reduce inter-hemispheric transfer time and increase information processing speed, resulting in more efficient cognitive functioning and faster reaction (73).
Moreover, many genetic correlations were uncovered for brain disorders, including schizophrenia, ADHD, ALS, Alzheimer's disease, and ASD. For example, significant genetic correlations were found between the posterior limb of internal capsule and Alzheimer's disease, the external capsule and ADHD, the fornix stria terminalis and ALS, the superior longitudinal fasciculus and schizophrenia, and the body of corpus callosum and ASD (∣gc∣ > 0.16, P < 2 × 10−3). Alterations of white matter integrity have been consistently observed in the corpus callosum among ASD patients (74). Fornix degeneration is involved in the pathology pathway of memory impairment in ALS, and the external capsule is related to ADHD symptoms (75). In addition, white matter changes in the posterior limb of internal capsule have been observed in people with Alzheimer's disease (76). For schizophrenia, case-control differences of FA were frequently reported in the superior longitudinal fasciculus (77).
We also found correlations for non-brain traits, many of which were risk factors for stroke (P < 2.4 × 10−3). For example, high blood pressure was genetically correlated with many white matter tracts including the superior longitudinal fasciculus, internal capsule, superior fronto-occipital fasciculus, uncinate fasciculus, and external capsule (P range = (5.7 × 10−11, 2.3 × 10−3)). Hypertension can result in vascular stiffness and impaired cerebral perfusion, leading to microstructural white matter disruption (78) and stroke. We also observed strong genetic correlations with white matter hyperintensities (P range = (3.6 × 10−12, 2.3 × 10−3)), which have been implicated in the connection between stroke and white matter (79). In addition, we found that BMI, bone mineral density, smoking, and T2D were all genetically correlated with the corticospinal tract (P range = (3 × 10−13, 2.3 × 10−3)). The corticospinal tract controls primary motor activity and is known to be closely related to stroke (80). Higher BMI, smoking, and T2D are risk factors for stroke and are associated with white matter changes (81, 82). Low bone mineral density may also have correlation with white matter changes that contribute to stroke (83). Moreover, genetic correlations were observed between drinking and the corpus callosum (P range = (6 × 10−5, 2.4 × 10−3)). whose atrophy is frequently observed in people with alcohol use disorders (84).
Next, we explored the genetic correlations between 215 DTI parameters and 101 regional brain volumes (85). After Bonferroni adjustment, there were 678 significant pairs between 132 DTI parameters and 62 regional brain volumes (215 × 101 tests, ∣gc∣ range = (0.18, 0.85), P < 2.2 × 10−6, Table S16). These results suggest the existence of widespread genetic overlaps between basic brain morphology and structural connectivity (Figs. S17-S18). More importantly, most of the significant genetic correlations were observed between white matter tracts and their neighboring brain regions, indicating the strong spatial colocalization of genetic co-variation.
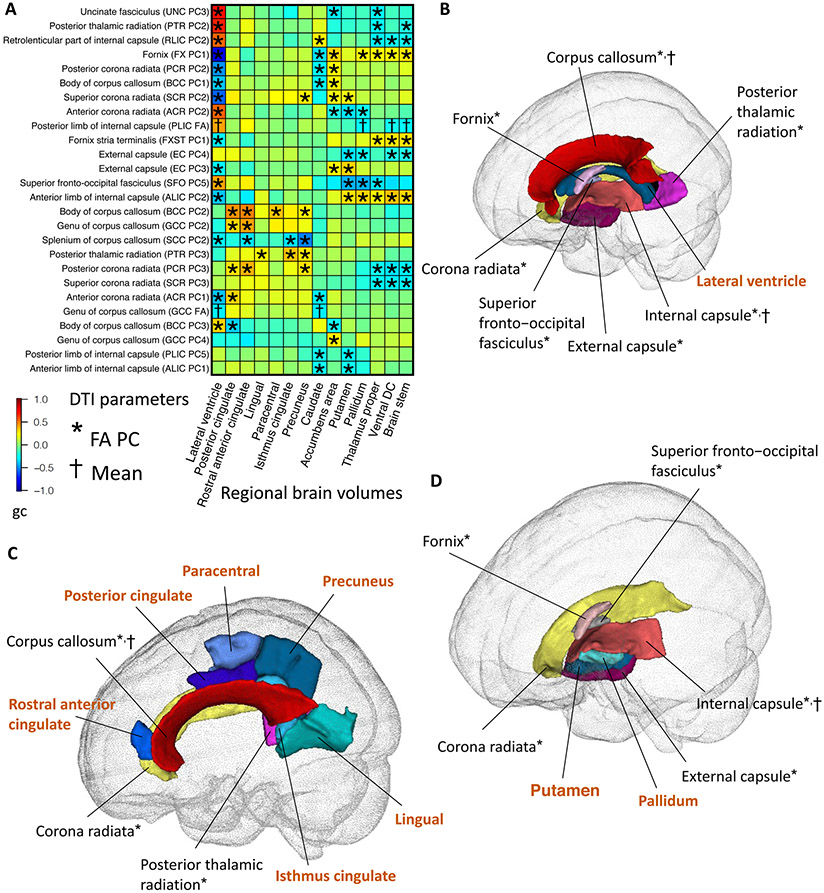
For example, lateral ventricle volumes were genetically correlated with the FA of white matter tracts surrounding the lateral ventricles, including the fornix, corona radiata, corpus callosum, posterior thalamic radiation, superior fronto-occipital fasciculus, internal capsule, and external capsule (Figs. 5A-5B). Both ventricular enlargement and altered white matter integrity were biomarkers of aging (86, 87) and brain disorders, such as normal pressure hydrocephalus (88, 89), Alzheimer's disease (90, 91), and Parkinson’s disease (87, 92). In addition, the FA of corpus callosum, corona radiata, and posterior thalamic radiation had genetic correlations with volumes of nearby cerebral cortex regions, such as posterior cingulate, rostral anterior cingulate, lingual, isthmus cingulate, paracentral, and precuneus (Fig. 5C). These cortex regions have connections with many other brain regions via white matter tracts to support a wide range of cognitive functions. For example, the cingulate gyrus overlies the corpus callosum and serves as a neural hub among emotion, action, and memory (93), and the corona radiata consists of prominent projection fibers carrying information between the cerebral cortex and brain stem. Moreover, putamen and pallidum volumes were correlated with the FA of internal capsule, external capsule, fornix, superior fronto-occipital fasciculus, and corona radiata. The putamen and pallidum lie on the lateral aspect of the internal capsule and separate internal capsule from external capsule (Fig. 5D), all of which are critical for human sensory motor functions (94). Similarly, volumes of other subcortical structures (such as the thalamus proper, accumbens area, and caudate) and the brain stem also had genetic correlations with adjacent white matter tracts, especially the corona radiata (Fig. S19).
Fig. 5. Selected pairwise genetic correlations between fractional anisotropy (FA) of white matter tracts and regional brain volumes.
(A) The asterisks (for FA PCs) and daggers (for mean parameters) highlight significant genetic correlations after Bonferroni adjustment for multiple testing. The y-axis lists the DTI parameters of white matter tracts and the x-axis provides the name of regional brain volumes. The colors represent genetic correlations (gc). PC, principal component of FA. (B) Location of the lateral ventricle region and its neighboring white matter tracts whose FA parameters were genetically correlated with the volume of lateral ventricle. The colors describe different brain regions (names highlighted in brown color) and white matter tracts. (C) Location of the cortex regions and their neighboring white matter tracts whose FA parameters were genetically correlated with the volume of these regions. (D) Location of the putamen and pallidum regions and their neighboring white matter tracts whose FA parameters were genetically correlated with the volume of these regions.
Gene-level analysis
We carried out MAGMA (95) gene-based association analysis for the 215 DTI parameters using our discovery GWAS summary statistics. There were 2,772 significant gene-level associations (P < 1.2 × 10−8, adjusted for 215 phenotypes) between 487 genes and 178 DTI parameters (Table S17), where 96 of the associated genes could only be discovered by PC parameters. We replicated 101 of 112 MAGMA genes reported in (19) and some other white matter-associated genes reported in previous studies (20-22, 44), such as SH3PXD2A (10q24.33), NBEAL1 (2q33.2), TRIM47 (17q25.1), VCAN (5q14.3), ALDH2 (12q24.12), PLEKHG1 (6q25.1), GNA12 (7p22.2), and GNA13 (17q24.1). Many of our MAGMA significant genes have been linked to other complex traits and brain disease (Table S18). For example, EGFR (7p11.2), OBFC1 (10q24.33), LMF1 (16p13.3), SLC16A8 (22q13.1), and CDKN2B (9p21.3) are risk genes for glioma (46). Colocalization analysis (96) found that EGFR and SLC16A8 had high colocalization probability (> 0.9) with expression quantitative trait loci (eQTL) signals in GTEx (v8) (97) brain tissues (Table S19). EGFR is critical for oligodendrocyte development and white matter recovery after neonatal hypoxia (98). It is known that EGFR plays an important role in the initiation of primary glioblastoma and progression of lower-grade glioma (46). In addition, many stroke risk genes, such as CDH11 (16q21), NTN4 (12q22), COL4A1 (13q34), FAT4 (4q28.1), CACNB2 (10p12.31), SUPT3H (6p21.1), ICA1L (2q33.2), CDKN2C (1p32.3), SH3PXD2A (10q24.33), ALDH2 (12q24.12), and ADAMTS7 (15q25.1) (50, 99), were associated with DTI parameters. These genes were also widely associated with hypertension, T2D, coronary artery disease, and BMI, suggesting the shared genetic components among stroke, white matter microstructure, and vascular risk factors.
Next, we mapped significant variants (P < 2.3 × 10−10) to genes according to physical position, eQTL association, and 3D chromatin (Hi-C) interaction via FUMA (38). FUMA yielded 1,183 associated genes (1,545 in total) that were not discovered in MAGMA analysis (Table S20), replicating 282 of the 292 FUMA genes identified in (19) and more genes from previous studies of white matter, such as CLDN23 (100) (8p23.1) and EFEMP1 (20, 21, 44) (2p16.1). In particular, 911 FUMA genes were solely mapped by significant Hi-C interactions in brain tissues (Table S21), demonstrating the power of integrating chromatin interaction profiles in GWAS of white matter. More overlapping genes were observed between white matter microstructure and other traits (Table S22). For example, glioma risk genes from five more regions were associated with white matter microstructure, including RAVER2 (1p31.3), MDM4 (1q32.1), ZBTB16 (11q23.2), AKT3 (1q44), and MAML2 (11q21) (46). AKT3 is hyper-expressed and activated in glioma (101) and is also associated with brain developmental disorders (102), learning and memory deficits (103), and disrupted integrity of white matter (104). Together, our variant-level and gene-level analyses uncovered the shared genetic influences with glioma in 11 different genomic regions. Considering the largest glioma GWAS (46) reported 25 risk regions, our results reveal the close genetic relationship between glioma and white matter integrity.
We performed drug target lookups in a recently established drug target network (105), which included 273 nervous system drugs (anatomical therapeutic chemical (ATC) code starts with “N”) and 241 targeted genes. We found that 14 white matter-associated genes were targets for 79 drugs, 23 of which were anti-psychotics (ATC: N05A, target such as CALM1) to manage psychosis like schizophrenia and bipolar disorder, 38 were anti-depressants (ATC: N06A, SLC6A4) to treat MDD and other conditions, 8 were used to treat addictive disorders (ATC: N07B, ALDH2), 6 were anti-Parkinson drugs (ATC: N04B, HTR2B), 4 were anti-dementia drugs (ATC: N06D, CHRNB4), and 3 were anti-convulsants (ATC: N03A, GABBR1) used in the treatment of epileptic seizures (Table S23). In addition, we treated white matter-associated genes as an annotation and performed partitioned heritability enrichment analysis (106) for the other 70 complex traits studied in our genetic correlation analysis. At FDR 5% level, heritability of 49 complex traits was significantly (P range = (1.2 × 10−8, 3.2 × 10−2)) enriched in regions influencing white matter microstructure, such as stroke, schizophrenia, ADHD, bipolar disorder, Alzheimer's disease, T2D, high blood pressure, and coronary artery disease (Fig. S20 and Table S24). Overall, these results suggest the potential clinical values of the genes identified for white matter microstructure.
Biological annotations
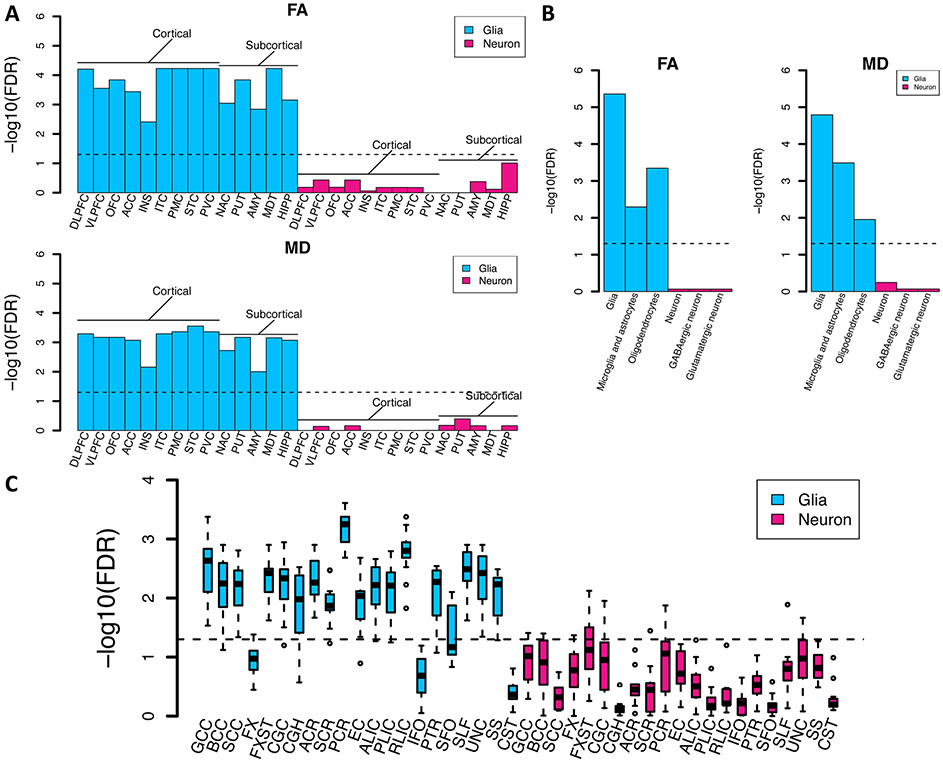
In order to identify tissues and cell types where genetic variation leads to changes in white matter microstructure, we performed partitioned heritability analyses (106) from the GWAS of global FA and MD within tissue type and cell type specific regulatory elements. First, we used regulatory elements across multiple adult and fetal tissues (107). As expected, both FA and MD had the strongest enrichment of heritability in active gene regulation regions of brain tissues (Fig. S21 and Table S25). To identify gross cell types, we again performed partitioned heritability analysis using chromatin accessibility data of two brain cell types, neurons (NeuN+) and glia (NeuN−) sampled from 14 brain regions, including both cortical and subcortical (108). For all regions, we found that enrichment of FA and MD heritability existed in glial, but not neuronal regulatory elements (Fig. 6A). These results are expected as white matter is largely composed of glial cell types. For further resolution on cell types, we tested partitioned heritability enrichment within differentially accessible chromatin of glial cell subtypes, oligodendrocyte (NeuN−/Sox10+) and microglia and astrocyte (NeuN−/Sox10−); and neuronal cell subtypes, GABAergic (NeuN+/Sox6+) and glutamatergic (NeuN+/Sox6−) neurons (109). Heritability of FA and MD was enriched in oligodendrocyte and microglia/astrocyte annotations. In contrast, no enrichment was observed in neurons (Fig. 6B). We also performed regional-specific enrichment analysis for FA in each white matter tract. Glial cell enrichment was observed in different white matter tracts and the strongest enrichments were found in the posterior corona radiata, posterior limb of internal capsule, and genu of corpus callosum (Fig. 6C). These analyses imply that common variants associated with white matter microstructure alter the function of regulatory elements in glial cells, particularly oligodendrocytes, the cell type expected to influence white matter microstructure, supporting the biological validity of the genetic associations.
Fig. 6. Partitioned heritability enrichment analysis in brain cell types.
(A) Heritability enrichment of global mean fractional anisotropy (FA) and mean diffusivity (MD) in regulatory elements of two brain cell types (neuron and glia) sampled from 14 brain cortical and subcortical regions. DLFPC, dorsolateral prefrontal cortex; VLPFC, ventrolateral prefrontal cortex; OFC, orbitofrontal cortex; ACC, anterior cingulate cortex; INS, insular cortex; ITC, inferior temporal cortex; STC, superior temporal cortex; PMC, primary motor cortex; PVC, primary visual cortex; AMY, amygdala; HIPP, hippocampus; MDT, mediodorsal thalamus; NAC, nucleus accumbens; PUT, putamen. (B) Heritability enrichment of global mean FA and MD in regulatory elements of glial cell subtypes (glia, including oligodendrocyte and microglia/ astrocyte) and neuronal cell subtypes (neurons, including GABAergic and glutamatergic neurons). (C) Heritability enrichment of mean FA of 21 white tracts in regulatory elements of two brain cell types (neuron and glia) sampled from 14 brain cortical and subcortical regions. Full names of the 21 white matter tracts can be found in Fig. 1 (B). The dashed lines indicate the significance level after controlling the FDR at 5% level.
Next, we performed MAGMA gene property (95) analysis for 13 GTEx (v8) brain tissues to examine whether tissue-specific gene expression levels were related to significance of the correlations between genes and DTI parameters. At FDR 5% level, 63 significant (P range = (7 × 10−16, 8.4 × 10−4)) associations were detected between DTI parameters and gene expression data sampled in GTEx v8 brain tissues (Table S26), suggesting that genes with higher transcription levels in brain tissues also had stronger genetic associations with DTI parameters. We also applied DEPICT (110) gene-set enrichment testing for 10,968 pre-constituted gene sets to prioritize enriched biological pathways. At FDR 5% levels, DEPICT found 381 significant gene sets (10,968 × 215 tests, P < 1 × 10−5, Table S27). Several gene sets of rat sarcoma (Ras) proteins, small guanosine triphosphate hydrolases (GTPases), and Rho family GTPases were prioritized by DEPICT, such as “small GTPase mediated signal transduction” (GO: 0007264), “Rho GTPase binding” (GO:0017048), “Ras protein signal transduction” (GO: 0007265), “reactome Rho GTPase cycle” (M27078), and “reactome signaling by Rho GTPases” (M501). The activity of Ras proteins is involved in developmental processes and abnormalities of neural cells in central nervous system (111); small and Rho family GTPases play crucial roles in basic cellular processes during the entire neurodevelopment process and are closely connected to several neurological disorders (112, 113). We also observed enrichment in pathways related to glial cells and nervous system, such as “glial cell differentiation” (GO:0010001), “abnormal radial glial cell morphology” (MP:0003648), “brain development” (GO:0007420), “abnormal forebrain morphology” (MP:0000783), “open neural tube” (MP:0000929), “regulation of neurogenesis” (GO:0050767), “abnormal neuronal migration” (MP:0006009), “central nervous system neuron development” (GO:0021954), and “abnormal neural fold formation” (MP:0004837).
Discussion
In this study, we analyzed the genetic architecture of brain white matter using dMRI scans of 43,802 subjects collected from five publicly accessible data resources. We identified 109 additional genomic regions for white matter microstructural differences. Many previously reported genetic hits were confirmed in our discovery GWAS, and we further validated our discovery GWAS in several replication cohorts. We evaluated the genetic relationships between white matter and a wide variety of complex traits in association lookups, genetic correlation estimations, and gene-level analyses. A large proportion of our findings were revealed by unconventional tract-specific PC parameters. Bioinformatics analyses found tissue and cell-specific functional enrichments and many enriched biological pathways. Together, these results suggest the value of large-scale neuroimaging data integration and the application of tract-specific FPCA in studying the genetics of human brain.
Many efforts have been made to study the gray matter abnormalities associated with brain diseases and cognition. Our results indicate that a better understanding of the etiopathogenesis and treatment of many brain disorders could be achieved from a more balanced perspective that includes both gray matter regions and white matter tracts. Moreover, we found that some commonly used centrally-acting medications have effects on genes with associations to white matter microstructure, suggesting that the neuropharmacology of brain disorders may potentially benefit from knowledge of the relationship between these medications and white matter. Many drugs for cognitive disorders have not been fully understood, and better knowledge of how these drugs work in the brain white matter could substantially improve patient care.
One limitation of the present study is that the majority of publicly available dMRI data are from subjects of European ancestry and our discovery GWAS focused on UKB British individuals. Such GWAS strategy can efficiently avoid false discoveries due to population stratifications and heterogeneities across studies (17, 114), but may raise questions as to the degree to which the research findings can be generalized and applied to global populations (115). In our analysis, the UKB British-derived PRS still showed prediction power in Hispanic, Asian, and Black/African American testing cohorts, but had reduced performance. This may indicate that the genetic architecture of white matter is similar, but not the same across different populations. Identifying the cross-population and population-specific components of genetic factors for human brain could be an interesting future topic. As more non-European neuroimaging data become available (for example, the ongoing CHIMGEN project (116) in Chinese population), global integration efforts are needed to study the comparative genetic architectures and to explore the multi-ethnic genetic relationships among brain and other human complex traits. Specifically, the anticipated non-European data will be relevant to non-European ancestry groups who often endure worse outcomes with respective to brain diseases that implicate white matter, such as stroke (117) and Alzheimer's disease (118). In addition, the present study focused on DTI parameters. Previous studies have found that some beyond-DTI parameters, such as white matter tract integrity (WMTI (119)), neurite orientation dispersion and density imaging (NODDI (120)), and diffusional kurtosis imaging (DKI (121)) may provide additional information of microstructural properties (122). The g-ratio measure (123, 124) is probably more directly correlated to neuronal conduction velocity and thus is of interest in assessing inter-hemispheric transfer time and information processing speed. Future work is needed to explore these beyond-DTI parameters in biobank-scale neuroimaging datasets. Finally, although we have harmonized the image processing pipelines to minimize the influences of imaging artifacts, confounding factors may still exist and limit the interpretation of our results. For example, the partial-volume effect (125) in DTI data could be a confounding factor that drives part of the observed genetic correlations between ventricle volumes and the FA of white matter tracts. More analysis is required to model and examine potential neuroimaging confounders (126).
Methods summary
We conducted a GWAS of white matter microstructure using dMRI data from 43,802 individuals. Measures of white matter microstructure were derived from DTI model using ENIGMA-DTI pipeline (30, 31). We analyzed five major DTI measures along 21 cerebral white matter tracts. For each individual, 215 DTI phenotypes were generated. We analyzed these phenotypes in subsequent genetic analyses.
Our discovery GWAS used data of UKB individuals of British ancestry. To replicate our results in UKB British GWAS, we performed additional GWAS with data from nine independent European and non-European datasets. We checked whether the associations detected in UKB British GWAS can be replicated in these independent GWAS. Additionally, we used polygenic risk scores to predict the proportion of variance of DTI phenotypes that can be explained by genetic variants in European and non-European cohorts.
SNP heritability explained by all autosomal variants was estimated using GCTA-GREML analysis (34). We used LDSC (36) to estimate genetic correlations between DTI parameters and other complex traits. Partitioned LDSC (106) was used to estimate the proportion of heritability explained by annotated regions of the genome. Heritability enrichment was estimated in multiple adult and fetal tissues (107), brain cell types (glia and neurons) (108), and glial and neuronal cell subtypes (109).
Gene-based association analysis was performed using MAGMA (95). We applied fastENLOC (96) to perform colocalization analysis with GTEx eQTL annotations. DEPICT (110) was used to explore the implicated biological pathways. Functional impact was investigated using FUMA (38), in which variants were linked to genes by a combination of positional, eQTL, and 3D chromatin interaction mappings.
Supplementary Material
Acknowledgments:
We thank Jinjie Lin for making panel D of the summary figure. We thank Sophia Cui, Xiaopeng Zong, and Peter Vandehaar for helpful conversations. We thank the individuals represented in the UKB, ABCD, HCP, PING, and PNC studies for their participation and the research teams for their work in collecting, processing and disseminating these datasets for analysis. We gratefully acknowledge all the studies and databases that made GWAS summary data available. This research has been conducted using the UK Biobank resource (application number 22783), subject to a data transfer agreement.
Funding:
This research was partially supported by NIH grants MH086633 (H.Z.), MH116527 (TF.L.), and HD079124 (Y.L.). Part of the data collection and sharing for this project was funded by the Pediatric Imaging, Neurocognition and Genetics Study (PING) (U.S. National Institutes of Health Grant RC2DA029475). PING is funded by the National Institute on Drug Abuse and the Eunice Kennedy Shriver National Institute of Child Health & Human Development. PING data are disseminated by the PING Coordinating Center at the Center for Human Development, University of California, San Diego. We thank the research team of PING, including Connor McCabe, Linda Chang, Natacha Akshoomoff, Erik Newman, Thomas Ernst, Peter Van Zijl, Joshua Kuperman, Sarah Murray, Cinnamon Bloss, Mark Appelbaum, Anthony Gamst, Wesley Thompson, and Hauke Bartsch. Support for the collection of the PNC datasets was provided by grant RC2MH089983 awarded to Raquel Gur and RC2MH089924 awarded to Hakon Hakonarson. All PNC subjects were recruited through the Center for Applied Genomics at The Children's Hospital in Philadelphia. Part of the data used in the preparation of this article were obtained from the Adolescent Brain Cognitive Development (ABCD) Study (https://abcdstudy.org), held in the NIMH Data Archive (NDA). This is a multisite, longitudinal study designed to recruit more than 10,000 children age 9-10 and follow them over 10 years into early adulthood. The ABCD Study is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041022, U01DA041028, U01DA041048, U01DA041089, U01DA041106, U01DA041117, U01DA041120, U01DA041134, U01DA041148, U01DA041156, U01DA041174, U24DA041123, U24DA041147, U01DA041093, and U01DA041025. A full list of supporters is available at https://abcdstudy.org/federal-partners.html. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/scientists/workgroups/. ABCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators. HCP data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: We made use of publicly available software and tools. The analysis code is freely available at https://doi.org/10.5281/zenodo.4549465. Our GWAS summary statistics are publicly available at https://doi.org/10.5281/zenodo.4549730 and https://doi.org/10.5281/zenodo.4549725. Our results can also be browsed through our BIG-KP knowledge portal https://bigkp.org/. The individual-level raw data used in this study can be obtained from five publicly accessible data resources.
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes:
- 1.van den Heuvel MP, Sporns O, A cross-disorder connectome landscape of brain dysconnectivity. Nature reviews neuroscience 20, 435–446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagmann P et al. , Mapping the structural core of human cerebral cortex. PLoS biology 6, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zielinski BA, Gennatas ED, Zhou J, Seeley WW, Network-level structural covariance in the developing brain. Proceedings of the National Academy of Sciences 107, 18191–18196 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filley CM, Fields RD, White matter and cognition: making the connection. Journal of neurophysiology 116, 2093–2104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koziol LF, Budding DE, Chidekel D, Adaptation, expertise, and giftedness: towards an understanding of cortical, subcortical, and cerebellar network contributions. The Cerebellum 9, 499–529 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Schmahmann JD, Smith EE, Eichler FS, Filley CM, Cerebral white matter: neuroanatomy, clinical neurology, and neurobehavioral correlates. Annals of the New York Academy of Sciences 1142, 266 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang EH et al. , The role of myelination in measures of white matter integrity: combination of diffusion tensor imaging and two-photon microscopy of CLARITY intact brains. Neuroimage 147, 253–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Z-M et al. , White matter microstructural alterations in children with ADHD: categorical and dimensional perspectives. Neuropsychopharmacology 42, 572–580 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou K et al. , Alterations of white matter integrity in adults with major depressive disorder: a magnetic resonance imaging study. Journal of psychiatry & neuroscience: JPN 33, 525 (2008). [PMC free article] [PubMed] [Google Scholar]
- 10.Cetin-Karayumak S et al. , White matter abnormalities across the lifespan of schizophrenia: a harmonized multi-site diffusion MRI study. Molecular psychiatry, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S et al. , White matter hyperintensities are a core feature of Alzheimer's disease: evidence from the dominantly inherited Alzheimer network. Annals of neurology 79, 929–939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basser PJ, Mattiello J, LeBihan D, Estimation of the effective self-diffusion tensor from the NMR spin echo. Journal of Magnetic Resonance, Series B 103, 247–254 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Grieve S, Williams L, Paul R, Clark C, Gordon E, Cognitive aging, executive function, and fractional anisotropy: a diffusion tensor MR imaging study. American Journal of Neuroradiology 28, 226–235 (2007). [PMC free article] [PubMed] [Google Scholar]
- 14.Tae W-S, Ham B-J, Pyun S-B, Kang S-H, Kim B-J, Current clinical applications of diffusion-tensor imaging in neurological disorders. Journal of Clinical Neurology 14, 129–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ennis DB, Kindlmann G, Orthogonal tensor invariants and the analysis of diffusion tensor magnetic resonance images. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 55, 136–146 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Vuoksimaa E et al. , Heritability of white matter microstructure in late middle age: A twin study of tract - based fractional anisotropy and absolute diffusivity indices. Human brain mapping 38, 2026–2036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott LT et al. , Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562, 210–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kochunov P et al. , Heritability of fractional anisotropy in human white matter: a comparison of Human Connectome Project and ENIGMA-DTI data. Neuroimage 111, 300–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao B et al. , Large-scale GWAS reveals genetic architecture of brain white matter microstructure and genetic overlap with cognitive and mental health traits (n= 17,706). Molecular Psychiatry, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rutten-Jacobs LC et al. , Genetic study of white matter integrity in UK Biobank (N= 8448) and the overlap with stroke, depression, and dementia. Stroke 49, 1340–1347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traylor M et al. , Genetic variation in PLEKHG1 is associated with white matter hyperintensities (n= 11,226). Neurology 92, e749–e757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fornage M et al. , Genome - wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Annals of neurology 69, 928–939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adib-Samii P et al. , 17q25 Locus is associated with white matter hyperintensity volume in ischemic stroke, but not with lacunar stroke status. Stroke 44, 1609–1615 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Safadi Z et al. , Functional segmentation of the anterior limb of the internal capsule: linking white matter abnormalities to specific connections. Journal of Neuroscience 38, 2106–2117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudlow C et al. , UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Medicine 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casey B et al. , The adolescent brain cognitive development (ABCD) study: imaging acquisition across 21 sites. Developmental cognitive neuroscience 32, 43–54 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Somerville LH et al. , The Lifespan Human Connectome Project in Development: A large-scale study of brain connectivity development in 5–21 year olds. NeuroImage 183, 456–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jernigan TL et al. , The pediatric imaging, neurocognition, and genetics (PING) data repository. Neuroimage 124, 1149–1154 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satterthwaite TD et al. , Neuroimaging of the Philadelphia neurodevelopmental cohort. Neuroimage 86, 544–553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jahanshad N et al. , Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: A pilot project of the ENIGMA–DTI working group. Neuroimage 81, 455–469 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kochunov P et al. , Multi-site study of additive genetic effects on fractional anisotropy of cerebral white matter: comparing meta and megaanalytical approaches for data pooling. Neuroimage 95, 136–150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall P, Müller H-G, Wang J-L, Properties of principal component methods for functional and longitudinal data analysis. The annals of statistics, 1493–1517 (2006). [Google Scholar]
- 33.Srivastava A, Klassen EP, Functional and shape data analysis. (Springer; ), vol. 1. [Google Scholar]
- 34.Yang J, Lee SH, Goddard ME, Visscher PM, GCTA: a tool for genome-wide complex trait analysis. The American Journal of Human Genetics 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kichaev G et al. , Leveraging polygenic functional enrichment to improve GWAS power. The American Journal of Human Genetics 104, 65–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bulik-Sullivan B et al. , An atlas of genetic correlations across human diseases and traits. Nature Genetics 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulik-Sullivan BK et al. , LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature genetics 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe K, Taskesen E, Bochoven A, Posthuma D, Functional mapping and annotation of genetic associations with FUMA. Nature Communications 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matoba N, Love M, Stein J, Evaluating brain structure traits as endophenotypes using polygenicity and discoverability. Human Brain Mapping, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jansen IE et al. , Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nature genetics 51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buniello A et al. , The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Research 47, D1005–D1012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikram MA et al. , Common variants at 6q22 and 17q21 are associated with intracranial volume. Nature Genetics 44, 539–544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vojinovic D et al. , Genome-wide association study of 23,500 individuals identifies 7 loci associated with brain ventricular volume. Nature communications 9, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verhaaren BF et al. , Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circulation: Cardiovascular Genetics 8, 398–409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van der Meer D et al. , Brain scans from 21,297 individuals reveal the genetic architecture of hippocampal subfield volumes. Molecular psychiatry 25, 3053–3065 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melin BS et al. , Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nature genetics 49, 789 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu D et al. , Alterations of white matter integrity associated with cognitive deficits in patients with glioma. Brain and Behavior, e01639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J et al. , Invasion of white matter tracts by glioma stem cells is regulated by a NOTCH1–SOX2 positive-feedback loop. Nature Neuroscience 22, 91–105 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Sharifi G, Pajavand AM, Nateghinia S, Meybodi TE, Hasooni H, Glioma Migration Through the Corpus Callosum and the Brainstem Detected by Diffusion and Magnetic Resonance Imaging: Initial Findings. Frontiers in Human Neuroscience 13, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malik R et al. , Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nature genetics 50, 524–537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hackett ML, Köhler S, T O'Brien J, Mead GE, Neuropsychiatric outcomes of stroke. The Lancet Neurology 13, 525–534 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Schlaug G, Siewert B, Benfield A, Edelman R, Warach S, Time course of the apparent diffusion coefficient (ADC) abnormality in human stroke. Neurology 49, 113–119 (1997). [DOI] [PubMed] [Google Scholar]
- 53.Ikeda M et al. , Genome-wide association study detected novel susceptibility genes for schizophrenia and shared trans-populations/diseases genetic effect. Schizophrenia bulletin 45, 824–834 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunkle BW et al. , Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nature genetics 51, 414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kouri N et al. , Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nature communications 6, 7247 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simon-Sanchez J et al. , Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nature genetics 41, 1308 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grove J et al. , Identification of common genetic risk variants for autism spectrum disorder. Nature genetics 51, 431 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez-Roige S et al. , Genome-wide association study meta-analysis of the Alcohol Use Disorders Identification Test (AUDIT) in two population-based cohorts. American Journal of Psychiatry 176, 107–118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyde CL et al. , Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nature genetics 48, 1031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JJ et al. , Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nature Genetics 50, 1112–1121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davies G et al. , Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nature Communications 9, 2098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luciano M et al. , Association analysis in over 329,000 individuals identifies 116 independent variants influencing neuroticism. Nature Genetics 50, 6–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baselmans BM et al. , Multivariate genome-wide analyses of the well-being spectrum. Nature genetics 51, 445–451 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Linnér RK et al. , Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nature Genetics 51, 245–257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu M et al. , Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature genetics 51, 237–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dashti HS et al. , Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nature communications 10, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones SE et al. , Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nature communications 10, 343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yasuno F et al. , Microstructural abnormality in white matter, regulatory T lymphocytes, and depressive symptoms after stroke. Psychogeriatrics 14, 213–221 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Liu Z et al. , Association between white matter impairment and cognitive dysfunction in patients with ischemic Moyamoya disease. BMC neurology 20, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li L et al. , Population-based case–control study of white matter changes on brain imaging in transient ischemic attack and ischemic stroke. Stroke 44, 3063–3070 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Ran S et al. , Atrophic Corpus Callosum Associated with Altered Functional Asymmetry in Major Depressive Disorder. Neuropsychiatric Disease and Treatment 16, 1473–1482 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao J, He Y, McWhinnie CM, Yao S, Altered white matter integrity in individuals with cognitive vulnerability to depression: a tract-based spatial statistics study. Scientific reports 5, 1–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Penke L et al. , Brain white matter tract integrity as a neural foundation for general intelligence. Molecular psychiatry 17, 1026–1030 (2012). [DOI] [PubMed] [Google Scholar]
- 74.Travers BG et al. , Diffusion tensor imaging in autism spectrum disorder: a review. Autism Research 5, 289–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gehricke J-G et al. , The brain anatomy of attention-deficit/hyperactivity disorder in young adults–a magnetic resonance imaging study. PloS one 12, e0175433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li X et al. , Impaired white matter connections of the limbic system networks associated with impaired emotional memory in Alzheimer's disease. Frontiers in aging neuroscience 8, 250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karlsgodt KH et al. , Diffusion tensor imaging of the superior longitudinal fasciculus and working memory in recent-onset schizophrenia. Biological psychiatry 63, 512–518 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Hannawi Y et al. , Hypertension is associated with white matter disruption in apparently healthy middle-aged individuals. American Journal of Neuroradiology 39, 2243–2248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Armstrong NJ et al. , Common genetic variation indicates separate causes for periventricular and deep white matter hyperintensities. Stroke 51, 2111–2121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maraka S et al. , Degree of corticospinal tract damage correlates with motor function after stroke. Annals of clinical and translational neurology 1, 891–899 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiong Y et al. , A diffusion tensor imaging study on white matter abnormalities in patients with type 2 diabetes using tract-based spatial statistics. American Journal of Neuroradiology 37, 1462–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wassenaar TM, Yaffe K, van der Werf YD, Sexton CE, Associations between modifiable risk factors and white matter of the aging brain: insights from diffusion tensor imaging studies. Neurobiology of aging 80, 56–70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Minn Y, Suk S, Do S, Osteoporosis as an independent risk factor for silent brain infarction and white matter changes in men and women: the PRESENT project. Osteoporosis International 25, 2465–2469 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Kashem MA, James G, Harper C, Wilce P, Matsumoto I, Differential protein expression in the corpus callosum (splenium) of human alcoholics: a proteomics study. Neurochemistry international 50, 450–459 (2007). [DOI] [PubMed] [Google Scholar]
- 85.Zhao B et al. , Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nature genetics 51, 1637–1644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hugenschmidt CE et al. , Relating imaging indices of white matter integrity and volume in healthy older adults. Cerebral Cortex 18, 433–442 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Apostolova LG et al. , Hippocampal atrophy and ventricular enlargement in normal aging, mild cognitive impairment and Alzheimer's disease. Alzheimer disease and associated disorders 26, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Damasceno BP, Neuroimaging in normal pressure hydrocephalus. Dementia & neuropsychologia 9, 350–355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teipel SJ et al. , Fractional anisotropy changes in Alzheimer's disease depend on the underlying fiber tract architecture: a multiparametric DTI study using joint independent component analysis. Journal of Alzheimer's Disease 41, 69–83 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Nestor SM et al. , Ventricular enlargement as a possible measure of Alzheimer's disease progression validated using the Alzheimer's disease neuroimaging initiative database. Brain 131, 2443–2454 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koyama T, Marumoto K, Domen K, Ohmura T, Miyake H, Diffusion tensor imaging of idiopathic normal pressure hydrocephalus: a voxel-based fractional anisotropy study. Neurologia medico-chirurgica 52, 68–74 (2012). [DOI] [PubMed] [Google Scholar]
- 92.Atkinson-Clement C, Pinto S, Eusebio A, Coulon O, Diffusion tensor imaging in Parkinson's disease: review and meta-analysis. Neuroimage: Clinical 16, 98–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rolls ET, The cingulate cortex and limbic systems for emotion, action, and memory. Brain Structure and Function 224, 3001–3018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stocco A, Lebiere C, Anderson JR, Conditional routing of information to the cortex: A model of the basal ganglia’s role in cognitive coordination. Psychological review 117, 541 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Leeuw CA, Mooij JM, Heskes T, Posthuma D, MAGMA: generalized gene-set analysis of GWAS data. PLoS Computational Biology 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wen X, Pique-Regi R, Luca F, Integrating molecular QTL data into genome-wide genetic association analysis: Probabilistic assessment of enrichment and colocalization. PLoS genetics 13, e1006646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Consortium G, The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scafidi J et al. , Intranasal epidermal growth factor treatment rescues neonatal brain injury. Nature 506, 230–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheng Y-C et al. , Genome-wide association analysis of young-onset stroke identifies a locus on chromosome 10q25 near HABP2. Stroke 47, 307–316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hofer E et al. , White matter lesion progression: genome-wide search for genetic influences. Stroke 46, 3048–3057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mure H et al. , Akt2 and Akt3 play a pivotal role in malignant gliomas. Neuro-oncology 12, 221–232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alcantara D et al. , Mutations of AKT3 are associated with a wide spectrum of developmental disorders including extreme megalencephaly. Brain 140, 2610–2622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Howell KR, Floyd K, Law AJ, PKBγ/AKT3 loss-of-function causes learning and memory deficits and deregulation of AKT/mTORC2 signaling: Relevance for schizophrenia. PLoS One 12, e0175993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang H et al. , Impaired spatial learning is associated with disrupted integrity of the white matter in Akt3 knockout mice. CNS Neuroscience & Therapeutics 23, 99 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang Q et al. , A Bayesian framework that integrates multi-omics data and gene networks predicts risk genes from schizophrenia GWAS data. Nature neuroscience 22, 691 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Finucane HK et al. , Partitioning heritability by functional annotation using genome-wide association summary statistics. Nature genetics 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kundaje A et al. , Integrative analysis of 111 reference human epigenomes. Nature 518, 317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fullard JF et al. , An atlas of chromatin accessibility in the adult human brain. Genome research 28, 1243–1252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hauberg ME et al. , Common schizophrenia risk variants are enriched in open chromatin regions of human glutamatergic neurons. Nature communications 11, 1–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pers TH et al. , Biological interpretation of genome-wide association studies using predicted gene functions. Nature Communications 6, 5890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kang M, Lee Y-S, The impact of RASopathy-associated mutations on CNS development in mice and humans. Molecular brain 12, 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Qu L et al. , The Ras Superfamily of Small GTPases in Non-neoplastic Cerebral Diseases. Frontiers in Molecular Neuroscience 12, 121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Govek E-E, Newey SE, Van Aelst L, The role of the Rho GTPases in neuronal development. Genes & development 19, 1–49 (2005). [DOI] [PubMed] [Google Scholar]
- 114.Smith SM, Nichols TE, Statistical challenges in “big data” human neuroimaging. Neuron 97, 263–268 (2018). [DOI] [PubMed] [Google Scholar]
- 115.Martin AR et al. , Clinical use of current polygenic risk scores may exacerbate health disparities. Nature genetics 51, 584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu Q et al. , CHIMGEN: a Chinese imaging genetics cohort to enhance cross-ethnic and cross-geographic brain research. Molecular Psychiatry, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gardener H et al. , Race and ethnic disparities in stroke incidence in the Northern Manhattan Study. Stroke 51, 1064–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen C, Zissimopoulos JM, Racial and ethnic differences in trends in dementia prevalence and risk factors in the United States. Alzheimer's & Dementia: Translational Research & Clinical Interventions 4, 510–520 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fieremans E, Jensen JH, Helpern JA, White matter characterization with diffusional kurtosis imaging. Neuroimage 58, 177–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC, NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016 (2012). [DOI] [PubMed] [Google Scholar]
- 121.Jensen JH, Helpern JA, Ramani A, Lu H, Kaczynski K, Diffusional kurtosis imaging: the quantification of non - gaussian water diffusion by means of magnetic resonance imaging. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 53, 1432–1440 (2005). [DOI] [PubMed] [Google Scholar]
- 122.Jelescu IO, Budde MD, Design and validation of diffusion MRI models of white matter. Frontiers in physics 5, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Campbell JS et al. , Promise and pitfalls of g-ratio estimation with MRI. Neuroimage 182, 80–96 (2018). [DOI] [PubMed] [Google Scholar]
- 124.Mohammadi S, Callaghan MF, Towards in vivo g-ratio mapping using MRI: Unifying myelin and diffusion imaging. Journal of Neuroscience Methods, 108990 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vos SB, Jones DK, Viergever MA, Leemans A, Partial volume effect as a hidden covariate in DTI analyses. Neuroimage 55, 1566–1576 (2011). [DOI] [PubMed] [Google Scholar]
- 126.Alfaro-Almagro F et al. , Confound modelling in UK Biobank brain imaging. NeuroImage 224, 117002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sotiropoulos SN et al. , Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage 80, 125–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miller KL et al. , Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nature neuroscience 19, 1523–1536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Alfaro-Almagro F et al. , Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 166, 400–424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Smith SM et al. , Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage 31, 1487–1505 (2006). [DOI] [PubMed] [Google Scholar]
- 131.Acheson A et al. , Reproducibility of tract - based white matter microstructural measures using the ENIGMA - DTI protocol. Brain and behavior 7, e00615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ramsay JO, Silverman BW, Functional data analysis. (Springer; 1997), vol.8. [Google Scholar]
- 133.Liu EY et al. , MaCH - Admix: genotype imputation for admixed populations. Genetic epidemiology 37, 25–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhao B et al. , Heritability of regional brain volumes in large-scale neuroimaging and genetic studies. Cerebral Cortex 29, 2904–2914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Das S et al. , Next-generation genotype imputation service and methods. Nature genetics 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.1000-Genomes-Project-Consortium et al. , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen CY et al. , Improved ancestry inference using weights from external reference panels. Bioinformatics 29, 1399–1406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Duncan LE et al. , Largest GWAS of PTSD (N= 20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Molecular psychiatry 23, 666–673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bycroft C et al. , The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jiang L et al. , A resource-efficient tool for mixed model association analysis of large-scale data. Nature genetics 51, 1749–1755 (2019). [DOI] [PubMed] [Google Scholar]
- 141.Purcell S et al. , PLINK: a tool set for whole-genome association and population-based linkage analyses. The American journal of human genetics 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Willer CJ et al. , METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.International HapMap 3 Consortium, Integrating common and rare genetic variation in diverse human populations. Nature 467, 52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Basser PJ et al. , MR diffusion tensor spectroscopy and imaging. Biophysical journal 66, 259–267 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mori S et al. , Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage 40, 570–582 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.