Abstract
Connections between distinct catalytic RNA motifs through networks of mutations that retain catalytic function (neutral networks) were likely central to the evolution of biocatalysis. Despite suggestions that functional RNAs collectively form an interconnected web of neutral networks, little evidence has emerged to demonstrate the existence of such intersecting networks in naturally occurring RNAs. Here we show that neutral networks of two naturally occurring, seemingly unrelated endonucleolytic ribozymes, the hammerhead (HH) and hairpin (HP), intersect. Sequences at the intersection of these networks exhibit catalytic functions corresponding to both ribozymes by potentially populating both catalytic folds and enable a smooth crossover between the two. Small and structurally simple endonucleolytic motifs like the HH ribozyme could, through mutational walks along their neutral networks, encounter novel catalytic phenotypes, and structurally flexible, bifunctional sequences at the intersection of these networks could have acted as nodes for evolutionary diversification in an RNA world. Considering the simplicity and small size of the HH ribozyme, we propose that this self-cleaving motif could have been a precursor to other more complex endonucleolytic ribozymes. More generally, our results suggest that RNAs that possess distinct sequences, structures, and catalytic functions, can potentially share evolutionary history through mutational connections in sequence space.
Keywords: ribozymes, RNA evolution, RNA world, neutral networks
INTRODUCTION
Early life is thought to have evolved via an RNA world, a hypothetical scenario where RNA served both as enzymes and as information carriers. Ribozymes still catalyze essential chemical processes in modern biology, and artificial ribozymes, relevant to metabolism in an RNA-based biology, have been identified by in vitro selection (Hirao and Ellington 1995; Chen et al. 2007; Martin et al. 2015). A major challenge for the RNA world hypothesis pertains to the emergence of complex ribozymes, as the assembly of ribonucleotides into oligoribonucleotides is slow, low yielding and thus unlikely to generate the diversity of catalytic RNAs required to sustain an RNA-based biology. However, small catalytic RNA motifs that potentially can be accessed by nonenzymatic assembly processes could serve as precursors to a diversity of more complex ribozymes via evolutionary walks along mutational pathways.
Acquisition of novel folds from preexisting functional folds has occurred during the evolution of protein enzymes and can occur in functional RNAs (Manrubia and Briones 2007; Petrov et al. 2015). For example, random mutagenesis of in vitro selected aptamers and ribozymes can yield new RNA folds from preexisting folds (Held et al. 2003; Huang and Szostak 2003; Curtis and Bartel 2005), and in some cases, simpler folds can produce new folds with increased structural complexity (Carothers et al. 2004). The metabolic sophistication ascribed to life in the RNA world (Benner and Ellington 1987; Benner et al. 1989) underscores the importance of identifying plausible evolutionary paths that could lead to the emergence of new, more complex catalytic folds from simple precursors. Mutational pathways connecting precursors to new ribozyme folds would be evolutionarily viable if they consisted of sequences that retain catalytic function, that is, if they are part of the neutral networks of both RNAs.
The first experimental demonstration for the existence of such networks in catalytic RNA was provided by Schultes and Bartel (2000), when they showed that the hepatitis delta virus (HDV) and an in vitro selected ligase ribozyme possess neutral networks that approach one another closely. The sequence at the intersection of the two networks could presumably adopt both ligase and HDV folds, although it catalyzed the respective reactions very slowly (rate of 10−5/min for cleavage and 10−8/min for ligation). Recent work using a combination of in vitro selection and high-throughput sequencing has identified numerous bifunctional sequences that exhibit both ligase and HDV functions supporting the “Schultes and Bartel” hypothesis that many ribozyme neutral networks closely approach one another (Bendixsen et al. 2019). Bifunctional sequences maximize the use of available nucleic acid sequence space and could have driven evolution in an RNA world. Kuhns and Joyce (2003) engineered complementary RNA and DNA sequences, each with the capacity to adopt a distinct structure and catalyze a particular chemical transformation. The ability of the heteroduplex to tolerate mismatches provides a pathway for one strand to retain function while the other can explore alternative folds. Nevertheless, in the nearly two decades following these initial suggestions, no subsequent reports describing networks connecting distinct ribozyme classes have emerged.
The HH ribozyme stands as the smallest endonucleolytic RNA motif in biology. This catalytic motif has emerged from in vitro selection experiments (Tang and Breaker 2000; Salehi-Ashtiani and Szostak 2001), including those that mimic prebiotic environments (Popovic et al. 2015) and in computational folding grids containing a pool of random RNA sequences (Knight et al. 2005). Moreover, HH self-cleaving motifs can be accessed in part through uncatalyzed template-directed polymerization of activated ribonucleotides (Prywes et al. 2016) or in its entirety through ribozyme-catalyzed transcription (Wochner et al. 2011; Tjhung et al. 2020). These observations suggest that HH-like RNA cleaving motifs could have been abundant in an RNA world. Variants of HH ribozymes that include circular permutations, mutations, deletions, and additions to the canonical HH sequence occur in all domains of life (Jimenez et al. 2015; de la Pena et al. 2017), underscoring the potential of a HH ribozyme sequence to embark on divergent trajectories via multiple sequence configurations. Therefore, the HH self-cleaving motif presents an attractive candidate for a primordial precursor to self-cleaving RNAs, and its intersection with other functional motifs in sequence space would provide a framework to explain the increase in complexity and diversity of endonucleolytic function. In this work, we demonstrate the existence of intersecting neutral networks between the HH ribozyme and the catalytic domain of the hairpin (HP) ribozyme comprising the conserved loop B, which is flanked by base-paired helix B (Supplemental Fig. S1) and contains residues important for function (Buzayan et al. 1986; Haseloff and Gerlach 1988). We identify a bifunctional sequence at the intersection of their neutral networks that performs HH cleavage in addition to cleaving a HP substrate, and outline neutral, stepwise mutational pathways that connect exclusive HH and HP ribozyme functions.
RESULTS
A single catalytic RNA sequence exhibits HH and HP functions simultaneously
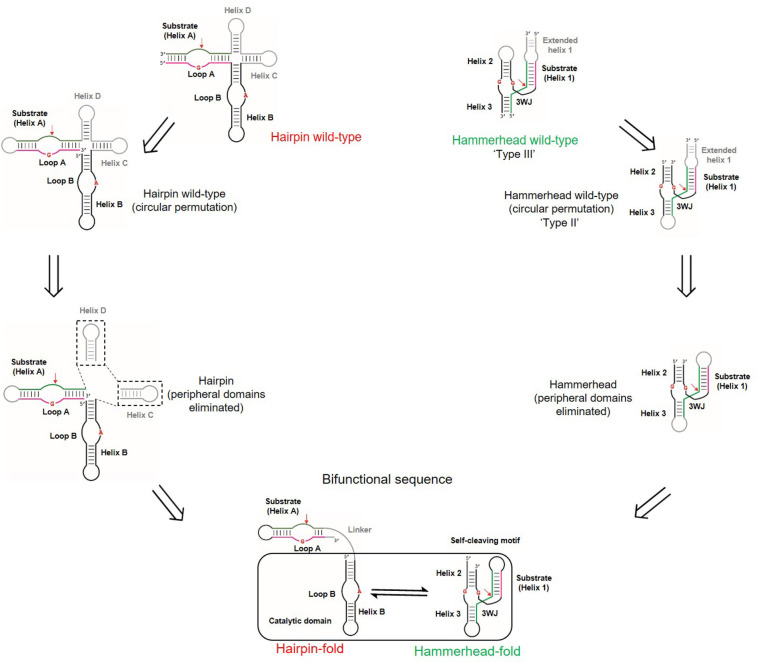
Intersecting neutral networks provide evolutionary mechanisms for functional diversification. As the HH catalytic motif does not contain a distinct separation of substrate and catalytic domains, unlike the larger HP ribozyme, we sought to identify a single sequence (boxed region in Fig. 1) that could exhibit HH self-cleavage and catalyze cleavage of a HP substrate when connected to it by a linker sequence. This rational design approach began with circular permutations to the wild-type HH (Supplemental Fig. S1A) and HP (Supplemental Fig. S1D) ribozymes so that the sequences had similar strand polarity (5′ to 3′). This operation was followed by the elimination of domains somewhat peripheral to their cleavage activities (Fig. 1). Extended helix 1 in the wild-type HH ribozyme was deleted and helix 1 was closed by a tetraloop. Similarly, relatively nonessential sequences from the four-way junction (helices C and D) of the HP ribozyme were deleted to yield HH and HP ribozyme constructs of comparable sizes (Fig. 1; Fedor 2000). Notably, these operations are plausible from an evolutionary perspective. Circularly permuted forms of ribozymes occur naturally and exhibit virtually indistinguishable catalytic functions (Perreault et al. 2011; Roth et al. 2014). Circular permutation of the “wild-type” HH ribozyme in this work converts a type III HH ribozyme to a type II HH ribozyme (Fig. 1; Supplemental Fig. S1B; Jimenez et al. 2011). Justification for the removal of peripheral domains comes from analyses of the primary, secondary, and tertiary structures of ribosomal RNA from bacteria, archea, and eukarya, which reveal evolutionary expansion through the addition of helical branches (junctions) to ancestral RNA stems while maintaining the structural integrity of the ancestral core (Petrov et al. 2015).
FIGURE 1.
Acquisition of a bifunctional intersection sequence from wild-type hammerhead (HH) and hairpin (HP) ribozymes. Circular permutations of wild-type ribozymes orient them so that they have similar strand polarities. Minimal catalytic cores corresponding to both ribozymes were obtained by deleting nonessential helices from wild-type ribozymes (helices C and D in HP and extended helix 1 in HP; shown in gray). These minimal catalytic sequences guided the design of the intersection sequence (Fig. 2A). Substrates and its complementary strands are shown in green and pink, respectively. Cleavage sites are indicated by red arrows and catalytic nucleotides are shown in red. The 17 nt linker connecting the substrate and catalytic domains of the HP ribozyme is shown in gray.
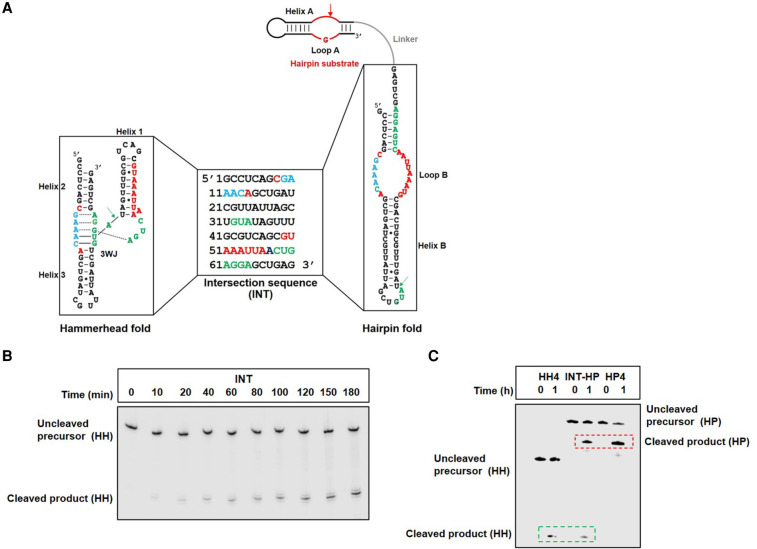
These simpler catalytic motifs resemble minimal forms of the HP ribozyme catalytic domain and the HH motif, which are widely used in the functional studies of these RNAs (Butcher et al. 1995; Komatsu et al. 1995, 1996; Blount and Uhlenbeck 2005). Guided by the consensus sequences of both HP (Fedor 2000) and HH ribozymes (Supplemental Fig. S1C,E; McCall et al. 2000), we designed sequences by hand that could be threaded through both the secondary structures of the HH ribozyme and the catalytic domain of the HP ribozyme and preserved the conserved structural features in each (Supplemental Fig. S1C,E). We screened these sequences computationally by calculating the relative stabilities of their HH and HP structures. Sequences predicted to misfold or predominantly adopt only one of the two ribozyme structures were discarded, and those with comparable predicted stabilities for both ribozyme structures were synthesized by in vitro transcription and tested for in vitro cleavage. While HH activity of these sequences was tested by assaying for self-cleavage, HP activity was tested by assaying for cleavage of a HP substrate physically linked to these sequences (Fig. 1). Most sequences that were expected to adopt HH or HP folds from folding algorithms exhibited barely detectable cleavage activities and were therefore discarded. Only a few sequences exhibited HH or HP ribozyme function and were optimized for dual function by introducing mutations. This workflow (Supplemental Fig. S2) generated an “intersection sequence,” INT (Fig. 2A), that underwent site-specific HH self-cleavage and cleaved a HP substrate in cis, when connected by a 17 nt linker (referred to as INT-HP), consistent with the capacity to adopt both HH and HP ribozyme folds (Fig. 2).
FIGURE 2.
Bifunctional intersection sequence, INT, that can be threaded through secondary structures of both the HH ribozyme and the catalytic domain of the HP ribozyme. (A) INT potentially adopts the secondary structures of both HH ribozyme and the catalytic domain of the HP ribozyme. Residues in INT that correspond to the essential nucleotides in the HH and HP ribozymes are colored in green and red, respectively, and residues important for both HH and HP functions are colored in blue. The HH and HP cleavage sites are denoted by green and red arrows, respectively. The HP substrate (shown in red) is connected to INT by a 17 nt RNA linker (shown in gray). (B) INT self-cleavage corresponding to HH activity. (C) The fusion of INT with the HP substrate (INT-HP) results in two cleavage products generated from HH and HP cleavage. HH4 and HP4 represent sequences that stabilize the HH and HP motifs, respectively, and are used here as positive controls for HH and HP cleavage (see Fig. 3; Supplemental Fig. S5).
Intersection sequence, INT, contains all the nucleotides critical for HH self-cleavage (Fig. 2A, in green and blue), and contains the important nucleotides of the HP catalytic domain (Fig. 2A, in red and blue). The HP catalytic domain is functional when fused to the HP substrate, which harbors conserved residues within an internal loop, referred to as loop A. The HH fold of INT harbors nucleotides essential for HH cleavage within a conserved three-way junction (3WJ); however, these residues are part of base-paired regions (helix B), an internal loop (loop B), and a terminal loop in its alternate fold corresponding to the HP catalytic domain. Similarly, essential HP nucleotides residing within loop B of the HP catalytic domain (HP fold of INT) constitute a major portion of helix 1 with two residues falling within helices 2 and 3 in the HH fold. A stretch of five nucleotides in INT (GAAAC, Fig. 2A in blue), important for both HH and HP function, is located within the conserved 3WJ of the HH fold and loop B of the HP fold. INT underwent HH self-cleavage and cleaved a HP substrate with rate acceleration of about three orders of magnitude in each case (Supplemental Fig. S3) (the rate of uncatalyzed RNA cleavage under similar conditions was measured as ∼0.00001 min−1). Although INT exhibits significant rate enhancements, INT-catalyzed cleavage rates were lower than wild-type HH or HP ribozymes (by about 100-fold), which could be a combined effect of conformational plasticity (which reduces the fraction of either fold) and the deletion of wild-type ribozyme domains. Nevertheless, these results support the hypothesis that INT partially populates the catalytically active fold of the HH ribozyme within the sequence context of a cis-cleaving HP ribozyme (Fig. 2) and underscore the robustness of the HH functional fold.
Mutational pathways connect the HH and HP ribozymes
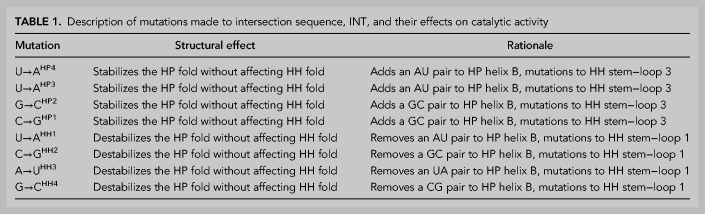
Intersection sequences with dual catalytic function can act as nodes from which two distinct catalytic motifs can be accessed via “neutral” mutational drifts (Lau and Ferre-D'Amare 2016). Mutations to INT designed to disrupt or stabilize the HP fold relative to the HH fold had a corresponding effect on the relative cleavage activity (Tables 1, 2; Fig. 3; Supplemental Figs. S4, S5). A set of four cumulative point mutations (Table 1; Fig. 3A) to residues corresponding to a loop region (terminal loop 3) in the HH fold of INT and expected to preferentially stabilize the HP fold of INT by introducing additional base pairs in its helix B, preserved HP activity while eliminating HH cleavage (Table 2; Fig. 3C; Supplemental Fig. S5). Conversely, a set of four cumulative point mutations (Table 1; Fig. 3B) to residues corresponding to a different loop (terminal loop 1) in the HH fold but expected to disrupt base pairs within the HP helix B (immediately under internal loop B), preserved HH cleavage while abolishing HP cleavage (Table 2; Fig. 3C; Supplemental Fig. S5). Among these four base pairs, disruption of the single A-U base pair distal from the conserved loop B of the HP fold (HH1A) still leaves a four base-paired helix flanking loop B and thus, has minimal effect on HH or HP cleavage. However, further mutations to base pairs proximal to loop B destabilize the HP stem and likely allow the HH fold to predominate. Accordingly, a slight reduction and eventual elimination of HP activity accompanies an increase in HH activity (Table 2; Fig. 3C; Supplemental Fig. S5).
TABLE 1.
Description of mutations made to intersection sequence, INT, and their effects on catalytic activity
TABLE 2.
Cleavage rate constants (min−1) of RNA sequences tested in this work
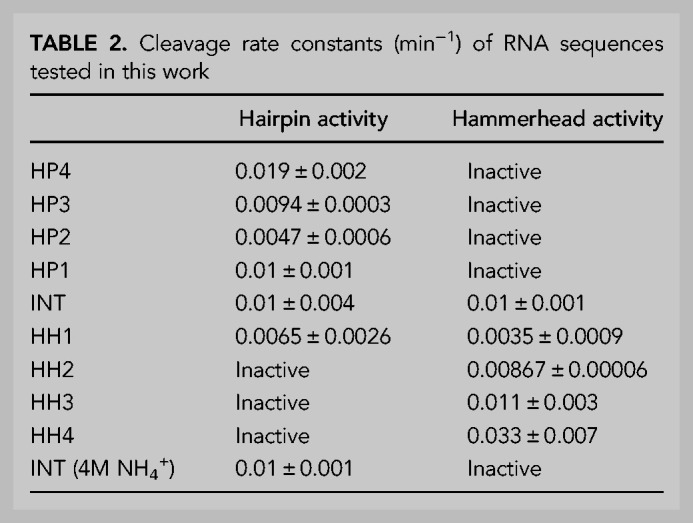
FIGURE 3.
Single point mutations connect HH and HP ribozyme functions. (A) Point mutations to the intersection sequence, INT is expected to stabilize the HP structure. This operation, which introduces new base pairs in helix B of its HP fold, corresponds to mutations in loop 3 of the HH structure, hence does not affect HH stability. This results in preferential stabilization of the HP fold. Mutations are indicated by arrows; for example, U→HP4A indicates a U to A mutation that produces the HP catalytic domain sequence, HP4 (see Table 1 for details). Catalytic residues (G8 and G12 in wild-type nomenclature) in the HH ribozyme are shown in green and catalytic residues (G8 and A38 in wild-type nomenclature) in the HP ribozyme are shown in red. (B) Point mutations to the intersection sequence, INT, expected to preferentially destabilize the HP structure. This operation, which disrupts base pairs in helix B (underneath loop B), correspond to mutations in loop 1 of the HH structure, hence do not affect HH stability. This results in preferential stabilization of the HH fold. (C) Smooth access to exclusive HH or HP ribozyme function is facilitated by intersecting neutral networks corresponding to both functions. Sequences, INT and HH1 retain dual function (dotted box); however, point mutations in either direction generate either HH or HP phenotype.
To illustrate that intersection sequence, INT, retains the catalytic mechanism of the wild-type HH ribozyme, we mutated either catalytic guanosine residue (“G8” or “G12” in wild-type nomenclature) to adenosine (Fig. 3A,B). Indicative of HH-mediated self-cleavage, mutations to either residue abolished activity (Supplemental Fig. S6A). In HH4, a sequence with exclusive HH function, these mutations also resulted in a loss of cleavage activity, suggesting a mechanistic continuity between the bifunctional INT and HH4 (Supplemental Fig. S6B). Similarly, mutations to residues that correspond to catalytic nucleotides in the HP fold (“G8” and “A38” in wild-type nomenclature) of INT-HP (Fig. 3A,B), eliminated cleavage of the HP substrate, and this result was recapitulated in HP4-HP, a sequence with exclusive HP function (Supplemental Fig. S6C,D). These results provide further evidence that INT harbors, within a single sequence, two distinct catalytic apparatuses.
Evolutionary innovation can occur in response to changing environmental conditions. In the context of bifunctional ribozymes, certain reaction conditions can favor one ribozyme function over the other. While both HH and HP ribozymes use Mg2+ to catalyze self-cleavage in vitro, replacing Mg2+ with 4 M NH4+ reduces wild-type HH ribozyme cleavage by ∼20- to 100-fold but enhances wild-type HP ribozyme cleavage by ∼40-fold (Murray et al. 1998). Consistent with this observation, INT lost its HH cleavage activity in the presence of 4 M NH4 (in the absence of Mg2+) but retained its ability to cleave a HP substrate (Supplemental Fig. S7), illustrating an additional path to evolutionary divergence.
DISCUSSION
Constructing neutral pathways connecting the HH and HP ribozymes presents an operational framework to examine the evolution of ribozymes from simpler precursors. With the active site of the HH ribozyme contained within a three-way junction that includes 13 conserved nucleotides, the robustness of the peripheral helix sequences can presumably allow exploration of mutational space. Accumulation of sufficient mutational perturbations to the original sequence can lead to a new functional fold, resulting in the sudden emergence of a new phenotype, while retaining the original phenotype (Fontana and Schuster 1998). We have demonstrated that sequence variants of the HH self-cleaving motif can adopt a new functional fold corresponding to the catalytic domain of the HP ribozyme. This functional plasticity endows the neutral networks of distinct catalytic RNAs with points of intersection that enable acquisition of new catalytic phenotypes through mutations or even changing environmental conditions. These evolutionary pathways could have been especially important in the evolution of viroid RNAs. For example, the HH and HP catalytic motifs occur on complementary strands of a satellite RNA in the tobacco ringspot virus (STobRV RNA) and both genomic and anti-genomic strands of the hepatitis delta virus harbor HDV ribozymes (Riccitelli and Luptak 2013). Recent studies have discovered type III HH ribozyme motifs within HDV-like circRNA genomes from metazoans like toads and termites (de la Pena et al. 2021). Since the evolutionary origins of ribozymes are lost to history, biochemical reconstruction of their neutral networks might help ascertain the fluidity with which these ribozyme motifs can be accessed from each other.
The importance of peripheral sequences for ribozyme folding has been widely recognized. Tertiary interactions mediated by peripheral regions in the HH and HP ribozymes facilitate the formation of their active folds, which exhibit the maximum catalytic power (Walter et al. 1998; Fedor 1999; De la Pena et al. 2003; Khvorova et al. 2003); therefore, eliminating these domains to generate minimal ribozymes result in compromised catalytic efficiencies as illustrated by the INT-derived sequences discussed here. Nevertheless, the present work illustrates how sacrificing some catalytic prowess through changes to their peripheral sequences may allow functional RNAs to escape existing functional folds and acquire novel structures and functions. Peripheral elements provide the freedom to explore vast expanses of sequence space without eliminating existing activity, consequently facilitating phenotypic “leaps” across intersecting neutral networks. The results presented here provide the first demonstration for the existence of intersecting neutral networks between naturally occurring functional RNAs. A simple HH-like self-cleaving motif could have evolved novel capabilities through adaptive walks across fitness landscapes corresponding to multiple catalytic phenotypes and could have served as precursors to structurally complex endonucleolytic ribozymes via common evolutionary processes such as insertions, deletions, base-pair covariation, and domain accretion (Fox et al. 1977; Bokov and Steinberg 2009; Petrov et al. 2015). Smaller, prefolded sequences like the HH could have been the catalytic core around which new ribozymes evolved. Although our results do not necessarily recapitulate evolutionary history, the apparent ease with which RNA sequences can access functional genotypes in sequence space, as illustrated here, further supports the plausibility of primordial RNA-based biocatalysis in the context of an RNA world.
MATERIALS AND METHODS
RNA synthesis, purification, and radiolabeling
DNA templates used to generate RNA constructs were prepared by PCR amplification of DNA Ultramers (for sequences between 100 and 200 nt) and custom oligos (for sequences under 100 nt), both purchased from Integrated DNA Technologies (IDT). RNA was prepared by in vitro transcription for 2 h at 37°C in buffer containing 40 mM Tris-HCl pH 7.9, 2 mM spermidine, 10 mM NaCl, 25 mM MgCl2, 10mM DTT, 30 U/mL RNase Inhibitor (NEB), 2.5 U/mL TIPPase (NEB), 4 mM of each NTPs, 30 pmol/mL DNA template, and 40 µg/mL T7 RNA polymerase (made in-house). Transcription reactions were quenched by the addition of 10 U/mL DNase I (Promega) and incubation at 37°C for 30 min. Transcribed RNAs were extracted by phenol-chloroform-isoamyl alcohol (pH 4.3) and purified by denaturing PAGE.
RNA obtained from transcription was subjected to phosphatase treatment (∼1 U shrimp alkaline phosphatase/50 pmol RNA to be labeled) to remove 5′ triphosphates. RNA sequences were 5′ 32P labeled in the presence of 25–50 pmol γ32-ATP (Perkin-Elmer) for 30 min at 37°C and PAGE purified. Purified, radiolabeled RNA sequences were ethanol precipitated in the presence of glycogen (Thermo-Fisher) to assist the formation of a visible pellet. Pellets were suspended in ddH2O prior to cleavage assays.
Ribozyme cleavage assays
Radiolabeled RNA in ddH2O was heated at 70°C for 1 min in the presence of 25 mM KCl, followed by incubation on ice for 3 min and at room temperature for 5 min. Tris-HCl (pH 8) and spermidine were added to final concentrations of 50 mM and 2 mM, respectively, and incubated at 37°C for 15 min. A total of 200 mM MgCl2 (or 4 M NH4Cl in Supplemental Fig. S7) was added to initiate cleavage and the reaction was incubated at 37°C. Aliquots at different time points were withdrawn from the reaction and quenched with stop dye containing excess EDTA and 90% (v/v) formamide, 0.05% (w/v) xylene cyanol, and 0.05% (w/v) bromophenol blue, and frozen on dry ice. Aliquots were loaded on to a 20% dPAGE gel for separating precursor and product bands. The gels were exposed to PhosphorImager screens (Amersham Biosciences) and scanned on a Typhoon Trio imager (GE Healthcare) in “Storage phosphor” mode. Bands were quantified in ImageQuant TL with an automated rolling ball algorithm for background subtraction. Fraction cleaved was calculated as the ratio of the intensities of the product and precursor bands. Fraction cleaved was plotted against reaction time points and data was fitted to a first-order exponential in GraphPad PRISM. Composite data were plotted in GraphPad PRISM.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Sandip Shelke for expression and purification of T7 RNA polymerase. We appreciate the insightful comments of Dr. Bryan Dickinson and Dr. Phoebe Rice on the overall project and Huw Rees and Michael Disare for careful review of the manuscript. This work was supported by grants from the National Institutes of Health (R01AI081987) to J.A.P.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078813.121.
REFERENCES
- Bendixsen DP, Collet J, Ostman B, Hayden EJ. 2019. Genotype network intersections promote evolutionary innovation. PLoS Biol 17: e3000300. 10.1371/journal.pbio.3000300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner SA, Ellington AD. 1987. The last ribo-organism. Nature 329: 295–296. 10.1038/329295a0 [DOI] [PubMed] [Google Scholar]
- Benner SA, Ellington AD, Tauer A. 1989. Modern metabolism as a palimpsest of the RNA world. Proc Natl Acad Sci 86: 7054–7058. 10.1073/pnas.86.18.7054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Uhlenbeck OC. 2005. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct 34: 415–440. 10.1146/annurev.biophys.34.122004.184428 [DOI] [PubMed] [Google Scholar]
- Bokov K, Steinberg SV. 2009. A hierarchical model for evolution of 23S ribosomal RNA. Nature 457: 977–980. 10.1038/nature07749 [DOI] [PubMed] [Google Scholar]
- Butcher SE, Heckman JE, Burke JM. 1995. Reconstitution of hairpin ribozyme activity following separation of functional domains. J Biol Chem 270: 29648–29651. 10.1074/jbc.270.50.29648 [DOI] [PubMed] [Google Scholar]
- Buzayan JM, Hampel A, Bruening G. 1986. Nucleotide sequence and newly formed phosphodiester bond of spontaneously ligated satellite tobacco ringspot virus RNA. Nucleic Acids Res 14: 9729–9743. 10.1093/nar/14.24.9729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers JM, Oestreich SC, Davis JH, Szostak JW. 2004. Informational complexity and functional activity of RNA structures. J Am Chem Soc 126: 5130–5137. 10.1021/ja031504a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li N, Ellington AD. 2007. Ribozyme catalysis of metabolism in the RNA world. Chem Biodivers 4: 633–655. 10.1002/cbdv.200790055 [DOI] [PubMed] [Google Scholar]
- Curtis EA, Bartel DP. 2005. New catalytic structures from an existing ribozyme. Nat Struct Mol Biol 12: 994–1000. 10.1038/nsmb1003 [DOI] [PubMed] [Google Scholar]
- de la Pena M, Gago S, Flores R. 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J 22: 5561–5570. 10.1093/emboj/cdg530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pena M, Garcia-Robles I, Cervera A. 2017. The hammerhead ribozyme: a long history for a short RNA. Molecules 22: 78. 10.3390/molecules22010078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pena M, Ceprian R, Casey JL, Cervera A. 2021. Hepatitis delta virus-like circular RNAs from diverse metazoans encode conserved hammerhead ribozymes. Virus Evol 7: veab016. 10.1093/ve/veab016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor MJ. 1999. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry 38: 11040–11050. 10.1021/bi991069q [DOI] [PubMed] [Google Scholar]
- Fedor MJ. 2000. Structure and function of the hairpin ribozyme. J Mol Biol 297: 269–291. 10.1006/jmbi.2000.3560 [DOI] [PubMed] [Google Scholar]
- Fontana W, Schuster P. 1998. Continuity in evolution: on the nature of transitions. Science 280: 1451–1455. 10.1126/science.280.5368.1451 [DOI] [PubMed] [Google Scholar]
- Fox GE, Magrum LJ, Balch WE, Wolfe RS, Woese CR. 1977. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc Natl Acad Sci 74: 4537–4541. 10.1073/pnas.74.10.4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Gerlach WL. 1988. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature 334: 585–591. 10.1038/334585a0 [DOI] [PubMed] [Google Scholar]
- Held DM, Greathouse ST, Agrawal A, Burke DH. 2003. Evolutionary landscapes for the acquisition of new ligand recognition by RNA aptamers. J Mol Evol 57: 299–308. 10.1007/s00239-003-2481-y [DOI] [PubMed] [Google Scholar]
- Hirao I, Ellington AD. 1995. Re-creating the RNA world. Curr Biol 5: 1017–1022. 10.1016/S0960-9822(95)00205-3 [DOI] [PubMed] [Google Scholar]
- Huang Z, Szostak JW. 2003. Evolution of aptamers with a new specificity and new secondary structures from an ATP aptamer. RNA 9: 1456–1463. 10.1261/rna.5990203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez RM, Delwart E, Luptak A. 2011. Structure-based search reveals hammerhead ribozymes in the human microbiome. J Biol Chem 286: 7737–7743. 10.1074/jbc.C110.209288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez RM, Polanco JA, Luptak A. 2015. Chemistry and biology of self-cleaving ribozymes. Trends Biochem Sci 40: 648–661. 10.1016/j.tibs.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD. 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 10: 708–712. 10.1038/nsb959 [DOI] [PubMed] [Google Scholar]
- Knight R, De Sterck H, Markel R, Smit S, Oshmyansky A, Yarus M. 2005. Abundance of correctly folded RNA motifs in sequence space, calculated on computational grids. Nucleic Acids Res 33: 5924–5935. 10.1093/nar/gki886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu Y, Kanzaki I, Koizumi M, Ohtsuka E. 1995. Modification of primary structures of hairpin ribozymes for probing active conformations. J Mol Biol 252: 296–304. 10.1006/jmbi.1995.0497 [DOI] [PubMed] [Google Scholar]
- Komatsu Y, Kanzaki I, Ohtsuka E. 1996. Enhanced folding of hairpin ribozymes with replaced domains. Biochemistry 35: 9815–9820. 10.1021/bi960627n [DOI] [PubMed] [Google Scholar]
- Kuhns ST, Joyce GF. 2003. Perfectly complementary nucleic acid enzymes. J Mol Evol 56: 711–717. 10.1007/s00239-002-2445-7 [DOI] [PubMed] [Google Scholar]
- Lau MW, Ferre-D'Amare AR. 2016. Many activities, one structure: functional plasticity of ribozyme folds. Molecules 21: 1570. 10.3390/molecules21111570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrubia SC, Briones C. 2007. Modular evolution and increase of functional complexity in replicating RNA molecules. RNA 13: 97–107. 10.1261/rna.203006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LL, Unrau PJ, Muller UF. 2015. RNA synthesis by in vitro selected ribozymes for recreating an RNA world. Life (Basel) 5: 247–268. 10.3390/life5010247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall MJ, Hendry P, Mir AA, Conaty J, Brown G, Lockett TJ. 2000. Small, efficient hammerhead ribozymes. Mol Biotechnol 14: 5–17. 10.1385/MB:14:1:5 [DOI] [PubMed] [Google Scholar]
- Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. 1998. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol 5: 587–595. 10.1016/S1074-5521(98)90116-8 [DOI] [PubMed] [Google Scholar]
- Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Ferbeyre G, Breaker RR. 2011. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol 7: e1002031. 10.1371/journal.pcbi.1002031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, et al. 2015. History of the ribosome and the origin of translation. Proc Natl Acad Sci 112: 15396–15401. 10.1073/pnas.1509761112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M, Fliss PS, Ditzler MA. 2015. In vitro evolution of distinct self-cleaving ribozymes in diverse environments. Nucleic Acids Res 43: 7070–7082. 10.1093/nar/gkv648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prywes N, Blain JC, Del Frate F, Szostak JW. 2016. Nonenzymatic copying of RNA templates containing all four letters is catalyzed by activated oligonucleotides. Elife 5: e17756. 10.7554/eLife.17756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccitelli N, Luptak A. 2013. HDV family of self-cleaving ribozymes. Prog Mol Biol Transl Sci 120: 123–171. 10.1016/B978-0-12-381286-5.00004-4 [DOI] [PubMed] [Google Scholar]
- Roth A, Weinberg Z, Chen AG, Kim PB, Ames TD, Breaker RR. 2014. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol 10: 56–60. 10.1038/nchembio.1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Szostak JW. 2001. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414: 82–84. 10.1038/35102081 [DOI] [PubMed] [Google Scholar]
- Schultes EA, Bartel DP. 2000. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science 289: 448–452. 10.1126/science.289.5478.448 [DOI] [PubMed] [Google Scholar]
- Tang J, Breaker RR. 2000. Structural diversity of self-cleaving ribozymes. Proc Natl Acad Sci 97: 5784–5789. 10.1073/pnas.97.11.5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjhung KF, Shokhirev MN, Horning DP, Joyce GF. 2020. An RNA polymerase ribozyme that synthesizes its own ancestor. Proc Natl Acad Sci 117: 2906–2913. 10.1073/pnas.1914282117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter F, Murchie AI, Lilley DM. 1998. Folding of the four-way RNA junction of the hairpin ribozyme. Biochemistry 37: 17629–17636. 10.1021/bi9821115 [DOI] [PubMed] [Google Scholar]
- Wochner A, Attwater J, Coulson A, Holliger P. 2011. Ribozyme-catalyzed transcription of an active ribozyme. Science 332: 209–212. 10.1126/science.1200752 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.