Abstract
SBI-0206965, originally identified as an inhibitor of the autophagy initiator kinase ULK1, has recently been reported as a more potent and selective AMP-activated protein kinase (AMPK) inhibitor relative to the widely used, but promiscuous inhibitor Compound C/Dorsomorphin. Here, we studied the effects of SBI-0206965 on AMPK signalling and metabolic readouts in multiple cell types, including hepatocytes, skeletal muscle cells and adipocytes. We observed SBI-0206965 dose dependently attenuated AMPK activator (991)-stimulated ACC phosphorylation and inhibition of lipogenesis in hepatocytes. SBI-0206965 (≥25 μM) modestly inhibited AMPK signalling in C2C12 myotubes, but also inhibited insulin signalling, insulin-mediated/AMPK-independent glucose uptake, and AICA-riboside uptake. We performed an extended screen of SBI-0206965 against a panel of 140 human protein kinases in vitro, which showed SBI-0206965 inhibits several kinases, including members of AMPK-related kinases (NUAK1, MARK3/4), equally or more potently than AMPK or ULK1. This screen, together with molecular modelling, revealed that most SBI-0206965-sensitive kinases contain a large gatekeeper residue with a preference for methionine at this position. We observed that mutation of the gatekeeper methionine to a smaller side chain amino acid (threonine) rendered AMPK and ULK1 resistant to SBI-0206965 inhibition. These results demonstrate that although SBI-0206965 has utility for delineating AMPK or ULK1 signalling and cellular functions, the compound potently inhibits several other kinases and critical cellular functions such as glucose and nucleoside uptake. Our study demonstrates a role for the gatekeeper residue as a determinant of the inhibitor sensitivity and inhibitor-resistant mutant forms could be exploited as potential controls to probe specific cellular effects of SBI-0206965.
Keywords: AMPK, AMPK-related kinase, glucose uptake, lipogenesis, SBI-0206965, ULK1
Introduction
AMP-activated protein kinase (AMPK) is an important regulator of energy balance [1,2]. AMPK has long held promise as a therapeutic target for metabolic syndrome, as its activation in multiple metabolic tissues resulted in amelioration of insulin resistance and reduction in hyperglycemia in preclinical studies [3–5]. For example, active AMPK inhibits fatty acid biosynthesis in the liver through phosphorylation and inactivation of acetyl-CoA carboxylase-1 (ACC1) [6,7]. The activation of AMPK also leads to increased fatty acid oxidation through phosphorylation of ACC2 [8], and glucose uptake in skeletal muscle, at least in part through phosphorylation of TBC1D1 [9,10], a Rab GTPase-activating protein (GAP) which modulates trafficking of GLUT4-containing vesicles [11]. AMPK exists as hetero-complexes of three subunits; a catalytic α and two regulatory β and γ subunits. Each exists as multiple isoforms (α1/α2, β1/β2, γ1/γ2/γ3), generating up to 12 possible combinations [2]. AMPK is activated through phosphorylation of threonine 172 (Thr172) within the activation segment of the α subunit kinase domain [12]. The major upstream kinases phosphorylating Thr172 are liver kinase B (LKB1) and Ca2+-calmodulin-dependent protein kinase kinase-2 (CaMKK2) [2]. The γ-subunits contain four tandem cystathionine β-synthase (CBS) motifs that bind adenine nucleotides. The binding of ADP and/or AMP to CBS motifs causes conformational changes that increase net Thr172 phosphorylation [13–15]. In addition, the binding of AMP, but not ADP, further increases AMPK activity through allosteric stimulation [13]. AMPK activity is, therefore, regulated in a multi-layered process, often involving a combination of posttranslational modifications and allosteric regulation [16].
To delineate cellular and physiological roles for AMPK, small-molecule activators have played important roles. The most widely used small-molecule activator of AMPK is 5-aminoimidazole-4-carboxamide riboside (AICAR), a prodrug of AMP-mimetic ZMP, which was used for probing the AMPK-dependent (and insulin-independent) stimulation of glucose uptake in skeletal muscle [17–19]. However, the specificity of AICAR has been challenged and numerous AMPK-independent actions of AICAR have been reported [20–23]. Specific allosteric activators and new mechanisms for their targeting of AMPK have recently been discovered. The crystallographic structures of AMPK trimeric complexes revealed that A-769662 (a member of the thienopyridone family [24]) and 991 (a benzimidazole derivative, also known as ex229) bind in an allosteric drug and metabolite (ADaM) pocket located at the interface of the α subunit (kinase domain N-lobe) and β subunit (carbohydrate-binding module) [25,26]. We and others have shown that 991, and its two related analogues with improved bioavailability (MK-8722, PF-739), are potent and highly specific AMPK activators [4,5,21,27]. Collectively, these activators and their analogues are exploited for drug development (e.g. for metabolic disorders) and have also been used extensively as research tools to delineate AMPK's molecular and physiological roles in cellular and organismal contexts [3].
In contrast, the availability of small-molecule AMPK inhibitors as research tools is vastly limited. The most widely utilised AMPK inhibitor, the pyrazolopyrimidine derivative Compound C (also known as Dorsomorphin), was originally discovered in a high-throughput screen and used to probe AMPK-dependent metabolic actions of metformin in hepatocytes [28]. Compound C is an ATP-competitive inhibitor and binds to the highly conserved active site of the AMPKα catalytic domain [29]. However, Compound C/Dorsomorphin inhibits several other kinases in vitro with similar or much greater potency than AMPK [30–32] and elicits numerous AMPK-independent biological effects [33].
SBI-0206965, a 2-aminopyrimidine derivative, was originally identified through screening a library of pyrimidine analogues as an ATP-competitive inhibitor of the autophagy initiator kinase ULK1, with the ability to inhibit ULK signalling and ULK1-mediated survival of lung cancer and glioblastoma cells when coupled with nutrient stress [34]. Interestingly, using cell-free phosphorylation of the AMPK and the ULK1 synthetic peptide substrate S108tide as a measure of kinase activity [35], it was recently reported that SBI-0206965 (used at 1 μM) was a more effective inhibitor of AMPK (80% inhibition) compared with ULK1 (63% inhibition) [32]. In addition, SBI-0206965 has demonstrated much higher potency and selectivity toward AMPK when directly compared with Compound C in cell-free assays, even though several other kinases (including JAK3, Src, FAK, Abl, Aurora A/B) were also selectively inhibited [32,34,36,37]. Mechanistically, a co-crystal structure of the AMPKα2 kinase domain-SBI-0206965 complex showed that the drug occupies a pocket that partially overlaps with the ATP-binding site; however, SBI-0206965 displayed mixed-competitive kinetics [32]. SBI-0206965 inhibits glucose deprivation-, AICAR-, A769662- or ionomycin-induced cellular AMPK activation, judged by phosphorylation of AMPK substrates, at doses ranging from 5 to 30 μM in multiple cell lines including HEK293, SH-SY5Y, mouse embryonic fibroblasts, or HUVECs [32,38,39] with no associated metabolic readouts of AMPK assessed.
In the current study, we initially sought to evaluate the effects of SBI-0206965 on AMPK signalling and metabolic readouts in multiple cell types, including hepatocytes, skeletal muscle cells, and adipocytes. We observed SBI-0206965 dose dependently attenuated AMPK-dependent stimulation of ACC phosphorylation and inhibition of lipogenesis with the direct AMPK activator 991 in mouse primary hepatocytes. SBI-0206965 (≥25 μM) only modestly inhibited AMPK signalling in C2C12 myotubes and adipocytes, but potently inhibited basal and AMPK-independent (or insulin-stimulated) glucose transport and nucleoside uptake. We next performed an extended (activity-based) screen of SBI-0206965 using a panel of 140 human protein kinases in vitro to test our hypothesis that SBI-0206965 would inhibit AMPK-related kinases, given the structural similarities of the kinase domains of AMPK and AMPK-related kinases [40,41]. This screen, together with molecular modelling and structure-guided mutagenesis, has identified that SBI-0206965 inhibits several kinases with equal or higher potency, including the AMPK-related kinases NUAK1, MARK3 and MARK4.
Materials and methods
Materials
5-Aminoimidazole-4-carboxamide ribonucleotide (AICAR, #OR1170T) was obtained from Apollo Scientific. 991 (#AOB8150) was purchased from Aobious. Insulin (#I9287) and SBI-0206965 (#SML1540) were obtained from MerckMilliporeSigma. A769662 (#3336) and S-(4-Nitrobenzyl)-6-thioinosine (NBMPR, #2924) were purchased from Tocris Bioscience. Deoxy-d-[1, 2-3H]-glucose, [5, 6-3H]-uridine, [1-14C]-Acetic acid and [U-14C]-d-glucose were purchased from PerkinElmer. Rotenone (#R8875) was obtained from MerckMilliporeSigma. Cell culture reagents were purchased from Thermo Fisher Scientific. All other materials, unless otherwise indicated, were from MerckMilliporeSigma.
Antibodies
AS160 (#07-741), Flag (#F7425), α-tubulin (#T6074), and phospho-TBC1D1 Ser237 (#07-2268) antibodies were purchased from MerckMilliporeSigma. ACC (#3676), phospho-ACC1 Ser79 (#3661), Akt (#4691), phospho-Akt Ser473 (#4060), phospho-Akt Thr308 (#9275) AMPKα (#2532), phospho-AMPKα Thr172 (#2535), phospho-AS160 Thr649 (#8881), ERK1/2 (#4695), phospho-ERK1/2 Thr202/Tyr204 (#4370), GSK3β (#9315), phospho-GSK3β Ser9 (#9322), HSP90 (#4874), p70S6K1 (#2708), phospho-p70S6K1 Thr389 (#9234), Raptor (#2280), phospho-Raptor Ser792 (#2083), TBC1D1 (#4629), ULK1 (#8054), phospho-ULK1 Ser555 (#5869), and vinculin (#13901) antibodies were purchased from Cell Signaling Technology.
Experimental animals
Animal experiments were performed in accordance with the European directive 2010/63/EU of the European Parliament and of the Council of the protection of animals used for scientific purposes. Ethical approval was given by the Danish Animal Experiments Inspectorate (#2015-15-0201-00796) or by the Regional Ethical Committee on Animal Experiments in Malmö/Lund (#5.8.18-18569/2018). Animal studies were conducted on C57BL/6NTac male mice (obtained from Taconic Biosciences) at 9–12 weeks of age in the animal facility at the Faculty of Health and Medical Sciences (University of Copenhagen) or 36- to 38-day-old Sprague-Dawley rats (obtained from Taconic Biosciences) in the animal facility at the Biomedical Centre (Lund University, Sweden). Animals were maintained on a standard chow diet and 12 : 12-h light–dark cycle.
Mouse primary hepatocytes isolation, culture, and lipogenesis assay
Mice were anesthetised via intraperitoneal injection of avertin, prepared from a stock of 1 g/ml tribromoethanol (#T48402, MerckMilliporeSigma) in 2-methyl-2-butanol (#152463, MerckMilliporeSigma), diluted 1 : 20 in saline, and dosed as 10 μl/g body weight. Hepatocytes were isolated by collagenase perfusion as previously described [42,43]. Hepatocytes were seeded in medium Eagle-199 (MEM-199) (#41150, Thermo Fisher Scientific) containing 100 U/ml penicillin G, 100 μg/ml streptomycin and 10% (vol/vol) FBS. Hepatocytes were left for attachment (3–4 h) and cultured overnight at 37°C with 5% CO2 in MEM-199 supplemented with 100 U/ml penicillin G, 100 μg/ml streptomycin, 10 nM insulin, and 100 nM dexamethasone. Twenty-four hours after seeding, primary hepatocytes were incubated for 1 h for signalling studies as described [42] or 3 h with MEM-199 supplemented with 0.6 μCi/ml [14C]-acetate in the presence of SBI-0206965 with 991 or vehicle (0.1% DMSO). Cells were harvested in 0.5 ml PBS, transferred into 1 ml 40% KOH and 2 ml methanol followed by 1 h incubation at 80°C. Lipids were saponified by acidifying the samples in 37% HCl and extracted with petroleum ether. Extracts were allowed to evaporate to dryness, then dissolved in Ultima Gold scintillation fluid for determination of [14C]-acetate incorporation into lipids.
C2C12 skeletal muscle cell culture, treatment, and glucose uptake measurement
C2C12 (#CRL-1772, ATCC) myoblasts were cultured and differentiated into myotubes as described [44]. Fully differentiated C2C12 myotubes (day 4–5) were serum starved in DMEM for 4 h followed by 1 h incubation with bicarbonate-free media (DMEM, 20 mM HEPES, 0.2% BSA, pH 7.4). Cells were then pre-incubated in Krebs-Ringer HEPES (KRH) buffer (140 mM NaCl, 4.7 mM KCl, 2.5 mM CaCl2, 1.25 mM MgSO4, 10 mM HEPES, 1.2 mM KH2PO4, and 0.2% BSA) with vehicle (0.1% DMSO) or SBI-0206965 for 30 min followed by an additional 1 h incubation with 991 (10 μM) or insulin (100 nM). Cells were incubated in KRH buffer containing 50 μM [3H]2-deoxyglucose (0.1 μCi/ml) for 5 min and the reaction was stopped by adding 25 mM glucose. The cells were washed in KRH buffer and harvested in ice-cold lysis buffer. Two-third of the lysate was used for immunoblot analysis and the remaining one-third was analysed by scintillation counting to measure 2-deoxyglucose uptake in the cells.
Ex vivo mouse skeletal muscle incubation
Mice were anesthetised by avertin (as described above) via intraperitoneal injection, and extensor digitorum longus (EDL) muscle was rapidly dissected and mounted in oxygenated (95% O2 and 5% CO2) and warmed (30°C) Krebs-Ringer buffer in a myograph system (820MS, Danish Myo Technology) as described [27,45]. The muscles were incubated in the presence of the indicated compounds or vehicle (0.1% DMSO) for 50 min before snap-frozen in liquid nitrogen.
Rat primary adipocyte isolation, culture, treatment, and glucose uptake assay
Adipocytes were isolated by collagenase digestion as described [46]. After isolation, primary adipocytes were stimulated directly (1 ml of 8–10% (v/v) cells per stimulation) as indicated in the figure legends. After stimulation, cells were washed in KRH buffer without BSA and lysed in lysis buffer. For glucose uptake, freshly isolated rat adipocytes were washed in glucose-free buffer containing 30 mM HEPES pH 7.4, 120 mM NaCl, 4 mM KH2OPO4, 1 mM MgSO4, 0.75 mM CaCl2, 10 mM NaHCO3, 200 nM adenosine and 1% (w/v) BSA (incubation buffer). Adipocyte suspensions (400 µl of 5% (v/v) cells) were pre-incubated with SBI-0206965 or 0.1% DMSO for 30 min before being stimulated with 10 nM insulin or 10 µM cytochalasin B for 30 min. Subsequently, 100 µl incubation buffer containing 0.25 µl (0.025 µCi) [14C]-glucose (275 mCi/mmol glucose; final glucose concentration = 0.18 µM) was added and cells were incubated for a further 30 min. Reactions were stopped by centrifuging 300 µl of each reaction in a Beckman microtube containing 75 µl dinonylphtalate. The adipocyte fraction was collected and subjected to liquid scintillation counting.
3T3-L1 adipocyte culture and glucose uptake
3T3-L1 fibroblasts were originally obtained from ATCC (#CL-173). The cells were cultured and differentiated into adipocytes as described previously [47] and used between passages 7 and 12. Experiments were performed at days 8–12 post differentiation. 2-deoxyglucose uptake assay and preparation of cell lysates were as described previously [46].
Cell and tissue extract preparation
Treated cells were washed once with PBS and lysed in ice-cold lysis buffer (containing 50 mM Tris–HCl, pH 7.5, 1 mM EGTA, 1 mM EDTA, 270 mM sucrose, 1% Triton X-100, 20 mM glycerol-2-phosphate, 50 mM NaF, 5 mM Na4P2O7, 1 mM DTT, 0.1 mM PMSF, 1 mM benzamidine, 1 μM microcystin-LR, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, and 1 mM Na3VO4). For mouse skeletal muscle tissue samples, ∼10 mg tissue was homogenised in 400 μl ice-cold lysis buffer using a Tissuelyser II (Qiagen) at 30 Hz for 30 s (x 2 sets). Cell or tissue lysates were clarified by centrifugation at 6000×g for 10 min at 4°C, and total protein concentration was determined using Bradford reagent (#23200, ThermoFisher) and BSA as standard.
Immunoblot analysis
Protein lysates were denatured in Laemmli buffer at 95°C for 5 min. Twenty micrograms of protein was separated by SDS–PAGE on home-made Tris-glycine or pre-cast Novex 4–12% Bis-Tris gels (ThermoFisher) and transferred onto nitrocellulose or Immobilon-FL polyvinylidene difluoride membranes. Membranes were blocked for 45 min in TBST [Tris-buffered saline (20 mM Tris–HCl, pH 7.6, 137 mM NaCl) with 0.1% Tween 20] containing 5% skimmed milk. Blocked membranes were incubated overnight at 4°C in primary antibody diluted in TBST containing 4% BSA. Protein detection was performed with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (ECL) substrate or LI-COR IRdye-conjugated secondary antibodies and a LI-COR Odyssey infrared imaging system (LI-COR Biosciences). ECL signals were visualised using a ChemiDoc XRS+ system followed by analysis of band intensities with the software Image Lab 6.0 (both from Bio-Rad). LI-COR signals were analysed and quantified using ImageStudioLite software (LI-COR Biosciences).
Protein expression constructs
AMPKα1 constructs (used in Figure 8) were made by the cloning team at Medical Research Council Protein Phosphorylation and Ubiquitylation Unit (MRC PPU) Reagents and Services (https://mrcppureagents.dundee.ac.uk/). The coding region of AMPKα1 (NM_006251.5) was amplified using oligonucleotides adding BglII and NotI restriction sites at the 5′ and 3′ end, respectively, and subcloned as a BglII/NotI insert into pcDNA5D-FRT/TO FLAG vector (DU41457) digested with BamHI and NotI to generate pcDNA5D-FRT/TO FLAG AMPKα1 (DU67861). The M104T mutant version of this plasmid was made according to the QuikChange method (Stratagene) using KOD polymerase (Novagen) to generate pcDNA5D-FRT/TO FLAG AMPKα1 M104T. Plasmid expression constructs for AMPK Flag-α1, β1-Myc, and HA-γ1 (used in Figure 7) were previously described [32]. Mutagenesis of Flag-α1 was performed as above using the QuikChange method. Note that the AMPKα1 construct used in Figure 7 is a variant of NM_006251.5 that starts at Met10 (the alternate initiation site), thus the gatekeeper methionine shown as M104 corresponds to M93. Plasmid constructs for full-length, N-terminal Flag-tagged human ULK1 (NM_003565.4) and the M92T mutant were generated by custom gene synthesis (Gene Universal) and cloned into pcDNA3.1(−) vector using XhoI/HindIII restriction sites. All constructs were sequence verified.
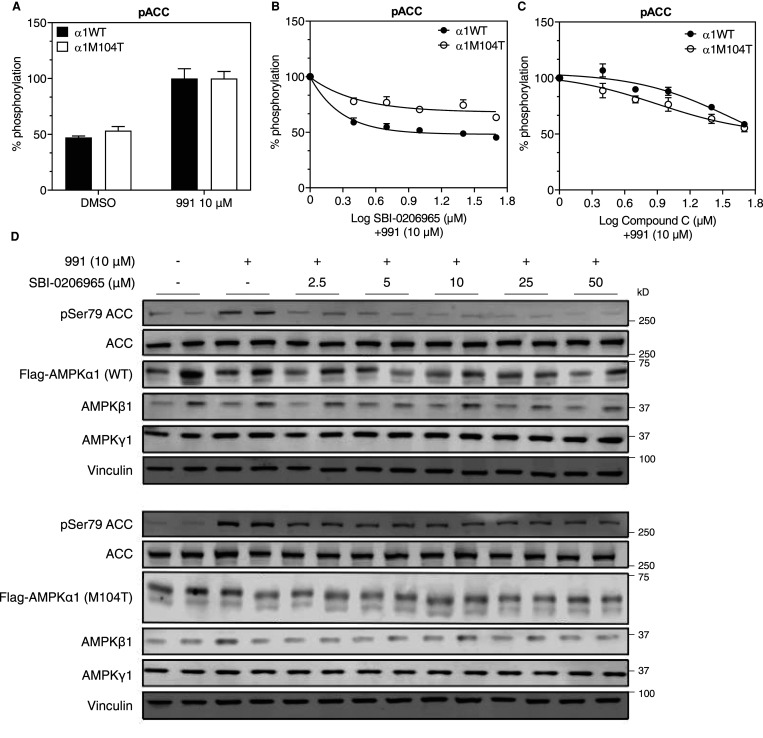
Figure 8. Mutation of gatekeeper methionine to threonine renders AMPK insensitive to SBI-0206965 in cells.
(A–D) cDNA constructs encoding Flag-AMPKα1 wild type (WT) or M104T mutant were transfected into AMPKα1/α2 double knockout (DKO) U2OS cells in 96-well plate. Forty-eight hours post transfection, cells were treated with indicated compounds for 1 h prior to cell lysis. Cell lysates were subjected to homogeneous time-resolved fluorescence (HTRF) assay, as described in the Materials and methods section, or immunoblot analysis. (A) Phosphorylation of ACC with vehicle (0.1% DMSO) or 991 (10 μM) using HTRF assay. Data are shown as % of 991-stimulated (100%) state. (B,C) % ACC phosphorylation (HTRF assay) relative to respective 991-stimulated value (100%) in response to increasing doses of SBI-0206965 or Compound C. (D) Immunoblot analysis of lysates from Flag-AMPKα1 WT (upper panel) or M104T mutant (lower panel) expressing cells using the indicated antibodies. Results are from three independent experiments (n = 6–9 per treatment) and are expressed as mean ± SEM.
Figure 7. Mutation of gatekeeper methionine to threonine renders AMPK and ULK1 insensitive to SBI-0206965 in cell-free assay.
(A–J) Immunopurified recombinant Flag-AMPKα1 (wild type (WT), M104T) complex or Flag-ULK1 (WT, M92T mutant) were subjected to immunoblot analysis with the indicated antibodies (A,B) or in vitro kinase assay without (C,D) or with the indicated compounds (E–J). Results are expressed as mean ± SEM.
Protein production and kinase assays
Heterotrimeric human AMPK Flag-α1β1γ1 and human Flag-ULK1 were produced in mammalian cells as described [32,35]. For AMPK expression, the cells were triply transfected at 60% confluency using FuGene HD (Roche Applied Science) and 1 μg of pcDNA3 plasmid expression constructs for AMPK Flag-α1, β1-Myc, and HA-γ1, whereas for ULK1 expression the cells were transfected with 2 μg of pcDNA3.1(−) Flag-ULK1 plasmid. After 48 h, the transfected cells were harvested by rinsing with ice-cold PBS, followed by rapid lysis using 500 μl of lysis buffer.
AMPK and ULK1 activity were determined by phosphorylation of synthetic peptide substrates SAMS (HMRSAMSGLHLVKRR) and S108tide (KLPLTRSHNNFVARRR), respectively [32,35]. Briefly, recombinant AMPK or ULK1 was immunoprecipitated from 10 μg of transfected cultured cell lysate using 10 μl of anti-FLAG M2 agarose beads (50% (v/v)) (MerckMilliporeSigma), washed and then added to a 25 μl reaction containing assay buffer (50 mM HEPES-NaOH, pH 7.4, 1 mM DTT, and 0.02% (v/v) Brij-35), 200 μM synthetic peptide substrate (SAMS or S108tide), 200 μM [γ-32P]-ATP (PerkinElmer), 5 mM MgCl2, in the presence of Compound C (0–400 μM) or SBI-0206965 (0–400 μM). Reactions were performed at 30°C and terminated after 10 min by spotting 15 μl onto P81 phosphocellulose paper (Whatman). Radioactivity was quantified by liquid scintillation counting.
Homogeneous time-resolved fluorescence (HTRF) assay using AMPKα1 and α2 double KO (DKO) U2OS cells
U2OS FRT Flp-In T-REx cell line was a kind gift from John Rouse (University of Dundee) and dual deletion of PRKAA1 and PRKAA2 (that encode AMPKα1 and α2, respectively) was performed by Horizon Discovery using CRISPR–CAS9 technology. HTRF was employed for a higher throughput application, as well as more sensitive and quantitative detection of ACC phosphorylation (using the phospho-ACC 1/2 (Ser79) kit, # 64ACCPET, Cisbio). Briefly, cells were seeded in a 96-well plate at a density of 5 × 104 cells per well. The next day, cells were treated with increasing concentration of the indicated compounds diluted in a serum-free medium. Post-treatment, cells were lysed in 50 μl per well of phospho-total protein lysis buffer 1 (#64KL1FDF, Cisbio) containing 1 : 100 blocking reagent (#64KB1ACC, Cisbio) with continuous vibration at room temperature for 30 min. ACC (Ser79) D2- and Cryptate-antibodies were diluted (1 : 40) in the detection buffer. In a 384-well plate, 16 μl lysate and 4 μl antibody solution were added per well. The plate was incubated at room temperature with light protection. The next day, FRET (665 nm/620 nm) was measured using Spectramax microplate reader (Molecular Devices) according to the manufacturer instruction.
Nucleotide and ZMP measurements
Nucleotides and ZMP were measured by liquid chromatography-mass spectrometry from perchlorate extracts of mouse primary hepatocytes, C2C12 myotubes, and mouse EDL muscle tissues as described previously [32,45]. Adenylate energy charge was calculated as:
Uridine uptake
Uridine uptake assay was performed as described [48]. Briefly, C2C12 myotubes were washed once in uridine uptake buffer (20 mM Tris–HCl, 3 mM KH2PO4, 1 mM MgCl2, 2 mM CaCl2, 5 mM glucose, 130 mM NaCl, pH 7.4) before incubating at room temperature in 400 μl uridine uptake buffer containing vehicle (0.1% DMSO), NBMPR, or SBI-0206965. Thirty minutes following incubation, the buffer was aspirated and the new 400 μl uridine uptake buffer containing the compounds (DMSO, NBMPR, or SBI-0206965) and 0.1 μM (2 μCi/ml) [3H]-uridine. After 1 min, uridine transport was terminated by washing the cells five times in ice-cold uridine uptake buffer containing cold 1 mM uridine. The cells were lysed in 200 μl 10% (w/v) SDS before scraping. The amount of uridine transported into the cells was determined by scintillation counting.
Adenosine kinase assay
Of human recombinant adenosine kinase (BioVision, #8024-AK), 0.5 μg was pre-incubated with non-nucleoside adenosine kinase inhibitor ABT-702 (Tocris, #2372) or vehicle (DMSO) for 30 min followed by a further incubation with or without increasing doses (0.1–10 μM) of SBI-0206965 for 60 min. Adenosine kinase activity was measured using the Universal Kinase Activity kit (R&D System, EA004) using adenosine (MerckMilliporeSigma, #A9251) as substrate according to the manufacturer protocol.
Protein kinase screen and IC50 determination
All protein kinases in the kinase panel were expressed, purified and assayed at the International Centre for Protein Kinase Profiling (http:// www.kinase-screen.mrc.ac.uk/), MRCPPU, University of Dundee, as previously described [30]. Briefly, all assays were carried out at room temperature. Assays were performed for 30 min (except Lck and PBK that were incubated for 15 min) using Multidrop Micro reagent dispensers (Thermo Electron Corporation) in a 96-well format. The concentration of magnesium acetate in the assays was 10 mM and [γ-33P]ATP (∼800 cpm/pmol) was used at either 5, 20, or 50 μM in order to be at or below the Km for ATP for each enzyme (described under ‘kinase panel’ in the webpage http://www.kinase-screen.mrc.ac.uk/services/premier-screen). The half-maximal inhibitory concentration (IC50) values of the selected kinases against SBI-0206965 were performed by the International Centre for Kinase Profiling team and determined after carrying out assays at ten different concentrations of the compound.
Molecular docking
Molecular docking was performed using the SwissDock server [49] available at http://www.swissdock.ch/ (last accessed on 20 August 2020). The kinase domain structures of AMPKα2 (PDBid: 6BX6), ULK2 (PDBid: 6YID), MARK3 (PDBid: 2QNJ), MARK4 (PDBid: 5ES1) were prepared for docking using the Chimera plugin DOCKPREP after removal of water and ligand molecules [50,51]. The ligand SBI-0206965 (ZINC database entry: 253387916) was docked to each kinase structure using the default settings for ‘accurate’ docking, allowing flexibility for side chains within 3 Å of any atom of the ligand in its reference binding mode.
Statistical analysis
Results are indicated as means ± standard error mean (SEM). Two-way analysis of variance (ANOVA) with Tukey's post hoc analysis was used to determine differences between multiple treatment groups. Significance was set at P < 0.05.
Results
SBI-0206965 dose dependently attenuates 991-induced phosphorylation of ACC and inhibition of lipogenesis in primary hepatocytes
SBI-0206965 was reported to inhibit cellular AMPK activation, assessed by phosphorylation of ACC, at doses ranging from 5 to 30 μM in multiple cell lines including HEK293, SH-SY5Y, or HUVECs [32,38]. We observed that SBI-0206965 attenuates basal or 991-stimulated phosphorylation of AMPK substrates (ACC and Raptor) in a dose-dependent manner in primary mouse hepatocytes (Figure 1A,B). Phosphorylation of AMPKα T172 was modestly elevated with 991, and was further increased with increasing doses of SBI-0206965 (Figure 1A,B). Since a previous study noted that cellular incubations at high concentrations of SBI-0206965 may induce AMPKα T172 phosphorylation through fluctuations in AMP/ATP and ADP/ATP ratios [32], we measured the effect of SBI-0206965 on adenylate energy charge. We indeed observed that SBI-0206965 promoted phosphorylation of AMPKα T172 at higher doses (Supplementary Figure S1A), however, there was no reduction in adenylate energy charge (Supplementary Figure S1B).
Figure 1. SBI-0206965 dose dependently attenuates 991-induced phosphorylation of ACC and inhibition of lipogenesis in mouse primary hepatocytes.
(A,B) Mouse primary hepatocytes were pre-incubated for 30 min with vehicle (0.1% DMSO) or the indicated concentrations of SBI-0206965 followed by an additional incubation for 1 h with 991 (10 μM) or vehicle. Cell lysates were subjected to immunoblot analysis using the indicated antibodies. Tubulin was used as a loading control. Representative images (A) and quantification of the indicated blots (B) from three independent experiments are shown (n = 6 per treatment). (C) Mouse hepatocytes were treated with the indicated concentrations of SBI-0206965 in the absence (vehicle) or presence of 991 (10 μM) and labelled with [14C]-acetate for 3 h. Rates of fatty acid synthesis were estimated from incorporation [14C]-acetate into saponifiable lipids. (D) Results from (B) and (C) (except vehicle and SBI-0206965 alone data) are plotted to visualise the inverse relationship between ACC phosphorylation and lipogenesis in response to increasing doses of SBI-0206965 in combination with 991 (10 μM). Results are expressed as percentage relative to the vehicle and represent the mean ± SEM for three independent experiments (n = 6–12 per treatment). *P < 0.05 (vehicle vs. indicated compound(s) treated). N.S.; not significant.
Since activation of AMPK leads to inhibition of lipogenesis through phosphorylation of ACC1 in hepatocytes [6,7,42], we tested if SBI-0206965 treatment could prevent 991-induced suppression of lipogenesis. As previously reported [7,27], 991 potently inhibited lipogenesis in hepatocytes (Figure 1C). Co-treatment with SBI-0206965 attenuated 991-induced inhibition of lipogenesis in a dose-dependent manner. When co-treated with a 25 μM dose of SBI-0206965, the inhibitory effect of 991 was fully restored to the level comparable to the vehicle-treated state (Figure 1C). As shown in Figure 1D, we observed an inverse relationship between ACC phosphorylation and lipogenesis in response to increasing doses of SBI-0206965 with 991 (10 μM). These results demonstrate that SBI-0206965 is a useful tool compound to probe AMPK-dependent regulation of lipogenesis by small-molecule activators in primary hepatocytes.
SBI-0206965 potently inhibits glucose uptake in C2C12 myotubes and adipocytes
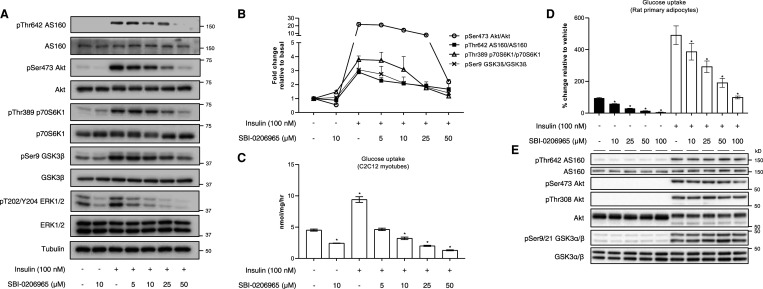
We next assessed the effects of SBI-0206965 on AMPK signalling and glucose uptake in C2C12 myotubes. SBI-0206965 (10 μM) alone modestly increased phosphorylation of AMPKα with no apparent effect on phosphorylation of AMPK substrates (ACC, Raptor, TBC1D1, ULK1) (Figure 2A,B and Supplementary Figure S1A). As anticipated, 991 (10 μM) enhanced phosphorylation of AMPK substrates to varying degrees. Treatment of C2C12 myotubes with increasing doses of SBI-0206965 in combination with 991 resulted in a dose-dependent decrease in Raptor phosphorylation, however, phosphorylation of ACC and ULK1 was only modestly decreased at doses up to 50 μM (Figure 2A,B). Notably, phosphorylation of TBC1D1, known to be involved in AMPK-mediated glucose uptake in skeletal muscle [9–11], was unchanged with SBI-0206965 (Figure 2A,B). In contrast, phosphorylation of extracellular signal-regulated kinase (ERK), used as control unrelated to AMPK signalling, was markedly inhibited with SBI-0206965 at 5 μM (Figure 2A). SBI-0206965 (10 μM) suppressed basal glucose uptake by more than 50%, whereas 991 stimulated glucose uptake by ∼2-fold and this was dose dependently reduced with SBI-0206965 (Figure 2C). Even though 5 μM SBI-0206965 had no apparent inhibitory effect on phosphorylation of AMPK substrates (Figure 2A,B), it was sufficient to fully inhibit 991-stimulated glucose uptake in C2C12 myotubes (Figure 2C). Contrary to the inhibitory effect of SBI-0206965 on phosphorylation of AMPK substrates, 991-stimulated phosphorylation of AMPKα T172 was further elevated with increasing doses of SBI-0206965 (Figure 2A). Consistent with the observation in hepatocytes (Supplementary Figure S1A) SBI-0206965 dose dependently promoted phosphorylation of AMPKα and there was no change in adenylate energy charge (Supplementary Figure 1C,D). Collectively, SBI-0206965 is a potent inhibitor for basal and 991-stimulated glucose uptake, although it promotes phosphorylation of AMPKα T172 and has only a modest inhibitory effect on phosphorylation of AMPK substrates in C2C12 myotubes.
Figure 2. SBI-0206965 inhibits unstimulated and 991-stimulated glucose uptake with no apparent inhibition of TBC1D1 Ser231 phosphorylation in C2C12 myotubes.
(A,B) C2C12 myotubes were pre-incubated for 30 min with vehicle (0.1% DMSO) or the indicated concentrations of SBI-0206965 followed by an additional 1 h incubation with 991 (10 μM) or vehicle. Cell lysates were subjected to immunoblot analysis using the indicated antibodies. Tubulin was used as a loading control. Representative images (A) and quantification of the indicated blots (B) from three independent experiments are shown (n = 3 per treatment). (C) C2C12 myotubes were incubated with vehicle or the indicated compounds for 1 h followed by addition of [3H]-2-deoxy-glucose and measurement of glucose uptake for 5 min (n = 8–9 per treatment). Results are expressed as mean ± SEM. *P < 0.05 (vehicle vs. indicated compound(s) treated).
Since we observed that SBI-0206965 potently inhibits basal and 991-stimulated glucose uptake with no detectable inhibition of TBC1D1 phosphorylation, we tested if SBI-0206965 suppresses insulin-stimulated (i.e. AMPK-independent) glucose uptake in C2C12 myotubes. SBI-0206965 doses ≥25 μM consistently inhibited insulin-stimulated phosphorylation of Akt and its downstream targets including AS160 (also known as TBC1D4), which plays a key role in regulating GLUT4 trafficking [11,52,53] (Figure 3A,B). We observed that basal and insulin-stimulated glucose uptake were robustly inhibited with SBI-0206965 (Figure 3C), which is consistent with a previous study using L6 myotubes and isolated mouse skeletal muscle ex vivo [54]. Collectively, these results demonstrate that SBI-0206965 potently inhibits insulin-stimulated glucose uptake and modestly inhibits insulin signalling intermediaries in C2C12 myotubes.
Figure 3. SBI-0206965 inhibits insulin-stimulated glucose uptake in C2C12 myotubes and rat primary adipocytes.
(A,B) C2C12 myotubes were pre-incubated for 30 min with vehicle (0.1% DMSO) or the indicated concentrations of SBI-0206965 followed by an additional incubation without (vehicle) or with insulin (100 nM) for 1 h. Cell lysates were subjected to immunoblot analysis using the indicated antibodies. Tubulin was used as a loading control. Representative images (A) and quantification of the indicated blots (B) from three independent experiments are shown (n = 3 per treatment). (C) C2C12 myotubes were incubated with vehicle or the indicated compounds for 1 h followed by addition of [3H]-2-deoxy-glucose and measurement of glucose uptake for 5 min (n = 6 per treatment). (D,E) Rat primary adipocytes were pre-incubated with vehicle (0.1% DMSO) or the indicated concentrations of SBI-0206965 for 30 min before being stimulated with insulin (10 nM) for 30 min. The adipocytes were either lysed for immunoblot analysis (E, n = 4 per treatment) or incubated further 30 min with an addition of [14C]-glucose for determination of glucose uptake (D, n = 4 per treatment). Results are expressed as mean ± SEM. *P < 0.05 (vehicle vs. indicated compound(s) treated).
To further examine if SBI-0206965 ubiquitously inhibits the glucose transport system in different cell types, we also treated rat primary adipocytes with SBI-0206965 in the presence or absence of insulin. Consistent with our observation in C2C12 myotubes, both basal and insulin-stimulated glucose uptake were dose dependently reduced in primary adipocytes (Figure 3D). However, contrary to the inhibitory effect on insulin signalling intermediaries in C2C12 myotubes (Figure 3A,B), SBI-0206965 had no effect on phosphorylation of Akt and its substrates (AS160 and GSK3α/β) in primary adipocytes (Figure 3E). We also assessed the effect of SBI-0206965 on 991-induced AMPK activation in primary adipocytes and observed that phosphorylation of Raptor, but not ACC, was only modestly reduced at 50 or 100 μM (Supplementary Figure S2A,B). We also used 3T3 L1 mouse adipocytes and observed that SBI-0206965 (30 μM) profoundly inhibited both basal and insulin-stimulated glucose uptake (Supplementary Figure S2C). Taken together, SBI-0206965 inhibits both stimulated and unstimulated glucose transport in multiple cell types.
SBI-0206965 inhibits the nucleoside transport system
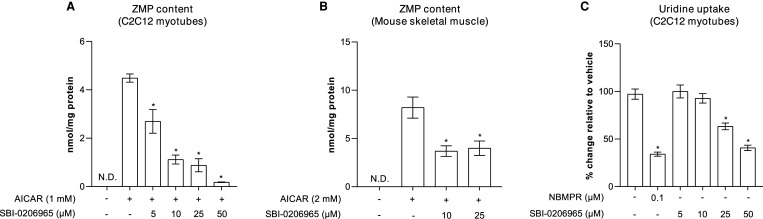
AICAR is the most widely used compound to pharmacologically activate AMPK and probe AMPK's cellular and physiological functions, although its specificity has recently been challenged [20–22]. Upon uptake into cells, AICAR is converted to the monophosphate derivative, ZMP, which can accumulate to high levels, and mimics the effects of AMP on the AMPK pathway. Consistent with previous findings using HEK293 cells [32], we observed that SBI-0206965 potently inhibits AICAR uptake (measured as cellular ZMP accumulation) in both C2C12 myotubes and isolated mouse skeletal muscle (EDL) tissue ex vivo (Figure 4A,B). To determine if the reduced accumulation of ZMP was due to inhibition of the transport or conversion of AICAR to ZMP through phosphorylation by adenosine kinase, we measured the effect of SBI-0206965 on the activity of human recombinant adenosine kinase in vitro. We observed that while a non-nucleoside adenosine kinase inhibitor (ABT-702) potently inhibited adenosine kinase activity at 0.01 μM, SBI-0206965 had no effect on adenosine kinase up to 10 μM (Supplementary Figure S3). In line with these observations, SBI-0206965 also inhibited the uptake of uridine (ribonucleoside), although the potency was much more modest compared with NBMPR, a known inhibitor of nucleoside transporters [55] (Figure 4C). These results, together with a previous report [32], establish that SBI-0206965 inhibits the nucleoside transport system when used at ≥5–10 μM in multiple cell types and skeletal muscle tissue.
Figure 4. SBI-0206965 inhibits AICAR and uridine uptake in C2C12 myotubes.
(A,B) C2C12 myotubes or isolated mouse skeletal muscle (extensor digitorum longus) were pre-incubated for 30 min with vehicle (0.1% DMSO) or the indicated concentrations of SBI-0206965 followed by an additional 1-h incubation without (vehicle) or with AICAR (1 mM). Cell or tissue extracts were subjected to ZMP analysis by liquid chromatography-mass spectrometry. (C) C2C12 myotubes were incubated in the absence (vehicle) or presence of the indicated compounds for 30 min followed by an addition of [3H]-uridine for 1 min for determination of uridine uptake. Results are expressed as mean ± SEM. *P < 0.05 (AICAR (A,B) or vehicle (C) vs. SBI-0206965 or NBMPR). N.D.; not detectable.
In vitro screen of SBI-0206965 against an extended panel of human protein kinases
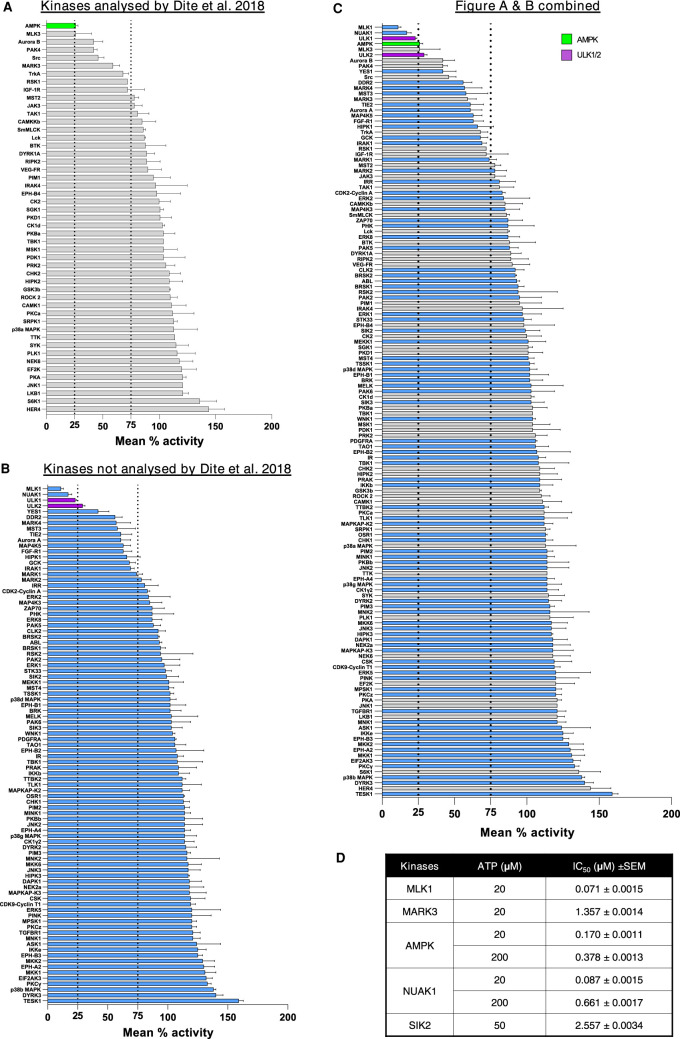
Given the similarity of the kinase domain structure between AMPK and AMPK-related kinases (BRSK1, BRSK2, MARK1, MARK2, MARK3, MARK4, MELK, NUAK1, NUAK2, SIK1, SIK2, SIK3, SNRK) [40,41], we hypothesised that SBI-0206965 also inhibits several members of the AMPK-related kinases. In a previous study [32], among 13 members of the AMPK-related kinases only MARK3 was included in the screen. Notably, while AMPK was most potently inhibited (∼80%) by SBI-0206965 (used at 0.25 μM), cross-reactivity with MARK3 (>40% inhibition) was also observed [32]. To gain a better understanding of SBI-0206965 selectivity we performed a screen against an expanded kinase panel consisting of 140 kinases, 49 of which overlap with a previous report [32]. Importantly, our panel includes 10 members of the AMPK-related kinases (BRSK1, BRSK2, MARK1, MARK2, MARK3, MARK4, MELK, NUAK1, SIK2, SIK3). Our results were similar to the results obtained in the previous screen [32] and SBI-0206965 robustly inhibited AMPK (∼80%) and to a lesser degree (∼30–50% inhibition) multiple other kinases (e.g. MARK3, MLK3, PAK4, Src, TrkA) (Figure 5A). Interestingly, our expanded screen revealed that SBI-0206965 also inhibits MLK1 and NUAK1 more potently than AMPK and ULK1/2, while an additional 25 kinases were inhibited by ≥25% (Figure 5B,C). We next determined the half-maximal inhibitory concentration (IC50) of SBI-0206965 against AMPK and MLK1, as well as selected AMPK-related kinases (MARK3/4, NUAK1, SIK2). In line with the results of our single concentration screen (Figure 5C), we observed that IC50 values (ATP concentration = 20 μM) for MLK1 (0.071 μM) and NUAK1 (0.087 μM) were noticeably lower than AMPK (0.170 μM) (Figure 5D).
Figure 5. Kinase selectivity profile for SBI-0206965.
(A–C) A screen of 140 human protein kinases (n = 2 per kinase, with or without 1 μM SBI-0206965) was performed in vitro using the MRC-PPU Premier Screen service as described in Materials and methods. Results from the current profiling (140 kinases) were shown as; kinases profiled previously (49 kinases) by Dite et al. [32] (A) or not profiled (91 kinases) (B) or combined data from A and B (C). (D) The half-maximal inhibitory concentration (IC50) values of the selected kinases against SBI-0206965 were performed using the indicated concentrations of ATP. Results are expressed as mean ± SEM.
SBI-0206965 inhibits kinases that contain methionine at the gatekeeper residue
Structures of SBI-0206965 (Figure 6A) bound to AMPKα2 and ULK2 kinase domains have provided a detailed view of how this inhibitor engages the ATP-binding cleft of protein kinases (Figure 6B). Both structures revealed a compound binding mode that is characteristic of type I kinase inhibitors, taking advantage of the ‘active-like’ conformation of the kinase domain. SBI-0206965 engages the hinge region, the canonical DFG motif and, importantly, the bromine atom fills the ‘back pocket’ next to the methionine ‘gatekeeper’ residue (Figure 6B). To gain a general overview of how SBI-0206965 may be interacting with MARK3 and MARK4 we performed molecular docking experiments using the SwissDock server [49]. As expected, SBI-0206965 engages MARK3 and MARK4 in a similar manner as AMPKα2 and ULK2 and binds in the ATP cleft with the bromine atom facing the methionine gatekeeper residue (Figure 6B). The position of docked SBI-0206965 was almost identical with the position of the bound compound to AMPKα2 and ULK2 (Figure 6B), thus validating our molecular docking approach for determining the general binding mode of SBI-0206965 to MARK3 and MARK4. Interestingly, we noticed that most SBI-0206965-sensitive kinases contain a large gatekeeper residue and a preference for methionine at this position (Figure 6C, Supplementary Figure S4).
Figure 6. SBI-0206965 inhibitor engagement with kinase ATP-binding clefts.
(A) Chemical structure of SBI-0206965. (B) Kinase domain structures with bound and/or docked SBI-0206965 in the active site. SBI-0206965 engages the ATP-binding cleft near the hinge region and gatekeeper residue. Docked compound structures are shown in grey with coloured heteroatoms (red, oxygen; blue, nitrogen). Accession PDBid codes for bound compound structures: 6BX6, AMPKα2; 6YID, ULK2. (C) Gatekeeper residues of AMPK-related kinases (top panel) and a select set of kinases sensitive to SBI-0206965 (middle panel). Gatekeeper residues of ULK3 and ULK4 are shown (bottom panel).
Mutation of the gatekeeper methionine to threonine renders AMPK and ULK1 resistant to SBI-0206965 in vitro
We wondered if converting the gatekeeper residue methionine to an amino acid with a smaller side chain (e.g. threonine) would lead to loss of SBI-0206965 inhibition. We mutated the gatekeeper residue of AMPKα1 (M104 → Thr; M104T) or ULK1 (M92T) and expressed the respective mutant or wild type (WT) in mammalian cells. Immunoblot analysis showed that levels of total (AMPKα1 and ULK1) and AMPK α1-pThr172 were comparable between WT and the respective gatekeeper mutant (Figure 7A,B). We observed that both AMPKα1 M104T and ULK1 M92T mutants had lower activity in vitro compared with respective WT forms (Figure 7C,D). Despite the reduced basal activity, AMPKα1 M93T mutant was equally responsive to allosteric AMPK activators AMP and A-769662 (that bind the nucleotide-binding domain of γ subunit and ADaM site located at the interface of the α and β subunit, respectively [3]) compared with WT (Figure 7E,F). Notably, we observed a marked increase in SBI-0206965 IC50 for AMPKα1 M104T (∼20 fold) and ULK1 M92T (∼7-fold) compared with their respective WT control suggesting the importance of a methionine gatekeeper for SBI-0206965 inhibition. In contrast, the IC50 was virtually unchanged (within 2-fold difference) when Compound C, an ATP-competitive inhibitor that binds to the highly conserved active site of AMPK [29], was tested for inhibition of AMPK and ULK1 WT and mutant forms (Figure 7G–J).
Mutation of the gatekeeper methionine to threonine renders AMPK resistant to SBI-0206965 in cells
We next wanted to verify if in vitro results of the SBI-0206965-insensitive AMPK mutant (Figure 7G,H) can be recapitulated in a cellular context. We generated AMPKα1/α2 double knockout (DKO) U2OS cells (Supplementary Figure S5) and transfected the DKO cells with AMPKα1 WT or M104T mutant, which allowed us to avoid the influence of endogenous AMPKα in the mutant-expressing cells. The transfected cells were treated with vehicle or SBI-0206965 followed by incubation with vehicle or 991. Using a sensitive and quantitative HTRF assay, phosphorylation of ACC (Ser79) was measured as surrogate readout of cellular AMPK activity. As shown in Figure 8A, 991 equally increased ACC phosphorylation ∼2.5-fold in AMPKα1 WT- and M104T-expressing cells. Consistent with in vitro kinase activity data, the inhibitory effect of SBI-0206965 on ACC phosphorylation was blunted in AMPKα1 M104T-expressing cells compared with WT-expressing cells (Figure 8B). In contrast, the inhibitory effect of Compound C on ACC phosphorylation was similar between WT- and M104T-expressing cells (Figure 8C). Immunoblot analysis confirmed differential effects of SBI-0206965 on ACC phosphorylation (measured by HTRF assay, Figure 8A, B) in WT- and M104T-expressing cells and also consistent/comparable expression of the transfected WT and M104T mutant (Figure 8D). Collectively, we demonstrate the key role of a methionine gatekeeper for SBI-0206965-mediated inhibition of AMPK and have designed an inhibitor-insensitive mutant form of AMPK that can be used as control.
Discussion
The use of protein kinase inhibitors as tool compounds have transformed our ability to study cell signalling and resulting biological responses. However, for chemical probes to be useful a thorough analysis of their off-target effects is paramount. We have demonstrated that SBI-0206965 is not a highly selective inhibitor for either ULK1 or AMPK. In cell-free assays, SBI-0206965 inhibits MLK1/3, NUAK1 and also FAK [34] at the same levels as, or more robustly than ULK1 or AMPK. We also show that in intact cells, SBI-0206965 potently inhibits glucose and nucleoside uptake independent of its inhibition of AMPK activity. Analysis of co-crystal structures of AMPK-SBI-0206965 or ULK2-SBI-0206965 complex and computational modelling has revealed the importance of the gatekeeper methionine as a key determinant of SBI-0206965 sensitivity.
A previous study reported superior selectivity and preferable characteristics of SBI-0206965 for AMPK relative to Compound C, and a co-crystal structure of the AMPKα2 kinase domain (residues 6–278)-SBI-0206965 complex showed that the drug occupies a pocket that partially overlaps with the ATP-binding site [32]. The ATP-binding site is composed of the adenine pocket (near the hinge region), the ribose-interacting region, the phosphate-coordinating region, the solvent accessible region, and the buried region [56,57]. The size and shape of the buried region is controlled by the gatekeeper residue, which serves as a molecular gate controlling the accessibility to this pocket and the nature of interactions with ligands (as seen in Figure 6C) [57,58]. In ∼73% of human kinases, a hydrophobic amino acid with a bulky side chain (Met, Phe, or Leu) is observed at that position, 22% have a small residue, such as Thr or Val and the remaining 5% have one of the remaining amino acids [57]. It has been reported that the amino acid distribution of the gatekeeper residue in some of the kinase groups differ significantly from the whole distribution. In particular, methionine is relatively more frequent (∼20 kinases) in the tyrosine kinase group than in the overall distribution [59]. We noticed that among the top 30 kinases inhibited by SBI-0206965 (>25% inhibition, Figure 5C) in the current screen, 25 kinases have medium (Met, Leu) or large gatekeepers (Phe, Tyr) and the other 4 kinases (YES, SRC, DDR2, TIE2, FGFR1) have a small gatekeeper (Thr, Ile, Val) (Supplementary Figure S4A). Notably, 19 out of 30 kinases (MLK1, NUAK1, ULK1, AMPK, MLK3, ULK2, Aurora B, PAK4, MARK4, MST3, MARK3, MAP4K5, GCK, IGF-1R, MARK1, MST2, MARK2, MARK2, JAK3, IRR, TAK1) have methionine in their gatekeeper residue. Consistent with this, out of the top 18 kinases whose binding/activity was shown to be inhibited by SBI-0206965 [34], seven of them (ULK1, ULK2, FAK, JAK3, JAK2, NUAK2, PAK1) have methionine in their gatekeeper residue. Nonetheless, it should also be noted that there are kinases with a methionine gatekeeper residue (e.g. JNKs), which are not selectively inhibited by SBI-0206965 [31,33].
We show here that converting the gatekeeper residue methionine of AMPKα1 (M104) or ULK1 (M92) to an amino acid with a smaller side chain (Thr) attenuated SBI-0206965 inhibition in a cell-free assay. Importantly, reintroducing WT or the M104T mutant of AMPKα1 into AMPKα1/α2 DKO U2O2 cells shows that the gatekeeper residue is also critical for efficient AMPK inhibition in cellulo. Interestingly, we observed that Compound C was not affected by mutations of the gatekeeper residue in AMPK and ULK1 in vitro or AMPK in cellulo. Structures of the AMPKα2 kinase domain bound to SBI-0206965 [32] and Compound C [29] show that SBI-0206965 contacts mainly the ATP pocket, hinge region and the gatekeeper region. In addition to the hinge and gatekeeper regions, Compound C interacts with the C-lobe αD helix and the activation segment, which has moved within the ATP-binding grove (Supplementary Figure S6). Therefore, Compound C is likely to be less reliant on interactions with the gatekeeper residue than SBI-0206965.
We and others [32,34] have observed that SBI-0206965 doses ≥25 μM increase phosphorylation of AMPKα T172 in multiple cell types, which may be due to off-target effects or cellular toxicity by reduced cellular adenylate energy charge as there was a trend toward reduced adenylate energy charge at 30 μM [32]. Contrary to this hypothesis, we did not observe reduced AMP/ATP, ADP/ATP ratio or adenylate energy charge in both C2C12 and primary hepatocytes with SBI-0206965 (up to 50 μM). We show that SBI-0206965 does not activate (>25%) upstream kinases (LKB1 or CaMKK2) that catalyse AMPKα T172 in the cell-free assay (Figure 5). It is unknown if SBI-0206965 inhibits a protein phosphatase that regulates AMPKα T172 or if inhibitor binding causes a conformational change that makes T172 less accessible to a phosphatase. Nonetheless, we observed that SBI-0206965-induced phosphorylation of AMPKα T172 was not accompanied by an increase but rather a decrease in AMPK substrate phosphorylation. This is presumably because SBI-0206965 binds the kinase domain and inhibits the catalytic activity of AMPK [32], negating the effect of increases in AMPKα T172 phosphorylation. This highlights the unreliability of using AMPKα T172 phosphorylation as a sole readout of cellular AMPK activity. In mouse embryonic fibroblasts (MEFs) and HEK293 cells, SBI-0206965 treatment at 10–50 μM (1 h) appeared to cause reduced activity of FAK, while mTOR, Akt, or ERK signalling were unaltered [34]. We observed that in C2C12 myotubes, but not adipocytes, Akt and ERK signalling were inhibited with ≥25 μM SBI-0206965.
In previous studies, SBI-0206965 was used to probe the effect of AMPK activators on glucose uptake in isolated rodent skeletal muscle ex vivo. It was reported that AICAR-mediated increase in phosphorylation of AMPK substrates (ACC, ULK1, or TBC1D1) was reduced with SBI-0206965 (used at 10 or 100 μM), which was accompanied by a profound reduction in glucose uptake in isolated rodent skeletal muscle ex vivo [54,60]. However, the results of these studies need to be cautiously interpreted as SBI-0206965 potently inhibits cellular uptake of AICAR (i.e. cellular accumulation of ZMP) at 10 μM in cultured cells and intact skeletal muscle tissue ([32] and the current study), and the above studies [54,60] did not report muscular ZMP content with AICAR in combination with SBI-0206965. Compound C also blocks cellular AICAR uptake through competition for adenosine transporter binding sites, which accounts for its inhibitory effects on AICAR-mediated AMPK activation [61]. Based on the results reported in the previous [32,61] and the current study, we would strongly advise against the use of SBI-0206965 or Compound C in combination with AICAR when investigating cellular AMPK signalling and function.
A recent study reported that SBI-0206965 inhibits unstimulated and insulin-stimulated glucose transport in isolated skeletal muscle tissue and cultured L6 myotubes without apparent effect on GLUT4 translocation in L6 muscle cells [54]. This is reminiscent of the p38 MAPK inhibitor SB-203580, which was shown to inhibit insulin-stimulated glucose transport in skeletal muscle tissue/cells and adipocytes [62]. Using a SB-203580-resistant mutant of p38 MAPK, it was unambiguously shown that the inhibitory effect of SB-203580 on glucose transport was independent of p38 MAPK [63]. Along the same lines, it would be useful to utilise SBI-0206965-resistant mutant AMPK or ULK1 to probe drug selectivity towards respective kinase signalling pathways. For this purpose, we generated AMPKα1α2 DKO U2OS cells and transfected the DKO cells with AMPKα1 WT or M104T mutant, which allowed us to avoid the effect of SBI-0206965 on endogenous AMPKα in the mutant-expressing cells. A similar approach may be needed for the ULK pathway (e.g. use of ULK1/2 DKO cells and introduction of WT or M92T mutant). It would also be interesting to assess if the SBI-0206965-resistant mutant can be exploited to delineate drug selectivity towards the functional/physiological outputs of the kinases. However, it might be technically challenging since the SBI-0206965-resistant mutants display only a partial resistance to the inhibitor action.
In conclusion, while SBI-0206965 was reported as a highly selective inhibitor against ULK1 [34] and AMPK [32], we and others demonstrate that in vitro the compound is a potent inhibitor of several other kinases, including NUAK1, FAK, MLK1/3, and MARK3/4, all of which contain a methionine in their gatekeeper position. In addition to these kinases, SBI-0206965 also inhibits glucose and nucleoside transport systems; therefore, the drug effect on cell growth, metabolism or survival needs to be carefully assessed not only when studying kinase signalling cascades, but also when investigating nutrient and metabolite levels. Considering the cross-talk in kinase signalling networks and the myriad of roles that AMPK has in regulating metabolism, the field is still in need of a more selective and potent tool compound. Until more specific AMPK inhibitors are discovered, cautious use of the current tools and careful interpretations of results are advised. Our study has revealed inhibitor-insensitive mutant forms of AMPK and ULK1 that could be exploited as potential controls. We hope that our conclusions stimulate a search for better tool compounds and improved experimental design to probe AMPK and ULK1/2 functions.
Acknowledgements
We thank Amy Ehrlich for her technical assistance and Sourav Banerjee for valuable input.
Abbreviations
- ACC1
acetyl-CoA carboxylase-1
- ADaM
allosteric drug and metabolite
- AMPK
AMP-activated protein kinase
- CaMKK2
Ca2+-calmodulin-dependent protein kinase kinase-2
- CBS
cystathionine β-synthase
- ECL
enhanced chemiluminescence
- EDL
extensor digitorum longus
- LKB1
liver kinase B
Data availability
Data and reagents are available upon request to the corresponding author.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was financially supported by The Swedish Research Council (project grant Dnr 2017-01295), The Swedish Diabetes Foundation, Royal Physiographic Society of Lund and The Påhlsson Foundation (to O.G.). J.W.S. was supported by National Health and Medical Research Council (NHMRC) project grant (GNT1138102). This project was also supported in part by the Victorian Government's Operational Infrastructure Support Programme. A.J.O. is supported by a PhD scholarship funded by the Australian Catholic University. M.D. is supported by a postdoc fellowship and A.S.H. by a PhD scholarship from the Danish Diabetes Academy, which is funded by the Novo Nordisk Foundation (NNF17SA0031406). P.G. was supported by the Erasmus+ programme. Y.A. was supported by a research scholarship from King Saud University. E.Z. was supported by a Sir Henry Dale Fellowship (Wellcome Trust and the Royal Society; 200523/Z/16/Z). The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent Research Center based at the University of Copenhagen, Denmark, and partially funded by an unconditional donation from the Novo Nordisk Foundation (Grant number NNF18CC0034900).
CRediT Author Contribution
Kei Sakamoto: Conceptualization, resources, supervision, funding acquisition, investigation, writing — original draft. Danial Ahwazi: Investigation, visualization, methodology, writing — review and editing. Katyayanee Neopane: Investigation, visualization, methodology, writing — review and editing. Greg R. Markby: Investigation, methodology, writing — review and editing. Franziska Kopietz: Investigation, methodology, writing — review and editing. Ashley J. Ovens: Investigation, methodology, writing — review and editing. Morten Dall: Investigation, writing — review and editing. Anna S. Hassing: Investigation, writing — review and editing. Pamina Graesle: Investigation, writing — review and editing. Yazeed Alshuweishi: Investigation, writing — review and editing. Jonas T. Treebak: Supervision, methodology, writing — review and editing. Ian P. Salt: Supervision, writing — review and editing. Olga Göransson: Resources, supervision, writing — review and editing. Elton Zeqiraj: Conceptualization, investigation, visualization, writing — original draft. John W. Scott: Conceptualization, resources, supervision, investigation, visualization, methodology, writing — review and editing.
Supplementary Material
References
- 1.Hardie, D.G. and Sakamoto, K. (2006) AMPK: a key sensor of fuel and energy status in skeletal muscle. Physiology (Bethesda) 21, 48–60 10.1152/physiol.00044.2005 [DOI] [PubMed] [Google Scholar]
- 2.Hardie, D.G. (2011) AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 25, 1895–1908 10.1101/gad.17420111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinberg, G.R. and Carling, D. (2019) AMP-activated protein kinase: the current landscape for drug development. Nat. Rev. Drug. Discov. 18, 527–551 10.1038/s41573-019-0019-2 [DOI] [PubMed] [Google Scholar]
- 4.Cokorinos, E.C., Delmore, J., Reyes, A.R., Albuquerque, B., Kjobsted, R., Jorgensen, N.O.et al. (2017) Activation of skeletal muscle AMPK promotes glucose disposal and glucose lowering in non-human primates and mice. Cell Metab. 25, 1147–1159.e1110 10.1016/j.cmet.2017.04.010 [DOI] [PubMed] [Google Scholar]
- 5.Myers, R.W., Guan, H.P., Ehrhart, J., Petrov, A., Prahalada, S., Tozzo, E.et al. (2017) Systemic pan-AMPK activator MK-8722 improves glucose homeostasis but induces cardiac hypertrophy. Science 357, 507–511 10.1126/science.aah5582 [DOI] [PubMed] [Google Scholar]
- 6.Fullerton, M.D., Galic, S., Marcinko, K., Sikkema, S., Pulinilkunnil, T., Chen, Z.P.et al. (2013) Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat. Med. 19, 1649–1654 10.1038/nm.3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudaba, N., Marion, A., Huet, C., Pierre, R., Viollet, B. and Foretz, M. (2018) AMPK Re-activation suppresses hepatic steatosis but its downregulation does not promote fatty liver development. EBioMedicine 28, 194–209 10.1016/j.ebiom.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Neill, H.M., Lally, J.S., Galic, S., Thomas, M., Azizi, P.D., Fullerton, M.D.et al. (2014) AMPK phosphorylation of ACC2 is required for skeletal muscle fatty acid oxidation and insulin sensitivity in mice. Diabetologia 57, 1693–1702 10.1007/s00125-014-3273-1 [DOI] [PubMed] [Google Scholar]
- 9.Chen, Q., Xie, B., Zhu, S., Rong, P., Sheng, Y., Ducommun, S.et al. (2017) A Tbc1d1 (Ser231Ala)-knockin mutation partially impairs AICAR- but not exercise-induced muscle glucose uptake in mice. Diabetologia 60, 336–345 10.1007/s00125-016-4151-9 [DOI] [PubMed] [Google Scholar]
- 10.Kjobsted, R., Roll, J.L.W., Jorgensen, N.O., Birk, J.B., Foretz, M., Viollet, B.et al. (2019) AMPK and TBC1D1 regulate muscle glucose uptake after, but not during, exercise and contraction. Diabetes 68, 1427–1440 10.2337/db19-0050 [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto, K. and Holman, G.D. (2008) Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am. J. Physiol. Endocrinol. Metab. 295, E29–E37 10.1152/ajpendo.90331.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawley, S.A., Davison, M., Woods, A., Davies, S.P., Beri, R.K., Carling, D.et al. (1996) Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 271, 27879–27887 10.1074/jbc.271.44.27879 [DOI] [PubMed] [Google Scholar]
- 13.Gowans, G.J., Hawley, S.A., Ross, F.A. and Hardie, D.G. (2013) AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 18, 556–566 10.1016/j.cmet.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao, B., Sanders, M.J., Underwood, E., Heath, R., Mayer, F.V., Carmena, D.et al. (2011) Structure of mammalian AMPK and its regulation by ADP. Nature 472, 230–233 10.1038/nature09932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakhill, J.S., Steel, R., Chen, Z.P., Scott, J.W., Ling, N., Tam, S.et al. (2011) AMPK is a direct adenylate charge-regulated protein kinase. Science 332, 1433–1435 10.1126/science.1200094 [DOI] [PubMed] [Google Scholar]
- 16.Ovens, A.J., Scott, J.W., Langendorf, C.G., Kemp, B.E., Oakhill, J.S. and Smiles, W.J. (2021) Post-translational modifications of the energy guardian AMP-activated protein kinase. Int. J. Mol. Sci 22, 1229 10.3390/ijms22031229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrill, G.F., Kurth, E.J., Hardie, D.G. and Winder, W.W. (1997) AICA riboside increases AMP-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am. J. Physiol. 273, E1107–E1112 10.1152/ajpendo.1997.273.6.E1107 [DOI] [PubMed] [Google Scholar]
- 18.Hayashi, T., Hirshman, M.F., Kurth, E.J., Winder, W.W. and Goodyear, L.J. (1998) Evidence for 5′ AMP-activated protein kinase mediation of the effect of muscle contraction on glucose transport. Diabetes 47, 1369–1373 10.2337/diab.47.8.1369 [DOI] [PubMed] [Google Scholar]
- 19.Mu, J., Brozinick, Jr, J.T., Valladares, O., Bucan, M. and Birnbaum, M.J. (2001) A role for AMP-activated protein kinase in contraction- and hypoxia-regulated glucose transport in skeletal muscle. Mol. Cell 7, 1085–1094 10.1016/S1097-2765(01)00251-9 [DOI] [PubMed] [Google Scholar]
- 20.Guigas, B., Sakamoto, K., Taleux, N., Reyna, S.M., Musi, N., Viollet, B.et al. (2009) Beyond AICA riboside: in search of new specific AMP-activated protein kinase activators. IUBMB Life 61, 18–26 10.1002/iub.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collodet, C., Foretz, M., Deak, M., Bultot, L., Metairon, S., Viollet, B.et al. (2019) AMPK promotes induction of the tumor suppressor FLCN through activation of TFEB independently of mTOR. FASEB J. 33, 12374–12391 10.1096/fj.201900841R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter, R.W., Hughey, C.C., Lantier, L., Sundelin, E.I., Peggie, M., Zeqiraj, E.et al. (2018) Metformin reduces liver glucose production by inhibition of fructose-1-6-bisphosphatase. Nat. Med. 24, 1395–1406 10.1038/s41591-018-0159-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foretz, M., Hebrard, S., Leclerc, J., Zarrinpashneh, E., Soty, M., Mithieux, G.et al. (2010) Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J. Clin. Invest. 120, 2355–2369 10.1172/JCI40671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cool, B., Zinker, B., Chiou, W., Kifle, L., Cao, N., Perham, M.et al. (2006) Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 3, 403–416 10.1016/j.cmet.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 25.Xiao, B., Sanders, M.J., Carmena, D., Bright, N.J., Haire, L.F., Underwood, E.et al. (2013) Structural basis of AMPK regulation by small molecule activators. Nat. Commun. 4, 3017 10.1038/ncomms4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calabrese, M.F., Rajamohan, F., Harris, M.S., Caspers, N.L., Magyar, R., Withka, J.M.et al. (2014) Structural basis for AMPK activation: natural and synthetic ligands regulate kinase activity from opposite poles by different molecular mechanisms. Structure 22, 1161–1172 10.1016/j.str.2014.06.009 [DOI] [PubMed] [Google Scholar]
- 27.Bultot, L., Jensen, T.E., Lai, Y.C., Madsen, A.L., Collodet, C., Kviklyte, S.et al. (2016) Benzimidazole derivative small-molecule 991 enhances AMPK activity and glucose uptake induced by AICAR or contraction in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 311, E706–E719 10.1152/ajpendo.00237.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou, G., Myers, R., Li, Y., Chen, Y., Shen, X., Fenyk-Melody, J.et al. (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108, 1167–1174 10.1172/JCI13505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handa, N., Takagi, T., Saijo, S., Kishishita, S., Takaya, D., Toyama, M.et al. (2011) Structural basis for compound C inhibition of the human AMP-activated protein kinase alpha2 subunit kinase domain. Acta Crystallogr. D Biol. Crystallogr. 67, 480–487 10.1107/S0907444911010201 [DOI] [PubMed] [Google Scholar]
- 30.Bain, J., Plater, L., Elliott, M., Shpiro, N., Hastie, C.J., McLauchlan, H.et al. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogt, J., Traynor, R. and Sapkota, G.P. (2011) The specificities of small molecule inhibitors of the TGFss and BMP pathways. Cell Signal. 23, 1831–1842 10.1016/j.cellsig.2011.06.019 [DOI] [PubMed] [Google Scholar]
- 32.Dite, T.A., Langendorf, C.G., Hoque, A., Galic, S., Rebello, R.J., Ovens, A.J.et al. (2018) AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965. J. Biol. Chem. 293, 8874–8885 10.1074/jbc.RA118.003547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dasgupta, B. and Seibel, W. (2018) Compound C/Dorsomorphin: its use and misuse as an AMPK inhibitor. Methods Mol. Biol. 1732, 195–202 10.1007/978-1-4939-7598-3_12 [DOI] [PubMed] [Google Scholar]
- 34.Egan, D.F., Chun, M.G., Vamos, M., Zou, H., Rong, J., Miller, C.J.et al. (2015) Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell 59, 285–297 10.1016/j.molcel.2015.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dite, T.A., Ling, N.X.Y., Scott, J.W., Hoque, A., Galic, S., Parker, B.L.et al. (2017) The autophagy initiator ULK1 sensitizes AMPK to allosteric drugs. Nat. Commun. 8, 571 10.1038/s41467-017-00628-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaikuad, A., Koschade, S.E., Stolz, A., Zivkovic, K., Pohl, C., Shaid, S.et al. (2019) Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2). Biochem. J. 476, 875–887 10.1042/BCJ20190038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji, X., Zhang, X. and Li, Z. (2020) ULK1 inhibitor induces spindle microtubule disorganization and inhibits phosphorylation of Ser10 of histone H3. FEBS Open Bio 10, 2452–2463 10.1002/2211-5463.13000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strembitska, A., Mancini, S.J., Gamwell, J.M., Palmer, T.M., Baillie, G.S. and Salt, I.P. (2018) A769662 inhibits insulin-stimulated Akt activation in human macrovascular endothelial cells independent of AMP-activated protein kinase. Int. J. Mol. Sci 19, 3886 10.3390/ijms19123886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zachari, M., Longo, M. and Ganley, I.G. (2020) Aberrant autophagosome formation occurs upon small molecule inhibition of ULK1 kinase activity. Life Sci. Alliance 3, e202000815 10.26508/lsa.202000815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alessi, D.R., Sakamoto, K. and Bayascas, J.R. (2006) LKB1-dependent signaling pathways. Annu. Rev. Biochem. 75, 137–163 10.1146/annurev.biochem.75.103004.142702 [DOI] [PubMed] [Google Scholar]
- 41.Lizcano, J.M., Goransson, O., Toth, R., Deak, M., Morrice, N.A., Boudeau, J.et al. (2004) LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833–843 10.1038/sj.emboj.7600110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter, R.W., Foretz, M., Bultot, L., Fullerton, M.D., Deak, M., Ross, F.A.et al. (2014) Mechanism of action of compound-13: an alpha1-selective small molecule activator of AMPK. Chem. Biol. 21, 866–879 10.1016/j.chembiol.2014.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dall, M., Trammell, S.A.J., Asping, M., Hassing, A.S., Agerholm, M., Vienberg, S.G.et al. (2019) Mitochondrial function in liver cells is resistant to perturbations in NAD(+) salvage capacity. J. Biol. Chem. 294, 13304–13326 10.1074/jbc.RA118.006756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ducommun, S., Ford, R.J., Bultot, L., Deak, M., Bertrand, L., Kemp, B.E.et al. (2014) Enhanced activation of cellular AMPK by dual-small molecule treatment: AICAR and A769662. Am. J. Physiol. Endocrinol. Metab. 306, E688–E696 10.1152/ajpendo.00672.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhein, P., Desjardins, E.M., Rong, P., Ahwazi, D., Bonhoure, N., Stolte, J.et al. (2021) Compound- and fiber type-selective requirement of AMPKgamma3 for insulin-independent glucose uptake in skeletal muscle. Mol. Metab. 101228 10.1016/j.molmet.2021.101228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kopietz, F., Alshuweish, Y., Bijland, S., Alghamdi, F., Degerman, E., Sakamoto, K.et al. (2021) A-769662 inhibits adipocyte glucose uptake in an AMPK-independent manner. Biochem. J. 478, 633–646 10.1042/BCJ20200659 [DOI] [PubMed] [Google Scholar]
- 47.Katwan, O.J., Alghamdi, F., Almabrouk, T.A., Mancini, S.J., Kennedy, S., Oakhill, J.S.et al. (2019) AMP-activated protein kinase complexes containing the beta2 regulatory subunit are up-regulated during and contribute to adipogenesis. Biochem. J. 476, 1725–1740 10.1042/BCJ20180714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Logie, L., Lees, Z., Allwood, J.W., McDougall, G., Beall, C. and Rena, G. (2018) Regulation of hepatic glucose production and AMPK by AICAR but not by metformin depends on drug uptake through the equilibrative nucleoside transporter 1 (ENT1). Diabetes Obes. Metab. 20, 2748–2758 10.1111/dom.13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grosdidier, A., Zoete, V. and Michielin, O. (2011) Swissdock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–W277 10.1093/nar/gkr366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lang, P.T., Brozell, S.R., Mukherjee, S., Pettersen, E.F., Meng, E.C., Thomas, V.et al. (2009) DOCK 6: combining techniques to model RNA-small molecule complexes. RNA 15, 1219–1230 10.1261/rna.1563609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C.et al. (2004) UCSF chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 52.Chen, S., Wasserman, D.H., MacKintosh, C. and Sakamoto, K. (2011) Mice with AS160/TBC1D4-Thr649Ala knockin mutation are glucose intolerant with reduced insulin sensitivity and altered GLUT4 trafficking. Cell Metab. 13, 68–79 10.1016/j.cmet.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sano, H., Kane, S., Sano, E., Miinea, C.P., Asara, J.M., Lane, W.S.et al. (2003) Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J. Biol. Chem. 278, 14599–14602 10.1074/jbc.C300063200 [DOI] [PubMed] [Google Scholar]
- 54.Knudsen, J.R., Madsen, A.B., Persson, K.W., Henriquez-Olguin, C., Li, Z. and Jensen, T.E. (2020) The ULK1/2 and AMPK inhibitor SBI-0206965 blocks AICAR and insulin-stimulated glucose transport. Int. J. Mol. Sci 21, 2344 10.3390/ijms21072344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright, N.J. and Lee, S.Y. (2019) Structures of human ENT1 in complex with adenosine reuptake inhibitors. Nat. Struct. Mol. Biol. 26, 599–606 10.1038/s41594-019-0245-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vulpetti, A. and Bosotti, R. (2004) Sequence and structural analysis of kinase ATP pocket residues. Farmaco 59, 759–765 10.1016/j.farmac.2004.05.010 [DOI] [PubMed] [Google Scholar]
- 57.Romano, V., de Beer, T.A. and Schwede, T. (2017) A computational protocol to evaluate the effects of protein mutants in the kinase gatekeeper position on the binding of ATP substrate analogues. BMC Res. Notes 10, 104 10.1186/s13104-017-2428-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elphick, L.M., Lee, S.E., Gouverneur, V. and Mann, D.J. (2007) Using chemical genetics and ATP analogues to dissect protein kinase function. ACS Chem. Biol. 2, 299–314 10.1021/cb700027u [DOI] [PubMed] [Google Scholar]
- 59.Huang, D., Zhou, T., Lafleur, K., Nevado, C. and Caflisch, A. (2010) Kinase selectivity potential for inhibitors targeting the ATP binding site: a network analysis. Bioinformatics 26, 198–204 10.1093/bioinformatics/btp650 [DOI] [PubMed] [Google Scholar]
- 60.Choumessi, A.T., Johanns, M., Beaufay, C., Herent, M.F., Stroobant, V., Vertommen, D.et al. (2019) Two isoprenylated flavonoids from Dorstenia psilurus activate AMPK, stimulate glucose uptake, inhibit glucose production and lower glycemia. Biochem. J. 476, 3687–3704 10.1042/BCJ20190326 [DOI] [PubMed] [Google Scholar]
- 61.Fryer, L.G., Parbu-Patel, A. and Carling, D. (2002) Protein kinase inhibitors block the stimulation of the AMP-activated protein kinase by 5-amino-4-imidazolecarboxamide riboside. FEBS Lett. 531, 189–192 10.1016/S0014-5793(02)03501-9 [DOI] [PubMed] [Google Scholar]
- 62.Turban, S., Beardmore, V.A., Carr, J.M., Sakamoto, K., Hajduch, E., Arthur, J.S.et al. (2005) Insulin-stimulated glucose uptake does not require p38 mitogen-activated protein kinase in adipose tissue or skeletal muscle. Diabetes 54, 3161–3168 10.2337/diabetes.54.11.3161 [DOI] [PubMed] [Google Scholar]
- 63.Antonescu, C.N., Huang, C., Niu, W., Liu, Z., Eyers, P.A., Heidenreich, K.A.et al. (2005) Reduction of insulin-stimulated glucose uptake in L6 myotubes by the protein kinase inhibitor SB203580 is independent of p38MAPK activity. Endocrinology 146, 3773–3781 10.1210/en.2005-0404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and reagents are available upon request to the corresponding author.