Abstract
The aim of this study was to investigate the effect of lncRNA KCNQ1OT1 on HCC and to explore the possible underlying mechanisms. The expression levels of KCNQ1OT1, miR-149 and S1PR1 were detected by qRT-PCR assay. A dual luciferase reporter assay was used to detect the interaction between KCNQ1OT1 and miR-149, as well as miR-149 and S1PR1. The interaction between KCNQ1OT1 and miR-149 was further investigated by RNA pull-down assay. Wound healing assays and Transwell assays were carried out to determine cell migration and invasion. A xenograft tumour assay was used to validate the role of KCNQ1OT1 in vivo. KCNQ1OT1 and S1PR1 were significantly increased, but miR-149 was decreased in HCC cells. Luciferase reporter assays and RNA pull-down assays revealed that KCNQ1OT1 directly targeted miR-149. In addition, miR-149 bound to the 3’-UTR of S1PR1. Knockdown of KCNQ1OT1 or overexpression of miR-149 inhibited the invasion and migration of HCC cells. However, suppression of miR-149 could abrogate the effect of KCNQ1OT1 knockdown on the invasion and migration abilities of HCC cells. In vivo assays showed that KCNQ1OT1 knockdown suppressed tumour growth. This work suggests that lncRNA KCNQ1OT1 might act as a potential therapeutic target in HCC.
Subject terms: Cancer, Medical research
Introduction
Hepatocellular carcinoma (HCC) is a common malignancy that is one of the most frequent causes of cancer-related deaths around the world. The treatments and prognosis of patients with HCC present challenges because they vary by the stage of disease, the specifics of tumour histology, the likelihood to tolerate surgery and the availability of liver transplants. Curative therapies such as liver transplantation or surgical resection are feasible in 20–40% of patients [1]. A comprehensive understanding of the molecular regulation of the carcinogenesis of HCC will help to improve the therapeutic strategy and survival rate of patients with HCC.
Long noncoding RNAs (lncRNAs) are involved in various aspects of gene regulation, such as mRNA splicing and translation, chromosome dosage compensation and nuclear and cytoplasmic trafficking [2–5]. KCNQ1OT1 is an lncRNA gene located within the KCNQ1 locus, and studies show that the dysregulation of KCNQ1OT1 can lead to deleterious physical and genetic effects. For example, Zhu and colleagues found that KCNQ1OT1 was increased in the plasma of patients with diabetic nephropathy and high-glucose (HG)-induced HK-2 cells and that KCNQ1OT1 interference could decrease the inflammation, oxidative stress and pyroptosis of HG-induced HK-2 cells [6]. The upregulated expression of KCNQ1OT1 has been shown to be correlated with acute myocardial infarction and atherosclerosis, which are high risk factors for stroke [7]. Zhang et al. found that KCNQ1OT1 acted as an oncogene, was highly expressed in tongue squamous cell carcinoma (TSCC) tissues, especially in chemoresistant TSCC patients, and played an important role in the regulation of TSCC chemoresistance [8]. The present study shows that a high level of KCNQ1OT1 is correlated with HCC [9]. However, the biological functions of KCNQ1OT1 and its mechanism of regulation in HCC are not clear. Bioinformatics prediction suggests that there is an interaction between lncRNA KCNQ1OT1 and miR-149. miR-149 has been reported as a potential prognostic marker for HCC [10], colorectal cancer [11], upper tract urothelial carcinoma [12] and gastric cancer [13].
Sphingosine-1-phosphate receptor 1 (S1PR1) has been identified as a G protein-coupled receptor engaged with S1P, which enhances cell proliferation and migration in normal and cancer cells. Extensive research has shown that S1PR1 is involved in immune cell regulation, vascular growth and development, motility of cancer cells and multiple sclerosis [14]. It has been demonstrated that the expression levels of S1PR1 are high in HCC cell lines [15]. Bioinformatics predictions indicate that miR-149 interacts with S1PR1. However, the regulatory mechanism of KCNQ1OT1/miR-149/S1PR1 in HCC progression is not clear.
Our research suggests that the KCNQ1OT1-modulated suppression of miR-149 is responsible for the activation of the S1PR1/phosphoinositide-3 kinase (PI3K)/AKT signalling cascade in the process of migration and invasion in HCC cells, providing novel insight into the diagnosis and treatment of HCC.
Materials and methods
Cell culture
The normal human healthy hepatic cell line WRL68 and the human HCC cell lines Bel-7402, HepG2, SMMC-7721 and Huh7 were obtained from the Chinese Academy of Sciences (Shanghai, China). The cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (purchased from Gibco, USA) supplemented with 10% foetal bovine serum (FBS; purchased from Gibco, USA), 100 μg/mL streptomycin and 100 units/mL penicillin at 37 °C in a humidified incubator with 5% CO2. The culture medium was replaced every 2–3 days.
Cell transfection
Sh-KCNQ1OT1, miR-149 mimics and miR-149 inhibitor were designed and synthesized by Shanghai GenePharma company. Sh-KCNQ1OT1 was cloned into the lentiviral vector psi-LVRH1GP, and the recombinant lentivirus was transfected into the HCC cell lines Huh7 and SMMC-7721. Moreover, the vector, miR-149 mimics and miR-149 inhibitor were transfected into Huh7 and SMMC-7221 cells at a concentration of 50 nM using Lipofectamine 3000 reagent (Life Technologies Corporation, Carlsbad, CA) in accordance to the manufacturer’s instructions.
Dual-luciferase reporter assay
The wild-type (WT) fragments from KCNQ1OT1 (KCNQ1OT1-WT) containing miR-149 potential binding sites and the corresponding mutant (MUT) fragments (KCNQ1OT1-MUT) were cloned into pMIR-basic vector (Promega, USA). Meanwhile, the WT fragment from the 3’-untranslated region (3’-UTR) of S1PR1 (S1PR1-WT) and the corresponding MUT fragment (S1PR1-MUT) were cloned into the pMIR-basic vector to generate the luciferase reporter construct of S1PR1. Then SMMC-7721 and Huh7 cells were cotransfected with the vectors and miR-149 mimics by using Lipofectamine 3000 reagent (Life Technologies Corporation). The cells were lysed, and the relative Rluc/Luc ratio was measured by the Dual-Luciferase reporter gene system (Promega, USA) following a published protocol [16].
Wound healing assay
After the cells were incubated to reach confluence, they were scratched with a sterile pipette tip. Then the plates were rinsed with fresh culture medium twice to remove the detached cells and incubated in an incubator with 5% CO2 at 37 °C. The cells migrated from the wound edge, and the distance between the two sides of the wound was monitored using a microscope. The degree of cell migration was quantified by the ratio of the measured gap distance at 24 h to that at 0 h.
Cell invasion assay
Matrigel-coated Transwell chamber models were used in the cell invasion assay [17]. After transfection for 48 h, cells resuspended in 50 μL serum-free DMEM culture medium were seeded on a polycarbonate membrane with fibronectin-coated insert with 8-μm pores in a Transwell apparatus. The Transwell inserts were placed in 24-well culture plates containing 10% FBS medium and solidified by incubating at 37 °C for 4 h. Then 5 × 104 cells in 200 μL medium were placed in the upper chambers. After adding mitomycin to stop mitosis, the membranes were fixed with 10% formalin at room temperature and stained with 0.05% crystal violet. Invasive cells were counted in five different fields of each filter.
Total RNA extraction and quantitative reverse transcriptase-PCR (qRT-PCR)
Total RNA was extracted from the indicated cultured cell lines using TRIzol reagent (Sigma, USA) following the manufacturer’s instructions. The optical density value at 260 nm was measured to determine the concentration and purity of RNA. Total RNA was transcribed into cDNAs using the M-MLV Kit (Takara, Japan) in accordance with the protocols provided by the manufacturer. The relative expression levels of KCNQ1OT1, miR-149 and S1PR1 were measured by qRT-PCR assay using the SYBR Green real-time PCR Master mix (Toyobo, Japan). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 small nuclear RNA served as endogenous controls [8, 18]. PCR amplification was carried out using a Light Cycler 2.0 instrument (Roche Applied Science), and the gene-specific primers for qRT-PCR analyses were used as previously described [9, 10, 15]. The sequences of the primers are listed in Table 1.
Table 1.
Primer sequences.
| Name | Sequence (5’ to 3’) |
|---|---|
| lncRNA KCNQ1OT1-F | GCAAGCCTCAGCATACACAA |
| lncRNA KCNQ1OT1-R | GGCTGTGTTGCTCTCTTTCC |
| miR-149-F | CGTCTGGCTCCGTGTCTTC |
| miR-149-R | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGGAGT |
| S1PR1-F | CCACAACGGGAGCAATAACT |
| S1PR1-R | CAGAATGACGATGGAGAGCA |
| GAPDH-F | CCAGGTGGTCTCCTCTGA |
| GAPDH-R | GCTGTAGCCAAATCGTTGT |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
Protein extraction and western blot assay
The cells were lysed in RIPA reagent (Beyotime, China), and protein levels were quantified using the BCA Protein Assay Kit (Bio-Rad Laboratories, USA). Forty micrograms of total protein for each sample was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride (PVDF) membranes (Invitrogen, USA). The PVDF membranes were incubated with 5% bovine serum albumin in Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBST buffer) at room temperature for 1 h to block non-specific bindings. The appropriate protein-specific primary antibodies were subsequently added and incubated at 4 °C overnight. The secondary horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit IgG antibodies (1:5000 dilution, Beyotime Biotechnology, China) were matched with the primary antibodies for 1 h at room temperature. Subsequently, the membranes were rinsed with TBST buffer 3 times for 10 min each. The target proteins were visualized using enhanced chemiluminescence reagent. The primary antibodies for the detection of corresponding proteins were used as follows: mouse anti-S1PR1 antibody (1:500 dilution, Santa Cruz Biotechnology, USA); rabbit anti-AKT1 (1:1000 dilution, Cell Signaling Technology, USA), anti-phospho-AKT1 (1:500 dilution, Cell Signaling Technology, USA), anti-PI3K (1:1000 dilution, Cell Signaling Technology, USA), anti-phospho-PI3K (1:1000 dilution, Cell Signaling Technology, USA), anti-E-cadherin (1:500 dilution, Cell Signaling Technology, USA), anti-N-cadherin (1:500 dilution, Cell Signaling Technology, USA), anti-Vimentin (1:500 dilution, Cell Signaling Technology, USA), anti-Snail (1:1000 dilution, Cell Signaling Technology, USA), anti-MMP-2 (1:1000 dilution, Cell Signaling Technology, USA), and anti-MMP-9 antibodies (1:1000 dilution, Cell Signaling Technology, USA). The rabbit anti-GAPDH antibody (1:1000 dilution, Bioss, China) was used as the internal control. The relative band density of each protein was analysed using the ImageJ software.
Mice xenograft assay
A total of 15 BALB/c male nude mice (5 weeks) were purchased from the Animal Center of the Chinese Academy of Science and housed in sterilized cages with a 12:12 h light:dark cycle and 45% humidity at 25 °C. The mice ate regularly and were provided free access to water. The mice were randomly divided into 3 groups with 5 mice in each group: control group, sh-NC group, and sh-KCNQ1OT1 group. The corresponding groups of SMMC-7721 cells were subcutaneously injected into the dorsal sides of the nude mice. The length and width of the tumour was measured every 7 days from injection to monitor the growth of the tumour. The tumour volume was calculated using the following formula: (length × width2)/2. Four weeks later, the mice were sacrificed, and the tumour tissue was imaged. Our protocol was approved by the Ethics Committee of Guangdong Medical University.
Immunohistochemistry assay
Tumours were embedded in paraffin and then cut into sections of 4 μm. Specific mouse anti-S1PR1 antibody (Santa Cruz Biotechnology, USA) and rabbit anti-Ki67 human primary antibody (Cell Signaling Technology, USA) were diluted with phosphate-buffered saline (PBS) buffer at 1:100 and incubated at 4 °C overnight. Then the tissues were incubated with goat anti-mouse secondary antibody or goat anti-rabbit secondary antibody, and the complexes were measured by using HRP–streptavidin conjugates and visualized using 3,3’-diaminobenzidine.
RNA pull-down assay
The interaction between KCNQ1OT1 and miR-149 was further determined by RNA pull-down assay using the Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, USA) in accordance with the manufacturer’s protocol. Protein extracts from SMMC-7721 and Huh7 cells were incubated with biotin-labelled miR-149-WT or miR-149-MUT and mixed with streptavidin agarose magnetic beads (Life Technologies Corporation, USA). The mixture was centrifuged at 1500 rpm for 1 h at 4 °C. Total RNA was extracted by TRIzol, and the relative expression of KCNQ1OT1 mRNA was detected by qRT-PCR.
Statistical methods
All of the experiments were repeated at least three times with similar results. The data of this study were statistically analysed by the SPSS 21.0 software, and all of the measured data are expressed as means ± standard deviations (SDs). Student’s t test was used for pairwise comparisons, and one-way analysis of variance was applied for multi-group comparisons. P < 0.05 indicated that the differences were statistically significant.
Results
Relative expression of KCNQ1OT1, miR-149 and S1PR1 in HCC cells
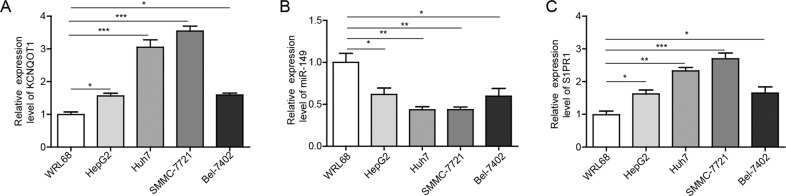
A qRT-PCR assay was performed to determine the mRNA expression of KCNQ1OT1, miR-149 and S1PR1 in normal liver cells and HCC cells. As shown in Fig. 1, the KCNQ1OT1 and S1PR1 transcription in HCC cells was higher than that in WRL68 cell line. Among the four HCC cells, the expression changes in SMMC-7721 and Huh7 cells are more obvious. In contrast, the level of miR-149 was lower in Bel-7402, HepG2, SMMC-7721 and Huh7 cells than in WRL68 cells. Moreover, the change in expression levels in SMMC-7721 and Huh7 cells was the most obvious.
Fig. 1. The expression levels of KCNQ1OT1, miR-149 and S1PR1 in normal human hepatocytes and hepatocellular carcinoma (HCC) cells.
a The normal human hepatic cell line WRL68 and the human HCC cell lines Bel-7402, HepG2, SMMC-7721 and Huh7 were used for qRT-PCR analysis. b qRT-PCR analysis of the expression of miR-149 was detected in HCC cell lines and the WRL68 cell line. c qRT-PCR analysis of S1PR1 expression was detected in HCC cell lines and the WRL68 cell line. *P < 0.05, **P < 0.01, ***P < 0.001.
The interactions between lncRNA KCNQ1OT1 and miR-149, miR-149 and S1PR1
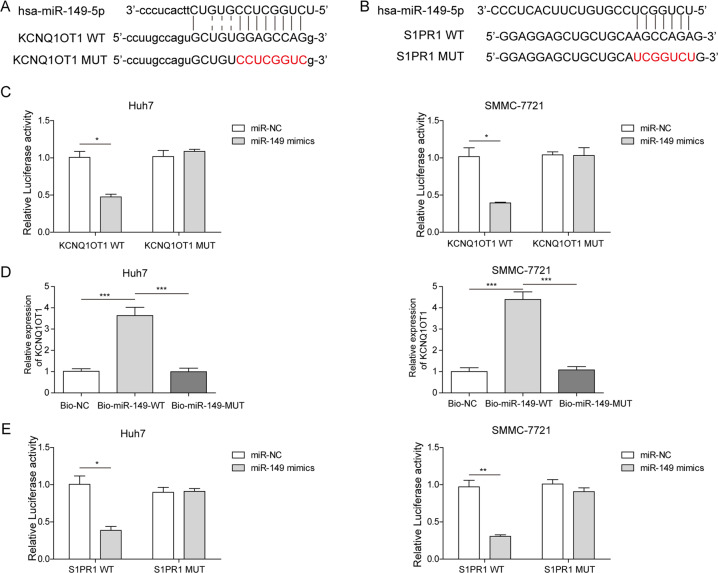
As shown in Fig. 2a, the region of miR-149 was identified to have putative binding sites for KCNQ1OT1. Using an online database analysis, we predicted that KCNQ1OT1 might act as a competing endogenous RNA to modulate the expression of miR-149. To determine whether miR-149 was regulated by the lncRNA KCNQ1OT1 in HCC, KCNQ1OT1-WT and KCNQ1OT1-MUT fragments were cloned into the psiCHECK-2 vector. A luciferase reporter assay was performed in SMMC-7721 and Huh7 cells. The results showed that the luciferase activity in KCNQ1OT1-WT cells was markedly inhibited by miR-149 mimics (P < 0.05, Fig. 2c). The relationship between KCNQ1OT1 and miR-149 was further confirmed by using the RNA pull-down assay. As shown in Fig. 2d, the level of KCNQ1OT1 in miR-149-WT pulled down pellets was higher than that in miR-149-MUT and negative control pellets. Overall, during the process of HCC, there is a putative interaction between KCNQ1OT1 and miR-149.
Fig. 2. The interactions between lncRNA KCNQ1OT1 and miR-149, miR-149 and S1PR1.
a The interaction region between KCNQ1OT1 and miR-149. b The interaction region between miR-149 and the S1PR1 3’-UTR. c A luciferase reporter assay was performed in SMMC-7721 and Huh7 cells to detect the interaction of KCNQ1OT1 and miR-149. d The relative expression level of KCNQ1OT1 was measured by RNA pull-down assay in extracts of SMMC-7721 and Huh7 cells. e The binding activity of miR-149 to the 3’-UTR of S1PR1 was measured by luciferase reporter assay in SMMC-7721 and Huh7 cells. *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate the potential mechanism of KCNQ1OT1/miR-149 in HCC regulation, the TargetScan, PicTar and MiRanda online algorithms were used to predict the possible binding sites. The results indicated that the 3’-UTR of S1PR1 contained a corresponding binding sequence to miR-149 (Fig. 2b). A luciferase reporter assay was performed in SMMC-7721 and Huh7 cells, and the luciferase activity was significantly inhibited when the S1PR1-WT reporter vector was co-transfected with miR-149 mimics (Fig. 2e). These results suggest that miR-149 binds to the 3’-UTR of S1PR1.
LncRNA KCNQ1OT1 drives HCC cell invasion and migration
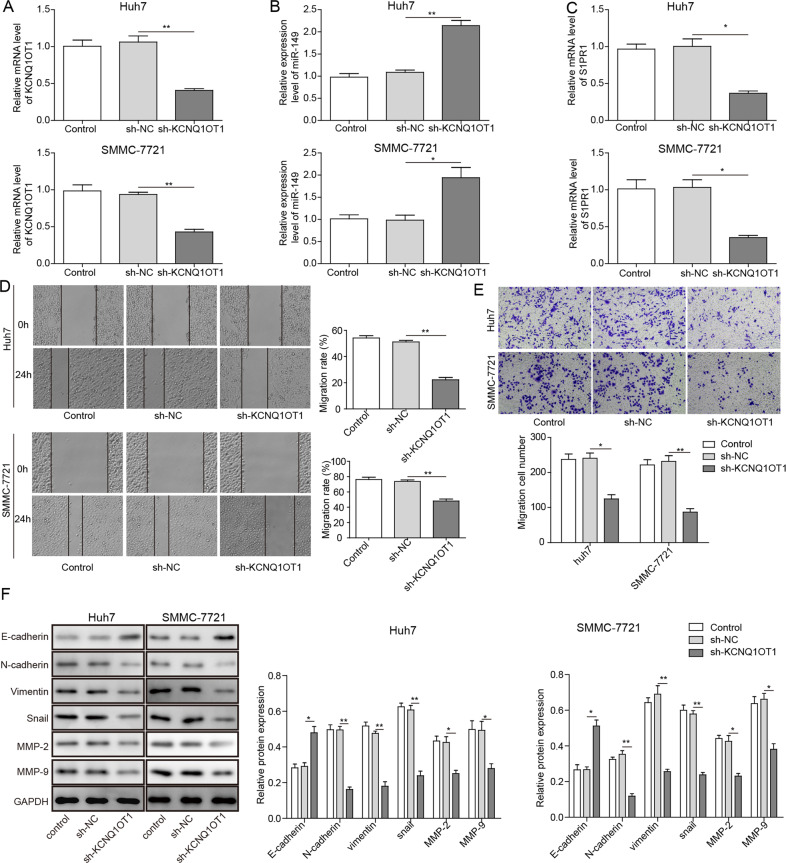
To further explore whether S1PR1 accumulation is affected by KCNQ1OT1 through the suppression of miR-149, qRT-PCR analysis was carried out to determine the expression of S1PR1 and miR-149 in SMMC-7721 and Huh7 cells with KCNQ1OT1 gene knockdown. The expression level of miR-149 was elevated when KCNQ1OT1 was knocked down and that of S1PR1 was downregulated (Fig. 3a–c). As shown in Fig. 3d, the wound healing motility assay indicated that knocking down the KCNQ1OT1 gene significantly reduced the migration activities of SMMC-7721 and Huh7 cells in comparison to those of control cells. Moreover, the Transwell assay revealed that knocking down the KCNQ1OT1 gene markedly reduced the invasion activities of SMMC-7721 and Huh7 cells in comparison to those of control cells (Fig. 3e). The epithelial–mesenchymal transition (EMT) process is highly related to the migration and invasion of malignant tumours, during which E-cadherin can limit peripheral infiltration, along with invasive migration, N-cadherin mainly inhibits cell apoptosis, Vimentin is expressed in mesenchymal tissues and is associated with tumour invasion and metastasis and the expression of Snail is high at the invasive edge of tumour cells. With disease progression, tumour cell invasion is aggravated, which may be reflected by the EMT process with reduced expression of the epithelial marker E-cadherin, as well as enhanced expression of the mesenchymal markers N-cadherin, Vimentin and Snail [19]. This study showed that the protein levels of E-cadherin were increased and those of N-cadherin, Vimentin and Snail were downregulated in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 (Fig. 3f). In addition, the expression of matrix metalloproteinase-2 (MMP-2) and MMP-9, which play vital roles in cell invasion and metastasis of cancer, was analysed. As shown in Fig. 3f, the protein levels of MMP-2 and MMP-9 were reduced in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1. The results demonstrate that KCNQ1OT1 is involved in regulating both the migration and invasion of HCC cells.
Fig. 3. LncRNA KCNQ1OT1 knockdown inhibits the migration and invasion ability of HCC cells.
a–c qRT-PCR analysis of KCNQ1OT1, miR-149 and S1PR1 expression levels in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1. d The wound healing assay was performed to detect the migration activities of SMMC-7721 and Huh7 cells. e The invasion activities of SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 were measured by the Transwell assay. f Western blot analyses of the EMT marker proteins E-cadherin, N-cadherin, Vimentin and Snail, as well as MMP-2 and MMP-9, which play vital roles in cell invasion and metastasis of cancer. *P < 0.05, **P < 0.01.
Overexpression of miR-149 inhibits the invasion and migration of HCC cells
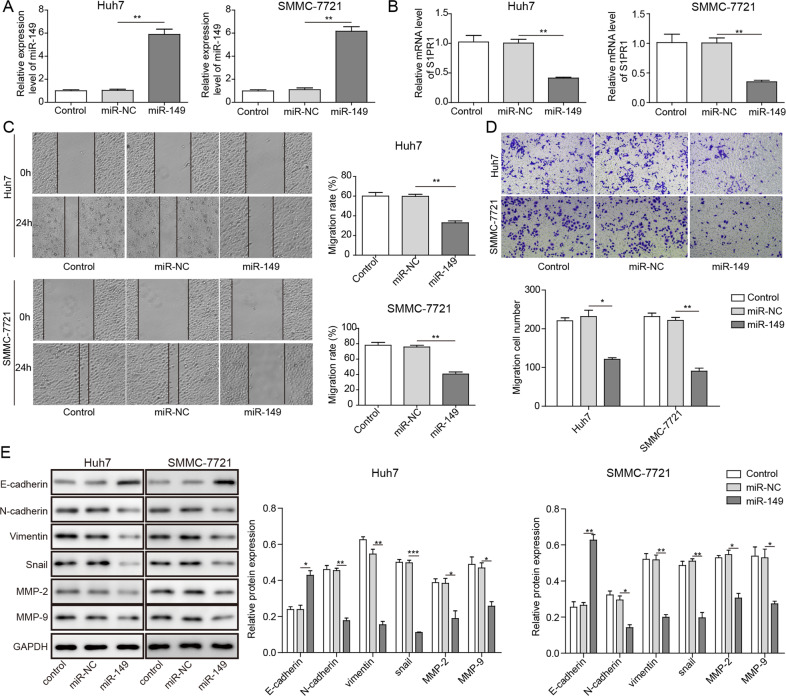
As shown above, the expression levels of miR-149 were downregulated in HCC cells, and miR-149 had binding sites for S1PR1. To confirm whether miR-149 plays a tumour-suppressor role in HCC by regulating the gene expression of S1PR1, we analysed the expression patterns of S1PR1 in SMMC-7721 and Huh7 cells transfected with miR-NC, miR-149 mimics, inhibitor NC and miR-149 inhibitor. In contrast to those in miR-NC-transfected SMMC-7721 and Huh7 cells, reduced S1PR1 transcriptional levels were detected in miR-149-overexpressing cell lines (Fig. 4a, b). Besides, the expression of S1PR1 was upregulated in SMMC-7721 and Huh7 cells transfected with the miR-149 inhibitor (Supplementary Fig. 1A, B). In addition, miR-149 overexpression remarkably reduced the migration and invasion activities of SMMC-7721 and Huh7 cells in comparison to those of the control cells, and the miR-149 inhibitor had the opposite effects (Fig. 4c, d; Supplementary Fig. 1C, D). It was also found that the protein expression levels of E-cadherin were elevated and those of N-cadherin, Vimentin, Snail, MMP-2 and MMP-9 were reduced in SMMC-7721 and Huh7 cells with miR-149 overexpression (Fig. 4e). In contrast, the levels of E-cadherin protein were decreased and those of N-cadherin, Vimentin, Snail, MMP-2 and MMP-9 were increased in SMMC-7721 and Huh7 cells transfected with the miR-149 inhibitor (Supplementary Fig. 1E). Based on these results, it is suggested that the tumour-inhibiting effect of miR-149 in HCC is partially mediated by targeting S1PR1.
Fig. 4. The overexpression of miR-149 inhibits the migration and invasion ability of HCC cells.
a, b The expression of miR-149 and S1PR1 was analysed by qRT-PCR assay in SMMC-7721 and Huh7 cells transfected with miR-149 mimics. c The migration activities of SMMC-7721 and Huh7 cells transfected with miR-149 mimics were determined by wound healing assay. d The invasion activities of SMMC-7721 and Huh7 cells overexpressing miR-149 were measured using the Transwell assay. e Western blot analyses of E-cadherin, N-cadherin, Vimentin, Snail, MMP-2 and MMP-9. *P < 0.05, **P < 0.01, ***P < 0.001.
KCNQ1OT1 regulates HCC tumorigenesis by regulating the PI3K/AKT signalling pathway
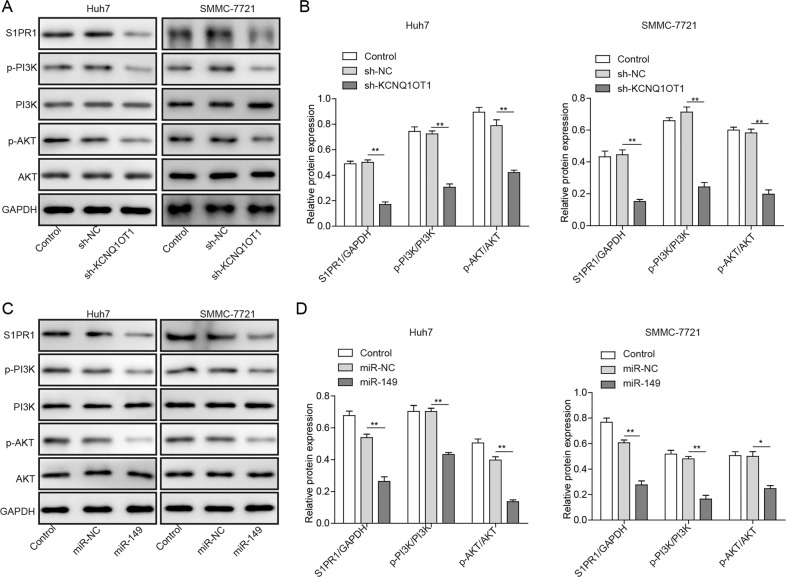
To investigate the underlying mechanisms of KCNQ1OT1/miR-149/S1PR1, the PI3K/AKT signalling pathway, which plays an important regulatory role in the progression of HCC, was evaluated. The expression of downstream S1PR1 protein and the key components in the abovementioned pathway, including PI3K, p-PI3K, AKT and p-AKT, in SMMC-7721 and Huh7 cells with or without KCNQ1OT1 knockdown and miR-149 overexpression was measured. The total levels of PI3K and AKT protein expression in SMMC-7721 and Huh7 cells were not changed and those of S1PR1 were markedly decreased when KCNQ1OT1 was knocked down or miR-149 was overexpressed (Fig. 5a, b). However, the phosphorylated PI3K and AKT protein expression levels were significantly reduced in SMMC-7721 and Huh7 cells with KCNQ1OT1 knockdown or miR-149 overexpression (Fig. 5c, d). These results indicate that KCNQ1OT1 is involved in the regulation of HCC tumorigenesis, possibly through the PI3K/AKT signalling pathway.
Fig. 5. KCNQ1OT1 regulates HCC tumorigenesis by regulating the PI3K/AKT signalling pathway.
a The protein expression of S1PR1, p-PI3K, PI3K, p-AKT and AKT in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 was measured by western blot assay. b Quantification of the results presented in a. c The protein expression of S1PR1, p-PI3K, PI3K, p-AKT and AKT in SMMC-7721 and Huh7 cells overexpressing miR-149 was analysed by western blot assay. d Greyscale analysis of the data in c. *P < 0.05, **P < 0.01.
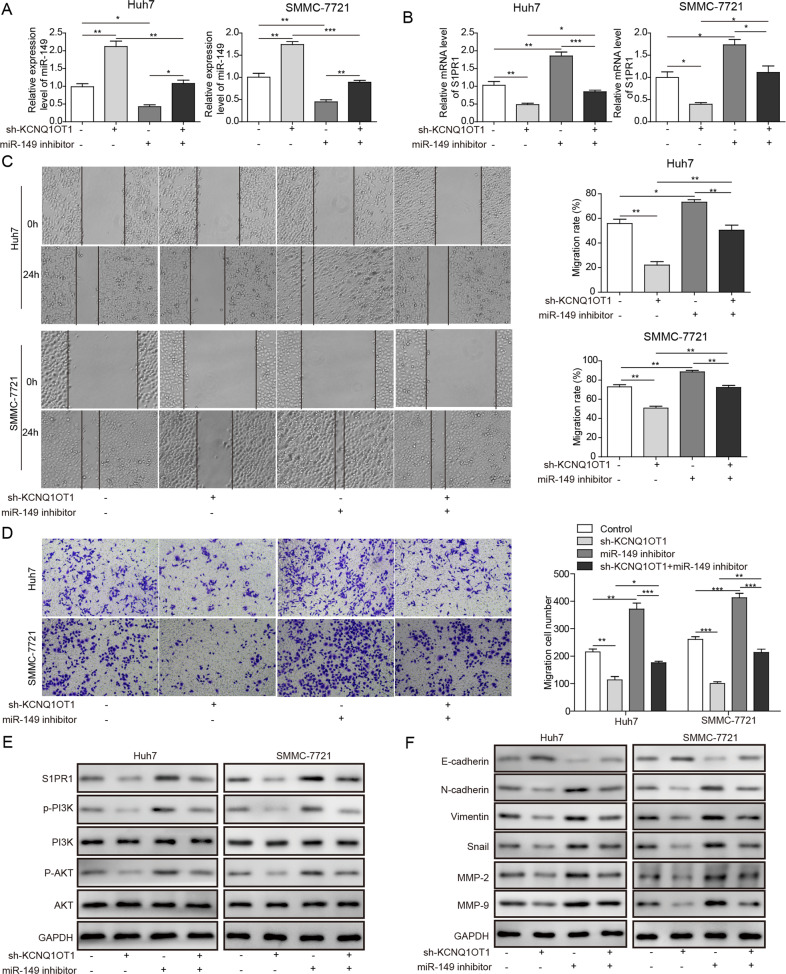
Inhibition of miR-149 abrogates the effect of KCNQ1OT1 knockdown on the invasion and migration abilities of HCC cells
To further explore the cross-talk between KCNQ1OT1 and miR-149 in HCC progression, the mRNA and protein expression of S1PR1 was determined following transfection with sh-KCNQ1OT1, miR-149 inhibitor or co-transfection of sh-KCNQ1OT1 and the miR-149 inhibitor. As expected, the expression levels of miR-149 were attenuated in SMMC-7721 and Huh7 cells co-transfected with sh-KCNQ1OT1 and the miR-149 inhibitor compared with those in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 (Fig. 6a). Moreover, the inhibition of miR-149 increased S1PR1 mRNA and protein levels and weakened the effect of KCNQ1OT1 knockdown on S1PR1 expression (Fig. 6b, e). As demonstrated above, KCNQ1OT1 knockdown attenuated the migration and invasion activities of SMMC-7721 and Huh7 cells, but this effect was partly abrogated by co-transfection with the miR-149 inhibitor (Fig. 6c, d). The migration and invasion activities were significantly increased in SMMC-7721 and Huh7 cells co-transfected with sh-KCNQ1OT1 and the miR-149 inhibitor compared with those in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 (Fig. 6c, d). Similarly, the levels of p-PI3K and p-AKT in SMMC-7721 and Huh7 cells co-transfected with sh-KCNQ1OT1 and the miR-149 inhibitor were higher than those in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 (Fig. 6e). The protein expression of E-cadherin was reduced, while that of N-cadherin, Vimentin, Snail, MMP-2 and MMP-9 was elevated in SMMC-7721 and Huh7 cells co-transfected with sh-KCNQ1OT1 and the miR-149 inhibitor compared with SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 (Fig. 6f). Taken together, these results lead to the conclusion that after sh-KCNQ1OT1 and miR-149 inhibitor co-transfection, the effect of sh-KCNQ1OT1 on the migration and invasion of HCC cells and the expression of related proteins is reversed.
Fig. 6. sh-KCNQ1OT1 was co-transfected with miR-149 inhibitor to reverse the effect of sh-KCNQ1OT1 on the migration and invasion of HCC cells.
a, b The expression of miR-149 and S1PR1 was determined in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 or miR-149 inhibitor or co-transfected with sh-KCNQ1OT1 and miR-149 inhibitor. c The migration activities of SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 or miR-149 inhibitor or co-transfected with sh-KCNQ1OT1 and miR-149 inhibitor were detected by the wound healing assay. d The invasion activities of SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 or miR-149 inhibitor or co-transfected with sh-KCNQ1OT1 and miR-149 inhibitor were detected by the Transwell assay. e Western blot analyses of S1PR1, p-PI3K, PI3K, p-AKT and AKT in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 or miR-149 inhibitor or co-transfected with sh-KCNQ1OT1 and miR-149 inhibitor. f The protein expression of the EMT marker proteins MMP-2 and MMP-9 was detected in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1 or miR-149 inhibitor or co-transfected with sh-KCNQ1OT1 and miR-149 inhibitor. *P < 0.05, **P < 0.01, ***P < 0.001.
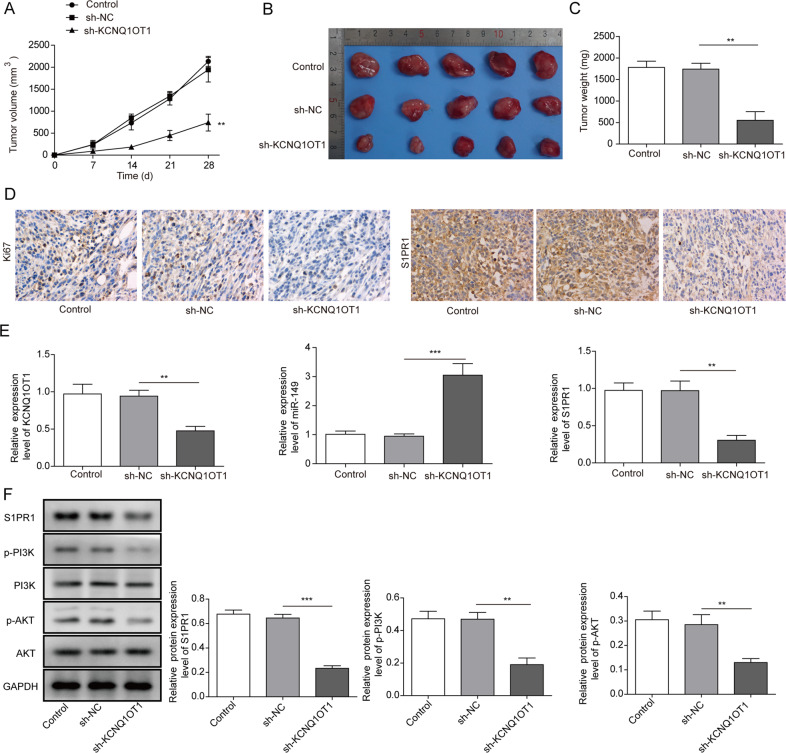
Knockdown of KCNQ1OT1 inhibits tumour growth in vivo
SMMC-7721 cells with KCNQ1OT1 knockdown were implanted into nude mice to determine the tumour growth of HCC. In comparison to the tumour volume in the sh-NC group, the volume in the sh-KCNQ1OT1 group grew more slowly during the 4-week observation (Fig. 7a–c). The expression levels of S1PR1 and the tumour cell proliferation marker Ki67 were decreased in the KCNQ1OT1 knockdown group compared to the negative control group (Fig. 7d). The expression levels of KCNQ1OT1, miR-149 and S1PR1 in tumour tissues of the sh-KCNQ1OT1 and sh-NC groups were detected by qPCR. The results indicated that the KCNQ1OT1 and S1PR1 expression was significantly reduced in KCNQ1OT1 knockdown cells and that the miR-149 expression level was obviously increased (Fig. 7e). Furthermore, the expression levels of S1PR1, p-PI3K, PI3K, p-AKT and AKT proteins in tumour tissues of the sh-KCNQ1OT1 and sh-NC groups were detected by western blot assay. S1PR1, p-PI3K and p-AKT protein levels were decreased in tumour tissues of the sh-KCNQ1OT1 group in comparison to those in the sh-NC group (Fig. 7f). These results suggest that knockdown of KCNQ1OT1 suppresses HCC tumour growth in vivo, downregulates the expression of S1PR1 and inhibits the PI3K/AKT pathway.
Fig. 7. The effect of KCNQ1OT1 knockdown on tumour growth in vivo.
a Tumour volume growth curve. b Tumour image with length scale. c Tumour weight histogram. d The expression of Ki67 and S1PR1 in tumour tissues was measured by immunohistochemistry. e KCNQ1OT1, miR-149 and S1PR1 expression levels in tumour tissues were measured by qPCR. f The protein expression levels of S1PR1, p-PI3K, PI3K, p-AKT and AKT were detected via western blotting. **P < 0.01, ***P < 0.001.
Discussion
HCC is regarded as one of the most common malignancies causing cancer-related death worldwide [20]. Five S1PRs, S1PR1–S1PR5, have been revealed to be involved in the progression and proliferation of a variety of cancers. Among them, S1PR1 has been shown to participate in HCC cell proliferation [15], but the upstream regulatory mechanisms of S1PR1 are still unclear. LncRNAs are a non-protein-coding RNA family that may play a vital role in regulating the cell proliferation of HCC [21, 22]. Previous studies have indicated that the lncRNA KCNQ1OT1 plays a key role in the proliferation and apoptosis of HCC cells and acts as a potential biomarker for the postsurgical surveillance of HCC [9]. miRNAs are a class of small non-coding RNAs with a length of 18–25 nucleotides that function as tumour suppressors or oncogenes during tumour progression [23, 24]. The prognostic value of multiple miRNAs has been shown in a variety of cancers, such as malignant pleural mesothelioma [25], colon cancer [26], ovarian cancer [27], lung cancer [28] and HCC [29–31]. It has been reported that miR-149 potentially acts as an important tumour suppressor by targeting the AKT/mTOR pathway [10]. Here we showed that KCNQ1OT1 and S1PR1 expression was higher in the human HCC cell lines Bel-7402, HepG2, SMMC-7721 and Huh7 than in the normal human hepatic cell line WRL68 (Fig. 1a, c). Conversely, the levels of miR-149 were lower in Bel-7402, HepG2, SMMC-7721 and Huh7 cells than in WRL68 cells (Fig. 1b). It was confirmed here that the transcriptional levels of KCNQ1OT1 and S1PR1 were upregulated in HCC cells and those of miR-149 were downregulated.
LncRNAs have been reported to sponge miRNA expression partially by interacting with the binding sites, and they may play a considerable role in tumour development [32]. In this work, KCNQ1OT1 was identified to directly bind to miR-149 by using a dual luciferase reporter assay and RNA pull-down assay (Fig. 2c, d). In addition, knockdown of KCNQ1OT1 upregulated the expression of miR-149 and weakened the migration and invasion abilities of HCC cells (Fig. 3a, b, d, e). The expression of MMP-2 and MMP-9, which play vital roles in cell invasion and metastasis of cancer, was analysed and showed reduced expression in SMMC-7721 and Huh7 cells transfected with sh-KCNQ1OT1. It has been demonstrated that EMT enhances the behaviours of HCC, including metastasis, invasion and vasculogenic mimicry formation [33, 34]. Other studies have shown that lncRNAs play important roles in the epigenetic regulation of EMT-related genes [35, 36]. As demonstrated in a previous study, we hypothesized that KCNQ1OT1 could promote HCC metastasis by regulating EMT. The protein expression levels of the EMT-associated marker E-cadherin were upregulated, while those of the EMT markers N-cadherin, Vimentin and Snail were decreased in KCNQ1OT1 knockdown HCC cells (Fig. 3f). These results indicate that KCNQ1OT1 knockdown exerts tumour-suppressive effects in HCC cells likely by regulating the expression of miR-149. An mRNA can be targeted by several miRNAs via different binding sites. As reported previously, the 3’-UTR of S1PR1 might contain a functional target site for certain miRNAs, such as miR-92a and miR-363 [15, 37, 38]. It was demonstrated here that miR-149 could downregulate S1PR1 expression levels by targeting the 3’-UTR of S1PR1 mRNA in HCC cells (Figs. 2e and 4b, Supplementary Fig. 1B). Moreover, overexpression of miR-149 induced decreased migration and invasion in HCC cells (Fig. 4c, d), while the miR-149 inhibitor had the opposite effects (Supplementary Fig. 1C, D), suggesting that miR-149 might act as a tumour suppressor in HCC cells. As shown in Fig. 4e, enhanced expression of E-cadherin protein and reduced expression of N-cadherin, Vimentin, Snail, MMP-2 and MMP-9 proteins were observed in SMMC-7721 and Huh7 cells with miR-149 overexpression. In contrast, the levels of E-cadherin protein were decreased and those of N-cadherin, Vimentin, Snail, MMP-2 and MMP-9 were increased in SMMC-7721 and Huh7 cells transfected with the miR-149 inhibitor (Supplementary Fig. 1E), providing stronger evidence that miR-149 is involved in inhibiting the behaviours of HCC. Furthermore, the molecular genetic data in this study suggested that KCNQ1OT1 silencing suppressed the expression of S1PR1 in HCC cells (Fig. 3c). A signalling cascade is presented in our analyses whereby by sponging miR-149 KCNQ1OT1 mediates S1PR1 expression, thus regulating the invasion and migration of HCC cells.
Evidence has shown that miR-300 inhibits EMT-mediated HCC cell migration and invasion via the downstream PI3K/AKT signalling pathway [39]. miR-149 has been reported to play a key role in inhibiting tumorigenesis of HCC by mediating the AKT/mTOR signalling pathway [10]. The overexpression of apolipoprotein M can promote cell invasion and proliferation of non-small cell lung carcinoma by upregulating S1PR1 and activating the PI3K/AKT signalling pathway. Here it was found that the phosphorylation levels of PI3K and AKT were reduced in both KCNQ1OT1 knockdown and miR-149-overexpressing HCC cells (Fig. 5). The upregulated expression levels of S1PR1 and enhanced migration and invasion were found in HCC cells with KCNQ1OT1 knockdown combined with miR-149 inhibition in comparison to those in sh-KCNQ1OT1-transfected HCC cells (Fig. 6a–d). In addition, the suppression of PI3K and AKT phosphorylation levels in HCC cells transfected with sh-KCNQ1OT1 was abrogated by miR-149 inhibition (Fig. 6e). In addition, the increased expression of E-cadherin protein and the decreased expression of N-cadherin, Vimentin, Snail, MMP-2 and MMP-9 by KCNQ1OT1 knockdown were also abrogated by miR-149 inhibition (Fig. 6f). Moreover, it was suggested in this study that KCNQ1OT1 knockdown inhibited HCC tumour growth, downregulated the expression of S1PR1 and suppressed the PI3K/AKT pathway (Fig. 7). These findings imply that lncRNA KCNQ1OT1 mediates S1PR1 expression in the regulation of the growth, invasion and migration of HCC cells by sponging miR-149 and activating the PI3K/AKT signalling pathway.
Conclusions
Our study shows the involvement of KCNQ1OT1 in miR-149 inhibition by interacting with each other, while miR-149 specifically binds to sequences in S1PR1 and increases its expression, which in turn regulates the PI3K/AKT signalling pathway. Taken together, these results suggest that the KCNQ1OT1-mediated signalling pathway is responsible for S1PR1 expression to promote the invasion and migration of HCC cells, providing new insight into the signalling cascades that regulate HCC tumorigenesis.
Supplementary information
Acknowledgements
This work was supported by the PhD Start-up Fund of Guangdong Medical University (No.: 2XB17025), Medical Scientific Research Foundation of Guangdong Province of China (No.: B2019024), Project of Traditional Chinese Medicine Bureau of Guangdong Province of China (No.: 20202095) and “Group-type” Special Support Project for Education Talents in Universities (No.: 4SG19045G).
Author contributions
Guarantor of integrity of the entire study and study concepts: A-HF. Study design: J-LC, D-JL, M-YL, LL. Definition of intellectual content: A-HF, D-JL. Literature research: D-JL, X-YL. Clinical studies: D-JL, M-YL. Experimental studies and data acquisition: J-LC, M-YL. Data analysis and statistical analysis: Y-JP. Manuscript preparation: J-LC, D-JL. Manuscript editing and manuscript review: X-JZ, A-HF.
Data availability
All data generated or analysed during this study are included in this published article.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
The informed consent was obtained from the study participants.
Ethics approval and consent to participate
The protocol has been approved by the Ethics Committee of Guangdong Medical University. All patients were informed of the study and signed the written consent.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41417-020-0203-x) contains supplementary material, which is available to authorized users.
References
- 1.Toro A, Ardiri A, Mannino M, Arcerito MC, Mannino G, Palermo F, et al. Effect of pre- and post-treatment α-fetoprotein levels and tumor size on survival of patients with hepatocellular carcinoma treated by resection, transarterial chemoembolization or radiofrequency ablation: a retrospective study. BMC Surg. 2014;14:40. doi: 10.1186/1471-2482-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–76. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 3.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, et al. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–7. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 4.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu B, Cheng XB, Jiang YL, Cheng M, Chen LP, Bao JJ, et al. Silencing of KCNQ1OT1 decreases oxidative stress and pyroptosis of renal tubular epithelial cells. Diabetes Metab Syndr Obes. 2020;13:365–75. doi: 10.2147/DMSO.S225791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren Y, Gao XP, Liang H, Zhang H, Hu CY. LncRNA KCNQ1OT1 contributes to oxygen-glucose-deprivation/reoxygenation-induced injury via sponging miR-9 in cultured neurons to regulate MMP8. Exp Mol Pathol. 2020;112:10435. doi: 10.1016/j.yexmp.2019.104356. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S, Ma H, Zhang D, Xie S, Wang W, Li Q, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9:742. doi: 10.1038/s41419-018-0793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, Miao R, Zhang J, Qu K, Liu C. Long non-coding RNA KCNQ1OT1 mediates the growth of hepatocellular carcinoma by functioning as a competing endogenous RNA of miR-504. Int J Oncol. 2018;52:1603–12. doi: 10.3892/ijo.2018.4313. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Guo X, Xiong L, Yu L, Li Z, Guo Q, et al. Comprehensive analysis of microRNA-regulated protein interaction network reveals the tumor suppressive role of microRNA-149 in human hepatocellular carcinoma via targeting Akt-mTOR pathway. Mol Cancer. 2014;13:253. doi: 10.1186/1476-4598-13-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu XX, Wang M, Xu D, Yang JH, Kang HF, Wang XJ, et al. Quantitative assessment of the association between genetic variants in microRNAs and colorectal cancer risk. Biomed Res Int. 2015;2015:1–10. doi: 10.1155/2015/276410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izquierdo L, Ingelmo-Torres M, Mallofrã C, Lozano JJ, Verhasselt-Crinquette M, Leroy X, et al. Prognostic value of microRNA expression pattern in upper tract urothelial carcinoma. BJU Int. 2014;113:813–21. doi: 10.1111/bju.12551. [DOI] [PubMed] [Google Scholar]
- 13.Xu Q, Liu JW, Yuan Y. Comprehensive assessment of the association between miRNA polymorphisms and gastric cancer risk. Mutat Res Rev Mutat Res. 2015;763:148–60. doi: 10.1016/j.mrrev.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, Mao J, Redfield S, Mo Y, Lage JM, Zhou X. Systemic distribution, subcellular localization and differential expression of sphingosine-1-phosphate receptors in benign and malignant human tissues. Exp Mol Pathol. 2014;97:259–65. doi: 10.1016/j.yexmp.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P, Huang G, Zhao Y, Zhong D, Xu Z, Zeng Y, et al. MicroRNA-363-mediated downregulation of S1PR1 suppresses the proliferation of hepatocellular carcinoma cells. Cell Signal. 2014;26:1347–54. doi: 10.1016/j.cellsig.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Guo X, Xiong L, Kong X, Xu Y, Liu C, et al. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett. 2012;586:4362–70. doi: 10.1016/j.febslet.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 17.Lin ZY, Huang YQ, Zhang YQ, Han ZD, He HC, Ling XH, et al. MicroRNA-224 inhibits progression of human prostate cancer by downregulating TRIB1. Int J Cancer. 2014;135:541–50. doi: 10.1002/ijc.28707. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Zhang Y, Sun XX, Ma X, Chen ZN. MicroRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol Cancer. 2015;14:5. doi: 10.1186/1476-4598-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang R, Zhao C, Wang X, Wang S, Sun X, Tian Y, et al. Resistin-like molecule-β promotes invasion and migration of gastric carcinoma cells. Med Sci Monit. 2016;22:937–42. doi: 10.12659/MSM.895598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen MF, Hou JL, Chutaputti A. Hepatocellular carcinoma in the Asia Pacific region. J Gastroenterol Hepatol. 2010;24:346–53. doi: 10.1111/j.1440-1746.2009.05784.x. [DOI] [PubMed] [Google Scholar]
- 21.Chi HC, Tsai CY, Tsai MM, Yeh CT, Lin KH. Roles of long noncoding RNAs in recurrence and metastasis of radiotherapy-resistant cancer stem cells. Int J Mol Sci. 2017;18:1903. doi: 10.3390/ijms18091903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 24.Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Sci. 2010;101:2309–15. doi: 10.1111/j.1349-7006.2010.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirschner MB, Cheng YY, Armstrong NJ, Lin RCY, Kao SC, Linton A, et al. MiR-score: a novel 6-microRNA signature that predicts survival outcomes in patients with malignant pleural mesothelioma. Mol Oncol. 2015;9:715–26. doi: 10.1016/j.molonc.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, et al. Prognostic and predictive value of a microRNA signature in stage Ii colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 27.Bagnoli M, Canevari S, Califano D, Losito S, Maio MD, Raspagliesi F, et al. Development and validation of a microRNA-based signature (MiROvaR) to predict early relapse or progression of epithelial ovarian cancer: a cohort study. Lancet Oncol. 2016;17:1137–46. doi: 10.1016/S1470-2045(16)30108-5. [DOI] [PubMed] [Google Scholar]
- 28.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–47. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong CC, Wong C, Tung EK, Au SL, Lee JM, Poon RT, et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140:322–31. doi: 10.1053/j.gastro.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor β receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205–17. doi: 10.1002/hep.26315. [DOI] [PubMed] [Google Scholar]
- 32.Qu J, Li M, Zhong W, Hu C. Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int J Clin Exp Med. 2015;8:17110. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao N, Sun H, Sun B, Zhu D, Zhao X, Wang Y, et al. Mir-27a-3p suppresses tumor metastasis and VM by down-regulating VE-cadherin expression and inhibiting EMT: an essential role for Twist-1 in HCC. Sci Rep. 2016;6:23091. doi: 10.1038/srep23091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tania M, Khan MA, Fu J, Tania M, Khan MA, Fu J. Epithelial to mesenchymal transition inducing transcription factors and metastatic cancer. Tumor Biol. 2014;35:7335–42. doi: 10.1007/s13277-014-2163-y. [DOI] [PubMed] [Google Scholar]
- 35.Battistelli C, Cicchini C, Santangelo L, Tramontano A, Grassi L, Gonzalez FJ, et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36:942–55. doi: 10.1038/onc.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang JW, Xie Y, Xu XL, Yin Y, Jiang RQ, Deng L, et al. Bidirectional transcription of Linc00441 and RB1 via H3K27 modification-dependent way promotes hepatocellular carcinoma. Cell Death Dis. 2017;8:e2675. doi: 10.1038/cddis.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ando H, Okamoto A, Yokota M, Shimizu K, Asai T, Dewa T, et al. Development of a miR-92a delivery system for anti-angiogenesis-based cancer therapy. J Gene Med. 2013;15:20–7. doi: 10.1002/jgm.2690. [DOI] [PubMed] [Google Scholar]
- 38.Shigoka M, Tsuchida A, Matsudo T, Nagakawa Y, Saito H, Suzuki Y, et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol Int. 2010;60:351–7. doi: 10.1111/j.1440-1827.2010.02526.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang R, Yu Z, Chen F, Xu H, Shen S, Chen W, et al. Mir-300 regulates the epithelial-mesenchymal transition and invasion of hepatocellular carcinoma by targeting the FAK/PI3K/AKT signaling pathway. Biomed Pharmacother. 2018;103:1632. doi: 10.1016/j.biopha.2018.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article.