Abstract
Merkel cell carcinoma (MCC) is an aggressive skin tumor with neuroendocrine differentiation, mainly affecting elderly population or immunocompromised individuals. As methylation of the human telomerase reverse transcriptase (mhTERT) has been shown to be a prognostic factor in different tumors, we investigated its role in MCC, in particular in intron 4–5 where rs10069690 has been mapped and recognized as a cancer susceptibility locus. DNA methylation analysis of hTERT gene was assessed retrospectively in a cohort of 69 MCC patients from the University of Bologna, University of Turin and University of Insubria. Overall mortality was evaluated with Kaplan-Meier curves and multivariable Royston-Parmar models. High levels of mhTERT (mhTERThigh) (HR = 2.500, p = 0.015) and p63 (HR = 2.659, p = 0.016) were the only two clinico-pathological features significantly associated with a higher overall mortality at the multivariate analysis. We did not find different levels of mhTERT between MCPyV (+) and (−) cases (21 vs 14, p = 0.554); furthermore, mhTERThigh was strongly associated with older age (80.5 vs 72 years, p = 0.026), no angioinvasion (40.7% vs 71.0%, p = 0.015), lower Ki67 (50 vs 70%, p = 0.005), and PD-L1 expressions in both tumor (0 vs 3%, p = 0.021) and immune cells (0 vs 10%, p = 0.002). mhTERT is a frequently involved epigenetic mechanism and a relevant prognostic factor in MCC. In addition, it belongs to the shared oncogenic pathways of MCC (MCPyV and UV-radiations) and it could be crucial, together with other epigenetic and genetic mechanisms as gene amplification, in determining the final levels of hTERT mRNA and telomerase activity in these patients.
Keywords: Merkel cell carcinoma, Merkel cell polyomavirus, Methylation, HTERT, HTERT intron 4–5, Rs10069690, Telomerase
Introduction
Merkel cell carcinoma (MCC) is an aggressive neuroendocrine tumor, predominantly affecting elderly population or immunocompromised individuals [1, 2]. Although MCC is rare, its annual incidence has significantly increased in the last decades, probably due to population aging and the increase of risk factors as chronic sun damage and immunosuppressive therapies [3, 4]. Despite aggressive therapeutic strategies, MCC has a 5-year overall mortality of about 33%, more than twice than that of cutaneous melanoma [4]. Therefore, deep insight into its genetic background could be crucial to improve targeted therapeutic strategies and outcomes of MCC patients. In the last years, several authors investigated the most commonly altered genes and genetic pathways (TP53, RB1, PIK3CA, KIT, PDGFRA, PDCD1, chromosomal abnormalities, miRNAs, and many others) in MCC [5–12]. They found that a number of oncogenes and tumor-suppressor genes, even if not mutated, are abnormally up- or down-regulated in MCC, through different epigenetic mechanisms [5–12]. Telomerase (TERT), an RNA-dependent DNA polymerase, represents one of the crucial steps in the malignant transformation of several tumors, stabilizing the telomere length and immortalizing cancer cells [9, 13–15]. The functional catalytic subunit of TERT (human telomerase reverse transcriptase-hTERT), is up-regulated through multiple genetic and epigenetic mechanisms, including hTERT promoter mutations, amplifications, promoter methylation, alternative splicing, and many others [9, 13–15]. Although hTERT has a well-established pathogenetic and prognostic role in several tumors, it has been poorly investigated in MCC and little is known about its function in this neoplasia [9, 16, 17]. Xie H. et al. showed that there is a widespread hTERT mRNA expression in MCC and that higher hTERT mRNA levels are associated with a significantly shorter overall survival [9]. Nevertheless, they only investigated hTERT promoter mutations and amplifications and, as clarified by the same authors, the complex genetic and epigenetic regulation system of hTERT expression in MCC is still largely unknown [9]. Genome-wide association studies (GWAS) have recently mapped risk alleles for at least 10 cancer types in a region of chromosome 5 (5p15.33), harboring hTERT gene [18]. Allele-specific effects on DNA methylation were seen in this region identifying the single-nucleotide polymorphism (SNP) biomarker rs10069690 as a possible effector on the methylation and subsequently on gene expression [18, 19]. In this work, we decided to evaluate epigenetic modifications with regard to DNA methylation in this precise region not located on the promoter but within the intron (mhTERT) [18, 19]. We previously characterized DNA methylation alterations in this locus in a large cohort of oral squamous cell carcinoma, identifying the most correlated CpG located at Chr5:1279643-1279644, with an area under the curve (AUC) of 0.92 [18, 19]. Since mhTERT could be a relevant epigenetic mechanism in hTERT expression and an independent prognostic factor in several tumors, we investigated whether mhTERT is associated with overall mortality and with the most relevant clinical-pathological features in a series of MCC patients [9, 13–32].
Materials and Methods
Ethics Statement
All clinical investigations were conducted according to the principles of the Declaration of Helsinki. The study was approved by the Institutional Review Board and the local ethics committee (study number CE18083, DIBINEM-UNIBO-rif.CE AVEC number 377/2018/OSS/AUSLBO). All information regarding the human material used in this study was managed using anonymous numerical codes.
Patients and Specimens
This is a multicentric study enrolling 69 subjects from three different Italian Universities: University of Turin (31 cases), University of Bologna (31 cases), and University of Insubria (7 cases). This cohort has been previously published by our group in a study on mPDCD1, and the follow-up has been updated [8]. All tumors were re-staged according to the 8th edition of the AJCC Cancer Staging Manual [4]. Tissues were fixed in 10% formalin, embedded in paraffin, and stained with hematoxylin and eosin (H&E). Slides from each case were reviewed by five pathologists (S.A., S.U., S.L.R., F.M., C.R.) to confirm the diagnosis and restage the tumors, to assess pathological parameters, and to choose one representative paraffin block for additional analyses. We evaluated the immune cells (ICs) rather than the tumor-infiltrating lymphocytes (TILs), as previously reported [33–35].
Immunohistochemistry
Immunohistochemistry (IHC) was performed on archival, formalin-fixed, paraffin-embedded tissues, using the following antibodies: PD-L1 (clone 123 C3SP142, Ventana-Diapath, dilution 1:50), CD3 (clone 2GV6, Ventana-Diapath, dilution RTU), Cytokeratin 20 (clone SP33, Ventana, dilution RTU), Chromogranin (clone LK2H10, Ventana, dilution RTU), TTF-1 (clone 847G3/1, Ventana, dilution RTU), and Ki67 (clone MIB-1, Dako, dilution 1:100). PD-L1 positivity was assessed on ICs and tumor cells (TCs), as previously described by Lipson et al. [34].
DNA Mutation Analysis of hTERT Promoter
DNA Purification
DNA from macrodissected tumor tissue was digested at 56 °C for 3 h to over-night using the Quick Extract™ FFPE DNA extraction kit (Epicentre, Madison, WI, USA). A denaturation step at 95 °C for 5 min, followed by centrifugation at 10,000×g at 4 °C for 5 min, was adopted to pellet the undigested tissue and solidify the paraffin afloat. DNA was collected from the interphase and stored at 4 °C. hTERT promoter mutation analysis was performed by next-generation sequencing (NGS) using a two-step PCR protocol, as previously described [36]. The region of interest spanned the two most common mutations including C228T (c.-124C>T) and C250T (c.-146C>T) (Chr5: 1295035-1295199 keeping hg38 as a reference genome version). Each NGS experiment was designed to allocate at least 2 k reads/region aimed to have a depth of coverage of at least 2000×. Reads were mapped in a Galaxy Project environment to the hg38 human reference genome with Bowtie2, GATK local realignment, HaplotypeCaller, and Picard MarkDuplicates [37]. Mutations were visualized using BAM files loaded onto the Integrative Genomic Viewer (IGV); only mutations with a variant allele frequency (VAF) threshold > 5% were reported.
Quantitative DNA Methylation Analysis of hTERT (mhTERT)
Bisulfite DNA Treatment
Bisulfite treatment of genomic DNA was carried out with the EZDNA Methylation-Lightning™ Kit (Zymo Research, cod. D5031) according to the manufacturer’s protocol.
Bisulfite Next-Generation Sequencing analysis
Quantitative DNA methylation analysis was performed by bisulfite-next-generation sequencing as previously described [19]. The library was prepared in two steps: a first multiplex PCR amplification for target enrichment and a second round of amplification for barcoding which was performed using the Nextera™ index kit. MethPrimer (http://www.urogene.org/cgi-in/methprimer/methprimer.cgi) was applied to identify CpGs and the best primers of choice for hTERT [38]. Three regions of interest were evaluated: the promoter region A as indicated previously by Svahn et al. with the following mapping coordinates: Chr5: 1295506-1295709, the promoter region B with mapping coordinates Chr5: 1295027-1295220, and an additional region previously described by Morandi et al. spanning from Chr5: 1.279.604 to 1.279.759 (intron-4–5) (keeping hg38 as a reference) (mhTERT) [19, 39]. Six different CpGs are located in this region: 1279732, 1279723, 1279714, 1279660, 1279643, 1279631. Amplicons were purified using MagSi-NGSPREP beads (Magnamedics, Geleen, The Netherlands), quantified with Fluorometer Quantus™ (Promega, Madison, WI, USA), and then pooled and loaded on Illumina MiSEQ (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol.
Bisulfite Sequencing Data Analysis
Each NGS experiment was designed to allocate at least 2 k reads/region, in order to have a depth of coverage of about 2000× to allow a good estimate of DNA methylation level. The methylation ratio of each CpG was calculated in a Galaxy Project environment (Europe) using BWA-meth for mapping and generating the bam file followed by the MethylDackel tool [37]. In parallel, DNA methylation level and quality control of various FASTQ files were evaluated using the web-tool EPIC-TABSAT [40].
Statistical Analysis
hTERT methylation levels (mhTERT) have been dichotomized according to the obtained median value (0.97) chosen as threshold (Fig. 1). In brief, the tumor was stratified in mhTERThigh if the methylation levels of the CpGs mapped on Chr5:1279643-1279642, Chr5:1279660-1279659, Chr5:1279714-1279713, Chr5:1279723-1279722 were all above the cut-off threshold (0.97); on the contrary, mhTERTlow was assigned if even just one was below the threshold (range of methylation = 0.33–1.000). Continuous variables were dichotomized according to sensible cutoffs in order to be used for survival analysis. Patients have been aggregated in two groups for age ≥ or < 75 years (corresponding to the population mean age), tumor size ≥ 2 cm, mitosis ≥ 10/high-power field (HPF), Ki67 ≥ 50%, tumor thickness ≥ 10 mm. Because Ki67 dichotomized according to cut-off of < 55%/≥ 55% was not an independent predictor of survival at the multivariate analysis as recently reported by our group, we adopted the cut-off of < 50%/≥ 50% according to our previous data [41–43]. Stage was dichotomized into two classes, I-II (low stages) and III-IV (high stages). Patients’ characteristics were compared across mhTERT status using chi-square or Mann-Whitney test. Overall survival (OS) was evaluated by means of incidence rate ratios and survival analysis using Kaplan-Meier curves and multivariable Royston-Parmar (RP) models. These models have the advantage, over the more traditional Cox regression approach, to allow a greater flexibility in the specification of the baseline log cumulative survival function, which provides a better fit to the data and, as a consequence, an improvement of model accuracy [44, 45]. RP models use restricted cubic splines with an appropriate number of knots to fit even complex mathematical functions that best represent the survival function inherent to the data [44, 45]. We selected the most appropriate functional form among proportional hazards models, proportional odds models, and probit models with 1 to 5 evenly spaced knots, based on the lowest values of the Akaike information criterion (AIC) and Bayesian information criterion (BIC) statistics, which denote the better fit to the data. After identifying the survival function, the multivariable fractional polynomial (MFP) procedure was used to select the variables influencing OS from a set of initial candidate predictors. MFP automatically performs backward selection by removing the weakly significant predictors and identifies the most appropriate functional form of the relationships between the outcome and the continuous covariates, thus improving model accuracy [46]. The initial model was estimated using mhTERT status as main exposure and age, gender, tumor size, tumor thickness, angioinvasion, p63, MCPyV, clinical stage III-IV, PD-L1 in TCs ≥ 5%, and infiltrative growth as covariates and obtaining robust standard errors for patients clustering into centers. In the backward selection process of MFP, mhTERT was imposed to be retained in the model, as well as covariates with p < 0.100. This confidence level was chosen in order to keep in the final model the variables that may not reach a p value lower than the standard 0.05 confidence level only because of the small sample size, although showing a remarkable effect on OS. Lastly, the model-adjusted survival curves of mhTERT and the retained clinical-pathological parameters were estimated. All analyses have been carried out using Stata v.15.1, specifically the stpm2 procedure was used to estimate RP models [47].
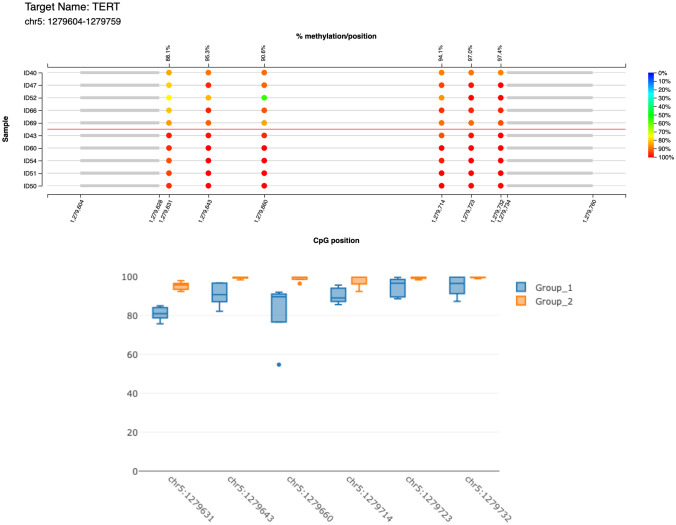
Fig. 1.
The lollipop graph displays the percent methylation per CpG position for 10 randomly selected samples, 5 mhTERThigh cases, and 5 mhTERTlow ones, respectively. The color of the circles represents the percent methylation as shown in the color legend on the right side of the plot. On the lower x-axis, the genomic positions of the CpGs are displayed, the upper x-axis shows the summarized average percent methylation for this CpG position
Results
Patients Characteristics
The clinico-pathological features of the cohort have already been described in a previous study [8]. In brief, the study cohort included a total of 69 patients, 38 (55.1%) males, and 31 (44.9%) females, with a median age of 77 years. According to the 8th edition of the AJCC Cancer Staging Manual, localized disease (AJCC Clinical Stage I-II) was present in 45 (65.2%) and advanced (AJCC Clinical Stage III-IV) in 24 (34.8%) patients, with only two (2.9%) patients displaying distant metastases (AJCC Clinical Stage IV) [4]. Pathological stage was available only for 28 patients (41%) because only after 2010, the SLNB has been recommended in all patients with localized disease [4, 48]. Patients were followed up for a maximum time of 192 months. At the end of follow-up, 39 (56.5%) patients died, 35 (50.7%) due to MCC (DOD) and 4 (5.8%) of other causes (DOC). The median follow-up time was 24 months (range 2–150) for DOD, 17 months (range 14–60) for DOC, and 46.5 months (range 6–192) for patients still alive at follow-up. At the end of follow-up, 9 patients (13.0%) were alive with disease progression (AWD) and 21 ones (30.4%) had no evidence of disease (NED).
hTERT Promoter Mutation Analysis
Only three cases showed the hTERT promoter mutation C250T (c.-146C>T) with a variant allele frequency below 7% for all.
DNA Methylation of Intron 4–5 (mhTERT) and Correlation with Clinico-pathological Features
The study population was composed by 30 (43.5%) patients with mhTERThigh and 39 (56.5%) patients with mhTERTlow, whose clinical-pathological features are summarized in Table 1. Patients with mhTERThigh differed significantly for an older median age (80.5 vs 72 years, p = 0.026), but a lower angioinvasion (40.7% vs 71.0%, p = 0.015), Ki67 expression (50 vs 70%, p = 0.005) and PD-L1 expressions in both TCs (0 vs 3%, p = 0.021 as continuous variable; 7 vs 17, p = 0.080 with cut-off of 5%), and ICs (0 vs 10%, p = 0.002 as continuous variable; 7 vs 23, p = 0.006 with cut-off of 5%). There was no difference in the distribution of mhTERT levels between MCPyV (+) and (−) cases (21 vs 14, p = 0.554). Among the 39 patients with a fatal outcome, 22 (56.4%) were in the mhTERThigh group and 17 (43.6%) in the mhTERTlow one. The mortality incidence ratio of mhTERThigh/mhTERTlow was 1.648 (95% CI: 0.836–3.305), suggesting higher, although not statistically significant, mortality in mhTERThigh group. The Kaplan-Meier survival analysis confirmed this result (log-rank test: t = 2.76, p = 0.097; Fig. 2) and showed that the curves diverge about 12 months after the surgery. Other clinical-pathological features associated to a higher mortality risk in the bivariate analysis were clinical stage III-IV (log-rank test: t = 4.16, p = 0.041), p63 (log-rank test: t = 13.12, p < 0.001), and absence of MCPyV (log-rank test: t = 11.62, p = 0.001). In the final multivariable RP model, mhTERThigh was confirmed as associated to a lower OS (HR = 2.500, p = 0.015) also after adjusting for p63 (HR = 2.659, p = 0.016) absence of MCPyV (HR = 0.478, p = 0.056) and angioinvasion (HR = 2.168, p = 0.060), while all the other covariates were removed because they did not show a remarkable influence on OS (p > 0.100) (Table 2 and Fig. 3).
Table 1.
Clinical-pathological data of the study population, divided in mhTERTlow and mhTERThigh groups
| n | Study population, n (% or range) | mhTERTlow, n (% or range) | mhTERThigh, n (% or range) | χ2 test; p | |
|---|---|---|---|---|---|
| Study population | 69 | 69(100.0) | 39(56.5) | 30(43.5) | |
| Age, median(range) | 69 | 77(41–95) | 72(41–95) | 80.5(59–89) | − 2.22; 0.026a |
| Age ≥ 75 years | 69 | 41(59.4) | 17(43.6) | 24(80.0) | 9.32; 0.002 |
| Males | 69 | 38(55.1) | 22(56.4) | 16(53.3) | 0.06; 0.799 |
| Size, median(range) | 65 | 2.5(0.3–8.5) | 2.5(0.3–6.5) | 2.4(0.6–8.5) | 0.440; 0.660a |
| Size ≥ 2 cm | 69 | 38(55.1) | 23(59.0) | 15(50.0) | 0.55; 0.458 |
| Clinical stage III-IV | 69 | 24(34.8) | 13(33.3) | 11(36.7) | 0.08; 0.773 |
| Tumor thickness, mm | 65 | 12(1–45) | 12.5(1–22) | 10(1–45) | 1.13; 0.257a |
| Tumor thickness ≥ 10 mm | 65 | 37(56.9) | 24(63.2) | 13(48.1) | 1.45; 0.228 |
| Angioinvasion | 65 | 38(58.5) | 27(71.0) | 11(40.7) | 5.97; 0.015 |
| Mitosis ≥ 10/HPF | 69 | 40(58.0) | 21(53.8) | 19(63.3) | 0.63; 0.429 |
| p63-IHC | 69 | 13(18.8) | 7(17.9) | 6(20.0) | 0.05; 0.829 |
| MCPyV | 69 | 35(50.7) | 21(53.8) | 14(46.7) | 0.35; 0.554 |
| Ki67, median(range) | 69 | 60(10–100) | 70(30–100) | 50(10–95) | 2.83; 0.005a |
| Ki67 ≥ 50% | 69 | 42(60.9) | 28(71.8) | 14(46.7) | 4.50; 0.034 |
| ICs | 65 | 34(52.3) | 23(60.5) | 11(40.7) | 2.48; 0.116 |
| Infiltrative growth | 65 | 22(33.8) | 16(42.1) | 6(22.2) | 2.79; 0.095 |
| PD-L1 ICs | 65 | 3(0–70) | 10(0–70) | 0(0–50) | 3.11; 0.002a |
| PD-L1 TCs | 69 | 1(0–30) | 3(0–30) | 0(0–20) | 2.31; 0.021a |
| Site | 69 | 1.36; 0.508 | |||
| Head and neck | 26(37.7) | 14(35.9) | 12(40.0) | ||
| Extremities | 28(40.6) | 18(46.2) | 10(33.3) | ||
| Trunk | 15(21.7) | 7(17.9) | 8(26.7) |
n number of patients for which clinical-pathological features were present and/or evaluated, ICs immune cells, TCs tumor cells, HPF high-power filed, p63 protein p63, IHC immunohistochemistry, MCPyV Merkel cell polyomavirus, Ki67 proliferation index Ki67/MIB1, PD-L1 programmed death-ligand 1, mhTERTlow low level of mhTERT, mhTERThigh high level of mhTERT
aMann-Whitney test
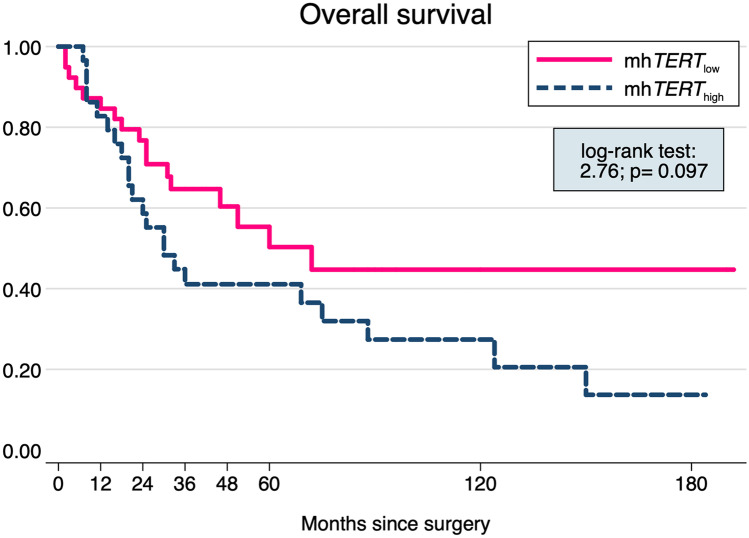
Fig. 2.
Kaplan-Meier survival curve shows a higher mortality in mhTERThigh group (log-rank test: t = 2.76, p = 0.097), with the curves diverging about 12 months after the surgery
Table 2.
Results of the multivariable Royston-Parmar model (n = 65)
| HR | 95% CI | p value | |
|---|---|---|---|
| mhTERThigh | 2.500 | 1.193–5.236 | 0.015 |
| Angioinvasion | 2.168 | 0.966–4.863 | 0.060 |
| p63-IHC | 2.659 | 1.203–5.877 | 0.016 |
| MCPyV | 0.478 | 0.224–1.020 | 0.056 |
CI confidence interval, HR hazard ratio, mhTERThigh high level of mhTERT, p63 protein p63, IHC immunohistochemistry, MCPyV Merkel cell polyomavirus
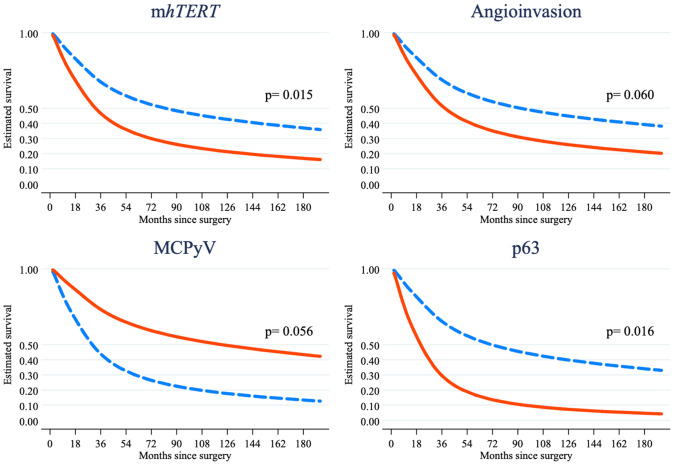
Fig. 3.
In the final multivariable RP model, mhTERThigh (p = 0.015) and p63 (p = 0.016) strongly influenced overall survival, whit borderline-significant associations for angioinvasion (p = 0.060) and absence of MCPyV (p = 0.056). Blue-dashed line: mhTERTlow or absence of examined parameter (p63, angioinvasion and MCPyV); red line: mhTERThigh or presence of examined parameter (p63, angioinvasion and MCPyV)
hTERT Promoter Methylation
DNA promoter methylation for regions A and B as previously described was available in only 13 cases [39]. This was due for overfixation problems, bisulfite treatment and for the amplicon length, which was higher than for intron 4–5 locus. Region A, spanning from Chr5: 1295506 to 1295709, revealed full methylation in all cases except for two, both showing mhTERTlow. Region B spanning from Chr5: 1295027 to 1295220 was found to be hypomethylated (ranged from 50 to 0% methylation) in all cases. Due to lack of data for most of cases, additional statistical analysis was not performed.
Discussion
Three main findings emerge from this study on mhTERT in MCC:
mhTERT is an epigenetic mechanism frequently activated in MCC, in particular in a locus mapped on intron 4–5 around the SNP variant rs10069690, which was previously described as correlated with several cancer types by genome-wide association studies [18];
mhTERT is an epigenetic mechanism involved in both MCPyV (+) and (−) MCC, but had a different distribution when compared with specific clinical-pathological features (age, angioinvasion, Ki67, and PD-L1 expression);
mhTERThigh was related to a higher mortality risk when adjusted for the main clinico-pathological features.
hTERT is the functional subunit of the enzyme TERT and it is crucial to erase senescence and immortalize tumor cells [13–15]. Numerous epigenetic mechanisms are involved in hTERT expression, and among these, one of the most studied is its promoter methylation [13–15]. For the majority of the genes, hypo-methylation of the promoter is fundamental to permit an active transcription [13–15]. Nevertheless, several studies showed that hypermethylation of hTERT was paradoxically associated with higher hTERT mRNA levels and telomerase activity depending on the region of interest investigated [13–32]. Although several mechanisms cooperate in determining the final telomerase activity, promoter methylation proved to be one of the most relevant ones, singly affecting survival, recurrence, and progression in pediatric brain tumors, meningiomas, head and neck squamous cell carcinoma, leukemias, prostatic adenocarcinoma, and several other tumors [17–32]. In our series, we investigated not only the promoter but also a region of intron 4–5, where the SNP rs10069690 has been mapped (mhTERT) [18, 19]. This locus was previously associated with estrogen- and progesterone receptor-negative breast cancer, glioma, prostate, testicular germ cell, pancreas, and urinary bladder [18, 19]. It is becoming clear that regions with variable methylation tend to be mapped also in the gene body, overlapping known regulatory elements enriched for disease-associated SNPs [18, 19, 49]. These results are promising from a therapeutic point of view, for the availability of therapies inducing demethylation and repression of hTERT expression, as just shown in carcinoma cell lines and other in vitro studies with the demethylating agent [50, 51]. This is the first study that investigated frequency, association with clinical-pathological features, and prognostic relevance of mhTERT within the gene body (intron 4–5) in MCC patients. Xie et al. showed that hTERT expression is widespread in MCC and higher hTERT mRNA levels are associated with a shorter OS [9]. Furthermore, the only two genetic mechanisms analyzed by the authors were promoter mutation (1/6 cell lines and 4/35 MCC cases) and gene amplification (1/6 cell lines and 11/14 MCC cases) [9]. As clarified by the same authors, these two genetic alterations could only partially explain the diffuse hTERT mRNA expression (6/6 cell lines and 41/43) and telomerase activity (6/6 cell lines and 11/11 MCC cases) [9]. Unfortunately, for tissue overfixation of several enrolled cases, we were not able to obtain good quality RNA to monitor hTERT gene expression level, so we could not correlate it with DNA methylation. In line with these findings, other studies showed that hTERT promoter mutation is infrequent in MCC and suggested that other epigenetic mechanisms are probably involved [16, 17]. Our series confirmed these data as only three cases showed the mutation C250T (c.-146C>T) with very low allele frequency below 7%. The first main finding coming out from our study is that mhTERThigh is an epigenetic mechanism frequently involved in MCC (30/69, 43.5%). Although this result needs to be validated in larger case series, it suggests that mhTERT could be one important epigenetic mechanism involved in hTERT regulation, cooperating with other ones to justify the high levels of hTERT mRNA and telomerase activity found in these patients. The second main finding is the prognostic impact of mhTERThigh, as just observed in other tumors [17–32]. In the multivariable model, we found that only mhTERThigh and p63 affected OS, with marginal significance for angioinvasion and absence of MCPyV. In addition, Kaplan-Meier analyses showed that mhTERThigh displays its effect on survival since about 12 months after the surgery. This finding suggests that mhTERThigh could be a middle- and long-term response predictor, as shown for other pathological features [8]. In addition, we found that mhTERThigh was a strong prognostic factor regardless its association with other unfavorable and favorable prognostic factors. Xie et al. found that, despite hTERT mRNA levels were associated with a shorter OS and they were strongly influenced by promoter mutation and gene amplification, these two genetic mechanisms did not affect the prognosis when singly evaluated [9]. This result is in line with what found by other authors in other tumors, where hTERT mRNA levels but not mhTERT promoter methylation affects the prognosis [30, 31]. A possible explanation, provided by the same authors and supported by us, is that mechanisms specifically regulating the levels of hTERT mRNA are differently involved in each tumor [9, 13–30]. This would explain why hTERT mRNA levels affect prognosis in almost all tumors but a singly evaluated mechanism could do it or not [9, 13–32]. Additionally, TERT promoter mutations have been shown to be spatially heterogeneous/sub-clonal in some tumors (follicular thyroid carcinoma, follicular thyroid tumors of uncertain malignant potential and meningiomas) and several genes (MLH1, MGMT, CDKN2B) could exhibit spatially heterogeneous/sub-clonal methylation patterns in different tumors (glioblastoma, breast carcinoma) [52–58]. Although these aspects have never been investigated in MCC, it is not possible to exclude that mhTERT and promoter methylation could be spatially heterogeneous/sub-clonal in this tumor and, as result, our data could be affected by the tissue/block selection. Future studies adopting single-cell sequencing will help to clarify if mhTERT and promoter methylation are heterogeneous/sub-clonal in MCC and if there are small fraction of tumor cells (i.e., tumoral stem cells) which could acquire hTERT demethylation in one allele causing small changes in overall DNA methylation. It is largely accepted that MCPyV and UV-radiations identify two divergent pathogenetic pathways in MCC, with many differences in the genetic landscape [5–8]. MCPyV (−) MCCs have a high mutation burden, frequent mutations in RB1, TP53, genes involved in chromatin remodeling (ASXL1, MLL2, and MLL3), and DNA damage [5–8]. By contrast, MCPyV (+) MCCs have a low mutation burden, reduced expression of H3K27me3, and no mutations in the previous listed genes [5–8]. Nevertheless, in both subgroups, there are involvement of common transcription factors (NFAT, P-CREB, and P-STAT3) and mutations in PI3K (HRAS, KRAS, PIK3CA, PTEN, and TSC1) and Notch pathways genes (Notch1 and Notch2), supporting the presence of shared oncogenic pathways in both MCPyV (+) and (−) MCCs [5]. We did not find different distribution of mhTERT levels between MCPyV (+) and (−) cases, suggesting that mhTERT belong to the shared oncogenic pathways. This hypothesis is supported by the absence of correlation between mhTERT and site (Table 1), with this latter known to be associated with the pathogenetic pathway [trunk: MCPyV (+); head-and-neck and extremities: MCPyV (−)] [3]. According to our findings, Chteinberg E et al. found that DNA methylation age (DNAmAge) is not associated with MCPyV in MCC, as previously described for other viral infections as HPV or EBV [59]. In addition, Xie et al. found that absence of virus was associated with promoter mutation (in line with the higher mutation burden of MCPyV (−) MCC) but none with gene amplification and hTERT mRNA levels, supporting that hTERT could be involved in both pathogenic pathways [9].
Conclusion
In conclusion, our study showed that mhTERT in intron 4–5 in proximity of the SNP rs10069690 strongly affects the prognosis of MCC patients, being involved in both MCPyV (+) and (−) tumors, cooperating with other epigenetic and genetic mechanisms as gene amplification in determining the final levels of hTERT mRNA and telomerase activity. Future studies with larger case series are needed to clarify the relationship between mhTERT, the other genetic and epigenetic mechanisms involved in the hTERT regulation, the pathogenesis (MCPyV and UV-radiations), and prognosis.
Acknowledgements
This work was supported by Ricerca Fondamentale Orientata (RFO) to Prof. Sofia Asioli, University of Bologna, Italy. The funding sources had no involvement/role (in study design, in the collection, analysis, and interpretation of data, in the writing of the report and in the decision to submit the article for publication); see also Funding section at the end of the manuscript.
Author Contribution
All authors contributed to the study conception and design. Laboratory analysis was performed by Luca Morandi. Material preparation, data collection, and analysis were performed by Costantino Ricci, Sofia Asioli, and Dino Gibertoni. The first draft of the manuscript was written by Costantino Ricci and Sofia Asioli and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript (see also Cover Letter and Authorship Statement attached as separate file).
Funding
Open access funding provided by Alma Mater Studiorum - Università di Bologna within the CRUI-CARE Agreement. This work was supported by Ricerca Fondamentale Orientata (RFO) to Prof. Sofia Asioli, University of Bologna, Italy. The funding sources had no involvement/role (in study design, in the collection, analysis and interpretation of data, in the writing of the report, and in the decision to submit the article for publication).
Availability of Data, Material (Data Transparency) and Code
The authors declare transparency and availability of data, material, and code (all data that support the findings of this research is deposited in a public repository).
Declarations
Informed Consent (Consent to Participate, Consent for Publication)
Informed consent (to participate and to publish) was obtained from all individual participants included in the study.
Research Involving Human Participants and/or Animals
All clinical investigations were conducted according to the principles of the Declaration of Helsinki. The study was approved by the Institutional Review Board and the local ethics committee (study number CE18083, DIBINEM-UNIBO-rif.CE AVEC number 377/2018/OSS/AUSLBO).
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Toker C. Trabecular carcinoma of the skin. Arch Dermatol. 1972;105:107–110. doi: 10.1001/archderm.1972.01620040075020. [DOI] [PubMed] [Google Scholar]
- 2.Sibley RK, Rosai J, Foucar E, Dehner LP, Boslet G. Neuroendocrine (Merkel cell) carcinoma of the skin. Am J Surg Pathol. 1980;4:211–221. doi: 10.1097/00000478-198006000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Busam KJ, Walsh N, Woods BA. Merkel cell carcinoma. In: Elder DE, Massi D, Scolyer RA, Willemze R, editors. World Health Organization classification of skin tumors. 4. Lyon: IARC Press; 2018. pp. 48–50. [Google Scholar]
- 4.Amin MB, Edge SB, Greene FL, et al. AJCC cancer staging manual. 8. New York: Springer; 2017. [Google Scholar]
- 5.González-Vela MD, Curiel-Olmo S, Derdak S, et al. Shared oncogenic pathways implicated in both virus-positive and UV-induced Merkel cell carcinomas. J Invest Dermatol. 2017;137:197–206. doi: 10.1016/j.jid.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Carter MD, Gaston D, Huang WY, Greer WL, Pasternak S, Ly TY, Walsh NM. Genetic profiles of different subsets of Merkel cell carcinoma show links between combined and pure MCPyV-negative tumors. Hum Pathol. 2018;71:117–125. doi: 10.1016/j.humpath.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Harms PW, Vats P, Verhaegen ME, et al. The distinctive mutational spectra of polyomavirus-negative Merkel cell carcinoma. Cancer Res. 2015;75:3720–3727. doi: 10.1158/0008-5472.CAN-15-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricci C, Morandi L, Righi A, et al. PD-1 (PDCD1) promoter methylation in Merkel cell carcinoma: prognostic relevance and relationship with clinico-pathological parameters. Mod Pathol. 2019;32:1359–1372. doi: 10.1038/s41379-019-0261-5. [DOI] [PubMed] [Google Scholar]
- 9.Xie H, Liu T, Wang N, et al. TERT promoter mutations and gene amplification: promoting TERT expression in Merkel cell carcinoma. Oncotarget. 2014;5:10048–10057. doi: 10.18632/oncotarget.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andea AA, Patel R, Ponnazhagan S, Kumar S, DeVilliers P, Jhala D, Eltoum IE, Siegal GP. Merkel cell carcinoma: correlation of KIT expression with survival and evaluation of KIT gene mutational status. Hum Pathol. 2010;41:1405–1412. doi: 10.1016/j.humpath.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waltari M, Sihto H, Kukko H, Koljonen V, Sankila R, Böhling T, Joensuu H. Association of Merkel cell polyomavirus infection with tumor p53, KIT, stem cell factor, PDGFR-alpha and survival in Merkel cell carcinoma. Int J Cancer. 2011;129:619–628. doi: 10.1002/ijc.25720. [DOI] [PubMed] [Google Scholar]
- 12.Fan K, Ritter C, Nghiem P, et al. Circulating cell-free miR-375 as surrogate marker of tumor burden in Merkel cell carcinoma. Clin Cancer Res. 2018;24:5873–5882. doi: 10.1158/1078-0432.CCR-18-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leão R, Apolónio JD, Lee D, Figueiredo A, Tabori U, Castelo-Branco P. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: clinical impacts in cancer. J Biomed Sci. 2018;25:22. doi: 10.1186/s12929-018-0422-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avin BA, Umbricht CB, Zeiger Martha A, MA, Human telomerase reverse transcriptase regulation by DNA methylation, transcription factor binding and alternative splicing (review) Int J Oncol. 2016;49:2199–2205. doi: 10.3892/ijo.2016.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Fang X, Ge Z, et al. The telomerase reverse transcriptase (hTERT) gene is a direct target of the histone methyltransferase SMYD3. Cancer Res. 2007;67:2626–2631. doi: 10.1158/0008-5472.CAN-06-4126. [DOI] [PubMed] [Google Scholar]
- 16.Chung HJ, Yang S, Succaria F, Bhawan J. Telomerase (TERT) Promoter mutations are infrequent in Merkel cell carcinomas. J Am Acad Dermatol. 2015;73:864–865. doi: 10.1016/j.jaad.2015.07.030. [DOI] [PubMed] [Google Scholar]
- 17.Zheng X, Zhuge J, Bezerra SM. High frequency of TERT promoter mutation in small cell carcinoma of bladder, but not in small cell carcinoma of other origins. J Hematol Oncol. 2014;7:47. doi: 10.1186/s13045-014-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Zhu B, Zhang M, et al. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the TERT-CLPTM1L region on chromosome 5p15.33. Hum Mol Genet. 2014;23:6616–6633. doi: 10.1093/hmg/ddu363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morandi L, Gissi D, Tarsitano A, et al. CpG location and methylation level are crucial factors for the early detection of oral squamous cell carcinoma in brushing samples using bisulfite sequencing of a 13-gene panel. Clin Epigenetics. 2017;9:85. doi: 10.1186/s13148-017-0386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gigek CO, Leal MF, Silva PN, et al. hTERT methylation and expression in gastric cancer. Biomarkers. 2009;14:630–636. doi: 10.3109/13547500903225912. [DOI] [PubMed] [Google Scholar]
- 21.Choi JH, Park SH, Park J, et al. Site-specific methylation of CpG nucleotides in the hTERT promoter region can control the expression of hTERT during malignant progression of colorectal carcinoma. Biochem Biophys Res Commun. 2007;361:615–620. doi: 10.1016/j.bbrc.2007.07.051. [DOI] [PubMed] [Google Scholar]
- 22.Haraguchi K, Yada N, Sato S, et al. The methylation status and expression of human telomerase reverse transcriptase is significantly high in oral carcinogenesis. APMIS. 2017;125:797–807. doi: 10.1111/apm.12723. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Weng X, Ye J, He L, Zhou D, Liu Y. Promoter hypermethylation of TERT is associated with hepatocellular carcinoma in the Han Chinese population. Clin Res Hepatol Gastroenterol. 2015;39:600–609. doi: 10.1016/j.clinre.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Ruland V, Hartung S, Kordes U, Wolff JE, Paulus W, Hasselblatt M. Methylation of the hTERT promoter is frequent in choroid plexus tumors but not of independent prognostic value. J Neurooncol. 2014;119:215–216. doi: 10.1007/s11060-014-1473-7. [DOI] [PubMed] [Google Scholar]
- 25.Sobecka A, Blaszczak W, Barczak W, et al. hTERT promoter methylation status in peripheral blood leukocytes as a molecular marker of head and neck cancer progression. J Appl Genet. 2018;59:453–461. doi: 10.1007/s13353-018-0458-1. [DOI] [PubMed] [Google Scholar]
- 26.Zhao X, Tian X, Kajigaya S, et al. Epigenetic landscape of the TERT promoter: a potential biomarker for high risk AML/MDS. Br J Haematol. 2016;175:427–439. doi: 10.1111/bjh.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castelo-Branco P, Choufani S, Mack S, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. 2013;14:534–542. doi: 10.1016/S1470-2045(13)70110-4. [DOI] [PubMed] [Google Scholar]
- 28.Fürtjes G, Köchling M, Peetz-Dienhart S, et al. hTERT promoter methylation in meningiomas and central nervous hemangiopericytomas. J Neurooncol. 2016;130:79–87. doi: 10.1007/s11060-016-2226-6. [DOI] [PubMed] [Google Scholar]
- 29.Castelo-Branco P, Leão R, Lipman T, et al. A cancer specific hypermethylation signature of the TERT promoter predicts biochemical relapse in prostate cancer: a retrospective cohort study. Oncotarget. 2016;7:57726–57736. doi: 10.18632/oncotarget.10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Köchling M, Ewelt C, Fürtjes G, et al. hTERT promoter methylation in pituitary adenomas. Brain Tumor Pathol. 2016;33:27–34. doi: 10.1007/s10014-015-0230-8. [DOI] [PubMed] [Google Scholar]
- 31.Kumari A, Srinivasan R, Vasishta RK, Wig JD. Positive regulation of human telomerase reverse transcriptase gene expression and telomerase activity by DNA methylation in pancreatic cancer. Ann Surg Oncol. 2009;16:1051–1059. doi: 10.1245/s10434-009-0333-8. [DOI] [PubMed] [Google Scholar]
- 32.Stögbauer L, Stummer W, Senner V, Brokinkel B. Telomerase activity, TERT expression, hTERT promoter alterations, and alternative lengthening of the telomeres (ALT) in meningiomas - a systematic review. Neurosurg Rev. 2020;43:903–910. doi: 10.1007/s10143-019-01087-3. [DOI] [PubMed] [Google Scholar]
- 33.Mitteldorf C, Berisha A, Tronnier M, Pfaltz MC, Kempf W. PD-1 and PD-L1 in neoplastic cells and the tumor microenvironment of Merkel cell carcinoma. J Cutan Pathol. 2017;44:740–746. doi: 10.1111/cup.12973. [DOI] [PubMed] [Google Scholar]
- 34.Lipson EJ, Vincent JG, Loyo M, et al. PD-L1 expression in the Merkel cell carcinoma microenvironment: association with inflammation, Merkel cell polyomavirus and overall survival. Cancer Immunol Res. 2013;1:54–63. doi: 10.1158/2326-6066.CIR-13-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricci C, Righi A, Ambrosi F, et al. Prognostic impact of MCPyV and TIL subtyping in Merkel cell carcinoma: evidence from a large European cohort of 95 patients. Endocr Pathol. 2020;31:21–32. doi: 10.1007/s12022-019-09601-5. [DOI] [PubMed] [Google Scholar]
- 36.Morandi L, Righi A, Maletta F, et al. Somatic mutation profiling of hobnail variant of papillary thyroid carcinoma. Endocr Relat Cancer. 2017;24:107–117. doi: 10.1530/ERC-16-0546. [DOI] [PubMed] [Google Scholar]
- 37.Afgan E, Baker D, Batut B, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 39.Svahn F, Paulsson JO, Stenman A. TERT promoter hypermethylation is associated with poor prognosis in adrenocortical carcinoma. Int J Mol Med. 2018;42:1675–1683. doi: 10.3892/ijmm.2018.3735. [DOI] [PubMed] [Google Scholar]
- 40.Krainer J, Weinhäusel A, Hanak K, Pulverer W, Özen S, Vierlinger K, Pabinger S. EPIC-TABSAT: analysis tool for targeted bisulfite sequencing experiments and array-based methylation studies. Nucleic Acids Res. 2019;47:W166–W170. doi: 10.1093/nar/gkz398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asioli S, Righi A, Volante M, Eusebi V, Bussolati G. p63 expression as a new prognostic marker in Merkel cell carcinoma. Cancer. 2007;110:640–647. doi: 10.1002/cncr.22828. [DOI] [PubMed] [Google Scholar]
- 42.La Rosa S, Bonzini M, Sciarra A, et al (2020) Exploring the Prognostic Role of Ki67 Proliferative Index in Merkel Cell Carcinoma of the Skin: Clinico-Pathologic Analysis of 84 Cases and Review of the Literature. Endocr Pathol 31(4):392-400. [DOI] [PMC free article] [PubMed]
- 43.Asioli S, Righi A, de Biase D, et al (2011) Expression of p63 is the sole independent marker of aggressiveness in localised (stage I-II) Merkel cell carcinomas. Mod Pathol 24:1451-61. [DOI] [PubMed]
- 44.Ng R, Kornas K, Sutradhar R, Wodchis WP, Rosella LC. The current application of the Royston-Parmar model for prognostic modeling in health research: a scoping review. Diagnostic Progn Res. 2018;2:4. doi: 10.1186/s41512-018-0026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crowther MJ, Lambert PC. A general framework for parametric survival analysis. Stat Med. 2014;33:5280–5297. doi: 10.1002/sim.6300. [DOI] [PubMed] [Google Scholar]
- 46.Sauerbrei W, Perperoglou A, Schmid M, et al. State of the art in selection of variables and functional forms in multivariable analysis-outstanding issues. Diagn Progn Res. 2020;4:3. doi: 10.1186/s41512-020-00074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambert PC, Royston P. Further development of flexible parametric models for survival analysis. Stata J. 2009;9:265–290. doi: 10.1177/1536867X0900900206. [DOI] [Google Scholar]
- 48.Lemos BD, Storer BE, Iyer JG, et al. Pathologic nodal evaluation improves prognostic accuracy in Merkel cell carcinoma: analysis of 5823 cases as the basis of the first consensus staging system. J Am Acad Dermatol. 2010;63:751–761. doi: 10.1016/j.jaad.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grundberg E, Meduri E, Sandling JK, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guilleret I, Benhattar J. Demethylation of the human telomerase catalytic subunit (hTERT) gene promoter reduced hTERT expression and telomerase activity and shortened telomeres. Exp Cell Res. 2003;289:326–334. doi: 10.1016/S0014-4827(03)00281-7. [DOI] [PubMed] [Google Scholar]
- 51.Tao SF, Zhang CS, Guo XL, et al. Anti-tumor effect of 5-aza-2'-deoxycytidine by inhibiting telomerase activity in hepatocellular carcinoma cells. World J Gastroenterol. 2012;18:2334–2343. doi: 10.3748/wjg.v18.i19.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stenman A, Hysek M, Jatta K, et al. TERT promoter mutation spatial heterogeneity in a metastatic follicular thyroid carcinoma: implications for clinical work-up. Endocr Pathol. 2019;30:246–248. doi: 10.1007/s12022-019-09580-7. [DOI] [PubMed] [Google Scholar]
- 53.Hysek M, Jatta K, Hellgren LS, Stenman A, Larsson C, Zedenius J, Juhlin CC. Spatial distribution patterns of clinically relevant TERT promoter mutations in follicular thyroid tumors of uncertain malignant potential (FT-UMPs): advantages of the digital droplet PCR (ddPCR) technique. J Mol Diagn. 2021;23:212–222. doi: 10.1016/j.jmoldx.2020.10.016. [DOI] [PubMed] [Google Scholar]
- 54.Theeler BJ. De novo and secondary anaplastic meningiomas: natural history, prognosis, and the TERT promoter. Neuro Oncol. 2018;20:1009–1010. doi: 10.1093/neuonc/noy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Juratli TA, Thiede C, Koerner MVA, et al. Intratumoral heterogeneity and TERT promoter mutations in progressive/higher-grade meningiomas. Oncotarget. 2017;8:109228–109237. doi: 10.18632/oncotarget.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varley KE, Mutch DG, Edmonston TB, Goodfellow PJ, Mitra RB. Intra-tumor heterogeneity of MLH1 promoter methylation revealed by deep single molecule bisulfite sequencing. Nucleic Acids Res. 2009;37:4603–4612. doi: 10.1093/nar/gkp457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moelans CB, de Groot JS, Pan X, van der Wall E, van Diest PJ. Clonal intratumor heterogeneity of promoter hypermethylation in breast cancer by MS-MLPA. Mod Pathol. 2014;27:869–874. doi: 10.1038/modpathol.2013.207. [DOI] [PubMed] [Google Scholar]
- 58.Wenger A, Ferreyra Vega S, Kling T, Bontell TO, Jakola AS, Carén H. Intratumor DNA methylation heterogeneity in glioblastoma: implications for DNA methylation-based classification. Neuro Oncol. 2019;21:616–627. doi: 10.1093/neuonc/noz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chteinberg E, Vogt J, Kolarova J, et al. The curious case of Merkel cell carcinoma: epigenetic youth and lack of pluripotency. Epigenetics. 2020;15:1319–1324. doi: 10.1080/15592294.2020.1773096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare transparency and availability of data, material, and code (all data that support the findings of this research is deposited in a public repository).