Abstract
Leucocytozoon sabrazesi is the intracellular protozoa of leucocytozoonosis, which is transmitted by the insect vectors and affects chickens in most subtropical and tropical regions of the globe, except South America, and causing enormous economic losses due to decreasing meat yield and egg production. In this study, L. sabrazesi gametocytes have been observed in the blood smears, and molecular methods have been used to analyse the occurrence and genetic diversity of L. sabrazesi in blood samples from 313 chickens raised in northern, western and southern parts of Thailand. The nested polymerase chain reaction (nested PCR) assay based on the cytb gene revealed that 80.51% (252/313) chickens were positive of L. sabrazesi. The phylogenetic analysis indicated that L. sabrazesi cytb gene is conserved in Thailand, showed 2 clades and 2 subclades with similarity ranged from 89.5 to 100%. The diversity analysis showed 13 and 18 haplotypes of the sequences from Thailand and from other countries, respectively. The entropy analyses of nucleic acid sequences showed 26 high entropy peaks with values ranging from 0.24493 to 1.21056, while those of amino acid sequences exhibited 5 high entropy peaks with values ranging from 0.39267 to 0.97012. The results; therefore, indicate a high molecular occurrence of L. sabrazesi in chicken blood samples with the associated factors that is statistically significant (p < 0.05). Hence, our results could be used to improve the immunodiagnostic methods and to find appropriate preventive control strategies or vaccination programs against leucocytozoonosis in order to mitigate or eliminate the harmful impact of this infection on chicken industry.
Subject terms: Parasitology, Environmental sciences
Introduction
Leucocytozoon is a haemosporidian protozoan that belongs to the phylum of Apicomplexa causing leucocytozoonosis in birds, including chickens. This parasite is found in blood cells of avian hosts, and transmitted by simuliid blackflies (Diptera: Simuliidae) or Culicoides midges (Diptera: Ceratopogonidae)1–3. The high pathogenic cases caused by L. sabrazesi exhibit chicken clinical mortality and subclinical decreased egg production with several symptoms including depression, weakness, anorexia, restlessness, anemia, pale comb, green feces and dead4–10. The infected chickens possess mortality rate more than 50% resulting in significant economic losses9. Outbreaks of Leucocytozoon infection have been reported in chicken (Asia and Africa), waterfowl (Asia, Europe and North America), turkeys (North America), and free-living and captive avian species throughout the globe7,11–16. In Thailand, the first case of leucocytozoonosis was reported by Campbell17 known as ‘Bangkok hemorhagic disease’ in chicken. There have been a few reports on Leucocytozoon sp. infections in both domestic chickens (42.86%) and wild birds (16.67–22.22%) in Chiang Mai and Phetchabun provinces, respectively18,19.
Conventional diagnostic method for Leucocytozoon sp. infection is direct microscopic observation of the circulating gametocytes in Giemsa-stained blood smear. In addition, detection of parasites DNA or RNA using polymerase chain reaction (PCR) method utilizing primers derived from parasites mitochondrial cytochrome b (cytb) gene could be more common reliable and widely used in diagnosing the infections particular in laboratory for high sensitivity and specificity even when blood smears are negative in case of low parasitemia or early stage of infection in animals9,12,20–22. Little is known about the prevalence of Leucocytozoon sabrazesi in Thailand. The aim of this study was to further investigate the occurrence and genetic diversity of this parasite in chickens from three regions (northern, western and southern) of Thailand. Likewise, haplotype diversity and entropy analysis among the isolated sequences discriminated in this study and those from other countries are investigated. These results could provide the information on the genetic diversity structure of this parasite’s population for further development of the immunodiagnostic methods and vaccine strategies.
Results
Occurrence of L. sabrazesi infection in chicken blood samples
Of the 313 chicken blood samples were examined under light microscope to observe the intra-erythrocytic L. sabrazesi gametocytes in blood smears (Fig. 1). Two hundred and fifty-two blood samples out of 313 (80.51%) were positive as investigated by the PCR analysis targeting cytb gene of L. sabrazesi (Table 1). The representative PCR products of parasite examined were 248 bp. The highest occurrence of L. sabrazesi infection (116/127; 91.34%) was found in Chiang Rai province, followed by Kanchanaburi province (72/86; 83.72%) and Phatthalung province (64/100; 64%) as shown in Table 1. The association between the occurence of L. sabrazesi infection and the factors, i.e., age (4, 4–12 and > 12 months), management system (backyard in households, free-range farms and non-evaporative cooling houses) and water sources (pond in the farm and river) nearby were considered to be statistically significant (p < 0.05), while the significant different was not found counting upon the chicken’s breed (native and layer), gender (male and female), type of feeds (commercial and natural) and insect control system (Table 2).
Figure 1.
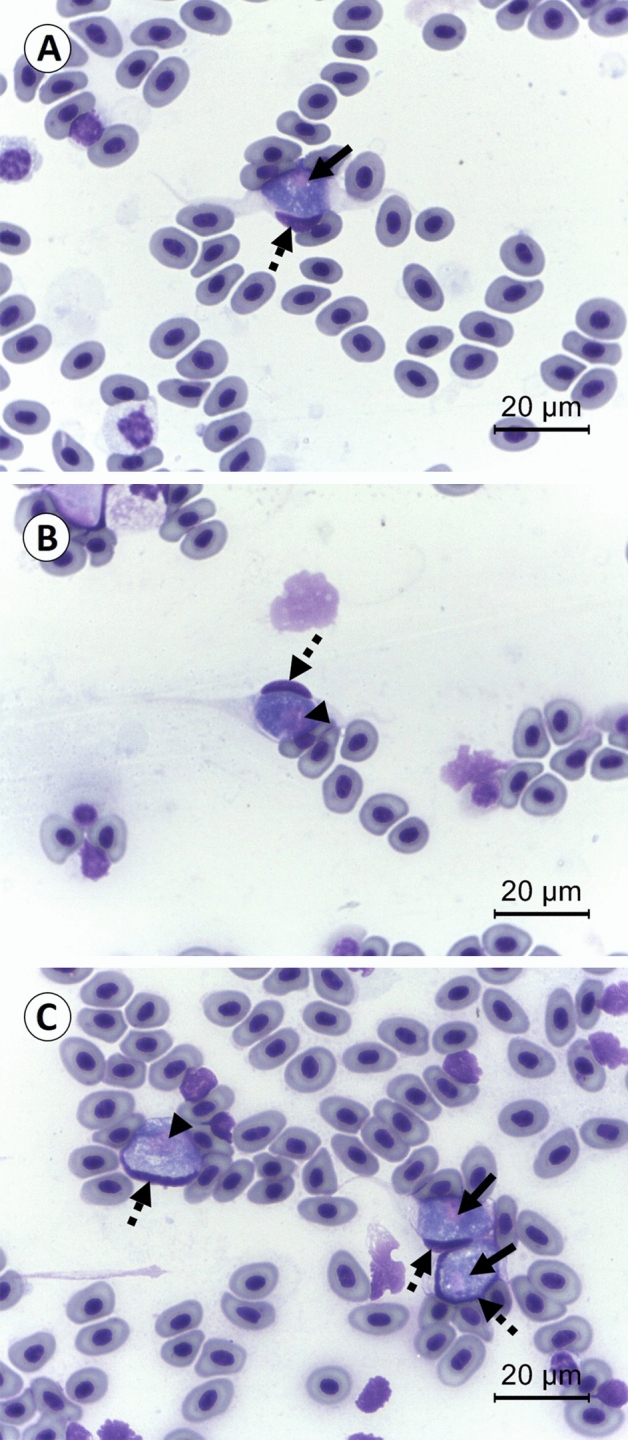
Gametocytes of L. sabrazesi in chicken’s cells identified from Geimsa stained blood smear. The high magnification micrographs showed the elongated macrogametocytes (A), elongated microgametocytes (B) and rounded microgametocyte (the left side) and rounded and elongated macrogametocytes (the right side) (C). The arrows present parasites’ nuclei of macrogametocytes. The arrowheads indicate parasites’ nuclei of microgametocytes. The dashed arrows demonstrate hosts’ nuclei.
Table 1.
Summary of L. sabrazesi infection in chickens from Chiang Rai, Kanchanaburi and Phatthalung provinces analyzed by PCR assay.
| Parameters | Number of examined samples | Number of positive samples | Total (%) |
|---|---|---|---|
| Chiang Rai province | 127 | 116 | 91.34 |
| Mae Suai district | |||
| Kanchanaburi province | 86 | 72 | 83.72 |
| Sai Yok district | |||
| Phatthalung province | 100 | 64 | 64.00 |
| Mueang Phatthalung district | 20 | 8 | 40.00 |
| Srinagarindra district | 16 | 13 | 81.25 |
| Khuan Khanun district | 42 | 31 | 73.81 |
| Kong Ra district | 22 | 12 | 54.55 |
| Total | 313 | 252 | 80.51 |
Table 2.
Factors associated with L. sabrazesi infection of chickens in Chiang Rai, Kanchanaburi and Phatthalung provinces.
| Parameters | Number of examined samples | Number of positive samples | Total (%) | χ2 (df) | p-value |
|---|---|---|---|---|---|
| Breed | 2.7132 (1) | 0 .099518 | |||
| Native | 150 | 115 | 76.67 | ||
| Layer | 163 | 137 | 84.05 | ||
| Total | 313 | 252 | 80.51 | ||
| Age | 53.9465 (1) | < 0.00001 | |||
| 0–4 months | 40 | 38 | 95.00 | ||
| 4–12 months | 182 | 164 | 90.11 | ||
| > 12 months | 91 | 50 | 54.95 | ||
| Total | 313 | 252 | 80.51 | ||
| Gender | 0.0383 (1) | 0.844754 | |||
| Male | 78 | 61 | 78.21 | ||
| Female | 235 | 191 | 81.28 | ||
| Total | 313 | 252 | 80.51 | ||
| Management system | 31.0085 (2) | < 0.00001 | |||
| Backyard in households | 186 | 138 | 74.19 | ||
| Free-range farms | 91 | 86 | 94.51 | ||
| Non-evaporative cooling houses | 36 | 28 | 77.78 | ||
| Total | 313 | 252 | 80.51 | ||
| Type of feeds | 0.6267 (2) | 0.731003 | |||
| Commercial | 36 | 28 | 77.78 | ||
| Natural | 156 | 124 | 79.49 | ||
| Mixed | 121 | 100 | 82.64 | ||
| Total | 313 | 252 | 80.51 | ||
| Water source | 18.4521 (1) | 0.000017 | |||
| Yes | 255 | 217 | 85.10 | ||
| No | 58 | 35 | 60.34 | ||
| Total | 313 | 252 | 80.51 | ||
| Insect control system | 1.2544 (1) | 0.262717 | |||
| Yes | 25 | 18 | 72.00 | ||
| No | 288 | 234 | 81.25 | ||
| Total | 313 | 252 | 80.51 | ||
χ2 = Chi-square; df = degree of freedom.
Phylogenetic and similarity analysis of L. sabrazesi cytb gene sequences
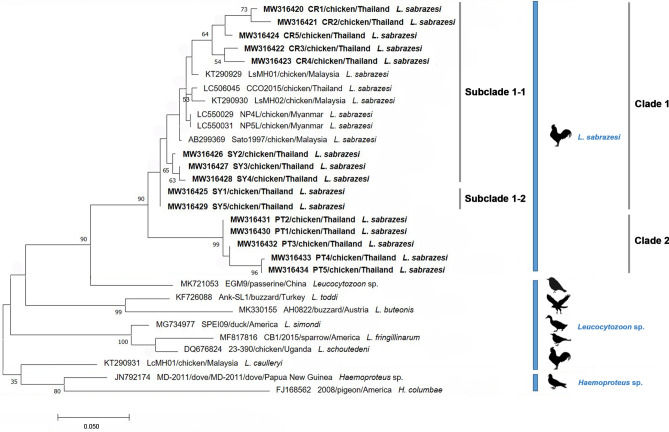
In this study, the phylogenetic tree based on the alignment of the 15 Thailand sequences of L. sabrazesi cytb gene with 6 other sequences taken from the GenBank were classified as 2 clades. Clade 1 was divided into two subclades (subclade 1–1 and subclade 1–2). The sequences assigned to subclade 1–1 exhibited the genetic variability of L. sabrazesi cytb gene sequences from Chiang Rai and Kanchanaburi provinces of Thailand together with sequences from Malaysia, Thailand and Myanmar obtained from the GenBank. Subclade 1–2 was composed of two sequences from Kanchanaburi province and showed phylogenetic proximity. Five sequences from Phatthalung province of Thailand comprised clade 2 and also showed phylogenetic proximity. The cytb gene sequences among L. sabrazesi were highly conserved when compared with other strains as outer groups (Fig. 2). In addition, the similarity ranged between 89.5 and 100% for Thailand cytb sequences. The similarity of subclade 1–1 and subclade 1–2 was in the range of 93.3–99.5% and 94.3—100% among the Thailand L. sabrazesi sequences, respectively, while the similarity of the sequences within 2nd clade ranged between 89.5 to 100% (Table 3). The nucleic acid substitution rate in cytb sequences among L. sabrazesi was estimated under the Tamura and Nei23 mode as shown in Table 4.
Figure 2.
A Maximum Likelihood (ML) phylogenetic tree of L. sabrazesi cytb gene sequences in this study (blodface) and those
taken from GenBank. The numbers on each node correspond to the bootstrap analysis of 1000 replicates (percentage more than 50% were listed). The GenBank assession numbers, the country and the parasite species name of the sequences are exhibited. Two gene sequences of Haemoproteus sp. were employed as the outer groups. The scale bar exhibits the number of substitutions per site.
Table 3.
Similarity of the L. sabrazesi cytb gene sequences as detected in chicken sampled in Thailand.
| Clade | Clade 1 | Clade 2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subclade | Subclade 1–1 | Subclade 1–2 | ||||||||||||||
| Acc. no | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| MW316420 | 1 | 100 | ||||||||||||||
| MW316421 | 2 | 98.1 | 100 | |||||||||||||
| MW316424 | 3 | 97.2 | 96.3 | 100 | ||||||||||||
| MW316422 | 4 | 96.8 | 95.8 | 96.7 | 100 | |||||||||||
| MW316423 | 5 | 95.3 | 96.3 | 96.2 | 97.7 | 100 | ||||||||||
| MW316426 | 6 | 94.3 | 94.8 | 95.8 | 95.3 | 95.3 | 100 | |||||||||
| MW316427 | 7 | 93.8 | 94.3 | 95.3 | 94.8 | 94.8 | 99.5 | 100 | ||||||||
| MW316428 | 8 | 93.3 | 94.3 | 94.8 | 94.3 | 94.7 | 99.1 | 99.5 | 100 | |||||||
| MW316425 | 9 | 94.3 | 94.3 | 95.8 | 96.2 | 94.7 | 99.1 | 98.6 | 98.2 | 100 | ||||||
| MW316429 | 10 | 94.3 | 94.3 | 95.8 | 96.2 | 94.7 | 99.1 | 98.6 | 98.2 | 100 | 100 | |||||
| MW316431 | 11 | 92.8 | 92.3 | 91.2 | 92.3 | 90.8 | 92.8 | 92.3 | 91.8 | 93.8 | 93.8 | 100 | ||||
| MW316430 | 12 | 92.8 | 92.3 | 91.2 | 92.3 | 90.8 | 92.8 | 92.3 | 91.8 | 93.8 | 93.8 | 100 | 100 | |||
| MW316432 | 13 | 92.3 | 91.8 | 90.7 | 91.8 | 90.2 | 92.3 | 92.8 | 92.4 | 93.3 | 93.3 | 99.5 | 99.5 | 100 | ||
| MW316433 | 14 | 90.2 | 90.2 | 89.5 | 90.7 | 89.7 | 91.3 | 91.8 | 91.8 | 92.3 | 92.3 | 96.7 | 96.7 | 97.2 | 100 | |
| MW316434 | 15 | 90.2 | 90.2 | 89.5 | 90.7 | 89.7 | 91.3 | 91.8 | 91.8 | 92.3 | 92.3 | 97.2 | 97.2 | 97.7 | 99.5 | 100 |
| Similarity (%) | ||||||||||||||||
Table 4.
The nucleic acid substitution rate in L. sabrazesi cytb sequences as detected in chickens in Thailand and other countries.
| Nucleic acid | A | T/U | C | G |
|---|---|---|---|---|
| Sequence within Thailand | ||||
| A | – | 4.27 | 1.80 | 10.57 |
| T/U | 3.35 | – | 12.13 | 1.30 |
| C | 3.35 | 28.70 | – | 1.30 |
| G | 27.15 | 4.27 | 1.80 | – |
| Sequence worldwide | ||||
| A | – | 3.96 | 1.68 | 11.41 |
| T/U | 3.15 | – | 11.49 | 1.19 |
| C | 3.15 | 27.04 | – | 1.19 |
| G | 30.10 | 3.96 | 1.68 | – |
Each entry is the probability of substitution from one base (row) to another base (column). Rates of different transitional substitutions are shown in bold and those of transversional substitutions are shown in italics. The maximum Log likelihood for this computation was − 554.346 and − 588.782 for the sequences within Thailand and worldwide, respectively.
Haplotype diversity
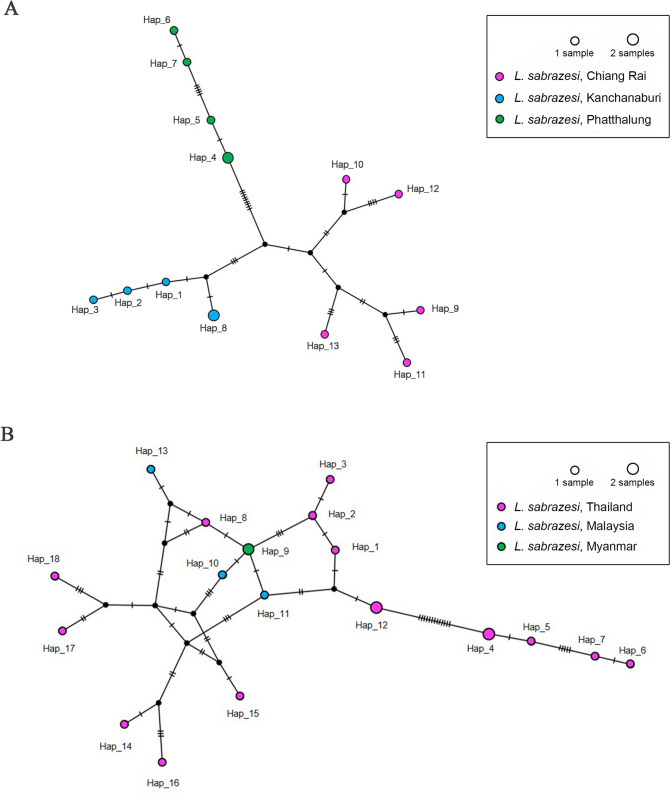
The TCS Network tool was employed to construct the haplotype network of L. sabrazesi cytb gene sequences. Haplotype diversity based on cytb gene found in Chiang Rai, Kanchanaburi and Phatthalung provinces of Thailand was diverse when compared to worldwide gene sequences (Fig. 3). In Thailand, a total of 13 different haplotypes were analyzed including the 5 sequences (haplotype #1 to #3 and #8), 5 sequences (haplotype #4 to #7) and 5 sequences (haplotype #9 to #13) taken from chickens in Kanchanaburi, Phatthalung and Chiang Rai provinces, respectively. Bearing in mind, haplotype #4 was composed of 2 identical sequences (PT1 and PT2) and haplotype #8 was also comprised of SY1 and SY5 sequences (Fig. 3A and Table 5). In addition, 18 haplotypes indicated in TCS network exhibited that haplotypes #1 to #8, #12, #14 to #18 were detected in chicken in three provinces of Thailand. The rest of haplotypes was detected in other countries worldwide (Fig. 3B and Table 5).
Figure 3.
TCS network of haplotypes based on the L. sabrazesi cytb gene sequences detected in Thailand (A) and other countries (B).
Table 5.
Polymorphism and genetic diversity of L. sabrazesi cytb gene sequences as detected in chickens in Thailand and other countries.
| Gene | Size (bp) | N | VS | GC% | h | Dh (mean ± SD) | π (mean ± SD) | K |
|---|---|---|---|---|---|---|---|---|
| Sequence within Thailand | ||||||||
| cytb | 222 | 15 | 26 | 29 | 13 | 0.981 ± 0.031 | 0.05393 ± 0.00428 | 11.9714 |
| Sequence worldwide | ||||||||
| cytb | 222 | 21 | 26 | 28.8 | 18 | 0.986 ± 0.019 | 0.04713 ± 0.00522 | 10.4619 |
N: number of analyzed sequences; VS: number of variable sites; GC: G × C content; h: number of haplotypes; Dh: diversity of haplotypes; SD: standard deviation; π: nucleotide diversity (per site); K: average number of nucleotide differences.
Entropy analysis
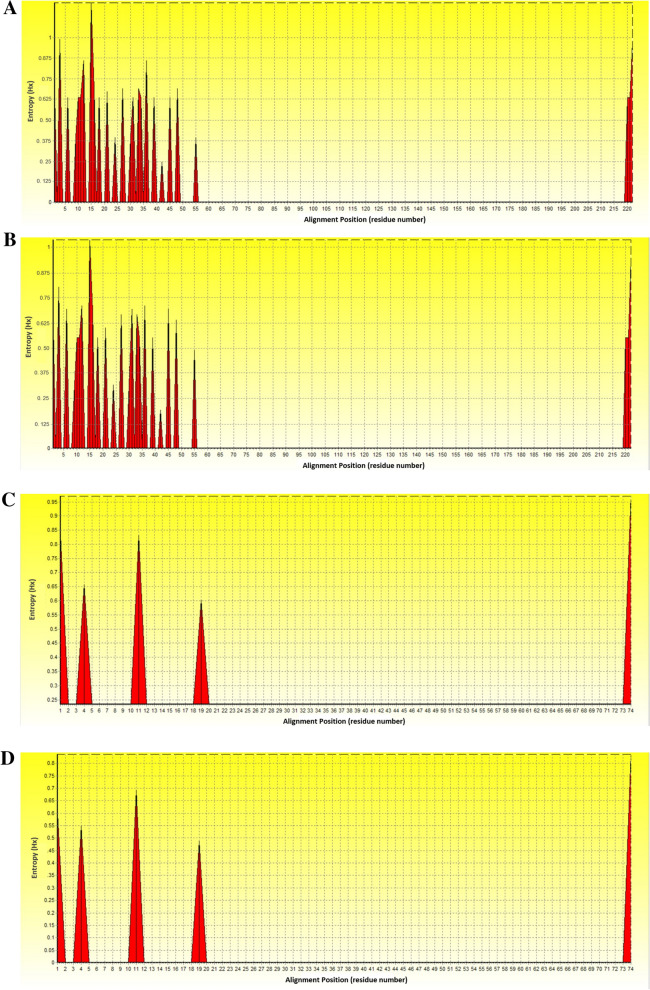
Nucleic acid entropy analysis of 15 L. sabrazesi cytb gene sequences obtained in this study showed 26 high entropy peaks at nucleic acid alignment position 1, 3, 6, 9–12, 15–16, 18, 21, 24, 27, 30–31, 33–34, 36, 39, 42, 45, 48, 55 and 220–222 with entropy values ranged between 0.24493 and 1.21056 (Fig. 4A). The entropy plot of our 15 Thailand sequences aligned with 6 other sequences obtained from the GenBank exhibited the same nucleic acid alignment position but entropy values (0.19144–1.03570) were different (Fig. 4B). In addition, amino acid entropy analysis was performed using 15 Thailand cytb sequence alignments. The chart showed 5 high entropy peaks at amino acid alignment position 1, 4, 11, 19 and 74 with entropy values ranged between 0.39267 and 0.97012 (Fig. 4C). The entropy chart of 15 Thailand sequences with 6 other sequences taken from the GenBank showed the same amino acid alignment position, but entropy value (0.39267–0.97012) were different (Fig. 4D).
Figure 4.
Nucleic and amino acid entropy plots obtained from L. sabrazesi cytb gene sequences. Entropy plot of nucleic acid sequences from Thailand (A) as well as Thailand and other countries (B). Entropy plot of amino acid sequences from Thailand (C) as well as Thailand and other countries (D).
Discussion
Leucocytozoonosis is an important disease causing clinical infection in chickens in many areas of the world, based on widespread distribution of the vector simuliid flies or culicoides midges. There are many conventional methods for detection of Leucocytozoon infection in chickens, such as clinical signs, hematological findings, microscopic examination of mature gametocytes in blood smears and serological tests. However, these methods require expertise since parasites are often missed when parasitemia are significantly low. Accurate diagnosis is needed to develop for appropriate treatment, transmission control and disease management. Thus, molecular assay, i.e., nested PCR, is a sensitive diagnostic method and used for detection and characterization of parasite DNA strains after gametocytes disappeared in host’s blood circulation.
Up to now, the detection of Leucocytozoon infection in chickens has only been reported in some areas of Thailand18,19. In addition, there is an obvious lack of relevant information on leucocytozoonosis isolated in chickens in Thailand. Our study is the first investigation that demonstrated a molecular occurrence of L. sabrazesi infection in chickens throughout Chiang Rai, Kanchanaburi and Phattalung provinces of Thailand.
In this study, our findings revealed a high molecular occurrence of L. sabrazesi in chicken blood samples in three regions (northern, western and southern) of Thailand. The molecular detection exhibited that of the animal sampled, 80.51% (252/313) were positive for L. sabrazesi based on the cytb gene (Table 1). In our work, the cytb gene has been used as a good marker for providing sufficient variation to establish phylogeographic patterns on the large scale and useful for epidemiological approaches of leucocytozoonosis24,25. The occurrence of L. sabrazesi infections was highest in chickens aged < 4 months (95%) compared to chickens aged 4–12 months (90.11%) and > 12 months (54.95%), which is in line with the previous studies by Naqvi et al.26, who showed the highest prevalence in chickens aged less than 6 months (24 weeks) was 78.8%. From our findings, it is possible that the intensive infection could be consistent with free-range farm system showing the higher statistically significant infection than other systems. Likewise, the climate condition, such as rainfall, are often involved with the occurrence of L. sabrazesi infection due to the longer survival of the parasite vectors (black flies and biting midges)27. Moreover, western and southern regions of Thailand have a long rainy season (about 3–4 months); this tropical area is a significant influence on the risk of L. sabrazesi infection and also seems to be a predisposing factor for the development of the parasite vectors which can survive in humid environment9,26,28–30.
Although the genetic diversity of Leucocytozoon sp. based on the sequences of cytb gene has been investigated in several countries8,12,14,16,22,25,31, little is known about the genetic diversity and the phylogeny of L. sabrazesi Thailand strain. In this study, the cytb gene in chicken population sampled in the northern, western and southern areas of Thailand was employed to determine the genetic diversity of L. sabrazesi in these regions. The phylogenetic analysis of chicken L. sabrazesi cytb gene Thailand isolate showed two clades together with the sequences from Malaysia and Myanmar. Our results exhibited that the genetic diversity observed in a phylogram was confirmed by the high similarity value for L. sabrazesi cytb gene (89.5–100%). This finding indicated high conserved sequences and phylogenetic proximity of cytb gene circulating in both Thailand and other countries.
In this work, the cytb gene sequence in blood samples of chickens in the northern, western and southern parts of Thailand were analyzed. Our results showed that the L. sabrazesi population was low diverse in Thailand, with the presence of probably more than one haplotype. The genotype of this gene was identified in haplotype networks. It was carried out with sequences detected in this study together with other sequences obtained from GenBank database that found in Malaysia and Myanmar. In this study, there has been mild different of morphological traits when compare the sequences from Chiang Rai haplotype to other haplotypes. Because Chiang Rai sequence has more prevalence and some chickens show pale comb or skinny correlate to amount of gametocytes in the blood smear. The severity of high prevalence is probably associated with areas where parasite’s vectors are abundant. The more prevalent haplotype is haplotype#12, which is 0–4 month age chickens. This indicated that there was some genetic diversity of cytb gene observed in the different haplotype networks in Thailand and other countries. Furthermore, the cytb sequence shared genetic traits with all sequences as ascertained previously from Malaysia and Myanmar. This finding exhibited that the genetic diversity among L. sabrazesi populations varied in accordance with the geographical area.
Regarding the analysis of L. sabrazesi nucleic acid sequences, our results approved the polymorphism with 26 entropy peaks reaching up to 1.21 (sequence within Thailand) and 1.04 (sequence worldwide). In addition, our results showed 5 entropy peaks of amino acid sequences reaching up to 0.97 (sequence within Thailand) and 0.84 (sequence worldwide). These indicated that different genotypes may involve being a genetic diversity of L. sabrazesi distribution in Thailand.
Conclusions
This study is the first report indicating a molecular occurrence and genetic diversity of L. sabrazesi in chicken blood samples in Thailand. Our findings showed that L. sabrazesi cytb gene is genetically conserved in Thailand and other countries. These could help to ameliorate the understanding of phylogeny and genetic diversity among cytb gene of L. sabrazesi Thailand strain. Therefore, the periodical assessment of the occurrence of leucocytozoonosis is necessitate to control the affectivity of the effective treatment and prevention throughout the country to reduce the infection of chicken vector-borne parasites.
Methods
Study locations and sample sizes
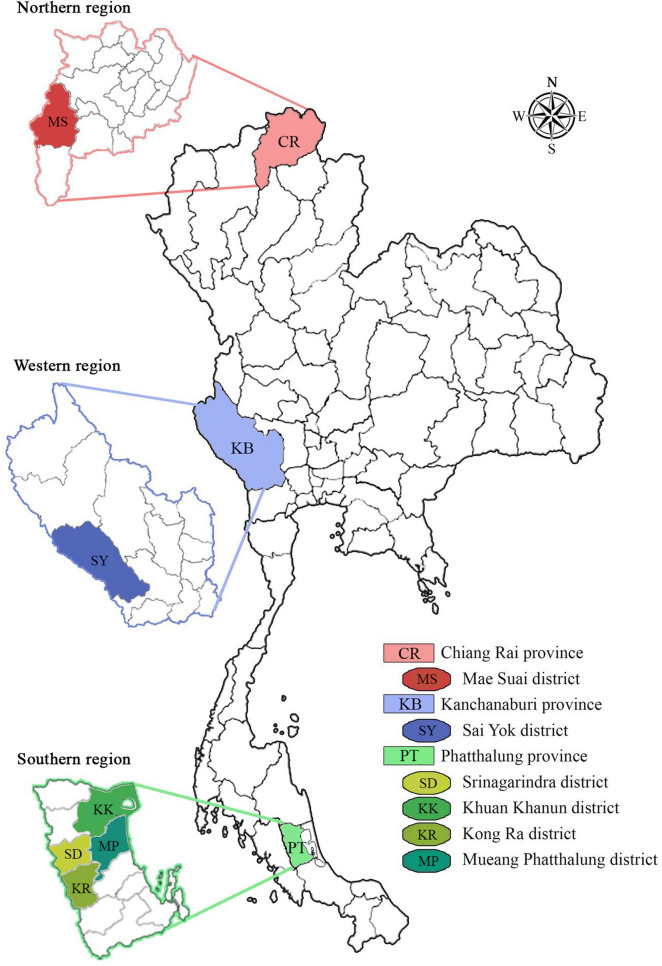
The present study was conducted in three regions of Thailand (Fig. 5). Chicken blood samples were collected in Mae Suai district (19°39′24″N, 99°32′30″E) of Chiang Rai province in northern region, in Sai Yok district (14°6′56″N, 99°8′40″E) of Kanchanaburi province in western region, and in four districts including Srinagarindra (7°34′24″N, 99°56′30″E), Khuan Khanun (7°44′6″N, 100°0′36″E), Kong Ra (7°24′12″N, 99°57′0″E) and Mueang Phatthalung (7°37′6″N, 100°4′24″E) of Phatthalung province in southern region. Total 313 blood samples were randomly collected from the chickens raised in both backyard in households, free-range farms and non-evaporative cooling houses during December 2019 and October 2020. The sample sizes were calculated using the formula based on an equation, n = t2 × p(1 − p)/m2, inserting the following values: the prevalence of L. sabrazesi infection among chickens in Thailand (p)18,19, a 95% confidence level (t) and 5% margin of error (m).
Figure 5.
Geographical location of northern, western and southern regions where chicken blood samples are collected and examined. Legends indicate the distribution of Leucocytozoon sabrazesi Thailand strains discriminated in chickens from Mae Suai district (MS) in Chiang Rai province (CR), Sai Yok district (SY) in Kanchanaburi province (KB) and Srinagarindra (SD), Khuan Khanun (KK), Kong Ra (KR) and Mueang Phatthalung (MP) districts in Phatthalung province (PL).
Collection of blood samples and microscopic examination
Between 0.3 and 2 ml of blood was collected via either the brachial wing vein or medial metatarsal vein of each chicken. They were kept into the centrifuge tube containing lithium heparin to preclude coagulation, and stored at − 80 °C until further used. In addition, some thin fresh blood smears were prepared for each chicken on glass slides, air-dried, fixed in 100% methanol for 1 min, and then stained with Giemsa stain (SIGMA-ALDISH, Germany) for 40 min.
DNA extraction
Genomic DNA of L. sabrazesi was extracted from all collected blood samples of chickens using Tissue DNA Extraction Kit (OMEGA, bio-tek, USA) following the protocol of Watthanadirek et al.32 and Junsiri et al.33 with some modifications. Briefly, 20 µl of blood samples were mixed with 25 µl of OB Protease solution and 250 µl of BL buffer and incubated at 70 °C for 1 min. After adding with 250 µl of 100% ethanol, samples were transferred to HIBIND DNA Mini Column. Finally, DNA samples were eluted in 50 µl Milli-Q water and kept at − 20 °C until used. The concentration and purity of DNA were defined with NANODROP 2000 Spectrophotometers (THERMO SCIENTIFIC) at 260/280 and 260/230 ratios.
Amplification and detection of L. sabrazesi DNA
The specific primer pairs designed from L. sabrazesi cytb sequence submitted in GenBank database under accession numbers AB299369.1, were utilized to amplify DNA fragments of the cytb gene. L. sabrazesi cytb gene was amplified by nested PCR using 2 pairs of specific primers. In the first step of amplification, the primers, namely LsF1 (5′-CATATATTAAGAGAATTATGGAG-3′) and LsR1 (5′-ATAAAATGYTAAGAAATACCATTC-3′) were used. In the second step, the primers, namely LsF2 (5′-TAATCACATGGGTTTGTGGA-3′) and LsR2 (5′-GCTTTGGGCTAAGAATAATACC-3′) were also used. The expected size of amplification products was 248 bp.
PCR reaction mixtures consisting of 50 ng DNA template, 0.2 µM each of the primers, 0.125 mM of each deoxynucleoside triphosphate (dNTPs), 3 mM MgCl2, 0.25 U Tag DNA polymerase (NEW ENGLAND BIOLABS, UK), 1× standard Tag reaction buffer and nuclease free water, were put through in a thermal cycle (BIO-RAD, USA) with the following condition: 40 cycles of denaturation at 94 °C for 1 min (1st step) and for 20 s (2nd step), annealing at 50 °C for 1 min (1st step) and 53 °C for 20 s (2nd step), extension at 68 °C for 1 min (1st step) and for 30 s (2nd step) as well as a final extension at 68 °C for 5 min (1st and 2nd steps). PCR product was observed by 1.2% agarose gel stained with Fluorostain DNA Fluorescent Staining Dye (SMOBIO, Taiwan) and viewed under ultraviolet (UV) transilluminator. A 100 bp Plus DNA Ladder (THERMO FISHER SCIENTIFIC, USA) was used as standard for defining the molecular mass of PCR products.
Cloning of the mitochondrial cytb gene from L. sabrazesi DNA
L. sabrazesi cytb gene was cloned into vector with following specific primers: LsF 5′-CACCTAATCACATGGGTTTGTGGA-3′ and LsR 5′-GCTTTGGGCTAAGAATAATACC-3′. The 4 nucleotides (CACC) were added at 5′ end of forward primer with the overhang sequence (GTGG) in pET100/D-TOPO® vector (INVITROGEN, USA) to enable directional cloning. The PCR reaction was conducted with the protocols as described in previous section. PCR products were purified using ULTRACLEAN 15 DNA Purification Kit (MO BIO LABORATORIES, USA) following the manufacturer’s instructions for cloning. The 20 ng of the blunt-end PCR products were inserted in the pET100/D-TOPO vector (INVITROGEN LIFE TECHNOLOGIES, USA). Then 3 µl of the cloning reactions were transformed into chemically competent Escherichia coli cells (INVITROGEN, USA). Subsequently, 200 µl of transformed bacterial culture was spread on the Luria Bertani (LB) agar plates containing 100 µg ampicillin and incubated for overnight at 37 ºC. The positive clones were selected and grown in LB medium containing ampicillin for overnight. Finally, the recombinant plasmids (pET100-cytb) were extracted from the competent cells using AXYPREP Plasmid Miniprep Kit (AXYGEN BIOSCIENCE, USA) following the manufacturer’s instructions and analyzed for correctly sized inserts by agarose gel electrophoresis32,33.
Sequencing analysis
Purified PCR products were confirmed by Sanger method of DNA sequencing. All DNA sequences were analyzed by BLAST (The National Center for Biotechnology Information, NCBI, http://www.ncbi.nlm.nih.gov/BLAST). All sequences were deposited in GenBank, accession numbers are provided in Table 6.
Table 6.
The L. sabrazesi cytb nucleotide sequences amplified in Thailand strains were deposited in GenBank database.
| Regions | Provinces | Districts | Animal ID | GenBank accession numbers |
|---|---|---|---|---|
| Northern | Chiang Rai | Mae Suai | CR1 | MW316420 |
| CR2 | MW316421 | |||
| CR3 | MW316422 | |||
| CR4 | MW316423 | |||
| CR5 | MW316424 | |||
| Western | Kanchanaburi | Sai Yok | SY1 | MW316425 |
| SY2 | MW316426 | |||
| SY3 | MW316427 | |||
| SY4 | MW316428 | |||
| SY5 | MW316429 | |||
| Southern | Phatthalung | Khuan Khanun | PT1 | MW316430 |
| PT2 | MW316431 | |||
| Kong Ra | PT3 | MW316432 | ||
| Mueang Phatthalung | PT4 | MW316433 | ||
| Srinagarindra | PT5 | MW316434 |
Phylogenetic sequence analysis
Leucocytozoon sabrazesi cytb gene sequences were used for sequence alignment and phylogenetic analysis as shown in Table 6 and two gene sequences of Haemoproteus sp. were employed as the outer groups (accession no. JN792174 and FJ168562). Multiple sequence alignment was proceeded with Clustal W algorithm and then genetic inference was carried out by Maximum Likelihood (ML) phylogenetic tree using MEGA software version 7.0.2.634,35. Bootstrap analysis with 1000 repetitions was employed to evaluate the confidence of the branching pattern of the trees36. The distance analyses were performed using the Kimura 2-parameter distance model37. The similarity was determined with the pairwise-distance method38.
Haplotype diversity
The obtained alignment of L. sabrazesi cytb gene sequences was used to estimate the nucleotide diversity (π), diversity of haplotypes (Dh), number of haplotypes, and the average number of nucleotide differences (K), using the DnaSP version 6.0 software39. Likewise, the nucleotide sequences were put through to the Population Analysis with the Reticulate Trees (popART) program40 in order to analyze the TCS Network construction41.
Entropy analysis
Entropy analysis was employed to verify the variability of the nucleic acid and amino acid sequences. L. sabrazesi cytb nucleotide sequences were translated into amino acids, aligned and then analyzed by the Entropy (H (x)) plot using Bioedit version 7.2.6.142.
Statistical analysis
The overall occurrence was calculated as a percent of number of L. sabrazesi-infected animals in the total number of examined animals. Relative occurrence of this parasite was calculated and proceeded by breed, age, gender, management system, type of feed, water source and insect control system. For all analyses, confidence level was obtained at 95% and p-value of ≤ 0.05 was considered to be the level of significance. Statistical analysis was defined using SPSS software for Window version 22.0 (SPSS Inc., Chicago, USA).
Ethic approval and permit
All experimental procedures regarding animals were carried out under the following approval and permit from the Animal Care and Use Committee (IMBMU-ACUC), Institute of Molecular Biosciences, Mahidol University, Thailand. All methods were performed in accordance with relevant guidelines and regulations.
Acknowledgements
This research project is financially supported by Mahidol University (Basic Research Fund: fiscal year 2021) and Research Grants from the National Research Council of Thailand (NRCT) and the Mid-Career Research Grant co-funded by National Research Council of Thailand (NRCT) and Mahidol University [grant number NRCT5-RSA63015-23] to Panat Anuracpreeda, and Royal Golden Jubilee Ph.D. (RGJ-PHD) Scholarship [grant number PHD/0050/2560] to Pacharaporn Khumpim.
Author contributions
P.K. was involved with all aspects of the planning and conducting of this research including the laboratory work, processing of data and drafting of manuscript. R.C., W.J., A.W., N.P., S.M. and N.S. contributed to study design, conducting of the laboratory work and manuscript revision. P.A. was involved in all aspects of the project from research design, laboratory work, data analysis, laboratory resources, funding acquisition, editing of manuscript, manuscript development and final revisions. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: Pacharaporn Khumpim was omitted from the author list in the original version of this Article. Modifications were made to the acknowledgement and author contribution sections. Full information regarding the corrections can be found in the correction of this article.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/9/2024
A Correction to this paper has been published: 10.1038/s41598-024-53903-6
References
- 1.Valkiunas G. Avian Malaria Parasites and Other Haemosporidia. CRC Press; 2005. [Google Scholar]
- 2.Forrester DJ, Greiner EC. Leucocytozoonosis. In: Atkinson CT, Thomas NJ, Hunter DB, editors. Parasitic Diseases of Wild Birds. Wiley-Blackwell; 2008. pp. 54–107. [Google Scholar]
- 3.Zhao W, Liu J, Xu R, Zhang C, Pang Q, Chen X, Su XZ. The gametocytes of Leucocytozoon sabrazesi infect chicken thrombocytes, not other blood cells. PLoS ONE. 2015;10:e0133478. doi: 10.1371/journal.pone.0133478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fallis AM, Jacobson RL, Raybould JN. Haematozoa in domestic chickens and guinea fowl in Tanzania and transmission of Leucocytozoon neavei and Leucocytozoon schoutedeni. J. Protozool. 1973;20:438–442. doi: 10.1111/j.1550-7408.1973.tb00918.x. [DOI] [PubMed] [Google Scholar]
- 5.Morii T, Matsui T, Iijima T, Fujinaga F. Infectivity of Leucocytozoon caulleryi sporozoites developed in vitro and in vivo. Int. J. Parasitol. 1984;14:135–139. doi: 10.1016/0020-7519(84)90040-7. [DOI] [PubMed] [Google Scholar]
- 6.Morii T, Nakamura K, Lee YC, Iijima T, Hoji K. Observations on the Taiwanese strain of Leucocytozoon caulleryi (Haemosporina) in chickens. J. Protozool. 1986;33:231–234. doi: 10.1111/j.1550-7408.1986.tb05597.x. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura K, Ogiso M, Shibahara T, Kasuga H, Isobe T. Pathogenicity of Leucocytozoon caulleryi for specific pathogen-free laying hens. J. Parasitol. 2001;87:1202–1204. doi: 10.1645/0022-3395(2001)087[1202:POLCFS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 8.Sehgal RN, Valkiunas G, Iezhova TA, Smith TB. Blood parasites of chickens in Uganda and Cameroon with molecular descriptions of Leucocytozoon schoutedeni and Trypanosoma gallinarum. J. Parasitol. 2006;92:1336–1343. doi: 10.1645/GE-927R.1. [DOI] [PubMed] [Google Scholar]
- 9.Zhao W, Pang Q, Xu R, Liu J, Liu S, Li J, Su XZ. Monitoring the Prevalence of Leucocytozoon sabrazesi in southern China and testing tricyclic compounds against gametocytes. PLoS ONE. 2016;11:e0161869. doi: 10.1371/journal.pone.0161869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donoghue P. Haemoprotozoa: Making biological sense of molecular phylogenies. Int. J. Parasitol. Parasites Wildl. 2017;6:241–256. doi: 10.1016/j.ijppaw.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishtiaq F, Gering E, Rappole JH, Rahmani AR, Jhala YV, Dove CJ. Prevalence and diversity of avian hematozoan parasites in Asia: A regional survey. J. Wild. Dis. 2007;43:382–398. doi: 10.7589/0090-3558-43.3.382. [DOI] [PubMed] [Google Scholar]
- 12.Sato Y, Tamada A, Mochizuki Y, Nakamura S, Okano E, Yoshida C, Ejiri H, Omori S, Yukawa M, Murata K. Molecular detection of Leucocytozoon lovati from probable vectors, black flies (Simuliudae) collected in the alpine regions of Japan. Parasitol. Res. 2009;104:251–255. doi: 10.1007/s00436-008-1183-1. [DOI] [PubMed] [Google Scholar]
- 13.Murdock, C. Studies on the ecology of avian malaria in an alpine ecosystem. Ph.D. thesis (University of Michigan, 2009).
- 14.Murdock C, Adler PH, Frank J, Perkins SL. Molecular analyses on host-seeking black flies (Diptera: Simuliidae) reveal a diverse assemblage of Leucocytozoon (Apicomplexa: Haemospororida) parasites in an alpine ecosystem. Parasit. Vectors. 2015;8:343–350. doi: 10.1186/s13071-015-0952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee DH, Jang JH, Kim BY, Kwon YK, Gomis S, Lee JB, Park SY, Choi IS, Song CS. Diagnosis of Leucocytozoon caulleryi infection in commercial broiler breeders in South Korea. Avian Dis. 2014;58:183–186. doi: 10.1637/10621-072913-Case.1. [DOI] [PubMed] [Google Scholar]
- 16.Lee HR, Koo BS, Jeon EO, Han MS, Min KC, Lee SB, Bae Y, Mo IP. Pathology and molecular characterization of recent Leucocytozoon caulleryi cases in layer flocks. J. Biomed. Res. 2016;30:517–524. doi: 10.7555/JBR.30.2016K0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell JG. Bangkok haemorrhagic disease of chickens; an unusual condition associated with an organism of uncertain taxonomy. J. Pathol. Bacteriol. 1954;68:423–429. doi: 10.1002/path.1700680216. [DOI] [PubMed] [Google Scholar]
- 18.Singjam, S. & Ruksachat, N. Case Report: Outbreak of Leucocytozoonosis in Captive Wild Birds, Khao Kor Wildlife Captive Breeding Center. Veterinary news: Lower northern region, veterinary research and development center, DLD Thailand, Report number. 8, 1–7 (2011).
- 19.Buranapim N, Chaiwisit P, Wangkawan A, Tiwananthagorn S. A survey on blood parasites of birds in Chiang Mai province. Vet. Integr. Sci. 2019;17:65–73. [Google Scholar]
- 20.Ortego J, Cordero PJ. PCR-based detection and genotyping of haematozoa (Protozoa) parasitizing eagle owls, Bubo bubo. Parasitol. Res. 2009;104:467–470. doi: 10.1007/s00436-008-1207-x. [DOI] [PubMed] [Google Scholar]
- 21.Ferraguti M, Martínez-de la Puente J, Ruiz S, Soriguer R, Figuerola J. On the study of the transmission networks of blood parasites from SW Spain: Diversity of avian haemosporidians in the biting midge Culicoides circumscriptus and wild birds. Parasit. Vectors. 2013;6:208. doi: 10.1186/1756-3305-6-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suprihati E, Yuniarti WM. The phylogenetics of Leucocytozoon caulleryi infecting broiler chickens in endemc areas in Indonesia. Vet. World. 2017;10:1324–1328. doi: 10.14202/vetworld.2017.1324-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 24.Clark NJ, Clegg SM, Lima MR. A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): New insights from molecular data. Int. J. Parasitol. 2014;44:329–338. doi: 10.1016/j.ijpara.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Galen SC, Nunes R, Sweet PR, Perkins SL. Integrating coalescent species delimitation with analysis of host specificity reveals extensive cryptic diversity despite minimal mitochondrial divergence in the malaria parasite genus Leucocytozoon. BMC Evol. Biol. 2018;18:128–143. doi: 10.1186/s12862-018-1242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naqvi MA, Khan MK, Iqbal Z, Rizwan HM, Khan MN, Naqvi SZ, Zafar A, Sindhu Z, Abbas RZ, Abbas A. Prevalence and associated risk factors of haemoparasites, and their effects on hematological profile in domesticated chickens in District Layyah, Punjab, Pakistan. Prev. Vet. Med. 2017;143:49–53. doi: 10.1016/j.prevetmed.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Paperna I, Soh MCK, Yap CAM, Sodhi NS, Lim SLH, Prawiradilaga DM, Nagata H. Blood parasite prevalence and abundance in the bird communities of several forested locations in Southeast Asia. Ornithol. Sci. 2005;4:129–138. doi: 10.2326/osj.4.129. [DOI] [Google Scholar]
- 28.Awa T, II, Khan PV, Tamungang Simon A, Marcel D. Effects of seasonal and site variations on the prevalence of parasites on Black-crowned waxbill (Estrilda nonnula) in Dschang and its environs, Cameroon. J. Appl. Biol. Biotechnol. 2014;2:012–016. doi: 10.7324/JABB.2014.2303. [DOI] [Google Scholar]
- 29.Elbers ARW, Koenraadt CJ, Meiswinkel R. Mosquitoes and Culicoides biting midges: Vector range and the influence of climate change. Rev. Sci. Tech. 2015;34:123–137. doi: 10.20506/rst.34.1.2349. [DOI] [PubMed] [Google Scholar]
- 30.Takang P, Pikulkaew S, Awaiwanont N, Numee S. Prevalence and risk factors of blood parasites infection in backyard chickens in Chiang Mai. Vet. Integr. Sci. 2017;15:157–167. [Google Scholar]
- 31.Argilla LS, Howe L, Gartrell BD, Alley MR. High prevalence of Leucocytozoon spp. in the endangered yellow-eyed penguin (Megadyptes antipodes) in the sub-Antarctic regions of New Zealand. Parasitology. 2013;140:672–682. doi: 10.1017/S0031182012002089. [DOI] [PubMed] [Google Scholar]
- 32.Watthanadirek A, Chawengkirttikul R, Poolsawat N, Junsiri W, Boonmekam D, Reamtong O, Anuracpreeda P. Recombinant expression and characterization of major surface protein 4 from Anaplasma marginale. Acta Trop. 2019;197:105047. doi: 10.1016/j.actatropica.2019.105047. [DOI] [PubMed] [Google Scholar]
- 33.Junsiri W, Watthanadirek A, Poolsawat N, Kaewmongkol S, Jittapalapong S, Chawengkiattikul R, Anuracpreeda P. Molecular detection and genetic diversity of Anaplasma marginale based on the major surface protein genes in Thailand. Acta Trop. 2020;205:105338. doi: 10.1016/j.actatropica.2020.105338. [DOI] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 37.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 38.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; 2000. [Google Scholar]
- 39.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 40.Leigh JW, Bryant D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 41.Clement, M., Snell, Q., Walker, P., Posada, D & Crandall, K. TCS: Estimating gene genealogies. In Parallel and Distributed Processing Symposium, International Proceedings Vol. 2, 184 (2002).
- 42.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41:95–98. doi: 10.14601/Phytopathol_Mediterr-14998u1.29. [DOI] [Google Scholar]