Abstract
Historically, the genetic analysis of mammalian cells entailed the isolation of randomly-arising mutant cell lines with altered properties, followed by laborious genetic mapping experiments to identify the mutant gene responsible for the phenotype. In recent years, somatic cell genetics has been revolutionized by functional genomics screens, in which expression of every protein-coding gene is systematically perturbed, and the phenotype of the perturbed cells is determined. We outline here a novel functional genomics screening strategy that differs fundamentally from commonly used approaches. In this strategy, we express libraries of artificial transmembrane proteins named traptamers and select rare cells with the desired phenotype because, by chance, a traptamer specifically perturbs the expression or activity of a target protein. Active traptamers are then recovered from cells and can be used as tools to dissect the biological process under study. We also briefly describe how we have used this new strategy to provide insights into the complex process by which human papillomaviruses (HPVs) enter cells.
Keywords: traptamers, HPV, human papillomavirus, BPV E5 protein, genetic screening, retromer, artificial proteins, transmembrane proteins, Rab7
Introduction
The vast majority of published functional genomics screens are based on knockdown of gene expression by siRNA- or shRNA-mediated RNA interference or on gene knockout by CRISPR-mediated gene disruption [1, 2]. The availability of complete genome sequences and advances in oligonucleotide synthesis allow the design and construction of libraries that target all known protein-coding sequences. These libraries can be used in two basic formats: arrayed screens and pooled selection screens. In arrayed screens, sequences that target each gene are tested individually. Once a phenotype is detected, the targeted gene is immediately known, based on the sequence of the active siRNA, shRNA, or sgRNA. In selection screens, complex libraries of thousands of sequences are introduced as a pool into cells, which are then subjected en mass to biological section to enrich for cells with the desired phenotype. Genomic DNA of the selected cell population is subjected to next-generation sequencing, and the target gene can be inferred from the identity of sequences that are enriched or depleted by selection. Transposon-mediated mutagenesis has been widely used as an alternate method to disrupt genes in prokaryotes and lower eukaryotes and has also been applied to mammalian cells and even mice (e.g., [3]).
Although knockdown and knockout screens have yielded a wealth of information and profoundly increased our understanding of many biological processes, they have some limitations. A general limitation of knockdown/knockout genomic screens is the difficulty of identifying relevant genes that are essential for cell viability. This problem can be circumvented in some situations by CRISPR-based activation and repression screens that modulate gene expression e.g., [4]. It is also difficult to identify genes that play redundant roles, because a process can tolerate the loss of one gene if a second gene can supply the missing function. In addition, off-target effects can occur in which repression of unknown non-targeted genes is responsible for the phenotype, so experiments should be performed to confirm the importance of the gene identified as the target of a particular RNA.
Traptamers
We have developed an alternative selection screening strategy that depends on protein interference or activation rather than gene knockdown or knockout. The screen is based on the expression of artificial proteins with the potential to bind to cellular proteins and modulate their activity, combined with strong biological selection for the desired phenotype. The overall strategy is to screen libraries expressing many artificial proteins, each containing a different, short (typically < 25 amino acid), randomized segment with the potential to bind specifically to a naturally occurring protein (Fig. 1A). To reduce the likelihood that randomized protein sequences will mis-fold or aggregate, we impose the additional structural criterion that the randomized segment is composed primarily of hydrophobic amino acids, so that the artificial proteins will spontaneously insert into cell membranes and fold into stable α-helices. By chance, a rare artificial transmembrane helix will bind to the transmembrane domain of a cellular (or viral) protein and modulate its activity. Because transmembrane protein-protein interactions are often highly specific [5, 6], these artificial proteins can have specific effects on cells. We refer to these artificial small hydrophobic proteins as traptamers, for transmembrane protein aptamers.
Fig. 1. Schematic diagram of traptamer library design and construction.
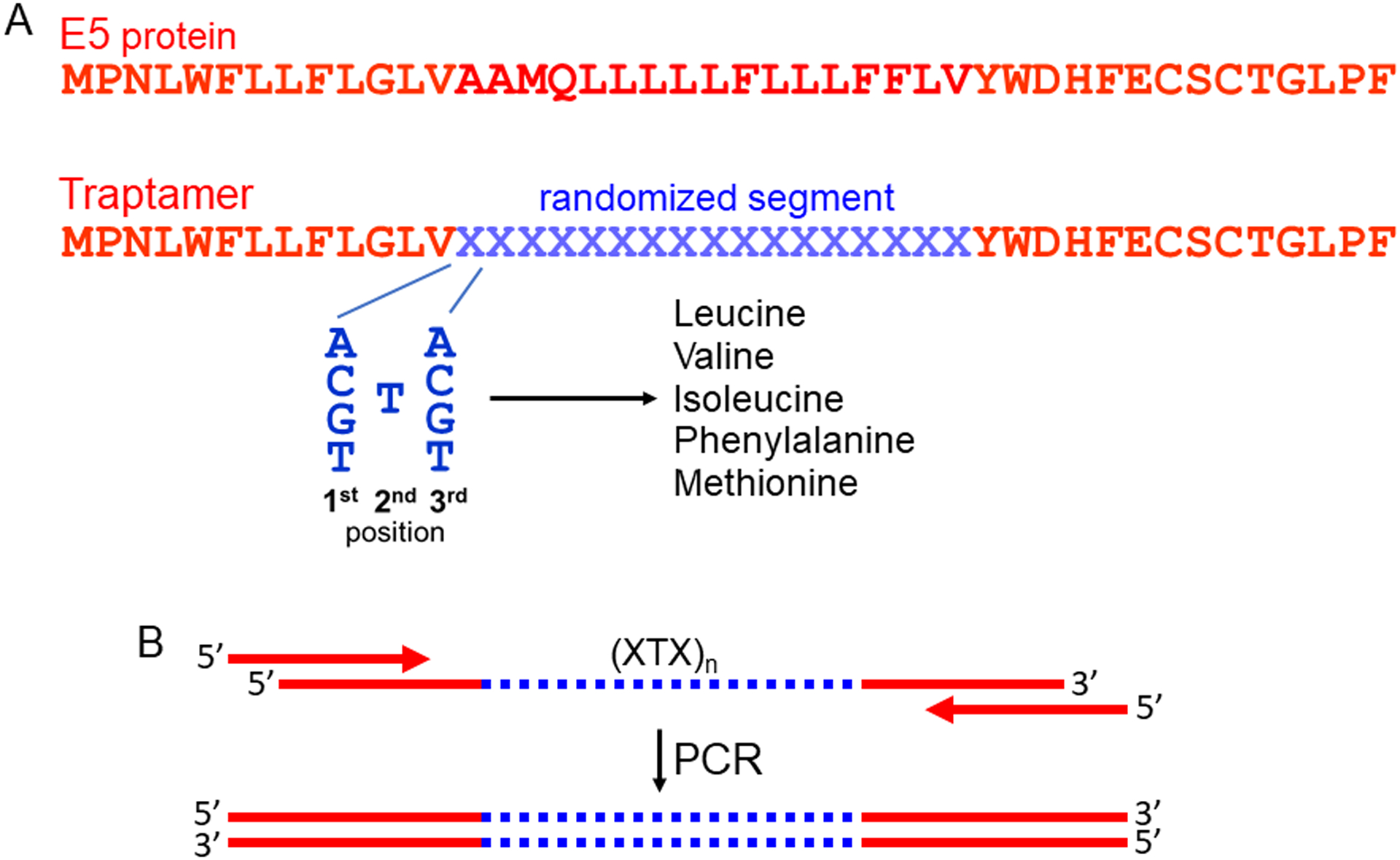
A. The top line shows the sequence of the 44-residue E5 protein of bovine papillomavirus, which has a central hydrophobic segment that serves as a transmembrane (TM) domain. E5 amino acids are shown in red in the single letter code. The second line shows the amino acid sequence of a traptamer library modeled on the E5 protein, with the randomized amino acids represented by the blue Xs. Each randomized codon consists of all four nucleotides in the first and third positions and a T at the second position. Codons of this composition will encode a segment consisting of the hydrophobic amino acids leucine, valine, isoleucine, phenylalanine, and methionine in random order at a 6:4:3:2:1 ratio. B. The top diagram shows a long, single-stranded DNA oligonucleotide containing a central segment (blue) randomized as in panel A [represented as (XTX)n]. This oligonucleotide is amplified by PCR using short primers that hybridize to the fixed sequences flanking the randomized segment. The resulting double-stranded amplification products are cloned as a pool into a retrovirus expression vector to generate the library used for selection.
The likelihood that any one traptamer will induce the desired phenotype is low, so traptamer screening requires the use of libraries expressing many thousands or even millions of traptamers combined with a genetic selection that can separate rare cells expressing the desired phenotype from a large excess of cells that do not express it. Degenerate oligonucleotides are used to construct libraries in which the randomized segment encodes exclusively or primarily hydrophobic amino acids. For example, by inserting a T at the second position in the codons in the randomized segment while all four bases are allowed at the first and third positions, the hydrophobic amino acids leucine, isoleucine, valine, phenylalanine, and methionine will be encoded in random order (Figs. 1A and 1B). Retrovirus vectors can be conveniently used to express the traptamer libraries in mammalian cells so that most cells express only one or a few traptamers, facilitating the eventual isolation of individual active traptamers (Fig. 2). Importantly, as is the case for all selection-based screens, the selected phenotype must be cell-autonomous, i.e., the desired phenotype is displayed only by cells expressing the active traptamer, otherwise inactive traptamers may be recovered from passenger cells. Positive growth selections in which cells displaying the desired phenotype are the only cells that proliferate are ideal, but we have also used fluorescent-activated cell sorting to isolate cells expressing active traptamers [7].
Fig. 2. Overview of traptamer screening.
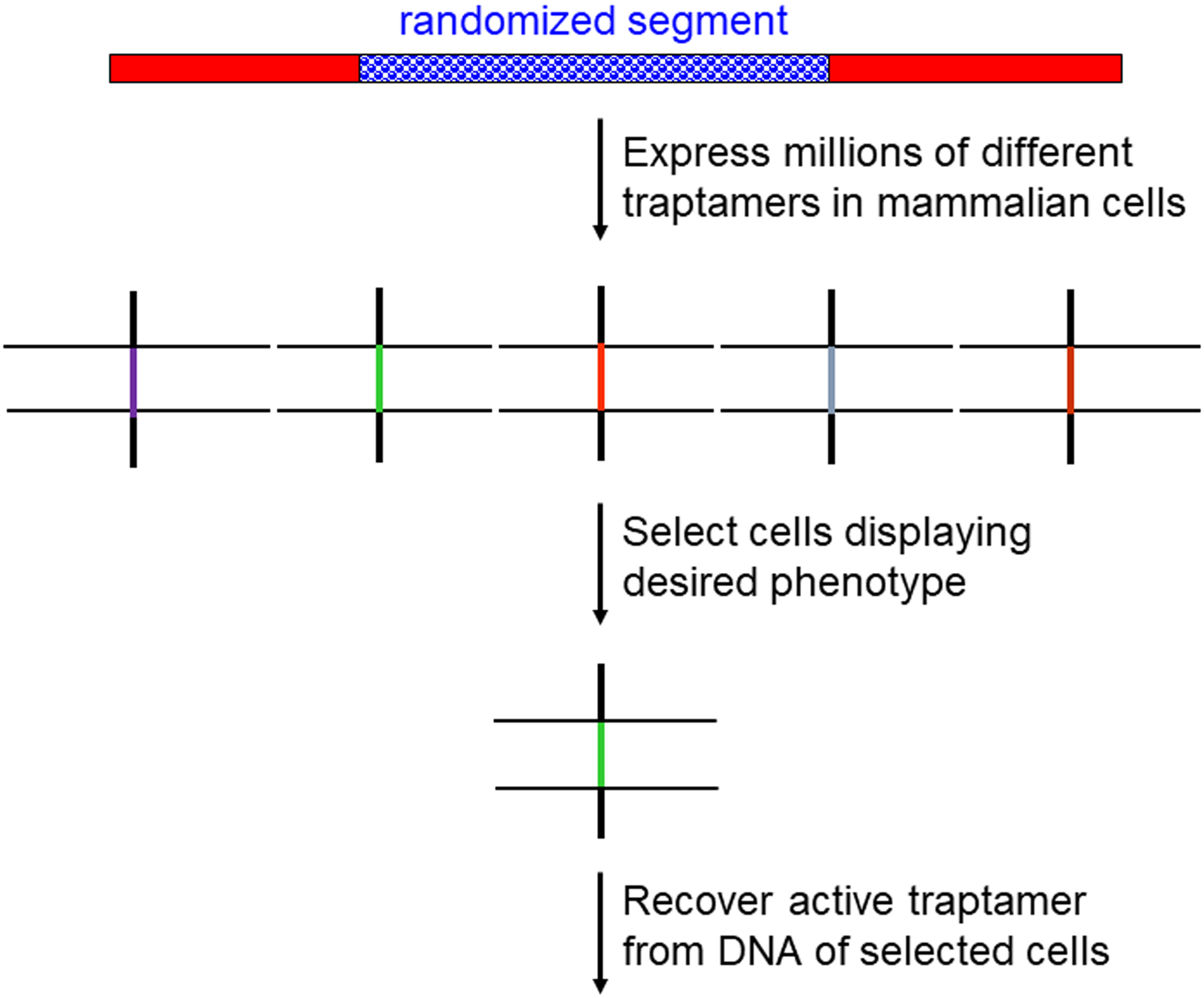
The top line shows the schematic structure of the traptamer library as in Figure 1A. Retroviruses are used to introduce the traptamer genes into cells so that most cells express a single traptamer, and genetic selection is imposed to isolate cells displaying the desired phenotype due the expression of a traptamer. PCR is then used to recover the gene encoding the active traptamer from the DNA of selected cells. The horizontal lines represent the lipid bilayer of cell membranes.
Genes encoding traptamers can be readily recovered from genomic DNA isolated from the selected cells by PCR amplification followed by molecular cloning. If a single round of selection is not sufficient to recover a unique traptamer from a population of selected cells, serial rounds of selection and recovery can be performed in naïve cells, a process which discards background cells surviving each round of selection. Once a unique traptamer sequence (or a few sequences) are recovered from a selected cell population, it is imperative to reintroduce it into cells as an individual clone and confirm activity. Alternatively, next-generation sequencing can be used to identify traptamer genes enriched or depleted in the selected cell population [8]. Traptamer screening bears some resemblance to Genetic Suppressor Element (GSE) screening described by Roninson and colleagues [5], in which fragments of naturally occurring proteins are expressed in cells and selected as inhibitors of protein function. Unlike GSEs, the active segments of traptamers are transmembrane domains and are not derived from naturally occurring sequences.
We have isolated traptamers that specifically activate the PDGF β receptor or the erythropoietin (EPO) receptor or inhibit the expression of the chemokine and human immunodeficiency virus receptor, CCR5 [7–15]. These traptamers appear to have diverse mechanisms of action. The PDGF receptor activators are thought to induce receptor homodimerization and autophosphorylation. In some cases, the EPO receptor activators appear to stabilize an active configuration of the homodimeric EPO receptor, whereas in another case a traptamer appears to activate a constitutive heterodimer of EPO receptor and the cytokine receptor β-common chain. Most but not all CCR5 inhibitors direct CCR5 to the lysosome for degradation. After active traptamers are isolated, they can be optimized by subjecting them to limited random mutagenesis and rescreening under more stringent conditions [7, 16].
Traptamers that inhibit Human papillomavirus entry
In the examples cited above, we selected traptamers that modulate a predetermined protein, but this approach can also be used in unbiased genetic screens for proteins that affect biological processes without knowledge of the relevant targets beforehand, an approach we term traptamer screening. We demonstrated the feasibility of this approach by isolating traptamers that inhibit infection by human papillomavirus (HPV). HPV is a non-enveloped DNA virus responsible for approximately 5% of human cancer. During virus entry, HPV traffics through the retrograde transport pathway to reach the nucleus where the viral genome replicates [17–20], but the details of HPV trafficking are not known.
For these screening experiments, we exploited the finding that ongoing expression of the integrated HPV18 E6 and E7 oncogenes is required for continued proliferation of HeLa cervical carcinoma cells. If the E6/E7 promoter is repressed by expression of the bovine papillomavirus (BPV) E2 transcription factor, the cells cease proliferation [21]. To isolate cells resistant to HPV infection, we constructed an HPV16 vector that expresses BPV E2. We then infected HeLa cells with this vector and selected cells that continued to proliferate because they were resistant to HPV16 infection and the resulting repression of E6 and E7 by BPV E2 [22] (Fig. 3). Traptamer genes were recovered from the DNA of proliferating cells and tested individually for their ability to inhibit expression of a reporter gene expressed from a different HPV16 virus. Details of library construction, selection, and traptamer recovery are described in [22].
Fig. 3. Scheme to isolate traptamers that inhibit HPV infection.
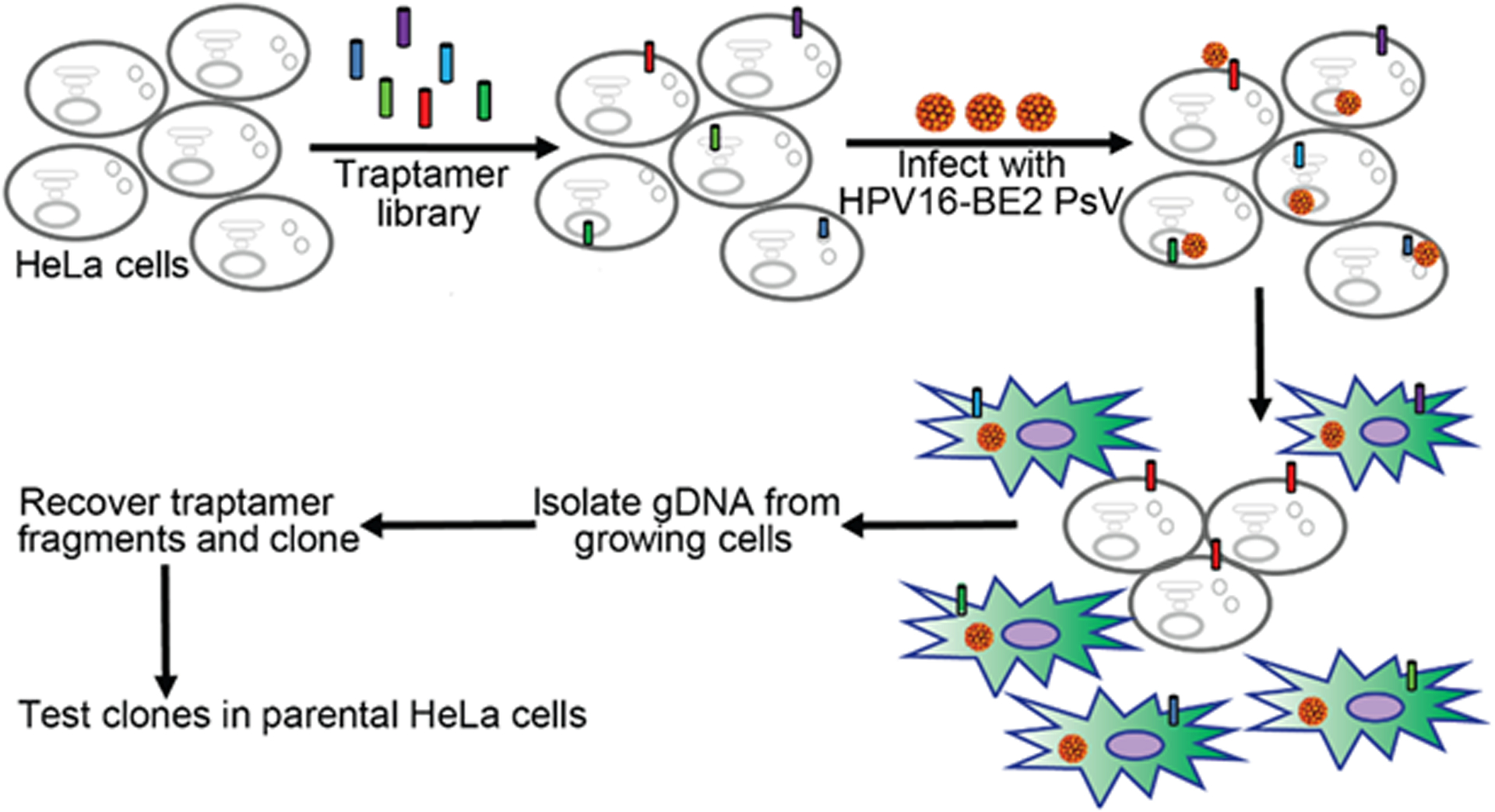
A traptamer library was introduced into HeLa cells, which were then infected with an HPV16 variant expressing the BPV E2 protein (designated HPV16-BE2), which represses expression of the HPV18 oncogenes in these cells. Oncogene repression causes the cells to cease proliferation and undergo replicative senescence, as indicated by the green cells. If a cell expresses a traptamer (red cylinder) that blocks infection by HPV16, the cell will continue to proliferate because the oncogenes are not repressed. PCR is used to recover traptamer genes from genomic DNA isolated from proliferating cells. The ability of individual cloned traptamers to inhibit HPV infection is tested to confirm activity. Figure reproduced from Xie et al. [22].
We used this approach to isolate four traptamers that inhibit infection by several oncogenic HPV types without affecting unrelated viruses [22, 23]. These inhibitory traptamers were then used as tools to dissect HPV entry by examining infection in cells expressing the traptamers. Intriguingly, all four traptamers contain unrelated hydrophobic sequences, display distinct intracellular localizations, and inhibit HPV entry at different steps [23]. By examining the localization of incoming virus in cells expressing individual traptamers or pairwise combinations of traptamers, we confirmed the importance of retrograde transport for HPV infection, placed the steps of HPV trafficking through retrograde compartments into a defined order, and established that none of the steps inhibited by the traptamers can be readily bypassed by alternative HPV entry pathways (Fig. 4A).
Fig. 4. A. Inhibitory traptamers act at different positions in the HPV entry pathway.
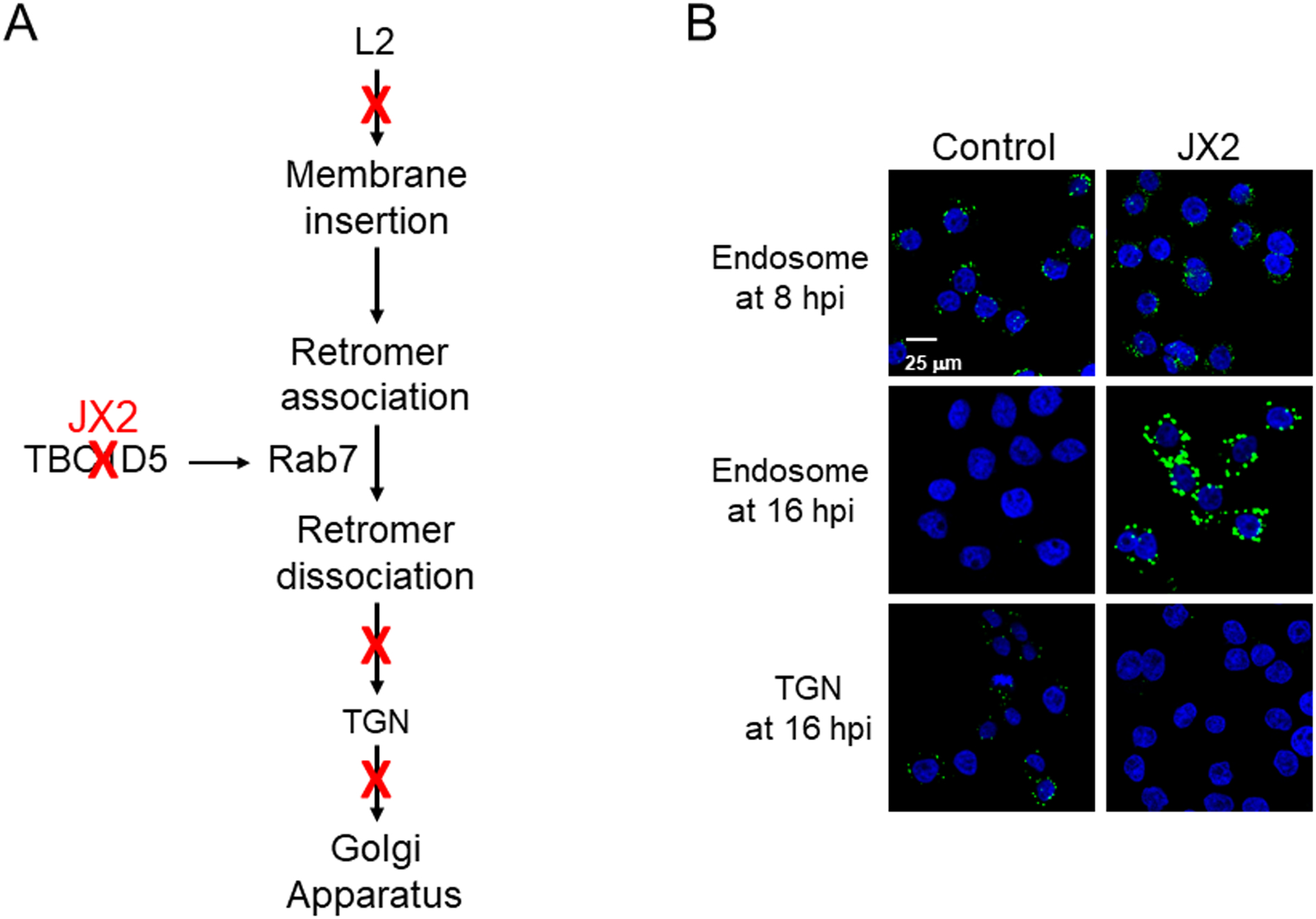
A. Summary of the effect of four traptamers on HPV entry. Sites of inhibition by different traptamers are shown by red Xs, with the inhibition of TBC1D5 by JX2 highlighted. B. Traptamer JX2 inhibits HPV exit from the endosome and arrival at the TGN. Images show proximity ligation assays (PLAs) for HPV capsid protein L1 and either the endosome marker EEA1 or the TGN marker TGN46 to assess the presence of incoming virus in these compartments. HPV16-infected cells expressing no traptamer (control) or expressing traptamer JX2 are shown. PLA signal is green; nuclei are stained blue. The scale bar shows 25 μM. Top row shows HPV in the endosome at 8 hours post-infection (h.p.i.), second row shows HPV in the endosome at 16 h.p.i., and bottom row shows HPV in the trans-Golgi network (TGN) at 16 h.p.i. Note that JX2 does not inhibit initial localization of HPV in the endosome but causes the virus to accumulate in the endosome at 16 h.p.i. and prevents its arrival in the TGN. Panel B modified from [22].
One of the four inhibitory traptamers, designated JX2, causes HPV to accumulate in the endosome without reaching the trans-Golgi network (TGN) during entry (Fig. 4A and B). The other traptamers do not cause endosomal accumulation [22, 24]. In order to exit the endosome during entry, HPV requires the action of a cellular trafficking protein complex named retromer, which itself is regulated by the small G protein, Rab7 [18, 25–27]. JX2 inhibits the activityTBC1D5, a GTPase-activating protein that regulates Rab7 activity [22, 28] and thereby causes the accumulation of GTP-bound Rab7, which inhibits the ability of retromer to dissociate from HPV and sort the virus from the endosome into the retrograde transport pathway (Figs. 4A and B). Not surprisingly, TBC1D5 knockdown has the same phenotype as JX2 expression and causes incoming HPV to accumulate in the endosome. Thus, by analyzing a traptamer selected for its ability to inhibit HPV infection, we identified TBC1D5 as a cell protein required for HPV endosome exit during virus entry and revealed the role of Rab7 in retromer function during HPV entry. Another traptamer directs incoming HPV to the lysosome, a non-productive destination, and still another causes the virus to accumulate in the TGN without progressing to the nucleus. The mechanisms by which these traptamers exert these diverse effects is under investigation. In preliminary results, we have also isolated traptamers that inhibit enveloped viruses.
Limitations and advantages of traptamer screening
There are several theoretical limitations and advantages of traptamer screening. One limitation is that only transmembrane proteins are directly targeted by this approach. However, since approximately 30% of all eukaryotic proteins appear to have transmembrane domains [29, 30], many if not most cellular processes are amenable to traptamer screening. A clear disadvantage of traptamer screening compared to gene knockdown/knockout screening is that the target of the active traptamers is not immediately obvious, whereas it can be directly deduced by the sequence of the active RNAs in knockdown and knockout experiments. However, it is possible to identify traptamer targets by co-immunoprecipitation followed by western blotting (as was the case for TBC1D5) or mass spectrometry. It might also be possible to functionalize traptamers by fusing them to a non-specific biotin ligase such as APEX2 [31] or to a ubiquitin ligase recruiting element to facilitate the identification of targets. As is the case for other screening strategies, off-target effects could occur. In limited surveys of candidate off-target proteins, traptamers appear to be highly specific [11, 12, 32], but once a potential traptamer target is identified, additional experiments are required to establish its importance in the studied process.
A theoretical advantage of traptamer screening follows from the fact that a traptamer can influence a process either by activating or by inhibiting a protein target. In contrast, knockdown/knockout screens only inhibit expression of their targets. Thus, target proteins that induce the desired phenotype when activated could be directly identified in a traptamer screen, but not in a knockdown screen. Second, traptamers have the potential to modulate the activity of their target proteins in subtler ways than merely affecting protein levels. This is illustrated most clearly by different traptamers that bind to the EPO receptor transmembrane domain and elicit distinct signaling patterns and cell phenotypes [8, 9]. Thus, traptamers may perturb an activity of a protein involved in the studied process while sparing other activities of the protein, including those required for cell viability. Therefore, it is possible that traptamer screening could identify hits that are proteins essential for cell growth, which are largely invisible to knockdown/knockout screens.
Unlike interfering RNAs or sgRNAs, the traptamer themselves can be readily localized in cells and can be directly used in affinity purification experiments. Finally, different hits are often identified when independent labs conduct knockdown/knockout screens of the same process. Presumably, the radically different design of traptamer screening is even more likely to yield divergent hits compared to conventional screens. Indeed, TBC1D5 was not identified in previous screens for HPV entry factors e.g., [18, 33–35], showing the utility of traptamer screening.
Conclusions
Functional genetics screens that modulate gene expression have revolutionized the study of mammalian cells. We describe an orthogonal screening strategy that is based on modulation of protein activity with artificial transmembrane proteins named traptamers. Our results indicate that traptamer screening will complement more standard screening strategies to allow the identification of new proteins required for a wide variety of cellular processes. In addition, the traptamers themselves will be valuable tools to dissect these processes in detail.
Acknowledgments
The authors thank Jan Zulkeski for assistance in preparing this manuscript. The research described in the DiMaio lab was supported by grants from the NIH to D.D. (R01 AI102876; R35 CA242462) and by a multi-PI grant from the NCI to B. Tsai (University of Michigan) and D.D. (R01 AI150897). The funding sources had no role in study design; in the collection, analysis or interpretation of data; in the writing of the report; and in the decision to submit the article for publication. The authors declare no conflicts of interest.
Abbreviations
- BPV
bovine papillomavirus
- HPV
human papillomavirus
- PLA
proximity ligation assay
- PDGF
platelet-derived growth factor
- h.p.i.
hours post-infection
- CCR5
C-C chemokine receptor 5
- TGN
trans-Golgi network
- PCR
polymerase chain reaction
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest. The funding sources had no role in study design; in the collection, analysis or interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
References
- 1.Shalem O, Sanjana NE & Zhang F (2015) High-throughput functional genomics using CRISPR-Cas9, Nat Rev Genet. 16, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva J, Chang K, Hannon GJ & Rivas FV (2004) RNA-interference-based functional genomics in mammalian cells: reverse genetics coming of age, Oncogene. 23, 8401–9. [DOI] [PubMed] [Google Scholar]
- 3.Chang H, Pan Y, Landrette S, Ding S, Yang D, Liu L, Tian L, Chai H, Li P, Li DM & Xu T (2019) Efficient genome-wide first-generation phenotypic screening system in mice using the piggyBac transposon, Proc Natl Acad Sci U S A. 116, 18507–18516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clements KE, Schleicher EM, Thakar T, Hale A, Dhoonmoon A, Tolman NJ, Sharma A, Liang X, Imamura Kawasawa Y, Nicolae CM, Wang HG, De S & Moldovan GL (2020) Identification of regulators of poly-ADP-ribose polymerase inhibitor response through complementary CRISPR knockout and activation screens, Nat Commun. 11, 6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudkov AV, Kazarov AR, Thimmapaya R, Axenovich SA, Mazo IA & Roninson IB (1994) Cloning mammalian genes by expression selection of genetic suppressor elements: association of kinesin with drug resistance and cell immortalization, Proc Natl Acad Sci U S A. 91, 3744–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore DT, Berger BW & DeGrado WF (2008) Protein-protein interactions in the membrane: sequence, structural, and biological motifs, Structure. 16, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheideman EH, Marlatt SA, Xie Y, Hu Y, Sutton RE & DiMaio D (2012) Transmembrane protein aptamers that inhibit CCR5 expression and HIV coreceptor function, J Virol. 86, 10281–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He L, Cohen EB, Edwards APB, Xavier-Ferrucio J, Bugge K, Federman RS, Absher D, Myers RM, Kragelund BB, Krause DS & DiMaio D (2019) Transmembrane Protein Aptamer Induces Cooperative Signaling by the EPO Receptor and the Cytokine Receptor beta-Common Subunit, iScience. 17, 167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cammett TJ, Jun SJ, Cohen EB, Barrera FN, Engelman DM & Dimaio D (2010) Construction and genetic selection of small transmembrane proteins that activate the human erythropoietin receptor, Proc Natl Acad Sci U S A. 107, 3447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman-Cook LL, Dixon AM, Frank JB, Xia Y, Ely L, Gerstein M, Engelman DM & DiMaio D (2004) Selection and characterization of small random transmembrane proteins that bind and activate the platelet-derived growth factor beta receptor, J Mol Biol. 338, 907–20. [DOI] [PubMed] [Google Scholar]
- 11.He L, Steinocher H, Shelar A, Cohen EB, Heim EN, Kragelund BB, Grigoryan G & DiMaio D (2017) Single methyl groups can act as toggle switches to specify transmembrane Protein-protein interactions, eLife. 6:e27701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heim EN, Marston JL, Federman RS, Edwards AP, Karabadzhak AG, Petti LM, Engelman DM & DiMaio D (2015) Biologically active LIL proteins built with minimal chemical diversity, Proc Natl Acad Sci U S A. 112, E4717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chacon KM, Petti LM, Scheideman EH, Pirazzoli V, Politi K & DiMaio D (2014) De novo selection of oncogenes, Proc Natl Acad Sci U S A. 111, E6–E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petti LM, Talbert-Slagle K, Hochstrasser ML & DiMaio D (2013) A single amino acid substitution converts a transmembrane protein activator of the platelet-derived growth factor beta receptor into an inhibitor, J Biol Chem. 288, 27273–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petti LM, Marlatt SA, Luo Y, Scheideman EH, Shelar A & DiMaio D (2018) Regulation of C-C chemokine receptor 5 (CCR5) stability by Lys-197 and by transmembrane protein aptamers that target it for lysosomal degradation (In press), J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen EB, Jun SJ, Bears Z, Barrera FN, Alonso M, Engelman DM & DiMaio D (2014) Mapping the homodimer interface of an optimized, artificial, transmembrane protein activator of the human erythropoietin receptor, PLoS One. 9, e95593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Day PM, Thompson CD, Schowalter RM, Lowy DR & Schiller JT (2013) Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection, J Virol. 87, 3862–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CD, Atwood WJ & DiMaio D (2013) Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus, Proc Natl Acad Sci USA. 110, 7452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie J, Zhang P, Crite M & DiMaio D (2020) Papillomaviruses Go Retro, Pathogens. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Kazakov T, Popa A & DiMaio D (2014) Vesicular trafficking of incoming human papillomavirus type 16 to the Golgi apparatus and endoplasmic reticulum requires gamma-secretase activity, mBio. 5, e01777–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodwin EC & DiMaio D (2000) Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways, Proc Natl Acad Sci USA 97, 12513–12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie J, Heim EN, Crite M & DiMaio D (2020) TBC1D5-Catalyzed Cycling of Rab7 Is Required for Retromer-Mediated Human Papillomavirus Trafficking during Virus Entry, Cell Rep. 31, 107750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie J, Zhang P, Crite M & DiMaio D (2020) High-resolution dissection of infectious HPV entry with artificial transmembrane proteins (Submitted).
- 24.Xie J & DiMaio D (2020) Traptamer screening: a new functional genomics approach to study virus entry and other cellular processes (In preparation). [DOI] [PMC free article] [PubMed]
- 25.Burd C & Cullen PJ (2014) Retromer: a master conductor of endosome sorting, Cold Spring Harbor perspectives in biology. 6, pii: a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popa A, Zhang W, Harrison MS, Goodner K, Kazakov T, Goodwin EC, Lipovsky A, Burd CG & DiMaio D (2015) Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during infection, PLoS Pathog. 11, e1004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu TT, Gomez TS, Sackey BK, Billadeau DD & Burd CG (2012) Rab GTPase regulation of retromer-mediated cargo export during endosome maturation, Mol Biol Cell. 23, 2505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borg Distefano M, Hofstad Haugen L, Wang Y, Perdreau-Dahl H, Kjos I, Jia D, Morth JP, Neefjes J, Bakke O & Progida C (2018) TBC1D5 controls the GTPase cycle of Rab7b, J Cell Sci. 131. [DOI] [PubMed] [Google Scholar]
- 29.Lehnert U, Xia Y, Royce TE, Goh CS, Liu Y, Senes A, Yu H, Zhang ZL, Engelman DM & Gerstein M (2004) Computational analysis of membrane proteins: genomic occurrence, structure prediction and helix interactions, Q Rev Biophys. 37, 121–46. [DOI] [PubMed] [Google Scholar]
- 30.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J & Ponten F (2015) Proteomics. Tissue-based map of the human proteome, Science. 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 31.Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA & Ting AY (2016) Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2, Nat Protoc. 11, 456–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Federman RS, Boguraev AS, Heim EN & DiMaio D (2019) Biologically Active Ultra-Simple Proteins Reveal Principles of Transmembrane Domain Interactions, J Mol Biol. 431, 3753–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aydin I, Weber S, Snijder B, Samperio Ventayol P, Kuhbacher A, Becker M, Day PM, Schiller JT, Kann M, Pelkmans L, Helenius A & Schelhaas M (2014) Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses, PLoS pathogens. 10, e1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipovsky A, Erden A, Kanaya E, Zhang W, Crite M, Bradfield C, MacMicking J, DiMaio D, Schoggins JW & Iwasaki A (2017) The cellular endosomal protein stannin inhibits intracellular trafficking of human papillomavirus during virus entry, J Gen Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergant Marusic M, Ozbun MA, Campos SK, Myers MP & Banks L (2012) Human papillomavirus L2 facilitates viral escape from late endosomes via sorting nexin 17, Traffic. 13, 455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]


