Dear Editor,
Currently, wild-type (WT) hamsters, ferrets, cats, and non-human primates are being used as COVID-19 animal models. However, no severe clinical symptoms develop in these animals1–5. Similarly, most of the human ACE2 (hACE2) transgenic mouse models develop only mild COVID-19 disease with only a few recent transgenic mouse models developing severe and fatal respiratory diseases6–10, calling for a better large animal model that could mimic the full spectrum of COVID-19 symptoms. Although the pig is thought to be a better model for human diseases in general11, due to its similarity to human anatomy, physiology, and immunology, previous studies have shown that WT pigs are not susceptible to SARS-CoV-22,12. Here we report our attempt to create the first humanized pig expressing the hACE2 receptor for COVID-19 research, speculating that humanization of the pig ACE2 receptor could make pigs susceptible to SARS-CoV-2.
To create a COVID-19 pig model with the targeted insertion of hACE2 at the pig ACE2 locus, we constructed a homologous recombination donor vector with homology arms of ~1 kb on each side. We used the CRISPR/Cas9 system to increase the chance of homologous recombination. Single-guide RNAs (sgRNAs) closest to the start codon of exon 1 were selected for the construction of sgRNA-expressing vector pX459 (Fig. 1a). To optimize the efficiency of the sgRNA, 9 sgRNAs were synthesized and assembled. IBRS-2 porcine kidney cells were electroporated with plasmids of pACE2-sgRNAs and Cas9. Sanger sequencing was used to identify the indels and evaluate the targeting efficiency for these sgRNAs. The cleavage efficiency of sgRNA3 was higher than that of other sgRNAs. Therefore, we chose sgRNA3 for subsequent experiments.
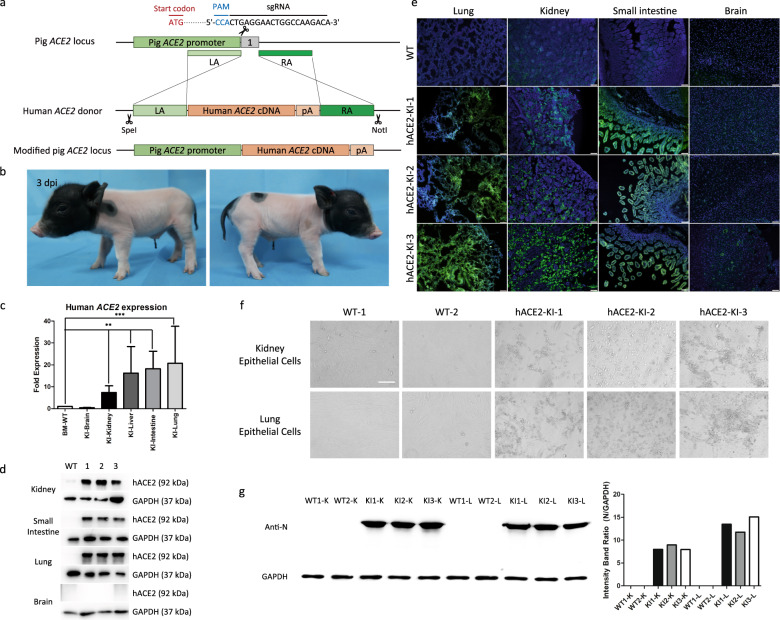
Fig. 1. Generation and characterization of a site‑specific hACE2 knock‑in pig model for COVID-19.
a Scheme for hACE2 integration in porcine fetal fibroblasts (PFFs) via CRISPR/Cas9-mediated homology-directed repair. b The physical appearance of representative cloned piglet. c Transcriptional analysis of hACE2 in cloned pigs showing mRNA expression levels of hACE2 in different tissues. Total RNA in different tissues of WT and knock-in pigs was extracted and subjected to reverse transcription, followed by qPCR to determine the expression of hACE2, with GAPDH as an internal reference gene. Data are presented as mean ± SD and are plotted from three independent experiments. **P < 0.01, ***P < 0.001, n = 3, unpaired one-tailed Student’s t-test. KI, knock-in. d Western blot showing expression of hACE2 protein in WT and knock-in (1, 2, 3) piglets. kDa, kilodalton. e Localization of hACE2 protein (green) in the lung, kidney, small intestine, and brain of WT and knock-in piglets indicated by immunofluorescent analysis. Nuclei are stained with DAPI (blue). Representative images were randomly chosen. Scale bars, 50 μm. KI, knock-in. f Primary epithelial cells of hACE2 knock-in piglets showed significant cytopathic effects after SARS-CoV-2 infection. Scale bars, 50 μm. KI, knock-in. g Western blot analysis for the expression of virus N protein in primary epithelial cells derived from WT and knock-in piglets. K, kidney epithelial cells; L, lung epithelial cells.
Porcine fetal fibroblasts (PFFs) isolated from embryos of Bama mini-pigs were electroporated with plasmids of pACE2-sgRNA3, Cas9, and donor linearized by in vitro cleavage of the donor vector by restriction enzymes (SpeI and NotI). We used puromycin selection (1 μg/mL for 2 days) to enrich positive cell colonies, and their genotype was identified using PCR and Sanger sequencing (Supplementary Fig. S1a, b). Among the 85 single cell colonies, three of them (3/85, 3.53%) were identified as positive for hACE2 insertion.
Next, the verified colonies were used as donor cells for somatic cell nuclear transfer (SCNT) into three surrogates. After about four months of pregnancy, these surrogates gave birth to nine genetically modified piglets (Fig. 1b). Genomic DNA was extracted from a variety of tissues of these piglets one day after birth, and their genotype was identified by PCR. The results showed that all of the piglets were positive for hACE2 insertion (Supplementary Fig. S1a).
The transcriptional level of hACE2 in several tissues of the knock-in piglets was quantified by qPCR, with GAPDH serving as the reference gene. The hACE2 mRNA levels in the kidney, liver, small intestine, and lung of the knock-in pigs were significantly higher than their WT littermates. In contrast, only low mRNA levels of hACE2 were detected in the brains of knock-in pigs (Fig. 1c).
To further determine whether the CRISPR/Cas9-mediated knock-in of hACE2 could increase the expression of hACE2, the protein was isolated from different organs in the WT and knock-in pigs. Western blot showed high expression of hACE2 in the knock-in pigs in all tissues except the brain (Fig. 1d).
We also performed immunofluorescent analysis to detect hACE2 in different organs of both WT and knock-in pigs. The results showed a high expression of hACE2 in the lung, kidney and small intestine of knock-in pigs, based on a strong green fluorescent signal. In contrast, very faint signal was seen in WT tissues and the brain of knock-in pigs, which was consistent with the results identified by qPCR and western blotting (Fig. 1e).
To determine the replication efficiency of SARS-CoV-2 in primary cells of the hACE2 knock-in pigs, lung and kidney epithelial cells were isolated and infected with SARS-CoV-2 (MOI = 0.01). After 72 h of infection, these primary cells of the hACE2 knock-in pigs showed significant cytopathic effects in contrast to wide-type swine cells (Fig. 1f).
At 72 h post-inoculation (hpi), the cells were collected for the detection of SARS-CoV-2 N protein by western blotting (Fig. 1g) and immunofluorescence assay (IFA) (Supplementary Fig. S2). Obvious fluorescent signal was detected in the cells of the knock-in pigs compared to the cells from WT pigs. As shown by western blot analysis, significant expression of viral N protein was detected in the cells from the knock-in pigs, but not in the cells from WT pigs. These results indicated that the primary epithelial cells from the hACE2 knock-in piglets are susceptible to SARS-CoV-2 infection.
In conclusion, we have shown that healthy hACE2 knock-in pigs can be generated by targeted insertion of hACE2 to the pig ACE2 locus. The expression of hACE2 protein is regulated by the endogenous swine promoter and appears to recapitulate the in vivo expression pattern. The expression patterns of hACE2 in the lung, kidney, testis, and intestine of our hACE2 pig models are similar to human and the humanized ACE2 mice8,13. In humanized ACE2 mice, the small intestine showed higher levels of hACE2 than other tissues by qPCR. In contrast, in our humanized hACE2 pigs the lungs show higher hACE2 expression levels than other organs.
Since the hACE2 knock-in pigs are created in this study by SCNT, they have a uniform genetic background and can be generated in large quantities in a short time. Also, severe cases of COVID-19 are often accompanied by underlying diseases, and pig models offer a unique opportunity to combine these existing diseases to reproduce severe cases of COVID-19. In addition to acute lung injury, some COVID-19 patients also experience organ damage, including acute kidney, heart, and liver dysfunction14–16, in particular for people with comorbidities such as hypertension, cardiovascular disease, and diabetes15. The humanized hACE2 pigs can be readily combined with other available pig models to accelerate the research of basic disease mechanisms, biomarkers, and treatment methods for high-risk people of COVID-19.
As SARS-CoV-2 mutants are rapidly emerging, scientists are scrambling to design and develop new vaccines, and are making continuous efforts to find antiviral drugs. The immediate urgency of treating severe disease and reining in viral escape variants calls for optimal animal models. Our hACE2 knock-in pigs have great potential to fulfill this need.
Supplementary information
Acknowledgements
We thank Dr. Lara Carroll (University of Utah) for the careful reading of the manuscript. We thank Xiaolan Zhang at the Institute of Microbiology, Chinese Academy of Sciences, for her technical support in confocal microscopy analysis. We are grateful for the help from the staff of the BSL-3 facility at the Institute of Microbiology, Chinese Academy of Sciences. This work was supported by the 2020 Research Program of Sanya Yazhou Bay Science and Technology City (202002011), the Open Project of Hainan Provincial Key Laboratory of Tropical Animal Reproduction & Breeding and Epidemic Disease Research (HKL20200301), the National Natural Science Foundation of China (32002180 and 32041010), the Strategic Priority Research Programme of the Chinese Academy of Sciences (XDB29010102), the NSFC Outstanding Young Scholars (31822055), and Youth Innovation Promotion Association of CAS (2017122 and 2021081).
Author contributions
S.W. conceived and designed the study. X.D., Z.G., C.C., and Y.Q. performed the cell culture and gene targeting experiments. F.G. and P.L. performed gene construction. W.F., P.J., and Y.B. performed virus challenge experiments. D.Y., T.H., X.D., C.C., Z.H., and J.R. performed nuclear transfer experiments. Z.G., W.F., P.J. and Y.Q. designed and performed histological experiments. S.W., D.Y., Y.B., and W.L. supervised the project. S.W., X.D., Z.G., and Y.B. analyzed the data and wrote the manuscript with input from all authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xuguang Du, Zihang Guo, Wenhui Fan, Tang Hai
Contributor Information
Yuhai Bi, Email: beeyh@im.ac.cn.
Dawei Yu, Email: ydw023@163.com.
Sen Wu, Email: swu@cau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41421-021-00313-x.
References
- 1.Sia SF, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi J, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YI, et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709.e702. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rockx B, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munster VJ, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 7.Jiang RD, et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58.e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, F.-L. et al. Rapid generation of ACE2 humanized inbred mouse model for COVID-19 with tetraploid complementation. Natl. Sci. Rev.8, (2020). [DOI] [PMC free article] [PubMed]
- 9.Zheng J, et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oladunni FS, et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020;11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walters EM, Wells KD, Bryda EC, Schommer S, Prather RS. Swine models, genomic tools and services to enhance our understanding of human health and diseases. Lab Anim. 2017;46:167–172. doi: 10.1038/laban.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlottau K, et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. 2020;1:e218–e225. doi: 10.1016/S2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman-Edelstein M, et al. Expression of the SARS-CoV-2 receptorACE2 in human heart is associated with uncontrolled diabetes, obesity, and activation of the renin angiotensin system. Cardiovasc. Diabetol. 2021;20:90. doi: 10.1186/s12933-021-01275-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen N, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.