Abstract
Slaughterhouse wastewater is considered a reservoir for antibiotic-resistant bacteria and antibiotic residues, which are not sufficiently removed by conventional treatment processes. This study focuses on the occurrence of ESKAPE bacteria (Enterococcus spp., S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, Enterobacter spp.), ESBL (extended-spectrum β-lactamase)-producing E. coli, antibiotic resistance genes (ARGs) and antibiotic residues in wastewater from a poultry slaughterhouse. The efficacy of conventional and advanced treatments (i.e., ozonation) of the in-house wastewater treatment plant regarding their removal was also evaluated. Target culturable bacteria were detected only in the influent and effluent after conventional treatment. High abundances of genes (e.g., blaTEM, blaCTX-M-15, blaCTX-M-32, blaOXA-48, blaCMY and mcr-1) of up to 1.48 × 106 copies/100 mL were detected in raw influent. All of them were already significantly reduced by 1–4.2 log units after conventional treatment. Following ozonation, mcr-1 and blaCTX-M-32 were further reduced below the limit of detection. Antibiotic residues were detected in 55.6% (n = 10/18) of the wastewater samples. Despite the significant reduction through conventional and advanced treatments, effluents still exhibited high concentrations of some ARGs (e.g., sul1, ermB and blaOXA-48), ranging from 1.75 × 102 to 3.44 × 103 copies/100 mL. Thus, a combination of oxidative, adsorptive and membrane-based technologies should be considered.
Subject terms: Applied microbiology, Environmental microbiology
Introduction
Currently, poultry represents one of the main sources of meat production worldwide1. In the first half of 2020, more than 800,000 tons of poultry were processed only in Germany2. Slaughtering, processing and cleaning and sanitizing of poultry production facilities are water-consuming processes. Depending on the slaughtering process, between 5,000 and 21,000 L of water per ton of meat is needed for processing3. This leads to high amounts of wastewater that arises along the slaughtering chain, which usually exhibits various enteric pathogens4. In addition to microbiological loads, slaughterhouse wastewater also exhibits a high organic content due to solved fibers, proteins, and fats5. Thus, the direct discharge of untreated livestock wastewater to surface waters is impractical and should be avoided because of environmental pollution and possible negative effects on human health. Moreover, after the prescribed withdrawal periods, it may further contain residues of antimicrobials commonly used in veterinary medicine as well as detergents and disinfectants6,7.
Interestingly, meat production has changed over the last 30 years, as it has doubled worldwide and is expected to double again until 20508. This implies a steadily increasing quantity of produced wastewater, which also forces an increase in the pollution loads discharged into the environment. Thus, mostly after pretreatment in on-site wastewater treatment plants (WWTPs), slaughterhouses commonly discharge their wastewater either directly into a river or other receiving body (direct dischargers) or to municipal WWTPs (indirect dischargers)9,10.
German slaughterhouses commonly treat their wastewater by different physicochemical and/or biological methods. This results in a moderate to high removal of nutrients and a reduction of the bacterial load by 1.1–3.4 log units6,11,12. Currently, advanced oxidation processes (AOPs) are becoming an interesting additional treatment option to the prevailing conventional methods. AOPs (e.g., ozone treatment) are currently discussed as effective technologies for inactivation of microorganisms, especially antibiotic-resistant bacteria and pathogens12. Ozone treatment has been shown to exhibit a high reduction efficacy of 98.4% against facultative pathogenic bacteria and their antibiotic resistance genes (ARGs)12. In German municipal WWTPs, microbial reduction to below the detection limit (101 cell equivalents per 100 mL) was recently reported in the case of additional advanced treatment technologies (ozonation and ultrafiltration)13. Thus, advanced treatment technologies could decrease the bacterial loads discharged into the aquatic environment, preventing the dissemination, inter alia, of clinically relevant antibiotic-resistant bacteria and their resistance determinants. However, upgrading WWTPs with advanced treatment technologies bears additional costs associated with high energy consumption, additional personnel and posttreatment of the effluents12,14.
Interestingly, in the EU, the use of advanced treatment technologies is not mandatory for operators of WWTPs in any sector (e.g., municipal, health care or industry), as no legal limits or reduction levels have been fitted for microbiological pollutants in wastewater. However, the occurrence of important pathogenic microorganisms in municipal and clinical wastewater is well documented15. Furthermore, antibiotic-resistant bacteria with zoonotic potential are prevalent in livestock wastewater16. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA), extended-spectrum β-lactamase (ESBL)-producing Escherichia coli/Klebsiella pneumoniae and mcr-1-carrying Enterobacterales, exhibiting resistance to highly and critically important antimicrobials, were already detected in biologically and physicochemically treated effluents of in-house WWTPs of German poultry slaughterhouses17,18. In addition, Savin et al. reported the detection of antimicrobial residues in wastewater from German pig slaughterhouses after conventional treatment in their on-site WWTPs19. This provides evidence on inadequate wastewater treatment by in-house WWTPs that enables further dissemination of clinically relevant bacteria of livestock origin and antimicrobial residues into surface waters and the environment.
However, comparable data on the occurrence of antibiotic-resistant facultative pathogenic bacteria and antimicrobial residues in livestock wastewater before and after treatment by advanced techniques such as ozonation are lacking. This study aimed to determine the occurrence of ESKAPE bacteria (Enterococcus spp., S. aureus, K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp.), ESBL-producing E. coli and antimicrobial residues in wastewater from a poultry slaughterhouse before and after conventional and advanced treatments. Furthermore, the reduction efficacy of the in-house WWTP upgraded with an ozone facility on clinically relevant ARGs and facultative pathogenic bacteria (i.e., E. coli, K. pneumoniae, A. baumannii, P. aeruginosa and enterococci) was evaluated.
Results
Detection of target ESKAPE bacteria and their phenotypic resistance
Overall, 97 isolates were recovered from water samples of the influent (n = 81) and effluent after physicochemical and biological treatments (n = 16). No target ESKAPE bacteria were detected in the effluent after subsequent ozone treatment. The majority of them belonged to the A. calcoaceticus-baumannii complex (ACB complex: 32.0%, 31/97), K. pneumoniae (24.7%, 24/97), E. coli (22.7%, 22/97) and S. aureus (14.4%, 14/97). The abundances of the E. cloacae complex and P. aeruginosa were low at 4.1% (4/97) and 2.1% (2/97), respectively. No VRE and CRE were detected.
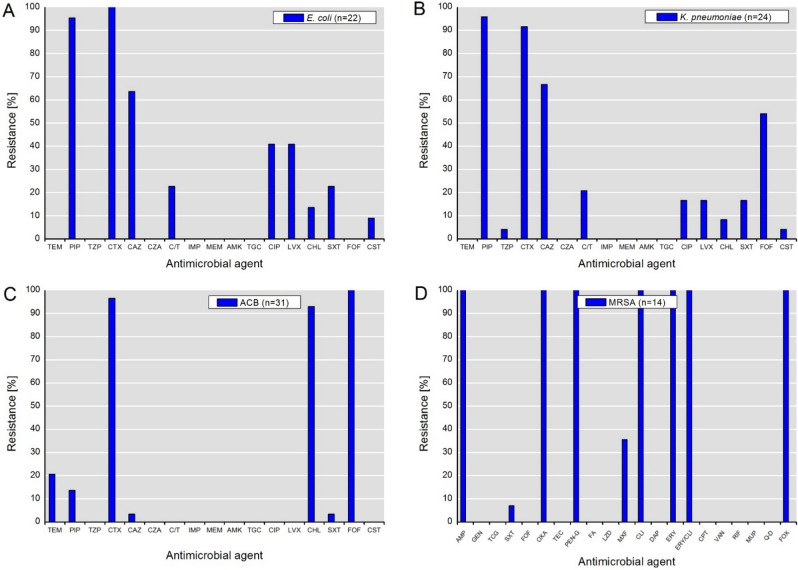
The phenotypic antimicrobial resistance of the recovered isolates is summarized in Fig. 1. Due to selective isolation, high rates of resistance to β-lactams (e.g., PIP and CTX) among isolates of E. coli and K. pneumoniae were expected. The highest 3MDRO rate (multidrug-resistant organisms) with combined resistance to PIP, CTX and CIP was exhibited by E. coli (40.9%), followed by K. pneumoniae (16.7%). Notably, all isolates were susceptible to carbapenems (IMP, MEM), ceftazidime-avibactam, amikacin and tigecycline. Interestingly, 22.7% and 20.8% of E. coli and K. pneumoniae isolates, respectively, exhibited resistance to the newly approved drug combination ceftolozane-tazobactam. Resistance to colistin was detected only at a low rate among E. coli (9.1%) and K. pneumoniae (4.2%) isolates. Furthermore, no 3MDRO phenotype was detected among isolates of the ACB complex. Moreover, considering the intrinsic resistance of ACB complex species to temocillin, cefotaxime, chloramphenicol and fosfomycin, only a minor percentage of isolates exhibited acquired resistance to ceftazidime and sulfamethoxazole-trimethoprim. Interestingly, in addition to the intrinsic resistance to ampicillin, oxacillin, penicillin-G, and cefoxitin, all MRSA isolates were resistant to clindamycin, erythromycin and the combination thereof.
Figure 1.
Phenotypic resistance to antimicrobial agents detected among isolates of (A) E. coli, (B) K. pneumoniae, (C*) ACB complex and (D**) MRSA. Abbreviations for antimicrobial agents: TEM temocillin, PIP piperacillin, TZP piperacillin-tazobactam, CTX cefotaxime; CAZ ceftazidime; CZA ceftazidime-avibactam; C/T ceftolozane-tazobactam; IMP imipenem, MEM meropenem, AMK amikacin, TGC tigecycline, CIP ciprofloxacin, LVX levofloxacin, CHL chloramphenicol, SXT sulfamethoxazole-trimethoprim, FOF fosfomycin, CST colistin, AMP ampicillin, GEN gentamicin; OXA oxacillin, TEC teicoplanin, PEN-G penicillin G, FA fusidic acid, LZD linezolid, DAP daptomycin, CPT ceftaroline,, VAN vancomycin, RIF rifampicin, MUP mupirocin, FOX cefoxitin; MXF moxifloxacin, CLI clindamycin; ERY erythromycin, Q-D synercid (quinupristin-dalfopristin). *Species of the ACB complex are considered intrinsically resistant to temocillin, cefotaxime, chloramphenicol and fosfomycin. **MRSA is considered intrinsically resistant to ampicillin, oxacillin, penicillin-G and cefoxitin.
Characterization of ARGs in target bacterial species
The majority of E. coli isolates (72.7%, 16/22) exhibited a blaTEM genotype, with blaTEM-116 accounting for 27.3%, as well as blaTEM-52 and blaTEM-1 accounting for 22.7% each, respectively. blaCTX-M-1 and blaSHV-12 were detected in 13.6% of E. coli isolates. In K. pneumoniae (n = 24), only genes of the blaSHV family were identified, with blaSHV-2 accounting for 66.7% and blaSHV-12 accounting for 8.3% of the isolates. Six K. pneumoniae isolates (25.0%) tested negative for bla genes encoding SHV, TEM, and CTX-M enzymes. Isolates of the E. cloacae complex (n = 4) carried blaSHV-12 (n = 2) and blaTEM-1 (n = 2).
No blaPER, blaGES, or blaVEB were detected among P. aeruginosa and isolates of the ACB complex. mcr-1.1 was detected in one colistin-resistant E. coli (1/2) isolate.
Molecular typing of ESBL-producing E. coli and MRSA
The majority of E. coli isolates were assigned to phylogroups B1 (50.0%) and E (27.3%), which are commonly associated with commensal strains. Less abundant phylogroups were represented by A and F (each 9.1%) as well as C (4.5%). Notably, extraintestinal pathogenic (ExPEC) groups B2 and D were not detected.
Recovered MRSA isolates (n = 14) belonged to the clonal complexes CC9 (t1430 and t13177, each 35.7%) and CC398 (t034, 28.6%).
Abundance of ARGs in water samples
According to Hembach et al.13, the abundance of “frequent” ARGs in influent, effluent after physicochemical and biological treatments, and subsequent ozone treatment is shown in Fig. 2. All measurements were performed after PMA treatment for living/dead discrimination. ermB and blaTEM exhibited the highest abundances of 5.61 × 106 and 1.48 × 106 gene copies per 100 mL, respectively, followed by sul1 (4.98 × 105) and tetM (9.25 × 104). After physicochemical and biological treatments, the highest significant reduction of 3.7 log units was achieved for blaTEM (p < 0.001), followed by ermB and tetM (each 2.7 log units) as well as sul1 (1 log unit). After the subsequent ozonation, blaTEM, sul1 (both p < 0.001) and ermB (p < 0.05) were further significantly reduced (6.6 – 34.4-fold).
Figure 2.
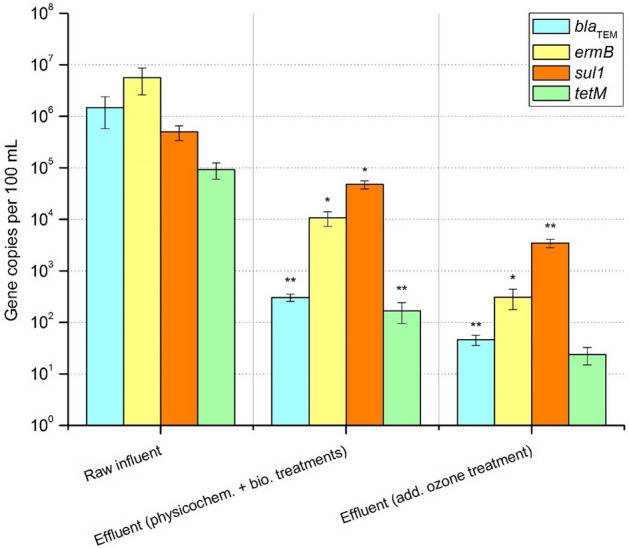
Abundance of “frequent” antibiotic resistance genes in influent and effluent after physicochemical and biological treatments and subsequent ozone treatment. Displayed are mean values with standard deviation. Significance is given by one-tailed nonparametric Mann–Whitney U test calculation and is shown by asterisks (*p ≤ 0.05, **p ≤ 0.001). Compared are gene copies per 100 mL in (i) raw influent with effluent after physicochemical and biological treatments and (ii) in effluent after physicochemical and biological treatments with effluent after subsequent ozone treatment.
The abundance of “intermediate and rare abundant” ARGs in the analysed samples is shown in Fig. 3. blaCTX-M-32 and mcr-1 were detected with the highest abundances of 3.58 × 104 and 3.17 × 104 gene copies per 100 mL, respectively, followed by blaOXA-48 (6.65 × 103), blaCMY-2 (4.07 × 103), blaCTX-M-15 (9.97 × 102), and vanA (1.30 × 102) gene copies per 100 mL. However, all of them were significantly reduced after physicochemical and biological treatments (p < 0.001), with reduction factors ranging from 2.2 log units in the case of carbapenemase blaOXA-48 to 4.2 log units in the case of blaCTX-M-32. Furthermore, blaCTX-M-15 and vanA were reduced to under the limit of detection (LOD). After subsequent ozonation, blaCTX-M-32 (p < 0.05) and mcr-1 (p < 0.001) were significantly reduced < LOD.
Figure 3.
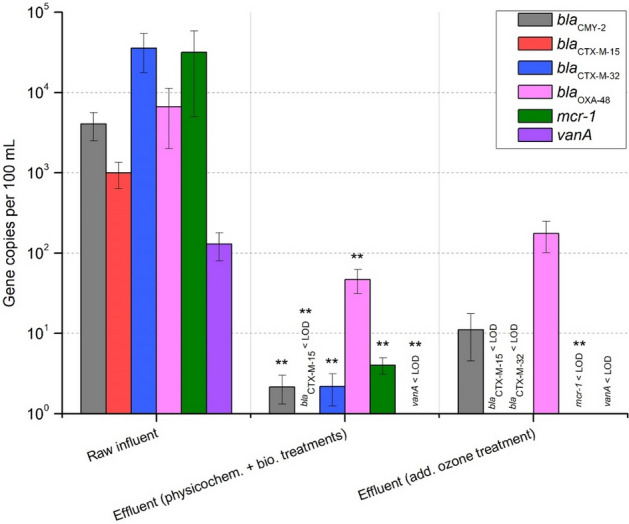
Abundance of “intermediate and rare abundant” antibiotic resistance genes in influent and effluent after physicochemical and biological treatments and subsequent ozone treatment. Displayed are mean values with standard deviation. Significance is given by one-tailed nonparametric Mann–Whitney U test calculation and is shown by asterisks (**p ≤ 0.001). Compared are gene copies per 100 mL in (i) raw influent with effluent after physicochemical and biological treatments and (ii) in effluent after physicochemical and biological treatments with effluent after subsequent ozone treatment. LOD – limit of detection.
Abundance of facultative pathogenic bacteria in water samples
E. coli, K. pneumoniae, A. baumannii, P. aeruginosa, and enterococci were quantified using species-specific gene markers after PMA treatment of the samples (Fig. 4). E. coli followed by enterococci and A. baumannii were the most predominant target species in the influent, with abundances of 6.90 × 104, 8.41 × 103 and 7.47 × 103 cell equivalents per 100 mL, respectively. P. aeruginosa exhibited the lowest concentration of 4.32 × 101 cell equivalents per 100 mL.
Figure 4.
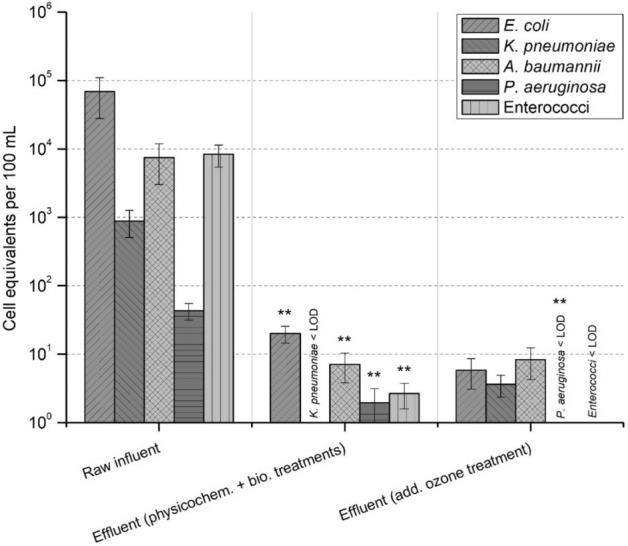
Abundance of facultative pathogenic bacteria in influent and effluent after physicochemical and biological treatments and subsequent ozone treatment. Displayed are mean values with standard deviation. Significance is given by one-tailed nonparametric Mann–Whitney U test calculation and is shown by asterisks (**p ≤ 0.001). Compared are gene copies per 100 mL in (i) raw influent with effluent after physicochemical and biological treatments and (ii) in effluent after physicochemical and biological treatments with effluent after subsequent ozone treatment. LOD – limit of detection.
After the physicochemical and biological treatments, all target species were reduced significantly (p < 0.001). Moreover, K. pneumoniae was reduced < LOD. After the subsequent ozonation, a nonsignificant reduction in E. coli and enterococci was observed, whereas P. aeruginosa was significantly reduced < LOD (p < 0.001).
Occurrence of antimicrobial residues in water samples
Of the wastewater samples, 55.6% (n = 10/18) were positive for antibiotic residues, which were detected only in the influent and effluent of the in-house WWTP after conventional treatment, indicating that antimicrobials were removed during ozonation (Table 1). The most prevalent antimicrobial classes detected were macrolides (38.9%, 7/18), sulfonamides (16.7%, 3/18), and fluoroquinolones (11.1%, 2/18).
Table 1.
Antimicrobial residues detected in wastewater samples from the in-house WWTP of the examined poultry slaughterhouse.
| Antimicrobiala [µg/L] | Influent (n = 6) | Effluent after physicochemical and biological treatments (n = 6) | Frequency c[%] | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nb1 | N2 | N3 | N4 | N6 | N1 | N2 | N3 | N4 | N5 | ||
| TYL | 3.7 | 0.83 | 12 | 0.27 | 0.24 | 0.08 | 0.11 | 38.9 | |||
| SXM | 0.06 | 0.04 | 11.1 | ||||||||
| SDD | 0.06 | 5.6 | |||||||||
| ENR | 3.9 | 0.47 | 11.1 | ||||||||
| AMP | < LOQ | - | |||||||||
aAbbreviations for antimicrobial agents: TYL tylosin, SXM sulfamethoxazole, SDD sulfadimidine, ENR enrofloxacin, AMP ampicillin.
bN—Sampling number.
cOnly samples with concentrations > LOQ were used for the evaluation.
Discussion
This study provides insights regarding the occurrence and impact of ESKAPE bacteria, including ESBL-producing E. coli, ARGs and antimicrobial residues, in wastewater samples from a German poultry slaughterhouse that might be disseminated to the environment. Furthermore, we evaluated the efficacy of ozonation treatment of wastewater for a reduction in bacterial loads and ARGs.
The high number of notified 3MDRO E. coli and K. pneumoniae isolates might be caused by the use of β-lactams (e.g., cephalosporines, penicillins) and fluoroquinolones for the treatment of animal infections in the veterinary medicine20. Cephalosporines and fluoroquinolones belong to the veterinary critically important antimicrobial agents (VCIAs) and are crucial for combating specific infectious diseases (e.g., septicaemias and respiratory and enteric diseases) in livestock. However, the use of VCIA also affects public health, as these antimicrobials are also considered the highest priority critically important antimicrobials (HPCIAs) for humans21.
Clinically associated fluoroquinolone resistance is mainly caused by chromosomal mutations in the quinolone resistance-determining regions (QRDRs) of gyrA and parC22,23. Furthermore, the cooccurrence of plasmid-mediated quinolone resistance (PMQR) genes and QRDR mutations can lead to high resistance (fluoro)quinolone phenotypes24. Recently, high percentages of ciprofloxacin resistance in ESBL-producing E. coli (50.5%) and K. pneumoniae (67.9%) from process and wastewater of poultry slaughterhouses have been reported17. Moreover, Savin et al. also notified a high number of PMQRs (31.4%) in 3MDRO extraintestinal pathogenic E. coli (ExPEC) and ESBL-producing K. pneumoniae (100%) from poultry slaughterhouses16.
The detected concentrations of enrofloxacin in untreated wastewater exceeded its Predicted No Effect Concentrations (PNECs) for resistance selection of 0.064 µg/L25, indicating that the withdrawal period of enrofloxacin might not be strictly adhered to. This selective pressure might further facilitate the development of fluoroquinolone resistance in clinically relevant bacteria. Thus, the use of fluoroquinolones should be restricted to individual treatments and no longer permitted for general medication of poultry herds. Thus, we strongly recommend forcing the development of alternative antimicrobial-free treatment strategies to avoid the use of similar antimicrobials in different compartments.
The high abundance of β-lactam (bla) resistance genes in the investigated cephalosporin-resistant E. coli correlates well with the detected prevalence of blaTEM-1, blaTEM-52, blaTEM-116, blaSHV-1, blaSHV-12 and blaCTX-M-1 in isolates of livestock wastewater and poultry products26,27. In general, ESBL-producing E. coli carrying diverse virulence determinants (i.e. for human colonization and infection) are widely disseminated in German poultry production16,28. While other reports have detected up to 16.2% ExPEC isolates from poultry wastewater, no isolate of this study was assigned to the ExPEC phylogroups B2 or D16.
The development and dissemination of ESBL-producing Enterobacteriaceae are not only promoted by the use of β-lactams in veterinary medicine but also facilitated by co- and cross-resistance through disinfectants, heavy metals, and other antimicrobials, since corresponding genes are often located on the same plasmid29. The cooccurrence of bla and mcr genes is of particular importance, since colistin was reintroduced into human therapy to treat infections caused by carbapenemase-producing Enterobacteriaceae (CPE) or multidrug-resistant A. baumannii and P. aeruginosa30. Moreover, mobile colistin resistance genes (esp. mcr-1) is considered to originate from livestock31. Colistin-resistant Enterobacteriaceae carrying mcr-1 on a wide variety of transmissible plasmids have already been reported from German process waters and wastewaters of livestock slaughterhouses, municipal WWTPs, and hospitals18,32.
Reports on the occurrence of ARGs in livestock wastewater conferring resistance to HPCIA have already been published33. However, there is a lack of qualitative and quantitative data for German slaughterhouses. We detected high abundances of the mcr-1, blaCMY-2 and blaCTX-M genes in untreated wastewater, which is worrying since they confer resistance to HPCIA in humans20. Their potential incorporation into clinically relevant bacteria might narrow antimicrobial treatment options and compromise their efficacy in cases of infection, especially if resistance against different last resort antibiotics (e.g., carbapenemases, mcr) occurs34,35. The human impact of livestock as a reservoir for combinations of antimicrobials of the last resort (i.e. blaNDM-1/mcr-1) have already been described36,37.
Of special concern is the occurrence of the carbapenemase gene blaOXA-48 among investigated wastewater samples detected by molecular-based methods. As selective cultivation did not yield any carbapenem-resistant isolates, culturable carbapenemase-producing Enterobacteriaceae can be excluded as predominant hosts for blaOXA-48. Thus, molecular detection of carbapenemase genes in environmental samples has only limited value, as the host species is not identified. However, other studies demonstrated different transferable plasmids carrying the blaOXA-48 gene in Enterobacteriaceae, which might not be cultivable or underrepresented in the analysed isolates of our study38,39. A probable reservoir for blaOXA-48 might be Shewanella spp., as this gene naturally occurs in these bacteria. As Shewanella spp. are not of particular clinical relevance, they might play a role as drivers of blaOXA-48 spread to clinically relevant bacteria (i.e. ESKAPE)40. The slight increase of blaOXA-48 in effluent samples treated with zone might indicate its selective properties regarding antibiotic-resistant, and more ozone-tolerant bacterial species41.
The presence of the vanA gene in untreated wastewater is worrying, as vancomycin also represents an antimicrobial of the last resort that is critically important for human medicine21. However, cultural analysis for vancomycin-resistant enterococci (VRE) was negative, which is in line with other studies reporting the absence or rare occurrence of VRE in livestock wastewater from German slaughterhouses17,19. This might indicate the presence of vancomycin-variable enterococci (VVE), which are vanA-positive but phenotypically susceptible to vancomycin42. Because of their susceptibility to vancomycin, traditional methods fail to detect VVE. vanA encodes inducible high-level resistance to glycopeptides, and VVE have the ability to revert into vancomycin-resistant phenotypes upon vancomycin exposure42. However, glycopeptides are not approved for the treatment of livestock in the EU, and the use of avoparcin in feed as a growth promoter was banned in Germany in 199643. Moreover, after treatments in WWTPs, vanA and enterococci were reduced below the detection limit. Thus, the role of agriculture in Germany in the development of glycopeptide-resistant enterococci is less significant than that in the general community and hospitals. Pseudomonas spp., Aeromonas spp. and Raoutella spp. isolated from surface water have also been reported to carry the vanA gene44.
However, coselection by macrolides, tetracycline, or copper might play an important role in maintaining the persistence of VRE in livestock45–47. Macrolides and tetracyclines together with penicillins and sulfonamides belong to the VCIA, for which there are fewer alternatives available48. In 2018, 571.7 tons of antimicrobials of these classes were sold to veterinarians in Germany, making up 79.1% of the total amount49. Interestingly, the supplied quantities of these antimicrobials correlate well with the abundances of particular ARGs in the investigated wastewater samples. Moreover, high abundances of ermB, sul1 and tetM genes are in line with other studies reporting a high prevalence of these resistance determinants in isolates recovered from livestock wastewater and poultry products, inter alia, in ExPEC and MDR E. coli isolates16.
The high abundance of ermB in wastewater is critical since ermB encodes resistance to macrolides, which are HPCIAs for human medicine and are crucial for the treatment of severe Campylobacter infections, particularly in children21. Furthermore, erm genes could be transferred to gram-positive pathogens (e.g., staphylococci, streptococci, enterococci), resulting in MLSB (macrolide, lincosamide, streptogramin B) cross-resistance, compromising the efficacy of further antibiotics such as erythromycin and clindamycin50. This might be the reason for the high resistance rates of MRSA to erythromycin and its combination with clindamycin. MRSA of CC9 and CC398, which were detected in this study, have already been reported in wastewater from poultry slaughterhouses and in retail poultry products17,51. They were also isolated from human infections, especially in regions with high livestock production, underlying their zoonotic potential52. Interestingly, the detected concentration of tylosin (12 µg/L) might exert selective pressure on species with resistance to macrolides, including MRSA, as it exceeded its PNEC of 4 µg/L. However, detected antimicrobials were eliminated after ozonation, which is in line with other studies reporting high degradation rates (70–100%) of macrolides as well as sulfonamides and fluoroquinolones depending on ozone dose and reaction time53. The removal of various organic micropollutants, which might not be reduced by other advanced treatment technologies, e.g., ultrafiltration, is an advantage of ozonation.
However, additional ozonation following conventional treatment did not significantly reduce the loads of facultative pathogenic bacteria, which might be due to different factors, such as ozonation time, concentration of the applied ozone, wastewater composition and physical characteristics of the examined organisms41. It is important to note that a high number of bacterial cells in wastewater might be aggregated in flocs, protecting the inner cells from the damaging effects of ozone. Comparable results regarding microbial reduction rates were observed in German municipal WWTPs after conventional treatment54.
Conventional treatment significantly reduced the abundances of ARGs conferring resistance to clinically relevant antimicrobials. Some of them (i.e., mcr-1, blaCTX-M-32) were further reduced below the limit of detection after subsequent additional treatment with ozone, underlying the importance of advanced treatment. However, despite the significant reduction through conventional and advanced treatments, effluents still exhibited relatively high concentrations of some ARGs, e.g., sul1, ermB and blaOXA-48 (> 102 copies/100 mL). Similar reduction rates were reported by Czekalski and colleagues41. Ozonation does not show the best effect on the reduction of ARGs in comparison to other methods, e.g., chlorination or ultrafiltration. This might be due to low ozone dosages frequently used in practice13,55. However, increasing the amount of ozone might lead to the release of harmful products to the environment and have an ecotoxicological effect56. Applying additional filtration steps (e.g., charcoal or sand filtration) after ozonation to remove remaining ozone and unwanted byproducts may result in bacterial regrowth13. Moreover, the receiving water bodies can support bacterial regrowth if they contain the necessary nutrients. Iakovides et al. reported the regrowth of total and antibiotic-resistant E. coli after the stress caused by ozone was relieved, indicating the importance of proper setting of ozonation parameters53. For wastewater exhibiting high concentrations of ARGs and antibiotic-resistant bacteria, a combination of oxidative, adsorptive (charcoal or sand filtration), and membrane-based technologies should be considered to interrupt dissemination of (facultative) pathogenic antibiotic-resistant bacteria to the environment.
Material and methods
Examined poultry slaughterhouse and its wastewater management
The investigated poultry slaughterhouse exhibited a capacity of > 100,000 slaughtered chickens per day by producing 3,600 m3 wastewater that was treated in an in-house wastewater treatment plant (WWTP) before being discharged into a preflooder and a receiving waterbody (i.e., river).
On-site treatment of wastewater is based on physicochemical, biological, and advanced oxidation processes. First, the wastewater was mechanically pretreated using screeners and sieves. Fat and greases are removed using grease traps. Afterwards, the wastewater is treated by dissolved air flotation with a subsequent biological treatment by activated sludge. Additionally, the in-house WWTP was equipped with an ozone system (Xylem Water Solutions Deutschland GmbH, Großostheim, Germany). The ozone dosage used was 75 g/m3, and the contact time varied between 15 and 30 min, depending on the water flow rate.
Sampling and sample preparation
In a two-month period, the in-house WWTP of a poultry slaughterhouse was sampled six times with a minimum time interval of 1 week. During sampling, a total of 18 samples were collected representing the influent (n = 6), the effluent after physicochemical and biological treatments (n = 6) and the effluent after ozone treatment (n = 6). The samples were taken as qualified samples according to the German standard methods for the examination of water, wastewater, and sludge (DIN 38402-11:2009-02)57. Therefore, five subsamples of 200 mL in two-minute sampling intervals were collected and mixed. The samples were transported to the laboratory in a Styrofoam box cooled to 5 ± 2 °C and were further processed within 24 h. Influent samples were manually filtered using stomacher strainer bags with a tissue filter (pore size, 0.5 mm; VWR, Radnor, PA, USA) to remove large particles.
Cultivation, identification and susceptibility testing of target bacteria
The samples were subjected to culturing on selective media to isolate gram-negative ESBL-producing and carbapenemase-producing Enterobacteriaceae (E. coli, Klebsiella spp., Enterobacter spp., Citrobacter spp. ), nonfermenting A. baumannii and P. aeruginosa, as well as MRSA and vancomycin-resistant enterococci (VRE). Detailed information on their cultivation procedures has already been published17.
Species identification was performed by MALDI-ToF MS (bioMérieux, Marcy-l'Étoile, France) equipped with Myla software. The isolates were purified on Columbia agar supplemented with 5% sheep blood, and cryopreservation at − 80 °C in cryotubes (Mast Diagnostics, Reinfeld, Germany) was used for storage. Isolated target bacteria were further subjected to antimicrobial susceptibility testing by the microdilution method according to protocols of the European Committee on Antimicrobial Susceptibility Testing (EUCAST v 11.0) using the Micronaut-S MDR MRGN-Screening system for Gram-negatives and the MICRONAUT-S MRSA/GP testing panel for Gram-positive bacteria (MERLIN, Gesellschaft für mikrobiologische Diagnostika GmbH, Bornheim-Hersel, Germany)58. The results were evaluated based on clinical cut-off values provided by EUCAST59. The multidrug-resistance phenotype (3MDRO) was defined based on combined resistance to piperacillin (PIP), cefotaxime (CTX) and/or ceftazidime (CAZ), and ciprofloxacin (CIP) as previously described60.
Detection and analysis of selected antibiotic resistance genes (ARGs) in target bacteria
Template DNA for PCRs was prepared by boiling bacterial suspensions in 10 mM Tris–EDTA pH 8.0 (Sigma-Aldrich, St. Louis, MO, USA) according to Aldous et al.61. Enterobacterales with phenotypic resistance to 3rd-generation cephalosporins were screened by PCR for β-lactamase (bla) genes encoding enzymes SHV, TEM, and CTX-M (groups 1, 2, 8 and 9) as previously described62–64. Isolates of the A. calcoaceticus-baumannii complex and P. aeruginosa were examined for the presence of blaPER, blaGES, and blaVEB by PCR as described65. Colistin-resistant isolates (MIC > 2 mg/L) were screened for mcr-1 to mcr-5 as well as mcr-6 to mcr-9 genes using multiplex PCR protocols as described by Rebelo et al.66 and Borowiak et al.67, respectively. The obtained PCR amplicons were purified with the innuPREP DOUBLEpure Kit (Analytik Jena AG, Jena, Germany) and subjected to Sanger sequencing at Microsynth Seqlab (Göttingen, Germany).
Molecular typing of resistant bacterial isolates
Phylotyping of E. coli isolates (A, B1, B2, C, D, E, F, clade I-V) was conducted as previously described68. MRSA isolates were spa-typed by amplifying and sequencing the Staphylococcus protein A repeat region according to Harmsen et al.69. The Ridom spa server database (http://www.spaserver.ridom.de) was used for assignment of spa types.
DNA extraction from water samples, quantification of antibiotic resistance genes (ARGs) and taxonomic gene markers
Volumes of 50 mL, 200 mL and 400 mL of influent, effluent after physicochemical and biological treatments and effluent after ozone treatment were subjected to DNA isolation after treatment with 0.25 mM propidium monoazide (PMA) (BLU-V viability PMA-kit, Qiagen GmbH, Hilden, Germany) as previously described12,14.
Antibiotic resistance genes (ARGs) that are most frequently detected in German urban wastewaters (sul1, ermB, blaTEM, tetM) as well as “intermediate and rare abundant”, which encode resistances to “Highest Priority Critically Important Antimicrobials” (blaCTX-M-15, blaCTX-M-32, blaCMY-2, mcr-1, blaOXA-48, blaNDM-1, vanA), were quantitatively amplified as previously published13,70. Furthermore, the facultative pathogenic bacteria E. coli (yccT), K. pneumoniae (gltA), A. baumannii (secE), P. aeruginosa (ecfX), and enterococci (23S rDNA) were quantified13,70. For qPCR analysis, a Bio-Rad Cycler CFX96 (CFX96 Touch Deep Well Real-Time PCR Detection System, Bio-Rad, Munich, Germany) and SYBR Green qPCR approach according to Hembach et al.70 and Jäger et al.12 were used. All samples were measured in technical triplicates. Cell equivalents were calculated according to Hembach et al.70 and normalized to 100 mL of water sample. In addition, a one-tailed nonparametric Mann–Whitney U test (Origin 8.1, OriginLab Corporation, Northampton, USA) was performed to identify the significance of the reduction in target genes during conventional and advanced wastewater treatments. Full primer sequences, qPCR reagent rations, thermal conditions and corresponding limit of detections (LODs) are listed in Supplemental Material (Tables S1, S2).
Determination of antimicrobial residues
Water samples were analysed for 45 antibiotics and two metabolites (N-acetylsulfamethoxazole and anhydroerythromycin) by high-performance liquid chromatography (HPLC) coupled to tandem mass spectrometry (MSMS) after dilution and filtration through hydrophilic PTFE filters (Macherey–Nagel, Düren, Germany) as previously described71. The analysed antibiotics belong to the following substance classes: β-lactams (i.e. penicillins, cephalosporins and carbapenems), tetracyclines, fluoroquinolones, sulfonamides (as well as their synergist trimethoprim), macrolides including tylosin and spiramycin, lincosamides, glycopeptides, oxazolidinones, and nitroimidazoles. All analysed antibiotics, including their limit of quantification (LOQ), are given in Supplemental Material Table S3. The LOD of each individual analyte was one-third of the respective LOQ.
Supplementary Information
Acknowledgements
We thank the staff of the participating slaughterhouse for their kind cooperation.
Author contributions
Project administration, conceptualization and data curation, M.S.; Funding acquisition, J.K. and T.S.; Investigation, M.S., J.A., G.B., J.A.H., N.H., R.M.S., E.S., A.V.; Methodology, M.S., J.A., N.H., A.V.; Writing—original draft, M.S.; Writing—review & editing, J.A., G.B., J.A.H., T.S., R.M.S., E.S., A.V. and J.K. All authors reviewed and approved the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was financed by the BMBF (Federal Ministry of Education and Research) funding measure HyReKA [02WRS1377]. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-96169-y.
References
- 1.Mottet A, Tempio G. Global poultry production: Current state and future outlook and challenges. Worlds Poult. Sci. J. 2017;73:245–256. doi: 10.1017/S0043933917000071. [DOI] [Google Scholar]
- 2.Statistisches Bundesamt (Destatis). Fleischproduktion im 1. Halbjahr 2020: -0,6 % gegenüber Vorjahr. Rückgang um 2,6 % im 2. Quartal 2020 sorgt für negatives Halbjahresergebnis. https://www.destatis.de/DE/Presse/Pressemitteilungen/2020/08/PD20_298_413.html (2021).
- 3.Genné I, Derden A. Handbook of Water and Energy Management in Food Processing. Elsevier; 2008. pp. 805–815. [Google Scholar]
- 4.Nielsen SS, et al. Welfare of pigs at slaughter. EFSA J. 2020;18:e06148. doi: 10.2903/j.efsa.2020.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustillo-Lecompte C, Mehrvar M. In: Physico-Chemical Wastewater Treatment and Resource Recovery. Farooq R, Ahmad Z, editors. InTech; 2017. [Google Scholar]
- 6.Bustillo-Lecompte CF, Mehrvar M. Slaughterhouse wastewater characteristics, treatment, and management in the meat processing industry: A review on trends and advances. J. Environ. Manage. 2015;161:287–302. doi: 10.1016/j.jenvman.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Chang X, et al. Determination of antibiotics in sewage from hospitals, nursery and slaughter house, wastewater treatment plant and source water in Chongqing region of Three Gorge Reservoir in China. Environ. Pollut. 2010;158:1444–1450. doi: 10.1016/j.envpol.2009.12.034. [DOI] [PubMed] [Google Scholar]
- 8.Shahbandeh, M. Global Meat Industry—Statistics & Facts. https://www.statista.com/topics/4880/global-meat-industry/ (2019).
- 9.COUNCIL DIRECTIVE of 21 May 1991 concerning urban waste water treatment (91/271/EEC) (2014).
- 10.DIRECTIVE 2010/75/EU OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL of 24 November 2010 on industrial emissions (integrated pollution prevention and control) (Recast) (2010).
- 11.Baker BR, Mohamed R, Al-Gheethi A, Aziz HA. Advanced technologies for poultry slaughterhouse wastewater treatment: A systematic review. J. Dispersion Sci. Technol. 2021;42:880–899. doi: 10.1080/01932691.2020.1721007. [DOI] [Google Scholar]
- 12.Jäger T, et al. Reduction of antibiotic resistant bacteria during conventional and advanced wastewater treatment, and the disseminated loads released to the environment. Front. Microbiol. 2018;9:2599. doi: 10.3389/fmicb.2018.02599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hembach N, Alexander J, Hiller C, Wieland A, Schwartz T. Dissemination prevention of antibiotic resistant and facultative pathogenic bacteria by ultrafiltration and ozone treatment at an urban wastewater treatment plant. Sci. Rep. 2019;9:12843. doi: 10.1038/s41598-019-49263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander J, Hembach N, Schwartz T. Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci. Rep. 2020;10:8952. doi: 10.1038/s41598-020-65635-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller H, et al. Dissemination of multi-resistant Gram-negative bacteria into German wastewater and surface waters. FEMS Microbiol. Ecol. 2018;94:fiy057. doi: 10.1093/femsec/fiy057. [DOI] [PubMed] [Google Scholar]
- 16.Savin M, et al. Clinically relevant Escherichia coli isolates from process waters and wastewater of poultry and pig slaughterhouses in Germany. Microorganisms. 2021;9:698. doi: 10.3390/microorganisms9040698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savin M, et al. ESKAPE bacteria and extended-spectrum-β-lactamase-producing Escherichia coli isolated from wastewater and process water from German poultry slaughterhouses. Appl. Environ. Microbiol. 2020;86:e02748. doi: 10.1128/AEM.02748-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savin M, et al. Colistin-resistant enterobacteriaceae isolated from process waters and wastewater from German Poultry and Pig Slaughterhouses. Front. Microbiol. 2020;11:575391. doi: 10.3389/fmicb.2020.575391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savin M, et al. Antibiotic-resistant bacteria and antimicrobial residues in wastewater and process water from German pig slaughterhouses and their receiving municipal wastewater treatment plants. Sci. Total Environ. 2020;727:138788. doi: 10.1016/j.scitotenv.2020.138788. [DOI] [PubMed] [Google Scholar]
- 20.European Food Safety Authority The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019;17:e05598. doi: 10.2903/j.efsa.2019.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization. Critically Important Antimicrobials for Human Medicine. Ranking of Medically Important Antimicrobials for Risk Management of Antimicrobial Resistance Due to Non-human Use. https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf?ua=1.
- 22.Ruiz J. Mechanisms of resistance to quinolones: Target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 2003;51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 23.Moon DC, et al. Emergence of a new mutation and its accumulation in the topoisomerase IV gene confers high levels of resistance to fluoroquinolones in Escherichia coli isolates. Int. J. Antimicrob. Agents. 2010;35:76–79. doi: 10.1016/j.ijantimicag.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Juraschek K, et al. Outcome of different sequencing and assembly approaches on the detection of plasmids and localization of antimicrobial resistance genes in commensal Escherichia coli. Microorganisms. 2021;9:598. doi: 10.3390/microorganisms9030598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bengtsson-Palme J, et al. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 2016;572:697–712. doi: 10.1016/j.scitotenv.2016.06.228. [DOI] [PubMed] [Google Scholar]
- 26.Laube H, et al. Longitudinal monitoring of extended-spectrum-beta-lactamase/AmpC-producing Escherichia coli at German broiler chicken fattening farms. Appl. Environ. Microbiol. 2013;79:4815–4820. doi: 10.1128/AEM.00856-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irrgang A, et al. Diversity of CTX-M-1-producing E. coli from German food samples and genetic diversity of the blaCTX-M-1 region on IncI1 ST3 plasmids. Vet. Microbiol. 2018;221:98–104. doi: 10.1016/j.vetmic.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Kola A, et al. High prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae in organic and conventional retail chicken meat, Germany. J. Antimicrob. Chemother. 2012;67:2631–2634. doi: 10.1093/jac/dks295. [DOI] [PubMed] [Google Scholar]
- 29.Cheng G, et al. Selection and dissemination of antimicrobial resistance in Agri-food production. Antimicrob. Resist. Infect. Control. 2019;8:158. doi: 10.1186/s13756-019-0623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azzopardi EA, Boyce DE, Thomas DW, Dickson WA. Colistin in burn intensive care: Back to the future? Burns. 2013;39:7–15. doi: 10.1016/j.burns.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y-Y, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet. Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 32.Zhao F, Feng Y, Lü X, McNally A, Zong Z. Remarkable diversity of Escherichia coli carrying mcr-1 from hospital sewage with the identification of two new mcr-1 variants. Front. Microbiol. 2017;8:2094. doi: 10.3389/fmicb.2017.02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diallo AA, et al. Persistence and prevalence of pathogenic and extended-spectrum beta-lactamase-producing Escherichia coli in municipal wastewater treatment plant receiving slaughterhouse wastewater. Water Res. 2013;47:4719–4729. doi: 10.1016/j.watres.2013.04.047. [DOI] [PubMed] [Google Scholar]
- 34.Tsui CKM, et al. Plasmid-mediated colistin resistance encoded by mcr-1 gene in Escherichia coli co-carrying blaCTX-M-15 and blaNDM-1 genes in pediatric patients in Qatar. J. Glob. Antimicrob. Resist. 2020;22:662–663. doi: 10.1016/j.jgar.2020.06.029. [DOI] [PubMed] [Google Scholar]
- 35.Chen, W. et al. Characterization of a clinical Enterobacter hormaechei strain belonging to epidemic clone ST418 co-carrying blaNDM-1, blaIMP-4 and mcr-9.1 (2020). [DOI] [PubMed]
- 36.Zheng B, et al. Coexistence of MCR-1 and NDM-1 in Clinical Escherichia coli Isolates. Clin. Infect. Dis. 2016;63:1393–1395. doi: 10.1093/cid/ciw553. [DOI] [PubMed] [Google Scholar]
- 37.Zheng B, et al. Complete genome sequencing and genomic characterization of two Escherichia coli strains co-producing MCR-1 and NDM-1 from bloodstream infection. Sci. Rep. 2017;7:17885. doi: 10.1038/s41598-017-18273-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger S, et al. Characterization of a new blaOXA-48-carrying plasmid in Enterobacteriaceae. Antimicrob. Agents Chemother. 2013;57:4064–4067. doi: 10.1128/AAC.02550-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludden C, et al. Sharing of carbapenemase-encoding plasmids between Enterobacteriaceae in UK sewage uncovered by MinION sequencing. Microb. Genom. 2017;3:e000114. doi: 10.1099/mgen.0.000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodford N, Wareham DW, Guerra B, Teale C. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: An emerging public health risk of our own making? J. Antimicrob. Chemother. 2014;69:287–291. doi: 10.1093/jac/dkt392. [DOI] [PubMed] [Google Scholar]
- 41.Czekalski N, et al. Inactivation of antibiotic resistant bacteria and resistance genes by ozone: From laboratory experiments to full-scale wastewater treatment. Environ. Sci. Technol. 2016;50:11862–11871. doi: 10.1021/acs.est.6b02640. [DOI] [PubMed] [Google Scholar]
- 42.Kohler P, et al. Prevalence of vancomycin-variable Enterococcus faecium (VVE) among vanA-positive sterile site isolates and patient factors associated with VVE bacteremia. PLoS ONE. 2018;13:e0193926. doi: 10.1371/journal.pone.0193926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casewell M, Friis C, Marco E, McMullin P, Phillips I. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J. Antimicrob. Chemother. 2003;52:159–161. doi: 10.1093/jac/dkg313. [DOI] [PubMed] [Google Scholar]
- 44.Nakipoğlu M, Yilmaz F, Icgen B. vanA gene harboring enterococcal and non-enterococcal isolates expressing high level vancomycin and teicoplanin resistance reservoired in surface waters. Bull. Environ. Contam. Toxicol. 2017;98:712–719. doi: 10.1007/s00128-016-1955-8. [DOI] [PubMed] [Google Scholar]
- 45.Aarestrup FM. Characterization of glycopeptide-resistant enterococcus faecium (GRE) from broilers and pigs in Denmark: Genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. J. Clin. Microbiol. 2000;38:2774–2777. doi: 10.1128/JCM.38.7.2774-2777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilsson O. Vancomycin resistant enterococci in farm animals - occurrence and importance. Infect. Ecol. Epidemiol. 2012;2:16959. doi: 10.3402/iee.v2i0.16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasman H, Aarestrup FM. Relationship between copper, glycopeptide, and macrolide resistance among Enterococcus faecium strains isolated from pigs in Denmark between 1997 and 2003. Antimicrob. Agents Chemother. 2005;49:454–456. doi: 10.1128/AAC.49.1.454-456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.World Organisation for Animal Health (OIE). OIE List of Antimicrobial Agents Of Veterinary Importance. https://www.oie.int/app/uploads/2021/03/a-oie-list-antimicrobials-may2018.pdf (2018).
- 49.Federal Ministry of Food and Agriculture. Report of the Federal Ministry of Food and Agriculture on the Evaluation of the Antibiotics Minimisation Concept introduced with the 16th Act to Amend the Medicinal Products Act (16th AMG Amendment). Evaluation based on section 58g of the Medicinal Products Act. https://www.bmel.de/SharedDocs/Downloads/EN/_Animals/Report-16thAMGAmendment.pdf?__blob=publicationFile&v=4 (2019).
- 50.Rosato A, Vicarini H, Leclercq R. Inducible or constitutive expression of resistance in clinical isolates of streptococci and enterococci cross-resistant to erythromycin and lincomycin. J. Antimicrob. Chemother. 1999;43:559–562. doi: 10.1093/jac/43.4.559. [DOI] [PubMed] [Google Scholar]
- 51.Tang Y, et al. Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int. J. Food Microbiol. 2017;249:72–76. doi: 10.1016/j.ijfoodmicro.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Köck R, et al. Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS ONE. 2013;8:e55040. doi: 10.1371/journal.pone.0055040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iakovides IC, et al. Continuous ozonation of urban wastewater: Removal of antibiotics, antibiotic-resistant Escherichia coli and antibiotic resistance genes and phytotoxicity. Water Res. 2019;159:333–347. doi: 10.1016/j.watres.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 54.Voigt AM, et al. Association between antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in anthropogenic wastewater: An evaluation of clinical influences. Chemosphere. 2020;241:125032. doi: 10.1016/j.chemosphere.2019.125032. [DOI] [PubMed] [Google Scholar]
- 55.Zhuang Y, et al. Inactivation of antibiotic resistance genes in municipal wastewater by chlorination, ultraviolet, and ozonation disinfection. Environ. Sci. Pollut. Res. Int. 2015;22:7037–7044. doi: 10.1007/s11356-014-3919-z. [DOI] [PubMed] [Google Scholar]
- 56.Wigh A, et al. Assessment of ozone or not-treated wastewater ecotoxicity using mechanism-based and zebrafish embryo bioassays. JEP. 2018;09:325–346. doi: 10.4236/jep.2018.94022. [DOI] [Google Scholar]
- 57.DIN. 38402-11:2009-02, German Standard Methods for the Examination of Water, Wastewater and Sludge: General Information (group A)—Part 11: Sampling of Waste Water (A 11).
- 58.European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters: Version 11.0. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf.
- 59.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf. (2021).
- 60.KRINKO Hygienemaßnahmen bei Infektionen oder Besiedlung mit multiresistenten gramnegativen Stäbchen. Empfehlung der Kommission für Kranken-haushygiene und Infektionsprävention (KRINKO) beim Robert Koch-Institut (RKI) Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012;55:1311–1354. doi: 10.1007/s00103-012-1549-5. [DOI] [PubMed] [Google Scholar]
- 61.Aldous WK, Pounder JI, Cloud JL, Woods GL. Comparison of six methods of extracting Mycobacterium tuberculosis DNA from processed sputum for testing by quantitative real-time PCR. J. Clin. Microbiol. 2005;43:2471–2473. doi: 10.1128/JCM.43.5.2471-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paterson DL, et al. Extended-spectrum beta-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: Dominance and widespread prevalence of SHV- and CTX-M-type beta-lactamases. Antimicrob. Agents Chemother. 2003;47:3554–3560. doi: 10.1128/AAC.47.11.3554-3560.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grimm V, et al. Use of DNA microarrays for rapid genotyping of TEM beta-lactamases that confer resistance. J. Clin. Microbiol. 2004;42:3766–3774. doi: 10.1128/JCM.42.8.3766-3774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gröbner S, et al. Emergence of carbapenem-non-susceptible extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tübingen, Germany. J. Med. Microbiol. 2009;58:912–922. doi: 10.1099/jmm.0.005850-0. [DOI] [PubMed] [Google Scholar]
- 65.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 66.Rebelo AR, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23:1700672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borowiak M, et al. Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011–2018) in Germany. Front. Microbiol. 2020;11:80. doi: 10.3389/fmicb.2020.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013;5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 69.Harmsen D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hembach N, et al. Occurrence of the mcr-1 Colistin resistance gene and other clinically relevant antibiotic resistance genes in microbial populations at different municipal wastewater treatment plants in Germany. Front. Microbiol. 2017;8:1282. doi: 10.3389/fmicb.2017.01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voigt AM, et al. The occurrence of antimicrobial substances in toilet, sink and shower drainpipes of clinical units: A neglected source of antibiotic residues. Int. J. Hyg. Environ. Health. 2019;222:455–467. doi: 10.1016/j.ijheh.2018.12.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.