Abstract
GM1 gangliosidosis is a rare lysosomal storage disease caused by a deficiency of β-galactosidase due to mutations in the GLB1 gene. We established a C57BL/6 mouse model with Glb1G455R mutation using CRISPR/Cas9 genome editing. The β-galactosidase enzyme activity of Glb1G455R mice measured by fluorometric assay was negligible throughout the whole body. Mutant mice displayed no marked phenotype at eight weeks. After 16 weeks, GM1 ganglioside accumulation in the brain of mutant mice was observed by immunohistochemical staining. Meanwhile, a declining performance in behavioral tests was observed among mutant mice from 16 to 32 weeks. As the disease progressed, the neurological symptoms of mutant mice worsened, and they then succumbed to the disease by 47 weeks of age. We also observed microglia activation and proliferation in the cerebral cortex of mutant mice at 16 and 32 weeks. In these activated microglia, the level of autophagy regulator LC3 was up-regulated but the mRNA level of LC3 was normal. In conclusion, we developed a novel murine model that mimicked the chronic phenotype of human GM1. This Glb1G455R murine model is a practical in vivo model for studying the pathogenesis of GM1 gangliosidosis and exploring potential therapies.
Keywords: GM1 gangliosidosis, mutation, mouse model, microglia
Impact statement
Lysosomal storage diseases are a group of 70 monogenic disorders characterized by the lysosomal accumulation of a substrate. As a group, LSDs affect ∼1 in 5000 live births; however, each individual storage disease is rare. GM1 gangliosidosis is due to a deficiency in lysosomal β-galactosidase resulting in the excessive accumulation of ganglioside GM1 in neuronal tissue and glycosaminoglycans or glycopeptides in visceral organs. Murine models have been critical in understanding the fundamentals of disease pathogenesis and for developing safe and effective therapies. In this work, we describe a new mutant mouse model of GM1 gangliosidosis, which mimicked the chronic phenotype of human GM1.
Introduction
GM1 gangliosidosis (GM1, MIM 230500) is a rare autosomal recessive lysosomal storage disorder (LSD) caused by a deficiency of β-galactosidase (GLB1; EC 3.2.1.23) due to mutations in the GLB1 gene. The deficiency of β-galactosidase leads to excessive accumulation of GM1ganglioside in neural and visceral tissue. Three clinical forms of GM1 gangliosidosis have been described. Type 1 is an infantile form with rapid cognitive decline, visceromegaly, skeletal abnormalities, and early death usually within two years of age. Type 2, the late-infantile/juvenile form, has a more heterogeneous phenotype, slowly progressing neurological defects, and fewer skeletal changes. The milder adult and chronic phenotype with moderate cognitive deterioration and progressive ataxia are termed type 3.1,2
The cellular pathogenesis of progressive neural degeneration in GM1 gangliosidosis remains unknown,3 and there are no approved treatments. Research is underway using animal models to evaluate gene therapy technologies, intravenous enzyme replacement therapies (ERT), and pharmacological chaperone therapies.4–6 We previously identified a GLB1 missense mutation c.1357G>A (p.G453R) (NM_000404.3) in a late-infantile patient with a genotype of p.S149F/p.G453R.7 In vitro, p.G453R was confirmed to be a pathogenic variant through its transient expression in COS-7 cells with the residual β-galactosidase activity being 0.55% of wild-type.
Inflammation of the central nervous system (CNS) is considered to significantly contribute to the pathogenesis and rapid progression of GM1 gangliosidosis.8 However, the initial cellular events leading to inflammation remain uncertain. Microglia are the resident macrophages of the CNS and play important roles in neurodegeneration. Microglial activation has been shown to precede the onset of neuron loss in many LSDs, which strongly implies that microglia play an important role in disease progression.9 In neurons and other cell types, the intracellular clearance of undesirable material predominantly takes place by autophagy.10 The whole process, from the formation of the autophagosome isolation membrane to the fusion with lysosomal vesicles, is referred to as autophagic flux.11 Malfunction of the lysosomal mechanism blocks flux across the entire autophagic-lysosomal network (ALN), which not only impacts neurons but also other glia that feed back to maintain a healthy CNS. ALN involves a number of vital cellular processes including inflammation, adaptive immunity, and mitochondrial homeostasis, which may be of significance in LSDs.
Human GLB1G453R is conserved to mouse Glb1G455R. In this study, we established a Glb1G455R knock-in (KI) mouse model using the CRISPR/Cas9 technique. We found an inflammatory response and abnormal autophagy in the brains of the GM1 mouse model; this suggests the possibility of new therapeutic targets.
Materials and methods
Generation of a C57BL/6 mouse model with Glb1G455R variant
Corresponding to human GLB1G453R, the p.G455R (NM_009752.2: c. 1363 G > A) at the murine Glb1 locus was edited using CRISPR/Cas9-mediated genome engineering at Cyagen. The G455R (GGA to AGA) mutation sites in the donor oligo were introduced into exon 14 by homology-directed repair (Figure 1(a)). One-cell stage zygotes were obtained by mating ICR males (Charles River Laboratories, Beijing, China) with C57BL/6N females (Charles River Laboratories). Cas9 mRNA and sgRNA were generated by in vitro transcription (AM1354+AM1908, Thermo, USA) and donor oligo harboring the G455R point mutation was synthesized (IDT, USA). A solution of 0.3 µM Cas9 mRNA, 0.75 µM sgRNA, and 0.75 µM donor oligo harboring the G455R point mutation was injected into the cytoplasm of pronuclear stage embryos. The injected embryos were cultured in KSOM medium (Millipore, Germany) overnight and those that developed to the two-cell stage were transferred into the oviducts of pseudopregnant ICR females (Charles River Laboratories). The sequences of sgRNA and ssDNA donor were as follows:
Figure 1.
Survival analysis and weight of Glb1G455R mice. (a) Schematic showing the generation of the Glb1 p.G455R knock-in mouse model. (b) Confirmation of the Glb1 p.G455R mutation by genomic DNA sequencing. (c) Decreased survival of KI/KI mice. For survival analysis n = 7 per group. (d) Serial body weights in female and male mice of the indicated genotype are shown as mean ± SEM. n for 4–13 mice per genotype. (A color version of this figure is available in the online journal.)
Glb1-sgRNA: 5′-GTTTCGATCAAGGATTCCTTGGG-3′
Donor oligo: 5′-CAGCTCCTTTTCTCTGATCTCACCCT CCATCCCCTCGGCTTTAACTCACAGGTCCCCCAAA GAATCCTTGATCGAAACCTCATGACAGCTCTGAA CATACGGGGGAAGGCTG GAGCCACGC TG-3′.
The potential founders that were born from microinjected embryos were genotyped and correct integration of the donor DNA carrying the G455R point mutation was confirmed by Sanger sequencing. The correctly integrated mutant founder mice were further back-crossed with wild type (WT) C57BL/6N mice for three additional generations before a detailed phenotype assessment was implemented. Male and female mice from the Glb1 KI/+ lines are fertile and produce normal litters. The WT, KI/+, and KI/KI mice produced by crossing heterozygous mice were subsequently genotyped by PCR and DNA sequencing analysis. WT littermates were used as controls. This study was approved by the Institutional Animal Care and Use Committee of the Guangzhou Medical University # 2018–242.
Animal care and experiments
All mice were housed in specific pathogen-free conditions and their genotypes were confirmed by PCR and DNA sequencing of tail tissue (Figure 1(b)). The following primers were employed: mGlb1-KI-F, GAAGGTTTCAAGT TCAAGGCAATCC and mGlb1-KI-R, AAGGAAAATAAA CTGCATTCCTTGGTAG. The coding and splicing regions of the GLB1 gene were amplified using Ex Taq DNA polymerase (TaKaRa Bio Inc., Japan). PCR was performed using 35 cycles of reaction at 94°C for 30 s, at 60°C for 30 s, and at 72°C for 30 s. The products were sequenced using an ABI 3730 sequencer (Applied Biosystems, USA) and the sequencing chromatograms were analyzed using Sequencher software (DNAMAN, Lynnon Biosoft, Inc., Canada). Humane endpoints for removal of an animal before the end of the planned experimental term were: the presence of limb paralysis, the inability to rear and feed normally, or weight loss greater than or equal to 15% of peak body weight. Homozygous mutant mice at 8, 16, and 32 weeks and age-matched WT, heterozygous littermates were anesthetized using sodium pentobarbital and perfused transcardially with saline followed by 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). The brains that were used for histological analysis were immersed in 4% PFA in PBS at 4°C overnight and then were processed into paraffin using standard histology techniques.
Paraffin sectioning
The fresh tissue was placed in fixative for more than 24 h. Tissues were removed from the fixative and the target tissue was trimmed with a scalpel in a ventilation cupboard, and then placed in a labeled dehydration box. Subsequently, the dehydration box was placed into a dehydrator (Donatello, DIAPATH, Italy) for dehydration with gradient alcohol. The protocol used included 75% alcohol for 4 h, 85% alcohol for 2 h, 90% alcohol for 2 h, 95% alcohol for 1 h, anhydrous ethanol I for 30 min, anhydrous ethanol II for 30 min, alcohol benzene for 5–10 min, xylene II for 5–10 min, 65°C melting paraffin I for 1 h, 65°C melting paraffin II for 1 h, and 65°C melting paraffin III for 1 h. The wax-soaked tissues were embedded in the embedding machine (JB-P5, Wuhan Junjie Electronics, China). First, the melted wax was placed into the embedding frame; before the wax solidified, the tissue was removed from the dewatering box and placed into the embedding frame according to the requirements of the embedding surface and a corresponding label was affixed. The frames were placed on a −20°C freezing table to cool. After the wax solidified, each block was removed from the embedding frame and repaired. Each trimmed wax block was placed in a paraffin slicer (RM2016, Leica, Germany) and slices were cut at a thickness of 4 μm. The slightly crumpled tissue sections were floated on the surface of a 40°C water bath from which the tissue was picked up onto a glass slide and dehydrated in an oven at 60°C. After the water-baked dried wax was melted, the slides were removed and stored at room temperature.
Assessment of mouse behavior at 8, 16, and 32 weeks
Forelimb suspension test
A metal bar elevated 40 cm above the floor was used for the gripping test.12 According to Feather-Schussler and Ferguson’s protocol, mice were allowed to grasp a bar with their forelimbs until they either fell from the bar or achieved the maximum trial duration of 10 min. The test was repeated three times for each mouse, and the maximum latency to fall from the bar was recorded.
Rotarod test
An accelerating rotarod with the capacity to test five mice simultaneously was used. Mice were placed on the rod for 3 min. For each session, mice were subjected to three rotarod trials. Testing was performed on a rotarod programmed to accelerate from 5 to 40 r/min. The trial was considered complete when either the mouse fell off the rotarod or after 180 s had elapsed. The time interval between trials was 15 min. We recorded the maximum latencies on the rotarod rod for three repetitions.
β-galactosidase activity assay
The β-galactosidase activities in tissue and dried blood spots from tails were measured using fluorogenic substrate 4-methylumbelliferyl-β-D-galactopyranoside (4-MU-β-D-Gal, Sigma, USA). For dried blood spots, briefly, a 3.2 mm dish was incubated with 100 μL substrate solution (0.75 mM 4-MU-β-D-Gal) for 2 h at 37°C. Then 120 μL of 0.2 M glycine buffer (pH 10.7) was added and fluorescence was measured using a fluorimeter (Perkin Elmer 1420 Multilabel Counter).
Tissue samples for β-galactosidase activity assay were homogenized using a VCX 130 Sonicator (Sonics & Materials Inc., USA). Protein homogenate samples of 10 μg were incubated with 20 μL of 0.75 mM 4-MUβ-D-Gal containing 0.1 M citrate acid and 0.2 M sodium phosphate at 37°C. After 30 min, 200 μL of 0.2 M glycine buffer (pH 10.7) was added to quench the reaction. This fluorescence was measured in a fluorimeter (Flx800, BioTek, USA) with an excitation of 360 nm and emission read of 460 nm. 4-MU (Sigma-Millipore) was used to generate a standard curve. β-gal enzyme activity was expressed in nanomoles of 4-MU released per hour, per milligram of protein (nmol/mg×h).
Histopathology and immunofluorescence
For the majority of the histopathology analyses, 4 µm thick coronal sections were cut, deparaffinized in xylene, and rehydrated with isopropyl alcohol, followed by 96%, 70%, and 60% ethanol for 10 min. Standard hematoxylin & eosin (H&E) staining was performed for analysis of brain histology. The H&E brain sections were assessed using a Leica microscope equipped with LAS X software.
For immunofluorescence staining, the sections were blocked in 10% normal goat serum in PBS for 30 min (at room temperature), incubated with primary antibodies (GM1, Abcam, ab23943, 1:50; IL-1β, Abcam, ab9722, 1:100; Iba1, Wako, 016–26721, 1:100; LC3B, CST, 3868S, 1:100), and diluted in 5% bovine serum albumin in phosphate-buffered saline (PBS) at 4°C overnight. After washing in PBS three times, the secondary antibodies were applied: Goat anti-rabbit IgG (H + L) cross-adsorbed secondary antibody conjugated to Alexa Fluor 488 (ThermoFisher, A11008) was used to detect anti-GM1 antibody and anti-IL-1β antibody. Anti-Iba1 antibody was detected by a goat anti-mouse IgG (H + L) cross-adsorbed secondary antibody conjugated to Alexa Fluor 594 (ThermoFisher, A11005). Nuclei were stained with DAPI. Immunofluorescence staining in the cerebral cortex was photographed at ×400 magnification using a fluorescence microscope (DM6000B, Leica, Germany). Eight randomly selected magnified fields of view were acquired for each section. For each group, six sections from different mice were viewed.
Transmission electron microscopy staining
Immediately after resection, cerebral cortical tissue was cut into 1 mm3 pieces. Tissues were post fixed with 1% OsO4 in 0.1 M PB (pH 7.4) for 2 h at room temperature in the dark. After being removed from the OsO4, the tissues were rinsed in 0.1 M PB (pH 7.4) for 15 min three times and were then dehydrated through the following sequence at room temperature: 30% ethanol for 20 min; 50% ethanol for 20 min; 70% ethanol for 20 min; 80% ethanol for 20 min; 95% ethanol for 20 min; two changes of 100% ethanol for 20 min per; finally, two changes of acetone for 15 min per. Resin penetration and embedding proceeded as follows. Acetone EMBed 812 = 1:1 for 2–4 h at 37°C; acetone: EMBed 812 = 1:2 overnight at 37°C; pure EMBed 812 for 5–8 h at 37°C. Tissues were then embedded in pure EMBed 812 at 37°C overnight and the embedding models with resin and sample resin were polymerized in an oven at 65°C for more than 48 h. Ultrathin sections (60–80 nm) were prepared and stained with 2% uranium acetate saturated alcohol solution and 2.6% lead citrate. The sections were observed under a transmission electron microscope (HT7700, Hitachi, Japan). To quantify autolysosomes in neurons, five cells per group were counted.
Quantitative PCR
Total RNA from the brain was extracted using TRIzol Reagent (Invitrogen, USA) and reverse transcribed with a PrimeScript RT Master Mix kit (TAKARA, Japan). Genomic DNA samples were measured in triplicate on a CFX96 Real-Time PCR system (Bio-RAD) using TB Green Premix Ex Taq II (TAKARA, Japan). Triplicate reactions for each sample were conducted. GAPDH had been used as the internal reference gene. The primers used for PCR are listed in Table 1.
Table 1.
Primer sequences for real time PCR assays.
| Gene | Forward primers | Reverse primers |
|---|---|---|
| Glb1 | TCCCACTGAACACTGAGGC | GGAGTATGAGGTCCGAAGAAT |
| IL1-β | TGGACCTTCCAGGATGAGGACA | GTTCATCTCGGAGCCTGTAGTG |
| TNF-α | GGTGCCTATGTCTCAGCCTCTT | GCCATAGAACTGATGAGAGGGAG |
| LC3 | CTGCCTGTCCTGGATAAGACCA | CTGGTTGACCAGCAGGAAGAAG |
Statistical analysis
GraphPad Prism 7 (v. 7.0a, GraphPad Software, Inc.) was used to display all data. For all experiments, WT, heterozygous, and homozygous mutant groups were compared by one-way factorial ANOVA combined with Scheffe’s test for all paired comparisons with SPSS 17.0 software. Data are presented as mean ± SEM. P < 0.05 indicates statistical significance.
Results
Phenotype of the Glb1G455R/G455R mouse model
The homozygous (KI/KI) mice were born in a Mendelian ratio and showed no gross morphologic defects. The lifespan of KI/KI mice was 47 weeks (mean ± SEM), which was significantly lower than that of WT and heterozygous (KI/+) mice (P < 0.001; Figure 1(c)). From 16 weeks, the body weight of KI/KI mice increased over that of the WT and KI/+ mice (Figure 1(d)). KI/KI males reached a maximum weight at an average of 36 weeks of age (50.7 ± 3.4 g compared to WT, 39.6 ± 1.6 g and KI/+, 46.9 ± 5.1 g) while female reached a maximum weight at an average of 32 weeks of age (32.2 ± 2.6 g compared to WT, 27.5 ± 2.0 g and KI/+, 26.6 ± 1.5 g). After peak weight, KI/KI mice began to lose weight, while WT and KI/+ mice continued to gain weight. Also, from 16 weeks, KI/KI mice began exhibiting ataxia and tremors; these symptoms were lifelong but gradually exacerbated with age (data not shown).
Assessment of mouse behavior
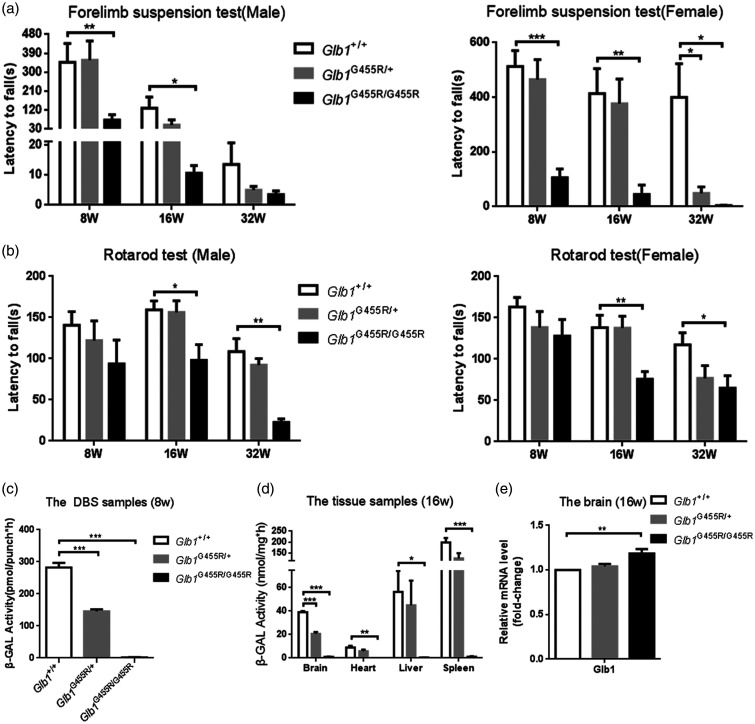
KI/KI mouse behavior in the forelimb grip strength test and rotarod test from 8 to 32 weeks showed progressive motor dysfunction compared to WT controls. Results of the mean of three maximum latencies in murine behavior tests are listed in Table 2. In the forelimb grip strength test, KI/KI mice from 8, 16, and 32 weeks were unable to hang from the metal bar for long, which indicated an impaired strength of the forelimbs (Figure 2(a)). There was no significant difference between KI/+ mice and WT controls, except for the female heterozygous mice at 32 weeks.
Table 2.
Data of the mouse behaviors.
|
Male |
Female |
|||||
|---|---|---|---|---|---|---|
| Latency to fall (s) | Glb1 +/+ | Glb1 G455R/+ | Glb1 G455R/G455R | Glb1 +/+ | Glb1 G455R/+ | Glb1 G455R/G455R |
| Forelimb grip strength test | ||||||
| 8 weeks | 347.2 ± 90.2 | 357.3 ± 91.9 | 72.9 ± 25.3 | 512.6 ± 58.7 | 465.4 ± 73.0 | 105.7 ± 31.7 |
| 16 weeks | 129.5 ± 52.7 | 49.5 ± 25.0 | 10.6 ± 2.5 | 413.0 ± 91.5 | 376.9 ± 89.9 | 44.3 ± 35.2 |
| 32 weeks | 13.5 ± 7.2 | 5.0 ± 1.2 | 3.5 ± 1.2 | 399.8 ± 122.8 | 48.8 ± 23.4 | 3.8 ± 1.0 |
| Accelerating rotarod test | ||||||
| 8 weeks | 140.6 ± 16.6 | 122.0 ± 23.9 | 93.6 ± 28.9 | 163.0 ± 11.6 | 138.3 ± 19.4 | 127.9 ± 20.1 |
| 16 weeks | 159.3 ± 10.8 | 156.3 ± 14.1 | 98.1 ± 19.0 | 138.1 ± 15.0 | 137.5 ± 14.3 | 75.5 ± 9.2 |
| 32 weeks | 108.6 ± 15.8 | 92.3 ± 7.9 | 22.5 ± 4.3 | 117.2 ± 14.7 | 76.8 ± 15.3 | 64.8 ± 15.2 |
Note: Data represent mean ± SEM.
Figure 2.
Progressive motor dysfunction and decreased β-galactosidase activity in KI/KI mice. (a) On the forelimb grip strength test, KI/KI mice (black bars) showed impaired strength of the forelimbs compared to wild type (WT) mice (white bars) and KI/+ mice (gray bars) (n = 4–11). (b) On the rotarod test of motor coordination and balance, the performance of KI/KI mice declined over time (n = 4–8). Results are expressed as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001. (c) The β-gal activity of DBS had decreased significantly in KI/KI mice (n = 24). (d) Significant deficiency of β-gal activity was observed in KI/KI mouse brain, heart, liver, and spleen tissues (n = 4). Results are expressed as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001. (e) qPCR analysis of Glb1 genes of WT, KI/+ and KI/KI mice (n = 3). The values are expressed as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
On the accelerating rotarod test, there was no marked difference in performance among the three groups of mice at eight weeks. However, a declining performance was observed among KI/KI mice from 16 to 32 weeks (Figure 2(b)).
β-galactosidase deficiency of KI/KI mice
The β-galactosidase activity in dried blood spots (DBS) collected from eight-week-old mouse tails was significantly decreased in KI/KI mice (1.9 ± 0.5 pmol/punch×h) as compared to WT mice (282.2 ± 14.7 pmol/punch×h) (P < 0.001) and KI/+ mice (145.4 ± 6.2 pmol/punch×h) (P < 0.001; Figure 2(c)). Similar findings were observed in brains, hearts, livers, and spleens of 16-week-old KI/KI mice indicating that the effects in these mice were sustained throughout the whole body (Table 3). The β-galactosidase activity in KI/KI mice was negligible in the brain, heart, liver, and spleen in comparison with WT mice and KI/+ mice (Figure 2(d)). We subsequently measured Glb1 mRNA expression by RT-qPCR and found that the p.G455R model retained mutant Glb1 mRNA (Figure 2(e)). Compared with WT mice, the mRNA levels of Glb1 were increased.
Table 3.
The β-galactosidase activity in the mice.
| The β-galactosidase activity (nmol/mg×h) | Glb1 +/+ | Glb1 G455R/+ | Glb1 G455R/G455R |
|---|---|---|---|
| Brain | 39.03 ± 0.95 | 20.64 ± 1.54 | 0.95 ± 0.32 |
| Heart | 8.94 ± 1.44 | 5.84 ± 1.44 | 0.04 ± 0.02 |
| Liver | 56.39 ± 17.75 | 45.08 ± 20.89 | 0.37 ± 0.14 |
| Spleen | 197.94 ± 21.25 | 125.42 ± 22.83 | 1.13 ± 0.44 |
Note: Data represent mean ± SEM.
GM1 ganglioside accumulation in KI/KI mouse brains
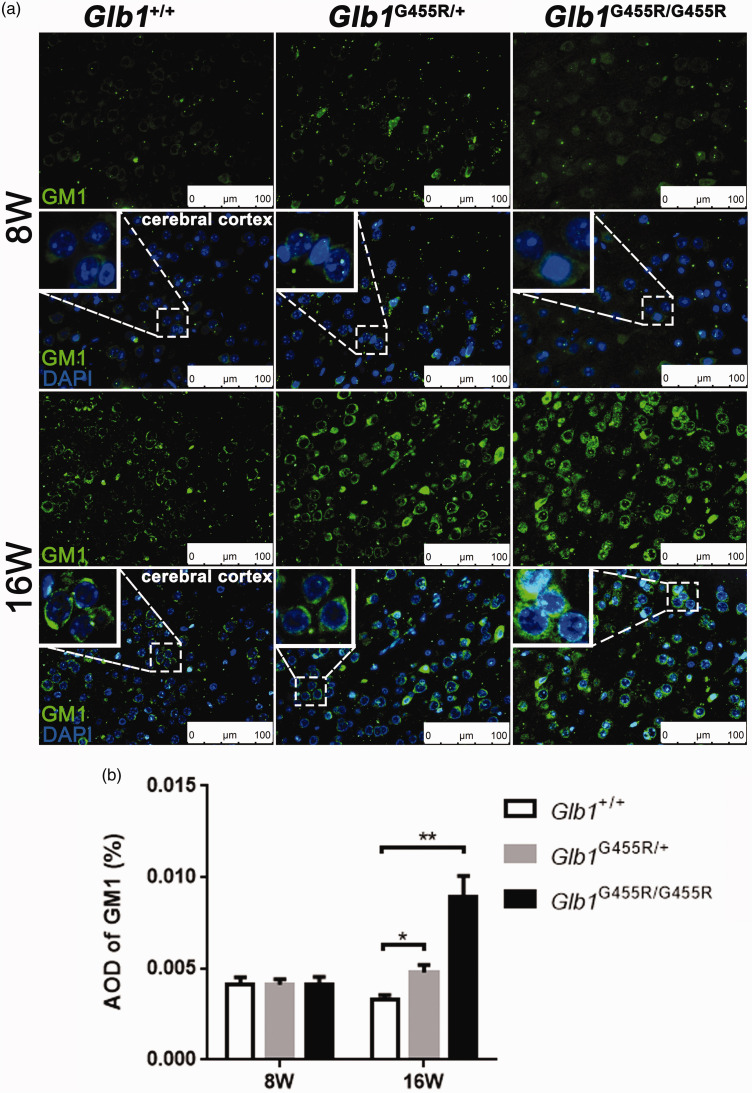
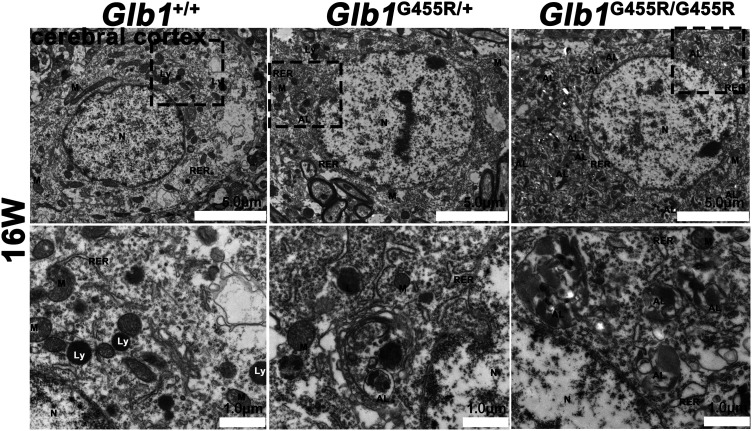
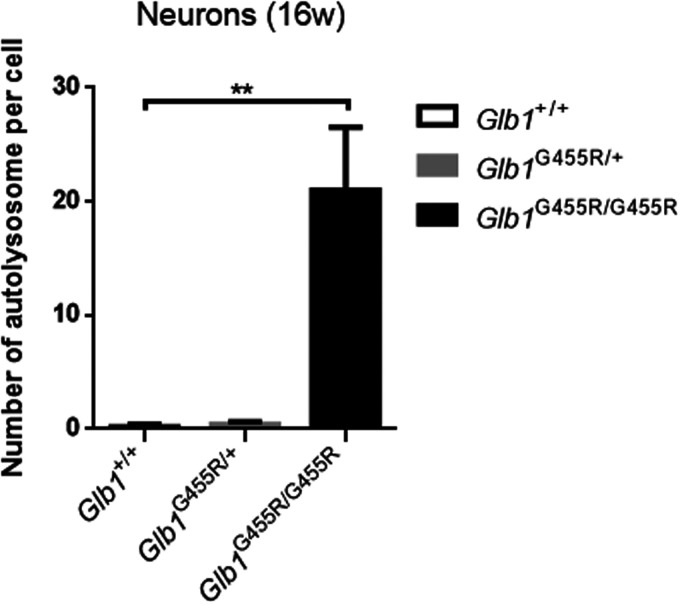
The GM1 immunohistochemistry staining of brain cortex from WT, KI/+, and KI/KI mice (at 8 and 16 weeks) showed that GM1 ganglioside accumulated in the brains of 16-week-old KI/KI mice but not in 8-week-old KI/KI mice (Figure 3(a)). Compared with WT mice, the average optical densities (AODs) of GM1 in 16-week-old KI/KI mice were markedly increased (P < 0.01; Figure 3(b)). We observed neuronal cytoplasmic vacuolation in the cerebral cortex and thalamus, where the Glb1 mutation triggered an accumulation of intralysosomal luminal vesicles and disrupted cytoplasmic structure (Figure 4). Similarly, in 16-week-old KI/KI mice, using electron microscopy, we observed the presence of a severe cytosolic vacuolization in cerebral cortical neurons (Figure 5). The cerebral cortical cells which appeared swollen were filled with autolysosomes (marked as AL; Figure 6). In addition to the AL, abnormal mitochondria with increased variation of size and disruption of the cristae (marked as M) and dilated rough endoplasmic reticulum (marked as RER) cisterns were noted in the cells.
Figure 3.
GM1 ganglioside accumulation in KI/KI mice. (a) The cerebral cortices of wild type (WT), KI/+, and KI/KI mice (at 8 and 16 weeks) were subjected to immunohistochemistry with GM1 antibody: GM1 (green), DAPI nuclear staining (blue), and merged signals. Objective ×40; scale bar indicates 100 μm. (b) AOD analysis of GM1 staining (n = 4). The values are expressed as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001. (A color version of this figure is available in the online journal.)
Figure 4.
Severe pathological change in KI/KI mice. Light micrographs of hematoxylin and eosin (H&E)-stained brain coronal sections from 16-week-old KI/KI, wild type (WT) and KI/+ mice. Neuronal cytoplasmic vacuolation (black arrowheads) is observed in KI/KI mice. Objective ×40; scale bar indicates 100 μm. (A color version of this figure is available in the online journal.)
Figure 5.
Numerous autolysosomes that were observed by electron microscopy in KI/KI mice. Electron microscopy images of cerebral cortex from 16-week-old KI/KI mice, wild type (WT), and KI/+ mice: objective ×2.5k; scale bar indicates 5 μm. Spinal cord control: objective ×8k; scale bar indicates 1 μm. Cell nuclei marked as N, autolysosomes marked as AL, lysosomes marked as Ly, cristae marked as M, and rough endoplasmic reticulum marked as RER.
Figure 6.
Quantification of autolysosomes in neurons (n = 5). The values are expressed as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
CNS inflammation in KI/KI mice
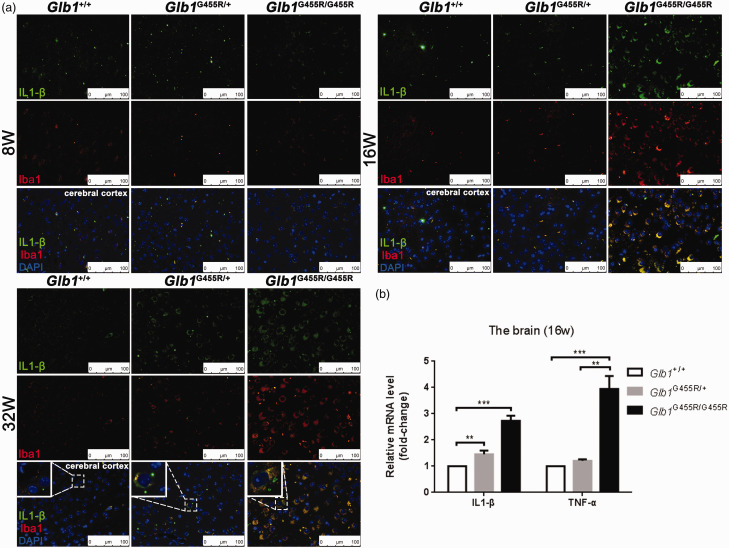
There were increased numbers of microglia in the brain tissues of 16-week-old KI/KI mice, as determined by increased Iba1 staining, a marker for microglial activation (Figure 7(a)). The majority of Iba1-positive cells appeared spherical with round cell bodies and were more numerous than those observed in the brains of eight-week-old mice. In parallel, the expression of IL-1β in the brains of KI/KI mice was increased and co-localized with Iba1 at 16 and 32 weeks (Figure 7(a)). We further measured the mRNA levels of IL-1β and TNF-α in the brains of 16-week-old mice (Figure 7(b)). The mRNA levels of IL-1β and TNF-α were significantly elevated in KI/KI mice over that of WT mice (2.7 and 3.9 fold higher, respectively). These microglia, which had an ameboid shape and secreted pro-inflammatory cytokines, suggested that activated microglia were polarized to proinflammatory M1 phenotypes in GM1 gangliosidosis.
Figure 7.
Activation of microglia in the brains of KI/KI mice. (a) Double staining anti IL1-β and anti Iba1 of cerebral cortex tissue in wild type (WT), KI/+, and KI/KI mice. IL1-β (green), Iba1 (red), and merged signals. Objective ×40; scale bar indicates 100 μm. (b) qPCR analysis of Glb1 and proinflammatory cytokine (IL-1β and TNF-α) genes of WT, KI/+, and KI/KI mice. Proinflammatory cytokines were highly enriched (n = 3). The values are expressed as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001. (A color version of this figure is available in the online journal.)
Disturbed autophagy of microglia in KI/KI mice
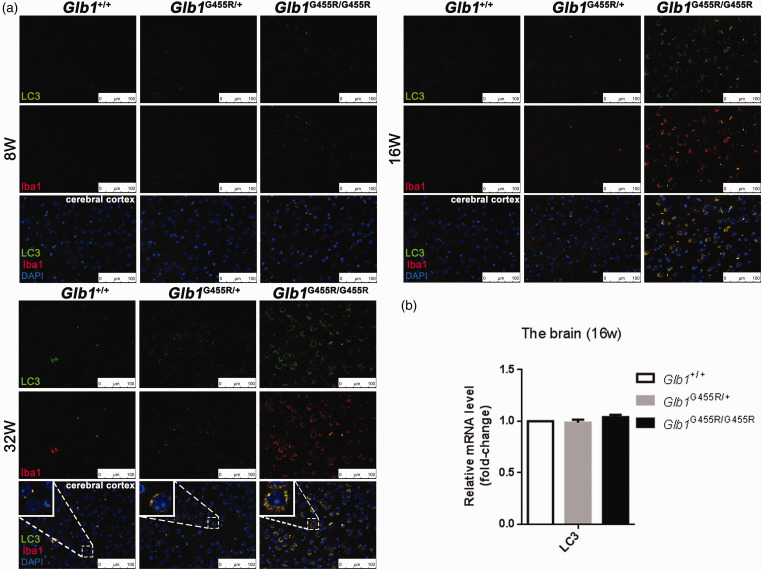
Furthermore, we assessed autophagy, a pathway that is commonly affected by lysosomal dysfunction in LSDs. We examined the autophagosomal marker LC3 in the cerebral cortex region at 8, 16, and 32 weeks, respectively. At eight weeks, the LC3 expression level was low and there was no significant difference among the three groups (Figure 8(a)). At 16 weeks, LC3 was present at significant levels in the cerebral cortices of KI/KI mice, while KI/+ and WT mice showed minor amounts of positive immunoreactivity. A strong LC3 signal was still detected in KI/KI mice at 32 weeks. However, the mRNA levels of LC3 showed no significant difference among the three groups at 16 weeks (Figure 8(b)), which indicated that the up-regulation of LC3 and accumulation of autolysosomes was not due to the increased initiation of autophagy, but rather to a blockade of autophagy flux. Additionally, immunofluorescence co-localization analysis showed that LC3 protein was co-localized with Iba1, indicating that disturbed autophagy occurred in microglia.
Figure 8.
Activation of autophagy of microglia in the brains of KI/KI mice. (a) Double staining anti LC3 and anti Iba1 of cerebral cortex tissue in wild type (WT), KI/+, and KI/KI mice. LC3 (green), Iba1 (red), and merged signals. Objective ×40; scale bar indicates 100 μm. (b) qPCR analysis of LC3 genes of WT, KI/+; and KI/KI mice. Proinflammatory cytokines were highly enriched (n = 3). The values are expressed as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001. (A color version of this figure is available in the online journal.)
Discussion
A faithful animal model is key to studying the pathogenesis of a disease and its novel therapies.13 We recently found a novel missense mutation (GLB1G453R) in a late-infantile form patient of GM1-gangliosidosis, which was confirmed to be related to a severe mutation in vitro.7 Here, we established a Glb1G455R mouse model carrying the human mutant GLB1G453R gene (corresponding to G455R in mice) using CRISPR-Cas9 genome editing. The KI/KI mice reflected the phenotypes observed in human GM1-gangliosidosis including β-galactosidase deficiency, GM1 ganglioside accumulation, neuromotor impairment, and shortened lifespan. The KI/+ mice showed no overt clinical phenotype, while their β-gal enzyme activity was slightly diminished compared to WT mice. All these observations demonstrated that our model system was reliable and the Glb1G455R mutation in mice was pathogenic.
The Glb1G455R mice were associated with a milder phenotype, though residual β-galactosidase activity in dried blood spots from mutant mice was only 0.6% of that in WT mice. The results were consistent with human patient and in vitro experiments.7 GM1 ganglioside accumulation was not conspicuous in the brain tissues until 16 weeks and a severe clinical phenotype was observed in mice at 32 weeks, similar to findings in the study of other Glb1 deficiency mouse models.14–16 The mechanisms of GM1 synthase regulation and alternative metabolic pathways may account for the different phenotypes between human and mouse GM1 gangliosidosis.17 GM1-ganglioside accumulation has an inhibitory effect on human but not on murine GM1 synthase. In addition, the murine GM1 ganglioside can be degraded by the removal of sialic acid from the corresponding asialo GA1, while GM1 ganglioside is only poorly degraded by sialidase in humans.14,18
Classic GM1 gangliosidosis mouse models, developed by Hahn et al.,14 who disrupted the gene by inserting the exogenous neomycin resistance gene in exon 6, and Matsuda et al.,16 who targeted exon 15, are currently the two most commonly used models for studying GM1. These mouse models were established through interrupting Glb1 gene expression to silence the expression of β-gal mRNA. The β-gal null mice showed obvious accumulation of GM1-gangliosidosis in the CNS at three weeks of age and started to display overt neurological problems beyond the age of five months. Furthermore, recent studies discovered another model of GM1-gangliosidosis developed based on a 20 bp deletion in the β-gal encoding gene.15 This β-gal−/− murine model detected that the deletion of catalytic nucleophiles and introduction of premature stop codons would result in loss of β-gal enzyme activity. The β-gal null mice exhibited neuromotor decline at four months of age and death by 10 months of age. In the same experimental report, the lab of Chester Whitley reported another novel model, which was produced by inducing Glb1W273L mutation.15 The mutation was commonly found in patients having Morquio syndrome type B. Experiments revealed that residual β-gal enzyme activity was detected in this mouse model and no change of phenotype was determined at 12 months. Our result was slightly different from the above results. The Glb1G455R mouse model showed accumulation of GM1-gangliosidosis in the CNS from 16 weeks and severe motor coordination deficits at 32 weeks as disease progressed.
Many of the mutations that cause human GM1 gangliosidosis are missense. Missense mutations can influence mRNA expression, protein folding, and protein stability. In feline GM1 gangliosidosis, unfolded protein response (UPR) in the ER, induced by accumulation of the mutant β-gal protein, plays a role in pathogenesis.19 The Glb1G455R mouse model retains mutant Glb1 mRNA in the brain, which can help us to explore the potential toxic consequences of aberrant mRNA and protein aggregation. Furthermore, the missense mutation model provides a valuable tool for testing β-gal enzyme stabilization and interaction with small molecules such as pharmacological chaperones.20
With the development of authentic animal models, we can investigate the mechanisms that show how the storage leads to CNS dysfunction in GM1-gangliosidosis. In eight-week-old mice, GM1 gangliosides did not accumulate and CNS homeostasis was not disrupted. At this time, microglia existed in a resting state and expressed little Iba1. At 16 weeks, levels of GM1 had markedly increased and microglia activation and proliferation were induced in the pathological condition. Microglia activation predated the onset of overt clinical symptoms at 32 weeks. We suggest that there is a correlation between microglial activation and disease progression in the Glb1G455R mutant model. Our hypothesis is supported by a study that showed that the severity of disease is positively correlated with the degree of inflammatory cytokine elevation in the cerebral spinal fluid of gangliosidoses patients.21 Actually, in most gangliosidoses lysosomal storage diseases, inflammatory responses are considered to contribute to pathogenesis and/or disease progression.9 In GM2 gangliosidosis mouse models, activation of inflammation predated the onset of overt clinical signs and its extent was correlated with disease progression.22 In Niemann-Pick diseased mice, lipid lysosomal overload of microglia leads to the polarization of microglia into pro-inflammatory phenotypes and worsened disease progression.23
Due to the vital role of lysosomes in autophagy, this pathway is an obvious candidate during LSD pathogenesis.24 We observed that levels of LC3 were increased in Glb1G455R mutant mice which was consistent with β-gal null mice.25 However, we did not observe the initiation of LC3 in RT-qPCR results, so it is more likely that autophagy flux was blocked in mutant mice. This was in agreement with studies in other LSDs, as shown by the following research. In fibroblasts of patients with Fabry disease, disease severity correlates with the extent of impaired autophagy.26 In Niemann-Pick Type C IPSc, autophagy inducers rescue the autophagy defect and improve cell viability.27 In Pompe mice, reducing autophagy levels reduces the formation of autophagic vesicles, while slowing down autophagic flux blockade-induced cell damage.28 Reversal of autophagy defects by rapamycin in a Gaucher disease drosophila model was able to partially ameliorate lifespan, locomotor, and oxidative stress phenotypes.29
Moreover, we noted that disturbed autophagy occurred in microglia. Recently, in different macromolecule storage disorders, it has been demonstrated that autophagy-lysosomal dysfunction in microglia contributes to cell damage through stress-related stimulation.30,31 In Alzheimer’s disease mice, a deficiency of LC3-associated endocytosis in microglia increases the production of pro-inflammatory cytokines in the hippocampus.32 Impaired autophagy in microglia exacerbates dopaminergic neuron loss in an MPTP-induced mouse model of Parkinson’s disease.33 While restoring autophagic capacity in mucopolysaccharidosis IIIB (MPS IIIB), a murine model reveals improved clearance of autophagic vacuoles in microglia and reduces inflammation in the brain.34 Taken together, we suggested that disturbed autophagy in microglia may play a role in GM1 gangliosidosis.
In conclusion, our murine GM1 model was reliable and closely reproduced the phenotype of human GM1. We suggest that disturbed autophagy in microglia may be involved in disease progression. This model provides a tool for exploring the potential toxic consequences of aberrant mRNA and pharmacological chaperone therapy for GM1 gangliosidosis.
Footnotes
AUTHORS’ CONTRIBUTIONS: All authors participated in the design, interpretation of the studies, analysis of the data, and review of the manuscript; SCL, XLJ, CFT, and FT conducted the experiments, SCL, YYF, and YLH wrote the manuscript.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: The author(s) disclose receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a fund from Guangzhou Institute of Pediatrics/Guangzhou Women Children’s Medical Center [No. Pre-NSFC-2019–013].
ORCID iD: Yonglan Huang https://orcid.org/0000-0002-7527-3754
References
- 1.Ferreira CR, Regier DS, Yoon R, Pan KS, Johnston JM, Yang S, Spranger JW, Tifft CJ.The skeletal phenotype of intermediate GM1 gangliosidosis: clinical, radiographic and densitometric features, and implications for clinical monitoring and intervention. Bone 2020; 131:115142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JS, Choi JM, Lee M, Kim SY, Lee S, Lim BC, Cheon JE, Kim IO, Kim KJ, Choi M, Seong MW, Chae JH.Diagnostic challenge for the rare lysosomal storage disease: late infantile GM1 gangliosidosis. Brain Dev 2018; 40:383–90 [DOI] [PubMed] [Google Scholar]
- 3.Poletti V, Biffi A.Gene-based approaches to inherited neurometabolic diseases. Hum Gene Ther 2019; 30:1222–35 [DOI] [PubMed] [Google Scholar]
- 4.Chen JC, Luu AR, Wise N, Angelis R, Agrawal V, Mangini L, Vincelette J, Handyside B, Sterling H, Lo MJ, Wong H, Galicia N, Pacheco G, Van Vleet J, Giaramita A, Fong S, Roy SM, Hague C, Lawrence R, Bullens S, Christianson TM, d'Azzo A, Crawford BE, Bunting S, LeBowitz JH, Yogalingam G.Intracerebroventricular enzyme replacement therapy with beta-galactosidase reverses brain pathologies due to GM1 gangliosidosis in mice. J Biol Chem 2020; 295:13532–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weismann CM, Ferreira J, Keeler AM, Su Q, Qui L, Shaffer SA, Xu Z, Gao G, Sena-Esteves M.Systemic AAV9 gene transfer in adult GM1 gangliosidosis mice reduces lysosomal storage in CNS and extends lifespan. Hum Mol Genet 2015; 24:4353–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yagci ZB, Esvap E, Ozkara HA, Ulgen KO, Olmez EO.Inflammatory response and its relation to sphingolipid metabolism proteins: chaperones as potential indirect anti-inflammatory agents. Adv Protein Chem Struct Biol 2019; 114:153–219 [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Huang Y, Zhao X, Sheng H, Feng Y, Zhang W, Liu L.Clinical and molecular characteristics of 11 Chinese probands with GM1 gangliosidosis. Metab Brain Dis 2018; 33:2051–7 [DOI] [PubMed] [Google Scholar]
- 8.Brunetti-Pierri N, Scaglia F.GM1 gangliosidosis: review of clinical, molecular, and therapeutic aspects. Mol Genet Metab 2008; 94:391–6 [DOI] [PubMed] [Google Scholar]
- 9.Bosch ME, Kielian T.Neuroinflammatory paradigms in lysosomal storage diseases. Front Neurosci 2015; 9:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palhegyi AM, Seranova E, Dimova S, Hoque S, Sarkar S.Biomedical implications of autophagy in macromolecule storage disorders. Front Cell Dev Biol 2019; 7:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballabio A, Bonifacino JS.Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol 2020; 21:101–18 [DOI] [PubMed] [Google Scholar]
- 12.Feather-Schussler DN, Ferguson TS.A battery of motor tests in a neonatal mouse model of cerebral palsy. J Vis Exp 2016; 117:53569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Favret JM, Weinstock NI, Feltri ML, Shin D.Pre-clinical mouse models of neurodegenerative lysosomal storage diseases. Front Mol Biosci 2020; 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn CN, del Pilar Martin M, Schroder M, Vanier MT, Hara Y, Suzuki K, Suzuki K, d'Azzo A.Generalized CNS disease and massive GM1-ganglioside accumulation in mice defective in lysosomal acid beta-galactosidase. Hum Mol Genet 1997; 6:205–11 [DOI] [PubMed] [Google Scholar]
- 15.Przybilla MJ, Ou L, Tabaran AF, Jiang X, Sidhu R, Kell PJ, Ory DS, O'Sullivan MG, Whitley CB.Comprehensive behavioral and biochemical outcomes of novel murine models of GM1-gangliosidosis and morquio syndrome type B. Mol Genet Metab 2019; 126:139–50 [DOI] [PubMed] [Google Scholar]
- 16.Matsuda J, Suzuki O, Oshima A, Ogura A, Noguchi Y, Yamamoto Y, Asano T, Takimoto K, Sukegawa K, Suzuki Y, Naiki M.Beta-galactosidase-deficient mouse as an animal model for GM1-gangliosidosis. Glycoconj J 1997; 14:729–36 [DOI] [PubMed] [Google Scholar]
- 17.Sano R, Trindade VM, Tessitore A, d'Azzo A, Vieira MB, Giugliani R, Coelho JC.G(M1)-ganglioside degradation and biosynthesis in human and murine G(M1)-gangliosidosis. Clin Chim Acta 2005; 354:131–9 [DOI] [PubMed] [Google Scholar]
- 18.Breiden B, Sandhoff K.Lysosomal glycosphingolipid storage diseases. Annu Rev Biochem 2019; 88:461–85 [DOI] [PubMed] [Google Scholar]
- 19.Martin DR, Rigat BA, Foureman P, Varadarajan GS, Hwang M, Krum BK, Smith BF, Callahan JW, Mahuran DJ, Baker HJ.Molecular consequences of the pathogenic mutation in feline GM1 gangliosidosis. Mol Genet Metab 2008; 94:212–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valenzano KJ, Khanna R, Powe AC, Boyd R, Lee G, Flanagan JJ, Benjamin ER.Identification and characterization of pharmacological chaperones to correct enzyme deficiencies in lysosomal storage disorders. Assay Drug Dev Technol 2011; 9:213–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Utz JR, Crutcher T, Schneider J, Sorgen P, Whitley CB.Biomarkers of Central nervous system inflammation in infantile and juvenile gangliosidoses. Mol Genet Metab 2015; 114:274–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeyakumar M.Central nervous system inflammation is a hallmark of pathogenesis in mouse models of GM1 and GM2 gangliosidosis. Brain 2003; 126:974–87 [DOI] [PubMed] [Google Scholar]
- 23.Gabande-Rodriguez E, Perez-Canamas A, Soto-Huelin B, Mitroi DN, Sanchez-Redondo S, Martinez-Saez E, Venero C, Peinado H, Ledesma MD.Lipid-induced lysosomal damage after demyelination corrupts microglia protective function in lysosomal storage disorders. EMBO J 2019; 38:e99553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren H, Wang G.Autophagy and lysosome storage disorders. Adv Exp Med Biol 2020; 1207:87–102 [DOI] [PubMed] [Google Scholar]
- 25.Takamura A, Higaki K, Kajimaki K, Otsuka S, Ninomiya H, Matsuda J, Ohno K, Suzuki Y, Nanba E.Enhanced autophagy and mitochondrial aberrations in murine G(M1)-gangliosidosis. Biochem Biophys Res Commun 2008; 367:616–22 [DOI] [PubMed] [Google Scholar]
- 26.Yanagisawa H, Hossain MA, Miyajima T, Nagao K, Miyashita T, Eto Y.Dysregulated DNA methylation of GLA gene was associated with dysfunction of autophagy. Mol Genet Metab 2019; 126:460–5 [DOI] [PubMed] [Google Scholar]
- 27.Maetzel D, Sarkar S, Wang H, Abi-Mosleh L, Xu P, Cheng AW, Gao Q, Mitalipova M, Jaenisch R.Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick type C patient-specific iPS cells. Stem Cell Rep 2014; 2:866–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim JA, Sun B, Puertollano R, Raben N.Therapeutic benefit of autophagy modulation in pompe disease. Mol Ther 2018; 26:1783–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinghorn KJ, Gronke S, Castillo-Quan JI, Woodling NS, Li L, Sirka E, Gegg M, Mills K, Hardy J, Bjedov I, Partridge L.A drosophila model of neuronopathic gaucher disease demonstrates lysosomal-autophagic defects and altered mTOR signalling and is functionally rescued by rapamycin. J Neurosci 2016; 36:11654–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cougnoux A, Drummond RA, Fellmeth M, Navid F, Collar AL, Iben J, Kulkarni AB, Pickel J, Schiffmann R, Wassif CA, Cawley NX, Lionakis MS, Porter FD.Unique molecular signature in mucolipidosis type IV microglia. J Neuroinflammation 2019; 16:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuil LE, Lopez Marti A, Carreras Mascaro A, van den Bosch JC, van den Berg P, van der Linde HC, Schoonderwoerd K, Ruijter GJG, van Ham TJ.Hexb enzyme deficiency leads to lysosomal abnormalities in radial glia and microglia in zebrafish brain development. Glia 2019; 67:1705–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, Guy CS, Zakharenko SS, Green DR.LC3-associated endocytosis facilitates beta-amyloid clearance and mitigates neurodegeneration in murine Alzheimer's disease. Cell 2019; 178:536–51.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue Q, Jingru Q, Ping W, Jia L, Yong Z, Fan J, Haiyan L.Impaired autophagy in microglia aggravates dopaminergic neurodegeneration by regulating NLRP3 inflammasome activation in experimental models of Parkinson's disease. Brain Behav Immun 2021; 91:324–38 [DOI] [PubMed] [Google Scholar]
- 34.Lotfi P, Tse DY, Di Ronza A, Seymour ML, Martano G, Cooper JD, Pereira FA, Passafaro M, Wu SM, Sardiello M.Trehalose reduces retinal degeneration, neuroinflammation and storage burden caused by a lysosomal hydrolase deficiency. Autophagy 2018; 14:1419–34 [DOI] [PMC free article] [PubMed] [Google Scholar]