Abstract
Abnormal lipid metabolism is regarded as a crucial cause of psoriasis. The specific mechanism of how phospholipase PLA2G4B mediates local immune dysfunction and skin lesions remains unclear. The aim of this study was to explore the mechanisms of anti-psoriasis and immune suppression effect by inhibiting PLA2G4B in psoriasis progression. We successfully transfected si-PLA2G4B in a murine keratinocyte cell-line PAM212 to verify the effect of progression by PLA2G4B. The Imiquimod psoriasis mouse model was then successfully constructed, followed by emulsion wrapped PLA2G4B-siRNA applied to the skin lesions. The phenotype, pathology, immunofluorescence staining of PLA2G4B, IL17, CD3, and CD1b, and bulk transcriptome analysis were performed to decipher the effect and mechanism of si-PLA2G4B. Interfering with PLA2G4B significantly inhibited the proliferation and migration of PAM212. The interference of PLA2G4B in vivo showed a therapeutic effect on psoriasis, comparable to that of betamethasone. The phenotype and pathology revealed reduced keratinocytes in the si-PLA2G4B group compared to the model mice. Immunofluorescence showed that CD1b, CD3+ T cells, and IL17 were suppressed in the skin lesions. RNA-seq and deconvolution revealed that immune cells such as myeloid dendritic cell and T cell CD8+ naive were inactivated. Th17 reduce the release of inflammatory factors such as IL17 and IL36. Pathway analysis revealed the potential therapeutic mechanism involved in the inhibition of sphingolipid or ceramide secretion. This study verified the anti-psoriatic effect of using si-PLA2G4B. The immune response was alleviated after administration. This phospholipase inhibition-based therapy sheds light on the pharmaceutical potential against psoriasis.
Keywords: Psoriasis, PLA2G4B, inflammatory factors, medicine, lipid metabolism disorder, transcriptome
Impact statement
PLA2G4B is significantly increased in the lesions of psoriasis patients and promotes abnormal lipid metabolism. The specific mechanism of how PLA2G4B mediates local immune dysfunction and skin lesions remains unclear. By constructing an animal model, cell biology experiments, and combining bioinformatics tools, we elucidated that increased PLA2G4B expression leads to abnormal lipid metabolism. The release of small molecules can be presented to CD1b by activating myeloid dendritic cells. This leads to an increase in the local concentration of inflammatory factors that participate in lipid metabolism-immunological dysregulation response, further exacerbating the inflammation of the skin lesion site and the excessive proliferation of keratinocytes. Inhibition of PLA2G4B may serve as a target for blocking the vicious cycle of abnormal lipid metabolism-lesion exacerbation. This provides a theoretical and experimental basis for a novel clinical approach to slow down psoriasis by correcting dyslipidemia and inhibiting inflammatory cell activation.
Introduction
Psoriasis is a chronic skin disease characterized by inflammation, rapid epidermal proliferation, and keratotic dysfunction, with approximately 2–3% in the human population. The pathogenesis is affected by various factors such as genes and the environment,1 and its pathogenesis is currently unclear. Many studies have shown that psoriasis also has abnormal lipid metabolism like other autoimmune diseases and autoinflammatory diseases.2 Changes in the psoriasis lesion’s phospholipid composition suggest that lipid deposition in the reticuloendothelial system can cause inflammation and dekeratinization of the skin.3,4 However, the causal relationship between psoriasis and lipid metabolism, and the mechanisms of lipid metabolism abnormalities and immune effects remain unclear.
The phospholipase A2 (PLA2) family consists of 11 members with distinct localizations and substrate specificities.5 PLA2 can catalyze the hydrolysis of cell membrane phospholipids, thereby releasing arachidonic acid, while psoriatic lesions also release arachidonic acid by activating sphingomyelinase in the cell surface.6 Arachidonic acid is further metabolized to a lipid hormone such as eicosanoid to regulate the inflammatory response. We found that PLA2 promotes lipids’ abnormal metabolism and causes pathological phenomena such as a hyperproliferative state, inflammatory secretion, keratosis dysfunction, and hypermigration. PLA2 is a promising predictor of comorbidities in patients with severe psoriasis.7 Combining previous literature studies8 and data analysis, we found that PLA2 can activate a cascade of downstream inflammatory factors and cause various pathological features of psoriasis. The exogenous PLA2 gene has recently been reported to promote the recruitment of CD1 to effector T cells to present new lipid antigens.9 The main function of CD1 protein is to mediate the presentation of lipid and glycolipid antigens derived from themselves or microorganisms to T cells. Van Rhijn et al. Found that CD1b has the characteristic of presenting both autophospholipids and pathogenic microorganism phospholipids to T cells without identification and then produces autoimmunity.10 PLA2G2F was expressed in terminally differentiated keratinocytes in the suprabasal epidermis. Blocking PLA2G2F may be a novel approach for specific treatment of psoriasis, skin cancer, or other conditions characterized by epidermal hyperplasia.11 Our previous research found that fatty acid and lipid metabolism pathways are involved in the process of psoriasis. The PLA2 family’s seven genes are significantly up-regulated, of which PLA2G4B is the most prominent (P value was significantly higher than that of PLA2G2F).12 By searching the human protein atlas database, we found that PLA2G4B was also expressed in keratinocytes. Furthermore, the down-regulation of PLA2G4B has been involved in phosphatidylcholine conversion to anti-inflammatory lipoxins.13 The current research on PLA2G4B is not thorough enough, and its specific mechanism of regulating skin homeostasis needs to be proved and extended experimentally.
In the process of downstream inflammatory reactions and keratinocyte dysplasia, innate immune-mediated skin cell inflammation plays a crucial role in developing psoriasis.14 Current studies have found that keratinocytes, macrophages, and dendritic cells release IL-1215 and promote T cell, Th1 cell, and Th22 cell release of IL-17.16,17 The release of such cytokines acts on keratinocytes, aggravating the skin inflammatory response of psoriasis. Th17 cells secreting IL-17 are among the first to be activated during immune responses,18 suggesting that IL-17 may play an essential role in the early stages of inflammation.
Therefore, we hypothesized that PLA2G4B could activate IL-17 production through the CD1b lipid antigen pathway as the key point. We used siRNA-505 to construct PLA2G4B targeted silencing cells, proving the role of PLA2G4B in psoriasis. Wrap siRNA-505 in liposomes to make an emulsifier, apply it to the Imiquimod (imq) psoriasis mouse model’s skin lesions, and compare the efficacy with betamethasone. The flow chart of the study design is shown in Figure 1. This study further demonstrates the importance of abnormal lipid metabolism in the occurrence of psoriasis. It also provides theoretical and experimental evidence that PLA2G4B can be used as a potential target to correct abnormal lipid metabolism and relieve psoriasis.
Figure 1.
Flow chart of the study design.
Materials and methods
Design and synthesis of PLA2G4B-siRNA interfering plasmid
According to the target design principles, the required siRNA core target sequences (one each in the forward and reverse directions) are obtained by calculation. The forward sequence is as follows:
siRNA-PLA2G4B-350: 5′-GGACCUGGUGACCGGAGAUTT-3′
siRNA-PLA2G4B-505: 5′-GCGAGUGGCUCGUCAGCAATT-3′
siRNA-PLA2G4B-1375: 5′-CCCUGAGUCAUGGCCAGAATT-3′
BbsI was selected as the endonuclease of the vector plasmid. The siRNA sequence plus the sticky end of the endonuclease BbsI is the final fragment of interest. Polymerase chain reaction (PCR) thermal cycling parameters: 37°C, 30 min; 95°C, 5 min; 25°C, 5 min. It was then diluted 200-fold with ddH2O. The vector plasmid and siRNA fragment were ligated in 1:1 molar T4 ligase, and the 10 µL system was left at 4°C overnight. Take the competent cells and melt them in ice. Add 5 µL of the ligation product to 50 µL of competent cells. Leave on ice for 30 min, 42°C, 90 s. Quickly transfer to ice and let stand for 3 min. Add 900 µL Luria-Bertani (LB) medium without Ampicillin (Amp), 180 r/min, 37°C, 1 h, and 20 µL coated LB selection plate medium overnight at 37°C. Pick single clones and expand culture, extract plasmids for sequencing and identification.
siRNA transfection and verification of interference efficiency
Murine keratinocyte cell line (PAM 212) was seeded in a cell culture plate with 50–90% cell density. Next, the siRNA-Hilymax complex (Shanghai GenePharma Co., Ltd) was added to the cell culture well, and the plate was incubated at 37°C in a CO2 incubator for 4 h. The medium was changed, and the plates were incubated for 48 h.
Total RNA from cells was extracted using the Cell Culture and Tissue Total RNA Extraction and Preparation Mini Kit according to the manufacturer’s instructions (Shanghai TIANGEN BIOTECH Co., Ltd). The quantity and quality of RNA were confirmed with a NanoDrop 1000. The primers were designed using Primer Premier 5.0 software and synthesized by General Biotech Co., Ltd. Quantitative real-time PCR was performed using the KAPA SYBR Green supermix PCR kit with the iCycler apparatus system (Bio-Rad). The forward sequence is as follows:
PLA2G4B: 5′-AGGAGACAGGAGACCAGAAGT-3′
GAPDH: 5′-GAGTCCACTGGCGTCTTCAC-3′
Scratch wound-healing assay
PAM 212 cells were seeded in 24-well plates at a density of 1 × 105 cells per well and incubated overnight. The cells were then scratched with a 10-µL tip at the bottom of the wells, and the floating cells were washed twice with the medium. Next, the cells were transfected with siRNA-505. Finally, each well was photographed by microscopy to calculate the healing change at 0 h and 16 h.
Proliferation assay
Cell proliferation was assessed by the Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan). Briefly, PAM 212 cells were seeded on a 96-well microplate at a density of 3 × 104 cells per well, and then the cells were transfected with siRNA-505. The cells were cultured for 0 h, 24 h, and 48 h. Next, 5 µL of CCK-8 solution was added to each well and incubated at 37°C for an additional 2 h. The optical density was determined at a wavelength of 450 nm.
siRNA liposome emulsifier formulation
Dilute siRNA-505 with buffer phosphate buffered solution (PBS) to prepare 50 µg/µL siRNA solution. Take 6 µL siRNA solution (300 µg) and mix with 100 µL vivo-jetPEI® reagent, vortex for 5 s, which is solution A. Use 50 µL sterile water to dilute 50 µL vivo-jetPEI® buffer to prepare B solution. Add diluted B solution to A solution, vortex gently, and incubate at room temperature for 15 min to prepare 200 µL transfection mixed solution (PLA2G4B-siRNA liposome emulsifier mixture).
Psoriasis model
Twenty BALB/C male mice (eight weeks old, specified pathogen free (SPF), purchased from Shanghai Lingchang Biological Technology Co., Ltd) were randomly divided into four groups, with five mice in each group. A 2 cm × 3 cm area of the back was selected for hair removal, and the corresponding therapeutic drugs were separately applied. The control group (NC) received an even application of vaseline. The psoriasis model group (OC) received an application of 5% imq cream. The betamethasone group (BT) received an application of 5% imq cream first and then an application of betamethasone ointment (Federal Beta) 2 h later. The si-PLA2G4B group (SR) received an application of 5% imq cream first and then application of PLA2G4B-siRNA liposome emulsifier mixture 2 h later. To avoid skin inflammation caused by male mice fighting when mixed, the mice were housed separately. For 10 consecutive days, the mice were sacrificed after the end of treatment. The dorsal lesions’ tissue was divided into two parts: one was combined with 4% paraformaldehyde for immunization, and the other was immediately stored in a refrigerator at –80°C for use. This model can be used to verify whether si-PLA2G4B emulsifier can delay the progression of psoriasis in the early stage. All experiments were performed according to the principles outlined in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Publication No. 86-23, revised 1985) and approved by the ethics committee of Shanghai Skin Disease Hospital.
Immunohistochemical staining
Mouse skin lesions were embedded and sliced for hematoxylin-eosin staining (HE) staining. Immunofluorescence experiments were performed to detect the expression of proteins such as CD3, CD1b, PLA2G4B, and IL-17 (antibodies purchased from Thermo Fisher Scientific). Images were scanned for image acquisition analysis.
cDNA library preparation and sequencing
The ribosomal RNA in the samples was removed using the Ribo-Zero Golden kit (Epicentre, Illumina, USA), and cDNA libraries were prepared by TruSeq total RNA sample preparation kit (Illumina, USA) according to the instructions. Fragments of 300–450 bp length were recovered using an agarose gel extraction kit (TIAGEN, China). All libraries were quantified using 2100 Bioanalyzer and pooled as 1:1 at 2 nM for sequencing in HiSeq 150 pair-end reads (Illumina, USA). Sequence quality was estimated using FastQC (v0.11.5).
Skin tissue RNA sequencing and functional analysis
RNA sequencing analysis was performed on NC, OC, and SR groups’ skin tissue, and heat maps were drawn. Path enrichment analysis of trend genes was performed using the “KEGG.db” and “KEGGprofile” packages in the R project (http://bioconductor.org/packages). The nCounter Mouse Immunology Panel includes 547 genes covering the core pathways and processes of the immune response and 14 internal reference genes for data normalization. We also intersected NC–OC–SR (expression increased first and then decreased) trend cytokines with nCounter Mouse Immunology Panel and presented it as a heat map.
RNAseq deconvolution to decipher microenvironment
We next calculated the fraction of the cell subsets using enrichment score-based algorithm xCell from RNA-seq data.19 Briefly, expression profile Trans Per Million (TPM) of all samples was employed as raw signatures. R package “immunedeconv” was applied to obtain an enrichment score of 26 immune cell types.
Results
Successfully transfected siRNA-505 and down-regulated PLA2G4B expression
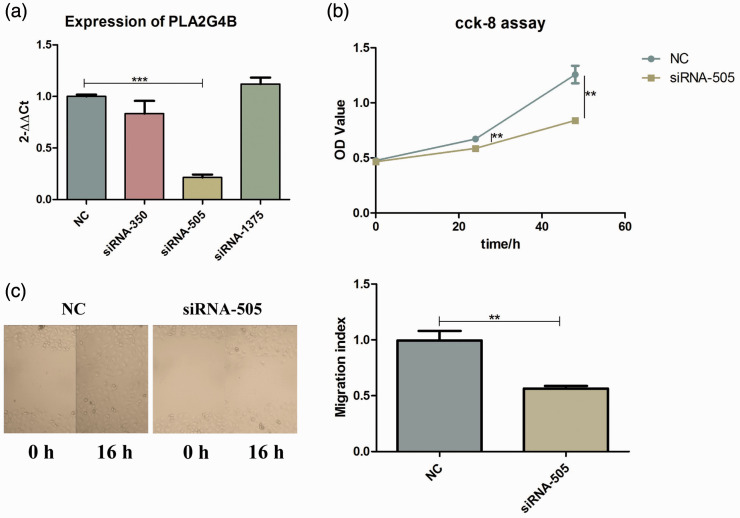
The quantitative real-time PCR (qPCR) results showed a successful knockdown of the PLA2G4B gene by siRNA-505 candidates. There is a statistically significant difference between the knockdown group and the NC group (P < 0.001), indicating that we successfully transfected the siRNA and down-regulated the expression of PLA2G4B (Figure 2(a)). CCK-8 results showed that the proliferation of murine keratinocyte cells was significantly inhibited after PLA2G4B was disturbed (P < 0.01) (Figure 2(b)). The wound healing experiment verified that PLA2G4B has the ability to make keratinocytes migrate. Compared with the control group, scratch healing ability will be affected after PLA2G4B is disturbed (Figure 2(c)).
Figure 2.
Verification of interference efficiency and PLA2G4B function. (a) siRNA-505 showed the highest inhibitory effect in PAM 212 cells. (b) CCK-8 analysis showed that the growth of PAM 212 cells was inhibited by siRNA-505. (c) Scratch wound-healing assay shows that siRNA-505 inhibits the migration ability of PAM 212 cells. (A color version of this figure is available in the online journal.)
Interfering with PLA2G4B has a therapeutic effect demonstrated by the mouse psoriasis model
We successfully constructed a mouse model of imq psoriasis. The phenotypic, molecular, and pathological assessments were combined with an overall psoriasis area and severity index (PASI) score to determine successful modeling. Combination of imq and drugs determines whether siRNA-505 PLA2G4B emulsifier can delay the progression of psoriasis in the early stage.
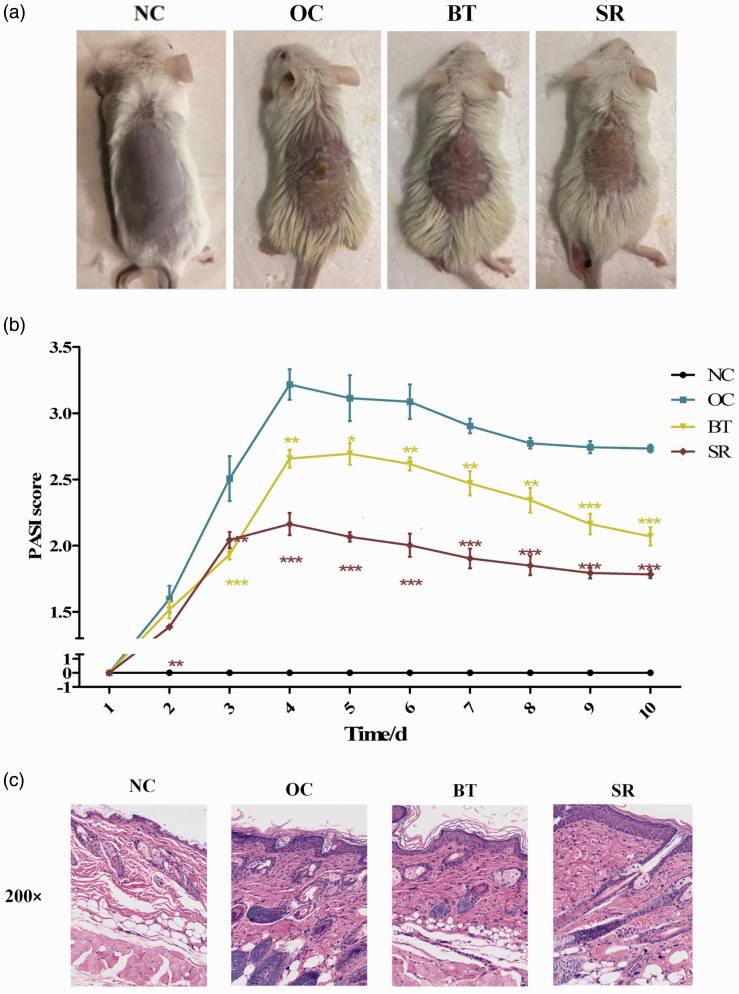
In the OC group, the skin lesions of the mice were increasingly severe. After one week of application, erythema, fine scales, skin folds, thickening, and scaly detachment were apparent. After a week of drug treatment, the degree of skin lesions was reduced. Compared with the OC group, the skin’s erythema color in the SR group was darker and thinner, and the silvery-white scales were reduced (Figure 3(a)). The PASI score of the BT group and the SR group decreased significantly, but the SR group better than the BT group (Figure 3(b)).
Figure 3.
Phenotype and pathology of skin lesions in mouse model of psoriasis. (a) Interference with PLA2G4B has a therapeutic effect on skin lesions in psoriatic model mice, and the effect is better than that in the BT group. (b) Significant decrease in PASI score after drug treatment. (c) HE staining showed that the degree of lesion repair after interference with PLA2G4B was better than that of the BT group. (A color version of this figure is available in the online journal.)
HE staining showed that compared with the NC group, the mice in the other groups had thicker epithelial layers, incomplete keratinization of the stratum corneum, and obvious inflammatory cell infiltration in the dermis, similar to the histological manifestations of psoriasis lesions. Compared with the OC group, pathological changes were confirmed in the BT and SR groups, the spinous layer became thinner than before, and the epidermal process gradually returned to normal. The SR group was better than the BT group (Figure 3(c)).
Immunofluorescence showed a positive correlation between PLA2G4B, CD1b, and IL17
PLA2G4B was significantly down-regulated in the SR group. The results of the immunofluorescence assay of IL-17 showed that the levels of IL-17 secreted by keratinocytes in the OC group were significantly higher than those in the other groups. Interference with PLA2G4B can significantly inhibit the production of inflammatory factors at the site of skin lesions. CD1b and CD3 immunofluorescence experiments also showed the same trend. These results indicate that PLA2G4B may induce overexpression of CD1b, a lipid antigen-presenting molecule. At the same time, the decrease of CD3 after treatment intimate that PLA2G4B may induce T cell activation (Figure 4).
Figure 4.
Immunofluorescence image of PLA2G4B, CD1b, CD3, and IL17. (A color version of this figure is available in the online journal.)
RNA sequencing and functional analysis of skin lesions
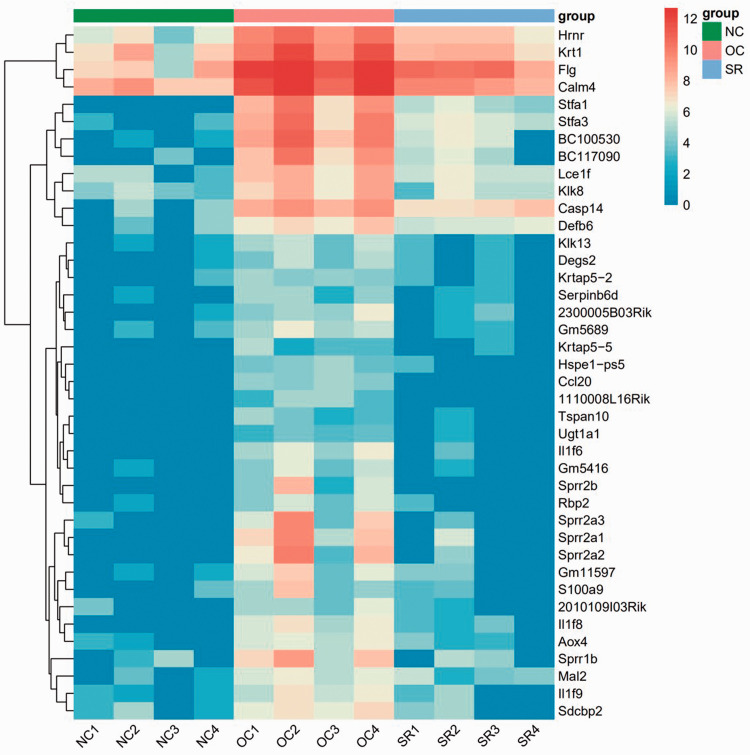
RNA sequencing analysis was performed on the skin lesions of each group. RNAseq deconvolution can decipher the microenvironment. The heatmap of the relative expression of immune cells is shown in Figure 5(a). Wilcox test analysis found that myeloid dendritic cell and T cell CD8+ naive were significantly activated in the OC group and recovered to a similar level to the NC group in the SR group (Figure 5(b)). The principal component analysis (PCA) diagram shows that the NC and SR groups are not distinctively different while having transcriptomic differences with the OC group. The dimensionality reduction positions of the SR transcriptome were between the NC and OC groups. This shows that the treatment in the SR group is effective (Figure 5(c)).
Figure 5.
Analysis of immune microenvironment in psoriasis lesions. (a, b) RNAseq deconvolution indicate that myeloid dendritic cell and T cell CD8+ naive were significantly activated in the OC group and recovered to a similar level to the NC group in the SR group. (c) PCA cluster analysis. (A color version of this figure is available in the online journal.)
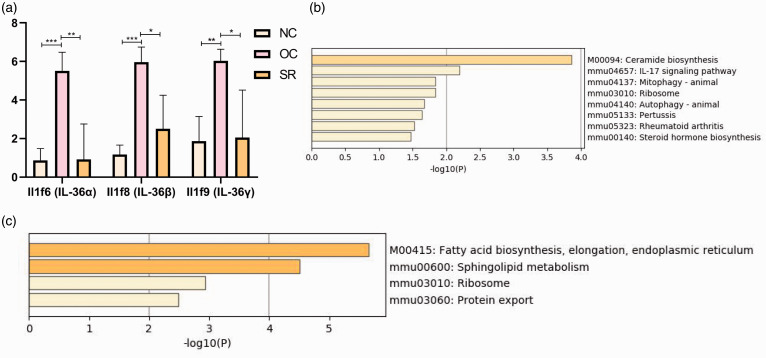
The expression level of trend gene increased first and then decreased (NC-OC-SR). It was found that Il1f6 (IL-36α), Il1f8 (IL-36β), and Il1f9 (IL-36γ) significantly increased in the OC group compared with the NC group. Gene expression in the SR group decreased to a similar level in the NC group after treatment (Figures 6 and 7(a)). Supplementary figure 1 summarized the changes in trend cytokines in NO–OC–SR (significantly different between groups), including Lce1f, Defb6, S100a9, 2010109l03Rik, Ccl20, Defb14, Cxcl1, ll1a, Tnfsf15, and Gp1bb.
Figure 6.
NC–OC–SR (expression increased first and then decreased) trend gene heat map. (A color version of this figure is available in the online journal.)
Figure 7.
Analysis of DEGs’ function in psoriasis lesions. (a) Changes in IL-36α, IL-36β, and IL-36γ expression trends. (b, c) Enrichment of the KEGG pathway of trend genes. (A color version of this figure is available in the online journal.)
The kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment of these trend genes indicates that the psoriasis treatment model mainly involves pathways such as ceramide biosynthesis, leukocyte cell–cell adhesion, and IL-17 signaling pathway (Figure 7(b)). A co-network was established for differentially expressed genes (DEGs) and nuclear transcription factors (TF) of OC vs. NC, and then KEGG pathway enrichment analysis was performed (Figure 7(c)). It was found that metabolic activation of sphingolipid will cause a high expression of ceramide. Lipids and amide transmitters are up-regulated in the inflammatory parts of the model and down-regulated after administration, accompanied by inhibition of IL-36α, IL-36β, IL-36γ, and IL-17 signaling pathways.
Discussion
Psoriasis is a kind of auto-inflammatory disease, and its mechanism is extremely complicated. Many patients are accompanied by abnormal lipid metabolism. When the lipid metabolism is corrected with drugs, psoriasis skin lesions are relieved accordingly.20 The skin lesions’ lipid component plays a crucial role in the vicious cycle of inflammation and abnormal lipid metabolism in skin lesions. However, the molecular regulatory mechanism that initiates this vicious cycle has not been reported. It has been found that the abnormal expression of PLA2 is involved in the occurrence of psoriasis, but the specific mechanism of how it mediates local skin lesions, lipid metabolism disorders, immunological disorders is unclear.
Our cell experiment proved the effective interference efficiency of siRNA-505 on PLA2G4B and found that PLA2G4B has the ability to promote the proliferation and migration of keratinocytes. We use imq ointment to construct mouse model of psoriasis, and use siRNA505 PLA2G4B liposome emulsifier and betamethasone for treatment. A clinical/molecular phenotypic evaluation was performed based on pathology and PASI score to verify whether the drug can inhibit the progression of psoriasis at an early stage. Compared with the control group, HE confirmed the pathological change recovery. Additionally, clinical phenotypic lesions decreased. From the immunofluorescence pictures of the three groups, it can be clearly seen that PLA2G4B increased significantly in the psoriasis lesions. After smearing with siRNA emulsifier, the fluorescence intensity of PLA2G4B decreased significantly, which means that the emulsifier can penetrate the skin surface to play a role. CD1b and IL17 accumulate in the skin lesions as we expected. A Th17 response commonly drives psoriasis at the molecular level, which serves as a major therapeutic target.21 The increase of CD3 means the activation of total T cells.
Using this treatment model, we also performed skin RNA sequencing of mouse lesions and treatment sites to obtain specific key molecules, downstream molecular pathways, and regulatory networks after PLA2G4B inhibition. RNA deconvolution can reveal the immune microenvironment of psoriasis lesions. In our analysis results, myeloid dendritic cell and T cell CD8+ naive are significantly activated in OC group. CD1b-restricted T cells can be activated by lipids presented through myeloid dendritic cells,22 Khasawneh et al. indicate that in psoriasis not only the skin but also the blood myeloid dendritic cells perform Th17 recruiting capacity.23 The study has now shown that T cell CD8+ is an important T cell cluster in psoriasis lesions.24 Using DEG for function and pathway enrichment, it was found that the model that interferes with PLA2G4B for psoriasis mainly involves ceramide biosynthesis, leukocyte cell–cell adhesion, and IL-17 signaling pathway. The activation of sphingolipid causes high expression of ceramide, and lipids and amide transmitters are up-regulated in the inflammatory parts of the model. Down-regulation after treatment was accompanied by suppression of IL-36α, IL-36β, IL-36γ, and IL-17 signaling pathways. IL-36α, IL-36β, and IL-36γ are agonists of IL36, and the role of IL-36 in keratinocytes has been widely discussed.25 A large amount of evidence shows that the serum level of IL-36 in patients with psoriasis is significantly higher than that in healthy people. Its expression is closely related to the disease.26,27 These results indicate that psoriatic models mainly involve sphingolipid metabolism-related pathways, and lipid metabolism plays an important role in psoriasis. Interference with PLA2G4B can significantly inhibit generally acknowledged targets of psoriasis, such as IL36 and IL17. The above results demonstrate the considerable potential of PLA2G4B in the clinical treatment of psoriasis.
Currently, calcipotriol is commonly used in combination with betamethasone as a compound preparation for the psoriasis treatment. Calcipotriol is a vitamin D derivative that inhibits the cell proliferation-associated antigen Ki-67, increases the activity of the differentiation antigen K-10, and reduces CD45RO-positive and CD8-positive T cells. Betamethasone, as a hormone receptor agonist, can enhance the activity of epidermal K-10 without activating Ki-67.28 This study demonstrates the potential of PLA2G4B to participate in clinical combination therapies.
This project explored the relationship between the abnormal expression of the PLA2G4B gene and the severity of lesions in psoriatic lesions. It clarified the abnormal proliferation and incomplete keratinization of epidermal cells caused by overexpression of the PLA2G4B gene. This study confirmed the abnormalities in small-molecule lipid metabolism caused by abnormal expression of the PLA2G4B gene and critical evidence related to the immune imbalance caused by the CD1–Th17 pathway. Target-specific PLA2G4B inhibition was produced by in vitro preparation, and lesion inhibition was achieved in the mouse psoriasis model. The treatment of psoriasis provides the basis for future research.
In summary, we speculate that in the early stages of psoriasis, PLA2G4B expression increased due to stress response, which caused abnormal lipid metabolism. The release of small molecules such as free fatty acids and lysophospholipids can be presented to CD1b by activating myeloid dendritic cells. This causes T cell CD8+ naive and Th17 to release inflammatory factors such as IL-17 and IL-36. The increase in the local concentration of inflammatory factors participates in lipid metabolism-immunological dysregulation response, further exacerbating the inflammation of the skin lesion site and the excessive proliferation of keratinocytes.
Supplemental Material
Supplemental material, sj-jpg-1-ebm-10.1177_1535370221993424 for Inhibition of phospholipases suppresses progression of psoriasis through modulation of inflammation by Yunlu Gao, Jiajing Lu, Xunxia Bao, Xuemei Yi, Chen Peng, Wenjuan Chen, Timing Zhen, Yong Shi, Kaichen Xing, Sibo Zhu and Yangfeng Ding in Experimental Biology and Medicine
Footnotes
AUTHORS’ CONTRIBUTIONS: All authors participated in the design, interpretation of the studies, and review of the manuscript. XMY, CP, WJC, and KCX collection and assembly of data. YLG, JJL, XXB, SBZ, and YFD analysis of the data; XXB, TMZ, and YS conducted the experiments, YLG, JJL, XXB, SBZ, and YFD wrote the manuscript.
ACKNOWLEDGMENTS: Special thanks to Dr Yiping Cao and Mr Guoxiang Zhan of Shenzhen Tika Biotech Co., Ltd for their technical support.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: This work was supported by the Natural Science Foundation of Shanghai (Grant No. 18ZR1433600).
ORCID iD: Jiajing Lu https://orcid.org/0000-0002-0827-0596
Xunxia Bao https://orcid.org/0000-0002-6085-7319
Sibo Zhu https://orcid.org/0000-0003-4800-8987
Yangfeng Ding https://orcid.org/0000-0002-4312-0446
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Boehncke WH, Schon MP.Psoriasis. Lancet 2015; 386:983–94 [DOI] [PubMed] [Google Scholar]
- 2.Ryu H, Kim J, Kim D, Lee JE, Chung Y.Cellular and molecular links between autoimmunity and lipid metabolism. Mol Cells 2019; 42:747–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietrzak A, Chodorowska G, Szepietowski J, Zalewska-Janowska A, Krasowska D, Hercogova J.Psoriasis and serum lipid abnormalities. Dermatol Ther 2010; 23:160–73 [DOI] [PubMed] [Google Scholar]
- 4.Holzer M, Wolf P, Curcic S, Birner-Gruenberger R, Weger W, Inzinger M, El-Gamal D, Wadsack C, Heinemann A, Marsche G.Psoriasis alters HDL composition and cholesterol efflux capacity. J Lipid Res 2012; 53:1618–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami M, Sato H, Miki Y, Yamamoto K, Taketomi Y.A new era of secreted phospholipase A(2). J Lipid Res 2015; 56:1248–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL.StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015; 33:290–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiluk P, Baran A, Swiderska M, Maciaszek M, Flisiak I.Lp-PLA2 as a promising predictor of comorbidities in patients with severe psoriasis. J Dermatolog Treat 2020; 5;524–30 [DOI] [PubMed] [Google Scholar]
- 8.Quaranta M, Knapp B, Garzorz N, Mattii M, Pullabhatla V, Pennino D, Andres C, Traidl-Hoffmann C, Cavani A, Theis FJ, Ring J, Schmidt-Weber CB, Eyerich S, Eyerich K.Intraindividual genome expression analysis reveals a specific molecular signature of psoriasis and eczema. Sci Transl Med 2014; 6:244–ra90 [DOI] [PubMed] [Google Scholar]
- 9.Cheung KL, Jarrett R, Subramaniam S, Salimi M, Gutowska-Owsiak D, Chen YL, Hardman C, Xue L, Cerundolo V, Ogg G.Psoriatic T cells recognize neolipid antigens generated by mast cell phospholipase delivered by exosomes and presented by CD1a. J Exp Med 2016; 213:2399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Rhijn I, van Berlo T, Hilmenyuk T, Cheng TY, Wolf BJ, Tatituri RV, Uldrich AP, Napolitani G, Cerundolo V, Altman JD, Willemsen P, Huang S, Rossjohn J, Besra GS, Brenner MB, Godfrey DI, Moody DB.Human autoreactive T cells recognize CD1b and phospholipids. Proc Natl Acad Sci U S A 2016; 113:380–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto K, Miki Y, Sato M, Taketomi Y, Nishito Y, Taya C, Muramatsu K, Ikeda K, Nakanishi H, Taguchi R, Kambe N, Kabashima K, Lambeau G, Gelb MH, Murakami M.The role of group IIF-secreted phospholipase A2 in epidermal homeostasis and hyperplasia. J Exp Med 2015; 212:1901–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang GY, Zhang YC, Zhang QL.PLA2 activity in serum and cutaneous tissues in patients with psoriasis vulgaris. Hunan Yi Ke Da Xue Xue Bao 2001; 26:455–6 [PubMed] [Google Scholar]
- 13.Saare M, Tserel L, Haljasmagi L, Taalberg E, Peet N, Eimre M, Vetik R, Kingo K, Saks K, Tamm R, Milani L, Kisand K, Peterson P.Monocytes present age-related changes in phospholipid concentration and decreased energy metabolism. Aging Cell 2020; 19:e13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowes MA, Suarez-Farinas M, Krueger JG.Immunology of psoriasis. Annu Rev Immunol 2014; 32:227–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Cesare A, Di Meglio P, Nestle FO.The IL-23/Th17 axis in the immunopathogenesis of psoriasis. J Invest Dermatol 2009; 129:1339–50 [DOI] [PubMed] [Google Scholar]
- 16.Lowes MA, Chamian F, Abello MV, Fuentes-Duculan J, Lin SL, Nussbaum R, Novitskaya I, Carbonaro H, Cardinale I, Kikuchi T, Gilleaudeau P, Sullivan-Whalen M, Wittkowski KM, Papp K, Garovoy M, Dummer W, Steinman RM, Krueger JG.Increase in TNF-alpha and inducible nitric oxide synthase-expressing dendritic cells in psoriasis and reduction with efalizumab (anti-CD11a). Proc Natl Acad Sci U S A 2005; 102:19057–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaba LC, Krueger JG, Lowes MA.Resident and “inflammatory” dendritic cells in human skin. J Invest Dermatol 2009; 129:302–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao Z, Painter SL, Fanslow WC, Ulrich D, Macduff BM, Spriggs MK, Armitage RJ.Human IL-17: a novel cytokine derived from T cells. J Immunol 1995; 155:5483–6 [PubMed] [Google Scholar]
- 19.Aran D, Hu Z, Butte AJ.xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017; 18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wojcik P, Biernacki M, Wronski A, Luczaj W, Waeg G, Zarkovic N, Skrzydlewska E.Altered lipid metabolism in blood mononuclear cells of psoriatic patients indicates differential changes in psoriasis vulgaris and psoriatic arthritis. Int J Mol Sci 2019; 20:4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zakostelska Z, Malkova J, Klimesova K, Rossmann P, Hornova M, Novosadova I, Stehlikova Z, Kostovcik M, Hudcovic T, Stepankova R, Juzlova K, Hercogova J, Tlaskalova-Hogenova H, Kverka M.Intestinal microbiota promotes Psoriasis-Like skin inflammation by enhancing Th17 response. PLoS One 2016; 11:e0159539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasaki N, Rillahan CD, Cheng TY, Van Rhijn I, Macauley MS, Moody DB, Paulson JC.Targeted delivery of mycobacterial antigens to human dendritic cells via siglec-7 induces robust T cell activation. J Immunol 2014; 193:1560–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khasawneh A, Barath S, Medgyesi B, Beke G, Dajnoki Z, Gaspar K, Jenei A, Pogacsas L, Pazmandi K, Gaal J, Bacsi A, Szegedi A, Kapitany A.Myeloid but not plasmacytoid blood DCs possess Th1 polarizing and Th1/Th17 recruiting capacity in psoriasis. Immunol Lett 2017; 189:109–13 [DOI] [PubMed] [Google Scholar]
- 24.Vugmeyster Y, Kikuchi T, Lowes MA, Chamian F, Kagen M, Gilleaudeau P, Lee E, Howell K, Bodary S, Dummer W, Krueger JG.Efalizumab (anti-CD11a)-induced increase in peripheral blood leukocytes in psoriasis patients is preferentially mediated by altered trafficking of memory CD8+ T cells into lesional skin. Clin Immunol 2004; 113:38–46 [DOI] [PubMed] [Google Scholar]
- 25.Dinarello C, Arend W, Sims J, Smith D, Blumberg H, O’Neill L, Goldbach-Mansky R, Pizarro T, Hoffman H, Bufler P, Nold M, Ghezzi P, Mantovani A, Garlanda C, Boraschi D, Rubartelli A, Netea M, van der Meer J, Joosten L, Mandrup-Poulsen T, Donath M, Lewis E, Pfeilschifter J, Martin M, Kracht M, Muehl H, Novick D, Lukic M, Conti B, Solinger A, Kelk P, van de Veerdonk F, Gabel C.IL-1 family nomenclature. Nat Immunol 2010; 11:973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachelez H, Choon SE, Marrakchi S, Burden AD, Tsai TF, Morita A, Turki H, Hall DB, Shear M, Baum P, Padula SJ, Thoma C.Inhibition of the interleukin-36 pathway for the treatment of generalized pustular psoriasis. N Engl J Med 2019; 380:981–3 [DOI] [PubMed] [Google Scholar]
- 27.Sehat M, Talaei R, Dadgostar E, Nikoueinejad H, Akbari H.Evaluating serum levels of IL-33, IL-36, IL-37 and gene expression of IL-37 in patients with psoriasis vulgaris. Iran J Allergy Asthma Immunol 2018; 17:179–87 [PubMed] [Google Scholar]
- 28.Uva L, Miguel D, Pinheiro C, Antunes J, Cruz D, Ferreira J, Filipe P.Mechanisms of action of topical corticosteroids in psoriasis. Int J Endocrinol 2012; 2012:561018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-jpg-1-ebm-10.1177_1535370221993424 for Inhibition of phospholipases suppresses progression of psoriasis through modulation of inflammation by Yunlu Gao, Jiajing Lu, Xunxia Bao, Xuemei Yi, Chen Peng, Wenjuan Chen, Timing Zhen, Yong Shi, Kaichen Xing, Sibo Zhu and Yangfeng Ding in Experimental Biology and Medicine