Abstract
14-3-3 proteins play a major role in the regulation of primary metabolism, protein transport, ion channel activity, signal transduction and biotic/abiotic stress responses. However, their involvement in petal growth and development is largely unknown. Here, we identified and characterized the expression patterns of seven genes of the 14-3-3 family in gerbera. While none of the genes showed any tissue or developmental specificity of spatiotemporal expression, all seven predicted proteins have the nine α-helices typical of 14-3-3 proteins. Following treatment with brassinolide, an endogenous brassinosteroid, the Gh14-3-3 genes displayed various response patterns; for example, Gh14-3-3b and Gh14-3-3f reached their highest expression level at early (2 h) and late (24 h) timepoints, respectively. Further study revealed that overexpression of Gh14-3-3b or Gh14-3-3f promoted cell elongation, leading to an increase in ray petal length. By contrast, silencing of Gh14-3-3b or Gh14-3-3f inhibited petal elongation, which was eliminated partly by brassinolide. Correspondingly, the expression of petal elongation-related and brassinosteroid signaling-related genes was modified in transgenic petals. Taken together, our research suggests that Gh14-3-3b and Gh14-3-3f are positive regulators of brassinosteroid-induced ray petal elongation and thus provides novel insights into the molecular mechanism of petal growth and development.
Keywords: 14-3-3 protein, ray petal elongation, cell elongation, BR, Gerbera hybrida
Introduction
14-3-3 proteins, with a molecular weight of 25∼32 kDa, are a class of highly conserved, acidic, soluble proteins that are present in almost all eukaryotes (Ferl, 1996; Mhawech, 2005). Each 14-3-3 isoform contains nine α-helices and shows a high degree of similarity to other 14-3-3 members (Aitken, 2006). As bridge proteins, they participate widely in the regulation of various physiological processes (including metabolism, hormone signaling and stress response) by interacting with numerous clients, such as metabolic enzymes, signaling proteins and transcription factors (Silva et al., 2001; Chevalier et al., 2009; Zhou et al., 2015; Li M. et al., 2016; Keicher et al., 2017; Camoni et al., 2018). Generally, 14-3-3 proteins, which are encoded by multiple genes in most species (Denison et al., 2011), bind to their clients via two sequence motifs (RSXpSXP and RXXXpSXP) to activate or inhibit the activity of their target proteins (Sehnke et al., 2002). The first plant 14-3-3 protein identified was cloned from maize (de Vetten et al., 1992). In Arabidopsis, there are thirteen functionally expressed 14-3-3 genes (Chevalier et al., 2009), while tobacco has seventeen potential isoforms, rather more than the eight genes in rice and seven in cotton (Konagaya et al., 2004; Chen et al., 2006; Zhou et al., 2015). According to their gene structure, 14-3-3 proteins can be divided into two groups, termed ε and non-ε (DeLille et al., 2001; Konagaya et al., 2004; Chen et al., 2006; Chevalier et al., 2009).
Brassinosteroids (BRs) are plant steroid hormones that play key roles in regulating a variety of physiological processes, including leaf expansion, flowering, senescence, stress resistance and cell expansion and elongation (Clouse et al., 1996; Clouse and Sasse, 1998; Yang et al., 2011; Kim et al., 2012; Kaneko-Suzuki et al., 2018; Oh et al., 2020). BR signal cascades are well characterized in Arabidopsis, in which BZR1 is the key transcription factor, affecting plant growth and development by modulating thousands of BR target genes and interacting with other hormone signaling components (He et al., 2005; Wang et al., 2012; Qiao et al., 2017; Zheng et al., 2019; Zhang et al., 2020). 14-3-3 proteins are also important regulatory components of the BR signaling pathway: they regulate plant growth by anchoring BZR1 in the cytoplasm (Gampala et al., 2007; Ryu et al., 2007; Kim and Wang, 2010). Mutants of the 14-3-3-binding site of BZR1, with a phenotype similar to that of the bzr1-1D mutant, show a constitutive BR response and an increase in BZR1 nuclear retention (Gampala et al., 2007; Ryu et al., 2007).
The 14-3-3 proteins involved in BR signaling participate in plant growth and flowering processes by modulating cell differentiation and elongation (Pertl et al., 2010; Zhang et al., 2010; Taoka et al., 2011; Zhou et al., 2015; Kaneko-Suzuki et al., 2018; Minami et al., 2019). Analysis of multiple 14-3-3 mutants revealed their specificity and functional redundancy in primary root elongation under different environmental conditions, in which these genes are positive regulators under control conditions and negative regulators during abiotic stress (van Kleeff et al., 2014). In lily (Lilium longiflorum), 14-3-3 proteins were shown to play a role in the germination and elongation of pollen (Pertl et al., 2010). Minami et al. (2019) reported that, during BR-induced hypocotyl elongation, a 14-3-3 protein interacts with the phosphorylated C-terminus, and thereby enhances the catalytic activity, of plasma membrane H+-ATPase. In addition, 14-3-3 proteins have a regulatory role in cotton fiber elongation (Zhou et al., 2015). Thus, overexpression of 14-3-3L promotes fiber elongation in cotton, while gene silencing of 14-3-3L results in a shortening of cotton fiber length (Zhou et al., 2015). Recently, Zuo et al. (2021) identified eighteen 14-3-3 genes in the apple genome and characterized their expression patterns, suggesting that some of them may participate in the regulation of the flowering process. These results all highlight the importance of 14-3-3 proteins in plant growth.
Gerbera hybrida, belonging to the Asteraceae family, is one of the mainstream cut flowers and its commercial and ornamental value depend on petal morphology and color (Bhatia et al., 2009; Mosqueda Frómeta et al., 2017). Thus, it is important to understand the regulatory mechanisms governing gerbera petal morphology. The research team of Prof. Elomaa has focused on the molecular mechanisms of flower development in Asteraceae, including G. hybrida, for many years (Kotilainen et al., 1999; Broholm et al., 2008; Tahtiharju et al., 2012; Juntheikki-Palovaara et al., 2014; Zhao et al., 2020; Zhang et al., 2021). Broholm et al. (2008) found that overexpression of GhCYC2 in gerbera results in conversion of disc florets into ray-like florets with elongated petals, as well as disruption of stamen development. Functional analysis of GhCYC proteins revealed redundant functions of GhCYC2, GhCYC3 and GhCYC4 in regulating ray floret identity and in promoting petal development (Juntheikki-Palovaara et al., 2014). Various hormones (gibberellin, abscisic acid, ethylene, and BRs) are involved in the regulation of late-stage petal development in gerbera (Zhang et al., 2012; Li et al., 2015; Han et al., 2017; Huang et al., 2017, 2020; Ren et al., 2018). Li et al. (2015) found that GA3 stimulates petal elongation in gerbera, while ABA inhibits it. Further research showed that GhWIP2, a WIP-type ZFP transcription factor, represses cell expansion during petal and leaf development by modulating crosstalk between gibberellin, abscisic acid and auxin (Ren et al., 2018). Another study found that exogenous brassinolide (BL) treatment can boost the elongation of ray floret petals, whereas BRZ (a BR synthesis inhibitor) reduces petal length (Huang et al., 2017). However, whether 14-3-3 proteins, as one of the BR signaling components, play a regulatory role in BR-induced petal elongation in gerbera, or indeed in any other flowering species, remains unknown.
Here, seven gerbera 14-3-3 genes were identified and their predicted proteins classified. The expression patterns of all seven genes were comprehensively investigated in various tissues and at different developmental stages. Overexpressing two of these genes, Gh14-3-3b and Gh14-3-3f, in ray florets increased petal length by promoting cell elongation, whereas gene silencing of Gh14-3-3b or Gh14-3-3f reduced petal growth. Further analysis found that several BR-related genes, such as BZR1 homologs (GhBEH1 and GhBEH2) and petal elongation-associated genes (like GhEXP1, GhEXP3, GhEXP10, GhXTH1, and GhXET), were modified in transgenic petals. These results demonstrate a positive regulatory role of Gh14-3-3b and Gh14-3-3f in BR-induced ray petal elongation.
Materials and Methods
Plant Materials and Growth Conditions
A variety of G. hybrida called “Shenzhen No. 5” was used in this work. The plants were cultured under greenhouse conditions at 26/18°C (day/night temperature) with a 16 h light/8 h dark photocycle and a relative humidity of 65∼80%. Three types of floret (ray floret, trans floret, and disc floret) at stage 6, as well as young leaf (leaf from plants transplanted into the soil for 10∼15 days), old leaf (basal leaf of plants transplanted into the soil for 3 months), young root (root of plants transplanted into the soil for 10∼15 days), old root (root of plants transplanted into the soil for 3 months), calyx, scape, and ray florets at different developmental stages, were sampled for quantitative real-time PCR (qRT-PCR) analysis. The development stages of ray florets (S1∼S6, “S” represents “stage”) are defined according to Meng and Wang (2004). Ray florets at stage 3 were used for transient transformation and hormone treatment assays.
Cloning and Sequence Analysis of Gh14-3-3 Genes
Using the sequences of 14-3-3 genes in Arabidopsis thaliana, BLAST was performed against the transcriptome shotgun assembly database (Accession: PRJNA179026) of G. hybrida cultivar “Shenzhen No. 5” (taxid: 18101) (Kuang et al., 2013), and seven Gh14-3-3 genes were identified. Seven full-length Gh14-3-3 cDNA sequences were amplified from a gerbera cDNA library by PCR using PrimeSTAR Max Premix (Takara, Cat. No. R045) with specific primers. Alignment of the deduced amino acid sequences with Gh14-3-3 homolog from different species was performed using DNAMAN 6.0. Conserved domain analysis was executed in the Conserved Domain Database1. Protein structure prediction was performed with SWISS-MODEL2. Phylogenetic analysis was performed in MEGA 6.0 using a neighbor-joining algorithm with 1,000 bootstrap replicates. The primers for the constructs in each experiment and 14-3-3 protein information for various species are listed in Supplementary Table 1.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from the samples using the Easystep® Super Total RNA Extraction Kit (Promega, Code No. LS1040) following the manufacturer’s protocol. First-strand cDNA was synthesized from 1 μg total RNA using the ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Code No. FSQ-301). qRT-PCR was performed using RealStar Green Fast Mixture (GenStar, Code No. A301-01). 1 μL cDNA was added as a qPCR template in a total reaction volume of 20 μL. The samples were amplified using the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., United States) as follows: melting at 95°C for 2 min and amplification with 40 cycles of 95°C for 5 s and 60°C for 30 s. All analyses used a housekeeping gene (GhACTIN, AJ763915) as a normalization control (Kuang et al., 2013). The expression level was calculated according to the 2–ΔΔCt method. The primers used for qRT-PCR are listed in Supplementary Table 1.
Transient Transformation of Ray Florets
Transient transformation of ray florets was performed as described by Han et al. (2017). Overexpression vectors (C17 and C17-Gh14-3-3b/f) and virus-induced gene silencing (VIGS) vectors (pTRV1, pTRV2, and pTRV2-Gh14-3-3b/f) were transformed into Agrobacterium tumefaciens strain C58C1. A. tumefaciens were cultured in Luria-Bertani medium containing 75 mg mL–1 kanamycin and 50 mg mL–1 rifampicin for 24 h at 28°C and then inoculated into 50 mL Luria-Bertani medium containing 20 μM acetosyringone (AS) and 10 mM 2-(N-morpholino) ethanesulfonic acid (MES) and shaken at 28°C overnight. When the absorbance (OD600) of A. tumefaciens reached approximately 1.5, the cells were resuspended in infiltration buffer (200 μM AS, 10 mM MES, 10 mM MgCl2, pH 5.6) to a final optical density at 600 nm (OD600) of 1.5. Agrobacterium tumefaciens cultures carrying C17-Gh14-3-3b and C17-Gh14-3-3f, and the empty C17 vector as a mock treatment control, were stored at 28°C for 4 h in the dark at room temperature. Agrobacterium tumefaciens cultures carrying pTRV2-Gh14-3-3b/f and pTRV1 at a ratio of 1:1 (v/v), and a mixture containing pTRV2/pTRV1 as a mock treatment control, were also stored under the same conditions for 4 h.
Detached ray petals from fresh inflorescences at stage 3 were cleaned, and then immersed in the various resuspension buffers mentioned above under a vacuum of −0.09 MPa for 5 min. After 2 min, the vacuum was slowly released and the petals were rinsed with sterile distilled water (dH2O) and placed in sterile Petri dishes with two layers of Whatman filter paper. After incubation at 4°C for 3 days, the transformed petals were grown at 23∼25°C for 9 days at 50∼60% humidity under long-day conditions (16 h light/8 h dark). At least 15 well-grown inflorescences were used for each treatment, and at least three biological replicates were used for each experiment.
Hormone Treatment of Ray Florets
Our previous study showed that detached petals can develop normally. The result of in vitro hormone and inhibitor experiments performed with detached petals were consistent with the result of in vivo experiments using intact inflorescences (Li et al., 2015; Huang et al., 2017). In this study, detached ray petals from inflorescences at stage 3 were used for BL treatments as described previously (Huang et al., 2017). Transiently transformed petals were placed in sterile Petri dishes with two layers of Whatman filter paper soaked in 10 μM BL or dH2O as control. Subsequently, the petals were cultured at 24∼26°C for 2 days. At least 15 well-grown inflorescences were used for each treatment, and at least three biological replicates were used for each experiment.
Measurement of Ray Petal and Cell Length
A total of 45 petals were selected to measure their length as previously described (Li et al., 2015). Petals were imaged with a Nikon camera D7200 (Japan) and measured using ImageJ software. To measure the petal cell length and number, the top, middle and basal region of each petal were stained with propidium iodide (0.1 mg mL–1) for 5 min. Next, images of adaxial epidermal cells were captured using a confocal laser scanning microscope (LSM710, Carl Zeiss, Germany) and more than 50 cells were analyzed using ImageJ software. At least three biological replicates were used for each observation. The elongation rate was calculated according Han et al. (2017).
Statistical Analysis
The data were analyzed with SPSS (version 13.0; IBM Corp., Armonk, NY, United States). Statistical significance between samples was investigated by Duncan’s new multiple range test. The data are presented as mean ± standard error (SE). Different lowercase letters above the bars or line charts indicate significantly different groups: ∗P < 0.05, ∗∗P < 0.01.
Results
Isolation and Characterization of Gh14-3-3 Genes in Gerbera
To identify 14-3-3 genes in gerbera, we performed a tBLASTn search against the gerbera transcriptome data using 14-3-3 protein sequences from Arabidopsis as queries (Kuang et al., 2013). Seven putative Gh14-3-3 genes (Gh14-3-3a, Gh14-3-3b, Gh14-3-3c, Gh14-3-3d, Gh14-3-3e, Gh14-3-3f, and Gh14-3-3g) were identified. As shown in Figure 1, sequence analysis predicted that the seven Gh14-3-3 genes encode 258, 259, 257, 261, 336, 254, and 258 amino acids, respectively. Sequence alignment of the gerbera protein sequences showed a high degree of identity, >65%, with the 14-3-3 proteins of Helianthus annuus, Lactuca sativa, and Cynara scolymus. All seven gerbera sequences contained the nine antiparallel α-helices that are highly conserved among 14-3-3 proteins (Figure 1A). These results, together with the conserved domain analysis shown in Supplementary Figure 1, revealed that the seven gerbera genes belong to the 14-3-3 family. Phylogenetic tree analysis showed that these seven sequences could be divided into either the ε group or the non-ε group of 14-3-3 proteins (Figure 1B). Gh14-3-3c is located in the ε branch of the tree, while the other six Gh14-3-3 proteins belong to the non-ε group.
FIGURE 1.
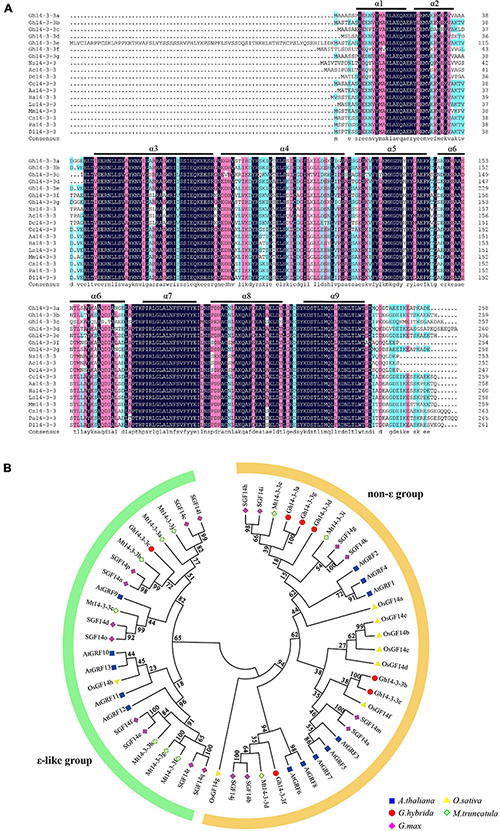
Sequence analysis and phylogenetic tree of Gh14-3-3 proteins. (A) Amino acid alignment of Gh14-3-3 proteins with those from other plants. Ns14-3-3 (KAA8528342.1) Nyssa sinensis; Ac14-3-3 (PSR99833.1) Actinidia chinensis; Dc14-3-3 (XP_017252199.1) Daucus carota; Cc14-3-3 (XP_024978074.1) Cynara cardunculus; Aa14-3-3 (PWA87588.1) Artemisia annua; Ha14-3-3 (XP_022009462.1) Helianthus annuus; Ls14-3-3 (XP_023753461.1) Lactuca sativa; Mm14-3-3 (KAD2805724.1) Mikania micrantha; Cs14-3-3 (XP_028084704.1) Camellia sinensis; Pa14-3-3 (PON78199.1) Parasponia andersonii; Dl14-3-3 (ACK76233.1) Dimocarpus longan. The regions of the nine conserved antiparallel α-helices (α1∼α9) are underlined in black. (B) Phylogenetic relationship showing two groups of 14-3-3 proteins in the five species. The minimum evolution tree was constructed using MEGA 6.0 from 1,000 bootstrap replicates. Protein designations consist of the prefixes Arabidopsis thaliana (At, blue squares), Oryza sativa (Os, yellow triangles), Gerbera hybrida (Gh, Red circles), Medicago truncatula (Mt, green rhombus), and Glycine max (Gm, pink rhombus). Detailed information for 14-3-3s from these plant species are listed in Supplementary Table 1.
Dimerization Patterns of Gh14-3-3 Proteins
The 14-3-3 proteins usually exist as homo- or heterodimers (Mhawech, 2005). To investigate protein-protein interactions among the seven Gh14-3-3s, a yeast two-hybrid assay was performed. The results showed that only three proteins (Gh14-3-3b, Gh14-3-3c, and Gh14-3-3f) can form homodimers, while the other protein interaction patterns varied. For example, Gh14-3-3b formed heterodimers with the remaining six Gh14-3-3 proteins, while Gh14-3-3e only interacted with Gh14-3-3b. The other five Gh14-3-3 proteins showed a variety of different dimerization behaviors (Figure 2 and Supplementary Figure 2). These results suggest that Gh14-3-3 protein interaction patterns vary according to isoform.
FIGURE 2.
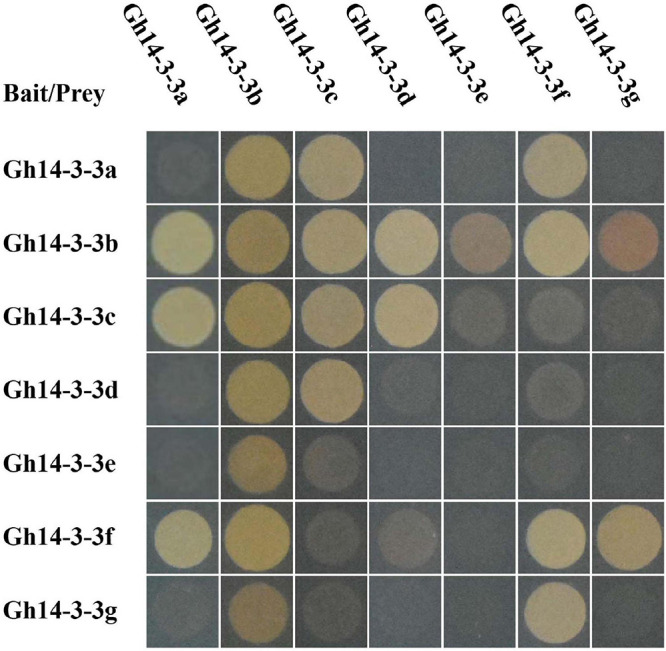
Specific interactions among Gh14-3-3 proteins. pGADT7-largeT7/pGBKT7-53 and pGADT7-largeT7/pGBKT7-laminC were used as positive and negative controls, respectively. The original figure is shown in Supplementary Figure 2.
Spatiotemporal Expression Patterns and Response of Gh14-3-3 Genes to BR
To explore the spatiotemporal expression patterns of the seven Gh14-3-3 family members in gerbera, qRT-PCR was performed. We first analyzed their expression in different tissues and found that each gene was expressed in various organs or tissues (Figure 3A). The highest expression levels appeared in young root, young leaf, disc floret, calyx and old leaf, while the lowest expression levels were mostly observed in old root. The different expression profiles in different tissues imply functional diversity in the Gh14-3-3 gene family.
FIGURE 3.
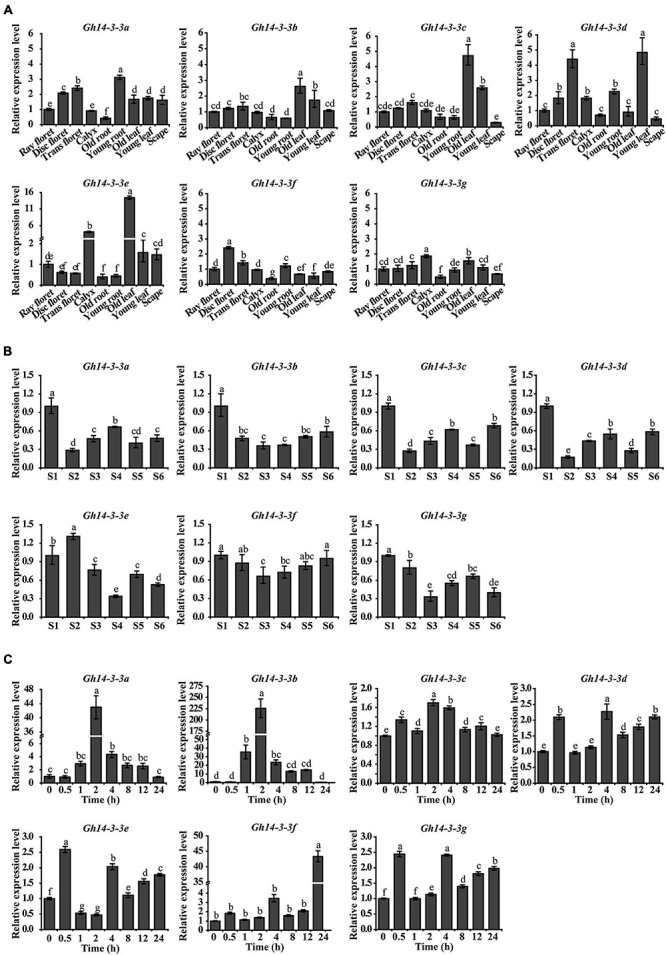
The spatiotemporal expression pattern of Gh14-3-3 genes. (A) The expression pattern of Gh14-3-3 genes in gerbera tissues and organs. Relative mRNA level of the Gh14-3-3 genes in gerbera tissues (ray floret, disc floret, trans floret, calyx, old root, young root, old leaf, young leaf, and scape) were detected by qRT-PCR. GhACTIN (AJ763915) is the reference gene (Kuang et al., 2013). Gene expression levels were set to 1 in ray floret. (B) The expression of Gh14-3-3 genes during different growth stages (S1∼S6, “S” represents “stage”) of ray floret in G. bybrida. The development stages of ray florets were defined according to Meng and Wang (2004). Gene expression levels were set to 1 in S1 ray floret petals. (C) The expression level of Gh14-3-3 genes in ray floret of G. bybrida under BL treatments. The expression levels of Gh14-3-3s in the ray floret of gerbera were detected within 0∼24 h after BL treatment. Gene expression levels were set to 1 in “0 h” and were calculated using the 2–ΔΔCt method. Values were the means ± SE from three biological replicates.
We next evaluated the expression pattern of Gh14-3-3 genes during ray floret developmental stages (S1∼S6, “S” represents “stage”) in gerbera. As ray floret petals developed, the expression levels of these genes changed in different ways (Figure 3B). However, some patterns were comparable: for example, Gh14-3-3b and Gh14-3-3f showed a similar expression pattern, such that their transcription levels declined from the highest level in S1 to the lowest level in S3, followed by a gradual increase. Similarly, the transcript abundance of three genes (Gh14-3-3a, Gh14-3-3c, and Gh14-3-3d) dropped to the lowest level from S1 to S2, and then fluctuated in a related manner. In addition, Gh14-3-3e and Gh14-3-3g showed the highest expression levels in S2 and S1, and the lowest expression levels in S4 and S3, respectively. These results suggest that the expression of Gh14-3-3 genes is developmentally regulated in petal cells of gerbera.
14-3-3 proteins play an essential role in the BR signaling pathway (Gampala et al., 2007; Ryu et al., 2007). To determine whether the expression of any of the seven Gh14-3-3 genes responds to BR, the transcript levels of these genes were evaluated following BL treatment. As shown in Figure 3C, Gh14-3-3a and Gh14-3-3b shared the same expression profile: both genes began to respond at 1 h after BL treatment, rising to the highest expression level at 2 h, and then gradually decreasing to the lowest level at 24 h. Specifically, Gh14-3-3b had the highest peak value (225) among all seven members in response to BL. Three other genes (Gh14-3-3d, Gh14-3-3e, and Gh14-3-3g) had a similar response pattern to BR with two comparable response peaks (2.0∼2.5) at 0.5 h and 4 h. The expression level of Gh14-3-3f increased over the study period to a maximum at 24 h, while Gh14-3-3c expression varied slightly within a narrow range in response to BR. These results indicate that all members of the Gh14-3-3 gene family responded to BR, with Gh14-3-3a and Gh14-3-3b both reaching the highest expression level at an early stage (2 h) after treatment and Gh14-3-3f at a late stage (24 h).
Gh14-3-3b and Gh14-3-3f Promote Ray Petal Elongation in Gerbera
Based on the above results, it is clear that Gh14-3-3b and Gh14-3-3f have the lowest expression level in S3 (the onset of cell elongation), compared to other developmental stages. The two genes reached their highest expression levels at early (2 h) and late stages (24 h) in response to BR, respectively. Thus, we chose to analyze the roles of Gh14-3-3b and Gh14-3-3f in ray petal elongation by transient overexpression and VIGS assays.
Overexpression of Gh14-3-3b significantly promoted petal length and elongation rate, while Gh14-3-3f-OE petals showed slightly increased petal length and elongation rate (Figures 4A,D–F). The elongation rate was 0.33 ± 0.01 in Gh14-3-3b-OE and 0.23 ± 0.01 in Gh14-3-3f-OE petals, which corresponds to increases of 57% and 10%, respectively, compared with an elongation rate of 0.21 ± 0.02 in the mock experiment (Figure 4F). To determine whether overexpression of Gh14-3-3b and Gh14-3-3f regulates petal length by promoting petal epidermal cell length, both cell length and number in the top, middle and basal regions of ray petals were measured (Figures 4B,C). The epidermal cell lengths of Gh14-3-3b-OE petals were markedly longer than in the mock controls in all three regions (Figures 4C,G). In addition, the epidermal cell numbers in Gh14-3-3b-OE petals were much smaller than in mock-treated equivalents (Figure 4H). For Gh14-3-3f-OE ray florets, epidermal cell lengths in the basal and middle regions were significantly longer than in the mock, while epidermal cell numbers in the basal and middle regions were lower. The results suggest that Gh14-3-3b and Gh14-3-3f promote ray petal elongation by regulating cell elongation.
FIGURE 4.
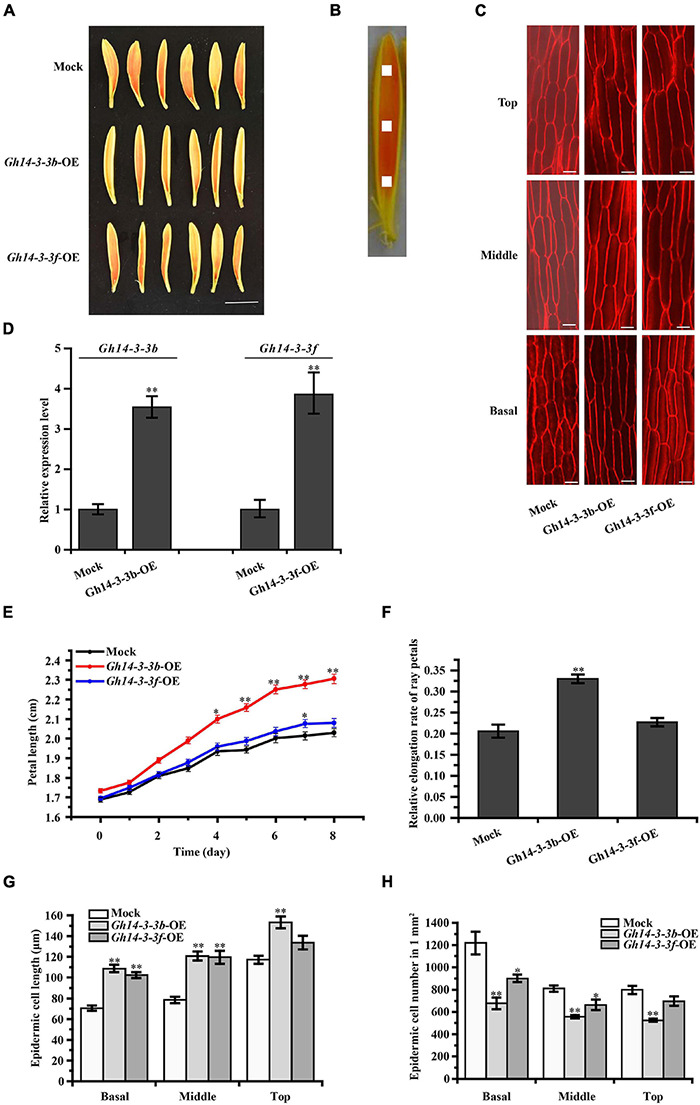
Petal elongation of ray florets were promoted by the overexpression of Gh14-3-3b and Gh14-3-3f. (A) Petal phenotype of the Gh14-3-3b, Gh14-3-3f transient overexpression and Mock (Gh14-3-3b-OE, Gh14-3-3f-OE, Mock) after 8 days. Bar = 1 cm. (B) Schematic diagram of the petal. The picture shows basal, middle, top regions of ray floret petals. The white area (1 mm2) shows the measured region for cell length. (C) The confocal microscope image of epidermal cells in top, middle and basal regions of petals. Bar = 20 μm. (D) The expression analysis of Gh14-3-3b and Gh14-3-3f in the Mock and transgenic ray petals by qRT-PCR. The length (E) and relative elongation rates (F) of mock, Gh14-3-3b-OE and Gh14-3-3f-OE petals (n = 45). The cell length (G) and cell number (H) in top, middle, and basal regions of transgenic ray petals. At least three biological replicates were used for each observation.
We further confirmed the role of Gh14-3-3b and Gh14-3-3f in ray petal elongation using the VIGS system. As shown in Figures 5A,E,F, gene silencing of both Gh14-3-3b and Gh14-3-3f significantly shortened the length and elongation rate of ray petals, relative to the mock. However, exogenous BL treatment eliminated partly this repression (Figures 5A,E). Gene silencing of Gh14-3-3b and Gh14-3-3f also reduced the epidermal cell lengths and boosted cell numbers compared to the mock, while BR reversed this phenotype to some extent (Figures 5B,G,H). In addition, we analyzed the expression of Gh14-3-3b and Gh14-3-3f in Gh14-3-3b-VIGS and Gh14-3-3f-VIGS petals without and with BL treatment (Figures 5C,D). Surprisingly, BL induced the expression of both Gh14-3-3b and Gh14-3-3f to levels that were approximately 10-fold and 60-fold greater, respectively, than those of Gh14-3-3b-VIGS and Gh14-3-3f-VIGS petals. Taken together, these results suggest that Gh14-3-3b and Gh14-3-3f promote BR-induced ray petal elongation.
FIGURE 5.
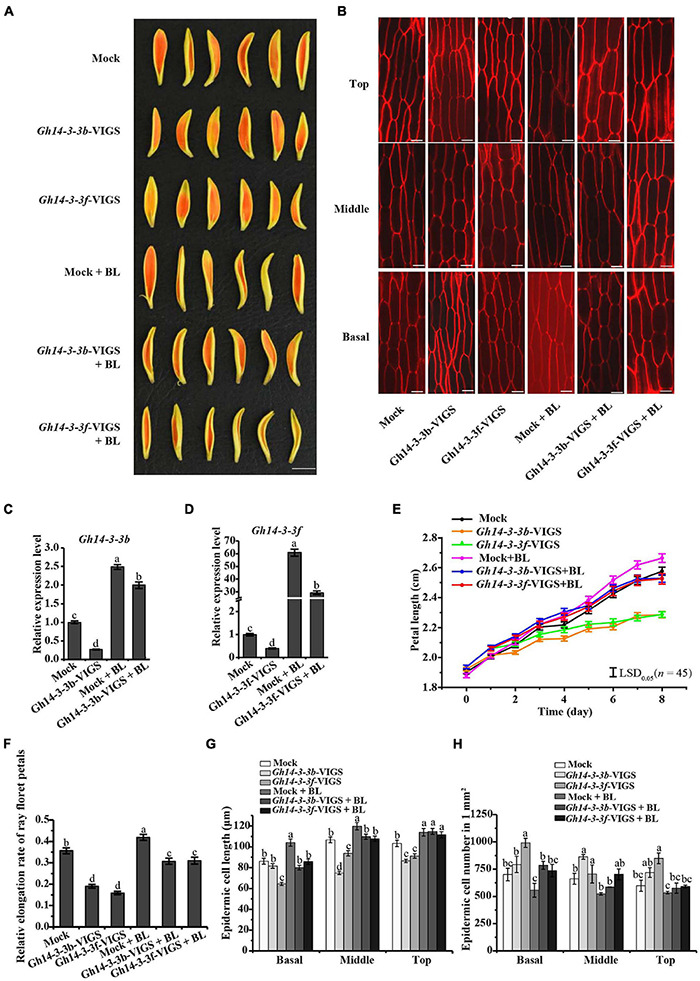
Petal elongation of ray florets was inhibited by transient silencing of Gh14-3-3b and Gh14-3-3f. (A) Petal phenotype of Mock, Gh14-3-3b transient silencing, Gh14-3-3f transient silencing, and those treated with 10 μM BL (Mock, Gh14-3-3b-VIGS, Gh14-3-3f-VIGS, Mock + BL, Gh14-3-3b-VIGS + BL, Gh14-3-3f-VIGS + BL). Bar = 1 cm. (B) The confocal microscope image of epidermal cells in top, middle and basal regions of petals. Bar = 20 μm. (C) The relative expression level of Gh14-3-3b in Mock, Gh14-3-3b-OE, Mock + BL and Gh14-3-3b-VIGS + BL. (D) The relative expression level of Gh14-3-3f in Mock, Gh14-3-3f-OE, Mock + BL, and Gh14-3-3f-VIGS + BL. The length (E) and relative elongation rate (F) of Mock, Gh14-3-3b-VIGS, Gh14-3-3f-VIGS, Mock + BL, Gh14-3-3b-VIGS + BL, and Gh14-3-3f-VIGS + BL petals (n = 45). The elongation rate was calculated at day 8. The cell length (G) and cell number (H) in top, middle, and basal regions in transgenic ray petals. At least three biological replicates were used for each observation.
Expression of Genes Involved in BR Signaling and Petal Elongation Is Altered in Gh14-3-3b and Gh14-3-3f Transgenic Ray Petals
To investigate the mechanism by which Gh14-3-3b and Gh14-3-3f regulate ray petal elongation, the expression levels of genes involved in BR signaling and petal elongation were determined. As shown in Figure 6A, the expression of BR signaling genes (GhBEH1, GhBEH2, and GhBIN2) was significantly increased in Gh14-3-3b-OE petals, but only marginally upregulated in Gh14-3-3f-OE petals. The expression levels of genes involved in petal elongation (GhEXP1, GhEXP2, GhEXP10, GhXTH1, and GhXET) were markedly boosted in Gh14-3-3b-OE and Gh14-3-3f-OE petals, especially in the former (Figure 6A).
FIGURE 6.
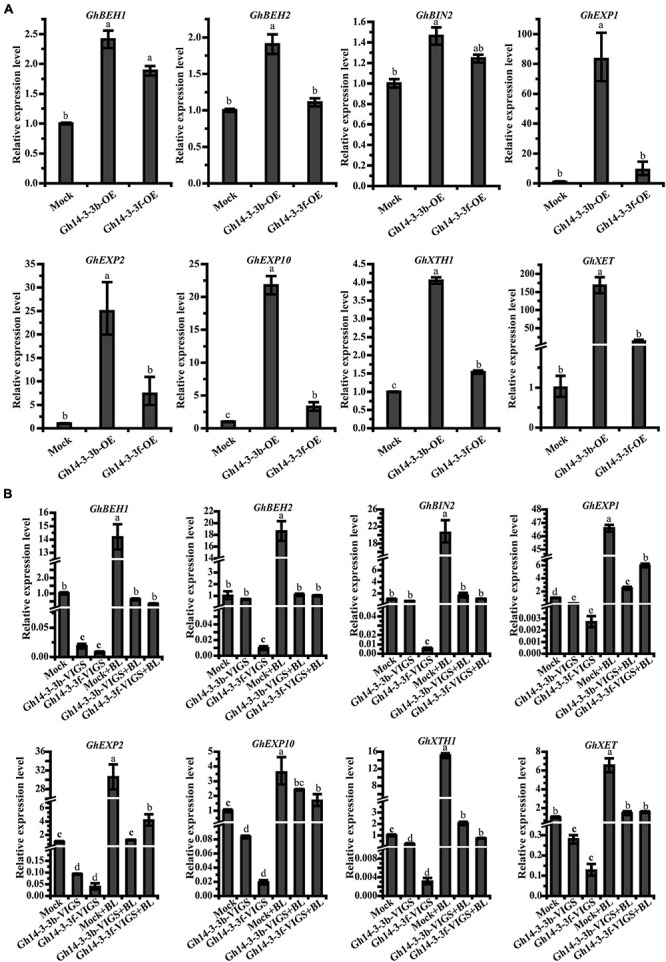
The expression level of related genes in Gh14-3-3b/f-OE ray floret petals (A) and Gh14-3-3b/f-VIGS ray floret petals (B). Gene expression levels were calculated using the 2–ΔΔCt method. The accession numbers of related genes in GenBank are: GhBEH1 (GACN01037487.1), GhBEH2 (GACN01006390.1), GhBIN2 (GACN01003499.1), GhEXP1 (GACN01041978.1), GhEXP2 (GACN01002748.1), GhEXP10 (GACN01039741.1), GhXTH1 (GACN01007419.1), and GhXET (GACN01007419.1).
On the other hand, the expression levels of the above genes showed, in many cases, a declining trend in Gh14-3-3b-VIGS and Gh14-3-3f-VIGS petals. In the Gh14-3-3f-VIGS petals, the expression of all eight genes was significantly reduced, compared to the mock (Figure 6B), while in Gh14-3-3b-VIGS petals, the expression of six genes (GhBEH1, GhEXP1, GhEXP2, GhEXP10, GhXTH1, and GhXET) was markedly downregulated, with two genes, GhBEH2 and GhBIN2, showing only a slightly decline. Notably, the expression of all eight genes was enhanced, albeit to different degrees, following BL treatment. These results suggest that Gh14-3-3b and Gh14-3-3f regulate BR-induced ray petal elongation by modulating genes associated with BR signaling and petal development.
Discussion
Gh14-3-3 Proteins, Which Fall Into Two Groups, May Possess Functional Diversity
Since the first plant 14-3-3 protein was cloned from maize (de Vetten et al., 1992), researchers have identified eight 14-3-3 proteins in rice (Chen et al., 2006; Denison et al., 2011), 18 in apple (Zuo et al., 2021), nine in common bean (Li M. et al., 2016) and seven in cotton (Zhou et al., 2015). In the present study, seven 14-3-3 isoforms were identified in the gerbera transcriptome. Sequence analysis showed that all isoforms share the conserved nine α-helical regions typical of the 14-3-3 family and have 254∼336 amino acids (Figure 1A). Phylogenetics classified the seven gerbera 14-3-3 proteins into two groups, the ε and non-ε groups, consistent with similar groupings in Arabidopsis, rice, and banana (Yaffe et al., 1997; Chevalier et al., 2009; Li M. et al., 2016). Furthermore, they show a high degree of identity with 14-3-3 proteins in Helianthus annuus and Lactuca sativa, both of which belong to Asteraceae family, hinting that these proteins have similar functions across the Asteraceae (Figure 1A).
Previous studies revealed that 14-3-3 proteins can form homodimers or, instead, can form heterodimers with different isoforms, which promotes functional diversity (Liu et al., 1995; Xiao et al., 1995; Aghazadeh et al., 2015; Ormancey et al., 2017). Each isoform displays a different propensity to dimerize with others, depending on the highly variable amino acid sequences in their N-terminal helices (Liu et al., 1995; Xiao et al., 1995). Consistent with this, the sequences of the N-terminal helices of gerbera 14-3-3 proteins are less well conserved than their C-terminal helices (Figure 1A) and only three of the gerbera proteins (Gh14-3-3b, Gh14-3-3c, and Gh14-3-3f) form homodimers (Figure 2 and Supplementary Figure 2). Among the seven Gh14-3-3 proteins, the number of heterodimers formed ranged from one for Gh14-3-3e to six for Gh14-3-3b, which demonstrates the selectivity of each isoform in protein-protein interactions.
Wilson et al. (2016) summarized the regulatory mechanisms of 14-3-3 proteins in several plants during the development of multiple organs, including seedling, leaf, root, flower, and developing seed. We found that the seven 14-3-3 isoforms are expressed in various organs in gerbera to different extents (Figure 3A). This implies a functional diversity among all seven members, similar to the 14-3-3 proteins of other species (Chen et al., 2006; Yao et al., 2007). In addition, we surveyed the expression patterns of the Gh14-3-3 genes during ray floret developmental phases (S1∼S6) (Figure 3B). As development progresses through stages S1 to S6, the Gh14-3-3 genes are expressed in various patterns. For example, Gh14-3-3b and Gh14-3-3f display a trend of earlier decrease and later increase from S1∼S6, and they both have the lowest expression level at S3 (Figure 3B). These genes also differ in their response to exogenous BL treatment (Figure 3C): Gh14-3-3b responds rapidly and reaches its highest expression level in the early phase of the experiment (2 h), while Gh14-3-3f shows a slow response pattern. These results suggest functional diversity of the Gh14-3-3 genes during BR-induced gerbera growth and development (Chen et al., 2006; Zhou et al., 2015; Zuo et al., 2021).
Gh14-3-3b and Gh14-3-3f Play a Positive Regulatory Role in BR-Induced Ray Petal Elongation
The roles of 14-3-3 proteins in plant growth and development have been reported (Zhou et al., 2015; Huang et al., 2018). At14-3-3λ and At14-3-3K are involved in shade-induced hypocotyl elongation via PHYTOCHROME-INTERACTING FACTOR 7 (Huang et al., 2018). In cotton, 14-3-3 proteins are involved in cotton fiber elongation by regulating GhBZR1 protein, which binds to the promoters of genes involved in fiber development (Zhou et al., 2015). In the present study, overexpression of Gh14-3-3b and Gh14-3-3f in gerbera increased the length of ray petals, whereas gene silencing of Gh14-3-3b and Gh14-3-3f shortened petal length (Figures 4E, 5E). Confocal images showed that this may be achieved by regulating epidermal cell length in petals (Figures 4G, 5G). Moreover, the qRT-PCR results revealed that the expression of genes involved in BR signaling (GhBEH1, GhBEH2, and GhBIN2) is enhanced in Gh14-3-3b-OE and Gh14-3-3f-OE petals and inhibited in Gh14-3-3b-VIGS and Gh14-3-3f-VIGS petals (Figures 6A,B). BL treatment promotes petal length in Gh14-3-3b-VIGS and Gh14-3-3f-VIGS petals, and enhances the expression of GhBEH1 in Gh14-3-3b-VIGS petals as well as the expressions of GhBEH1, GhBEH2, and GhBIN2 in Gh14-3-3f-VIGS petals (Figures 5E, 6B). These results suggest that Gh14-3-3b and Gh14-3-3f play a role in BR-induced ray petal elongation. In a previous study, BL was shown to promote ray petal elongation and GhBEH1 expression in gerbera (Huang et al., 2017). In Arabidopsis, AtBZR1 is one of the most important transcription factors in BR signaling and promotes cell elongation in response to BR (Oh et al., 2012; Chaiwanon and Wang, 2015). Therefore, it is possible that Gh14-3-3b and Gh14-3-3f modulate ray petal elongation by regulating GhBEH1 and GhBEH2. The specific molecular mechanisms will be investigated further in future studies.
Expansins (EXPs), xyloglucan endotransglycosylases (XETs), and xylan transferases/hydrolases (XTHs) are involved in cell wall remodeling and cell elongation (Takeda et al., 2002; Harada et al., 2011; Che et al., 2016; Li Y. et al., 2016; Rao and Dixon, 2017; Ackerman-Lavert and Savaldi-Goldstein, 2020). Two XTH genes (DcXTH2 and DcXTH3) and two expansin genes (DcEXPA1 and DcEXPA2) are associated with petal growth and development during flower opening in carnation (Harada et al., 2011). OsEXPA10 is expressed in the root tips and is necessary for cell elongation in rice (Che et al., 2016). Additionally, the expression of some XTH and EXP genes is markedly enhanced by BL treatment in Arabidopsis (Park et al., 2010) and soybean (Rao and Dixon, 2017). Our previous study also showed that BR promotes ray petal elongation and initiates the expression of a number of genes, including those encoding two putative cell wall proteins (Huang et al., 2017). In this study, the expression of a number of petal elongation-related genes (including GhEXP1, GhEXP2, GhEXP10, GhXET, and GhXEH) were altered in Gh14-3-3b-OE and Gh14-3-3f-OE, as well as Gh14-3-3b-VIGS and Gh14-3-3f-VIGS petals (Figure 6B). This suggests that Gh14-3-3b and Gh14-3-3f modulate petal elongation-related gene transcription, thereby mediating petal cell elongation and petal development.
As one of the mainstream cut flowers, gerbera has a high demand in the market. However, it has fewer flower types compared to chrysanthemum. Thus, obtaining a variety of flower types is one of the main goals of gerbera breeding, which requires an understanding of the regulatory mechanism of gerbera flower development. In this study, seven Gh14-3-3 protein genes were identified and their expression patterns were characterized. These genes share a conserved structure, but display different dimerization patterns, which implies they are functionally diverse. Transient transformation assays demonstrated that Gh14-3-3b and Gh14-3-3f play a positive regulatory role in BR-induced ray petal elongation. Thus, as well as providing novel insights into the role of 14-3-3 proteins in ray petal elongation, this study also highlights a number of candidate genes for flower type breeding of gerbera.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
XL carried out the experiments, drafted the manuscript, and revised manuscript. SH conducted the experiments, analyzed the data, and prepared the figures. GH participated in part of the experiments. YC and XW revised the manuscript. YW conceived the study, participated in its design, and revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Ling Li for providing the overexpression vector C17. We are grateful to Xiaoyan Tang and Cambridge Academic Manuscripts (www.CambridgeAcademicManuscripts.com) for manuscript editing.
Funding. This work was supported by the National Key R&D Program of China (2018YFD1000404), Research Team Project of Natural Science Foundation of Guangdong Province (2017A030312004), National Natural Science Foundation of China (31672188), and Natural Science Foundation of Guangdong Province (2021A1515012479, 2021A1515011315, and 201904010127).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.718091/full#supplementary-material
Conserved domains analysis of seven Gh14-3-3s in the Conserved Domain Database.
The interactions among Gh14-3-3 proteins. Gh14-3-3s were introduced into pGADT7-AD and pGBKT7-BD. pGADT7-largeT7/pGBKT7-53 and pGADT7-largeT7/pGBKT7-laminC were used as positive and negative controls, respectively.
Primers used in this study and amino acid sequences of 14-3-3s in different species.
References
- Ackerman-Lavert M., Savaldi-Goldstein S. (2020). Growth models from a brassinosteroid perspective. Curr. Opin. Plant Biol. 53 90–97. 10.1016/j.pbi.2019.10.008 [DOI] [PubMed] [Google Scholar]
- Aghazadeh Y., Zirkin B. R., Papadopoulos V. (2015). Pharmacological regulation of the cholesterol transport machinery in steroidogenic cells of the testis. Vitam. Hormon. 98 189–227. 10.1016/bs.vh.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Aitken A. (2006). 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 16 162–172. 10.1016/j.semcancer.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Bhatia R., Singh K. P., Jhang T., Sharma T. R. (2009). Assessment of clonal fidelity of micropropagated gerbera plants by ISSR markers. Sci. Hortic. 119 208–211. 10.1016/j.scienta.2008.07.024 [DOI] [Google Scholar]
- Broholm S. K., Tähtiharju S., Laitinen R. A. E., Albert V. A., Teeri T. H., Elomaa P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the gerbera (Asteraceae) inflorescence. PNAS 105 9117–9122. 10.1073/pnas.0801359105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camoni L., Visconti S., Aducci P., Marra M. (2018). 14-3-3 proteins in plant hormone signaling: doing several things at once. Front. Plant Sci. 9:297. 10.3389/fpls.2018.00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwanon J., Wang Z. Y. (2015). Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 25 1031–1042. 10.1016/j.cub.2015.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che J., Yamaji N., Shen R. F., Ma J. F. (2016). An Al-inducible expansin gene, OsEXPA10 is involved in root cell elongation of rice. Plant J. 88 132–142. 10.1111/tpj.13237 [DOI] [PubMed] [Google Scholar]
- Chen F., Li Q., Sun L., He Z. (2006). The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 13 53–63. 10.1093/dnares/dsl001 [DOI] [PubMed] [Google Scholar]
- Chevalier D., Morris E. R., Walker J. C. (2009). 14-3-3 and FHA domains mediate phosphoprotein interactions. Annu. Rev. Plant Biol. 60 67–91. 10.1146/annurev.arplant.59.032607.092844 [DOI] [PubMed] [Google Scholar]
- Clouse S. D., Langford M., McMorris T. C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111 671–678. 10.1104/pp.111.3.671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S. D., Sasse J. M. (1998). BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu. Rev. Plant Biol. 49 427–451. 10.1146/annurev.arplant.49.1.427 [DOI] [PubMed] [Google Scholar]
- de Vetten N. C., Lu G., Feri R. J. (1992). A maize protein associated with the G-box binding complex has homology to brain regulatory proteins. Plant Cell 4 1295–1307. 10.1105/tpc.4.10.1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLille J. M., Sehnke P. C., Ferl R. J. (2001). The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 126 35–38. 10.1104/pp.126.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison F. C., Paul A. L., Zupanska A. K., Ferl R. J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22 720–727. 10.1016/j.semcdb.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Ferl R. J. (1996). 14-3-3 Proteins and signal transduction. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 47 49–73. 10.1146/annurev.arplant.47.1.49 [DOI] [PubMed] [Google Scholar]
- Gampala S. S., Kim T. W., He J., Tang W., Deng Z., Bai M., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in arabidopsis. Dev. Cell 13 177–189. 10.1016/j.devcel.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Jin X., Yao W., Kong L., Huang G., Tao Y., et al. (2017). A mini zinc-finger protein (MIF) from Gerbera hybrida activates the GASA protein family gene, GEG, to inhibit ray petal elongation. Front. Plant Sci. 8:1649. 10.3389/fpls.2017.01649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T., Torii Y., Morita S., Onodera R., Hara Y., Yokoyama R., et al. (2011). Cloning, characterization, and expression of xyloglucan endotransglucosylase/hydrolase and expansin genes associated with petal growth and development during carnation flower opening. J. Exp. Bot. 62 815–823. 10.1093/jxb/erq319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. X., Gendron J. M., Sun Y., Gampala S. S. L., Gendron N., Sun C. Q., et al. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307 1634–1638. 10.1126/science.1107580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Han M., Jian L., Chen Y., Sun S., Wang X., et al. (2020). An ETHYLENE INSENSITIVE3-LIKE1 protein directly targets the GEG promoter and mediates ethylene-induced ray petal elongation in Gerbera hybrida. Front. Plant Sci. 10:1737. 10.3389/fpls.2019.01737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Han M., Yao W., Wang Y. (2017). Transcriptome analysis reveals the regulation of brassinosteroids on petal growth in Gerbera hybrida. PeerJ 5:e3382. 10.7717/peerj.3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Zhang Q., Jiang Y., Yang C., Wang Q., Li L. (2018). Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. Plant Biol. 7:17. 10.7554/eLife.31636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntheikki-Palovaara I., Tähtiharju S., Lan T., Broholm S. K., Rijpkema A. S., Ruonala R., et al. (2014). Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J. 79 783–796. 10.1111/tpj.12583 [DOI] [PubMed] [Google Scholar]
- Kaneko-Suzuki M., Kurihara-Ishikawa R., Okushita-Terakawa C., Kojima C., Nagano-Fujiwara M., Ohki I., et al. (2018). TFL1-Like proteins in rice antagonize rice FT-Like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol. 59 458–468. 10.1093/pcp/pcy021 [DOI] [PubMed] [Google Scholar]
- Keicher J., Jaspert N., Weckermann K., Möller C., Throm C., Kintzi A., et al. (2017). Arabidopsis 14-3-3 epsilon members contribute to polarity of PIN auxin carrier and auxin transport-related development. eLife 6:e24336. 10.7554/eLife.24336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. W., Michniewicz M., Bergmann D. C., Wang Z. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482 419–422. 10.1038/nature10794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. W., Wang Z. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61 681–704. 10.1146/annurev.arplant.043008.092057 [DOI] [PubMed] [Google Scholar]
- Konagaya K., Matsushita Y., Kasahara M., Nyunoya H. (2004). Members of 14-3-3 protein isoforms interacting with the resistance gene product N and the elicitor of Tobacco mosaic virus. J. Gen. Plant Pathol. 70 221–231. 10.1007/s10327-003-0113-4 [DOI] [Google Scholar]
- Kotilainen M., Helariutta Y., Mehto M., Pöllänen E., Albert V. A., Elomaa P., et al. (1999). GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell 11 1093–1104. 10.1105/tpc.11.6.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Q., Li L., Peng J., Sun S., Wang X. (2013). Transcriptome analysis of Gerbera hybrida ray florets: putative genes associated with gibberellin metabolism and signal transduction. PLoS One 8:e57715. 10.1371/journal.pone.0057715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang W., Zhang L., Li N., Peng J., Wang Y., et al. (2015). Transcriptomic insights into antagonistic effects of gibberellin and abscisic acid on petal growth in Gerbera hybrida. Front. Plant Sci. 6:168. 10.3389/fpls.2015.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Ren L., Xu B., Yang X., Xia Q., He P., et al. (2016). Genome-wide identification, phylogeny, and expression analyses of the 14-3-3 family reveal their involvement in the development, ripening, and abiotic stress response in banana. Front. Plant Sci. 7:1442. 10.3389/fpls.2016.01442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tu L., Pettolino F. A., Ji S., Hao J., Yuan D., et al. (2016). GbEXPATR, a species-specific expansin, enhances cotton fiber elongation through cell wall restructuring. Plant Biotechnol. J. 14 951–963. 10.1111/pbi.12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Bienkowska J., Petosa C., Collier R. J., Fu H., Liddington R. (1995). Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376 191–194. 10.1038/376191a0 [DOI] [PubMed] [Google Scholar]
- Meng X., Wang X. (2004). Regulation of flower development and anthocyanin accumulation in Gerbera hybrida. J. Hortic. Sci. Biotechnol. 79 131–137. 10.1080/14620316.2004.11511725 [DOI] [Google Scholar]
- Mhawech P. (2005). 14-3-3 proteins—an update. Cell Res. 15 228–236. 10.1038/sj.cr.7290291 [DOI] [PubMed] [Google Scholar]
- Minami A., Takahashi K., Inoue S., Tada Y., Kinoshita T. (2019). Brassinosteroid induces phosphorylation of the plasma membrane H+-ATPase during hypocotyl elongation in Arabidopsis thaliana. Plant Cell Physiol. 60 935–944. 10.1093/pcp/pcz005 [DOI] [PubMed] [Google Scholar]
- Mosqueda Frómeta O., Escalona Morgado M. M., Teixeira da Silva J. A., Pina Morgado D. T., Daquinta Gradaille M. A. (2017). In vitro propagation of Gerbera jamesonii Bolus ex Hooker f. in a temporary immersion bioreactor. Plant Cell Tissure Organ Cult. 129 543–551. 10.1007/s11240-017-1186-7 [DOI] [Google Scholar]
- Oh E., Zhu J. Y., Wang Z. Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14 802–809. 10.1038/ncb2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M. H., Honey S. H., Tax F. E. (2020). The control of cell expansion, cell division, and vascular development by brassinosteroids: a historical perspective. Int. J. Mol. Sci. 21:1743. 10.3390/ijms21051743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormancey M., Thuleau P., Mazars C., Cotelle V. (2017). CDPKs and 14-3-3 proteins: emerging duo in signaling. Trends Plant Sci. 22 263–272. 10.1016/j.tplants.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Park C. H., Kim T. W., Son S. H., Hwang J. Y., Lee S. C., Chang S. C., et al. (2010). Brassinosteroids control AtEXPA5 gene expression in Arabidopsis thaliana. Phytochemistry 71 380–387. 10.1016/j.phytochem.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Pertl H., Pöckl M., Blaschke C., Obermeyer G. (2010). Osmoregulation in lilium pollen grains occurs via modulation of the plasma membrane H+ ATPase activity by 14-3-3 proteins. Plant Physiol. 154 1921–1928. 10.1104/pp.110.165696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S., Sun S., Wang L., Wu Z., Li C., Li X., et al. (2017). The RLA1/SMOS1 transcription factor functions with OsBZR1 to regulate brassinosteroid signaling and rice architecture. Plant Cell 29 292–309. 10.1105/tpc.16.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X., Dixon R. A. (2017). Brassinosteroid mediated cell wall remodeling in grasses under abiotic stress. Front. Plant Sci. 8:806. 10.3389/fpls.2017.00806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G., Li L., Huang Y., Wang Y., Zhang W., Zheng R., et al. (2018). GhWIP2, a WIP zinc finger protein, suppresses cell expansion in Gerbera hybrida by mediating crosstalk between gibberellin, abscisic acid, and auxin. New Phytol. 219 728–742. 10.1111/nph.15175 [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19 2749–2762. 10.1105/tpc.107.053728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke P. C., DeLille J. M., Ferl R. J. (2002). Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell 14(Suppl.) S339–S354. 10.1105/tpc.010430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N. F., Christie L. N., Mazzurco M., Goring D. R. (2001). Characterization of a novel Brassica napus kinase, BNK1. Plant Sci. 160 611–620. 10.1016/s0168-9452(00)00426-x [DOI] [PubMed] [Google Scholar]
- Tahtiharju S., Rijpkema A. S., Vetterli A., Albert V. A., Teeri T. H., Elomaa P. (2012). Evolution and diversification of the CYC/TB1 gene family in asteraceae—a comparative study in gerbera (Mutisieae) and sunflower (Heliantheae). Mol. Biol. Evol. 29 1155–1166. 10.1093/molbev/msr283 [DOI] [PubMed] [Google Scholar]
- Takeda T., Furuta Y., Awano T., Mizuno K., Mitsuishi Y., Hayashi T. (2002). Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. PNAS 99 9055–9060. 10.1073/pnas.132080299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K., Ohki I., Tsuji H., Furuita K., Hayashi K., Yanase T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476 332–335. 10.1038/nature10272 [DOI] [PubMed] [Google Scholar]
- van Kleeff P. J. M., Jaspert N., Li K. W., Rauch S., Oecking C., de Boer A. H. (2014). Higher order Arabidopsis 14-3-3 mutants show 14-3-3 involvement in primary root growth both under control and abiotic stress conditions. J. Exp. Bot. 65 5877–5888. 10.1093/jxb/eru338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-Y., Bai M. Y., Oh E., Zhu J. Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46 701–724. 10.1146/annurev-genet-102209-163450 [DOI] [PubMed] [Google Scholar]
- Wilson R. S., Swatek K. N., Thelen J. J. (2016). Regulation of the regulators: post-translational modifications, subcellular, and spatiotemporal distribution of plant 14-3-3 proteins. Front. Plant Sci. 7:611. 10.3389/fpls.2016.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B., Smerdon S. J., Jones D. H., Dodson G. G., Soneji Y., Aitken A., et al. (1995). Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376 188–191. 10.1038/376188a0 [DOI] [PubMed] [Google Scholar]
- Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., et al. (1997). The structural basis for 14-3-3: phosphopeptide binding specificity. Cell 91 961–971. 10.1016/S0092-8674(00)80487-0 [DOI] [PubMed] [Google Scholar]
- Yang C., Zhang C., Lu Y., Jin J., Wang X. (2011). The mechanisms of brassinosteroids’ action: from signal transduction to plant development. Mol. Plant 4 588–600. 10.1093/mp/ssr020 [DOI] [PubMed] [Google Scholar]
- Yao Y., Du Y., Jiang L., Liu J. (2007). Molecular analysis and expression patterns of the 14-3-3 gene family from Oryza sativa. J. Biochem. Mol. Biol. 40 349–357. 10.5483/bmbrep.2007.40.3.349 [DOI] [PubMed] [Google Scholar]
- Zhang L., Li L., Wu J., Peng J., Zhang L., Wang X. (2012). Cell expansion and microtubule behavior in ray floret petals of Gerbera hybrida: responses to light and gibberellic acid. Photochem. Photobiol. Sci. 11 279–288. 10.1039/C1PP05218G [DOI] [PubMed] [Google Scholar]
- Zhang T., Cieslak M., Owens A., Wang F., Broholm S. K., Teeri T. H., et al. (2021). Phyllotactic patterning of gerbera flower heads. PNAS 118:e2016304118. 10.1073/pnas.2016304118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Guo W., Du D., Pu L., Zhang C. (2020). Overexpression of a maize BR transcription factor ZmBZR1 in Arabidopsis enlarges organ and seed size of the transgenic plants. Plant Sci. 292:110378. 10.1016/j.plantsci.2019.110378 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhou Y., Li Y., Shao S., Li B., Shi H., et al. (2010). Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibres and involved in cell elongation. J. Exp. Bot. 61 3331–3344. 10.1093/jxb/erq155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Broholm S. K., Wang F., Rijpkema A. S., Lan T., Albert V. A., et al. (2020). TCP and MADS-box transcription factor networks regulate heteromorphic flower. Plant Physiol. 184 1455–1468. 10.1104/pp.20.00702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Hu M., Yang H., Tang M., Zhang L., Liu H., et al. (2019). Three BnaIAA7 homologs are involved in auxin/brassinosteroid-mediated plant morphogenesis in rapeseed (Brassica napus L.). Plant Cell Rep. 38 883–897. 10.1007/s00299-019-02410-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhang Z., Li M., Wei X., Li X., Li B., et al. (2015). Cotton (Gossypium hirsutum) 14-3-3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signalling. Plant Biotechnol. J. 13 269–280. 10.1111/pbi.12275 [DOI] [PubMed] [Google Scholar]
- Zuo X., Wang S., Xiang W., Yang H., Tahir M. M., Zheng S., et al. (2021). Genome-wide identification of the 14-3-3 gene family and its participation in floral transition by interacting with TFL1/FT in apple. BMC Genom. 22:41. 10.1186/s12864-020-07330-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conserved domains analysis of seven Gh14-3-3s in the Conserved Domain Database.
The interactions among Gh14-3-3 proteins. Gh14-3-3s were introduced into pGADT7-AD and pGBKT7-BD. pGADT7-largeT7/pGBKT7-53 and pGADT7-largeT7/pGBKT7-laminC were used as positive and negative controls, respectively.
Primers used in this study and amino acid sequences of 14-3-3s in different species.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.


