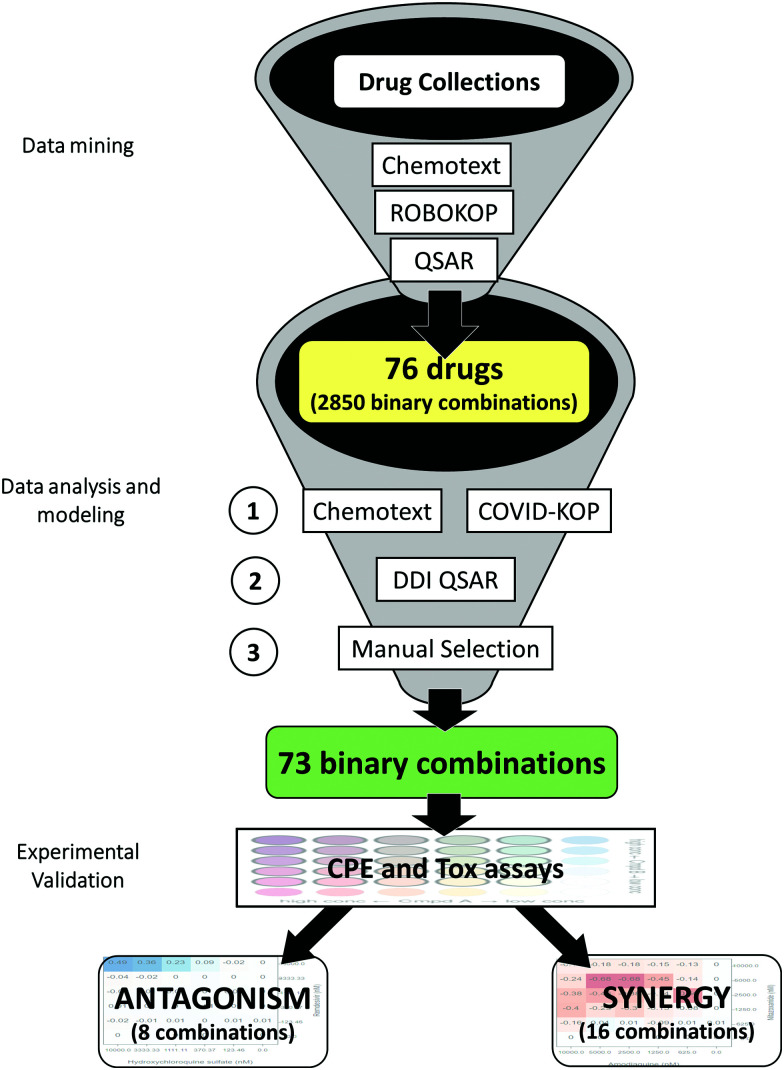
Abstract
COVID-19 has resulted in huge numbers of infections and deaths worldwide and brought the most severe disruptions to societies and economies since the Great Depression. Massive experimental and computational research effort to understand and characterize the disease and rapidly develop diagnostics, vaccines, and drugs has emerged in response to this devastating pandemic and more than 130 000 COVID-19-related research papers have been published in peer-reviewed journals or deposited in preprint servers. Much of the research effort has focused on the discovery of novel drug candidates or repurposing of existing drugs against COVID-19, and many such projects have been either exclusively computational or computer-aided experimental studies. Herein, we provide an expert overview of the key computational methods and their applications for the discovery of COVID-19 small-molecule therapeutics that have been reported in the research literature. We further outline that, after the first year the COVID-19 pandemic, it appears that drug repurposing has not produced rapid and global solutions. However, several known drugs have been used in the clinic to cure COVID-19 patients, and a few repurposed drugs continue to be considered in clinical trials, along with several novel clinical candidates. We posit that truly impactful computational tools must deliver actionable, experimentally testable hypotheses enabling the discovery of novel drugs and drug combinations, and that open science and rapid sharing of research results are critical to accelerate the development of novel, much needed therapeutics for COVID-19.
We cover diverse methodologies, computational approaches, and case studies illustrating the ongoing efforts to develop viable drug candidates for treatment of COVID-19.
Introduction. A brief survey of COVID-19 drug discovery landscape
With almost 180 million cases and 4 million deaths worldwide (June 2021),1 the COVID-19 pandemic generated a need for a rapid, massive and effective therapeutic response. Since the emergence of COVID-19 in late December 2019, both its causative agent, SARS-CoV-2 virus, and the host response to the virus have been extensively studied to understand the disease pathogenesis, the structure of the constituent viral proteins, and the viral-host interactome to guide rapid development of both direct-acting antiviral (DAA) and host-directed agents. Clearly, an immediate emergence of new, effective drugs for COVID-19 has not been an option because the usual extensive drug development and clinical testing takes 10–15 years. Thus, along with immense, and fortunately, successful efforts to develop vaccines against COVID-19, many scientists and clinicians have pursued the repurposing of existing drugs, clinical trials candidates, and approved natural products that have already been in man and whose toxicity and preliminary pharmacokinetics have been known.
The response by the research community to the pandemic measured by the number of publications has been substantial. As of April 2021, nearly 125 000 research papers on COVID-19 have been annotated in Pubmed2 and more than 14 500 preprints have been deposited by the scientific community in MedRxiv or BioRxiv,3 with many more appearing in other preprint servers. Many of these publications reported on extensive structural and proteomic studies of SARS-CoV-2 components, biological screening of chemical libraries, and other experimental investigations that provided valuable data to support multiple computational approaches to COVID-19 drug discovery. Conversely, many computational studies proposed candidates for drug repurposing as well as novel drug candidates, but the overwhelming majority of respective publications reported no supporting experimental evidence. The number of such manuscripts has become so overwhelming that even preprint servers have stopped accepting manuscripts describing purely computational submissions.4 However, comprehensive studies combining computational investigations with experimental validations have emerged as well.
Due to this unprecedented number of studies by both specialists and novices in computer-aided drug discovery (CADD) who embarked on virtual searches for COVID-19 drug candidates, we considered it extremely timely to critically review computational approaches employed in CADD for COVID-19 and the results of their application. We felt it was important to summarize the strategies and best practices of computational drug discovery that have emerged from the analysis of the most impactful publications. We have focused on small molecule drugs, as vaccine development has been reviewed elsewhere.5,6 It worth noting that although multiple effective COVID-19 vaccines have been developed, tested, and distributed with unprecedented speed, their long term efficacy, side effects, and coverage of rapidly emerging SARS-CoV-2 variants are not fully understood. In addition, none of the vaccines developed thus far offered 100% protection to all vaccinated people. It is also important to state that small-molecule DAA agents and vaccines correspond to fully complementary therapy- and prevention-oriented approaches, both aiming to contain COVID-19 pandemics. Thus, as emphasized in the recent Nature editorial7 and argued in a recent historical survey on antiviral drug discovery,8 efforts to develop new antiviral medications should not only continue but accelerate.
In this review, we provide a critical summary of research efforts that emerged in the CADD community in response to the pandemic. The overall flow of this review is shown in Fig. 1.
Fig. 1. Summary of key developments in CADD for COVID-19.
We start by providing brief overview of small molecule drug discovery and repurposing efforts and key data-rich resources that have been developed in the last year with the focus on SARS-CoV-2 and COVID-19. We follow with the detailed consideration of SARS-CoV-2 proteins critical to the virus’ life cycle and a critical overview of the computational drug discovery studies that can be classified into three major categories: structure-based approaches including molecular docking, molecular dynamics (MD) and free energy perturbations (FEP) (reviewed, in part, recently9); ligand-based methods such as Quantitative Structure–Activity Relationship (QSAR) modeling; and knowledge-mining approaches, including Artificial Intelligence (AI), that led to data-supported nomination and testing of several repurposed drug candidates and drug combinations. In reviewing these approaches and their applications, we emphasize the importance of reliable experimental validation of computational hits and describe the advantages of open drug discovery to accelerate the discovery of novel therapeutics against both the current and possible future pandemics.
We can summarize our analysis of the CADD research literature for COVID-19 as follows:
– The magnitude and urgency of the research response to COVID-19 pandemics highlights the ability of CADD to capture and transform both pre-existing and new data of relevance to the pandemic into actionable drug discovery hypotheses.
– CADD provides a robust framework for open science including knowledge exchange, open-source software implementation, and data sharing, as the nature of the field embodies collaboration between computational, experimental, and clinical scientists, and convergence of multi-disciplinary, goal-oriented approaches toward discovery and development of novel and powerful medicines.
– The expert use of methods and adherence to the best practices of CADD catalyze faster experimental success and enable rapid emergence of valid, experimentally confirmed drug candidates.
We trust that our observations and summaries of the best practice approaches to CADD in the times of pandemic are helpful to all investigators working on COVID-19 as well as other important drug targets. We hope this critical review will prove valuable not only for researchers but also for journal editors by helping them to assess quality and impact of manuscript submissions and media stories on COVID-19 drug discovery.
Critical assessment of early experimental, clinical, and computational studies on drug repurposing against COVID-19
The emergence of COVID-19 generated a sense of urgency among scientists from around the world. Many scientists with diverse educational and professional backgrounds have refocused their computational or experimental research toward the discovery of drug candidates for COVID-19. In the earliest stages of the outbreak, several publications reported compounds with low micromolar in vitro activity against SARS-CoV-2. Most of these studies involved FDA approved drugs with limited assessment of novel chemical entities. Larger screens were subsequently performed, and many hits were screened with human, or animal cells infected with the virus. To date, hundreds of structurally diverse small molecules have been assessed for their activity in virus-infected cells (Table 1). We briefly review some of these studies below as many of them have provided data to empower computational model development and hit validation.
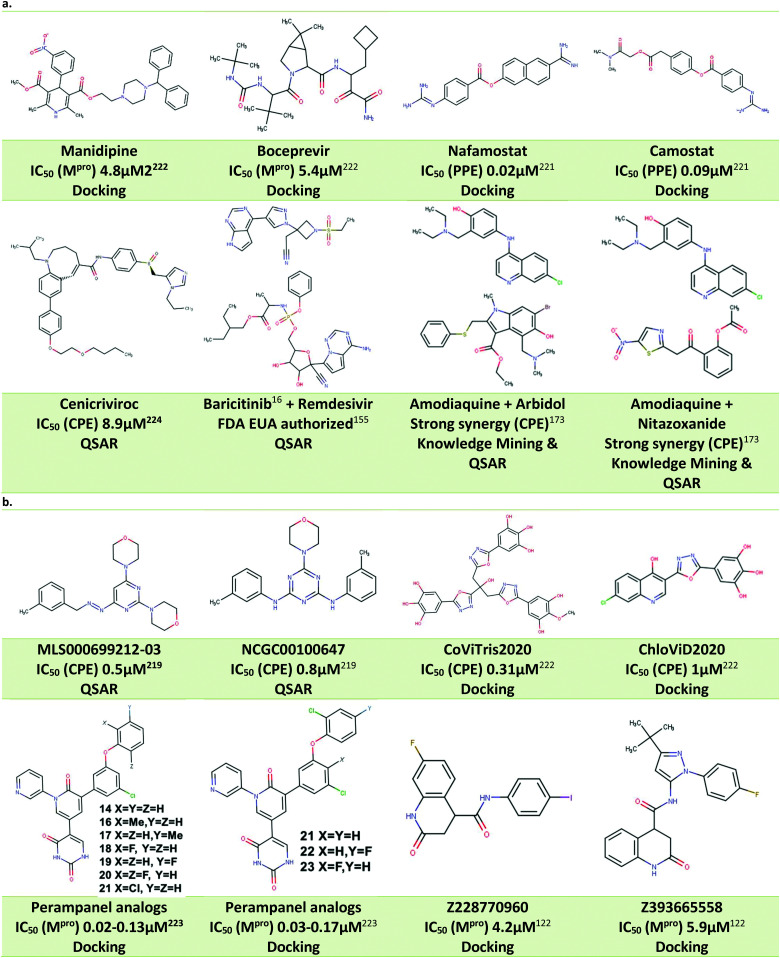
Examples of actives derived from drug repurposing for SARS-CoV-2.
| Molecule | Name | Target | SARS-CoV-2 activity in Vero cells | SARS-CoV-2 activity on other cell types |
|---|---|---|---|---|
|
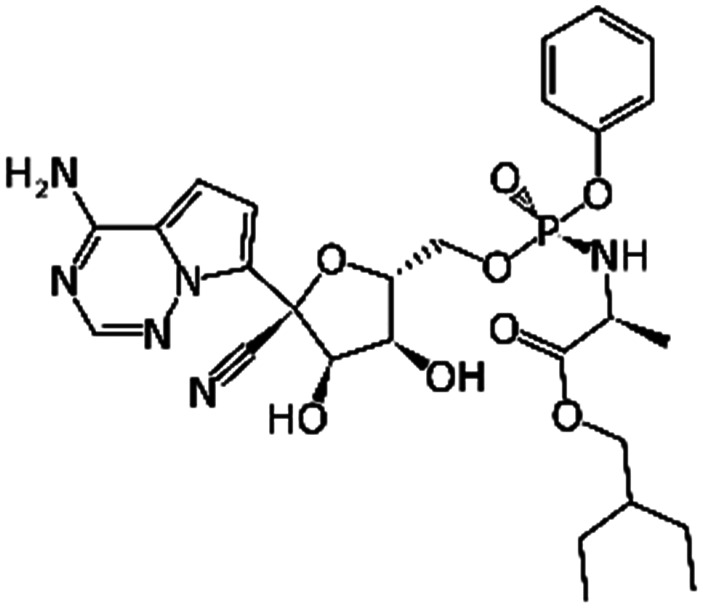
Remdesivir | RNA-dependent RNA polymerase | EC50 0.77 μM2 | Human epithelial cell culture (EC50 0.01 μM); Calu3 (EC50 0.28 μM)10 |
| EC50 1.65 μM5 | ||||
|
Apilimod | PIKfyve | EC50 0.023 μM11 | 293T cells (EC50 0.012 μM)12 |
| IC50 < 0.08 μM13 | Huh-7 cells (0.088 μM)12 | |||
| A549 cells (IC50 0.007 μM)14 | ||||
|
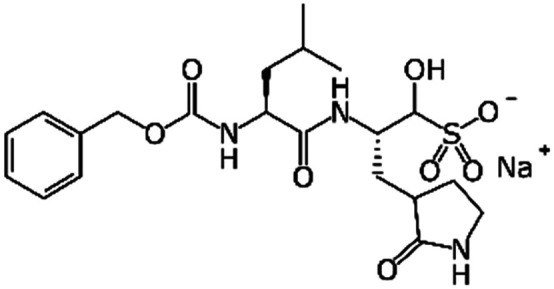
GC376 | Mpro (Ki 12 nM)15 | EC50 0.91 μM15 | Not tested |
|
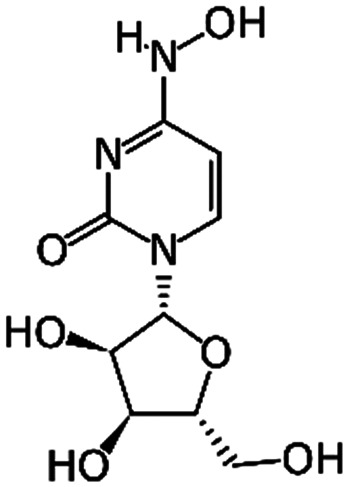
EIDD-1931 | RNA-dependent RNA polymerase | IC50 0.3 μM16 | Calu-3 (IC50 0.08 μM)17 |
One of the earliest drug repurposing studies18 identified several previously known antivirals with low μM activity against SARS-CoV-2 virus in Vero cells and possessing a selectivity index (SI) greater than 10,. Those included FDA-approved drugs nitazoxanide (EC50 2.12 μM), remdesivir (EC50 0.77 μM), and chloroquine (EC50 1.13 μM). Although subsequent clinical trials did not deliver a ‘silver bullet’ for COVID-19, remdesivir was eventually authorized for the clinical use.19 Other notable repurposing examples included lumefantrine (EC50 23.50 μM), the natural products lycorine (EC50 0.31 μM) and oxysophoridine (EC50 0.18 μM), where the latter two demonstrated Vero cell activity superior to gemcitabine (EC50 1.24 μM) and chloroquine (EC50 1.38 μM). Another repurposing screen identified niclosamide (IC50 0.28 μM), ciclesonide (IC50 4.33 μM), and tilorone (IC50 4 μM), previously shown to be active against MERS and Ebola. Pyronaridine (IC50 31 μM) was also identified as a SARS-CoV-2 candidate inhibitor, and both tilorone and pyronaridine have progressed into clinical trials.20 The FDA approved antiparasitic, ivermectin (IC50 2.8 μM) also demonstrated significant in vitro activity in Vero cells leading to broad discussions in the literature21 and eventual nomination for clinical trials.22
Progressive growth of assay- and robotic capabilities has enabled large-scale screening campaigns against SARS-CoV-2. For example, a recent study23 used biological activity-based modeling to identify 311 chemicals, of which 99 demonstrated in vitro activity against the virus. In another notable large-scale study, 12 000 clinical stage or FDA approved compounds from the ReFRAME library were evaluated in a Vero cell assay.12 As the result, twenty-one hits were identified with promising dose–response readouts. Of those, clofazimine (EC50 0.31 μM) and the kinase inhibitor apilimod (EC50 0.023 μM) were of particular interest. Apilimod was subsequently tested in 293T and Huh-7 infected cells where it demonstrated striking potency (12 and 88 nM, respectively);12 the drug entered clinical trials for COVID-19 in June 2020.24 In July 2020, clofazimine has also advanced into clinical trials as a part of a combination therapy.25
In another study,26 authors demonstrated that SARS-CoV-2 virus can rewire phosphorylation signaling in infected Vero and A549 cells, also suggesting the use of kinase inhibitors, including apilimod. A drug repurposing study14 used a protein interaction map to identify approved and experimental drugs that bind to sigma-1 and 2 receptors (acting as host factors), where the most potent compound, PB28 demonstrated IC90 of 280 nM in Vero cells.
According to DrugBank, more than 680 medications have been in over 3300 clinical trials, including remdesivir, hydroxychloroquine, chloroquine, lopinavir, ritonavir, camostat, ivermectin and baricitinib, among others.27 Unfortunately, despite significant effort toward finding COVID-19 drugs among approved therapeutics, most repurposing studies (including clinical trials) have proved unsuccessful. A recent summary of trends observed across several thousand of COVID-19 therapeutic clinical trials of drug products and antibody-based agents with the total enrolment of over 500 000 patients was recently published by the FDA.11 The study came to rather unenthusiastic finding that “the vast majority of trials of therapeutics for COVID-19 are not designed to yield actionable information; low randomization rates and underpowered outcome data render matters of safety and efficacy generally uninterpretable”. This observation, however, does not obviate the need for carefully designed and executed trials involving evidence-supported drug candidates. For instance, Pfizer's SARS-CoV-2 Main protease (MPro) inhibitor PF-0083523113 continues to be clinically evaluated and still provides hope. Moreover, Pfizer recently announced that the company started clinical trials of another, new oral antiviral agent PF-07321332, designed as specific SARS-CoV-2 MPro inhibitor in less than a year.
Along with experimental repurposing screening campaigns, there has been an avalanche of computational drug repurposing studies, especially against SARS-CoV-2 main protease (Mpro) that was the first viral protein with X-ray resolved structure.28 Shortly after the first structure Mpro was deposited into the Protein Data Bank,29 numerous research groups from all around the world started submitting manuscripts describing docking experiments with SARS-CoV-2 Mpro and various drugs, natural products, nutraceuticals, etc., have been annotated as putative hits.
In our observation, many researchers started to use molecular modeling and cheminformatics tools for the first time. Consequently, many were unaware of the best practices of CADD and rigorous protocols required for data preparation, curation, and proper validation of predictions. Arguably, the most common issue was the absence of chemical standardization and curation, leading to the use of incorrect protonation states in the ligands, missing hydrogen atoms, presence of salts, duplicates, inconsistent representations of chemical moieties and tautomers, etc.30 Additionally, some studies employing molecular docking, omitted key steps of protein structure preparation, including removal of water molecules, addition of explicit hydrogens and assignment of accurate protonation states for residues, identification and addition of missing side chains or loops, removal of overlapping atoms and energy-minimization of sidechains among others. Some papers apparently docked their library “directly from SMILES strings”, strongly suggesting neglect of proper compound curation and preparation, which are critical.30 Another common shortcoming was the use of rigid docking, which has significant limitations and may require additional post-processing steps.31 Unfortunately, as mentioned above, many of such papers (frequently accompanied by press releases) made misleading claims about the discovery of COVID-19 cures based solely on computational model predictions.32 Clearly, such statements can only be made after robust experimental and, ideally, clinical validation of computer-generated drug candidates.
Most promising drug candidates that were not FDA approved drugs have been previously known or well-advanced experimental DAA agents. For example, the SARS-CoV Mpro inhibitor GC376 also showed excellent potency against SARS-CoV-2 Mpro (Ki 12 nM) and demonstrated significant activity in Vero cells (EC50 0.91 μM). Another important example is EIDD-1931, a broad-spectrum antiviral, targeting RNA viruses and causing mutations to accumulate in viral RNA. It was shown to inhibit SARS-CoV-2 in Vero (IC50 0.3 μM) and Calu-3 cells (IC50 0.08 μM) and a prodrug version of this molecule was previously reported active against SARS-CoV and MERS-CoV in mouse models. Notably, recent clinical trials33 of the Pfizer's SARS-CoV Mpro inhibitor PF-07304814 (a prodrug form of the aforementioned PF-00835231) generated promising initial results warranting the continuation of the study.13 There is growing understanding that future computational and experimental studies need to place greater focus on the development of novel chemical entities with targeted, tailored activity against SARS-CoV-2 virus. As mentioned above, clinical studies of another Pfizer compound, PF-07321332, have begun: if approved, it could become the first DAA drug developed specifically against SARS-CoV-2. This compound is an example of the focused drug discovery approach enabled by the knowledge of the specific viral target. Thus, continuously evolving knowledge of these targets along with the expert use of current and novel computational approaches to antiviral drug discovery using constantly emerging SARS-CoV-2 and COVID-19 knowledge bases is critical for guiding DAA efforts as discussed in the next sections of this review.
Databases and research resources that support COVID-19 drug discovery
In response to the pandemic, many established research resources have created focused COVID-19 data and publication collections, and several new resources have appeared as well. To name a few, the world's premier biomedical and life sciences literature collection, PubMed, has created a special SARS-CoV-2 Data Resource,2 providing linkages to the respective collections of publications annotated in both Pubmed and Pubmed Central, clinical trials described in ClinicalTrials.gov, and other information summaries. One of the most important general collections of biochemical endpoints – the ChEMBL database – released a special edition including COVID-19 relevant screening results for more than 20 000 compounds.34 The European Bioinformatics Institute (EBI) that hosts ChEMBL established a comprehensive COVID-19 Data Portal35 that integrates data on both viral and host protein sequences, interacting viral-host proteins, and several other information sources. An important chemical genomics resource, providing data on biological screening of chemical libraries in SARS-CoV-2 target-specific as well as phenotypic assays, has been established by the National Center for Advancing Translational Studied (NCATS) at the NIH.36
In support of structure based drug discovery, the Diamond Synchrotron source has made available a set of ∼1500 resolved crystal structures of low-molecular weight fragments bound to SARS-CoV-2 Mpro, along with their experimentally estimated binding affinities.37 This and similar efforts resulted in more than 1100 protein structures deposited into the Protein Data Bank (PDB) to date, covering most of SARS-CoV-2 RNA translates.29 Furthermore, the Diamond fragments collection was used as a starting point for collaborative, community-sourced de novo ligand design led by PostEra.38 As the result, more than 1800 of specifically designed compounds have been proposed, synthesized, and screened to date and the results were publicly disclosed.
Unstructured data depositories offer another source of valuable information on the virus and the infection. Thus, most scientific publishers agreed to freely disclose all COVID-19 related papers to the public. Kaggle has made available the COVID-19 Open Research Dataset (CORD-19) containing about 200 ,00 scholarly articles on new and related coronaviruses, including over 100 000 full-text items.15 Similarly, Elsevier, released the free Coronavirus Information Center encompassing more than 30 000 papers and book chapters.39
The experimental information of protein–protein interaction (PPI) in SARS-CoV-2-virus represents another invaluable knowledge source. Such PPI networks have been reconstructed for proteins encoded by genes, which expression is altered in SARS-CoV-2-infected human cells organs, model organoids and cell lines. These networks enable to identify hubs (highly connected protein nodes), and bottlenecks (proteins exclusively connecting distinct modules), that represent potentially valuable drug targets for COVID-19.16 A powerful Coronavirus Discovery Resource to visualize such network was developed by the Institute of Cancer Research in the UK.40
Similar approach have been used to construct drug–protein interaction networks, such as Connectivity Map, which has been extensively employed to flag potential COVID-19 therapeutics.16 This approach identifies compounds (including known drugs), which upregulate human genes that are suppressed in cells invaded by SARS-CoV-2. These chemically induced gene expression profiles can be obtained from LINCS L1000 database, which contains information on thousands of perturbed genes at various time points, doses, and cell lines. This approach can be used separately or together with network-based applications to identify possible anti-COVID drugs.16 Examples of such studies are summarized in Table 2 illustrating that COVID-19 targets can be identified from PPI networks, from compound-target interactions, at transcription levels, as well as from pathways and biological processes.
Examples of PPI network-based analysis for COVID-19.
| Study | Cava et al.41 | Hazra et al.42 | Karakurt et al.43 | Zhou et al.44 |
|---|---|---|---|---|
| Source of network | Human PPI network subnetwork from the genes, which are co-expressed with ACE2. Human PPIs were obtained using SpidermiR tool (PMID: 28134831) | Human PPI network from STRING (https://string-db.org) | Metabolic network of bronchus respiratory epithelial cell based on Recon2 (PMID: 23455439), human PPI network from STRING (https://string-db.org) | SARS-CoV-2-human PPIs,41 viral-human PPIs for other coronaviruses, human PPIs from 18 public databases. |
| Source of compound-target interactions | Drug-target interactions were obtained from Matador (http://matador.embl.de) and DGIdb (https://www.dgidb.org) databases | STITCH (http://stitch.embl.de) | NAa | Drug–target associations from DrugBank (https://www.drugbank.com), Therapeutic Target Database (http://db.idrblab.net/ttd), ChEMBL (https://www.ebi.ac.uk/chembl), PharmGKB (https://www.pharmgkb.org), BindingDB (https://www.bindingdb.org/bind/index.jsp), Guide To Pharmacology (https://www.guidetopharmacology.org) |
| Transcription dataset | Data on transcription in normal lungs was obtained from Cancer Genome Atlas (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) and Genotype-Tissue Expression (https://gtexportal.org) databases. | Transcription profiles of peripheral blood mononuclear cells from SARS-CoV-1 infected patients (GEO ID: GSE1739) | Transcription profiles from SARS-CoV-2 infected human lung epithelial cells (GEO ID: GSE147507) | Transcription profiles from SARS-CoV-2 infected human lung epithelial cells (GEO ID: GSE147507). Protein expression profile from human Caco-2 cells infected with SARS-CoV-2 (PRIDE ID: PXD017710) |
| Pathways and biological processes | Genes correlated with ACE2 are mainly enriched in the sterol biosynthetic process, aryldialkylphosphatase activity, adenosylhomocysteinase activity, trialkylsulfonium hydrolase activity, acetate-CoA and CoA ligase activity | MMP9 showed functional annotations associated with neutrophil mediated immune-inflammation | Matrix metalloproteinase 2 (MMP2) and matrix metalloproteinase 9 (MMP9) with keratan sulfate synthesis pathway may play a key role in the infection. | Co-expression of ACE2 and TMPRSS2 was elevated in absorptive enterocytes from the inflamed ileal tissues of Crohn's disease patients compared to uninflamed tissues, revealing shared pathobiology by COVID-19 and inflammatory bowel disease. COVID-19 shared intermediate inflammatory endophenotypes with asthma (including IRAK3 and ADRB2) |
| Potential targets | NAa | Hub-bottleneck node MMP9 | IL-6, IL6R, IL6ST, MMP2, MMP9 | NAa |
| Potential drugs | 36 potential anti-COVID drugs. Among possible interesting 36 drugs for COVID-19 treatment, the authors found Nimesulide, Fluticasone Propionate, Thiabendazole, Photofrin, Didanosine and Flutamide | Chloroquine and melatonin targeting MMP9. Melatonin appears to be more promising repurposed drug against MMP9 for better immune-compromising action in COVID-19 | MMP9 inhibitors may have potential to prevent “cytokine storm” in severely affected patients | 34 potential anti-COVID drugs. Among them melatonin was confirmed by observational study of 18,118 patients from a COVID-19 registry. Melatonin was associated with 64% reduced likelihood of a positive laboratory test result for SARS-CoV-2 |
NA – Not applicable.
In summary, data accumulated in multiple databases and repositories enable the application of ligand based, structure based, and knowledge mining approaches in support of COVID-19 drug discovery that we discuss below. The role of SARS-CoV-2 targets in guiding DAA drug discovery efforts is discussed in the next section of this review.
Targets for antiviral drug discovery for SARS-CoV-2
SARS-CoV-2 is a member of the same single positive-stranded RNA enveloped virus Coronaviridae family responsible for the 2002 severe acute respiratory syndrome (SARS) and 2012 Middle East respiratory syndrome (MERS) epidemics. Notably, the number of potentially harmful pathogens is very large, while resources for anti-infective research are limited and, in fact, have been diminishing over recent years. In April 2018, a World Health Organization (WHO) panel of scientists and public health experts listed nine highly pathogenic viruses likely to cause major epidemics, including Ebola, Zika and Lassa viruses, as well as MERS and SARS coronaviruses. Although none of them are new, there are no DAA agents or vaccines capable to address these life-threatening pathogens.45 Remarkably, the WHO panel also considered a likely-to-emerge “Disease X” with epidemic or pandemic potential caused by a previously undisclosed pathogen.46 In hindsight, COVID-19 became the first such “Disease X”, and there is a significant likelihood that similar pandemics will emerge in the future, unless the need for “disease preparedness” is recognized and properly resourced. We anticipate that rapid CADD methodology should become an integral part of future integrated pathogen-defense systems, and the current efforts on targeting SARS-CoV-2 could be used as a practical road map.
The relatively small SARS-CoV-2 genome suggests that most of its 29 encoded proteins should play important roles in host invasion and/or viral replication. Hence, successful inhibition of many of them could lead to useful therapeutics. An insightful recent study has examined the variability of these targets across 58 coronaviruses (CoVs) to support the search for broad spectrum antivirals.47 The authors have also established an interactive web portal48 displaying the 3D structures available for 15 of the SARS-CoV-2 proteins with 19 putative drug binding sites mapped on these structures; this set of binding sites was collectively called a SARS-CoV-2 pocketome. This portal is very useful for scientists interested in analyzing these binding sites as part of the future structure based drug discovery efforts. Computer-aided discovery of drug candidates targeting key coronaviral proteins is discussed in subsequent sections. Herein, we summarize relevant information about the SARS-CoV-2 protein targets that can be explored by computational modeling.
At the whole-genome level SARS-CoV-2 exhibits 79% sequence identity to SARS-CoV and about 50% identity to MERS-CoV. In spite of the relatively modest levels of sequence conservation, CoVs share essential (more conserved) genomic targets. This suggests that repurposing of existing antivirals and, or rational development of novel DAAs using the wealth of information collected from previous drug discovery efforts, both represent promising avenues; both approaches are discussed below in greater details.
Viral proteins can be grouped into three main functional categories: attachment and penetration into host cells; viral replication and transcription; and suppression of the host immune response. Although the SARS-CoV-2 replicative and host invasion mechanisms are not yet fully understood, rapid structure determination of many SARS-CoV-2 proteins from all three groups enables structure-based drug discovery. Table 3 summarizes druggable sites in experimental structures of SARS-CoV-2 proteins that can be exploited using a wide range of CADD methods. We provide brief functional description of SARS-CoV-2 proteins listed in Table 3.
Potential targetable sites identified in structurally resolved SARS-CoV-2 proteins.
| ID | Protein | Target | PDB | Ligand | Ref. |
|---|---|---|---|---|---|
| 1 | Nsp1 | Nsp1/ribosome 40S interaction interface | 6ZLW | NA | 49 |
| 2 | Phosphatase | ADP-ribose binding site | 6W02 | ADP-ribose | 50 |
| 3 | PLpro (nsp3) | Active site | 7JIW | PLP_Snyder530 | 51 |
| 4 | Mpro (nsp5) | Active site | 6W63 | X77 | 52 |
| 5 | Mpro (nsp5) | Dimerization interface | 5RFA | Fragment x1187 | 53 |
| 6 | Primase (nsp7) | nsp7/nsp8 interaction interface | 6XIP | NA | |
| 7 | Nsp9 | Peptide binding site | 6W9Q | NA | 54 |
| 8 | Nsp10 | Predicted pocket, not annotated | 6ZCT | NA | 55 |
| 9 | RdRp (nsp12) | NiRAN domain | 6XEZ | ADP-Mg2+ | 56 |
| 10 | RdRp (nsp12) | Active site | 7BV1 | NA | 57 |
| 11 | RdRp (nsp12) | NTP entry site | 7CTT | NA | 57 |
| 12 | RdRp (nsp12) | Nsp12-Nsp7/Nsp8 interaction site | 7BV1 | NA | 58 |
| 13 | Helicase (nsp13) | ATP/ADP binding site | 6XEZ | NA | 56 |
| 14 | Helicase (nsp13) | DNA/RNA binding site | 6ZSL | NA | 59 |
| 15 | Endoribonuclease (nsp15) | Catalytic site | 6WXC | Tipiracil | 60 |
| 16 | 2′-O methyltransferase (nsp16) | RNA binding site | 6WKS | RNA cap | 55,61 |
| 17 | 2′-O methyltransferase (nsp16) | Active site | 6YZ1 | Sinefungin | 55 |
| 18 | 2′-O methyltransferase (nsp16) | Allosteric site | 6WKS | Adenosine | 61 |
| 19 | Spike (post-fusion) | HR2 linker motif | 6M3W | HR2 motif | 62 |
| 20 | Spike (post-fusion) | S2 HR1/HR2 bundle fold | 6M3W | NA | 62 |
| 21 | Spike (pre-fusion) | S2 U-turn loop | 6NB6 | NA | 62 |
| 22 | ORF3a | Predicted pocket, not annotated | 6XDC | NA | 63 |
| 23 | ORF8 | Predicted pocket, not annotated | 7JTL | NA | 64 |
| 24 | ORF9b | Lipid binding site | 6Z4U | PEG lipid | 65 |
| 25 | Nucleoprotein | RNA binding site | 6M3M | NA | 66 |
Following release of the viral RNA into the host cytoplasm, two open-reading frames translate the viral RNA into two overlapping co-terminal polyproteins, pp1a and pp1ab, containing the non-structural proteins (nsps) (1–16), involved in immune suppression, replication, and transcription of the RNA.
Nsp1 suppresses host gene expression, thus weakening cellular antiviral defense mechanisms, including the interferon response.49 A recently resolved crystal structure of nsp1 bound tightly to the mRNA entry channel of the 40S ribosomal subunit (PDB: 6ZLW) suggests that blocking this interaction (ID = 1 in Table 3) could help reactivate the host immune response against SARS-CoV-2.
Nsp3 is a multifunctional protein comprised of several distinct domains, some with papain-like protease (PLpro), activity while others play important complementary roles. Thus, the phosphatase domain of nsp3, also referred as MacroD (ID = 2), is believed to interfere with the immune response by acting as a ADP-ribose phosphatase to remove ADP-ribose from host proteins and RNAs.50 The recently reported crystal structure of a liganded MacroD (PDB: 6W02) provides an important avenue for rational development of MacroD-directed inhibitors that could restore host immune capabilities.
PLpro (nsp3) (ID = 3) and main protease Mpro (nsp5) (ID = 4) are enzymes that carry the critical upstream function of cleaving mature nsps from the pp1a and pp1ab polyproteins, following their initial translation. Both protease targets are under intensive investigation as discussed in great detail below. It is important to note that Mpro is a stable homodimer and its dimerization interface (ID = 5) may be an important site for targeting this critical viral enzyme.53
Nsp7 and nsp8 form a primase complex, involved in the RNA synthesis pathway and required for enhanced functionality of RNA-dependent RNA polymerase RdRp.67 A recently published crystal structure of nsp7 complexed with the C-terminus of nsp8 (PDB: 6XIP) identified several potentially druggable pockets in the dimerization interface (ID = 6) that could potentially be used to design interaction inhibitors capable of suppressing SARS-CoV-2 replication.
The exact role of nsp9 in SARS-CoV-2 biology is not yet fully defined, but its structural homolog in SARS-CoV species suggests that the protein may be essential for viral replication. To be functional, nsp9 needs to form an obligate homodimer via its conserved “GxxxG” motif (ID = 7). Notably, disruption of key residues in this motif in related coronaviruses resulted in reduction of viral replication.54
The replication-transcription complex (RTC) represents the major viral assembly responsible for RNA synthesis, replication, and transcription. The RTC consists of RNA-dependent RNA polymerase (RdRp, nsp12), the primase complex (nsp7–nsp8), and helicase (nsp13) that combine to maintain optimal functioning of the replication machinery. The RTC provides numerous opportunities to inhibit SARS-CoV-2 replication. In particular, RTC activity relies heavily on RdRp, an indispensable enzyme in the life cycle of all RNA viruses.68 RdRp supports the transcription and replication of viral RNA genome by catalyzing the synthesis of viral RNA templates to produce genomic and subgenomic RNAs.69 SARS-CoV-2 RdRp contains an extended N-terminal nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain (ID = 9), and, although its exact role is still unknown, its enzymatic activity is considered critical for viral propagation.56 Recent cryo-EM structures of the NiRAN domain (PDB: 6XEZ) revealed a potential allosteric site that may be a suitable target for drugs that disrupt the function of NiRAN and RdRp. In the resolved structure of the complex, ADP is located in the active site of the NiRAN domain, highlighting a potentially druggable area, although further investigations will be required to determine the exact NiRAN activity as well as its preferred substrate.
The core and main structural motifs of RdRp are highly conserved between SARS-CoV-2 and SARS-CoV species (96% sequence identity) including sharing key residues in their active sites.70 An apo-structure of RdRp has pockets in the catalytic chamber (active site) (ID = 10) where the RNA template needs to bind for replication (PDB: 7BV1). These pockets could be targeted by small molecules to impede RNA binding and disrupt RNA replication by the RTC. Situated next to the catalytic chamber, the NTP entry tunnel (ID = 11) guides new NTP into the extending RNA primer. Recently resolved structure of the RdRp (PDB: 7BW4) demonstrates that the tunnel could be blocked to interrupt elongation of the RNA duplex.68 As previously mentioned, the primase complex (nsp7–nsp8) also interacts with RdRp to significantly enhance polymerase activity of the RTC.58 Thus, the critical interaction between nsp7–nsp8 complex and RdRp (ID = 12) (PDB: 7BV1) represents another rational drug target. In the early phase of the pandemic, remdesivir (RDV) was considered a potential treatment for COVID-19, as studies reported that it was highly effective in inhibiting growth of SARS-CoV-2.18 RDV targets the RdRp to arrest RNA synthesis, thus highlighting the key role of RdRp in replication of SARS-CoV-2 and its potential as a druggable target.71
The helicase (nsp13) facilitates unwinding of RNA helices to prepare a template strand for replication and to hydrolyze various NTPs.72 There are two major functionalities of helicase that could be therapeutic targeted: the ATP binding site (PDB: 6XEZ) and RNA binding site (PDB: 6ZSL).56
The function of a nidoviral RNA uridylate-specific endoribonuclease NendoU (nsp15) in the viral replication cycle is also not fully understood. Nsp15 may be involved in interfering with host response and/or in viral replication by processing RNA. Nonetheless, the role of NendoU protein is considered essential,73 and its crystal structure (with the active site occupied by citrate, PDB: 6WXC) provides an attractive starting point for structure-based drug design.
2′-O-RNA methyltransferase protein (MTase, nsp16) is also involved in viral RNA replication. MTase ensures the integrity of viral RNA by adding to its 5′end a cap fragment consisting of a N-methylated GTP and C2′-O-methyl-ribosyladenine moiety. The cap ensures adequate RNA integrity and stability for translation.55 Based on the available crystal structure of MTase, several potentially targetable sites have been identified: a positively charged RNA binding canyon (capping site, PDB: 6WKS), S-adenosylmethionine binding site (ID = 17, PDB: 6YZ1) and a unique allosteric site (ID = 18) found to be occupied by adenosine in the 6WKS crystal structure.61
The spike glycoprotein (S protein) initiates the attachment and penetration of SARS-CoV-2 into host cells and consists of two subunits: S1 and S2. The former is responsible for binding the virus particle to the host's angiotensin-converting enzyme 2 (ACE2) receptor, while the latter facilitates the fusion of the viral and host cellular membranes. The receptor-binding domain (RBD) of S1 is the only exposed part of the virus and, therefore represents an exceptional targeting opportunity (described in detail in the following sections).
The S2 subunit of S protein also presents opportunities to inhibit attachment of SARS-CoV-2 to host cells. S protein undergoes significant structural rearrangements upon binding to ACE2 to allow fusion of host and viral membranes. Thus, disrupting S protein from reaching its stable fusion conformation could be a viable therapeutic approach. A linker needs to bind in a cavity upstream of the heptad repeat 2 (HR2) in S2, and this positively charged cavity represents a rational surface target (ID = 19). Moreover, small pockets along the HR1–HR2 six-helix bundle in the post-fusion state (ID = 20) could be targeted to prevent S protein from forming its fusion core (PDB: 6M3W). Similarly, the S2 U-turn loop in the pre-fusion state (ID = 21) could also be targeted by small molecules to hamper S protein's appropriate refolding (PDB: 6NB6).62
The SARS-CoV-2 virus evades the host immune system through an intricate network of interfering proteins. Thus, accessory protein 9b (ORF9b) is another virulence factor that may suppress type I interferon responses by associating with TOM70 human protein (translocase of outer membrane 70). This reduces the development of innate and adaptive immunity.65 A recently resolved crystal structure of ORF9b (PDB: 6Z4U) revealed the presence of a lipid binding site (ID = 24) that could be relevant for targeting with small molecules.
Finally, the nucleocapsid protein (N protein) of SARS-CoV-2 virus plays a structural role in protecting viral RNA. The N protein enhances the efficiency of virion assembly by binding to viral RNA to form functional ribonucleocapsid (PDB: 6M3M).74 Therefore, blocking RNA binding to the N protein may disrupt the critical RNA packing event.66
The valuable structural information on SARS-CoV-2 proteins generated to date has identified up to 25 potential target sites for rational drug discovery campaigns. Notably, this list is constantly evolving with more viral proteins and protein complexes qualifying as potential targets. Additional potentially targetable sites in SARS-CoV-2 proteins and complexes are being discovered and researched by CADD methods. Furthermore, various cryptic target sites on SARS-CoV-2 proteins represent another important targets for structure-based drug discovery.75 So far, these has been identified in nsp10, ORF3a, and ORF8 proteins from Table 3 (ID= 8, 22, 23) that do not exhibit distinct druggable sites on their surfaces.63 By combining Molecular Dynamics (MD) simulations with target site prediction tools, one could identify such cryptic protein pockets and develop inhibition strategies for SARS-CoV-2 proteins that are otherwise deemed non targetable. A more detailed discussion on this topic will be presented in a later section on molecular dynamics simulations for discovery of cryptic pockets.
Although this section has focused on SARS-CoV-2 protein targets for drug discovery, substantial efforts are underway to repurpose or discover drugs acting on human proteins that play significant roles in SARS-CoV-2 infection. Thus, in a seminal work by Gordon et al.,14 UCSF researchers expressed 26 of the 29 SARS-CoV-2 proteins and used them as baits in a mass-spectral proteomics experiment to identify 332 critical interactions with human proteins. Subsequently, cheminformatics and text-mining tools identified 66 human proteins that could targeted by 69 approved and experimental drugs. A subset of those identified by docking experiments was assessed in multiple viral assays. Ultimately, two series of host-directed pharmacological agents (inhibitors of mRNA translation and the sigma-1,2 receptor regulators) demonstrated significant antiviral activity.14
In summary, detailed structural information on both static and dynamic pockets in viral proteins and PPIs provide significant opportunities for structure-based drug discovery.
Structure-based drug discovery approaches
Computational methods of structure-based drug discovery (SBDD) simulate how potential ligands can interact with the putative binding (target) site under investigation. The ultimate objective of SBDD is to rank known or de novo designed chemicals for desired biological activity and, most importantly, to translate computer-generated hypotheses into actionable experimental steps. Along with the use of conventional molecular docking and scoring protocols, recent SBDD studies for COVID-19 have begun to exploit novel DL and AI methodologies.
While no repurposed or novel SARS-CoV-2 inhibitors have yet been identified with SBDD tools, an important trend has emerged that involves applying supercomputing resources to COVID-19 drug discovery. Early work by Smith and Smith76 employed the world's largest supercomputer – the IBM SUMMMIT to screen the SWEETLAND library consisting of 8000 drugs and natural products against the complex of SARS-CoV-2 Spike protein and human ACE2 receptor. The computationally demanding replica-exchange MD simulations were combined with ensemble docking and resulted in identification of 77 candidate drugs, of which five were approved therapeutics (pemirolast, isoniazid pyruvate, nitrofurantoin, ergoloid, and cepharanthine) that constituted putative treatment options for COVID-19. While the work by Smith and Smith has received broad coverage,32 the proposed repurposing candidates have not been properly validated nor confirmed by experiments. Moreover, ergoloid is a mixture of three different compounds but there was no indication which of them was identified. Such modest outcome from 200 petaflops of computational power, together with a notable lack of validation of the repurposing hits, might suggest that it would be more effective to simply screen relatively small drug libraries (e.g., a few thousand compounds) in a wet lab. In support of this notion, recent high-throughput repurposing campaigns conducted by NCATS and leading academic groups12 resulted in a number of attractive repurposing candidates that demonstrate potent inhibition of SARS-CoV-2 virus, as described in earlier sections of this review. However, computational resources are still very important in virtual screening campaigns that aim to identify novel chemical entities as potential COVID-19 therapeutics. This scenario, which promises to design or discover bespoke drugs with greater potency than repurposed drugs, needs to work in much larger chemical spaces that are currently inaccessible to experimental screening methods.
Below we summarize expert SBDD approaches that have been applied to SARS-CoV-2 targets and discuss recent trends in SBDD that aim at more rigorous, computationally efficient, and affective COVID-19 drug discovery.
SBDD studies with key SARS-CoV-2 targets
Most of the SBDD research involving SARS-CoV-2 proteome has been focused on three main targets: the Spike glycoprotein (S-protein); papain-like protease (PLpro); and prominently, main protease Mpro (that has already been extensively highlighted in previous sections). More than 1100 structures have been deposited to date in the RCSB's COVID19/SARS-CoV-2 Special collection,29 and an important recent study mapped binding pockets of all major SARS-CoV-2 proteins.48 Practically all major docking programs and molecular databases have been used to identify approved, pre-clinical or experimental drugs, natural products, or nutraceutics (among others) that could be rapidly repurposed. The main docking tools used are AutoDock, AutoDock Vina, SMNA, PLANTS, Glide, DOCK, and ICM. These have largely screened the DrugBank, ZINC, SuperDRUG2, Selleckchem, TargetMol, Drug Target Commons (DTC), BindingDB, Supernatural II, Drugs-lib, SWEETLAND and several other repurposing databases. In accordance with best practices, the docking campaigns were often followed by more rigorous determination of binding poses and free energies estimations using MD packages AMBER, MOE, MM-PBSA-WSAS, SOMD, GROMACS and MM-GBSA/MM-PBSA, among others. These studies aimed to find a ‘silver bullet’ that will either halt the pandemic or at least provide effective treatment for those severely affected by SARS-CoV-2.
On the other hand, the use of rigorous SBDD tools significantly facilitated our knowledge about SARS-CoV-2 target proteins including their dynamic behavior, induced ionization states and plasticity among other major factors potentially influencing ligand binding. The SARS-CoV-2 pocketome portal48 mentioned above can be used to visualize the details of the binding sites within individual target structures described below.
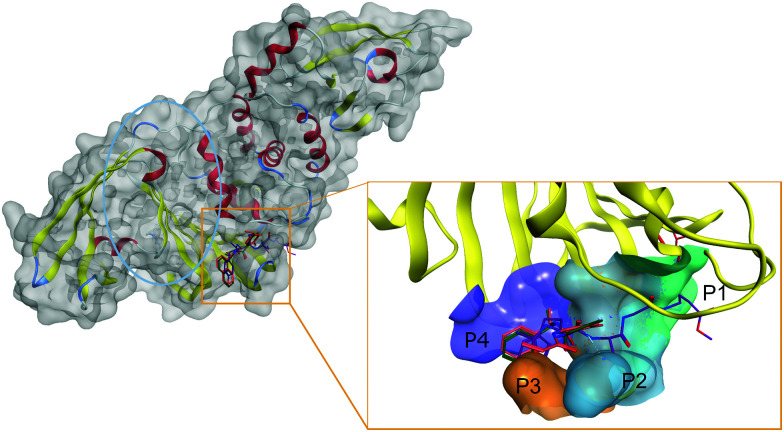
Thus, SARS-CoV-2 PLpro active site is centered on the catalytic triad of C111-H272-D286, which cleaves the replicase polyproteins at three specific sites featuring a conserved LXGG motif.77 The motif residues are labeled based on their relative position within the cleavage site. Position P1 is closest to the cleavage site, followed by P2, P3, and P4 at the end of the site, as shown in Fig. 2.
Fig. 2. Structure of SARS-CoV-2 PLpro and inhibitors in its catalytic site. In PLpro (left), the proximity of the ubiquitin binding site (circled in blue) to the catalytic site (squared in red) offers unique inhibition opportunities to target both activities of PLpro. The active site can be divided into subpockets (right) to guide drug design against SARS-CoV-2 PLpro. The four main pockets P1, P2, P3, and P4 (colored teal, blue, orange, and purple, respectively) need to be occupied for optimal inhibition. Ligands are represented in colored sticks. VIR251: purple; PLP_Snyder530: pink; GRL-0617: green. Parts of the pocket's surface were omitted for easier visualization.
The catalytic site of PLpro can be divided into different sub pockets identified by the residue recognized at each position. Flexibility in the PLpro active site complicates rational SBDD. Notably, the loop formed by Tyr268/Gln269 is highly flexible and adopts a closed conformation via an induced-fit mechanism by interaction with specific inhibitors.78 Thus, the active site cavity can change from an open to closed state depending on the co-crystallized ligand.
Detailed structural information on active site of PLpro has been used to design a potent inhibitor GRL-0617 (PDB: 7JIW), which binds to the active site of PLpro and inhibits its enzymatic activity. Due to the proximity of the active site to the S1 ubiquitin binding site, it was suggested that GRL-0617 could also inhibit the interaction of PLpro with ubiquitin-like protein ISG15 responsible for regulating host innate immune response.51 Other inhibitors with similar modes of action, such as the PLP_Snyder series and VIR250/VIR251, are also currently under investigation.79 Although GRL-0617 and the PLP_Snyder series do not explicitly interact with the P1 catalytic site, both compounds bound to the active site at P3-P4 and GRL-0617, exhibited potent activity (IC50 of 2.2 μM).
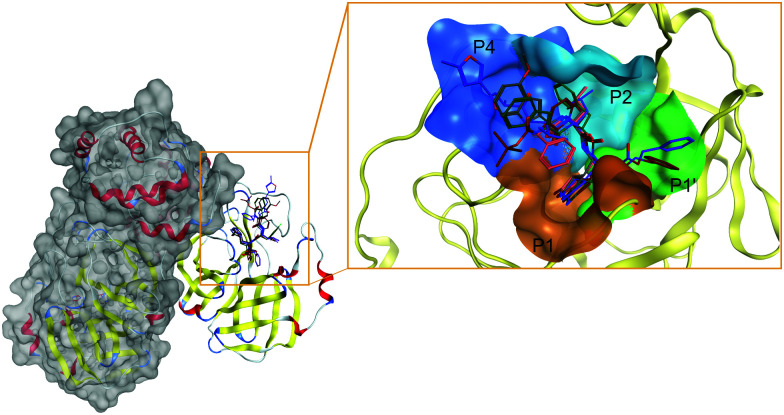
The active site of, perhaps, the most prominent SARS-CoV-2 target protein Mpro is centered on the catalytic dyad of Cys145-His41. It cleaves the replicase polyproteins at 11 specific positions, using core sequences in the polyproteins to determine the cleavage sites.80 The recognized residues on the polyproteins are named depending on their relative position to the cleavage site (see Fig. 3). Position P1 corresponds to the residue before the cleavage site up until the N-terminal (P2, P3, P4, P5), while position P1′ corresponds to the residue immediately after the cleavage site up until the C-terminal (P2′, P3′, P4′, P5′, etc.).81 Therefore, the active site of Mpro can be partitioned into different pockets, depending on the residue occupancy at each position. Rational drug design must again take into consideration the flexible nature of Mpro active site. Structural rearrangements of Met49 and Gln189 in the P2 position affects the size of the pocket for optimal occupancy. Therefore, special scrutiny must be taken when designing Mpro-specific inhibitors to fit the highly flexible P2 pocket in either the open or closed state. The latest study also indicated that the ionization state of Mpro active site residues could also be context-dependent which further complicated SBDD efforts with this protein.82
Fig. 3. Structure of SARS-CoV-2 Mpro and inhibitors in its active site. The unique dimer structure of Mpro (left) offers one distinct path to block its catalytic activity through the substrate-binding pocket. The active site can be partitioned into subpockets (right) to rationalize the design strategy against SARS-CoV-2 Mpro. The four main pockets P4, P2, P1, and P1′ (colored blue, teal, orange, and green, respectively) need to be occupied for optimal inhibition of Mpro. Ligands are represented in colored sticks in the active site. PF-00835231: grey; GC376: orange; 11b: green; N3: purple; 13: red.
Nonetheless, based on the recognition sequence of the polyproteins, ligands can be effectively designed to occupy the same pockets in SARS-CoV-2 Mpro active site. Recent X-ray structures of Mpro with potent inhibitors revealed key features of various different binding modes to the enzyme. Thus, P1 and P2 pockets must be occupied to inhibit Mpro activity, as all current ligands interact with Mprovia those two sites. Thus, early SBDD efforts with Mpro target site allowed the development of a potent covalent inhibitor, 11b (partly occupying the P1′ pocket) that exhibited IC50 values of ∼50 nM in vitro.83 The same molecular scaffold was later combined with different covalent warheads to generate more soluble Mpro inhibitors such as GC376, occupying pockets P1 and P2. This candidate had an in vitro IC50 ∼ 500 nM and is undergoing more extensive evaluation. More recently, a more potent derivative, PF-00835231 (with a prodrug PF-07304814), was developed with nanomolar potency against Mpro. Conspicuously, it also exhibited potent in vitro suppression of SARS-CoV-2 as a single agent and in combination with remdesivir.33
De novo drug design efforts exploiting the Mpro active site received a significant boost in early 2020 when scientists in the UK Diamond Center made available >70 experimentally resolved structures with diverse chemical fragments non-covalently bound to the Mpro active site.37 Importantly, these structures have been crowdsourced for de novo design of fragment-based Mpro inhibitors. This resulted in >10 000 submissions from all around the world.84 Of these, ∼ 1000 compounds have been synthesized and experimentally tested, resulting in several low- to sub-micromolar hits that await rigorous evaluation. The most active fragment-derived derivatives are being further refined using another computational crowdsourcing campaign, folding@home.85
A recent seminal work by Lyu et al.86 demonstrated that expanding virtual screening to include large ‘make-on-demand’ chemical libraries yields highly potent compounds and new scaffolds not present in available chemicals libraries. Importantly, this study used extensive computational resources but could only process 170 million molecules. However, the number of accessible small molecules to date is numbered in the billions. Potential synergy between massive chemical libraries, such as ZINC15,87 and supercomputing facilities, such as SUMMIT at the Oak Ridge National Laboratory, have been identified in a recent study.88 Enhanced sampling MD and ensemble docking with AutoDock-GPU was applied to eight SARS-CoV-2 target proteins. This achieved exhaustive docking of 1 billion compounds against the 8 targets in under 24 hours. Unfortunately, as noted above, this extensive computational study was not followed by the experimental evaluation, so the value of the practical value of the identified hits is yet to be determined. However, this study highlights the previously unattainable boundaries of molecular docking that have emerged in the time of pandemics.
Ligand docking with template-based approaches, shown to often outperform conventional docking, were used to discover novel inhibitors of SARS-CoV-2 Mpro. Thus, LigTBM89 was employed to obtain a model of SARS-CoV-2 Mpro active site in complex with a low μM noncovalent inhibitor characterized crystallographically in the COVID Moonshot initiative.89 Unlike conventional docking, template-based methods are particularly useful because they do not require detailed binding site information. They also provide measures of model quality based on the similarity between the target and the template. Template-based approaches can be readily applied to modeling interactions between inhibitors and SARS-CoV-2 viral and human targets relevant to COVID-19, as their structural coverage in PDB is on exponential trajectory.
Molecular dynamics simulations and the discovery of cryptic target pockets
Drug targets are complex, dynamic entities and no experiments can currently deduce all possible aspects of their biological function. Hence, molecular dynamics (MD) simulations are the only way to obtain detailed information on drug target dynamics and their interactions with potential ligands. Although MD simulations are computationally intensive, the COVID-19 High Performance Computing Consortium established in March 2020, provides rapid access to powerful computational resources for teams studying SARS-CoV-2 targets. Furthermore, the MD simulation community collectively committed to a set of principles governing methodologies and data sharing practices for COVID-19 related MD models.90
The already outlined, the massive increase in SARS-CoV-2 structural information provides valuable inputs for MD simulations. In mid-February, just when cases in the US were very low, the McLellan group developed the first cryoEM dataset of the SARS-CoV-2 main infection machinery, the spike protein.91 Its early release set the stage for the first SBDD efforts using that key target. Subsequent work by several groups established strong methodological frameworks for the construction and simulation of the glycosylated spike protein,92 including the need for long MD runs (μS) in order to reveal the active participation of glycans in the spike opening motions.93 Additionally, a large-scale simulation of a patch of viral membrane containing four spikes, coupled with data from cryoEM, indicated that the spike stalk has joints that enable it to undergo hinge bending motions.94
In addition to the ability to explore orthosteric and allosteric binding pockets, an interesting recent application of MD simulations is the analysis of hidden (cryptic) binding sites. These sites are particularly useful for the design of compounds that have enhanced selectivity or resistance profiles.95 Because MD simulations explore the low lying energy landscape around the minimum energy, high-resolution static structure from x-ray crystallography or cryoEM, simulations are increasingly being used to discover these cryptic pockets.9697 MD has identified cryptic pockets for both SARS-CoV-2 spike protein (at or near joints or hinges in the protein)98 and Mpro (near the active site and at the allosteric site), though these have not yet been experimentally validated.
Finally, MD simulations can identify potentially useful pharmacophores or targetable epitopes. For example, simulations of truncated human ACE2 in complex with the spike receptor binding domain generated a topological map of the key interactions. It further suggested the importance of rigidity at the binding interface,99 molecular details that can inform on the design of peptidomimetics, for example. MD simulations of the full length ACE2 embedded in the host cell membrane indicated an unexpectedly large degree of flexibility in the linker domain. This may provide another avenue of exploration for small molecules that disrupt mechanical processes related to the virus-cell fusion.100
Another potentially impactful study used MD to explore details of molecular complexes between the Spike protein and nicotinic acetylcholine receptors in the muscle and brain.101 These simulations provided support for the nicotinic hypothesis and provided a molecular basis for receptor subtype specificity. These findings may facilitate development of compounds selective for the α7 subtype as a way of blocking the interaction. Undoubtedly there will be many additional simulation-based studies that contribute to therapeutic programs against COVID-19.
Machine learning methods of scoring protein–ligand interactions at quantum-mechanical level
The development of fast and accurate methods to predict protein–ligand binding affinities represents a key challenge of SBDD because of two bottlenecks: statistical sampling; and the scoring problem. The former involves protein and ligand flexibility, solvent effects, and overall complexity of the protein–ligand interaction (particularly challenging in such cases as Mpro active site mentioned throughout above sections). The latter deals with accurate estimation of the interaction energy of the ligand with the target protein in the complex.102
The explicit use of quantum mechanical (QM) methods can aid solving the scoring problem and can provide more accurate estimates of binding affinity.102 This is especially important in cases involving metal ions, covalent bond formation, strong polarization and charge transfer effects, halogen bonding, etc.102 However, accurate QM calculations are very computationally demanding. Conventional Density Functional Theory (DFT) method scales nominally as O(N3), N being a measure of the system size. Wave-function based post-Hartree–Fock methods could scale even worse: O(N4–N7). The most popular strategies for addressing this challenge include hybrid QM/MM methods that partition the protein–ligand system such as only small most important region is treated with QM (e.g., ONIOM or QM/MM) and semiempirical and tight-binding methods that are applicable to thousands of atoms but need parametrization to overcome their inaccuracies.103
To date QM studies related to COVID19 have focused on reaction mechanisms and substrate specificity of the SARS-CoV-2 Mpro enzyme. Thus, Ramos-Guzmán et al.104 presented a detailed QM/MM analysis of the proteolysis reaction catalyzed by Mpro, modelling different states along the reaction pathway. These calculations were consistent with recently reported kinetic data for SARS-CoV-2 Mpro. Both studies presented a detailed analysis of key protein interactions and the critical importance of the P1/P1′ pockets in the design of potent and specific inhibitors.
Hatada et al.105 employed a fragment molecular orbital (FMO) interaction analysis of the complex between the SARS-CoV-2 Mpro and its peptide-like inhibitor N3 (PDB ID: 6LU7). They computed the contributions of different residues and elucidated the nature of interactions in this complex. Similarly, Ramos-Guzmán et al.104 identified the important role of His41 and Cys145 in the design of covalent inhibitors of SARS-CoV-2 Mpro. Furthermore, Khrenova et al.106 used hybrid QM/MM MD simulations to derive a simple descriptor, based on the Laplacian of the electron density and the electron localization function, that discriminated between covalent and non-covalent complexes. Cavasotto et al.107 used semiempirical PM7 calculations to rescore docking to the SARS-CoV-2 Mpro, PLpro, and spike glycoprotein, while Adhikari et al.108 used large-scale DFT calculations to analyze interactions in the RBD domain of the spike protein.
The modest contribution of QM studies to the body of literature covered in this review highlights the slow pace of these approaches. Therefore, the ability of QM methods to contribute to the development of therapies for COVID-19, under the time constraints of the pandemic, is quite limited. However, very recent, and exciting developments in AI and ML have the potential to greatly enhance the role of QM methods in drug discovery and development. Substantial progress has been made in the development of general-purpose atomistic potentials using ML, in particular, using deep neural networks (DNN).109
The ANAKIN-ME (or ANI for short) method110 is one example of transferable DNN-based molecular potentials. The key components of ANI models include the selection of diverse training data with active learning, non-equilibrium sampling of 3D conformations, and atom-centered descriptors to represent molecules for learning.111 The ANI-1ccx model was built from energies and forces of ∼60 000 small organic molecules (constituted of C, H, N and O atoms), considering non-equilibrium molecular conformations, using 5 million DFT (wB97x-D/DZ) and 0.5 million DLPNO-CCSD(T)/CBS calculations. These benchmark studies demonstrated the ANI-1ccx model to be within 1–2 kcal mol−1 of the reference (and extremely computationally demanding) Coupled Cluster calculations and to exceed the accuracy of DFT in multiple applications.112 The Atoms-In-Molecules neural Network or AIMNet improves the performance of ANI models for charged states and continuum solvent effects.113
The recently-developed ANI-2x model supports three additional chemical elements: S, F, and Cl. ANI-2x underwent torsional refinement training to better predict molecular torsion profiles.114 These new features open a wide range of new applications, including receptor–ligand systems, as they now cover 90% of drug-like molecules. Consequently, Lahey et al.115 demonstrated that by using the ANI potential to represent intramolecular interactions of ligands in protein pockets, both binding poses and conformational energies could be accurately calculated.
The NSF Molecular Sciences Software Institute (MolSSI), in collaboration with BioExcel, has set up a centralized hub and file sharing service for COVID-19 applications. It will connect scientists across the global biomolecular simulation community. The COVID-19 Molecular Structure and Therapeutics Hub also improves connection and communication between simulation, experimental, and clinical data investigators.90
The ANI-2x model was used to generate two public datasets, ANI–FDA Drugs and ANI–CAS Antiviral, for SBDD research for COVID-19.116 ANI–FDA Drugs contains low-energy conformers, tautomers, and dipole-consistent partial atomic charges for 6433 FDA approved and investigational drugs. It consists of 32 036 tautomeric structures and approximately 3 million conformers. ANI-CAS Antiviral contains 67 167 tautomeric structures and ∼6.6M conformers for 20 306 molecules from the CAS Antiviral database.117 Axelrod and Gomez-Bombarelli118 used semi-empirical tight-binding density functional theory (GFN2-xTB) to compute minimal conformers for 278 622 molecules that have been tested for in-vitro inhibition of SARS-CoV-related assays in PubChem. These recent developments are bound to improve the accuracy of both ligand representation and scoring functions used in virtual screening of chemical libraries against SARS-CoV-2 targets.
Deep learning approaches for SBDD
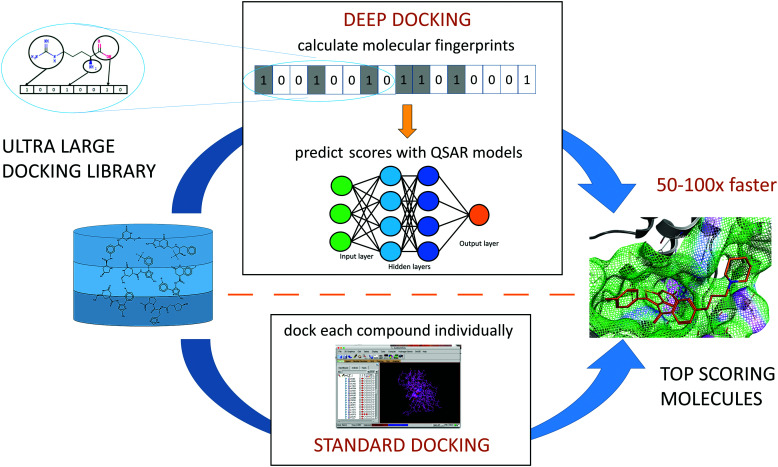
While none of supercomputer-driven campaigns have yet delivered validated, therapeutic candidates, the ultra-large chemical libraries can provide novel leads for COVID-19 drug discovery. An virtual screening campaign on the active site of Mpro, used deep learning (DL) methods to address the mismatch between the size of available chemical databases (the ZINC15 contains > 1.3B molecules) and conventional docking resources.119 The Deep Docking platform generated QSAR models trained on docking scores (Fig. 4). This approach takes a full advantage of all docking results (both favorable and negative) in contrast to conventional docking that does a complete screening run and selects only a small set of favorably docked molecules (hits). In the pilot study, Deep Docking rapidly and accurately predicted docking scores for 1.36 billion molecules from ZINC15 library against 12 prominent target proteins. It demonstrated up to 100-fold higher computational efficiency of virtual screening and up to 6000-fold enrichment for high scoring molecules.119 When used for virtual screening against Mpro,52 Deep Docking enabled the filtering of >1.4B molecules (the ZINC15 database, plus the Enamine PPI and Life Chemical antiviral libraries) down to 1000 potential hits in just one week. It used 640 CPU and 40 GPU units (running GLIDE docking and DL computations, respectively). Remarkably, without DL augmentation, the conventional docking programs would take years of continuous computation on this hardware.
Fig. 4. Schematic representation of a Deep Docking (DD) workflow.
Consistent with the Best Practices of CADD,120 consensus filtering and post-processing of GLIDE hits with a 3-feature pharmacophore generated from the active site of Mpro was employed (see Fig. 4).
This protocol enabled the identifications of 211 compounds highly ranked by both GLIDE and pharmacophore model that were selected for experimental evaluation. A continuous fluorescence resonance energy transfer (FRET)-based assay using recombinant Mpro allowed reliable and fast identification of small molecule inhibitors.121 Recombinant his-tagged protein was purified from E. coli lysates by Ni2q binding chromatography following protocols for SARS-CoV-2 Mpro108 >95% pure samples of the 211 selected compounds were acquired from vendors and tested in the FRET assay with serial dilutions. Ultimately, 25 molecules were confirmed as active, with IC50 values in the range 10–100 μM, a respectable 12% hit rate for the Deep Docking method.
Notably, eight top-scoring ZINC compounds from the original Deep Docking paper were also evaluated by a third-party group resulted in identification of two low micromolar hits for SARS-CoV-2 Mpro.122
Importantly, these results identify the need for the use of stringent methods and consensus protocols, relying on a larger number of more diverse CADD and experimental approaches discussed below.
Ligand-based antiviral drug discovery approaches
Here, we discuss application of traditional ligand-based methods, sometimes combined with knowledge mining approaches, that not only leverage but guide the experimental drug discovery for COVID-19. We highlight the utility of some recent innovative techniques such as Generative Topographic Mapping (GTM) and deep learning (DL) for the discovery of novel DAA agents as well as for COVID-19 drug repurposing.
REDIAL-2020 machine learning platform
The utility of predictive models is ultimately judged by their ability to guide the experiments. This objective formed a part of REDIAL-2020 – a suite of ML models aiming to predict anti-SARS-CoV-2 activities from chemical structure.123 It utilizes ML algorithms from the scikit-learn package, combined with cheminformatics protocols from RDKit.124 The platform was used to generate six best-in-class models for the following assays: viral entry (cytopathic effect, CPE, and host cell cytotoxicity counter-screen); viral replication (Mpro inhibition); and live virus infectivity (a spike-ACE2 interaction (AlphaLISA) assay, its TruHit counter-screen, and an ACE2 inhibition counter-screen). The corresponding data for 11 SARS-CoV-2 related assays were made openly available at NCATS COVID-19 portal.125
These NCATS datasets were consequently processed within this workflow using three different families of descriptors: fingerprints; pharmacophores; and physico-chemical properties. Starting from 22 different ML algorithms, six best performing algorithm/descriptor/assay combinations were selected, and voting-based consensus models were implemented on the REDIAL-2020 server for most of the assays (except AlphaLISA and ACE2). When tested on the external data, the REDIAL-2020 models correctly predicted 24 out of 39 published compounds for the CPE assay,126 15 out of 21 CPE actives from the ReFRAME library,12 and four out of the six Mpro inhibitors.127
Comparisons of a large number of anti-SARS-CoV-2 active compounds from the literature126 highlight frequent inconsistencies and discrepancies between different experimental measurements. For instance, out of 9 compounds tested in 6 published CPE assays,20,128,129 only remdesivir was active across all studies. As noted in the beginning of this review, the rush to publish initiated by the urgency of the COVID-19 pandemic has resulted in an unprecedented number of communications in peer-reviewed sources and media.130 Hence, it is particularly important to obtain an independent confirmation of anti-SARS-CoV-2 activities using alternative approaches. A recent study131 provides an example of such confirmatory evaluation of SARS-CoV-2 DAAs predicted by REDIAL-2020 with independent ligand-based virtual screen. From an initial set of 9 “chloroquine-like” drugs, zuclopenthixol, a typical antipsychotic, and nebivolol, an antihypertensive beta-adrenergic blocker, were identified as efficient inhibitors of SARS-CoV-2 infection with EC50 values in low micromolar range (see Table 4). The anti-SARS-CoV-2 activity of the antimalarial drug amodiaquine20,129 was also confirmed, and its metabolite, N-mono-desethyl amodiaquine, also appeared active and had a notable half-life of 21 days. Furthermore, two additional independent experimental evaluations were conducted, both of which confirmed zuclopenthixol and nebivolol as potential therapeutic agents for the treatment of incubation and early stage COVID-19 infections. The REDIAL-2020 platform123 can be accessed from any web browser; it accepts SMILES, drug names (e.g., generic or trade names), or PubChem IDs as an input, and generates predictions against 11 assays, with the top compounds from the NCATS training set, ranked by the corresponding chemical similarity; applicability domain was estimated for each assay.
Anti-SARS-CoV-2 activity values from two separate experiments, and pharmacokinetic properties for amodiaquine, its active metabolite, nebivolol, and zuclopenthixol. First column, EC50 CPE measured at UTHSC; 2nd EC50 values were determined at UNM. Pharmacokinetic properties were extracted from literature.
| Compound | EC50 (μM) | EC50 (μM) | C max (μM) | % oral | t 1/2 (hours) |
|---|---|---|---|---|---|
| Amodiaquine | 5.4 | 0.13 | 0.13 | 29 | 7.9 |
| N-Mono desethyl amodiaquine | 4 | N/A | 2.5 | N/A | 500 |
| Nebivolol | 2.8 | 2.72 | 0.02 | 12 | 10 |
| Zuclopenthixol | 0.015 | 1.35 | 0.03 | ∼50 | 20 |
Exploring chemical space of DAA candidates by chemography
Methods of chemical cartography, or chemography, enable visual analysis of an ensemble of chemical structures encoded by vectors of molecular descriptors enabling the projection of very complex data onto a two-dimensional chemical space maps.132 This approach exploits the ‘neighborhood behavior’ principle implying that close-proximity compounds possess similar properties, and hence chemical space maps can reflects relevant SARs. One of the most widely used chemical space mapping (chemography) approaches is Generative Topographic Mapping (GTM), a nonlinear grid-based method where the manifold is fitted into a high-dimensional descriptor space followed by projections of the chemical entities onto a grid of nodes superposed with the manifold.132 In such representation each compound is fuzzily associated to one or more of such nodes with certain probabilities (responsibilities) and, therefore, can be characterized by its responsibility vector. A distinctive feature of the GTM method is the combination of intuitive visualization and significant predictive ability. Any biological endpoint can be associated with a map via activity or classification landscapes that can visualize particular areas populated by molecules with a given activity and therefore enabling proximity-based classification of untested compounds.133
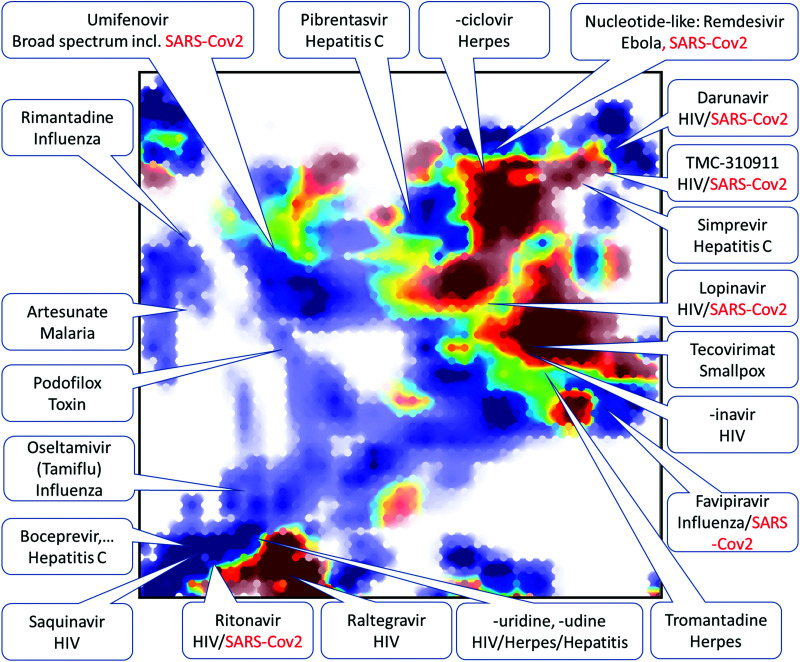
In the early days of drug discovery for SARS-CoV-2 no experimental data were available, and therefore, the initial studies were based on the prior data for related pathogens and hence chemography helped developing a global overview of the coronaviral DAA agent landscape. For instance, Horvath et al.134 have prepared several GTMs representing previous medicinal chemistry efforts to target CoVs. All CoV-associated molecules and antiviral DrugBank135 entries were projected onto seven maps hosting over 700 predictive activity landscapes.136 The list of approved or pending drugs associated with an “antiviral” label in DrugBank annotated the maps and fixed specific residence areas corresponding to compounds under clinical evaluation against SARS-CoV-2 (see Fig. 5). This framework, presenting the density distribution of CoV DAA agents, helped to highlight structural relatedness between compounds of different categories. Thus, similarity between umifenovir and SARS-CoV Mpro-inhibiting indole esters raised a new hypothesis that umifenovir might also act on viral proteases.
Fig. 5. Pool of 1000 compounds predicted to inhibit the 3CL proteinase of the novel SARS-CoV-2 (red) mapped against the SARS-CoV (betacoronavirus) compounds (blue). Location of several “antiviral” DrugBank molecules color-coded by their approval status (not-yet approved in red) is shown. Reproduced from ref. 134 with permission from the WILEY, copyright 2021.
Generative neural network models for de novo drug discovery
In contrast to virtual screening of available chemical libraries, de novo molecule construction provides access to a virtually infinite chemical space and offers innovative molecular architecture with desired properties.137 Recent advances of molecular design with the use of AI include so-called ‘generative’ (or ‘constructive’) models,138 which support augmented design of innovative therapeutics, including DAA agents.
Contemporary generative approaches usually build on deep neural networks (DNN),139 aiming to model the underlying distribution of a given set of molecules and, by sampling from the modelled distribution, construct novel chemical entities.140 Recurrent neural networks (RNNs) with long short-term memory (LSTM),141 as well as variational autoencoders,142 generative adversarial networks (GANs),143 graph neural networks (GNNs),144 and other network architectures145 have been explored. These methods are trained using algorithms that are successful for language analysis. Accordingly, for the purpose of molecular design, the training molecules are represented in terms of string notations, most often as simplified molecular input line entry systems (SMILES strings). Importantly, generative DL models automatically derive internal representations of SMILES, without relying on human-engineered molecular descriptors or reaction schemes. The generative model captures the syntax of these training molecules and generates new SMILES-encoded molecules that satisfy the constraints of the training set. This RNN-LSTM approach previously resulted in prospective discovery of novel compounds with desired bioactivities.146
As an example of generative de novo design, RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2147 was targeted, aiming to obtain new potent DAA agents. An RNN-LSTM model was employed for molecule generation,141 that was trained in two steps. Firstly, a generalized model (‘virtual medicinal chemist’) was developed by learning the syntax of approximately 400 000 SMILES strings of known bioactive compounds.148 Secondly, the model was fine-tuned with four nucleoside analogues that were effective against SARS-CoV-2 RdRp: approved favipiravir, and ribavirin; investigational galidesivir; and the active component GS-5734 of remdesivir prodrug. These four template compounds biased the model toward nucleoside analogues. Consequently, new SMILES were sampled by the tuned model, and the computer-generated molecules were ranked according to their topological pharmacophore similarity to the four RdRp inhibitor templates. Notably, the de novo generated structures contained several substructures of known RdRp inhibitors, but also carried novel chemical moieties, especially among the lower ranking designs (data not shown). We anticipate that these computer-generated molecules could serve as prospective templates rather than elaborated DAA designs because of limitations of the approach. For example, no background information about nucleoside interaction in RNA was considered during RNN-LSTM training. Neither target selectivity, pharmacokinetic and -dynamic properties, nor the synthesizability of the designs were explicitly considered. Consequently, the suggested molecules will benefit from careful checking by human experts and other computational tools. The selected designs then have to be synthesized and tested before any claim of pharmacological activity can be made. Nonetheless, some of the de novo generated molecules appear chemically feasible and attractive, contain innovative molecular scaffolds and deserve further consideration, illustrating the potential of generative models for rapid delivery of testable chemical designs and concepts.149
Knowledge mining tools for COVID-19 drug discovery
The severity of coronaviral pandemics prompted open science and FAIR (Findable, Accessible, Interpretable, Reusable) data initiatives150 to be embraced by researchers, institutions, publishers, companies and regulators to better understand the disease and to rapidly find an effective cure. Various structured and unstructured COVID-19 data sources have been made publicly available, enabling broader use of knowledge mining approaches and Artificial Intelligence (AI) – accelerated tools for COVID-19 drug discovery,151 with some notable examples discussed in the following section.
The use of knowledge graph approaches for COVID-19 drug repurposing
Biomedical Knowledge Graphs (KG) aim to provide a high level overview of the association between diseases (symptoms, ontologies, etc.), biological targets (genes, proteins, protein complexes, nucleic acids), and chemical entities (clinical and investigational drugs, tool compounds, etc.).152 These associations may be extracted directly from structured data sources such as medical and biochemical databases, or unstructured data such as a corpus of scientific articles and patents using text mining techniques assisted by ML methods. Building a KG from unstructured data can be achieved by NLP algorithms called entity recognition. This identify which objects in the text refer to the same underlying entities; relation extraction, finding relevant subject – predicate – object triplets in the text; and relation ranking, assessing the reliability of the information extracted algorithmically or with human supervision).153 Once such a KG is built, experts can explore the associations to discover important new drug targets or chemicals implicated in a disease mechanism. Other ML algorithms, such as tensor factorization, graph convolutional neural networks, logical inference algorithms, can be used for graph completion, to predict novel links between objects in the graph.153
One of the most notable examples of a KG was developed for drug repurposing against COVID-19 by BenevolentAI. This KG integrated a vast repository of structured medical information, including numerous connections extracted from scientific literature by various ML algorithms.154 To find a drug effective against COVID-19, a custom graph was created and a subgraph relating to SARS-CoV-2 extracted to permit inspection by experts.16 This KG revealed that the virus binds the host cells via the ACE2 receptor expressed on the surface of lung AT2 alveolar epithelial cells. ACE2 is involved in clathrin-mediated endocytosis, which in turn is promoted by members of the numb-associated kinase (NAK) family, including AAK1 and GAK. Baricitinib, a drug approved for the treatment of rheumatoid arthritis, was identified as a NAK inhibitor with sufficient plasma concentration to inhibit AAK1. It was therefore submitted for clinical testing.16 Furthermore, baricitinib is a JAK–STAT signaling inhibitor and was predicted to be effective against the elevated levels of cytokines (cytokine storm) observed in people with COVID-19. It was also predicted to have a tolerable side effect profile and low risk of interactions with other drugs based on the KG.154
These predictions were verified in vitro: baricitinib inhibited signaling of cytokines implicated in COVID-19 infection, it showed high affinity to several members of the NAK family, and it showed reduced viral infectivity in human primary liver spheroids.16 Initial clinical data has shown that baricitinib treatment was associated with clinical and radiologic signs of recovery, and a rapid decline in viral load and inflammatory markers in patients with bilateral COVID-19 pneumonia. A randomized clinical trial, ACTT-II, has been initiated by Eli Lilly and NIAID to study the effectiveness of baricitinib for serious COVID-19 infections and resulted in drug's approval for emergency use in combination with remdesivir.155
Taking advantage of publicly available information, a network of universities and biotechnology companies in China have created a KG for target-drug interactions, protein–protein interactions, drug molecular similarities, and protein sequence similarities. The KG was queried, using a network-based knowledge mining algorithm, for suitable drugs. These were identified as hit candidates if another NLP relation extraction model found a bag of sentences from the PubMed abstracts corpus describing a relation between the drug and a target in the coronavirus of interest. This method identified a PARP1 inhibitor, CVL218, which subsequently exhibited effective inhibitory activity against viral replication with no apparent signs of toxicity in rats and monkeys. It also possessed anti-inflammatory effects.
Researchers from Amazon Web Services (AWS) and a network of organizations in China and the USA have created a KG with 15 million edges (interactions) across 39 types of relationships connecting drugs, diseases, genes, pathways, and expressions, from a large scientific corpus of 24 million PubMed publications, the GNBR data set, and the DrugBank database.156 The RotatE algorithm was used to generate a low dimensional embedding of the KG that suggested 41 drug candidates for repurposing. These were supported by a high score in the treatment space, their proximity in the low dimensional embedding, and gene-set enrichment analysis from transcriptomic and proteomic data. AWS has also generated a similar biological knowledge graph, called DRKG, to fight COVID-19. It included information from six databases (DrugBank, Hetionet, GNBR, String, IntAct and DGIdb), and data collected from recent publications particularly related to COVID-19, containing nearly 6 million edges between 100 thousand entities of 13 entity types.157
Other open source COVID-19 KGs include the extension of ROBOKOP to COVID-KOP by researchers at the University of North Carolina,158 and KG-COVID-19 by investigators at Berkeley, California.159 The ROBOKOP biomedical KG was enriched with information from recent biomedical literature on COVID-19 annotated in the CORD-19 collection. Sentence-by-sentence co-occurrence analysis added 800 000 new edges to the COVID-KOP graph, and co-occurrence counts at the paper level led to 4.5 million new edges. Gene ontology data for viral proteins and symptom data was also added to the KG. The authors demonstrated the utility of the new KG by retrieving the pathway serving as a rationale for the linagliptin clinical trial against COVID-19 and suggesting new inferences.158 Thus, KG-COVID-19 was created by incorporating the latest data extracted from several biomedical databases and literature, including drug, protein–protein interactions, SARS-CoV-2 gene annotations, concept, and publication data from the CORD19 data set in an ontology-aware way. it contains about 16 million edges between nearly 300 thousand entities. The KG can be queried using SPARQL and the authors provide example queries to ease entry.
Another recent example of relevant KG construction is provided by Neo4COVID-19,160 a knowledge mining workflow inspired by SmartGraph161 and Hetionet,162 which served to assemble a Neo4j network with essential ingredients such as virus-host protein–protein interactions (VHPPIs), human protein–protein interactions (hPPIs), and drug-target interactions (DTIs). Its purpose is to better evaluate network-pharmacology-driven hypotheses and accelerate anti-SARS-CoV-2 drug repositioning. VHPPI sources included two proteomic studies,14,127 the SARS-CoV-2 subset from the viral-human interactions atlas163 and a genome-wide CRISPR screen for host genes related to SARS-CoV-2 infection.164 To streamline these non-overlapping VHPPIs with hPPIs,14,127,164 the authors used a KG based machine learning step (described elsewhere in the context of autophagy),165 by using the “positive” (known) interactions against true negatives (from the above experiments) in the context of data aggregated from 17 distinct machine-learning ready sets from TCRD/Pharos.166 For the pharmacology component of the network, DTIs were extracted from the DrugCentral database.167 DrugCentral currently includes 4642 drugs, of which 2549 have regulatory approval dates. DrugCentral DTI annotations include 19 959 human DTIs and 2570 non-human DTIs; of these 2752 are mode-of-action DTIs.167
In summary, recent efforts in knowledge graph construction and data mining illustrate the immense amount of research already performed on COVID-19, and the utility of KG approaches for the drug repurposing is outlined by the stellar example of baricitinib.
Open-source implementations are also available for anyone wanting to extend this work. It is important to note, however, that proper clinical validation of suggested candidates will require strong collaborations between academic, industrial, and government partners, and will take much longer than a KG query. It is a testament to the urgency of the pandemic that such a huge amount of data has been released to the community, and a vast array of AI and ML approaches have been brought to bear in the challenge of discovering effective treatments for COVID-19.
Knowledge-based discovery of synergistic drug combinations for COVID-19
Many valuable therapeutic opportunities arise from the synergistic action of drugs. The great success of anti-HIV drug combinations, and synergism of many other DAA agents highlight the importance of exploration of combination therapy for COVID-19. For that modern AI technologies can be used as powerful analytics tools for exploring drug combinations with synergistic action against SARS-CoV2.168
A detailed description of such study design is outlined in Fig. 6, where the initial step corresponds to the use of the combination of text mining (using Chemotext),169 knowledge mining (using ROBOKOP/COVID-KOP knowledge graphs),158,170 and machine learning (QSAR)171 tools to identify existing drugs with possible activities against SARS-CoV-2.172 Based on the initial findings, 76 individual drug candidates were identified as components of possible combinations.
Fig. 6. Study design for identifying drug combinations. Reproduced from ref. 173 with permission from the Cell Press, copyright 2021.
These drugs can generate 2850 unique-component combinations; to increase their synergetic probability, pairs of drugs with different mechanisms of action, and/or targeting virus at different lifecycle stages174 were prioritized. Consequently, 281 binary combinations of 38 drugs, and 95 ternary combinations of 15 drugs were chosen for further consideration. The in silico pipeline incorporating Chemotext169 along with recently developed COVID-KOP,158 and QSAR models of major drug-drug interactions175 was then used to determine whether selected compounds had been previously tested together and whether negative drug-drug interactions could be anticipated. The resulting prioritized list included 32 drugs and their 73 selected binary combinations for testing in vitro against SARS-CoV-2.173
Selected combinations were then experimentally screened in a 6 × 6 dose matrix format, involving two biological batches (cell and SARS-CoV-2 virus) and two assays (cytopathic effect and cytotoxicity against Vero-E6 cells) across 42 384-well plates, including replicates. Each batch was then assessed with five known DAAs used as a positive control. The batch readouts were highly reproducible, emphasizing the importance of using a dose matrix, instead of a single dose combination, to enhance the confidence of synergism/antagonism findings. The highest single agent (HSA) synergy model was subsequently applied to the screening outcomes and revealed that within 73 binary combinations of 32 compounds, there were 16 synergistic and 8 antagonistic pairs, with 4 displaying both synergistic and antagonistic interactions at different concentrations.176
Notably, these results demonstrated a strong antagonistic effect between remdesivir and the antimalarial drugs hydroxychloroquine, mefloquine, and amodiaquine (Fig. 7). Remarkably, the most striking antagonism was observed in the combination of the only two drugs approved with FDA Emergency Use Authorization (EUA) to treat COVID-19: hydroxychloroquine and remdesivir (the EUA for hydroxychloroquine has since been withdrawn by the FDA).177
Fig. 7. Activity and synergy/antagonism matrices for selected drug combinations (A: Remdesivir + Hydroxychloroquine; B: Remdesivir + Amodiaquine; C: Nitazoxanide + Remdesivir; D; Nitazoxanide + Amodiaquine). Reproduced from ref. 173 with permission from the Cell Press, copyright 2021.
Among the identified 16 synergistic combinations, a significant enrichment for nitazoxanide (FDA-approved broad-spectrum antiviral and antiparasitic drug) was also observed. The three most synergistic combinations were: nitazoxanide with remdesivir; nitazoxanide with umifenovir; and nitazoxanide with amodiaquine. A complete rescue of CPE was observed when 0.6–5 μM of nitazoxanide combined with remdesivir/umifenovir/amodiaquine, while any of these drugs alone only achieved 40–60% rescue. Important to note that amodiaquine, one of 32 drugs identified as descibed above and found active in CPE assay,172 subsequenty was found to have antiviral activity against SARS-CoV-2 in vitro131 and in vivo.178
These findings demonstrate the importance of preclinical research on antiviral drug combinations, as well as the utility of data and text mining approaches to explore modes of action (MoA) underlying synergism/antagonism in the context of COVID-19. These results also signal that the paucity of preclinical studies on drug combinations, prior to their use in patients, may significantly increase risks of undesirable side effects and poor outcomes. Furthermore, the developed matrix screening platform173 represents an efficient, data-driven means for prioritizing synergistic combinations of COVID-19 therapies and flagging undesirable drug interactions. All the results were made publicly available via NCATS Open Data Platform.125
The importance of rigor in computational and experimental approaches to COVID-19 drug discovery
As we repeatedly outlined in this review, rigor and best practices must be executed in all major elements of the modeling workflow: data curation; model development and virtual screening; and experimental analysis of computational hits. Best data curation and model development practices have been extensively covered in the scientific literature. Specific approaches to both chemical179 and biological30,180 data curation have been discussed elsewhere. Similarly, best practices of computational model validation have been discussed in multiple well-known papers and reports.171,181,182 The importance of rigor in data curation and model validation was additionally publicized in highly cited reviews.183,184 However, we felt it was important to highlight here best practices that should be followed in nominating and testing compounds that emerge from in silico studies as high-confidence tentative hits, especially in regard to molecular docking.
Consensus approaches to SBDD screening
Typically, only one scoring function from a single software would be employed in docking papers. Best CADD practice places particular emphasis on consensus-based voting schemes as generating the most enriched hit lists.120 This allows comparisons between the results from each scoring function and should better prioritize the docked compounds and should significantly increase confidence in the hits identified. However, relatively few academic groups have access to more sophisticated, commercial docking solutions such as Schrodinger's Glide (SP and XP scoring functions), ICM or CCG's MOE, among others. Additionally, very small datasets of virtual compounds with limited structural diversity were used in some of these docking studies. This puts comprehensive ensemble docking using multiple conformations of the target protein and multiple scoring functions out of the reach of many research groups.
Consensus approaches and/or post-docking processing have been used to a greater or lesser extent in the majority of VS campaigns on SARS-CoV-2 targets reported to date. As we have mentioned, the vast majority of these studies have not provided experimental validation for predictions.107,185–187 Only a few reported VS campaigns on Mpro resulted in identification of confirmed hits.28,52,188 While these represent rare cases of experimentally validated inhibitors of SARS-CoV-2 targets, the levels of activity achieved were not sufficient for direct therapeutic use. As we have described above, such hits may conceivably be improved by conventional medicinal chemistry-driven hit to lead optimization, however any NCEs that arise would have to follow the lengthy drug development pipeline. It is likely, therefore, that the flexible and chemically active enzymatic site of the Mpro requires use of more diverse and accurate CADD tools and more stringent and sophisticated consensus protocols.
To address the question of whether more rigorous scoring schemes could lead to more accurate VS performance, various consensus docking approaches were investigated using four major programs Autodock-GPU,189 FRED,190 GLIDE,191 and ICM.192 They were applied to the consensus protocol in sequential order (noting the decrease in the respective program's efficiency). A closely related SARS-CoV main protease (PDB: 4MDS193) was employed, for which many validated, diverse non-covalent inhibitors have been reported.194 From the literature, 81 such non-redundant inhibitors were identified,194 and for each, up to 50 molecular decoys were generated using the Directory of Useful Decoys–Enhanced (DUD-E) server.195 The resulting test set included 81 active and about 4000 inactive molecules, corresponding to a rather optimistic 2% background (random) hit rate.
For all poses generated by different docking programs for the same ligand, their pairwise RMSD values were calculated. Molecules that were docked by different programs with and RMSD < 2A were then considered to have been predicted by the consensus. The generated docking scored were consequently ranked by the last docking protocol used. The performance of this consensus approach was evaluated by the Enrichment Factor:196 Other common scoring criteria used were the receiver operating curve (ROC) and the area under the curve (AUC)120 metrics that illustrate the general quality of the ranking schemes. The resulting EF and ROC metrics estimated for the four VS strategies are presented in Fig. 8.
Fig. 8. (A) Enrichment factors for different consensus docking schemes applied to SARS-CoV-1 Mpro test set. (B) Receiver operating curves (ROC) for virtual screening using one software for docking and ranking (Autodock-GPU, AD, in green), and consensus docking using two (gold), three (grey) and four (purple) programs followed by ranking using the scoring function of the last indicate program for each strategy. Area under the curve (AUC) values are reported in brackets.
These results demonstrate that consensus prediction by two or more docking programs results in significantly better ROC statistics (with improvements in both initial slope and AUC values). In cases when all four docking programs were used, the AUC value was as high as 0.96 using ICM scoring function, indicating a very significant capability to distinguish between active and inactive compounds in the test set (Fig. 8). Similarly, EF values consistently increased with the number of programs combined in the consensus strategy, clearly indicating that consensus discarded decoys at a significant higher rate than active molecules.
In another conceptually similar study by Ghahremanpour et al.,188 consensus docking approaches also led to notable success. The authors concurrently employed Glide, AutoDock Vina, and two protocols with AutoDock 4.2 for concurrent virtual screening of ∼2000 existing drugs against the Mpro active site to arrive at 42 top-scoring consensus hit compounds. Then, taking into account intermolecular contacts, conformation, stability in molecular dynamics (MD) simulations, and potential for synthetic modification, 17 compounds were selected for purchasing. Remarkably, 14 out of these 17 tested compounds were found to be micromolar inhibitors of Mpro with IC50 values of 5–10 μM. This investigation suggests that rigorous approaches to molecular docking and consensus hit selection afford very high experimental hit rates. While compounds demonstrating micromolar activities in vitro are unlikely to be potent enough to be stand-alone drug candidates, these compounds were expected to be very useful for conventional hit-to-lead medicinal chemistry optimization. Indeed, in a recent exciting sequel to the aforementioned study,188 using Free Energy Perturbation (FEP approach, Zhang et al. redesigned the weak hit perampanel to yield multiple noncovalent, nonpeptidic inhibitors with ca. 20 nM IC50 values in a kinetic assay.
In summary, examples of studies described in this section, demonstrate that rational reduction of a molecular database through consensus VS could represent a rational strategy to find elusive, potent noncovalent SARS-2-CoV Mpro inhibitors. They also show the importance of rigor in evaluating computational hits and the power of the experimental confirmation of hits selected by computational protocols to increase the impact and recognition of CADD methods. We additionally reflect on the importance of rigorous execution of both molecular simulations and confirmatory experimental bioactivity testing in the next sections of this review.
On the importance of ligand entropy in SBDD for COVID-19
Studies described above188 illustrate that given the limitations of the use of rigid ligands and/or receptors in docking it is essential that the ligand:receptor complexes of molecules with the best docking scores be simulated by subsequent MD calculations. This allows more realistic contributions of ligand entropy to binding free energy to obtained, and also eliminates much of the errors resulting from the use of rigid receptors or ligands in docking. Guterres and Im showed how substantial improvements in protein–ligand docking results could be achieved using high-throughput MD simulations.197 They employed AutoDock Vina for docking followed by MD simulation using CHARMM. Over 56 protein targets (of 7 different protein classes) and 560 ligands they demonstrated a 22% improvement in the area under receiver operating characteristics curve, from an initial value of 0.68 using AutoDock Vina alone to a final value of 0.83 when the Vina results were refined by MD.
Experienced CADD users know that very flexible ligands suffer from entropic penalties that can affect their binding affinities. Thus, some important candidate hits that have emerged from virtual screening against the SARS-CoV-2 Mpro and RdRp are very flexible, with large numbers of rotatable bonds that make significant conformational entropy contributions to the ligand binding free energies. Studies that combine docking calculations with MD simulations of the best scoring hits often employ the Poisson–Boltzmann or Generalized Born and surface area continuum solvation (MM/PBSA and MM/GBSA) methods to estimate the free energy of the binding of small ligands to their targets. These popular methods are intermediate in accuracy and computational effort between empirical docking scores and strict alchemical perturbation methods. While they do a reasonable job of accounting for the entropic contributions of solvent, they ignore or approximate the conformational entropy of ligands due to the high computational cost of normal mode analysis.198 For example, Alamri et al. reported the results of a combined AutoDock Vina and MMGBSA study of the binding of libraries of covalent inhibitors and antiviral compounds against the SARS-Cov-2 Mpro.199
Researchers using MD simulations of molecules with the most favorable docking scores to calculate absolute binding energies need to be aware of the approximations inherent in the popular MMPBSA and MMGBSA methods and in the use thermodynamic cycle methods with insufficient conformational sampling. There are several recent developments that allow ligand entropies to the accounted for in more computational efficient ways.200 We expect that the use of the such corrections will improve the accuracy of docking calculations as applied to SARS-CoV-2 targets whereas approaches considered in following sections will help improve their computational efficiency.
Best practices of experimental validation of computational hits
Rapid accumulation of computational hit compounds has driven a demand for their proper experimental validation. Thus, many industrial and academic groups have established a large number of SARS-CoV-2 assays, broadly classified by (i) the type of assay – biochemical, biophysical, cell-based, proximity (immunoassays), (ii) the assay category – viral entry, viral replication, live virus infectivity, in vitro infectivity, and (iii) the detection method – fluorescence, microscale thermophoresis, high-content imaging, luminescence, AlphaLISA, bio-layer interferometry, etc.
Different types of assays can assess activities in different ways and can be used orthogonally to increase the confidence in hits. For example, Hanson et al.201 developed a proximity-based AlphaLISA assay to measure binding of SARS-CoV-2 spike RBD protein to the ACE2 receptor that can be used to find small molecules disrupting this critical interaction. These researchers screened 3384 drugs and pre-clinical candidates and identified 25 hits with IC50 values ranging from 0.1 to 29 μM.
Identification of false positives during any HTS campaign is similarly crucial. There are many assay components that can cause non-specific compound interference, such as readout type, signal generation or detection, platform automation, assay conditions, etc. To eliminate such potential false positives, Hanson et al. used the AlphaLISA TruHits kit as a counter-screen. This kit identifies inner filters, light scatterers (insoluble compounds), singlet oxygen quenchers and biotin mimetics interfering with the assay signal, thus eliminating false positives and helping to improve HTS outcomes.
Several cell-based live virus assays have been developed for SARS-CoV-2.125 One measures the ability of compounds to reverse the viral induced cytopathic effect (CPE) in infected Vero E6 host cells. The CPE reduction assay,202 indirectly detects the ability of a compound of interest to inhibit viral replication and/or infection through mechanisms such as direct inhibition of a viral entry, suppression of enzymatic processes, and action on host pathways that modulate viral replication. The CPE reduction assay was used in many studies of individual DAA agents or drug combinations that have been discussed in the previous sections.
To summarize, a significant number of cell-based and biochemical assays have been developed to aid drug discovery for SARS-CoV-2.203 Sharing and dissemination of such assays, along with screening results and successful CADD protocols, is in high demand. To address this need, the NCATS has developed open science data portal125 offering real-time results of various SARS-CoV-2 screening campaigns. This online resource contains readouts for more than 10 000 compounds, when possible, evaluated over full dose–response ranges. This portal stimulates multi-faceted collaboration between groups from different fields and represents the best practice scenario for drug discovery research against COVID-19 as described in the final section of this review.
COVID-19 and Open Science
This contribution was conceived and developed by a group of scientists who have dedicated their professional careers to molecular modelling and drug discovery. Many have worked on the comprehensive reviews of QSAR and CADD best practices.183,184 Here, we provided an overview of the broad landscape of CADD approaches used to target SARS-CoV-2, one of the most dangerous pathogens known to mankind. We reviewed known and emerging computational methods and described best practices for data processing and algorithm execution that will achieve meaningful and impactful drug discovery for COVID-19. We also stressed the importance of collaborative efforts and open science.
Conspicuously, the COVID-19 crisis has done much to stimulate collaboration and greater openness in science,204 driven by the assumption that openness accelerates the research. Sharing data and ideas with minimal restrictions allows all parties to capitalize on new knowledge more quickly and effectively, avoiding unnecessary duplication. Our willingness and efforts to bypass traditional scientific restrictions (e.g., the need to patent, secure research funding, or boost our academic profile) is encapsulated in three initiatives: Open access, Open data and Open source.
(1) Open access to computational research
Broad dissemination of peer-reviewed research is crucial for rapid drug discovery. In the wake of a global pandemics, all major publishers made COVID-related papers freely available in an unprecedented move.205 Moreover, recent explosion in use of free (unreviewed) preprint services has contributed significantly to Open access COVID-19 research. However, the extraordinary surge of preprints on computational discovery of potential COVID-19 therapies that providing no experimental verification has led to bans being placed on such unsubstantiated submissions.4
(2) Open data in COVID-19 computational research
The deposition of open-license datasets has been crucial for science, as illustrated by the high- profile example of the Human Genome Project that fueled the life sciences for the last two decades. The value of the Open data has also been clearly seen through numerous COVID-19 related initiatives, including the following: –
(i) Proteins and fragments
Central to all computational work is free access to data on protein targets. Thus, the Protein Data Bank developed a resource dedicated to SARS-CoV-2.29
The Diamond Light Source has also placed many of its structures, and associated fragment screening results, into public domain,206 preparing the ground for the community-based Moonshot initiative.
(ii) Assays and target screening results
The NCATS conducted a large number of screens with most important SARS-CoV-2 drug targets and disclosed all results in a real time, providing a wealth of information for CADD efforts.125
(iii) Data management
Tools for data analysis and aggregation are invaluable in defining drug design strategies. Thus, the Institute of Cancer Research has repurposed its cancer data tool, CanSAR,207 for coronavirus targets. CORD-1915 represents an integrated textual data platform for COVID-19 research. Another notable project, Nextstrain, is tracking viral evolution on the daily basis.208
(3) Open source COVID-19 CADD
The philosophy of Open source is participatory than observational.209 It has been implemented in many areas of research including discovery and synthesis of drugs for neglected and tropical diseases,210 for sharing physical samples for biomedical research,211 for collective optimization of drug candidates,212 among many others. In all those cases the underlying belief is that the goal is achieved more efficiently if there are “more eyeballs on the problem.” Such a belief is particularly widespread in the time of the crisis, with a corresponding number of COVID-19 research initiatives on a rise: –
(i) Collaborative drug design
The COVID Moonshot initiative mentioned above is hosted by a for-profit startup and contributors can submit candidate Mpro inhibitors built from crystallographic fragments from the Diamond Center.38 Following triage, the proposed compounds are synthesized by a contract research organization and evaluated in two open source orthogonal Mpro assays that provide new data into the next design round. Other distributed, participatory projects have also been launched for COVID-19 CADD campaigns.15
(ii) Competitions of approaches
The Joint European Disruptive Initiative (JEDI) is hosting a grand challenge project aimed at identifying the most promising computational approaches in compound design by combining the predictions with experimental validation of the best proposed structures. A prize is offered following a three-stage competition.213
(iii) Sample sharing
The sharing of physical proteins samples or potential drug/tool compounds can catalyze both computational and experimental research efforts; several public sources of both antiviral compounds214 and coronaviral target proteins215 have been developed in this way.
(iv) Shared computational resources
The Folding@Home project, a long-standing initiative for pooling computational powers, has been adapted for rigorous MD simulations of SARS-2-CoV target proteins, and major protein-host interactions.216
A generalized consensus on Open science has been agreed on by a number of global and national coalitions,217 research, business and regulatory consortia218 and progress-tracking initiatives committed to supporting open research, and collectively battling the deadly pandemic.
Concluding remarks
To conclude this review, we would like to state that along with the emphasis on openness and data sharing, we continue to stress the importance of best practices, rigor, and experimental validation in computer-aided drug discovery. The greater accessibility of computational resources and software has made it easy for non-experts to employ CADD tools to datasets and biological targets relevant to SARS-CoV-2 drug discovery. Hundreds of drugs have been computationally repurposed as putative treatments for COVID-19, many have gone into clinical trials with little or no supporting data or rationale. A few (e.g., remdesivir and baricitinib) have won regulatory approval, and yet, none has emerged yet as a curative treatment for the disease. This observation suggests that even massive computational resources cannot replace experimental methods for identifying promising drug candidates. However, the use of carefully processed prior experimental data and rigorous computational tools can enable successful experimental discovery of viable drug candidates.
To illustrate this assertion, selected examples of de novo designed chemical compounds, drugs, or drug combinations discovered or repurposed using computational approaches are shown in Table 5. Compounds MLS000699212-03 and NCGC00100647 were discovered using biological activity-based modeling approach, in which compound activity profiles established across multiple assays were used as signatures to predict compound activity in other assays or against a new target.219 Although the idea of using activity data in the modeling is not new,220 the authors validated its utility by achieving ∼30% success rate in the discovery of novel antivirals. Consensus of docking techniques resulted in repurposing of nafamostat and camostat,221 manidipine and boceprevir,222 and subsequent discovery of two antioxidant polyhydroxy-1,3,4-oxadiazole compounds CoViTris2020 and ChloViD2020 with high activities in vitro,222 and several perampanel analogs223 (identified using free-energy perturbation method) as novel, non-covalent Mpro inhibitors with 20 nM–5 μM IC50 values in a kinetic assay for Mpro. Several Deep Docking52 and QSAR224 hits were selected and confirmed experimentally by independent research groups that led to discovery of several potent Mpro inhibitors122 and repurposing of cenicriviroc and two other drugs, among others.125 In case of mixtures, AI-derived hypothesis of baricitinib as a potential treatment for COVID-1916 resulted in Emergency Use Authorization (EUA) by the U.S. Food and Drug Administration (FDA) granted for its combination with Remdesivir.155 Sixteen synergistic and eight antagonistic drug combinations, including most notable nitazoxanide – umifenovir for synergy and remdesivir – (hydroxy)chloroquine for antagonism, were identified using knowledge mining approaches and QSAR and then confirmed experimentally. Importantly, amodiaquine, identified as potential anti-COVID-19 repurposing candidate by knowledge-mining approaches,172 was confirmed to have experimental antiviral activity in CPE172 and titer reduction131 assays as well as in animal studies.178 Given its half-life of 3 weeks, amodiaquine could be a great solution, particularly for countries lacking access to Remdesivir, Favipiravir and other antivirals.
Selected repurposed drugs or drug combinations (a), and de novo designed compounds (b) identified by computational approaches and successfully confirmed in experimental studies.
Finally, Open Science and data sharing can go far in helping computational modelers discover new therapies and computational scientists must routinely seek experimental validation of their “digital dreams” before promoting computational results. Adhering to rigorous practices of modern research may dramatically reduce the number of publications but also dramatically improve the number of computer-assisted, experimentally validated potent antivirals discovered. We hope this collective contribution will be useful for data modelling and experimental researchers wishing to expand their toolkits to include rigorous computational approaches in their efforts to combat current and future pandemics.
Conflicts of interest
G. S. declares a potential financial conflict of interest as a founder of inSili.com GmbH, Zurich, and in his role as consultant to the pharmaceutical industry.
Supplementary Material
Acknowledgments
This review combined a series of separately written, invited contributions from the various coauthors (some sections with multiple coauthors). Primary attributions for the various contributed sections are as follows: introduction – A. Tropsha, A. Cherkasov, D. Fourches, S. Ekins, C. Andrade, and V. Poroikov; knowledge mining, AI, and ligand based approaches to drug discovery against COVID-19 – E. Muratov, A. Zakharov, T. Oprea, N. Brown, A. Varnek, G. Schneider, and A. Cherkasov; structure-based drug discovery approaches – D. Kozakov, R. Amaro, K. Merz, O. Isayev, D. Winkler, and A. Cherkasov; best practices of COVID-19 drug discovery – A. Zakharov, A. Cherkasov, and A. Tropsha; COVID-19 and open science – M. Todd, J. Medina-Franco, A. Cherkasov, and A. Tropsha; concluding Remarks – A. Cherkasov and A. Tropsha. Final editing was accomplished by A. Cherkasov and A. Tropsha, with the help of D. Winkler and E. Muratov. A. Cherkasov and A. Tropsha also take primary responsibility for the final content. Mentioning of trade names or commercial products does not constitute endorsement or recommendation for use. The authors acknowledge many fruitful discussions with members of their groups and other colleagues. TIO acknowledge NIH funding support (U24 CA224370, U24 TR002278, and U01CA239108). AT and EM acknowledge NIH funding support (U01CA207160). REA acknowledges support from NIH GM 132826, NSF RAPID MCB-2032054, UCSD Moores Cancer Center 2020 SARS-CoV-2 seed grant, and RCSA COVID Initiative Grant #27350. JLMF thanks DGAPA (UNAM) for grant no. IV200121. O. I. acknowledges support from NSF CHE-1802789 and CHE-2041108, DSF Charitable Foundation and the COVID-19 HPC Consortium. GS acknowledges RETHINK initiative at ETH Zurich, the Novartis Forschungsstiftung (FreeNovation: AI in Drug Discovery), and the Swiss National Science Foundation (grant no. 205321_182176) and inSili.com LLC, Zurich, for providing access to CopyCATS software. VP would like to acknowledge the support by the Russian Foundation of Basic Research grant No. 20-04-60285 and useful discussions with Dr Sergey Ivanov (Institute of Biomedical Chemistry, Moscow, Russia). AZ's contribution has been supported in part by the Intramural Research Program of the National Center for Advancing Translational Sciences, National Institutes of Health. SE kindly acknowledges support from R44GM122196-02A1 and the efforts of Mr Victor Gawriljuk, Dr Ana Puhl, and other colleagues and collaborators for their assistance with our SARS-CoV-2 research. AC acknowledges significant contribution of Mr Anh-Tien Ton and Dr Francesco Gentle to the writing of several sections of the review. E. M. is grateful to Dr Vinicius Alves (NIEHS) for his kind help with the figures.
Biographies
Biography
Artem Cherkasov .

Artem Cherkasov is a Professor of Medicine at the University of British Columbia (Vancouver, Canada) and a Director of Therapeutics Development at Vancouver Prostate Centre. Research interests include computer-aided drug discovery (CADD), QSAR modeling, drug reprofiling and development of new cancer therapies. Dr Cherkasov co-authored more than 200 research papers, 80 patent filings and several book chapters. During his tenure at the UBC, Dr Cherkasov has been a principal applicant or co-applicant on a number of successful grants totalling over 80M dollars, and licenced 8 drug candidates to big pharma companies, major international venture funds and spin off companies.
Biography
Alexander Tropsha .

Alexander Tropsha, PhD is K. H. Lee Distinguished Professor and Associate Dean for Data and Data Science at the UNC Eshelman School of Pharmacy (ranked #1 in the country by US News & World Report), UNC-Chapel Hill. Prof. Tropsha obtained his PhD in Chemical Enzymology in 1986 from Moscow State University, Russia and came to UNC-Chapel Hill in 1989 as a postdoctoral fellow. He joined the School of Pharmacy in 1991 as an Assistant Professor and became full professor in 2002. His research interests are in the areas of Computer-Assisted Drug Design, Computational Toxicology, Cheminformatics, (Nano)Materials Informatics, and Structural Bioinformatics. His has authored or co-authored more than 250 peer-reviewed research papers, reviews and book chapters and co-edited two monographs. He is an Associate Editor of the ACS Journal of Chemical Information and Modeling. His research has been supported by multiple grants from the NIH, NSF, EPA, DOD, foundations, and private companies.
Notes and references
- Coronavirus Update (Live): 87,457,764 Cases and 1,887,064 Deaths from COVID-19 Virus Pandemic – Worldometer, https://www.worldometers.info/coronavirus/, accessed 17 June 2021
- SARS-CoV-2 Resources – NCBI, https://www.ncbi.nlm.nih.gov/sars-cov-2/, accessed 17 April 2021
- bioRxiv COVID-19 SARS-CoV-2 preprints from medRxiv and bioRxiv, https://connect.biorxiv.org/relate/content/181, accessed 3 June 2021
- Kwon D. Nature. 2020;581:130–131. doi: 10.1038/d41586-020-01394-6. [DOI] [PubMed] [Google Scholar]
- Dong Y. Dai T. Wei Y. Zhang L. Zheng M. Zhou F. Signal Transduction Targeted Ther. 2020;5:1–14. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Editorial Nature. 2021;592:326. [Google Scholar]
- Bobrowski T. Melo-Filho C. C. Korn D. Alves V. M. Popov K. I. Auerbach S. Schmitt C. Moorman N. J. Muratov E. N. Tropsha A. Drug Discovery Today. 2020;25:1604–1613. doi: 10.1016/j.drudis.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindez G. Matschinske J. Rose T. D. Sadegh S. Salgado-Albarrán M. Späth J. Baumbach J. Pauling J. K. Nat. Comput. Sci. 2021;1:33–41. doi: 10.1038/s43588-020-00007-6. [DOI] [PubMed] [Google Scholar]
- Pruijssers A. J. George A. S. Schäfer A. Leist S. R. Gralinksi L. E. Dinnon K. H. Yount B. L. Agostini M. L. Stevens L. J. Chappell J. D. Lu X. Hughes T. M. Gully K. Martinez D. R. Brown A. J. Graham R. L. Perry J. K. Du Pont V. Pitts J. Ma B. Babusis D. Murakami E. Feng J. Y. Bilello J. P. Porter D. P. Cihlar T. Baric R. S. Denison M. R. Sheahan T. P. Cell Rep. 2020;32:107940. doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugin K. Woodcock J. Nat. Rev. Drug Discovery. 2021;20:254–255. doi: 10.1038/d41573-021-00037-3. [DOI] [PubMed] [Google Scholar]
- Riva L. Yuan S. Yin X. Martin-Sancho L. Matsunaga N. Pache L. Burgstaller-Muehlbacher S. De Jesus P. D. Teriete P. Hull M. V. Chang M. W. Chan J. F. W. Cao J. Poon V. K. M. Herbert K. M. Cheng K. Nguyen T. T. H. Rubanov A. Pu Y. Nguyen C. Choi A. Rathnasinghe R. Schotsaert M. Miorin L. Dejosez M. Zwaka T. P. Sit K. Y. Martinez-Sobrido L. Liu W. C. White K. M. Chapman M. E. Lendy E. K. Glynne R. J. Albrecht R. Ruppin E. Mesecar A. D. Johnson J. R. Benner C. Sun R. Schultz P. G. Su A. I. García-Sastre A. Chatterjee A. K. Yuen K. Y. Chanda S. K. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First-In-Human Study To Evaluate Safety, Tolerability, And Pharmacokinetics Following Single Ascending And Multiple Ascending Doses of PF-07304814 In Hospitalized Participants With COVID-19. – Full Text View – ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT04535167?term=pfizer&cond=covid-19&cntry=US&draw=2&rank=9, accessed 30 March 2021
- Gordon D. E. Jang G. M. Bouhaddou M. Xu J. Obernier K. White K. M. O’Meara M. J. Rezelj V. V. Guo J. Z. Swaney D. L. Tummino T. A. Hüttenhain R. Kaake R. M. Richards A. L. Tutuncuoglu B. Foussard H. Batra J. Haas K. Modak M. Kim M. Haas P. Polacco B. J. Braberg H. Fabius J. M. Eckhardt M. Soucheray M. Bennett M. J. Cakir M. McGregor M. J. Li Q. Meyer B. Roesch F. Vallet T. Mac Kain A. Miorin L. Moreno E. Naing Z. Z. C. Zhou Y. Peng S. Shi Y. Zhang Z. Shen W. Kirby I. T. Melnyk J. E. Chorba J. S. Lou K. Dai S. A. Barrio-Hernandez I. Memon D. Hernandez-Armenta C. Lyu J. Mathy C. J. P. Perica T. Pilla K. B. Ganesan S. J. Saltzberg D. J. Rakesh R. Liu X. Rosenthal S. B. Calviello L. Venkataramanan S. Liboy-Lugo J. Lin Y. Huang X.-P. Liu Y. Wankowicz S. A. Bohn M. Safari M. Ugur F. S. Koh C. Savar N. S. Tran Q. D. Shengjuler D. Fletcher S. J. O’Neal M. C. Cai Y. Chang J. C. J. Broadhurst D. J. Klippsten S. Sharp P. P. Wenzell N. A. Kuzuoglu-Ozturk D. Wang H.-Y. Trenker R. Young J. M. Cavero D. A. Hiatt J. Roth T. L. Rathore U. Subramanian A. Noack J. Hubert M. Stroud R. M. Frankel A. D. Rosenberg O. S. Verba K. A. Agard D. A. Ott M. Emerman M. Jura N. von Zastrow M. Verdin E. Ashworth A. Schwartz O. D’Enfert C. Mukherjee S. Jacobson M. Malik H. S. Fujimori D. G. Ideker T. Craik C. S. Floor S. N. Fraser J. S. Gross J. D. Sali A. Roth B. L. Ruggero D. Taunton J. Kortemme T. Beltrao P. Vignuzzi M. García-Sastre A. Shokat K. M. Shoichet B. K. Krogan N. J. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. L., Lo K., Chandrasekhar Y., Reas R., Yang J., Burdick D., Eide D., Funk K., Katsis Y., Kinney R., Li Y., Liu Z., Merrill W., Mooney P., Murdick D., Rishi D., Sheehan J., Shen Z., Stilson B., Wade A., Wang K., Wang N. X. R., Wilhelm C., Xie B., Raymond D., Weld D. S., Etzioni O. and Kohlmeier S., CORD-19: The COVID-19 Open Research Dataset, http://arxiv.org/abs/2004.10706, accessed 20 May 2021
- Richardson P. Griffin I. Tucker C. Smith D. Oechsle O. Phelan A. Stebbing J. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P. Sims A. C. Zhou S. Graham R. L. Pruijssers A. J. Agostini M. L. Leist S. R. Schafer A. Dinnon K. H. Stevens L. J. Chappell J. D. Lu X. Hughes T. M. George A. S. Hill C. S. Montgomery S. A. Brown A. J. Bluemling G. R. Natchus M. G. Saindane M. Kolykhalov A. A. Painter G. Harcourt J. Tamin A. Thornburg N. J. Swanstrom R. Denison M. R. Baric R. S. Sci. Transl. Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Cao R. Zhang L. Yang X. Liu J. Xu M. Shi Z. Hu Z. Zhong W. Xiao G. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA Approves First Treatment for COVID-19|FDA, https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19, accessed 20 May 2021
- Jeon S. Ko M. Lee J. Choi I. Byun S. Y. Park S. Shum D. Kim S. Antimicrob. Agents Chemother. 2020;64:e00819–e00820. doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L. Druce J. D. Catton M. G. Jans D. A. Wagstaff K. M. Antiviral Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivermectin in Adults With Severe COVID-19., https://clinicaltrials.gov/ct2/show/NCT04602507, accessed 17 January 2021
- Huang R. Xu M. Zhu H. Chen C. Z. Lee E. M. He S. Shamim K. Bougie D. Huang W. Hall M. D. Lo D. Simeonov A. Austin C. P. Qiu X. Tang H. Zheng W. Nat. Biotechnol. 2021;39:747–753. doi: 10.1038/s41587-021-00839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A Study of LAM-002A for the Prevention of Progression of COVID-19 – Full Text View – ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT04446377, accessed 22 April 2021
- Dual Therapy With Interferon Beta-1b and Clofazimine for COVID-19 – Full Text View – ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT04465695, accessed 22 April 2021
- Bouhaddou M. Memon D. Meyer B. White K. M. Rezelj V. V. Marrero M. C. Polacco B. J. Melnyk J. E. Ulferts S. Kaake R. M. Batra J. Richards A. L. Stevenson E. Gordon D. E. Rojc A. Obernier K. Fabius J. M. Soucheray M. Miorin L. Moreno E. Koh C. Tran Q. D. Hardy A. Robinot R. Vallet T. Nilsson-Payant B. E. Hernandez-Armenta C. Dunham A. Weigang S. Knerr J. Modak M. Quintero D. Zhou Y. Dugourd A. Valdeolivas A. Patil T. Li Q. Hüttenhain R. Cakir M. Muralidharan M. Kim M. Jang G. Tutuncuoglu B. Hiatt J. Guo J. Z. Xu J. Bouhaddou S. Mathy C. J. P. Gaulton A. Manners E. J. Félix E. Shi Y. Goff M. Lim J. K. McBride T. O’Neal M. C. Cai Y. Chang J. C. J. Broadhurst D. J. Klippsten S. De wit E. Leach A. R. Kortemme T. Shoichet B. Ott M. Saez-Rodriguez J. TenOever B. R. Mullins D. Fischer E. R. Kochs G. Grosse R. García-Sastre A. Vignuzzi M. Johnson J. R. Shokat K. M. Swaney D. L. Beltrao P. Krogan N. J. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Dashboard | DrugBank Online, https://go.drugbank.com/covid-19#clinical-trials, accessed 25 January 2021
- Jin Z. Du X. Xu Y. Deng Y. Liu M. Zhao Y. Zhang B. Li X. Zhang L. Peng C. Duan Y. Yu J. Wang L. Yang K. Liu F. Jiang R. Yang X. You T. Liu X. Yang X. Bai F. Liu H. Liu X. Guddat L. W. Xu W. Xiao G. Qin C. Shi Z. Jiang H. Rao Z. Yang H. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- COVID-19/SARS-CoV-2 Resources, https://www.rcsb.org/news?year=2020&article=5e74d55d2d410731e9944f52&feature=true, accessed 31 March 2021
- Fourches D. Muratov E. Tropsha A. Nat. Chem. Biol. 2015;11:535. doi: 10.1038/nchembio.1881. [DOI] [PubMed] [Google Scholar]
- Knegtel R. M. Wagener M. Proteins. 1999;37:334–345. doi: 10.1002/(sici)1097-0134(19991115)37:3<334::aid-prot3>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Parks J. M. Smith J. C. N. Engl. J. Med. 2020;382:2261–2264. doi: 10.1056/NEJMcibr2007042. [DOI] [PubMed] [Google Scholar]
- Boras B. Jones R. M. Anson B. J. Arenson D. Aschenbrenner L. Bakowski M. A. Beutler N. Binder J. Chen E. Eng H. Hammond J. Hoffman R. Kadar E. P. Kania R. Kimoto E. Kirkpatrick M. G. Lanyon L. Lendy E. K. Lillis J. R. Luthra S. A. Ma C. Noell S. Obach R. S. O’Brien M. N. O’Connor R. Ogilvie K. Owen D. Pettersson M. Reese M. R. Rogers T. Rossulek M. I. Sathish J. G. Steppan C. Ticehurst M. Updyke L. W. Zhu Y. Wang J. Chatterjee A. K. Mesecar A. D. Anderson A. S. Allerton C. bioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.09.12.293498. [DOI] [Google Scholar]
- ChEMBL, ChEMBL_27 SARS-CoV-2 release, http://chembl.blogspot.com/2020/05/chembl27-sars-cov-2-release.html, accessed 18 January 2021
- COVID-19 Data Portal – accelerating scientific research through data, https://www.covid19dataportal.org/, accessed 17 April 2021
- NCATS, Open Science Data Portal, https://opendata.ncats.nih.gov/covid19/databrowser, accessed 17 April 2021
- Main protease structure and XChem fragment screen – Diamond Light Source, https://www.diamond.ac.uk/covid-19/for-scientists/Main-protease-structure-and-XChem.html, accessed 26 January 2021
- PostEra|COVID-19, https://covid.postera.ai/covid, accessed 31 March 2021
- Novel Coronavirus Information Center, https://www.elsevier.com/connect/coronavirus-information-center, accessed 18 January 2021
- canSAR Coronavirus Research Tool|ICR CRUK Cancer Therapeutics Unit, https://corona.cansar.icr.ac.uk/, accessed 8 July 2020
- Cava C. Bertoli G. Castiglioni I. Viruses. 2020;12:404. doi: 10.3390/v12040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra S. Chaudhuri A. G. Tiwary B. K. Chakrabarti N. Life Sci. 2020;257:118096. doi: 10.1016/j.lfs.2020.118096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakurt H. Pir P. Turk. J. Biol. 2020;44:168–177. doi: 10.3906/biy-2005-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunakaran K. Ganapathiraju M. Res. Sq. 2020 doi: 10.21203/rs.3.rs-30363/v1. [DOI] [Google Scholar]
- Ekins S. Mottin M. Ramos P. De Paula Sousa B. K. junior Neves B. Foil D. Zorn K. Campos Braga R. Coffee M. Southan C. Puhl A. Horta Andrade C. OSF Prepr. 2020:1–50. doi: 10.1016/j.drudis.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioritizing diseases for research and development in emergency contexts, https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts, accessed 20 May 2021
- Yazdani S. De Maio N. Ding Y. Shahani V. Goldman N. Schapira M. bioRxiv. 2021 doi: 10.1101/2021.03.23.436637. [DOI] [PubMed] [Google Scholar]
- Genetic variability of SARS-CoV-2 drug binding sites, https://apps.thesgc.org/SARSCoV2_pocketome/, accessed 3 April 2021
- Thoms M. Buschauer R. Ameismeier M. Koepke L. Denk T. Hirschenberger M. Kratzat H. Hayn M. Mackens-Kiani T. Cheng J. Straub J. H. Stürzel C. M. Fröhlich T. Berninghausen O. Becker T. Kirchhoff F. Sparrer K. M. J. Beckmann R. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska K. Kim Y. Jedrzejczak R. Maltseva N. I. Stols L. Endres M. Joachimiak A. IUCrJ. 2020;7:814–824. doi: 10.1107/S2052252520009653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D. Mukherjee R. Grewe D. Bojkova D. Baek K. Bhattacharya A. Schulz L. Widera M. Mehdipour A. R. Tascher G. Geurink P. P. Wilhelm A. van der Heden van Noort G. J. Ovaa H. Müller S. Knobeloch K. P. Rajalingam K. Schulman B. A. Cinatl J. Hummer G. Ciesek S. Dikic I. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A.-T. Gentile F. Hsing M. Ban F. Cherkasov A. Mol. Inform. 2020;39:e2000028. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. C. Chang G. G. Chou C. Y. Biophys. J. 2010;98:1327–1336. doi: 10.1016/j.bpj.2009.12.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littler D. R. Gully B. S. Colson R. N. Rossjohn J. iScience. 2020;23:101258. doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafcikova P. Silhan J. Nencka R. Boura E. Nat. Commun. 2020;11:1–7. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Wu J. Wang H. Gao Y. Liu Q. Mu A. Ji W. Yan L. Zhu Y. Zhu C. Fang X. Yang X. Huang Y. Gao H. Liu F. Ge J. Sun Q. Yang X. Xu W. Liu Z. Yang H. Lou Z. Jiang B. Guddat L. W. Gong P. Rao Z. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W. Mao C. Luan X. Shen D. D. Shen Q. Su H. Wang X. Zhou F. Zhao W. Gao M. Chang S. Xie Y. C. Tian G. Jiang H. W. Tao S. C. Shen J. Jiang Y. Jiang H. Xu Y. Zhang S. Zhang Y. Xu H. E. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H. S. Kokic G. Farnung L. Dienemann C. Tegunov D. Cramer P. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Jia Z. Yan L. Ren Z. Wu L. Wang J. Guo J. Zheng L. Ming Z. Zhang L. Lou Z. Rao Z. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. Jedrzejczak R. Maltseva N. I. Wilamowski M. Endres M. Godzik A. Michalska K. Joachimiak A. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan T. Arya S. Chan S. H. Qi S. Dai N. Misra A. Park J. G. Oladunni F. Kovalskyy D. Hromas R. A. Martinez-Sobrido L. Gupta Y. K. Nat. Commun. 2020;11:1–7. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X. Cao D. Kong L. Zhang X. Nat. Commun. 2020;11:3618. doi: 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y. Shu T. Wu D. Mu J. Wang C. Huang M. Han Y. Zhang X. Y. Zhou W. Qiu Y. Zhou X. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower T. G. Buffalo C. Z. Hooy R. M. Allaire M. Ren X. Hurley J. H. Proc. Natl. Acad. Sci. U. S. A. 2021;118:e2021785118. doi: 10.1073/pnas.2021785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Jiang H. Nan Zhang H. Feng Meng Q. Xie J. Li Y. Chen H. Xiao Zheng Y. Ning Wang X. Qi H. Zhang J. Wang P. H. Han Z. G. Ce Tao S. Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. Yang M. Hong Z. Zhang L. Huang Z. Chen X. He S. Zhou Z. Zhou Z. Chen Q. Yan Y. Zhang C. Shan H. Chen S. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkolova E. Klima M. Nencka R. Boura E. J. Struct. Biol. 2020;211:107548. doi: 10.1016/j.jsb.2020.107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman S. Prasad B. V. L. S. Selvarajan R. Viruses. 2018:10. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A. Chan J. F. W. Azhar E. I. Hui D. S. C. Yuen K. Y. Nat. Rev. Drug Discovery. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko W. C. Rolain J. M. Lee N. Y. Chen P. L. Huang C. T. Lee P. I. Hsueh P. R. Int. J. Antimicrob. Agents. 2020;55:105933. doi: 10.1016/j.ijantimicag.2020.105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C. J. Tchesnokov E. P. Feng J. Y. Porter D. P. Götte M. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T. Huang M. Wu D. Ren Y. Zhang X. Han Y. Mu J. Wang R. Qiu Y. Zhang D. Y. Zhou X. Virol. Sin. 2020;35:321–329. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devillers J. Pandard P. Richard B. SAR QSAR Environ. Res. 2013;24:979–993. doi: 10.1080/1062936X.2013.848632. [DOI] [PubMed] [Google Scholar]
- McBride R. van Zyl M. Fielding B. C. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda S. Beglov D. Wakefield A. E. Egbert M. Whitty A. Curr. Opin. Chem. Biol. 2018;44:1–8. doi: 10.1016/j.cbpa.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. Smith J. C. ChemRxiv. 2020 doi: 10.26434/CHEMRXIV.11871402.V3. [DOI] [Google Scholar]
- Gao X. Qin B. Chen P. Zhu K. Hou P. Wojdyla J. A. Wang M. Cui S. Acta Pharm. Sin. B. 2020;11:237–245. doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y. M. Barraza S. J. Wilson M. W. Agius M. P. Mielech A. M. Davis N. M. Baker S. C. Larsen S. D. Mesecar A. D. J. Med. Chem. 2014;57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rut W. Lv Z. Zmudzinski M. Patchett S. Nayak D. Snipas S. J. El Oualid F. Huang T. T. Bekes M. Drag M. Olsen S. K. Sci. Adv. 2020;6:eabd4596. doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T. Takemoto C. Kim Y. T. Wang H. Nishii W. Terada T. Shirouzu M. Yokoyama S. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiemer L. Lund O. Brunak S. Blom N. BMC Bioinf. 2004;5:72. doi: 10.1186/1471-2105-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller D. W. Phillips G. Weiss K. L. Zhang Q. Coates L. Kovalevsky A. J. Med. Chem. 2021;64:4991–5000. doi: 10.1021/acs.jmedchem.1c00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W. Zhang B. Jiang X. M. Su H. Li J. Zhao Y. Xie X. Jin Z. Peng J. Liu F. Li C. Li Y. Bai F. Wang H. Cheng X. Cen X. Hu S. Yang X. Wang J. Liu X. Xiao G. Jiang H. Rao Z. Zhang L. K. Xu Y. Yang H. Liu H. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodera J. Lee A. A. London N. von Delft F. Nat. Chem. 2020;12:581. doi: 10.1038/s41557-020-0496-2. [DOI] [PubMed] [Google Scholar]
- Together We Are Powerful – Folding@home, https://foldingathome.org/, accessed 22 April 2021
- Lyu J. Wang S. Balius T. E. Singh I. Levit A. Moroz Y. S. O’Meara M. J. Che T. Algaa E. Tolmachova K. Tolmachev A. A. Shoichet B. K. Roth B. L. Irwin J. J. Nature. 2019;566:224–229. doi: 10.1038/s41586-019-0917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling T. Irwin J. J. J. Chem. Inf. Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya A. Agarwal R. Baker M. B. Baudry J. Bhowmik D. Boehm S. Byler K. G. Chen S. Y. Coates L. Cooper C. J. Demerdash O. Daidone I. Eblen J. D. Ellingson S. Forli S. Glaser J. Gumbart J. C. Gunnels J. Hernandez O. Irle S. Kneller D. W. Kovalevsky A. Larkin J. Lawrence T. J. LeGrand S. Liu S.-H. Mitchell J. C. Park G. Parks J. M. Pavlova A. Petridis L. Poole D. Pouchard L. Ramanathan A. Rogers D. M. Santos-Martins D. Scheinberg A. Sedova A. Shen Y. Smith J. C. Smith M. D. Soto C. Tsaris A. Thavappiragasam M. Tillack A. F. Vermaas J. V. Vuong V. Q. Yin J. Yoo S. Zahran M. Zanetti-Polzi L. J. Chem. Inf. Model. 2020;60:5832–5852. doi: 10.1021/acs.jcim.0c01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse J. S. Lalonde T. Xu S. Liu W. R. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro R. E. Mulholland A. J. J. Chem. Inf. Model. 2020;60:2653–2656. doi: 10.1021/acs.jcim.0c00319. [DOI] [PubMed] [Google Scholar]
- Wrapp D. Wang N. Corbett K. S. Goldsmith J. A. Hsieh C.-L. Abiona O. Graham B. S. McLellan J. S. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. Park S. Yeol C. Park T. Maham T. Cao Y. Kern N. Lee J. Min Y. Croll T. Seok C. Im W. J. Phys. Chem. B. 2020;124:7128–7137. doi: 10.1021/acs.jpcb.0c04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalino L. Gaieb Z. Goldsmith J. Hjorth C. Dommer A. Harbison A. Fogarthy C. Barros E. Taylor B. McLellan J. Fadda E. Amaro R. ACS Cenral Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoňová B. Sikora M. Schürmann C. Hagen W. J. H. Welsch S. Blanc F. E. C. von Bülow S. Gecht M. Bagola K. Hörner C. van Zandbergen G. Landry J. de Azevedo N. T. D. Mosalaganti S. Schwarz A. Covino R. Mühlebach M. D. Hummer G. Locker J. K. Beck M. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. Tsai C. J. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- Wassman C. D. Baronio R. Demir Ö. Wallentine B. D. Chen C. K. Hall L. V. Salehi F. Lin D. W. Chung B. P. Wesley Hatfield G. Richard Chamberlin A. Luecke H. Lathrop R. H. Kaiser P. Amaro R. E. Nat. Commun. 2013;4:1–9. doi: 10.1038/ncomms2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztain T. Amaro R. McCammon J. J. Chem. Inf. Model. 2021 doi: 10.1021/acs.jcim.1c00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon L. Belfon K. Raquette L. Wang Y. Corbo C. Stepanenko D. Cuomo A. Guerra J. Budhan S. Varghese S. Rizzo R. Simmerling C. ChemRxiv. 2020 doi: 10.26434/chemrxiv.13502646.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinello A. Saltalamacchia A. Magistrato A. J. Phys. Chem. Lett. 2020;11:4785–4790. doi: 10.1021/acs.jpclett.0c01148. [DOI] [PubMed] [Google Scholar]
- Barros E. P. Casalino L. Gaieb Z. Dommer A. C. Wang Y. Fallon L. Raguette L. Belfon K. Simmerling C. Amaro R. E. Biophys. J. 2020;120:1072–1084. doi: 10.1016/j.bpj.2020.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A. S. F. Ibarra A. A. Bermudez I. Casalino L. Gaieb Z. Shoemark D. Gallagher T. Sessions R. Amaro R. Mulholland A. Biophys. Lett. 2021;120(6):983–993. doi: 10.1016/j.bpj.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha K. Peters M. B. Wang B. Yu N. Wollacott A. M. Westerhoff L. M. Merz K. M. Drug Discovery Today. 2007;12:725–731. doi: 10.1016/j.drudis.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Cavasotto C. N. Aucar M. G. Front. Chem. 2020;8:246. doi: 10.3389/fchem.2020.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Guzmán C. A. Ruiz-Pernía J. J. Tuñón I. ACS Catal. 2020;10:12544–12554. doi: 10.1021/acscatal.0c03420. [DOI] [PubMed] [Google Scholar]
- Hatada R. Okuwaki K. Mochizuki Y. Handa Y. Fukuzawa K. Komeiji Y. Okiyama Y. Tanaka S. J. Chem. Inf. Model. 2020;60:3593–3602. doi: 10.1021/acs.jcim.0c00283. [DOI] [PubMed] [Google Scholar]
- Khrenova M. G. Tsirelson V. G. Nemukhin A. V. Phys. Chem. Chem. Phys. 2020;22:19069–19079. doi: 10.1039/d0cp03560b. [DOI] [PubMed] [Google Scholar]
- Cavasotto C. N. Di Filippo J. I. Mol. Inform. 2021;40:2000–2115. doi: 10.1002/minf.202000115. [DOI] [PubMed] [Google Scholar]
- Adhikari P. Li N. Shin M. Steinmetz N. F. Twarock R. Podgornik R. Ching W. Y. Phys. Chem. Chem. Phys. 2020;22:18272–18283. doi: 10.1039/d0cp03145c. [DOI] [PubMed] [Google Scholar]
- Behler J. J. Chem. Phys. 2016;145:170901. doi: 10.1063/1.4966192. [DOI] [PubMed] [Google Scholar]
- Smith J. S. Isayev O. Roitberg A. E. Chem. Sci. 2017;8:3192–3203. doi: 10.1039/c6sc05720a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. S. Nebgen B. Lubbers N. Isayev O. Roitberg A. E. A. E. J. Chem. Phys. 2018;148:241733. doi: 10.1063/1.5023802. [DOI] [PubMed] [Google Scholar]
- Smith J. S. Nebgen B. T. Zubatyuk R. Lubbers N. Devereux C. Barros K. Tretiak S. Isayev O. Roitberg A. E. Nat. Commun. 2019;10:2903. doi: 10.1038/s41467-019-10827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubatyuk R. Smith J. S. Leszczynski J. Isayev O. Sci. Adv. 2019;5:eaav6490. doi: 10.1126/sciadv.aav6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux C. Smith J. Davis K. Barros K. Zubatyuk R. Isayev O. Roitberg A. J. Chem. Theory Comput. 2020;16:4192–4202. doi: 10.1021/acs.jctc.0c00121. [DOI] [PubMed] [Google Scholar]
- Lahey S.-L. J. Rowley C. N. Chem. Sci. 2020;11:2362–2368. doi: 10.1039/c9sc06017k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therapeutics and Small Molecules, https://covid.molssi.org/therapeutics/#res_therapeutics, accessed 3 April 2021
- Download CAS COVID-19 Antiviral Candidate Compounds Dataset|CAS, https://www.cas.org/covid-19-antiviral-compounds-dataset, accessed 22 January 2021
- Axelrod S. and Gomez-Bombarelli R., GEOM: Energy-annotated molecular conformations for property prediction and molecular generation, http://arxiv.org/abs/2006.05531, accessed 22 January 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile F. Agrawal V. Hsing M. Ton A. T. Ban F. Norinder U. Gleave M. E. Cherkasov A. ACS Cent. Sci. 2020;6:939–949. doi: 10.1021/acscentsci.0c00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban F. Dalal K. Li H. LeBlanc E. Rennie P. S. Cherkasov A. J. Chem. Inf. Model. 2017;57:1018–1028. doi: 10.1021/acs.jcim.7b00137. [DOI] [PubMed] [Google Scholar]
- Hamill P. Hudson D. Kao R. Y. Chow P. Raj M. Xu H. Richer M. J. Jean F. Biol. Chem. 2006;387:1063–1074. doi: 10.1515/BC.2006.131. [DOI] [PubMed] [Google Scholar]
- Bocci G. B. K. C. Verma S. Hasan M. Holmes J. Sirimulla S. Oprea T. I. Nature Machine Intel. 2021;3:527–535. [Google Scholar]
- Drug Central -REDIAL, http://drugcentral.org/Redial, accessed 14 March 2021
- Landrum G., RDKit: Open-Source Cheminformatics Software, http://rdkit.org/, accessed 24 March 2021 [Google Scholar]
- Brimacombe K. R. Zhao T. Eastman R. T. Hu X. Wang K. Backus M. Baljinnyam B. Chen C. Z. Chen L. Eicher T. Ferrer M. Fu Y. Gorshkov K. Guo H. Hanson Q. M. Itkin Z. Kales S. C. Klumpp-Thomas C. Lee E. M. Michael S. Mierzwa T. Patt A. Pradhan M. Renn A. Shinn P. Shrimp J. H. Viraktamath A. Wilson K. M. Xu M. Zakharov A. V. Zhu W. Zheng W. Simeonov A. Mathé E. A. Lo D. C. Hall M. D. Shen M. bioRxiv. 2020 doi: 10.1101/2020.06.04.135046. [DOI] [Google Scholar]
- Kuleshov M. Clarke D. Kropiwnicki E. Jagodnik K. Bartal A. Evangelista J. E. Zhou A. Ferguson L. Lachmann A. Ma’ayan A. Patterns. 2020;1:100090. doi: 10.1016/j.patter.2020.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger B. Bojkova D. Zaliani A. Cinatl J. Claussen C. Westhaus S. Reinshagen J. Kuzikov M. Wolf M. Geisslinger G. Gribbon P. Ciesek S. Res. Sq. 2020 doi: 10.21203/RS.3.RS-23951/V1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touret F. Gilles M. Barral K. Nougairède A. Decroly E. de Lamballerie X. Coutard B. bioRxiv. 2020 doi: 10.1101/2020.04.03.023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S. Coleman C. M. Haupt R. Logue J. Matthews K. Frieman M. bioRxiv. 2020 doi: 10.1101/2020.03.25.008482. [DOI] [Google Scholar]
- Levin J. M. Oprea T. I. Davidovich S. Clozel T. Overington J. P. Vanhaelen Q. Cantor C. R. Bischof E. Zhavoronkov A. Nat. Biotechnol. 2020;38:1127–1131. doi: 10.1038/s41587-020-0686-x. [DOI] [PubMed] [Google Scholar]
- Bocci G. Bradfute S. B. Ye C. Garcia M. J. Parvathareddy J. Reichard W. Surendranathan S. Bansal S. Bologa C. G. Perkins D. J. Jonsson C. B. Sklar L. A. Oprea T. I. ACS Pharmacol. Transl. Sci. 2020;3:1278–1292. doi: 10.1021/acsptsci.0c00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar H. A. Baskin I. I. Marcou G. Horvath D. Varnek A. J. Chem. Inf. Model. 2015;55:84–94. doi: 10.1021/ci500575y. [DOI] [PubMed] [Google Scholar]
- Zabolotna Y. Lin A. Horvath D. Marcou G. Volochnyuk D. M. Varnek A. J. Chem. Inf. Model. 2021;61:179–188. doi: 10.1021/acs.jcim.0c00936. [DOI] [PubMed] [Google Scholar]
- Horvath D. Orlov A. Osolodkin D. I. Ishmukhametov A. A. Marcou G. Varnek A. Mol. Inform. 2020;39:e2000080. doi: 10.1002/minf.202000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S. Knox C. Guo A. C. Cheng D. Shrivastava S. Tzur D. Gautam B. Hassanali M. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casciuc I. Zabolotna Y. Horvath D. Marcou G. Bajorath J. Varnek A. J. Chem. Inf. Model. 2019;59:564–572. doi: 10.1021/acs.jcim.8b00650. [DOI] [PubMed] [Google Scholar]
- Yang X. Wang Y. Byrne R. Schneider G. Yang S. Chem. Rev. 2019;119:10520–10594. doi: 10.1021/acs.chemrev.8b00728. [DOI] [PubMed] [Google Scholar]
- Schneider G. Clark D. E. Angew. Chem., Int. Ed. 2019;58:10792–10803. doi: 10.1002/anie.201814681. [DOI] [PubMed] [Google Scholar]
- Chen H. Engkvist O. Wang Y. Olivecrona M. Blaschke T. Drug Discovery Today. 2018;23:1241–1250. doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- Generative Deep Learning: Teaching Machines to Paint, Write, Compose, and Play: Foster, David: 9781492041948: Amazon.com: Books, https://www.amazon.com/Generative-Deep-Learning-Teaching-Machines/dp/1492041947, accessed 21 January 2021
- Grisoni F. Schneider G. Chimia. 2019;73:1006–1011. doi: 10.2533/chimia.2019.1006. [DOI] [PubMed] [Google Scholar]
- Blaschke T. Olivecrona M. Engkvist O. Bajorath J. Chen H. Mol. Inform. 2018;37:1700123. doi: 10.1002/minf.201700123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E. Lin C. H. Lane H. Y. Molecules. 2020:25. doi: 10.3390/molecules25143250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogány P. Arad N. Genway S. Pickett S. D. J. Chem. Inf. Model. 2019;59:1136–1146. doi: 10.1021/acs.jcim.8b00626. [DOI] [PubMed] [Google Scholar]
- Sattarov B. Baskin I. I. Horvath D. Marcou G. Bjerrum E. J. Varnek A. J. Chem. Inf. Model. 2019;59:1182–1196. doi: 10.1021/acs.jcim.8b00751. [DOI] [PubMed] [Google Scholar]
- Merk D. Friedrich L. Grisoni F. Schneider G. Mol. Inf. 2018;37:1700153. doi: 10.1002/minf.201700153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. Clercq E. De. Nat. Rev. Drug Discovery. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Schneider P. Welin M. Svensson B. Walse B. Schneider G. Mol. Inform. 2020;39:e2000109. doi: 10.1002/minf.202000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Luna J. Grisoni F. Weskamp N. Schneider G. Expert Opin. Drug Discovery. 2021;2:1–11. doi: 10.1080/17460441.2021.1909567. [DOI] [PubMed] [Google Scholar]
- Wilkinson M. D. Dumontier M. Aalbersberg I. J. J. Appleton G. Axton M. Baak A. Blomberg N. Boiten J.-W. da Silva Santos L. B. Bourne P. E. Bouwman J. Brookes A. J. Clark T. Crosas M. Dillo I. Dumon O. Edmunds S. Evelo C. T. Finkers R. Gonzalez-Beltran A. Gray A. J. G. Groth P. Goble C. Grethe J. S. Heringa J. C.’t Hoen P. A. Hooft R. Kuhn T. Kok R. Kok J. Lusher S. J. Martone M. E. Mons A. Packer A. L. Persson B. Rocca-Serra P. Roos M. van Schaik R. Sansone S.-A. Schultes E. Sengstag T. Slater T. Strawn G. Swertz M. A. Thompson M. van der Lei J. van Mulligen E. Velterop J. Waagmeester A. Wittenburg P. Wolstencroft K. Zhao J. Mons B. Sci. data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock J. Luccioni A. Pham K. H. Lam C. S. N. Luengo-Oroz M. J. Artif. Intell. Res. 2020;69:807–845. [Google Scholar]
- Biomedical Data Translator Consortium Clin. Transl. Sci. 2019;12:86–90. doi: 10.1111/cts.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton K. Wang P. Bizon C. Cox S. Balhoff J. Kebede Y. Fecho K. Tropsha A. Bioinformatics. 2019;35:5382–5384. doi: 10.1093/bioinformatics/btz604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J. Phelan A. Griffin I. Tucker C. Oechsle O. Smith D. Richardson P. Lancet Infect. Dis. 2020;20:400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA, Coronavirus (COVID-19) Update: FDA Authorizes Drug Combination for Treatment of COVID-19|FDA, https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-drug-combination-treatment-covid-19, accessed 7 January 2021
- Zeng X. Song X. Ma T. Pan X. Zhou Y. Hou Y. Zhang Z. Li K. Karypis G. Cheng F. J. Proteome Res. 2020;19:4624–4636. doi: 10.1021/acs.jproteome.0c00316. [DOI] [PubMed] [Google Scholar]
- Amazon Web Services open-sources biological knowledge graph to fight COVID-19, https://www.amazon.science/blog/amazon-web-services-open-sources-biological-knowledge-graph-to-fight-covid-19, accessed 18 January 2021
- Korn D. Bobrowski T. Li M. Kebede Y. Wang P. Owen P. Vaidya G. Muratov E. Chirkova R. Bizon C. Tropsha A. Bioinformatics. 2021;37:586–587. doi: 10.1093/bioinformatics/btaa718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GitHub – Knowledge-Graph-Hub/kg-covid-19: An instance of KG Hub to produce a knowledge graph for COVID-19 response, https://github.com/Knowledge-Graph-Hub/kg-covid-19, accessed 18 January 2021
- Neo4COVID-19, https://neo4covid19.ncats.io/, accessed 26 January 2021
- Zahoránszky-Kohalmi G. Sheils T. Oprea T. I. J. Cheminform. 2020;12:5. doi: 10.1186/s13321-020-0409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelstein D. S. Lizee A. Hessler C. Brueggeman L. Chen S. L. Hadley D. Green A. Khankhanian P. Baranzini S. E. eLife. 2017;6:e26726. doi: 10.7554/eLife.26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasso G. Mayer S. V. Winkelmann E. R. Chu T. Elliot O. Patino-Galindo J. A. Park K. Rabadan R. Honig B. Shapira S. D. Cell. 2019;178:1526–1541. doi: 10.1016/j.cell.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J. Alfajaro M. M. DeWeirdt P. C. Hanna R. E. Lu-Culligan W. J. Cai W. L. Strine M. S. Zhang S. M. Graziano V. R. Schmitz C. O. Chen J. S. Mankowski M. C. Filler R. B. Ravindra N. G. Gasque V. de Miguel F. J. Patil A. Chen H. Oguntuyo K. Y. Abriola L. Surovtseva Y. V. Orchard R. C. Lee B. Lindenbach B. D. Politi K. van Dijk D. Kadoch C. Simon M. D. Yan Q. Doench J. G. Wilen C. B. Cell. 2021;184:76–91. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea T. I. Yang J. J. Byrd D. R. Deretic V. bioRxiv. 2019 doi: 10.1101/715037. [DOI] [Google Scholar]
- Sheils T. Mathias D. Kelleher K. Siramshetty V. Nguyen D. T. Bologa C. Jensen L. Vidović D. Koleti A. Schürer S. Waller A. Yang J. Holmes J. Bocci J. Southall N. Dharkar P. Mathé M. Simeonov A. Oprea T. Nucleic Acids Res. 2021;49:D1160–D1169. doi: 10.1093/nar/gkaa997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram S. Bologa C. Holmes J. Bocci G. Wilson T. Nguyen D. T. Curpan R. Halip L. Bora A. Yang J. Knockel J. Sirimulla S. Ursu O. Oprea T. Nucleic Acids Res. 2021;49:D1160–D1169. doi: 10.1093/nar/gkaa997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratov E. Zakharov A. chemRxiv. 2020 doi: 10.26434/chemrxiv.12143355.v1. [DOI] [Google Scholar]
- Capuzzi S. J. Thornton T. E. Liu K. Baker N. Lam W. I. O’Banion C. Muratov E. N. Pozefsky D. Tropsha A. J. Chem. Inf. Model. 2018;58:212–218. doi: 10.1021/acs.jcim.7b00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizon C. Cox S. Balhoff J. Kebede Y. Wang P. Morton K. Fecho K. Tropsha A. J. Chem. Inf. Model. 2019;59:4968–4973. doi: 10.1021/acs.jcim.9b00683. [DOI] [PubMed] [Google Scholar]
- Tropsha A. Mol. Inform. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- Bobrowski T. Chen L. Eastman R. T. Itkin Z. Shinn P. Chen C. Guo H. Zheng W. Michael S. Simeonov A. Hall M. D. Zakharov A. V. Muratov E. N. bioRxiv. 2020 doi: 10.1101/2020.06.29.178889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowski T. Chen L. Eastman R. T. Itkin Z. Shinn P. Chen C. Z. Guo H. Zheng W. Michael S. Simeonov A. Hall M. D. Zakharov A. V. Muratov E. N. Mol. Ther. 2021;29:873–885. doi: 10.1016/j.ymthe.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratov E. N. Varlamova E. V. Artemenko A. G. Polishchuk P. G. Nikolaeva-Glomb L. Galabov A. S. Kuz’min V. E. Struct. Chem. 2013;24:1665–1679. [Google Scholar]
- Zakharov A. V. Varlamova E. V. Lagunin A. A. Dmitriev A. V. Muratov E. N. Fourches D. Kuz’min V. E. Poroikov V. V. Tropsha A. Nicklaus M. C. Mol. Pharm. 2016;13:545–556. doi: 10.1021/acs.molpharmaceut.5b00762. [DOI] [PubMed] [Google Scholar]
- Foucquier J. Guedj M. Pharmacol. Res. Perspect. 2015;3:e00149. doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Monoclonal Antibody Bamlanivimab|FDA, https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-monoclonal-antibody-bamlanivimab, accessed 21 May 2021
- Si L. Bai H. Rodas M. Cao W. Oh C. Jay A. Moller R. Hoagland D. Oishi K. Shu H. Uhl S. Blanco-Melo D. Albrecht R. Liu W. Jordan T. Payant B. Golynker I. Frere J. Logue J. Haupt R. McGrath M. Weston S. Zhang T. Plebani R. Soong M. Nurani A. Kim S. Zhu D. Benam K. Goyal G. Gilpin S. Baun R. Gygi S. Powers R. Carlson K. Frieman M. tenOever B. Ingber D. Nat. Biomed. Eng. 2021 doi: 10.1038/s41551-021-00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourches D. Muratov E. Tropsha A. J. Chem. Inf. Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourches D. Muratov E. Tropsha A. J. Chem. Inf. Model. 2016;56:1243–1252. doi: 10.1021/acs.jcim.6b00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearden J. C. Cronin M. T. D. Kaiser K. L. E. SAR QSAR Environ. Res. 2009;20:241–266. doi: 10.1080/10629360902949567. [DOI] [PubMed] [Google Scholar]
- Organisation for Economic Co-operation and Development and OECD, OECD principles for the validation, for regulatory purposes, of (Quantitative) Structure–Activity Relationship models, http://europa.eu.int/comm/environment/chemicals/reach.htm, accessed 17 February 2017
- Cherkasov A. Muratov E. N. Fourches D. Varnek A. Baskin I. I. Cronin M. Dearden J. C. Gramatica P. Martin Y. C. Todeschini R. Consonni V. Kuz’min V. E. V. E. Cramer R. D. Benigni R. Yang C. Rathman J. F. Terfloth L. Gasteiger J. Richard A. M. Tropsha A. J. Med. Chem. 2014;57:4977–5010. doi: 10.1021/jm4004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratov E. N. Bajorath J. Sheridan R. P. Tetko I. V. Filimonov D. Poroikov V. Oprea T. I. Baskin I. I. Varnek A. Roitberg A. Isayev O. Curtalolo S. Fourches D. Cohen Y. Aspuru-Guzik A. Winkler D. A. Agrafiotis D. Cherkasov A. Tropsha A. Chem. Soc. Rev. 2020;49:3525–3564. doi: 10.1039/d0cs00098a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabato B. Falchi F. Cavalli A. ChemRxiv Prepr. Serv. 2020 doi: 10.26434/CHEMRXIV.12264503.V1. [DOI] [Google Scholar]
- Santibáñez-Morán M. G. López-López E. Prieto-Martínez F. D. Sánchez-Cruz N. Medina-Franco J. L. RSC Adv. 2020;10:25089–25099. doi: 10.1039/d0ra04922k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno A. Mestres-Truyol J. Ojeda-Montes M. J. Macip G. Saldivar-Espinoza B. Cereto-Massagué A. Pujadas G. Garcia-Vallvé S. Int. J. Mol. Sci. 2020;21:3793. doi: 10.3390/ijms21113793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahremanpour M. M. Tirado-Rives J. Deshmukh M. Ippolito J. A. Zhang C.-H. Cabeza De Vaca I. Liosi M.-E. Anderson K. S. Jorgensen W. L. bioRxiv. 2020 doi: 10.1101/2020.08.28.271957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand S., Scheinberg A., Tillack A. F., Thavappiragasam M., Vermaas J. V., Agarwal R., Larkin J., Poole D., Santos-Martins D., Solis-Vasquez L., Koch A., Forli S., Hernandez O., Smith J. C. and Sedova A., in Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, BCB 2020, Association for Computing Machinery, Inc, 2020
- McGann M. J. Chem. Inf. Model. 2011;51:578–596. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- Friesner R. A. Banks J. L. Murphy R. B. Halgren T. A. Klicic J. J. Mainz D. T. Repasky M. P. Knoll E. H. Shelley M. Perry J. K. Shaw D. E. Francis P. Shenkin P. S. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Neves M. A. C. Totrov M. Abagyan R. J. Comput. Aided. Mol. Des. 2012;26:675–686. doi: 10.1007/s10822-012-9547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlington M. Chun A. Tomar S. Eggler A. Grum-Tokars V. Jacobs J. Daniels J. S. Dawson E. Saldanha A. Chase P. Baez-Santos Y. M. Lindsley C. W. Hodder P. Mesecar A. D. Stauffer S. R. Bioorganic Med. Chem. Lett. 2013;23:6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T. Manickam M. Namasivayam V. Hayashi Y. Jung S.-H. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysinger M. M. Carchia M. Irwin J. J. Shoichet B. K. J. Med. Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A. Glen R. C. J. Chem. Inf. Model. 2005;45:1369–1375. doi: 10.1021/ci0500177. [DOI] [PubMed] [Google Scholar]
- Guterres H. Im W. J. Chem. Inf. Model. 2020;60:2189–2198. doi: 10.1021/acs.jcim.0c00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden S. Ryde U. Expert Opin. Drug Discovery. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamri M. A. Altharawi A. Alabbas A. B. Alossaimi M. A. Alqahtani S. M. Arab. J. Chem. 2020;13:7224–7234. doi: 10.1016/j.arabjc.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler D. A. J. Chem. Inf. Model. 2020;60:4421–4423. doi: 10.1021/acs.jcim.0c01146. [DOI] [PubMed] [Google Scholar]
- Hanson Q. Wilson K. Shen M. Itkin Z. Eastman R. Shinn P. Hall M. bioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.06.16.154708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Z. Shinn P. Itkin Z. Eastman R. T. Bostwick R. Rasmussen L. Huang R. Shen M. Hu X. Wilson K. M. Brooks B. M. Guo H. Zhao T. Klump-Thomas C. Simeonov A. Michael S. G. Lo D. C. Hall M. D. Zheng W. Front. Pharmacol. 2021;11:2005. doi: 10.3389/fphar.2020.592737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyr Z. A. Gorshkov K. Chen C. Z. Zheng W. J. Pharmacol. Exp. Ther. 2020;375:127–138. doi: 10.1124/jpet.120.000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial, Nature, 2020, 578, 7 [Google Scholar]
- Burrows C. J. Wang S. Kim H. J. Meyer G. J. Schanze K. Lee T. R. Lutkenhaus J. L. Kaplan D. Jones C. Bertozzi C. Kiessling L. Mulcahy M. B. Lindsley C. W. Finn M. G. Blum J. D. Kamat P. Aldrich C. C. Rowan S. Liu B. Liotta D. Weiss P. S. Zhang D. Ganesh K. N. Sexton P. Atwater H. A. Gooding J. J. Allen D. T. Voigt C. A. Sweedler J. Schepartz A. Rotello V. Lecommandoux S. Sturla S. J. Hammes-Schiffer S. Buriak J. Steed J. W. Wu H. Zimmerman J. Brooks B. Savage P. Tolman W. Hofmann T. F. Brennecke J. F. Holme T. A. Merz K. M. Scuseria G. Jorgensen W. Georg G. I. Wang S. Proteau P. Yates J. R. Stang P. Walker G. C. Hillmyer M. Taylor L. S. Odom T. W. Carreira E. Rossen K. Chirik P. Miller S. J. McCoy A. Shea J. E. Zanni M. Murphy C. Scholes G. Loo J. A. J. Am. Chem. Soc. 2020;142:8059–8060. doi: 10.1021/jacs.0c04253. [DOI] [PubMed] [Google Scholar]
- Walsh M. A. Douangamath A. Fearon D. Gehrtz P. Krojer T. Lukacik P. Owen D. Resnick E. Strain-Damerell C. Aimon A. Ábrányi P. Brandaõ-Neto J. Carbery A. Davison G. Dias A. Dunnett L. Fairhead M. Firth J. D. Jones S. Paul. Keely A. Keserü G. M. Klein H. F. Martin M. P. Noble M. E. M. Powell A. Reddi R. Skyner R. Snee M. Waring M. J. London N. von Delft F. Walsh M. A. bioRxiv. 2020 doi: 10.1101/2020.05.27.118117. [DOI] [Google Scholar]
- Mitsopoulos C. Di Micco P. Fernandez E. V. Dolciami D. Holt E. Mica I. L. Coker E. A. Tym J. E. Campbell J. Che K. H. Ozer B. Kannas C. Antolin A. A. Workman P. Al-Lazikani B. Nucleic Acids Res. 2021;49:D1074–D1082. doi: 10.1093/nar/gkaa1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J. Megill C. Bell S. M. Huddleston J. Potter B. Callender C. Sagulenko P. Bedford T. Neher R. A. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd M. H. ChemMedChem. 2019;14:1804–1809. doi: 10.1002/cmdc.201900565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfle M. Olliaro P. Todd M. H. Nat. Chem. 2011;3:745–748. doi: 10.1038/nchem.1149. [DOI] [PubMed] [Google Scholar]
- Arrowsmith C. H. Audia J. E. Austin C. Baell J. Bennett J. Blagg J. Bountra C. Brennan P. E. Brown P. J. Bunnage M. E. Buser-Doepner C. Campbell R. M. Carter A. J. Cohen P. Copeland R. A. Cravatt B. Dahlin J. L. Dhanak D. Edwards A. M. Frederiksen M. Frye S. V. Gray N. Grimshaw C. E. Hepworth D. Howe T. Huber K. V. M. Jin J. Knapp S. Kotz J. D. Kruger R. G. Lowe D. Mader M. M. Marsden B. Mueller-Fahrnow A. Müller S. O’Hagan R. C. Overington J. P. Owen D. R. Rosenberg S. H. Ross R. Roth B. Schapira M. Schreiber S. L. Shoichet B. Sundström M. Superti-Furga G. Taunton J. Toledo-Sherman L. Walpole C. Walters M. A. Willson T. M. Workman P. Young R. N. Zuercher W. J. Nat. Chem. Biol. 2015;11:536–541. doi: 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smil D. Wong J. F. Williams E. P. Adamson R. J. Howarth A. McLeod D. A. Mamai A. Kim S. Wilson B. J. Kiyota T. Aman A. Owen J. Poda G. Horiuchi K. Y. Kuznetsova E. Ma H. Hamblin J. N. Cramp S. Roberts O. G. Edwards A. M. Uehling D. Al-Awar R. Bullock A. N. O’Meara J. A. Isaac M. B. J. Med. Chem. 2020;63:10061–10085. doi: 10.1021/acs.jmedchem.0c01199. [DOI] [PubMed] [Google Scholar]
- JEDI – Joint European Disruptive Initiative – The European Darpa, https://www.jedi.foundation/, accessed 24 January 2021
- The COVID Box|Medicines for Malaria Venture, https://www.mmv.org/mmv-open/covid-box, accessed 24 January 2021
- COVID-19 Protein Portal, https://covid19proteinportal.org/index.html, accessed 24 January 2021
- CAPTURING THE COVID-19 DEMOGORGON (AKA SPIKE) IN ACTION – Folding@home, https://foldingathome.org/2020/04/03/capturing-the-covid-19-demogorgon-aka-spike-in-action/, accessed 24 January 2021
- Todd M. H. Tse E. G. Klug D. M. F1000Research. 2020;9:1043. doi: 10.12688/f1000research.26084.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Open COVID Pledge – Open Covid Pledge, https://opencovidpledge.org/, accessed 24 January 2021
- Huang R. Xu M. Zhu H. Chen C. Z. Zhu W. Lee E. M. He S. Zhang L. Zhao J. Shamim K. Bougie D. Huang W. Xia M. Hall M. D. Lo D. Simeonov A. Austin C. P. Qiu X. Tang H. Zheng W. Nat. Biotechnol. 2021;39:747–753. doi: 10.1038/s41587-021-00839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geronikaki A. Babaev E. Dearden J. Dehaen W. Filimonov D. Galaeva I. Krajneva V. Lagunin A. Macaev F. Molodavkin G. Poroikov V. Pogrebnoi S. Saloutin V. Stepanchikova A. Stingaci E. Tkach N. Vlad L. Voronina T. Bioorg. Med. Chem. 2004;12:6559–6568. doi: 10.1016/j.bmc.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Hu X. Shrimp J. H. Guo H. Zakharov A. Jain S. Shinn P. Simeonov A. Hall M. D. Shen M. ACS Pharmacol. Transl. Sci. 2021;4:1124–1135. doi: 10.1021/acsptsci.0c00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie A. M. New J. Chem. 2021;45:761–771. [Google Scholar]
- Zhang C. H. Stone E. A. Deshmukh M. Ippolito J. A. Ghahremanpour M. M. Tirado-Rives J. Spasov K. A. Zhang S. Takeo Y. Kudalkar S. N. Liang Z. Isaacs F. Lindenbach B. Miller S. J. Anderson K. S. Jorgensen W. L. ACS Cent. Sci. 2021;7:467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves V. M. Bobrowski T. Melo-Filho C. C. Korn D. Auerbach S. Schmitt C. Muratov E. N. Tropsha A. Mol. Inf. 2021;40:2000113. doi: 10.1002/minf.202000113. [DOI] [PubMed] [Google Scholar]