Background
Micronuclei is a cytogenetic term describing small nuclei composed of one to a few chromosomes or chromosomal fragments, which often result from abnormal cell divisions (Fenech et al., 2016). Micronuclei were discovered by Henry Howell and Justin Marie Jolly in erythrocytes over a century ago (Sears and Udden, 2012). Initially, Howell-Jolly bodies were used to describe the visible DNA fragments following the expulsion of the nucleus. Later on, the term “micronuclei” was adapted to describe all smaller fragments of the nucleus (1/5−1/20 the size of a normal nucleus) (Evans et al., 1959; Schmid, 1975). Micronuclei were initially correlated with conditions such as vitamin B12 and folate deficiency (Dawson and Bury, 1961). Soon thereafter, they were linked to many other genotoxic factors such as chemical exposure and radiation (Fenech et al., 2011). The presence of micronuclei is usually considered as an indicator of DNA damage and defects in mitosis.
Several different assays have been developed for the use of micronuclei. The cytokinesis-block, peripheral lymphocytic, and buccal MN assays have all been developed for use in mammalian cells in vivo (Sommer et al., 2020). Particularly, the development of the peripheral lymphocytic MN assay pioneered the use of micronuclei as a biomarker in mammalian cells with higher efficiency, along with a more rigorous definition for scoring (<1/3 of a nucleus, similar staining, no overlap, within 3–4 nuclear diameters from the main nucleus) (Countryman and Heddle, 1976).
Recently, enthusiasm for micronuclei studies has reemerged (Xie et al., 2016; Guo et al., 2019, 2020a,b; Fenech, 2020; Lepage et al., 2020; Mirzayans and Murray, 2020). First, the linkage to chromosomal instability, cancer, and other aging-related diseases has made micronuclei a potential potent biomarker (Aranda et al., 2018). Secondly, micronuclei, among many other previously ignored chromosomal abnormalities (most of which belong to the non-clonal chromosome aberrations or NCCAs), are key contributing factors for cancer by re-organizing the karyotype coding (Iourov et al., 2010, 2020; Heng et al., 2013; Ye et al., 2019a). Thirdly, micronuclei involve the activation of the immune system, further broadening the utility of micronuclei (Bartsch et al., 2017; Mackenzie et al., 2017; Kirsch-Volders et al., 2020). Finally, micronuclei are linked to stress response-mediated genome chaos, a driving force for cancer evolution (Heng, 2019; Ye et al., 2019b, 2021; Shoshani et al., 2020).
Micronuclei Clusters: Changing the System Inheritance
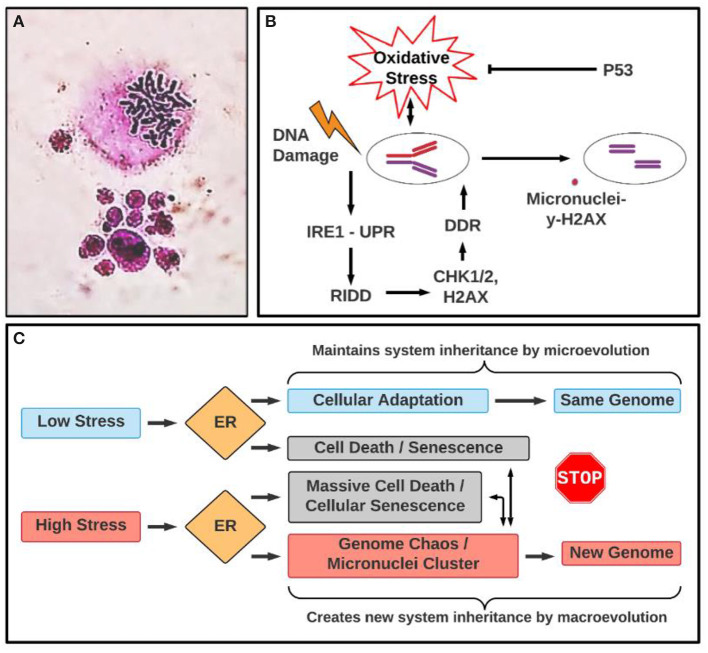
Micronuclei clusters are a group of micronuclei with variable size, which are often generated from one cell (either diploid or polyploid cell) (Figure 1A). These structures have been largely ignored, based on the reasoning that they represented cells that would soon be dead regardless. However, recent studies have demonstrated that micronuclei clusters belong to a type of chaotic genome, some of which can continue to evolve by cellular fusion and fission, representing important transitional structures for cellular macroevolution (Heng, 2015, 2019). In fact, the micronuclei cluster is most commonly observed in cancer samples, especially following drug treatment (Heng et al., 2008). Moreover, polyploid giant cancer cells (PGCCs) have generated excitement in cancer research lately, due to their contribution to rapid drug resistance and induced cancer lethality (Mirzayans et al., 2018; Erenpreisa et al., 2020; Mannan et al., 2020; Pienta et al., 2020a,b). PGCCs belong to numerical genome chaos, which can generate many aggressive near-diploid cancer cells with newly rearranged karyotypes (Heng et al., 2008; Liu et al., 2014). During this transition, PGCCs also can generate polyploid types of micronuclei clusters (Heng et al., 2013; Zhang et al., 2014). A model of how PGCCs contribute to drug resistance and aggressive cancer growth has been proposed, which involves high levels of cellular stress-induced abnormal developmental processes (dedifferentiation), genome chaos-mediated macroevolution (creation of new genome systems by PGCCs in smaller cell populations), and microevolution (growth of stable cancer populations) (Niu et al., 2016, 2017; Liu, 2018, 2020). This series of studies has also highlighted the importance of micronuclei, as they have now been shown to be initiators of genome instability and macroevolution, rather than just a reflection of genotoxic conditions. Along these lines, micronuclei clusters are often detected from drug treatment-induced genome chaos, including PGCCs. Clearly, the micronuclei cluster represents a means to change the karyotype coding, and by extension, the system inheritance (Ye et al., 2019b).
Figure 1.
Micronuclei cluster: morphology and involved pathways in ER stress. (A) Example of micronuclei cluster. The upper image is a mitotic figure and the lower image is a micronuclei cluster where different sized nuclei clustered with each other. Mitotic figure and micronuclei cluster was stained by Giemsa. (B) Illustrating a link between oxidative stress, unfolded protein response (UPR) and micronuclei formation under genotoxic stress. DDR, DNA damage response; RIDD: IRE1α-dependent decay. (C) A proposed conceptual relationship between different types of stresses, ER stress response, the types of evolution (macroevolution vs. microevolution). Even though ER response acts differently according to low and high stress, which are linked to microevolution and macroevolution, respectively, they are overlapping in terms of leading to senescence and genome chaos. For example, while high stress often can induce genome chaos including micronuclei clusters and senescence, coupling with massive death, low stress also could lead to death and senescence, which can lead to genome reorganization, albeit at a much lower rate. The association between senescence and ER stress response (UPR) is context-dependent. Indeed, senescence and ER stress response (UPR) make an interconnected network (UPR is activated in consequence to cell senescence or UPR is a driver of senescence) in which oxidative stress (ROS) acts as a central element responsible for an auto-amplification loop (Pluquet et al., 2015). The conceptual basis of this model is stress-induced genome reorganization that is essential for somatic evolution. Cellular stress in general, and ER stress in particular, can serve as an informational code that determines the cell's fate (Heng and Heng, 2021). Depending on the stress intensity or duration, UPR can trigger diverse cellular responses, including apoptotic pathways, which can be linked to genome chaos with the involvement of micronuclei clusters. This model also predicts the complex relationship between the UPR, genome reorganization, and functional relevance in cancer immunobalance. Even though the UPR may promote genome reorganization under acute or severe stress conditions, newly formed genomes can either be favorable or unfavorable for cancer cell malignancy depending on the genomic context. For example, the newly formed genomes, including micronuclei clusters, could either activate the immune system to fight against cancer or instead help cancer unexpectedly. It was reported that cell fusion can occur between cancer cells and immune cells (such as macrophages), promoting cancer cells to become more aggressive (Gast et al., 2018). In fact, a German gynecologist, Otto Aichel, first introduced this idea in 1911. It is likely that fused hybrid cells change their genomes through genome chaos including micronuclei clusters. According to the Genome Architecture Theory, such a mechanism of genome information creation under stress should be a universal phenomenon. However, for future research direction, quantitative studies are needed to predict the clinical odds (beneficial or harmful) under different treatment conditions.
Search For The Link Between Endoplasmic Reticulum (ER) Stress, Micronuclei, and Giant Cells
Interestingly, the cellular stress response machinery controls both the switching of genome stability and the dynamics leading to a spectrum of numerical and structural karyotypic variants (normal to chaotic) (Beaupere and Labunskyy, 2019; Heng, 2019; Limia et al., 2019). It is thus necessary to investigate how ER stress response impacts micronuclei and their triggered genome chaos (Heng et al., 2013; Zhang et al., 2015). The ER is responsible for the majority of cellular protein synthesis and folding while playing a key role in sensing cellular stress. For example, ER stress response or unfolded protein response (UPR) can either promote cellular survival or commit the cell to a pathway of apoptosis under different stress conditions. There are already two types of established linkages between intracellular stress and micronuclei: (1). Linking ER stress and oxidative stress responses to micronuclei/chromosome instability; (2). Linking ER stress to micronuclei clusters and polyploidy.
Most publications belong to the first category. For example, many studies suggest that micronuclei not only serve as an index of genotoxic effects and chromosomal instability (Guo et al., 2020b), but also are associated with the cellular stress response and immune activation following DNA damage (Chatterjee et al., 2018). Many biochemical and pathophysiological conditions, such as double-stranded DNA breaks, impaired DNA repair response, improper DNA replication, treatment of DNA adduct-forming chemicals, inhibition of microtubule polymerization, and centromere interference (Ye et al., 2019b; Guo et al., 2020a) can all directly or indirectly cause the formation of micronuclei. Studies further suggest that micronuclei can result from natural processes, such as metabolism and aging, and can be induced by environmental factors or irregular lifestyles (Luzhna et al., 2013; Nefic and Handzic, 2013). Over the past decade, mounting evidence suggests that micronucleus-causing genotoxicity is intrinsically linked to intracellular stress responses, particularly oxidative stress and ER stress responses (Luzhna et al., 2013; Horne et al., 2014; Hetz et al., 2020).
During mitotic exit, mis-segregated chromosomes can recruit their own nuclear envelope to form micronuclei. Over 60% of micronuclei undergo an irreversible loss of compartmentalization during interphase due to the collapse of the nuclear envelope (Hatch et al., 2013). This disruption of micronuclei reduces nuclear functions and can trigger massive DNA damage. Micronuclei disruption is associated with chromatin compaction and invasion of ER tubules into the chromatin. Disrupted micronuclei were identified in both major subtypes of human non-small cell lung cancer, solidifying evidence that disrupted micronuclei are useful objective biomarkers for genomic instability in cancer.
The process leading to the formation of micronuclei is associated with intracellular stress response and genotoxicity. It has been demonstrated that oxidative stress preferentially induces a subtype of micronuclei, specifically, the subclass marked by pan-staining of γ-H2AX or γ-H2AX (+), and mediates genomic instability caused by p53 dysfunction (Xu et al., 2014) (Figure 1B). Furthermore, reactive oxygen species (ROS) are known to cause many types of DNA lesions that can be converted into cancer-promoting genetic alterations. However, the tumor suppressor p53 plays an important role in regulating the generation of cellular ROS, by reducing oxidative stress under physiological or stress conditions. Indeed, in human and mouse cells that are deficient in p53, the frequencies of γ-H2AX (+) micronuclei are significantly elevated but can be attenuated by the antioxidant N-acetylcysteine (NAC). These findings implicate the importance of p53-regulated redox levels in the maintenance of genomic stability by preventing the formation of micronuclei.
Genotoxic stress-induced micronuclei formation is associated with the activation of ER stress response or UPR that modulates DNA damage repair programs and sustains cell survival (Dufey et al., 2020). DNA damage triggers the UPR signaling pathway mediated through the ER stress sensor inositol-requiring enzyme 1α (IRE1α), leading to the activation of regulated IRE1α-dependent decay (RIDD). The RIDD pathway sustains the activity of the key factors involved in the DNA damage response, including checkpoint kinase (CHK) 1, CHK2, and H2A.X Variant Histone (H2AX), therefore boosting the DNA damage response (DDR) (Figure 1B) (Dufey et al., 2020). Through modulation of DNA damage repair, cell cycle arrest, and apoptosis, UPR signaling through IRE1α may preserve genome stability and therefore protect the disruption of micronuclei under genotoxic stress. Additionally, ER stress and inflammatory responses are linked to genomic instability induced by gamma radiation (Chatterjee et al., 2018).
As of late, an increasing number of reports belonging to the second category that links ER stress to micronuclei clusters and polyploidy are being produced. Micronuclei clusters have been frequently observed in cancer, but many images of them were left unpublished due to their supposed lack of scientific worth (Hatch et al., 2013; Ye et al., 2019b). As the interaction of PGCCs and genome chaos-mediated macroevolution represents a newly emerging field in cancer research, we anticipate many studies of the same type to soon follow (Heng and Heng, 2020; Ye et al., 2021).
ER stress response has been linked to drug-induced-chromosome fragmentation, a type of mitotic cell death, and micronuclei clusters (Stevens et al., 2007, 2011). Similarly, ER stress response has been linked to chromosome instability in GWI (unpublished observation). Another noted study used the ER stress response to study mechanisms of genomic instability in polyploidization. Ploidy can activate ER stress response, resulting in anti-cancer immune responses. Immunoselection can then reduce ploidy, ER stress, and calreticulin exposure (Senovilla et al., 2012, 2017). It is thus interesting to study the complex relationship between ER stress, PGCCs, micronuclei clusters, and immune responses, which may yield new discoveries.
Future Perspective
The ER stress response represents a major regulating mechanism for cellular function under stress. Since both ER stress and micronuclei clusters can be linked to numerical molecular pathways, and the micronuclei are linked to genome chaos (Zhang et al., 2015; Ye et al., 2019b), we expect a sizable number of diverse molecular mechanisms to be published in the near future. It is thus essential to research the common mechanisms rather than focus on specific, individual ones, as in complex systems, there will always be too many genomic and environmental factors involved, with most contributing only moderately. Since CIN can be used to unify the triggering factors of cancer evolution including drug resistance (Ye et al., 2020, 2021), establishing the quantitative link between the ER stress response and CIN is a promising starting point. The quantitative data of micronuclei, especially of micronuclei clusters, is of importance when integrating into this platform.
Depending on the context, the ER stress response can play paradoxical roles. Under normal circumstances, it maintains system stability by repairing and eliminating unrepairable cellular elements or cells; under crisis, it might promote changes by incomplete cell death, and genome chaos (Heng, 2015, 2019). It is known that during cellular death, some outliers can form survivable genomes, and form new cellular populations. Furthermore, according to the Genome Architecture Theory, karyotype coding is maintained via the function of sexual reproduction and somatic genome instability (Gorelick and Heng, 2011; Ye et al., 2019a). Under crisis, however, the process of genome chaos can create new karyotype coding systems for speciation. It is thus timely to investigate the role played by ER stress response in this process. Similar opposing functions can be found in cancer immune surveillance as well. As mentioned previously, while ploidy can activate anti-cancer immune responses via the ER stress response (Senovilla et al., 2012), a high level of chromosomal structural variations can also suppress the immune response to cancer (Minton, 2012; Zanetti and Mahadevan, 2012; Zanetti, 2017). In other words, chromosomal chaos may promote or silence immune surveillance depending on different environmental and informational contexts. Consequently, how ER stress plays a role in the interplay between chromosomal abnormalities and immune surveillance is of importance in cancer research.
During tumorigenesis, high proliferation rates of cancer cells demand increased activities of ER protein folding and transport, a condition that triggers ER stress. As tumors grow, cancer cells experience nutrient starvation and hypoxia, which can induce the accumulation of unfolded or misfolded proteins in the ER and activation of the ER stress response (Hetz et al., 2020). It has been demonstrated that ER stress response is an important mechanism required for cancer cells to adapt to and survive from oncogenic stress conditions (Wang et al., 2010). Recently, the cancer problem, traditionally considered as an issue of out-of-control growth, has been rephrased as different phase transitions (from normal cells to transformed cells, from non-invasive tumor to cancer, and from drug-sensitive cancer cells to drug-resistant cells) (Heng and Heng, 2020). Accordingly, it would be interesting to investigate the ER homeostasis during these various phase transitions. Such types of information, including different ER stress response pathways, levels of overall stress, and types of genomic information involved (gene, epigenetic, and karyotype changes), are essential to understand the stress-information relationship in somatic and organismal evolution (Kültz, 2005, 2020; Heng, 2019). To initiate such an effort, a model that illustrates the relationship between stress levels, cellular responses, and types of evolution is proposed for future study (Figure 1C).
Author Contributions
EH, KZ, and HH drafted the manuscript. AM and GL participated in the initial observations and discussion. AM was a Summer Undergraduate Research Student of Center for Molecular Medicine and Genomics, Wayne State University. EH was a summer student. All authors involved in literature search and editing of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding. This work was partially supported by the Summer Undergraduate Research Program of CMMG, and by National Institutes of Health (NIH) grant DK090313.
References
- Aranda F., Chaba K., Bloy N., Garcia P., Bordenave C., Martins I., et al. (2018). Immune effectors responsible for the elimination of hyperploid cancer cells. Oncoimmunology 7:e1463947. 10.1080/2162402X.2018.1463947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch K., Knittler K., Borowski C., Rudnik S., Damme M., Aden K., et al. (2017). Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum. Mol. Genet. 26, 3960–3972. 10.1093/hmg/ddx283 [DOI] [PubMed] [Google Scholar]
- Beaupere C., Labunskyy V. M. (2019). (Un)folding mechanisms of adaptation to ER stress: lessons from aneuploidy. Curr. Genet. 65, 467–471. 10.1007/s00294-018-0914-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee J., Nairy R. K., Langhnoja J., Tripathi A., Patil R. K., Pillai P. P., et al. (2018). ER stress and genomic instability induced by gamma radiation in mice primary cultured glial cells. Metab Brain Dis. 33, 855–868. 10.1007/s11011-018-0183-9 [DOI] [PubMed] [Google Scholar]
- Countryman P. I., Heddle J. A. (1976). The production of micronuclei from chromosome aberrations in irradiated cultures of human lymphocytes. Mutat. Res. 41, 321–332. 10.1016/0027-5107(76)90105-6 [DOI] [PubMed] [Google Scholar]
- Dawson D. W., Bury H. P. (1961). The significance of Howell-Jolly bodies and giant metamyelocytes in marrow smears. J. Clin. Pathol. 14, 374–380. 10.1136/jcp.14.4.374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufey E., Bravo-San Pedro J. M., Eggers C., González-Quiroz M., Urra H., Sagredo A. I., et al. (2020). Genotoxic stress triggers the activation of IRE1α-dependent RNA decay to modulate the DNA damage response. Nat. Commun. 11:2401. 10.1038/s41467-020-15694-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erenpreisa J., Salmina K., Anatskaya O., Cragg M. S. (2020). Paradoxes of cancer: survival at the brink. Semin. Cancer Biol. 10.1016/j.semcancer.2020.12.009. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Evans H. J., Neary G. J., Williamson F. S. (1959) The relative biological efficiency of single doses of fast neutrons gamma-rays on vicia faba roots the effect of oxygen. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 3, 216–229. 10.1080/09553005914550311 [DOI] [PubMed] [Google Scholar]
- Fenech M. (2020). Cytokinesis-block micronucleus cytome assay evolution into a more comprehensive method to measure chromosomal instability. Genes. 11:1203. 10.3390/genes11101203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech M., Kirsch-Volders M., Natarajan A. T., Surralles J., Crott J. W., Parry J., et al. (2011). Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 26, 125–132. 10.1093/mutage/geq052 [DOI] [PubMed] [Google Scholar]
- Fenech M., Knasmueller S., Bolognesi C., Bonassi S., Holland N., Migliore L., et al. (2016). Molecular mechanisms by which in vivo exposure to exogenous chemical genotoxic agents can lead to micronucleus formation in lymphocytes in vivo and ex vivo in humans. Mutat Res. 770, 12–25. 10.1016/j.mrrev.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Gast C. E., Silk A. D., Zarour L., Riegler L., Burkhart J. G., Gustafson K. T., et al. (2018). Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci Adv. 4:eaat7828. 10.1126/sciadv.aat7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick R., Heng H. H. (2011). Sex reduces genetic variation: a multidisciplinary review. Evolution 65, 1088–1098. 10.1111/j.1558-5646.2010.01173.x [DOI] [PubMed] [Google Scholar]
- Guo X., Dai X., Wu X., Zhou T., Ni J., Xue J., et al. (2020b). Understanding the birth of rupture-prone and irreparable micronuclei. Chromosoma 129, 181–200. 10.1007/s00412-020-00741-w [DOI] [PubMed] [Google Scholar]
- Guo X., Dai X., Zhou T., Wang H., Ni J., Xue J., et al. (2020a). Mosaic loss of human Y chromosome: what, how and why. Human Genet. 139, 421–446. 10.1007/s00439-020-02114-w [DOI] [PubMed] [Google Scholar]
- Guo X., Ni J., Liang Z., Xue J., Fenech M. F., Wang X. (2019). The molecular origins and pathophysiological consequences of micronuclei: new insights into an age-old problem. Mutat. Res. 779, 1–35. 10.1016/j.mrrev.2018.11.001 [DOI] [PubMed] [Google Scholar]
- Hatch E. M., Fischer A. H., Deerinck T. J., Hetzer M. W. (2013). Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60. 10.1016/j.cell.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng H. H. (2015). Debating Cancer: The Paradox in Cancer Research. Singapore: World Scientific Publishing Co. ISBN 978-981-4520-84-3. 10.1142/8879 [DOI] [Google Scholar]
- Heng H. H. (2019). Genome Chaos: Rethinking Genetics, Evolution, and Molecular Medicine. Cambridge, MA: Academic Press Elsevier. ISBN 978-012-8136-35-5. [Google Scholar]
- Heng H. H., Liu G., Stevens J. B., Abdallah B. Y., Horne S. D., Ye K. J., et al. (2013). Karyotype heterogeneity and unclassified chromosomal abnormalities. Cytogenet. Genome Res. 139, 144–157. 10.1159/000348682 [DOI] [PubMed] [Google Scholar]
- Heng H. H., Stevens J. B., Lawrenson L., Liu G., Ye K. J., Bremer S. W., et al. (2008). Patterns of genome dynamics and cancer evolution. Cell. Oncol. 30, 513–514. 10.1155/2008/267326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng J., Heng H. H. (2020). Genome chaos: creating new genomic information essential for cancer macroevolution. Semin. Cancer Biol. 10.1016/j.semcancer.2020.11.003. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Heng J., Heng H. H. (2021). Karyotype coding: the creation and maintenance of system information for complexity and biodiversity. Biosystem 208:104476. 10.1016/j.biosystems.2021.104476 [DOI] [PubMed] [Google Scholar]
- Hetz C., Zhang K., Kaufman R. J. (2020). Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21, 421–438. 10.1038/s41580-020-0250-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne S. D., Chowdhury S. K., Heng H. H. (2014). Stress, genomic adaptation, and the evolutionary trade-off. Front. Genet. 5:92. 10.3389/fgene.2014.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iourov I. Y., Vorsanova S. G., Yurov Y. B. (2010). Somatic genome variations in health and disease. Curr. Genomics 11, 387–396. 10.2174/138920210793176065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iourov I. Y., Vorsanova S. G., Yurov Y. B., Zelenova M. A., Kurinnaia O. S., Vasin K. S., et al. (2020). The cytogenomic “Theory of Everything”: chromohelkosis may underlie chromosomal instability and mosaicism in disease and aging. Int. J. Mol. Sci. 21:8328. 10.3390/ijms21218328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch-Volders M., Bolognesi C., Ceppi M., Bruzzone M., Fenech M. (2020). Micronuclei, inflammation and auto-immune disease. Mutat. Res. 786:108335. 10.1016/j.mrrev.2020.108335 [DOI] [PubMed] [Google Scholar]
- Kültz D. (2005). Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 67, 225–257. 10.1146/annurev.physiol.67.040403.103635 [DOI] [PubMed] [Google Scholar]
- Kültz D. (2020). Evolution of cellular stress response mechanisms. J. Exp. Zool. A Ecol. Integrative Physiol. 333, 359–378. 10.1002/jez.2347 [DOI] [PubMed] [Google Scholar]
- Lepage C. C., Thompson L. L., Larson B., McManus K. J. (2020). An automated, single cell quantitative imaging microscopy approach to assess micronucleus formation, genotoxicity and chromosome instability. Cells 9:344. 10.3390/cells9020344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limia C. M., Sauzay C., Urra H., Hetz C., Chevet E., Avril T. (2019). Emerging roles of the endoplasmic reticulum associated unfolded protein response in cancer cell migration and invasion. Cancers 11:631. 10.3390/cancers11050631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Stevens J., Horne S., Abdallah B. Y., Ye K. J., Bremer S. W., et al. (2014). Genome chaos: Survival strategy during crisis. Cell Cycle 13, 528–537. 10.4161/cc.27378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. (2018). The dualistic origin of human tumors. Semin. Cancer Biol. 53, 1–16. 10.1016/j.semcancer.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. (2020). The “life code”: a theory that unifies the human life cycle and the origin of human tumors. Semin. Cancer Biol. 60, 380–397. 10.1016/j.semcancer.2019.09.005 [DOI] [PubMed] [Google Scholar]
- Luzhna L., Kathiria P., Kovalchuk O. (2013). Micronuclei in genotoxicity assessment: from genetics to epigenetics and beyond. Front. Genet. 4:131. 10.3389/fgene.2013.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie K. J., Carroll P., Martin C. A., Murina O., Fluteau A., Simpson D. J., et al. (2017). cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465. 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannan R., Wang X., Bawa P. S., Spratt D. E., Wilson A., Jentzen J., et al. (2020). Polypoidal giant cancer cells in metastatic castration-resistant prostate cancer: observations from the Michigan Legacy Tissue Program. Med. Oncol. 37:16. 10.1007/s12032-020-1341-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton K. (2012). Tumour immunology: chromosome overload. Nat. Rev. Immunol. 12:745. 10.1038/nri3326 [DOI] [PubMed] [Google Scholar]
- Mirzayans R., Andrais B., Murray D. (2018). Roles of polyploid/multinucleated giant cancer cells in metastasis and disease relapse following anticancer treatment. Cancers 10:118. 10.3390/cancers10040118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayans R., Murray D. (2020). Do TUNEL and other apoptosis assays detect cell death in preclinical studies?. Int. J. Mol. Sci. 21:9090. 10.3390/ijms21239090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefic H., Handzic I. (2013). The effect of age, sex, and lifestyle factors on micronucleus frequency in peripheral blood lymphocytes of the Bosnian population. Mutat. Res. 753, 1–11. 10.1016/j.mrgentox.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Niu N., Mercado-Uribe I., Liu J. (2017). Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells. Oncogene 36, 4887–4900. 10.1038/onc.2017.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N., Zhang J., Zhang N., Mercado-Uribe I., Tao F., Han Z., et al. (2016). Linking genomic reorganization to tumor initiation via the giant cell cycle. Oncogenesis 5:e281. 10.1038/oncsis.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienta K. J., Hammarlund E. U., Austin R. H., Axelrod R., Brown J. S., Amend S. R. (2020a). Cancer cells employ an evolutionarily conserved polyploidization program to resist therapy. Semin. Cancer Biol. 10.1016/j.semcancer.2020.11.016. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Pienta K. J., Hammarlund E. U., Axelrod R., Amend S. R., Brown J. S. (2020b). Convergent evolution, evolving evolvability, and the origins of lethal cancer. Mol. Cancer Res. 18, 801–810. 10.1158/1541-7786.MCR-19-1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluquet O., Pourtier A., Abbadie C. (2015). The unfolded protein response and cellular senescence. A review in the theme: cellular mechanisms of endoplasmic reticulum stress signaling in health and disease. Am. J. Physiol. Cell Physiol. 308, C415–C425. 10.1152/ajpcell.00334.2014 [DOI] [PubMed] [Google Scholar]
- Schmid W. (1975). The micronucleus test. Mutat. Res. 31, 9–15. 10.1016/0165-1161(75)90058-8 [DOI] [PubMed] [Google Scholar]
- Sears D. A., Udden M. M. (2012). Howell-Jolly bodies: a brief historical review. Am. J. Med. Sci. 343, 407–409. 10.1097/MAJ.0b013e31823020d1 [DOI] [PubMed] [Google Scholar]
- Senovilla L., Demont Y., Humeau J., Bloy N., Kroemer G. (2017). Image cytofluorometry for the quantification of ploidy and endoplasmic reticulum stress in cancer cells. Methods Mol. Biol. 1524, 53–64. 10.1007/978-1-4939-6603-5_3 [DOI] [PubMed] [Google Scholar]
- Senovilla L., Vitale I., Martins I., Tailler M., Pailleret C., Michaud M., et al. (2012). An immunosurveillance mechanism controls cancer cell ploidy. Science 337, 1678–1684. 10.1126/science.1224922 [DOI] [PubMed] [Google Scholar]
- Shoshani O., Brunner S. F., Yaeger R., Ly P., Nechemia-Arbely Y., Kim D. H., et al. (2020). Chromothripsis drives the evolution of gene amplification in cancer. Nature 591, 137–141. 10.1038/s41586-020-03064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S., Buraczewska I., Kruszewski M. (2020). Micronucleus assay: the state of art, and future directions. Int. J. Mol. Sci. 21:1534. 10.3390/ijms21041534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. B., Abdallah B. Y., Liu G., Ye C. J., Horne S. D., Wang G., et al. (2011). Diverse system stresses: common mechanisms of chromosome fragmentation. Cell Death Dis. 2:e178. 10.1038/cddis.2011.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. B., Liu G., Bremer S. W., Ye K. J., Xu W., Xu J., et al. (2007). Mitotic cell death by chromosome fragmentation. Cancer Res. 67, 7686–7694. 10.1158/0008-5472.CAN-07-0472 [DOI] [PubMed] [Google Scholar]
- Wang G., Yang Z. Q., Zhang K. (2010). Endoplasmic reticulum stress response in cancer: molecular mechanism and therapeutic potential. Am. J. Transl. Res. 2, 65–74. [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Ye S., Zhang J., He M., Dong C., Tu W., et al. (2016). Protective effect of mild endoplasmic reticulum stress on radiation-induced bystander effects in hepatocyte cells. Sci. Rep. 6:38832. 10.1038/srep38832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Wang W., Guo H., Sun Z., Wei Z., Zhang X., et al. (2014). Oxidative stress preferentially induces a subtype of micronuclei and mediates the genomic instability caused by p53 dysfunction. Mutat. Res. 770, 1–8. 10.1016/j.mrfmmm.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C. J., Sharpe Z., Alemara S., Mackenzie S., Liu G., Abdallah B., et al. (2019b). Micronuclei cluster and genome chaos: changing the system inheritance. Genes 10:366. 10.3390/genes10050366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C. J., Sharpe Z., Heng H. H. (2020). Origins and consequences of chromosomal instability: from cellular adaptation to genome chaos-mediated system survival. Genes 11:1162. 10.3390/genes11101162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C. J., Stilgenbauer L., Moy A., Liu G., Heng H. H. (2019a). What is karyotype coding and why is genomic topology important for cancer and evolution?. Front. Genet. 10:1082. 10.3389/fgene.2019.01082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. C., Horne S., Zhang J. Z., Jackson L., Heng H. H. (2021). Therapy induced genome chaos: a novel mechanism of rapid cancer drug resistance. Front. Cell Dev. Biol. 9:676344. 10.3389/fcell.2021.676344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M. (2017). Chromosomal chaos silences immune surveillance. Science, New York, NY. 355, 249–250. 10.1126/science.aam5331 [DOI] [PubMed] [Google Scholar]
- Zanetti M., Mahadevan N. R. (2012). Cancer. immune surveillance from chromosomal chaos? Science 337, 1616–1617. 10.1126/science.1228464 [DOI] [PubMed] [Google Scholar]
- Zhang C. Z., Spektor A., Cornils H., Francis J. M., Jackson E. K., Liu S., et al. (2015). Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184. 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Mercado-Uribe I., Xing Z., Sun B., Kuang J., Liu J. (2014). Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene 33, 116–128. 10.1038/onc.2013.96 [DOI] [PMC free article] [PubMed] [Google Scholar]