Abstract
Blocking the expression of programmed cell death ligand 1 (PD-L1) is a promising approach for the treatment of colon cancer. The binding of PD-L1 to its receptor programmed cell death 1 (PD-1) on immune cells leads to the apoptosis of activated T cells and causes immune escape. However, there is a limited number of patients with colon cancer that can benefit from the inhibition of PD-L1, and the regulation of PD-L1 expression is poorly understood in colon cancer. The present study demonstrated that interleukin-22 (IL-22) and PD-L1 were upregulated in colon cancer tissues and there was a positive correlation between IL-22 expression and PD-L1 expression. In the present study, exogenous IL-22 was found to upregulate PD-L1 expression via the signal transducer and activator of transcription 3 signaling pathway in human colon cancer cells (DLD-1 and primary colon cancer cells). The results of the present study revealed a novel regulatory mechanism of PD-L1 expression in colon cancer, which provides a theoretical basis for decreasing the immune tolerance of colon cancer via IL-22 overexpression.
Keywords: PD-L1, IL-22, colon cancer, STAT3
Introduction
Colon cancer is a common gastrointestinal tumor, which ranks fourth in terms of incidence and third in terms of mortality worldwide (1). The incidence and mortality rates of colon cancer are rapidly increasing worldwide, with the exception of a few developed countries (1). To date, the common and effective treatment for colon cancer includes radical excision at the early stages, followed by chemotherapy and/or radiotherapy for patients at advanced stages, which are invasive treatments associated with adverse side effects (2). Recently, immune checkpoint molecules programmed cell death-1 (PD-1) and programmed cell death ligand 1 (PD-L1) have been identified as promising targets for immunotherapy in colon cancer (3).
PD-L1 is widely expressed in the human body, and PD-L1 can be expressed by tumor cells and tumor stroma as a type I transmembrane protein (4). As a co-inhibitory molecule on T cells, PD-1 binds to PD-L1 on the surface of tumor cells or stromal cells, which leads to the apoptosis of activated T cells and causes immune escape (5). The expression of PD-L1 is significantly elevated in colon cancer tissues (6,7), and positive PD-L1 expression is an independent risk factor for poor prognosis (8). Anti-PD-1 or anti-PD-L1 therapy recovers the anti-tumor activity of immune cells, which have been proven effective in various types of solid tumors, including colon cancer with high PD-L1 expression and microsatellite instability (9). Nevertheless, the majority of patients with colon cancer are in mismatch repair proficient or microsatellite stability subtypes, and thus fail to respond to anti-PD-1 or anti-PD-L1 therapy (10). Therefore, further exploration of the regulatory mechanism of PD-1/PD-L1 is a required for the application of immune checkpoint protein inhibitors in colon cancer.
As a member of the interleukin (IL)-10 cytokine family, IL-22 plays an important role in the occurrence and development of colon cancer. IL-22 is mainly distributed in cytoplasm and stroma, which is secreted by various immune cells and binds to the IL-22 receptor complex that leads to the activation of signal transducer and activator of transcription 3 (STAT3) (11). IL-22 promotes the proliferation, migration, chemotherapy resistance and stemness of colon cancer by activating the STAT3 pathway (12–14). In a previous study, it was found that IL-22 was involved in the aerobic glycolysis of colon cancer through STAT3 phosphorylation (15). STAT3 is an important signaling mechanism that regulates PD-L1 expression in tumor cells (16). Meanwhile, Seki et al (17) reported that IL-22 was associated with the regulation of PD-L1 expression in airway epithelial cells via a STAT3-dependent mechanism. However, as an activator of the STAT3 signaling pathway, the effect of IL-22 on PD-L1 expression in colon cancer is still unclear. The aim of the present study was to preliminary explore the association between IL-22 and PD-L1 in vivo and in vitro, and briefly elucidate the mechanism.
Materials and methods
Clinical samples
A total of 23 fresh tissue specimens were obtained from patients who had received a pathological diagnosis of colon cancer between August 2019 and November 2019. The patients included 13 males and 10 females, ranging between 43 and 75 years with a mean age of 58.6 years. Tumor tissues and adjacent normal tissues (2 cm away from the tumor) were collected with RNase-free centrifuge tubes, and immediately placed into liquid nitrogen. Written informed consent was obtained from all patients who provided the samples for the present study. This study was approved by the Ethics Committee of The First Affiliated Hospital of Shandong First Medical University.
Cell culture
Two primary colon cancer cell lines (WRCA and JRCA) were provided by Professor Weiping Zou (University of Michigan, USA) (14). The colon cancer cell line DLD-1 was obtained from the American Type Culture Collection. Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) in a humidified incubator with 5% CO2 atmosphere at 37°C. To test the role of STAT3 in the expression of PD-L1 induced by IL-22, DLD-1 cells were stimulated with IL-22 (10 ng/ml; PeproTech, Inc.) for 24 h after pre-stimulation with Sttatic (5 µM; Selleck Chemicals) for 12 h. The optimal Sttatic concentration (5 µM) was selected from the concentration gradient (0, 2.5, 5 and 10 µM).
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from clinical tissues and colon cancer cell DLD-1 with RNAiso Plus reagent (Takara Bio, Inc.) and reverse transcribed into cDNA with PrimeScript™ RT Master Mix kit (Takara Bio, Inc.), according to the manufacturer's instructions. qPCR was performed using the SYBR green method (Takara Bio, Inc.) on the StepOnePlus™ Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). Primer sequences obtained from PrimerBank and were as follows: PD-L1 forward, GATACAAACTCAAAGAAGCAAAG and reverse, CAAAATAAATAGGAAAAACTCAT; IL-22 forward, GCAGGCTTGACAAGTCCAACT and reverse, GCCTCCTTAGCCAGCATGAA; β-actin forward, TGGCACCCAGCACAATGAA and reverse CTAAGTCATAGTCCGCCTAGAAGC A. The thermocycling conditions were as follows: Initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 62°C for 20 sec and 72°C for 10 sec. The mRNA expression of PD-L1 and IL-22 were normalized to the expression of β-actin. The relative expression of mRNA was calculated using the2−∆∆Cq method (18).
Flow cytometry analysis
Cells were digested with pancreatin for single-cell suspension and incubated with APC-A-conjugated mouse anti-human PD-L1 antibody (cat. no. 329708; 1:100; BioLegend, Inc.) or isotype control antibody (cat. no. 401210; 1:100; BioLegend, Inc.) for 30 min at 4°C. Cells were washed twice with PBS and resuspended to detect PD-L1 expression on the surface of tumor cells using FACS Aria II (BD Biosciences) and analyzed using FlowJo software (version 7.6.1; FlowJo LLC).
Western blotting analysis
Protein was extracted from cells with RIPA lysis buffer (Beyotime Institute of Biotechnology), and a BCA assay kit (Beyotime Institute of Biotechnology) was used to determine the protein concentration. Protein extracts (10 µg/lane) were separated via 10% SDS-PAGE, and subsequently transferred to polyvinylidene fluoride membranes. Membranes were blocked with 5% non-fat dry milk for 1 h at room temperature, and then incubated overnight at 4°C with the following primary antibodies: Anti-PD-L1 antibody (cat. no. 13684T), anti-phosphorylated STAT3 (Tyr705) antibody (cat. no. 9145T), anti-STAT3 antibody (cat. no. 12640S) (all 1:1,000; Cell Signaling Technology, Inc.) and anti-β-actin (cat. no. A3853; 1:4,000; Sigma-Aldrich; Merck KGaA). The membranes were incubated with a horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature and visualized with electrochemiluminescence (cat. no. A38555; Thermo Fisher Scientific, Inc.). Protein levels were normalized to the level of β-actin.
Statistical analysis
A paired Student's t-test was performed to compare data from clinical samples. ANOVA and Tukey's post hoc tests were applied to identify significant differences between the indicated cell groups. The correlation between IL-22 and PD-L1 mRNA expression levels was calculated with a Pearson's correlation analysis. The data are expressed as the mean ± standard deviation (SD). P<0.05 was considered to indicate a statistically significant difference.
Results
mRNA expression of PD-L1 is positively correlated with IL-22 expression in colon cancer tissues
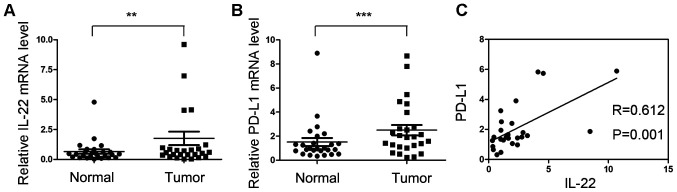
The mRNA expression of IL-22 and PD-L1 were investigated in 23 colon cancer tissues and adjacent normal tissues. The mRNA expression of IL-22 in tumor tissues was significantly higher compared with that of normal tissues (P<0.05; Fig. 1A). The relative mRNA level of PD-L1 was significantly upregulated in tumor tissues (P<0.05; Fig. 1B). Correlation analysis indicated that the mRNA expression of IL-22 was positively correlated with PD-L1 (r=0.612; P=0.001; Fig. 1C).
Figure 1.
Correlation analysis between IL-22 and PD-L1 mRNA expression in colon cancer tissues. (A and B) Total RNA was extracted from clinical tissues and the mRNA expression of IL-22 and PD-L1 were analyzed by reverse transcription-quantitative PCR in colon cancer tissues and adjacent normal tissues. (C) The correlation between IL-22 and PD-L1 mRNA expression was calculated by Pearson's correlation analysis. **P<0.01, ***P<0.001. IL-22, interleukin-22; PD-L1 programmed cell death ligand 1.
IL-22 promotes the expression of PD-L1 in colon cancer cells
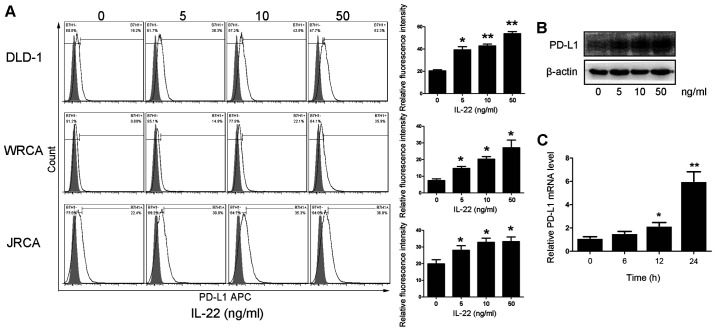
In order to evaluate the effect of IL-22 on the expression level of PD-L1 in colon cancer cells, primary colon cancer cells and DLD-1 cells were treated with different concentrations of IL-22 (0, 5, 10 and 50 ng/ml) for 24 h. The protein expression of PD-L1 was significantly upregulated in colon cancer cells in a dose-dependent manner (Fig. 2A and B). At the transcription level, IL-22 promoted the mRNA expression of PD-L1 in DLD-1 cells in a dose-dependent manner (Fig. 2C).
Figure 2.
Effect of IL-22 on the expression of PD-L1 in colon cancer cells. Primary colon cancer cells and DLD-1 cells were stimulated with IL-22 at the indicated concentration (5–50 ng/ml) for 24 h. The expression of PD-L1 was tested by (A) flow cytometry, (B) western blotting and (C) reverse transcription-quantitative PCR. *P<0.05 and **P<0.01. PD-L1, programmed cell death ligand 1; IL-22, interleukin-22.
IL-22 activates STAT3 in colon cancer cells
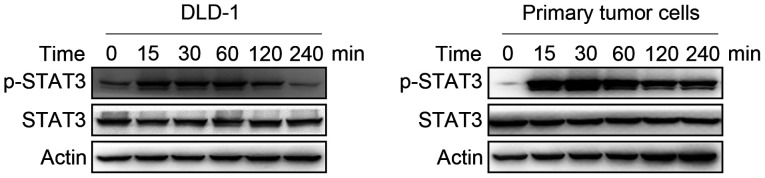
It is commonly known that STAT3 is an important component of the signaling pathway induced by IL-22. Primary colon cancer cells and DLD-1 cells were treated with IL-22 (10 ng/ml) for 15, 30, 60, 120 and 240 min. STAT3 phosphorylated was increased by IL-22 in colon cancer cells up to 4 h (Fig. 3).
Figure 3.
Effect of IL-22 on the phosphorylation of STAT3 in colon cancer cells. Cells were stimulated with IL-22 (10 ng/ml) for 15, 30, 60, 120 and 240 min. The expression of total and p-STAT3 was examined by western blotting in primary colon cancer cells and DLD-1 cells. IL-22, interleukin-22; p-, phosphorylated.
STAT3 is involved in IL-22-induced PD-L1 expression
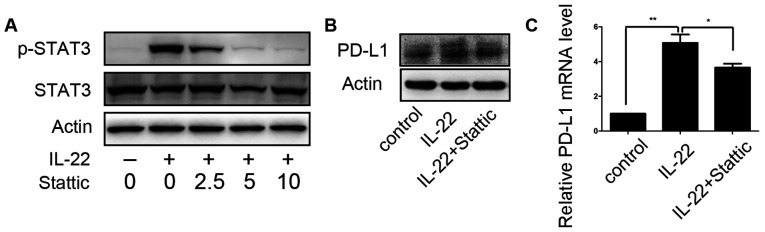
STAT3 small-molecule inhibitor Stattic was employed to assess the role of STAT3 activation in the regulation of PD-L1 expression of colon cancer cells by IL-22. After DLD-1 cells were pretreated with different concentrations of Stattic (0, 2.5, 5 and 10 µM) for 12 h, IL-22 (10 ng/ml) was added to the culture medium for 60 min. At a concentration of 5.0 µM, Stattic efficiently inhibited STAT3 phosphorylation, as shown in Fig. 4A. Thus, DLD-1 cells were stimulated with IL-22 (10 ng/ml) for 24 h after pre-stimulation on with Sttatic (5 µM) for 12 h. IL-22 induced upregulation of PD-L1 expression was significantly attenuated (Fig. 4B).
Figure 4.
Suppression of IL-22-induced PD-L1 upregulation by STAT3 small-molecule inhibitor Stattic. (A) DLD-1 cells were stimulated with IL-22 (10 ng/ml) for 30 min after pre-stimulation with Sttatic at the indicated concentrations (2.5–10 µM) for 12 h. The expression levels of total and p-STAT3 were examined by western blotting. (B) DLD-1 cells were stimulated with IL-22 (10 ng/ml) for 24 h after pre-stimulation with Sttatic (5 µM) for 12 h. The expression of PD-L1 was examined by western blotting. (C) DLD-1 cells were stimulated with IL-22 (10 ng/ml) for 24 h after pre-stimulation with Sttatic (5 µM) for 12 h. The mRNA expression of PD-L1 was examined by reverse transcription-quantitative PCR. *P<0.05 and **P<0.01. IL-22, interleukin-22; p-, phosphorylated; PD-L1, programmed cell death ligand 1.
Discussion
Tumor occurrence is often accompanied by the failure of the immune surveillance system, namely immune escape of tumors. Immune checkpoints act as a central mediator of immunosuppression in the tumor microenvironment. PD-1 is an important immune checkpoint. PD-L1 is the principal ligand of PD-1, which is not only expressed on immune cells but also expressed on tumor cells (19). Therefore, the present study examined the mRNA expression of PD-L1 in colon cancer tissues. The expression of PD-L1 was elevated in cancer tissues, which was in line with the previous studies (6). It is commonly known that a number of cytokines can induce PD-L1 expression on tumor cells, especially interferon-γ (20–22). In the present study, it was found that the mRNA expression of PD-L1 was increased and positively correlated with IL-22 expression in colon cancer tissues. Meanwhile, experiments in vitro confirmed that exogenous IL-22 could induce an increase in the mRNA and protein expression level of PD-L1 in colon cancer cells. To the best of our knowledge, the present study is the first to investigate the effect of IL-22 on the expression of PD-L1 in colon cancer cells.
IL-22 is a unique cytokine that is produced by immune cells, but only acts on non-lymphoid cells, epithelial cells in particular (23). Interestingly, IL-22 always plays a protective role on epithelial cells regardless of whether they have gone bad. It has been demonstrated that IL-22 modulates the expression of numerous genes that encode proteins involved in tissue protection and the remodeling of normal colon epithelial cell (24). Furthermore, IL-22 facilitates the migration of immune cells to attack the pathogen by supporting the release of metalloproteinases (25). Numerous studies have indicated that IL-22 is involved in the occurrence and development of colon cancer via various different pathways. IL-22 could not only promote the proliferation, migration and invasion of colon cancer cells (12), but also maintain colon cancer stemness (14). PD-L1 acts as an immunosuppressor on colon cancer cells, which prevents surveillance and elimination by immune cells. Hence, there are theoretical foundations to support the notion that IL-22 promotes the expression of PD-L1, which plays a role in facilitating the development of colon cancer. However, the direct effect of IL-22 on immune cells is absence, which will be the focus of the future study.
The pro-tumorigenic potential of IL-22 is mostly mediated by STAT3, a well-established oncogene that induces the expression of a large number of genes involved in tumor development (26–28). STAT3 is also an important regulator of PD-L1 expression in tumors. A previous report indicated that fibroblast growth factor receptor 2 induces the expression of PD-L1 via the JAK/STAT3 signaling pathway in human colon cancer cells to increase the apoptosis of Jurkat T cells (29). A recent study reported that STAT3 inhibition activates an efficient immune response by decreasing PD-L1 expression in colon cancer cells (30). The present study also confirmed that STAT3 inhibition impaired IL-22-induced upregulation of PD-L1 expression in colon cancer cells. Of course, the regulatory mechanism of PD-L1 expression calls for further study.
In brief, the present findings revealed that IL-22 promoted the expression of PD-L1 in colon cancer cells by activating the STAT3 signaling pathway, which may attenuate anti-tumor immunity and thus promote tumor development.
Acknowledgements
The authors would like to thank Professor Fan Xiang (Department of Gastrointestinal Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China) for their technical assistance.
Glossary
Abbreviations
- PD-L1
programmed cell death ligand 1
- PD-1
programmed cell death 1
- IL-22
interleukin-22
- STAT3
signal transducer and activator of transcription 3
Funding Statement
This work was supported by the National Natural Science Foundation (grant no. 81903044), Jinan Science and Technology Innovation Development Program (grant no. 202019083) and the Dean Foundation of The First Affiliated Hospital of Shandong First Medical University (grant no. QYPY2019NSFC0805).
Funding
This work was supported by the National Natural Science Foundation (grant no. 81903044), Jinan Science and Technology Innovation Development Program (grant no. 202019083) and the Dean Foundation of The First Affiliated Hospital of Shandong First Medical University (grant no. QYPY2019NSFC0805).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
MT and LX contributed to the conception of the project. JC and YL contributed towards project design. Acquisition of data was by KX and YZ. XX, RH and QW conducted the molecular experiments. HY and ZC analyzed and interpreted the data. XX and RH wrote the original draft of the manuscript. JC and YL reviewed and editing the manuscript. XX and YL confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of The First Affiliated Hospital of Shandong First Medical University (approval no. 2020S003). Written informed consent was obtained from all patients who provided the samples for the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Body A, Prenen H, Latham S, Lam M, Tipping-Smith S, Raghunath A, Segelov E. The Role of Neoadjuvant Chemotherapy in Locally Advanced Colon Cancer. Cancer Manag Res. 2021;13:2567–2579. doi: 10.2147/CMAR.S262870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yaghoubi N, Soltani A, Ghazvini K, Hassanian SM, Hashemy SI. PD-1/ PD-L1 blockade as a novel treatment for colorectal cancer. Biomed Pharmacother. 2019;110:312–318. doi: 10.1016/j.biopha.2018.11.105. [DOI] [PubMed] [Google Scholar]
- 4.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, Freeman GJ, Sharpe AH. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med. 2017;214:895–904. doi: 10.1084/jem.20160801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masugi Y, Nishihara R, Yang J, Mima K, da Silva A, Shi Y, Inamura K, Cao Y, Song M, Nowak JA, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut. 2017;66:1463–1473. doi: 10.1136/gutjnl-2016-311421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyo JS, Ko SH, Ko YS, Kim NY. Clinicopathological significance of PD-L1 expression in colorectal cancer: Impact of PD-L1 expression on pFOXO1 expression. Pathol Res Pract. 2020;216:152764. doi: 10.1016/j.prp.2019.152764. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, He M, Zhou Y, Yang C, Wei S, Bian X, Christopher O, Xie L. The prognostic and clinicopathological roles of PD-L1 expression in colorectal cancer: A systematic review and meta-analysis. Front Pharmacol. 2019;10:139. doi: 10.3389/fphar.2019.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinicrope FA, Foster NR, Thibodeau SN, Marsoni S, Monges G, Labianca R, Kim GP, Yothers G, Allegra C, Moore MJ, et al. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103:863–875. doi: 10.1093/jnci/djr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolk K, Witte E, Witte K, Warszawska K, Sabat R. Biology of interleukin-22. Semin Immunopathol. 2010;32:17–31. doi: 10.1007/s00281-009-0188-x. [DOI] [PubMed] [Google Scholar]
- 12.Jiang R, Wang H, Deng L, Hou J, Shi R, Yao M, Gao Y, Yao A, Wang X, Yu L, Sun B. IL-22 is related to development of human colon cancer by activation of STAT3. BMC Cancer. 2013;13:59. doi: 10.1186/1471-2407-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun D, Lin Y, Hong J, Chen H, Nagarsheth N, Peng D, Wei S, Huang E, Fang J, Kryczek I, Zou W. Th22 cells control colon tumorigenesis through STAT3 and Polycomb Repression complex 2 signaling. Oncoimmunology. 2015;5:e1082704. doi: 10.1080/2162402X.2015.1082704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, Vatan L, Szeliga W, Dou Y, Owens S, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. 2014;40:772–784. doi: 10.1016/j.immuni.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Xiang F, Huang Y, Shi L, Hu C, Yang Y, Wang D, He N, Tao K, Wu K, Wang G. Interleukin-22 promotes aerobic glycolysis associated with tumor progression via targeting hexokinase-2 in human colon cancer cells. Oncotarget. 2017;8:25372–25383. doi: 10.18632/oncotarget.15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Crabill GA, Pritchard TS, McMiller TL, Wei P, Pardoll DM, Pan F, Topalian SL. Mechanisms regulating PD-L1 expression on tumor and immune cells. J Immunother Cancer. 2019;7:305. doi: 10.1186/s40425-019-0770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seki N, Kan-O K, Matsumoto K, Fukuyama S, Hamano S, Tonai K, Ota K, Inoue H, Nakanishi Y. Interleukin-22 attenuates double-stranded RNA-induced upregulation of PD-L1 in airway epithelial cells via a STAT3-dependent mechanism. Biochem Biophys Res Comm. 2017;494:242–248. doi: 10.1016/j.bbrc.2017.10.045. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Eppihimer MJ, Gunn J, Freeman GJ, Greenfield EA, Chernova T, Erickson J, Leonard JP. Expression and regulation of the PD-L1 immunoinhibitory molecule on microvascular endothelial cells. Microcirculation. 2002;9:133–145. doi: 10.1080/713774061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Yang L, Huang F, Zhang Q, Liu S, Ma L, You Z. Inflammatory cytokines IL-17 and TNF-α up-regulate PD-L1 expression in human prostate and colon cancer cells. Immunol Lett. 2017;184:7–14. doi: 10.1016/j.imlet.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian J, Wang C, Wang B, Yang J, Wang Y, Luo F, Xu J, Zhao C, Liu R, Chu Y. The IFN-γ/PD-L1 axis between T cells and tumor microenvironment: Hints for glioma anti-PD-1/PD-L1 therapy. J Neuroinflammation. 2018;15:290. doi: 10.1186/s12974-018-1330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha JH, Wang HL, Yang WH, Yen EY, Chang WC, et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest. 2019;129:3324–3338. doi: 10.1172/JCI126022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez P, Gronke K, Diefenbach A. A catch-22: Interleukin-22 and cancer. Eur J Immunol. 2018;48:15–31. doi: 10.1002/eji.201747183. [DOI] [PubMed] [Google Scholar]
- 24.Wolk K, Witte E, Wallace E, Döcke WD, Kunz S, Asadullah K, Volk HD, Sterry W, Sabat R. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: A potential role in psoriasis. Eur J Immunol. 2006;36:1309–1323. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- 25.Eyerich S, Eyerich K, Pennino D, Carbone T, Nasorri F, Pallotta S, Cianfarani F, Odorisio T, Traidl-Hoffmann C, Behrendt H, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119:3573–3585. doi: 10.1172/JCI40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galoczova M, Coates P, Vojtesek B. STAT3, stem cells, cancer stem cells and p63. Cell Mol Biol Lett. 2018;23:12. doi: 10.1186/s11658-018-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 28.Fathi N, Rashidi G, Khodadadi A, Shahi S, Sharifi S. STAT3 and apoptosis challenges in cancer. Int J Biol Macromol. 2018;117:993–1001. doi: 10.1016/j.ijbiomac.2018.05.121. [DOI] [PubMed] [Google Scholar]
- 29.Li P, Huang T, Zou Q. FGFR2 promotes expression of PD-L1 in colorectal cancer via the JAK/STAT3 signaling pathway. J Immunol. 2019;202:3065–3075. doi: 10.4049/jimmunol.1801199. [DOI] [PubMed] [Google Scholar]
- 30.Jahangiri A, Dadmanesh M. STAT3 inhibition reduced PD-L1 expression and enhanced antitumor immune responses. J Cell Physiol. 2020;235:9457–9463. doi: 10.1002/jcp.29750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.