Summary
Machine learning (ML) for classification and prediction based on a set of features is used to make decisions in healthcare, economics, criminal justice and more. However, implementing an ML pipeline including preprocessing, model selection, and evaluation can be time-consuming, confusing, and difficult. Here, we present mikropml (prononced “meek-ROPE em el”), an easy-to-use R package that implements ML pipelines using regression, support vector machines, decision trees, random forest, or gradient-boosted trees. The package is available on GitHub, CRAN, and conda.
Graphical Abstract
Statement of need
Most applications of machine learning (ML) require reproducible steps for data pre-processing, cross-validation, testing, model evaluation, and often interpretation of why the model makes particular predictions. Performing these steps is important, as failure to implement them can result in incorrect and misleading results (Teschendorff, 2019; Wiens et al., 2019).
Supervised ML is widely used to recognize patterns in large datasets and to make predictions about outcomes of interest. Several packages including caret (Kuhn, 2008) and tidymod els (Kuhn et al., 2020) in R, scikitlearn (Pedregosa et al., 2011) in Python, and the H2O autoML platform (H2O.ai, 2020) allow scientists to train ML models with a variety of algorithms. While these packages provide the tools necessary for each ML step, they do not implement a complete ML pipeline according to good practices in the literature. This makes it difficult for practitioners new to ML to easily begin to perform ML analyses.
To enable a broader range of researchers to apply ML to their problem domains, we created mikropml, an easy-to-use R package (R Core Team, 2020) that implements the ML pipeline created by Topçuoğlu et al. (Topçuoğlu et al., 2020) in a single function that returns a trained model, model performance metrics and feature importance. mikropml leverages the caret package to support several ML algorithms: linear regression, logistic regression, support vector machines with a radial basis kernel, decision trees, random forest, and gradient boosted trees. It incorporates good practices in ML training, testing, and model evaluation (Teschendorff, 2019; Topçuoğlu et al., 2020). Furthermore, it provides data preprocessing steps based on the FIDDLE (FlexIble Data-Driven pipeLinE) framework outlined in Tang et al. (Tang et al., 2020) and post-training permutation importance steps to estimate the importance of each feature in the models trained (Breiman, 2001; Fisher et al., 2018).
mikropml can be used as a starting point in the application of ML to datasets from many different fields. It has already been applied to microbiome data to categorize patients with colorectal cancer (Topçuoğlu et al., 2020), to identify differences in genomic and clinical features associated with bacterial infections (Lapp et al., 2020), and to predict gender-based biases in academic publishing (Hagan et al., 2020).
mikropml package
The mikropml package includes functionality to preprocess the data, train ML models, evaluate model performance, and quantify feature importance (Figure 1). We also provide vignettes and an example Snakemake workflow (Köster & Rahmann, 2012) to showcase how to run an ideal ML pipeline with multiple different train/test data splits. The results can be visualized using helper functions that use ggplot2 (Wickham, 2016).
Figure 1:
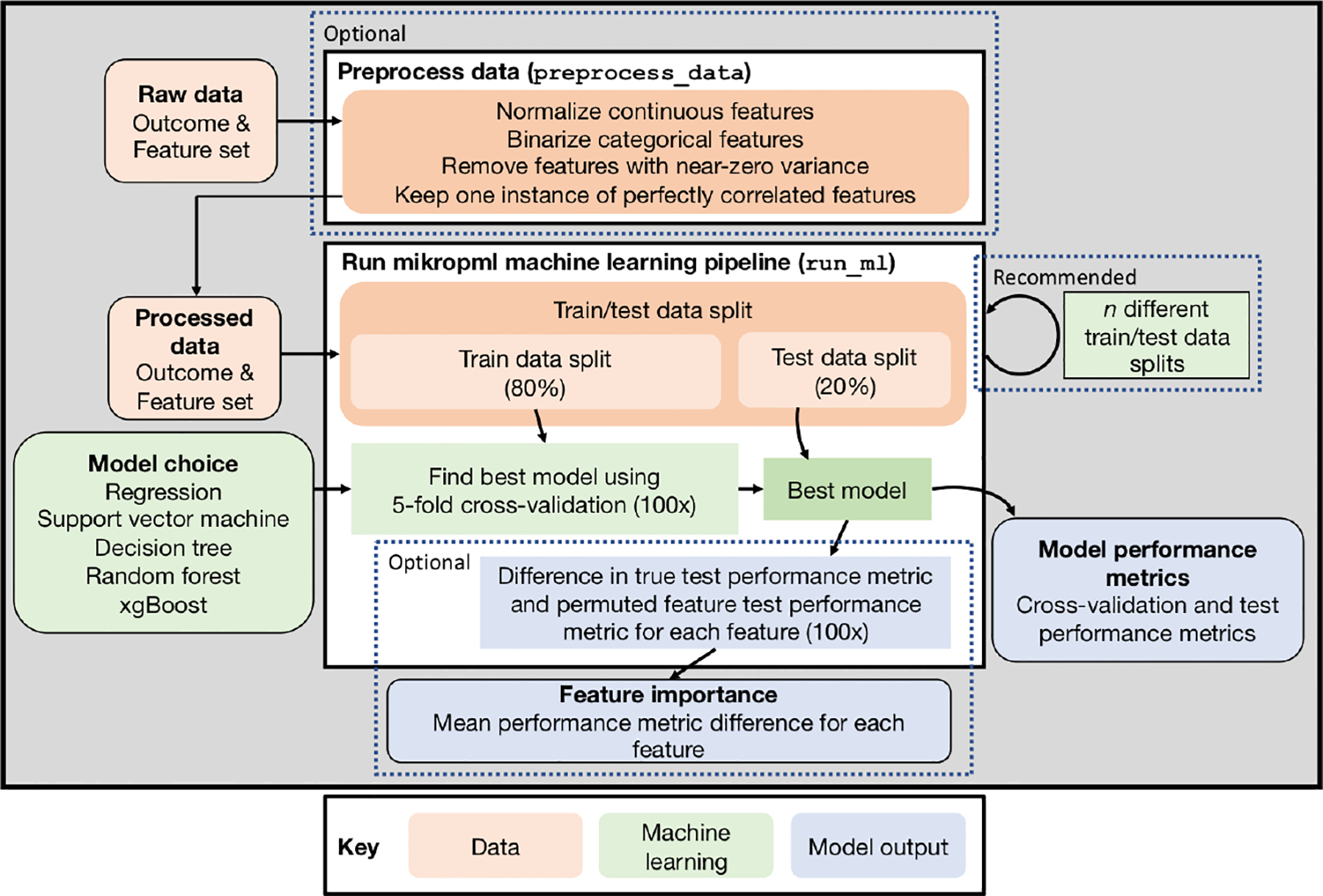
mikropml pipeline
While mikropml allows users to get started quickly and facilitates reproducibility, it is not a replacement for understanding the ML workflow which is still necessary when interpreting results (Pollard et al., 2019). To facilitate understanding and enable one to tailor the code to their application, we have heavily commented the code and have provided supporting documentation which can be read online.
Preprocessing data
We provide the function preprocess_data() to preprocess features using several different functions from the caret package. preprocess_data() takes continuous and categorical data, re-factors categorical data into binary features, and provides options to normalize continuous data, remove features with near-zero variance, and keep only one instance of perfectly correlated features. We set the default options based on those implemented in FIDDLE (Tang et al., 2020). More details on how to use preprocess_data() can be found in the accompanying vignette.
Running ML
The main function in mikropml, run_ml(), minimally takes in the model choice and a data frame with an outcome column and feature columns. For model choice, mikropml currently supports logistic and linear regression (glmnet: Friedman et al., 2010), support vector machines with a radial basis kernel (kernlab: Karatzoglou et al., 2004), decision trees (rpart: Therneau et al., 2019), random forest (randomForest: Liaw & Wiener, 2002), and gradient-boosted trees (xgboost: Chen et al., 2020). run_ml() randomly splits the data into train and test sets while maintaining the distribution of the outcomes found in the full dataset. It also provides the option to split the data into train and test sets based on categorical variables (e.g. batch, geographic location, etc.). mikropml uses the caret package (Kuhn, 2008) to train and evaluate the models, and optionally quantifies feature importance. The output includes the best model built based on tuning hyperparameters in an internal and repeated cross-validation step, model evaluation metrics, and optional feature importances. Feature importances are calculated using a permutation test, which breaks the relationship between the feature and the true outcome in the test data, and measures the change in model performance. This provides an intuitive metric of how individual features influence model performance and is comparable across model types, which is particularly useful for model interpretation (Topçuoğlu et al., 2020). Our introductory vignette contains a comprehensive tutorial on how to use run_ml().
Ideal workflow for running mikropml with many different train/test splits
To investigate the variation in model performance depending on the train and test set used (Lapp et al., 2020; Topçuoğlu et al., 2020), we provide examples of how to run_ml() many times with different train/test splits and how to get summary information about model performance on a local computer or on a high-performance computing cluster using a Snakemake workflow.
Tuning & visualization
One particularly important aspect of ML is hyperparameter tuning. We provide a reasonable range of default hyperparameters for each model type. However practitioners should explore whether that range is appropriate for their data, or if they should customize the hyperparameter range. Therefore, we provide a function plot_hp_performance() to plot the cross-validation performance metric of a single model or models built using different train/test splits. This helps evaluate if the hyperparameter range is being searched exhaustively and allows the user to pick the ideal set. We also provide summary plots of test performance metrics for the many train/test splits with different models using plot_model_performance(). Examples are described in the accompanying vignette on hyperparameter tuning.
Dependencies
mikropml is written in R (R Core Team, 2020) and depends on several packages: dplyr (Wickham et al., 2020), rlang (Henry et al., 2020) and caret (Kuhn, 2008). The ML algorithms supported by mikropml require: glmnet (Friedman et al., 2010), e1071 (Meyer et al., 2020), and MLmetrics (Yan, 2016) for logistic regression, rpart2 (Therneau et al., 2019) for decision trees, randomForest (Liaw & Wiener, 2002) for random forest, xgboost (Chen et al., 2020) for xgboost, and kernlab (Karatzoglou et al., 2004) for support vector machines. We also allow for parallelization of cross-validation and other steps using the foreach, doFuture, future.apply, and future packages (Bengtsson & Team, 2020). Finally, we use ggplot2 for plotting (Wickham, 2016).
Acknowledgments
We thank members of the Schloss Lab who participated in code clubs related to the initial development of the pipeline, made documentation improvements, and provided general feedback. We also thank Nick Lesniak for designing the mikropml logo.
We thank the US Research Software Sustainability Institute (NSF #1743188) for providing training to KLS at the Winter School in Research Software Engineering.
Funding
Salary support for PDS came from NIH grant 1R01CA215574. KLS received support from the NIH Training Program in Bioinformatics (T32 GM070449). ZL received support from the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE 1256260. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Conflicts of interest
None.
References
- Bengtsson H, & Team RC (2020). Future.apply: Apply Function to Elements in Parallel using Futures.
- Breiman L (2001). Random forests. Machine Learning, 45(1), 5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- Chen T, He T, Benesty M, Khotilovich V, Tang Y, Cho H, Chen K, Mitchell R, Cano I, Zhou T, Li M, Xie J, Lin M, Geng Y, Li Y, & implementation), X. contributors (base. X. (2020). Xgboost: Extreme Gradient Boosting.
- Fisher A, Rudin C, & Dominici F (2018). All models are wrong, but many are useful: Learning a variable’s importance by studying an entire class of prediction models simultaneously. [PMC free article] [PubMed]
- Friedman JH, Hastie T, & Tibshirani R (2010). Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software, 33(1), 1–22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- H2O.ai. (2020). H2O: Scalable machine learning platform [Manual].
- Hagan AK, Topçuoğlu BD, Gregory ME, Barton HA, & Schloss PD (2020). Women Are Underrepresented and Receive Differential Outcomes at ASM Journals: A Six-Year Retrospective Analysis. mBio, 11(6). 10.1128/mBio.01680-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry L, Wickham H, & RStudio. (2020). Rlang: Functions for Base Types and Core R and ‘Tidyverse’ Features.
- Karatzoglou A, Smola A, Hornik K, & Zeileis A (2004). Kernlab - An S4 Package for Kernel Methods in R. Journal of Statistical Software, 11(1), 1–20. 10.18637/jss.v011.i09 [DOI] [Google Scholar]
- Köster J, & Rahmann S (2012). Snakemakea scalable bioinformatics workflow engine.Bioinformatics, 28(19), 2520–2522. 10.1093/bioinformatics/bts480 [DOI] [PubMed] [Google Scholar]
- Kuhn M (2008). Building Predictive Models in R Using the caret Package. Journal of Statistical Software, 28(1), 1–26. 10.18637/jss.v028.i0527774042 [DOI] [Google Scholar]
- Kuhn M, Wickham H, & RStudio. (2020). Tidymodels: Easily Install and Load the ‘Tidymodels’ Packages.
- Lapp Z, Han J, Wiens J, Goldstein EJ, Lautenbach E, & Snitkin E (2020). Machine learning models to identify patient and microbial genetic factors associated with carbapenem-resistant Klebsiella pneumoniae infection. medRxiv, 2020.07.06.20147306. 10.1101/2020.07.06.20147306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, & Wiener M (2002). Classification and Regression by randomForest. 2, 5. [Google Scholar]
- Meyer D, Dimitriadou E, Hornik K, Weingessel A, Leisch F, C++-code), C.-C. C. (libsvm., & C++-code), C.-C. L. (libsvm. (2020). E1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien.
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, & Duchesnay É (2011). Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research, 12(85), 2825–2830. [Google Scholar]
- Pollard TJ, Chen I, Wiens J, Horng S, Wong D, Ghassemi M, Mattie H, Lindemer E, & Panch T (2019). Turning the crank for machine learning: Ease, at what expense? The Lancet Digital Health, 1(5), e198–e199. 10.1016/S2589-7500(19)30112-8 [DOI] [PubMed] [Google Scholar]
- R Core Team. (2020). R: A Language and Environment for Statistical Computing.
- Tang S, Davarmanesh P, Song Y, Koutra D, Sjoding MW, & Wiens J (2020). Democratizing EHR analyses with FIDDLE: A flexible data-driven preprocessing pipeline for structured clinical data. J Am Med Inform Assoc. 10.1093/jamia/ocaa139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE (2019). Avoiding common pitfalls in machine learning omic data science. Nature Materials, 18(5), 422–427. 10.1038/s41563-018-0241-z [DOI] [PubMed] [Google Scholar]
- Therneau T, Atkinson B, port BR (producer. of the initial R., & 1999–2017), maintainer. (2019). Rpart: Recursive Partitioning and Regression Trees.
- Topçuoğlu BD, Lesniak NA, Ruffin MT, Wiens J, & Schloss PD (2020). A Framework for Effective Application of Machine Learning to Microbiome-Based Classification Problems. mBio, 11(3). 10.1128/mBio.00434-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016). Ggplot2: Elegant Graphics for Data Analysis. Springer International Publishing. 10.1007/978-3-319-24277-4 [DOI] [Google Scholar]
- Wickham H, François R, Henry L, Müller K, & RStudio. (2020). Dplyr: A Grammar of Data Manipulation.
- Wiens J, Saria S, Sendak M, Ghassemi M, Liu VX, Doshi-Velez F, Jung K, Heller K, Kale D, Saeed M, Ossorio PN, Thadaney-Israni S, & Goldenberg A (2019). Do no harm: A roadmap for responsible machine learning for health care. Nat. Med, 25(9), 1337–1340. 10.1038/s41591-019-0548-6 [DOI] [PubMed] [Google Scholar]
- Yan Y (2016). MLmetrics: Machine Learning Evaluation Metrics.


