Abstract
Emerging tissue transformation technologies provide an unprecedented opportunity to investigate system-level molecular and anatomical features in situ. Hydrogel-based methods engineer physicochemical tissue properties to render intact organs optically transparent, size- and shape-adjustable while preserving biomolecules at their physiological locations. When combined with advanced molecular tools, labeling, and imaging techniques, tissue transformation enables three-dimensional (3D) mapping of molecules, cells, and their interrelationships at increasing speeds and resolutions. In this review, we discuss the basic engineering principles of tissue transformation and labeling techniques, as well as their broad applications, current challenges, and future potential.
Graphical Abstract
The principles, applications, challenges and future of hydrogel-based tissue transformation technologies, which have expanded the limits of biomolecular imaging in intact organs.
INTRODUCTION
Mammalian organs encompass a vast number of functionally specialized cells that are highly interconnected with each other. For a comprehensive understanding of organ function and dysfunction, it is imperative to interrogate cells at multiple scales (from the nanoscopic organization of biomolecular constituents to organ-scale cellular connectivity) through various lenses (e.g., transcriptome, proteome, projectome). Recent advances in single-cell sequencing, tissue processing, and imaging technologies have allowed researchers to extract unprecedented levels of information about cellular features and their spatial relationships, enabling discoveries of new cell-types and their functions (Darmanis et al., 2015; Frei et al., 2016; Zeisel et al., 2015). By harnessing the power of these technologies in a synergistic way, large-scale initiatives— namely, the Human Cell Atlas Initiative, Human Biomolecular Atlas Program, and Brain Initiative Cell Census Network (Corsortium, 2019; Insel et al., 2013; Regev et al., 2013)—aim to create comprehensive reference atlases of all human cell types to accelerate the pace of discovery. However, probing the extraordinary complexity of biological systems in their spatial context still requires the development of new technologies and approaches.
Classical histology has been the gold standard in probing biological tissues. Various chemical and mechanical processes have been developed to preserve tissue and extract its structural and molecular information. One of the most commonly used tissue processing methods is aldehyde-based chemical fixation and mechanical tissue sectioning. Aldehyde fixation minimizes tissue degradation by crosslinking biomolecules in situ, hardening tissue architecture, and inactivating proteins (e.g., enzymes) that can degrade biomolecules. The fixed tissues are then mechanically sliced into thin sections (10-200 μm) to expose cells to molecular probes and photons for labeling and imaging, respectively. In some advanced technologies (e.g., array tomography), plastic resin is used to embed fixed tissues for ultrathin sectioning (100-200 nm), followed by high-resolution imaging and precise 3D reconstruction (Micheva & Smith, 2007). Although these approaches have been widely used, their limited scalability to organ-scale tissues due to the laborious sectioning and 3D reconstruction processes, and the loss of information such as neural connectivity led to the development of large-scale intact tissue processing technologies.
Over the past decade, next-generation optical tissue clearing methods and hydrogel-based tissue transformation technologies have emerged as new platforms for molecular and structural interrogation of various biological systems. These methods engineer tissue physicochemical properties, such as optical transparency, molecular permeability, tissue size, and mechanical properties, while preserving the spatial context of cells and molecules. Such transmuted tissue platforms, combined with rapidly evolving chemical and optical tools (reviewed in ref. Ueda et al., 2020), provide ready access to the rich 3D information preserved within intact tissues without mechanical sectioning. For example, since the pioneering attempts in the early 1900s to render tissues transparent (Spalteholz, 1914), many researchers have developed next-generation tissue clearing methods that enable 3D cellular-resolution imaging of intact biological systems including those of plants, invertebrates, mice, primates, and human organs (Chiang et al., 2001; Chung et al., 2013; Dodt et al., 2007; Ertürk, Becker, et al., 2012; Ertürk, Mauch, et al., 2012; Hama et al., 2011, 2015; M. T. Ke et al., 2013; Kuwajima et al., 2013; Palmer et al., 2015; Schwarz et al., 2015; Susaki et al., 2014; Yang et al., 2014). Together with powerful genetic labeling tools, molecular probes, and microscopy technologies, optical tissue clearing has enabled neuronal circuit mapping (Belle et al., 2017; Chung et al., 2013; Ertürk, Mauch, et al., 2012; Hama et al., 2011, 2015; M. T. Ke et al., 2013; Murray et al., 2015; Y. Park et al., 2018; Renier et al., 2014; Schwarz et al., 2015), neural activity mapping (Renier et al., 2014; Susaki et al., 2014), and cellular phenotyping (Belle et al., 2017; Chung et al., 2013; Ku et al., 2016; Murray et al., 2015; Y. Park et al., 2018; Renier et al., 2014; Yang et al., 2014).
The advent of hydrogel-based tissue transformation methods led to a paradigm shift in the ways we can engineer the physicochemical properties of biological tissues beyond optical clearing. The main challenge in volumetric tissue phenotyping is the lipid content of the tissue that physically hinders probe and light penetration. Hydrogel-tissue fusion overcomes this challenge by enabling complete lipid removal while preserving the spatial context of cells and molecules in intact tissues. Hydrogel-based techniques further allowed utilization of various chemical modifiers and synthetic hydrogels to enhance tissue properties, resulting in heat- and chemical-resistant tissue-gels (Chung et al., 2013; Murray et al., 2015; Y. Park et al., 2018), size-adjustable tissue-gels (F. Chen et al., 2015; Ku et al., 2016; H. E. Park et al., 2019), and elastic tissue-gels (Ku et al., 2020). These unique tissue-gel properties enable multi-scale imaging (ranging from nanoscale to macroscale) of diverse features of intact biological tissues, including molecular (e.g., transcriptomic, proteomic) and anatomical (e.g., projectomic, connectomic) features. For example, these techniques have enabled the analysis of nanoscale synaptic structures (Chang et al., 2017; F. Chen et al., 2015; Chozinski et al., 2016; Ku et al., 2016; H. E. Park et al., 2019), molecular profiles of individual cells (F. Chen et al., 2016; Chozinski et al., 2016; Chung et al., 2013; Murray et al., 2015; Y. Park et al., 2018; Sylwestrak et al., 2016; Tillberg et al., 2016; G. Wang et al., 2018; Yang et al., 2014), and intercellular connectivity (Murray et al., 2015; Y. Park et al., 2018).
In this review, we assess the current state of hydrogel-based tissue transformation techniques and discuss their underlying engineering principles. By facilitating the understanding of the factors determining the physicochemical properties of tissue-gel hybrids and the major challenges involved in scaling these technologies, we aim to assist the research community in adopting these techniques and synergistically combining them with other pioneering tools to holistically assess biological systems.
HYDROGEL-BASED TISSUE TRANSFORMATION TECHNOLOGIES AND PROPERTIES
Hydrogel-based tissue transformation methods alter the physicochemical properties of tissue-gel hybrids to allow efficient preservation and extraction of biological information across multiple scales, uncovering information that ranges from macroscopic organ-wide properties to nanoscopic subcellular architectures (Figure 1A–E). To effectively manipulate tissue-gel properties, it is crucial to have deeper understanding of the basic chemical and physical principles behind these techniques. In this section, we introduce two broad categories of hydrogel techniques: the inter-biomolecular fixation method, which creates an internal mesh of crosslinked biomolecules, and the synthetic-hydrogel-based method, which creates an exogenous hydrogel mesh scaffold with integrated biomolecules. Understanding how each group of methods allows enhanced preservation and manipulation of physicochemical properties will enable researchers to rationally select and optimize the protocols for their desired application.
Figure 1. Multi-scale molecular and structural interrogation enabled by tissue-hydrogel transformation.

The gray bars below show the scales covered by hydrogel-based tissue transformation technologies. (A) Whole-organ scale. (B) Neural circuits and inter-regional connectivity. (C) Tissue micro-environment. (D) Single cell. (E) Subcellular and nanoscopic architectures.
1. Inter-biomolecular fixation-based methods
In inter-biomolecular fixation-based methods, chemical additives are added to form chemical bonds between biomolecules within the tissue to stabilize the tissue architecture and the biomolecules from degradation (Qidwai et al., 2014). Formaldehyde, a monomeric form of paraformaldehyde (PFA), is the most widely used tissue fixative in commercial, medical, and academic settings. Formaldehyde penetrates tissue rapidly and is slow to react with biomolecules, rendering the chemical uniquely suited for uniform fixation of large-scale tissues. Formaldehyde reacts with amine groups of nucleic acids and proteins to form reversible methylol adducts, which then irreversibly react with the nucleophiles of other biomolecules to form an intermolecular mesh connected by methylene bridges (Figure 2A, B). However, the formaldehyde molecule’s small size and monofunctionality limit the degree of intermolecular crosslinking, resulting in a loss of biomolecules and tissue architecture upon disruption of cell membranes during the tissue delipidation process required by most tissue transformation methods (Chung et al., 2013; Murray et al., 2015; Y. Park et al., 2018).
Figure 2. Overview of common tissue-hydrogel techniques.
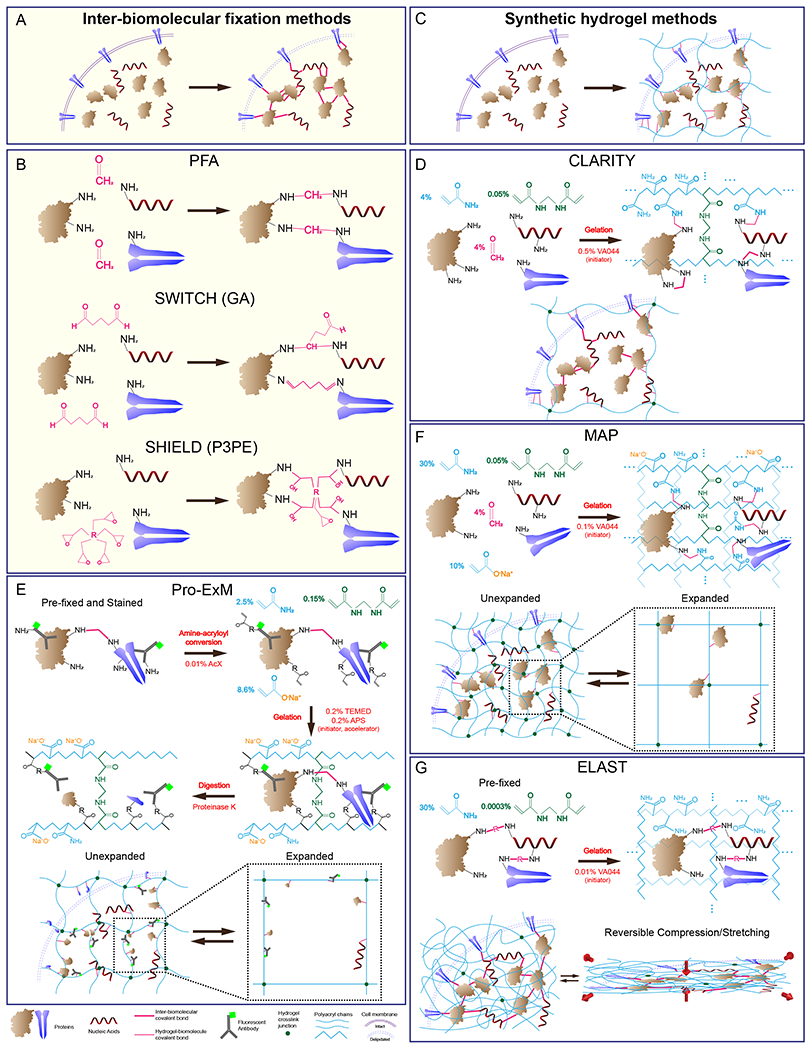
(A) Inter-biomolecular-fixation-based methods use covalently linked inter- and intra-molecular biomolecule meshes. (B) Schematics of PFA, GA, and P3PE fixation chemistries. (C) Synthetic-hydrogel-based techniques generate external polymeric meshes to which biomolecules are anchored. (D) CLARITY uses PFA and 4% acrylamide to covalently crosslink biomolecules to each other and to the pAAm mesh. (E) In Pro-ExM, PFA-fixed and antibody-stained biomolecules undergo amine-acryloyl conversion to enable their covalent anchoring to a polymeric mesh. This is followed by protease digestion to produce isotropic expansion in DI water. (F) MAP uses PFA and a dense heteropolymer composed of high acrylamide and sodium acrylate monomeric units to promote covalent crosslinking of biomolecules exclusively to the mesh (and not to each other), allowing isotropic expansion in DI water. (G) In ELAST, high acrylamide, and low crosslinker and initiator are used without PFA to physically entangle a pre-fixed tissue with a dense pAAm mesh that enables reversible compression and stretching.
Another commonly used fixative is glutaraldehyde (GA), a chemical widely used for EM sample processing. GA is a linear 5-carbon dialdehyde with two reactive groups and a rigid backbone. While the aldehyde-amine chemistry is the same between GA and PFA, GA reacts significantly faster than PFA (Hopwood, 1969) and enables a higher degree of intermolecular crosslinking (Figure 2B). To improve upon the tissue biomolecular preservation of PFA-based tissue-gels, Murray and colleagues developed a GA-based fixation protocol known as system-wide control of interaction time and kinetics of chemicals (SWITCH, Murray et al., 2015). Due to its larger size and faster reaction kinetics, however, GA diffuses into tissue more slowly than does PFA, resulting in a heterogeneous fixation profile (Hopwood, 1970). To overcome this limitation, the SWITCH technique uses a low-pH “OFF” buffer to allow diffusion of fixatives evenly throughout the tissue: the buffer inhibits the fixation reaction through the protonation of endogenous biomolecules. Subsequent incubation in a physiological pH “ON” buffer triggers the tissue-wide crosslinking reaction, providing uniform fixation of large-scale tissues such as the intact adult rat brain and young marmoset brain. The resulting GA-tissue-gels retain similar antigenicity compared to the PFA-fixed tissues (Murray et al., 2015). Furthermore, SWITCH tissues can endure full delipidation and repeated antibody destaining processes with low protein and structural loss, enabling highly multiplexed protein mapping. SWITCH also allows uniform fixation within the bulk specimen without perfusion, which is especially useful for formalin-banked human tissues. Due to its wide applicability on tissues of varying sizes, researchers have adapted SWITCH for application to various biological samples, including stem cell-derived cerebral organoids (Renner et al., 2017) and explanted liver seed grafts (Stevens et al., 2017).
GA-tissue-gels, however, suffer from high autofluorescence and chemical damage of mRNAs. To improve upon these limitations, Park and colleagues developed a polyepoxide-mediated technique, called stabilization under harsh conditions via intramolecular epoxide linkages to prevent degradation (SHIELD, Y. Park et al., 2018). The method’s primary fixative is polyglycerol-3-polyglycidyl ether (P3PE), which has five epoxy groups connected through a flexible backbone. Each epoxy group can undergo a ring opening reaction via nucleophilic attack by the amine groups of biomolecules to form a covalent bond (Figure 2B). Molecular dynamics simulations and experiments have confirmed that P3PE can react with multiple amine groups within a single protein. Such intramolecular crosslinking through the flexible P3PE backbone protects the protein’s tertiary structure against chemical and heat stressors, enabling maximal preservation of protein fluorescence. In addition, a small amount of P3PE can crosslink multiple biomolecules with minimal chemical modification and without adding autofluorescing conjugated π-electron systems. As a result, SHIELD offers higher mechanical stability, lower autofluorescence, and almost identical antigenicity compared to PFA-fixed tissues as well as better preservation of biomolecules compared to the GA and PFA-based preservation methods (Y. Park et al., 2018). Importantly, by applying the SWITCH approach, the SHIELD protocol can also be adapted to non-perfusion-based post-fixation of PFA-fixed tissues—the largest portion of human clinical samples. With SHIELD, Park and colleagues successfully processed 2-mm-thick human brain tissue slabs as well as whole mouse brain hemispheres.
2. Synthetic-hydrogel-based methods
In synthetic-hydrogel-based techniques, biomolecules are either covalently bonded to or physically entrapped inside an external polymer mesh (Figure 2C–G). Like the inter-biomolecular fixation methods, synthetic-hydrogel-based techniques were originally developed to provide a means of robust preservation of biomolecules and tissue architecture; however, researchers have since explored novel ways to harness hydrogel properties (e.g., expandability and elasticity) to overcome unique challenges in tissue phenotyping. We categorize synthetic-hydrogel techniques into three types: (1) CLARITY-based, (2) expansion-based, and (3) physical-entanglement-based methods.
1). CLARITY-based
CLARITY is the first tissue-gel technique to introduce a synthetic hydrogel mesh as a covalently linked scaffold for anchoring biomolecules in situ (Figure 2D, Chung et al., 2013). In CLARITY, endogenous biomolecules form covalent bonds with acrylamide monomers by first reacting with PFA to form methylols, which subsequently react with acrylamides’ amide side chains (Figure 2D) to form methylene bridges. The polyacrylamide (pAAm) mesh is synthesized in situ by the chain polymerization reaction of acrylamide molecules, initiated by the decomposition of an azo-initiator (such as VA044) at 37°C into free radicals. Another component of the hydrogel, bis-acrylamide (Bis), contains two terminal acryloyl groups that can crosslink separate pAAm chains to form a mechanically stable mesh framework. Such covalent linking of biomolecules significantly decreases protein loss upon delipidation compared to PFA-fixed tissues. CLARITY samples delipidated with sodium dodecyl sulfate (SDS) can then be rendered optically transparent through immersion in a medium with a matching refractive index (RI). In the original CLARITY paper, Chung and colleagues demonstrated its use on whole mouse brains and 0.5-mm-thick human brain slabs. Owing to the good mechanical and molecular stability, and the optical transparency of CLARITY-treated samples, researchers have applied variants of CLARITY to a wide range of organisms and tissue types, including post-mortem human brain blocks (Chung et al., 2013), mouse brains (Chung et al., 2013; Menegas et al., 2015, 2017; Yang et al., 2014), mouse whole bodies (Yang et al., 2014), plant leaves (PEA-CLARITY, Palmer et al., 2015), and animal bones (Bone-CLARITY, Greenbaum et al., 2017).
To improve the rate of delipidation and probe penetration in CLARITY, researchers developed the perfusion assisted release in situ (PARS) method (Yang et al., 2014). PARS harnesses existing vasculature for active transport of fixatives, monomers, and clearing solution to enable the clearing of a whole mouse brain as well as a whole mouse body. Aside from its extended use of perfusion, the PARS synthetic tissue-hydrogel formation protocol further improves chemical diffusivity by excluding PFA and Bis crosslinkers when forming the hydrogel mesh to increase gel pore size. This approach allows faster antibody penetration compared to the original CLARITY technique, but it sacrifices some mechanochemical stability by eliminating key fixative and crosslinker chemicals from the hydrogel monomer solution.
Although delipidation enables higher chemical and light penetration into thick tissues, many tissue-hydrogels still require immersion in RI matching solutions to achieve optical transparency. Minimizing RI mismatch between the tissue-gel network and the surrounding medium reduces light diffraction and increases optical transparency. Previous studies of the RIs of human and mammalian tissue showed that depending on the light wavelength and tissue type, tissue RIs could fall in the range 1.3-1.5, with many samples having a RI around the 1.38-1.46 range (Bolin et al., 1989; Dirckx et al., 2005; Zhou et al., 2013). The original CLARITY protocol rendered samples transparent through immersion in a 85% glycerol or FocusClear solution possessing RIs of around 1.45 (Chung et al., 2013), while PARS utilized RIMS (Refractive Index Matching Solution), a Histodenz-based solution (RI = 1.46) to achieve optical transparency (Yang et al., 2014). High RI media (RI > 1.5) based on iohexol and iodixanol are also used in various inter-biomolecular fixation methods like SWITCH or SHIELD (Murray et al., 2015; Y. Park et al., 2018). Using these RI-matching media, high-quality images with minimal light scattering artifacts can be achieved in thick, delipidated tissue-hydrogels.
2). Expansion-based hydrogel embedding
By amplifying the natural tendency of hydrogels to swell in aqueous solutions, researchers have physically expanded tissue-gels to many times their original size, allowing for super-resolution imaging using conventional fluorescent microscopes. Since expansion techniques are frequently applied to visualize nanoscale components, expansion isotropy on both macro- and microscopic scales is necessary to ensure accurate spatial interrogation. Expansion ratio is also important in enhancing and determining the effective imaging resolution.
Expansion Microscopy (ExM), developed by Chen and colleagues, utilizes a superabsorbent pAAm-acrylate gel to enable physical tissue expansion and super-resolution imaging of nanoscopic biological structures with diffraction-limited microscopy (F. Chen et al., 2015). In the original ExM protocol, in situ superabsorbent pAAm-acrylate gel formation is induced with an accelerated polymerization scheme using the free radical initiator ammonium persulfate (APS) and the accelerator TEMED. Unlike CLARITY, conventional ExM does not use PFA to covalently anchor biomolecules to the mesh (although the tissue is initially PFA-fixed). Instead, oligo-bearing antibodies bound to the desired biomolecular epitopes are linked to the mesh through hybridization to trifunctional tags containing complementary oligonucleotide sequences, fluorophores for visualization, and reactive methacrylol moieties for incorporation into the polymer mesh. Then, a harsh proteinase K step digests endogenous proteins to homogenize the gel, rendering the tissue-gel transparent and increasing its expansion isotropy while keeping anchored fluorophores intact (Figure 2E). The post-gelation digestion step and the charge of fixed acrylate groups enable a 4.5-fold linear expansion of ExM tissue-gels, which was demonstrated on single cells and 100-μm-thick mouse brain slices. ExM’s pre-expansion labeling of non-delipidated PFA tissue overcomes various issues of protein loss, macroscopic structural integrity, and antigenicity common in tissue processing. However, low antibody permeability on ExM samples due to a lack of pre-expansion delipidation as well as the need for custom-designed oligonucleotide sequences for antibody-gel hybridization limits the applicability of the method.
Variants of ExM have been developed to widen the applicability of ExM. Instead of anchoring the fluorescent probes to the mesh as in ExM, Pro-ExM directly incorporates proteins into the pAAm mesh using acryloyl-X, which converts biomolecule amine moieties to acrylamides (Figure 2E, Tillberg et al., 2016). A similar variant of Pro-ExM was reported independently by Chozinski and colleagues (Chozinski et al., 2016). The direct anchoring of the protein to the mesh enables immunostaining with antibodies after gel embedding and expansion, allowing faster delivery of labels into digested and expanded tissue-gels. Unlike ExM, Pro-ExM does not require custom-designed oligonucleotides and allows the use of commercially available antibodies for labelling, broadening its usability to more labs. However, Pro-ExM still results in protein loss due to protease treatment, and expansion renders ExM tissue (>99% water) mechanically weak, making it difficult to conduct multi-round immunolabeling, and to mount and unmount samples for repeated imaging without damage.
Other ExM variants have been developed for transcriptomic visualization. Expansion fluorescence in situ hybridization (ExFISH) anchors RNA molecules to the hydrogel mesh, which is followed by FISH (F. Chen et al., 2016). ExFISH can accommodate single-molecule FISH (smFISH) as well as hybridization chain reaction (HCR) to amplify the signals. Following ExFISH, MERFISH-ExM combines the error-robust mRNA barcoding scheme with ExM for multiplexing and de-crowding of optical signals (G. Wang et al., 2018).
Other variants of ExM have focused on extending expansion capacity, improving isotropy, reducing the spatial errors, and broadening the applicability of the original ExM protocol. For example, iterative ExM (iExM) enables 20-fold linear expansion by reembedding a cleavable ExM gel (by replacing Bis crosslinker with DHEBA, a cleavable crosslinker) into a non-cleavable ExM gel (Chang et al., 2017). The first cleavable gel’s fluorophores are anchored to the new non-cleavable gel using an additional oligonucleotide sequence, and the old mesh is cleaved at a high pH to allow another round of expansion. Recently, Pan-ExM added protein retention to the iExM concept, enabling the visualization of cellular ultrastructure with light microscopy (M’Saad & Bewersdorf, 2020). As an alternative way to interrogate nanostructures, expansion single-molecule localization microscopy (Ex-SMLM) adapted ExM to be compatible with super-resolution microscopy while reducing the linkage error (i.e., the spatial imaging error of resolving a given antigen due to the size of its bound primary-secondary antibody complex) of standard ExM protocols from 17.5 nm to 5 nm (Zwettler et al., 2020). Ultrastructure-ExM (U-ExM) demonstrated enhanced tissue-gel isotropy compared to Pro-ExM by using lower degrees of PFA and acrylamide crosslinking (Gambarotto et al., 2019). The utility of ExM has been further expanded to anchoring amino-modified sphingolipid ceramide, enabling lipid expansion and the visualization of membrane structures (Götz et al., 2020), while Click-ExM introduced click chemistry to retain many biomolecules that could not previously be expanded (Sun et al., 2021). As shown from these versatile applications of ExM variants, hydrogel-based tissue transformation techniques enable engineering of tissue size while preserving desired biomolecules for super-resolution imaging.
Ku and colleagues developed a new class of expansion techniques, termed magnified analysis of proteome (MAP, Ku et al., 2016), based upon the CLARITY chemistry. MAP takes advantage of the in situ thermal polymerization system developed in CLARITY, but it uses significantly higher acrylamide concentration (30%) than does CLARITY (4%). At a high monomer concentration, each biomolecule is more likely to covalently bond to the hydrogel mesh rather than to nearby biomolecules, preventing intra- and inter-protein crosslinking and promoting isotropic expansion (Figure 2F). Once a densely packed hydrogel is formed in situ, it naturally expands after tissue delipidation. A denaturation step dissociates protein complexes to further enhance the expansion ratio. Thermally controlled initiator system used in both MAP and CLARITY enables uniform in situ polymerization throughout organ-scale samples despite the exothermic nature of the radical polymerization process, which could cause overheating and uneven gelling especially when high concentrations of monomers (40% in MAP) are used. MAP-processed tissues can withstand harsh antibody destaining conditions, allowing multiplexed protein mapping. Furthermore, MAP is compatible with active antibody staining using stochastic electrotransport (Kim et al., 2015), enabling highly scalable, multiplexed, and super-resolution imaging of subcellular structures in large-scale tissues, such as whole mouse brains and other organs (Ku et al., 2016).
To allow for easy, flexible, and application-dependent expansion adjustment, Park and colleagues built on MAP and recently developed a technique called ZOOM that enables tuning of expansion ratio (H. E. Park et al., 2019). The authors omitted sodium acrylate from the MAP monomer solution and introduced heating steps immediately after gelation to hydrolyze a portion of the acrylamide groups to acrylate moieties, which enabled flexible modulation of the expansion ratio simply by varying the hydrolysis duration. ZOOM-processed 500-μm-thick mouse tissue can undergo a maximum of 8-fold isotropic linear expansion, enabling detection of fine dendritic spine architectures. Due to the methods’ similar monomer concentration and processing steps, ZOOM and MAP-processed tissues have similar properties; however, ZOOM-processed tissue-gels, which are formed purely from acrylamide monomer units, were shown to have higher compressive strength than tissue-gels formed from acrylamide—sodium-acrylate mixtures and tissue-gels formed purely from sodium acrylate monomer units (H. E. Park et al., 2019). Thus, pAAm gels seem to exhibit mechanical properties superior to pAAm-acrylate gels even after partial hydrolysis of the pAAm gels, demonstrating that downstream tissue-gel qualities are sensitive to monomer composition and gelation conditions.
3). Physical entanglement techniques
Unlike covalent crosslinking methods, physical entanglement techniques secure endogenous biomolecules at their physiological locations purely through physical entanglements between the dense synthetic hydrogel mesh and endogenous biomolecular networks (Ku et al., 2020). The lack of covalent bonding between the pAAm gel and biomolecules enhances tissue elasticity, resulting in tough and stretchable tissue-gels.
Ku and colleagues recently developed a physical entanglement technique termed entangled link-augmented stretchable tissue hydrogel (ELAST, Ku et al., 2020). Although its dense monomer composition is similar to that of MAP, ELAST uses a 300-fold lower crosslinker concentration and a 10-fold lower thermal initiator (VA-044) concentration without PFA. The high acrylamide monomer concentration and low initiator concentration promote the synthesis of longer pAAm chains, while the low Bis concentration enables more physical entanglement of the long pAAm chains by preventing extensive covalent crosslinking. The resulting interpenetrating polymer network (IPN) contains slip-link connections that allow elastic movements, rendering the tissue-gel tough as well as reversibly compressible and stretchable (Figure 2G). Not only do ELAST’s toughness and compressibility prevent tissue damage and enhance reproducibility, but these properties also enable researchers to apply cyclic compression to decrease sample thickness, which shortens the time required for immunostaining. Another key difference between ELAST and other synthetic hydrogel technologies is the former’s use of delipidated SHIELD-fixed tissue, as opposed to uncleared tissue, for gel embedding. The SHIELD process effectively preserves biomolecules during lipid removal, and delipidated tissues permit higher penetration of the pAAm mesh for more homogeneous physical entanglement. Since ELAST does not introduce additional covalent bonding, epitopes are not further masked, resulting in the same antigenicity between SHIELD and ELAST tissues. Using ELAST and cyclic compression, Ku and colleagues demonstrated rapid immunolabeling of mm-thick human and mouse brain samples that is 100 times faster than labeling of CLARITY-processed tissues using passive diffusion.
HYDROGEL-BASED TISSUE TRANSFORMATION OPTIMIZATION CRITERIA
We have reviewed various hydrogel-based techniques that specialize in probing biological systems of desired scope and scale. Here, we discuss essential hydrogel properties and explain how they can be engineered to help researchers select or optimize tissue transformation methods for their own specific systems and goals.
1. Essential hydrogel properties
Hydrogel properties are important in maximizing the amount of biological information that can be extracted from the specimen of interest. There are four essential hydrogel properties to consider while optimizing tissue-gel hybrids (Figure 3A).
Figure 3. Optimization of tissue-gel properties.

(A) Qualitative trends of four criteria for optimizing tissue-gel properties as well as their dependence on five experimental parameters. (B) GFP signal retention after 70°C, 24h SDS treatment. SHIELD (P3PE) outperforms other methods (adapted with permission from Y. Park et al., 2018). (C) GFP signal retention is worse in harsher delipidation conditions, but higher degree of delipidation offers better permeability and transparency (adapted with permission from Y. Park et al., 2018). (D) Stress-relaxation curves for three regimes. Strong but brittle materials can withstand high stress but fracture at low strains (orange), while ductile materials are permanently deformed after small stresses (red). Tough tissue-gels can withstand high strain and stress (green). (E) Rigidity is a measure of strength, which is desirable in tissue-hydrogels (adapted with permission from Y. Park et al., 2018). (F) Low crosslinker (Bis) concentration confers high compressibility and toughness to pAAm gels by encouraging more chain entanglement (adapted with permission from Ku et al., 2020). (G) A graph showing pre-expansion vs. post-expansion effective monomer density. High pre-expansion monomer density does not necessarily correlate with high post-expansion gel density (i.e., low permeability) due to hydrogel expansion (H) A graph showing monomer:crosslinker ratios for each hydrogel-based technique (Blue: expansion-based, Green: CLARITY-based, Magenta: physical entanglement-based). (I)-(K) Factors that influence expansion ratios of tissue-hydrogels. (I) Πel is the elastic pressure contribution to overall swelling pressure. High delipidation/digestion, low crosslinker %, low fixation strength (bottom) results in higher Πel (elastic pressure) and swelling. (J) Low monomer % (top) yields lower Πmix (polymer-solvent mixing) than high monomer % (bottom), resulting in lower expansion. (K) Higher concentration of fixed charges (bottom, i.e., sodium acrylate) results in higher Πion (ion exchange osmotic pressure) and more expansion.
1). Probe compatibility.
This is a measure of preserved sites for molecular probes to bind. Good probe compatibility is required for unbiased labeling of biological specimens.
2). Biomolecular stability.
This is the quality of biomolecule preservation and it can be measured by molecular loss assays and fluorescent protein signal retention assays (Figure 3B, C). Effectively transformed tissue-hydrogels have high biomolecular stability even under harsh delipidation conditions.
3). Mechanical toughness.
This is a material’s ability to withstand fracture at high stress (a measure of strength), even when deformed to high strains (a measure of deformability), and it is often quantified as the area underneath a material’s stress-strain curve (Figure 3D). For most tissue transformation applications, rigid tissue-gels, that are strong but not deformable (i.e., moderate toughness), are desirable (Figure 3E). However, a tougher tissue-gel that maintains a high level of strength while also being highly stretchable or compressible is significantly less brittle, more damage-resistant, and less prone to tears (Figure 3F).
4). Permeability and transparency.
Permeability refers to the tissue-hydrogel pore size and tortuosity, which directly influence molecular probe diffusivity. High permeability allows deeper penetration of molecular probes in a given time frame. Optical transparency enables deep, non-scattered imaging of thick specimens, and is often closely related to permeability.
Next, we will discuss how these parameters could be modulated.
2. Modulation parameters for hydrogels
Although there are many established hydrogel-based protocols available, understanding the ways to alter hydrogel properties will grant researchers flexibility in adopting hydrogel-based techniques to their systems. To alter the essential hydrogel properties, five parameters (listed below) can be modulated. General trends for how tissue-hydrogel properties vary based on these are outlined in Figure 3A. However, it is important to note that parameters are not always completely independent, and exceptions likely exist for these broad generalizations.
1). Fixation strength.
This can be increased by lengthening fixation time, increasing chemical fixative concentration, increasing temperature, using reactive pH, or using more potent crosslinker chemicals (e.g. GA vs. PFA).
By increasing the degree of tissue fixation, the covalent bonds between biomolecules are strengthened and tightened. This has the benefit of sample-wide tissue strengthening, which increases its rigidity and mechanical toughness (as shown in Figure 3E), while also enhancing biomolecule preservation. Furthermore, the tissue and its biomolecules become more resilient to downstream processing and sample handling. For example, P3PE, the primary fixative molecule in SHIELD, has a larger number of amine-reactive functional groups than PFA, GA, and CLARITY monomers, allowing it to better preserve the fluorescence of fluorescent proteins (Figure 3B). However, tightened inter- and intramolecular bonds decrease pore size and permeability. Furthermore, increased participation of biomolecular chemical moieties in covalent bonding results in more extensive epitope masking of fluorescent probe binding sites, decreasing the degree of probe compatibility.
2). Delipidation/digestion harshness.
The delipidation/digestion (clearing) harshness can be increased by lengthening clearing time, using harsher chemicals (e.g., using Proteinase K for digestion vs. using SDS for delipidation), using higher concentrations of clearing agents, and/or increasing the temperature.
The delipidation/digestion harshness of a protocol affects the degree of lipid and protein loss in a tissue sample. Harsher treatments can remove physical barriers and light-scattering molecules more effectively, resulting in higher permeability and transparency. However, this tends to come at the cost of weaker tissue mechanical properties, higher loss of proteins and other biomolecules (Figure 3C), and higher damage to epitopes to which fluorescent probes bind.
The following three parameters are only relevant in synthetic-hydrogel-based methods.
3). Monomer concentration.
Generally, this refers to the concentration of chemical units used as building blocks for the bulk of the polymer mesh in synthetic-hydrogel-based techniques. Often, monomer concentration refers to the combined concentration of acrylamide and sodium acrylate.
Monomer concentration has a wide range of effects on synthetic-hydrogel-based tissue-gel properties. At higher monomer concentrations, the biomolecular stability may be marginally enhanced either because of physical entrapment of biomolecules or due to more effective biomolecular anchoring to the hydrogel mesh when used in conjunction with PFA. Similarly, the overall mechanical toughness of the tissue may be increased because of a denser hydrogel scaffold. Finally, the permeability and transparency have the potential to increase at high monomer concentrations because of the tendency for dense hydrogel polymers to swell and expand, thus lowering the overall post-expansion density of the tissue-gel (Figure 3G).
4). Crosslinker concentration.
This refers to the concentration of chemicals used for crosslinking individual polymer chains into a covalently-bonded polymer mesh. For tissue transformation methods, this generally refers to Bis-acrylamide concentration.
By increasing crosslinker concentration in a synthetic-tissue-hydrogel, the bonds between polymeric acrylamide chains in the hydrogel mesh are tightened and strengthened, lowering the overall pore size (which decreases the permeability and transparency) while physically entrapping and stabilizing biomolecules. At a lower crosslinker concentration, the hydrogel mesh relies more heavily on physical polymeric chain entanglement rather than on covalent crosslinking for gel strength. Extremely low crosslinker concentrations can lead to a mechanically weak tissue-gel after hydration, but high crosslinker concentrations can lead to rigid, brittle gels. Thus, the optimal crosslinker concentration for mechanical toughness is low but non-zero, especially when combined with a high monomer concentration (Figure 3H). This promotes long polymeric chain growth which can form a dense, physically entangled gel with sparse covalent crosslinking support for both mechanical strength and reversible deformability (Figure 3F).
5). Initiator concentration.
This refers to the concentration of the chemical used for initiation of the polymerization reaction that leads to formation of the synthetic hydrogel mesh. For ExM, the initiator is the TEMED/APS system, whereas in most other acrylamide-based techniques, the initiator is a thermal azo-initiator (e.g., VA-044).
At extremely low initiator concentrations, polymerization is often too slow, resulting in incomplete gelation and a weak, amorphous gel. When combined with high acrylamide concentration (such as in ELAST), higher initiator concentration increases the number of pAAm chains generated, which shortens the average chain length. Shorter polymeric chains lead to a lower degree of entanglement and higher permeability at the expense of lower mechanical toughness and biomolecular stability (Figure 3A). However, in protocols with low acrylamide concentration (such as CLARITY), the monomer concentration is insufficient to form long chains for entanglement. Thus, a high initiator concentration in this regime leads to higher incorporation of acrylamide monomers, increasing gel density and resulting in lower permeability.
3. Additional criteria and properties of expansion-based tissue-hydrogels
When optimizing for expansion-based techniques, expansion ratio and expansion isotropy need to be considered. The expansion ratio (and transparency) can be modulated by understanding that tissue-gel expansion occurs due to swelling pressure described by the following equation (Horkay et al., 2000; H. E. Park et al., 2019).
Here, Πel is the elastic pressure, Πmix is the osmotic pressure due to polymer-solvent mixing, and Πion is the osmotic pressure due to ion exchange to satisfy the electroneutrality condition, in addition to the contributions from solvent-ion and polymerion interactions. Lower crosslinking (increasing Πel), higher monomer concentration (increasing Πmix), higher fixed charged moieties in the gel (increasing Πion), and decreased solution ion concentrations (increasing Πion) lead to an overall increase in swelling pressure Π and expansion. Consequently, we can think of the parameters that affect expansion ratio to be crosslinker concentration and fixation strength (decreases Πel, Figure 3I), delipidation/digestion harshness (increases Πel, Figure 3I), monomer concentration (increases Πmix , Figure 3J), and sodium acrylate concentration (increases Πion, Figure 3K).
Isotropy is generally improved by increasing the tissue-gel homogeneity. In ExM and many of its variants, proteinase digestion was used to decrease tissue contents and increase homogeneity. In MAP and its variants, denaturation-mediated dissociation of protein complexes and high monomer concentration were used to preserve the 3D proteome while increasing tissue-gel homogeneity. Synthesizing a dense polymeric mesh in situ using 40 wt% acrylic monomers (vs 10 wt% endogenous proteins) effectively lowers tissue-to-hydrogel density and increases overall homogeneity.
Depending on the tissue size, the necessity for multi-round labeling, and the desired imaging resolution, the requirements for expansion ratio may vary. For instance, for multi-round staining of thick tissues at synaptic resolution, MAP would be preferable due to its high mechanical stability and biomolecular preservation. For single round staining of thin tissues with a requirement for a high degree of expansion, iExM would be suitable, since mechanical toughness and biomolecular preservation are not as important.
Optimizing all factors is not trivial, especially when considering complex correlations among parameters. Depending on the goal of a given biological study, one must select or modify methods based on the tradeoffs involved, or develop new techniques to mitigate these tradeoffs. Regardless of the tissue transformation method, however, volumetric sample processing may require additional considerations of scalability when applying it to an organ-size tissues. We will discuss these challenges and offer necessary insights in the following section.
CHALLENGES IN SCALING UP HYDROGEL-BASED METHODS TO VOLUMETRIC SAMPLES
An important role of the hydrogel mesh is to secure biomolecules (proteins and transcripts) at their physiological locations during the lipid removal step (delipidation). Endogenous lipid layers act as both physical and optical barriers that prevent intracellular delivery of molecular probes and photons, respectively (Figure 4A, Richardson & Lichtman, 2015). Varying degrees of delipidation, therefore, have been used in tissue clearing and transformation methods to increase both molecular permeability and optical transparency of biological tissues. The synthetic-gel-based methods, in particular, allow complete delipidation using strong detergents because the hydrogel mesh can prevent loss of biomolecules and their spatial organization even after complete removal of lipid bilayers (Figure 4A).
Figure 4. Basic principles of tissue clearing and molecular deliveries.
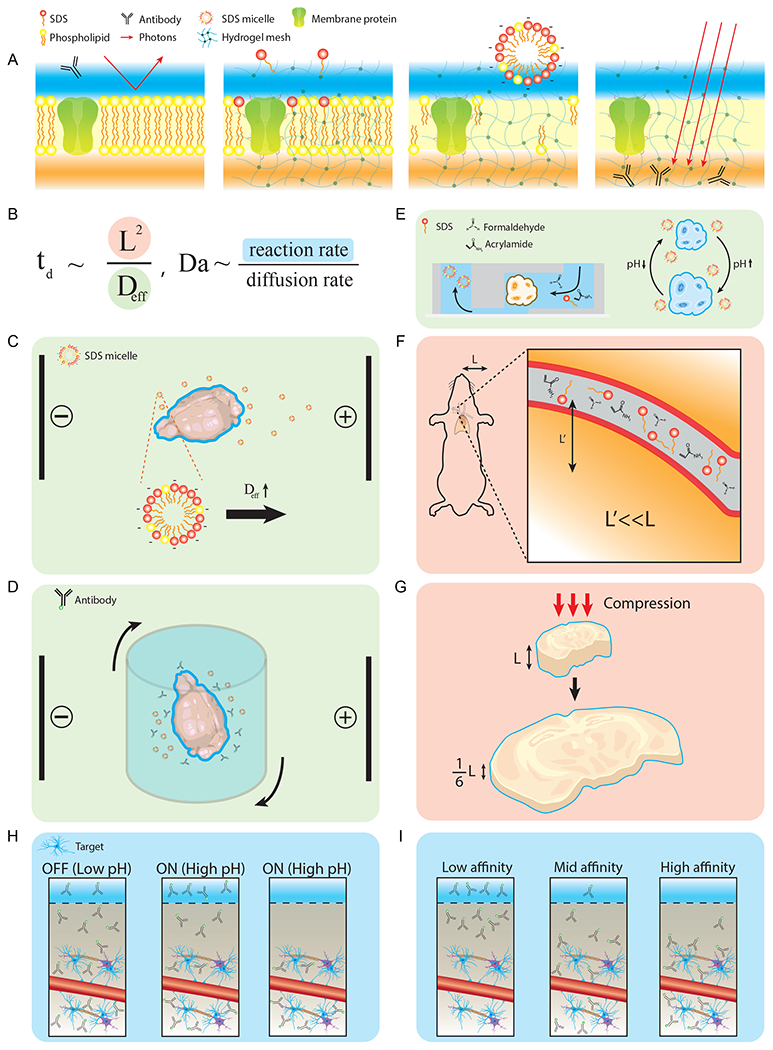
(A) A schematic of the delipidation process and the molecular preservation by hydrogel-based techniques. (B) Diffusion time scale, td, is proportional to the characteristic length (L) squared and inversely proportional to the effective diffusivity (Deff). Damköhler number, Da, dictates the relationship between the reaction rate and the diffusion rate. (C) CLARITY increases the value of Deff by applying an electric field on thick tissues for active delipidation. (D) Stochastic electrotransport applies a rotational electric field to selectively accelerate highly electromobile species (micelles/antibodies). (E) Use of convective flow and osmotic pumping to increase Deff in delivering CLARITY solution. (F) Repurposing of existing vasculature to deliver fixatives and detergents by effective reduction of the characteristic length (L’≪L). (G) Reversibly compressible tissue-gel hybrid by ELAST to modify the characteristic length scale. (H) SWITCH overcomes fast reaction time scales by suppressing reactions at low pH and by triggering reactions at physiological pH. (I) In eFLASH, a gradual modulation of antibody affinity allows steady change of Da, improving scalability over SWITCH.
The delipidation mechanism involves the transport of detergent micelles carrying endogenous lipid molecules (Figure 4A, Lichtenberg et al., 2013). While preserving biomolecules at their respective locations, hydrogels can decrease the permeability of tissue-gel hybrids by forming crosslinked meshes that decrease their effective pore sizes. This trade-off adversely affects the delipidation process since small pores hinder the movement of the detergent micelles. In the case of organ-scale samples, passive delipidation can take weeks. The relationship between the tissue size and the time required for full delipidation can be estimated by deriving the following expression for diffusion time scale (td, Box 1).
Box 1.
Derivation of diffusion time scale
We set up a chemical conservation equation for a control volume containing a biological sample:
Here, C denotes the concentration of chemicals (in this case micelle), and Deff is the effective diffusivity. We are not considering any chemical reaction here, since the removal of lipid-containing micelle is solely dependent on passive diffusion. If we perform scaling analysis, which is to simplify the equation by considering only the dominant terms using their order of magnitude, we can simplify the equation to:
Here C0 denotes the initial concentration of micelles, td represents the diffusion time scale (from the core to the surface), and L is the characteristic length that the micelles have to travel. Rearranging the terms, we derive an expression for the diffusion time scale:
Derivation of Damköhler number expression
We set up a chemical conservation equation for a control volume containing a biological sample, but this time we include the reaction term. Assuming steady state (no time-dependent term):
Here, RV denotes the volumetric reaction rate. Assuming we are modeling the reaction between the antibody (ab) and the target antigen (g), we have the following reaction rate expression:
Here, k denotes the reaction rate coefficient. Performing scaling analysis again:
In this expression, δ is the antibody penetration length, which represents the antibody travel distance before a reaction happens. If we compare this penetration length to the characteristic length L, we get the following expression for the Damköhler number:
From this relationship, we can see that the removal time of micelles is proportional to the square of the characteristic length (L2) of biological tissues and inversely proportional to the effective diffusivity (Deff, Figure 4B). Therefore, the time (td) for complete lipid removal in volumetric samples increases in a quadratic manner with their length scales (L), meaning the process can take weeks to complete in the case of whole organs. Various technologies have been developed to accelerate the rate of molecular transport in hydrogel-based techniques. We will categorize these methods into two groups: techniques that increase Deff and techniques that decrease L.
1. Increasing the effective diffusivity (Deff)
CLARITY was the first hydrogel-based technique that pioneered the Electrophoretic Tissue Clearing (ETC) method to increase the effective diffusivity (Figure 4C, Chung et al., 2013). By applying an electric field, ETC accelerates the extraction of highly charged micelles from tissue-gel, shortening the delipidation time from weeks to days. Applying higher electric fields can further reduce the processing time, but the electric field strength is limited to avoid tissue damage caused by the electrophoretic displacement of the charged tissue matrix. Kim and colleagues addressed this challenge by developing stochastic electrotransport that selectively disperses highly electromobile species (micelles and antibodies) while keeping the species with low electromobility (endogenous biomolecules tethered in the hydrogel) stationary (Figure 4D, Kim et al., 2015). This selective acceleration allows further reduction in the diffusion time scale, as it enables application of a higher electric field without causing tissue damage.
Another application of CLARITY utilized osmotic pumping to increase Deff (Silva Santisteban et al., 2018). At a basic pH, the CLARITY-processed tissue-gel swells due to deprotonation of crosslinked amino acids, and it shrinks at an acidic pH due to protonation. Repetition of this swelling-shrinkage cycle enables osmotic pumping, which acts as an external force to increase effective diffusivity. By applying this technique within a microfluidic device, the authors achieved highly parallelized and rapid delipidation of CLARITY-processed spheroids (Figure 4E).
2. Decreasing the characteristic length (L)
Decreasing the characteristic length (L) of tissues can yield a similar effect in accelerating molecular transport. For example, tissue sectioning is one way that directly benefits from decreased characteristic length. To achieve similar effects in intact volumetric tissues, PARS, CUBIC-perfusion (CB-perfusion), and uDISCO repurposed the existing vasculature system in animals to deliver both fixatives and detergents through densely distributed blood vessels (Figure 4F, Pan et al., 2016; Tainaka et al., 2014; Yang et al., 2014). However, these methods require intact vasculature for rapid delivery, limiting their use on extracted tissue samples. Recently, ELAST leveraged the elastic properties of hydrogels to enable reversible compression and stretching of tissue-gel hybrids (Ku et al., 2020). Like thinly sectioned tissue, compressed tissue has a small thickness and characteristic length scale, allowing fast labeling of volumetric tissues (Figure 2G, 4G). Using ELAST and cyclic compression, the authors demonstrated rapid staining of 5-mm-thick human brain samples within one day, which is 100 times faster than the passive staining demonstrated in CLARITY. As exemplified by these techniques, the unique mechanical properties of tissue-gel hybrids allow the application of various strategies to expedite chemical transport for rapid and scalable tissue processing.
3. Diffusion-reaction kinetics considerations
Molecular imaging and phenotyping often require labeling of delipidated tissues with appropriate probes (e.g., antibodies, FISH probes). Like lipid removal, probe delivery follows similar rules for the diffusion time scale. Besides the probe transport to the tissue core, however, there are also chemical reactions (e.g., antibody-antigen binding) between probes and biomolecules. Therefore, a balance between diffusion and reaction of molecular probes must be considered to achieve optimal labeling of large-scale tissues. In chemical engineering, the Damköhler number (Da, Figure 4B, Box 1) has been widely used to relate the chemical reaction timescale to the diffusion timescale.
Here, k represents the reaction rate coefficient, and Cp0 is the initial concentration of the reactive endogenous molecules. In the case of Da > 1, where the reaction is faster than the diffusion, most of the probes react with target molecules along the outer edge of the tissue, causing surface saturation and poor probe penetration. However, when Da ≤ 1, where the diffusion is faster than the reaction, the probes can penetrate deep into the tissue before the reaction occurs, allowing more uniform and complete staining (although extremely low Da can lead to an under-labeled/under-reacted tissue). In intact tissue processing, most reactions (e.g., tissue fixation, probe-target binding) are significantly faster than passive diffusion, making Da ≫ 1. For example, passive labeling of thick mouse brain tissue with antibodies results in an uneven signal profile, with only surface targets labeled (Ku et al., 2020). Therefore, the key parameters (k, Cpo, L, Deff) need to be carefully modulated to achieve Da ≤ 1 and optimal labeling.
The first method to take these parameters into account was SWITCH, which used SWITCH OFF and ON buffers to modulate the reaction rate, k (Murray et al., 2015). To achieve uniform fixation of large-scale tissues with GA, the GA-biomolecule reaction rate (k) was first significantly decreased using the low-pH SWITCH-OFF buffer, allowing even distribution of GA throughout the tissue prior to any reaction (Da ≪ 1). Once GA was uniformly dispersed, the sample was moved to high-pH SWITCH-ON buffer to increase the reaction rate (k) and achieve adequate degrees of fixation (Figure 4H). Analogously, to achieve more uniform molecular labeling, probe-target reactions were first suppressed (low k and Da) using the SWITCH-OFF buffers containing a small amount of SDS. Once the probes were uniformly dispersed, the tissue was immersed in the SWITCH-ON buffer containing non-ionic detergents that neutralize SDS and allow the binding reaction to occur (high k and Da). SWITCH-labeling protocols, however, employ two extreme ends of Da (Da ≪1 in the OFF step and Da≫1 in the ON step), and thus suffer from non-uniform staining. For example, during the ON step, probes dispersed inside the tissue quickly bind to nearby target molecules. However, the probes near the surface will diffuse out of the tissue into the ON buffer, leading to an under-labeling profile on the surface. In addition, like other passive staining methods, SWITCH requires long incubation times because the diffusion rate (Deff) remains slow.
A technique termed eFLASH (unpublished) overcame this challenge and achieved uniform staining of organ-scale tissues within one day by simultaneously modulating both the reaction rate (k) and the diffusion rate (Deff). In this method, the reaction rate (k) was gradually increased over the course of one day while applying stochastic electrotransport to increase the diffusion rate (Deff) of labeling molecules (Figure 4I). The initially unfavorable binding condition (Da ≪ 1) allows the antibodies to travel through biological tissues instead of binding on the surface. A progressive increase in binding affinity and rapid transport of chemicals ensure all biomolecules in the organ-scale tissue experience the same reaction conditions at any given time. Using this approach, a complete and uniform staining of organ-scale tissues was achieved within one day with a large library of antibodies.
As exemplified by various techniques, scalable processing of large-scale samples requires the consideration of additional parameters: td and Da. Although we only discuss hydrogel-based transformation methods, recent CUBIC-HV protocols modeled fixed biological tissues as a gel and optimized whole-organ staining conditions based on diffusion-reaction kinetics (Susaki et al., 2020) which underscores the necessity of careful consideration of these parameters. When these considerations are combined with hydrogel-based tissue transformation methods, we can fully apply the benefits of tissue-gel hybrids to volumetric samples in a scalable manner.
APPLICATIONS OF TISSUE TRANSFORMATION TECHNOLOGIES
Tissue transformation technologies provide unprecedented opportunities to investigate multi-omic (transcriptomic, proteomic, and connectomic) information at multiple scales (from nanoscopic to organ-level). Here, we will review how this synergy has been at the forefront of multimodal molecular phenotyping of biological systems.
1. Spatial transcriptome
Analyzing the spatial organization of various cell types and their gene expressions is crucial to understand the structure-function relationships in complex biological systems. Recent advances in in situ transcriptomics technologies have enabled spatial mapping of gene expression with increasingly high resolution and multiplexing. For instance, smFISH can map individual RNA molecules at single-cell resolution in their native tissue context (Figure 5A,B, Femino et al., 1998; Codeluppi et al., 2018). Spatial, spectral, and sequential barcoding approaches in smFISH have enabled mapping of many RNA species in individual cells (Figure 5C, D, Lubeck et al., 2014; Lubeck & Cai, 2012). Among these barcoding schemes, multiplexed error-robust FISH (MERFISH, Figure 5E, F, K. H. Chen et al., 2015) substantially increased the number of RNA species that can be simultaneously imaged in single cells to ~1,000 RNA species using error-robust encoding schemes. Together with single-cell RNA sequencing, MERFISH enabled identification of ~70 distinct neuronal populations in the hypothalamic preoptic region in mice (Moffitt et al., 2018). Many of these spatial-transcriptomic methods, however, were demonstrated to be effective on monolayer cell cultures or thin tissue sections. Hydrogel-based methods can offer a unique opportunity to extend the utility of these technologies to large-scale intact tissues by effectively preserving mRNA transcripts while allowing efficient tagging and 3D visualization of multiple mRNA species.
Figure 5. Overview of spatial transcriptome analysis.
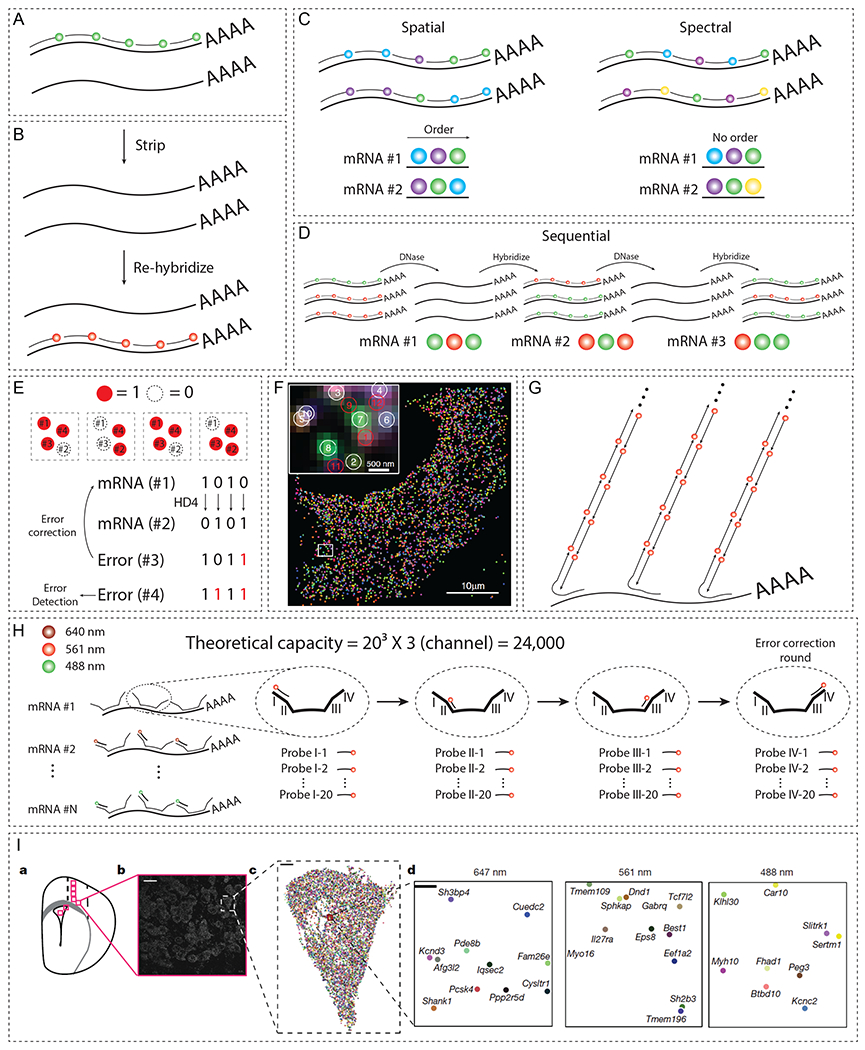
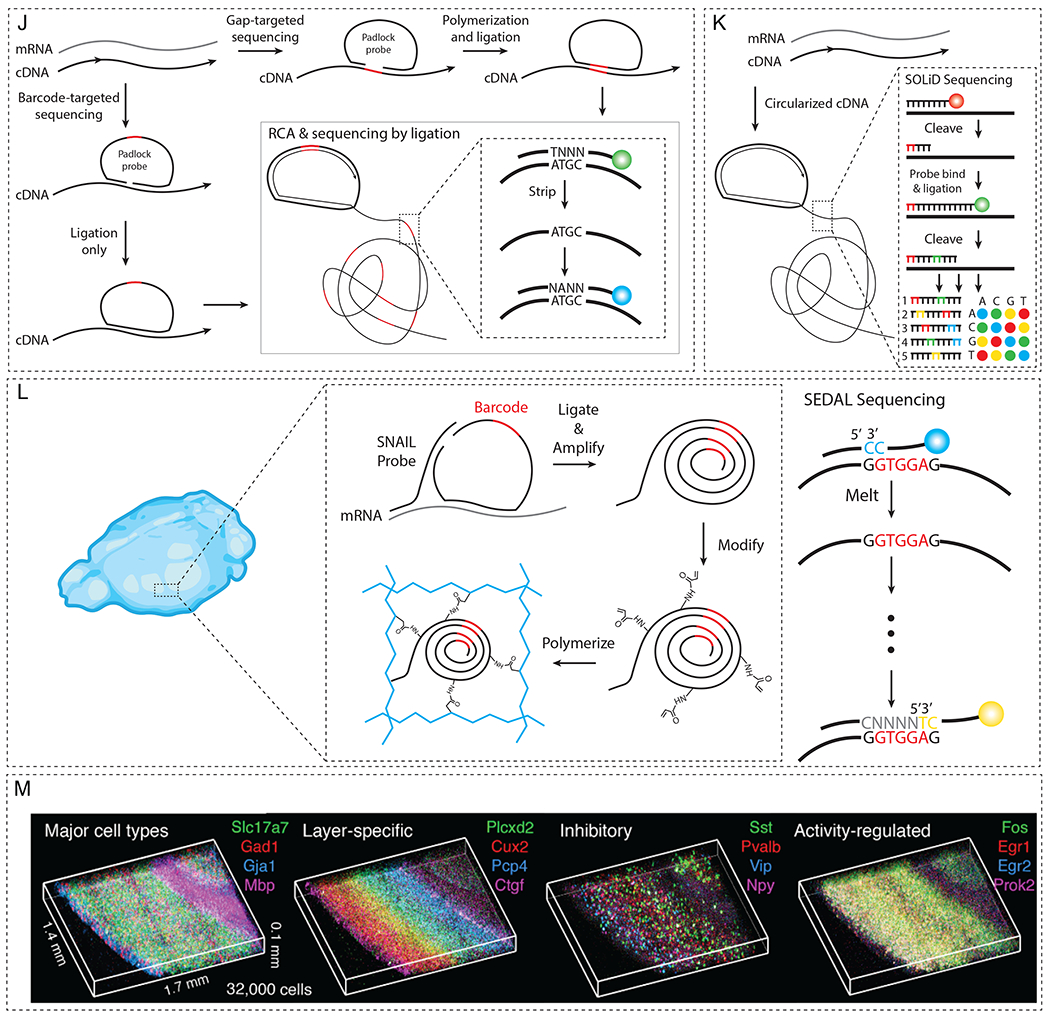
(A) smFISH hybridizes multiple fluorescently labeled DNA probes to the target mRNA. (B) FISH probes can be repeatedly hybridized and stripped off for the multiplexed detection of RNA molecules. (C) Spatial (left) and spectral (right) barcoding schemes that allow exponential increase in the number of targets with a given set of fluorophores. (D) A schematic of sequential barcoding, relying on multiple rounds of hybridization. (E) Concept of MERFISH to decrease the error rates by utilizing HD4 coding scheme. (F) Simultaneous measurements of 1,001 RNA species in single cells using MERFISH (adapted with permission from K. H. Chen et al., 2015). (G) HCR amplification for improving SNR. (H) Schematic of SeqFISH+, which extends multiplexing capacity to ~104. (I) SeqFISH+ detecting 9,418 mRNAs in a single cell (adapted with permission from Eng et al., 2019). (J) Schematic of in situ sequencing, in which target mRNA is reverse transcribed to cDNA using padlock probes and amplified via RCA. (K) FISSEQ circularizes cDNA into an amplicon, allowing untargeted analyses of mRNA profiles. (L) STARmap co-polymerizes DNA nanoballs into a hydrogel mesh, preserving the spatial information of the mRNA sequence. (M) STARmap on 100-μm-thick mouse visual cortex (adapted with permission from X. Wang et al., 2018).
Covalent tethering of mRNA to a porous hydrogel mesh can effectively preserve the integrity of mRNAs in a chemically and optically accessible 3D framework for spatial transcriptomics. CLARITY first succeeded in the preservation of mRNA within tissue-gel hybrids and demonstrated its compatibility with FISH using mouse brain sections (Chung et al., 2013; Yang et al., 2014). Hydrogel chemistry has been further improved for optimal RNA preservation. For example, Sylwestrak and colleagues crosslinked RNA transcripts to the hydrogel mesh through 1-Ethyl-3-3-dimethyl-aminopropyl carbodiimide (EDC) to enhance the preservation of messenger, non-coding, and micro RNAs (Sylwestrak et al., 2016). Using this approach, the authors successfully conducted hybridization-based 3D visualization of RNA transcripts in mm-thick tissues. Park and colleagues showed that SHIELD combined with SWITCH can effectively and uniformly preserve mRNAs in organ-scale tissues while simultaneously protecting protein fluorescence, antigenicity, and tissue architecture (Y. Park et al., 2018). In addition, Chen and colleagues conducted serial rounds of hybridization on an expandable tissue gel, confirming that the hydrogel mesh can stabilize mRNA transcripts through repeated rounds of hybridization and washing (Figure 5B, F. Chen et al., 2016). These techniques have proven that robust preservation of RNA transcripts can be achieved with hydrogel-tissue hybrid frameworks.
Unique physical and optical properties of the synthetic tissue gel platforms can improve the detection of labeled RNA molecules in 3D. Various factors including optical crowding and autofluorescence can decrease the signal-to-noise ratio (SNR) and increase error rates (false positive/negative signals) in RNA detection. To address these, ExFISH was combined with MERFISH (MERFISH-ExM) to physically de-crowd FISH signals which enabled the detection of high-density RNA species (F. Chen et al., 2016; G. Wang et al., 2018). In addition to physical expansion, delipidation and optical homogenization in hydrogel-based methods can substantially decrease autofluorescence. For instance, SeqFISH employed PACT-delipidation to achieve low autofluorescence and applied HCR amplification of smFISH signals to reliably perform imaging of mRNA signals in 0.5-mm-thick tissue sections (Figure 5G, Shah et al., 2016). This improved SNR is a shared benefit of many hydrogel-based transcriptomic methods, enabling the visualization of bacterial rRNA (DePas et al., 2016) and enhancing the multiplexing capacity by better resolving barcoded signals (Moffitt et al., 2016). Notably, seqFISH+ reported a multiplexing scheme that is capable of visualizing ~10,000 genes on hydrogel-embedded samples (Figure 5H, I, Eng et al., 2019), showing how hydrogel-tissue techniques can synergize with the pioneering in situ transcriptomics approaches to extend their utility.
Another in situ transcriptomics approach sequences mRNA directly within the cell/tissue of interest. For example, in situ sequencing (Figure 5J, Ke et al., 2013) and fluorescent in situ RNA sequencing (FISSEQ, Figure 5K, J. H. Lee et al., 2014) generate complementary DNA (cDNA) amplicons within a biological sample and sequence the amplicons by repeatedly hybridizing and imaging fluorescent probes. Similarly, a hydrogel-assisted in situ sequencing method, Spatially-resolved Transcript Amplicon Readout Mapping (STARmap), generates cDNA nanoballs (amplicons) directly inside thick tissues (Figure 5L, M, X. Wang et al., 2018) and reads out the sequence using serial probe hybridization. STARmap amplifies the cDNA into DNA nanoballs, which contain amine-modified nucleotides that are functionalized with acrylic acid N-hydroxysuccinimide ester. After delipidation and protein digestion, volumetric gene expression profiles secured inside the hydrogel-DNA amplicon network are mapped by reading the sequence information of the amplicons using fluorescent probes. Using this method, Wang and colleagues mapped 160 to 1,020 genes and characterized the distribution of excitatory and inhibitory neurons in the mouse brain cortex. In summary, hydrogel-based tissue transformation technologies can synergize with various spatial transcriptomics approaches to enable both targeted and untargeted analysis of transcriptomic profiles in thick tissues in a scalable and comprehensive manner.
2. Proteome
Imaging proteins in situ can provide rich molecular, morphological, and functional details of individual cells. Proteomic imaging can also complement transcriptomic analysis by directly probing protein expression and post-translational modifications (Sharma et al., 2015; Uhlén et al., 2015; Thul et al., 2017). A large library of antibodies could enable the detection of more than 70% of the human proteins and their subcellular localizations (Uhlen et al., 2010). However, in traditional immunohistology, only up to five fluorophore conjugated antibodies can be differentially excited due to spectral overlap. Furthermore, given that commercially available antibodies are mostly unconjugated, the secondary antibody-based signal amplification (indirect immunofluorescence) further limits multiplexing since the cross-reactivity imposed by the host animals requires host-matching labeling schemes.
Recent advances in proteomic imaging technologies have overcome these challenges and allowed researchers to extract an unprecedented level of proteomic details from tissue sections. For instance, Angelo and colleagues studied cellular heterogeneity in tumor tissues by simultaneously imaging 10 metal-tagged antibodies (scalable to 100 targets) in 4-μm-thick tissue sections using multiplexed ion beam imaging (MIBI, Angelo et al., 2014) and mass-spectrometry-based detection (Figure 6A, B). These metal-labeling methods only require a single round of antibody incubation, and mass-spectrometry-based methods are free from autofluorescence, rendering them attractive options for mapping two-dimensional protein distributions in thin tissue sections. Similarly, barcoding methods including CODEX (Goltsev et al., 2018), diffusible probes (Guo et al., 2019), and Immuno-SABER (Saka et al., 2019) enable single round labeling of tissue sections with a library of oligonucleotide-conjugated antibodies, coding each target with a unique nucleotide sequence (Figure 6C, D). Repeated cycles of fluorescent nucleotide hybridization are then used to image the tagged antibodies to enable visualization of dozens of targets within the same thin tissue section. For 3D proteomic tissue profiling, array tomography has been widely used (Micheva & Smith, 2007). Array tomography embeds tissues in a plastic resin, followed by serial sectioning, multi-round staining, and imaging of the section arrays (Figure 6E). Although it is powerful, array tomography has been limited to relatively small tissue volumes due to the difficulties in 3D image reconstruction, requiring alternative protocols for organ-scale proteomic studies. In this section, we will discuss the current state of proteomic phenotyping and how hydrogel-based techniques synergize with other technologies to expand the boundaries of in situ proteomics.
Figure 6. Overview of spatial proteome and connectome analysis.
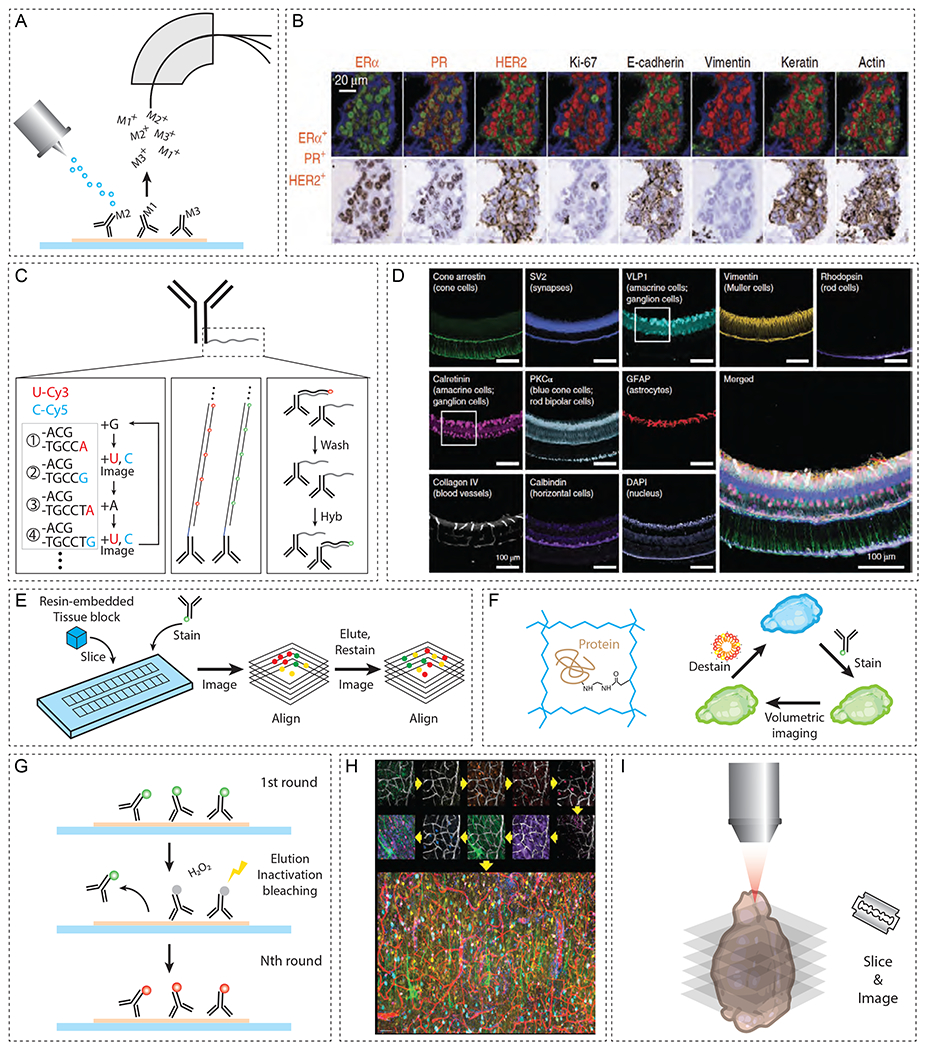
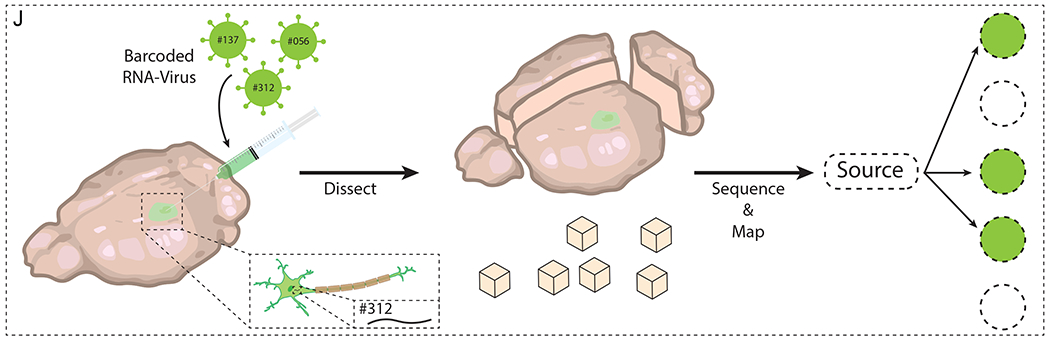
(A) MS-based multiplexing of antibody signals by tagging each antibody with different metals. (B) MIBI of patient FFPE tissue sections (adapted with permission from Angelo et al., 2014). (C) Antibodies can be tagged with oligonucleotide barcode sequences for multiplexing. (D) Immuno-SABER on 40-μm-thick mouse retina labeling ten different targets (adapted with permission from Saka et al., 2019). (E) Array tomography images of serially sectioned, resin-embedded bulk tissue to reconstruct a volumetric dataset. (F) CLARITY embeds tissues into a hydrogel mesh, reinforcing mechanical/chemical stability to allow delipidation, multi-round labeling, and volumetric imaging. (G) Repeated cycles of antibody staining with various methods of antibody removal (photobleaching, fluorophore inactivation, and physical elution). (H) Multi-round antibody staining and lectin-based co-registration in SWITCH (adapted from Murray et al., 2015). (I) Basic principle behind connectome approach relying on serial imaging of sliced planes. (J) MAPseq-based methods to multiplex connectomic analysis. Barcoded mRNAs are injected into the volumetric sample, which is then dissected and sequenced in a region-specific manner to reveal the projectome.
Hydrogel-based tissue transformation methods (e.g. CLARITY, SWITCH, SHIELD, MAP, ELAST, and ExM variants) allow volumetric visualization of a proteome in large-scale tissues (Chung et al., 2013; Murray et al., 2015; Y. Park et al., 2018; Tillberg et al., 2016). Tissue-gel hybrids (Figure 6F) can withstand antibody stripping conditions, allowing multiple-rounds of protein labeling and imaging using commercially available antibodies without any custom tagging. Removal of imaged antibodies or fluorophores can be performed in various ways, including photobleaching, chemical inactivation, and detergent-mediated dissociation of imaged antibodies (Figure 6G, Chung et al., 2013; Gut et al., 2018; J. R. Lin et al., 2015; Murray et al., 2015; Schubert et al., 2006). Detergent-based antibody stripping has been widely used in hydrogel-based methods owing to the permanent anchoring of endogenous proteins within the gel matrix. Notably, Murray and colleagues demonstrated >20 rounds of antibody labeling and imaging using SWITCH (Figure 6H, Murray et al., 2015). Hydrogel-based methods can also synergize with various antibody tagging approaches to improve throughput and multiplexing. For instance, Immuno-SABER combines ExM and SABER to enable highly multiplexed, high-resolution imaging of amplified proteomic signals (Saka et al., 2019).
Hydrogel-based tissue transformation technologies allow multi-scale proteomic interrogation of cellular features ranging from fine subcellular architectures to long-range intercellular connectivity. For example, expansion-based hydrogel transformation techniques, such as ExM and its variants (Pro-ExM, iExM) and MAP, can visualize nm-scale structures such as synapses and dendritic spines (Chang et al., 2017; F. Chen et al., 2015; Ku et al., 2016; Tillberg et al., 2016). Further improvements in expansion isotropy and compatibility with existing microscopy technologies enabled the imaging of subcellular ultrastructure such as microtubules and centrioles (Gambarotto et al., 2019; Zwettler et al., 2020). Owing to the versatile properties of hydrogels, researchers can now perform multi-scale proteomic analysis of biological systems, ranging from cm to nm.
With the aid of hydrogel-based tissue transformation methods, we can now envisage the highly multiplexed, unbiased proteomic profiles of human organs. At the moment, however, several multiplexing methods utilizing oligonucleotide-conjugated antibodies (Immuno-SABER and CODEX) have demonstrated successful application only on thin tissue sections (Goltsev et al., 2018; Saka et al., 2019). To scale up these approaches to study organs, we can employ volumetric antibody staining techniques (e.g. SWITCH, eFLASH (unpublished), ELAST) to produce homogeneous labeling profiles (Ku et al., 2020; Murray et al., 2015). These antibodies could be tagged with oligonucleotides for multiplexing, and such signals can be amplified for higher SNR, analogously to the primer exchange reactions (PER) in Immuno-SABER. In addition, expansion techniques could de-crowd the signals (F. Chen et al., 2015; Ku et al., 2016), and light sheet fluorescence microscopy (LSFM) can shorten the image acquisition time.
These futuristic suggestions must be accompanied by rigorous validation and optimization. However, we already have numerous techniques that leverage the versatile nature of hydrogels to facilitate the 3D phenotyping of biological systems. Along with transcriptomic approaches, proteomic analysis can complement our understanding of the functions and structures of mammalian organs, and hydrogel-based techniques are optimal for these applications.
3. Connectome
Intricate cellular connectivity serves as a structural foundation underlying the function of biological systems. Determining cellular connectivity is particularly important in understanding mammalian brains, which contain extensively intertwined networks organized at multiple levels. The mammalian brain consists of billions of neurons connected to local and brain-wide circuits through trillions of synapses. To map local wiring at synaptic resolution, electron microscopy (EM) is an ideal technology. EM can be used to trace all neurites and synaptic connections within small volumes without sparse labeling of individual neurons. Combined with functional imaging and electrophysiological analysis, synaptic-level network reconstruction has enabled the study of structure-function relationships of individual cells within local circuits. To map long-range connectivity between brain regions, however, light-based techniques are more appropriate. Advancements in light microscopy and genetic labeling methods have enabled imaging of sparsely labeled neurons in whole rodent brains to characterize areal connectivity. This so-called projectome provides an anatomical framework to understand system-level computations. In this section, we will discuss how tissue transformation technologies synergize with light-based approaches to facilitate connectomic studies.
Optical tissue clearing enables rapid characterization of brain-wide bulk projection patterns when combined with genetic labeling and light-sheet imaging techniques. Cell-type- and region-specific projections can be labeled by expressing FPs in a subset of neurons using genetic targeting tools. Advanced tissue clearing technologies, such as CLARITY, SHIELD, CUBIC, and uDISCO, can effectively preserve FP signal while rendering whole mouse brains transparent (Chung et al., 2013; Pan et al., 2016; Y. Park et al., 2018; Susaki et al., 2014). The cleared brains can be rapidly imaged using a light sheet microscope at single-cell resolution (1-20 μm). Intermingled axons and dendrites, however, cannot be resolved and individually traced at this resolution unless sparse labeling schemes are employed (Kebschull et al., 2016), but this approach is sufficient to visualize bulk brain-wide projection patterns. For instance, Renier and colleagues were able to use 3DISCO to visualize the bulk-projections of retinal ganglion cells after injecting cholera toxin beta (CTb) (Renier et al., 2014). In hydrogel-based tissue transformation methods, Menegas and colleagues combined CLARITY with light-sheet microscopy to map out monosynaptic input to dopaminergic neurons in the mouse midbrain that project to different brain regions (Menegas et al., 2015), and to locate the optogenetic fibers within the whole mouse brain (Menegas et al., 2017). Also, Park and colleagues used SHIELD to map brain-wide project patterns of parvalbumin-positive (PV+) globus pallidus externa (GPe) neurons and to identify their downstream target regions (Y. Park et al., 2018).
When combined with high-resolution fluorescence microscopy, tissue transformation techniques can provide sufficient resolution to map connections between individual cells. For instance, Economo and colleagues combined aqueous-based tissue clearing methods with serial two-photon (STP) tomography (Figure 6I, Economo et al., 2016; Ragan et al., 2012) to image cleared whole mouse brains with higher throughput and improved lateral (0.45 μm) and axial (1.33 μm) resolution (Economo et al., 2016). Recently, STP tomography combined with tissue clearing reconstructed axonal projection patterns of ~1,000 individual neurons (Winnubst et al., 2019), generating a database of new cell types with previously unreported connectivity in mouse brains. Taking advantage of its capability to engineer tissue size and chemical properties of biomolecules, hydrogel-based tissue transformation methods have been used to probe connections in cellular levels. For instance, Park and colleagues combined SHIELD and MAP to image sparsely labeled PV+ neurons in GPe and reconstruct their axonal projection patterns at single-cell resolution (Y. Park et al., 2018). Furthermore, Gao and colleagues combined expansion microscopy with lattice light-sheet microscopy to study neuronal projections in the Drosophila brain (Gao et al., 2019).
Although tissue transformation techniques can aid in the volumetric interrogation of a connectome, the inherent bottlenecks in connectome studies (sparse labeling and the challenge of resolving individual neurons) require higher-throughput methods of neuronal labeling. Existing high-throughput neuronal labeling methods, such as mosaic gene expression encoding various FPs (Brainbow, Livet et al., 2007) or barcoding methods (MAPseq, Han et al., 2018; Kebschull et al., 2016), can offer solutions for improving labeling efficiency. For instance, multiplexed analysis of projections by sequencing, or MAPseq, improves multiplexing by introducing barcoded RNAs into specific brain regions via viral vectors (Figure 6J, Han et al., 2018; Kebschull et al., 2016). After a certain period of viral incubation, each brain area is dissected and sequenced, revealing the destination of neuronal fibers. Compared to the tracer-based methods that are limited by neuronal density and spectral overlap, MAPseq possesses much higher multiplexing capacity since the number of barcodes increases exponentially with the sequence length. However, MAPseq is destructive, which inherently limits post hoc analysis and spatial resolution. In the future, these limitations may be overcome by combining MAPseq with hydrogel-based tissue transformation techniques and in situ sequencing. Instead of relying on post-lysis sequencing, one could decipher barcodes from MAPseq in situ, an approach that can benefit further from hydrogel-mediated expansion (higher resolution) and LSFM (fast acquisition rate). Recently, ExSeq demonstrated in situ sequencing within expanded tissues, addressing its potential use in connectome studies (Alon et al., 2021). Combined with other spatial - omics approaches, connectomics can help us better understand the communications within biological systems as well as how such communications are altered in diseased systems.
CONCLUSION
The advantage of hydrogel-based tissue transformation techniques is that researchers can readily modify physicochemical properties to preserve and extract rich molecular, anatomical, and functional information from intact tissues. In this review, we have explored the current status of hydrogel-based tissue transformation methods and outlined their basic principles to help researchers choose, optimize, and strategically combine the technologies for their studies. Furthermore, we have discussed the difficulties in scaling up 3D in situ molecular phenotyping and introduced emerging technologies that directly tackle the challenge.
The field of tissue transformation technologies has certainly made great strides over the course of the last decade; these advances have also synergized with the parallel evolution of microscopy techniques that allow rapid and high-resolution imaging of volumetric samples. There is, however, still much to be done to maximize these technologies’ impact for the research community. Continuous improvements at each step along the pipeline, from tissue transformation, labeling, and imaging to automated image analysis, will be essential, as many of the established protocols are restricted to specialized labs, often the labs of origin. During this process of improvement, the community will benefit from setting rigorous validation criteria that can be applied across different technology platforms. Considering the closely interconnected nature of each component within the pipeline, establishing an effective workflow remains challenging. For instance, one must carefully choose labeling and imaging technologies based on the tissue transformation technology used. A community-wide effort to provide detailed guidelines and standards for various applications will enable effective sharing of the technologies and protocols across laboratories.
One of the most exciting future directions is an integrative omics approach. Recently, spatial transcriptomics was highlighted as Method of the Year 2020 (Close et al., 2021; Larsson et al., 2021; “Method of the Year 2020: Spatially Resolved Transcriptomics,” 2021; Zhuang, 2021), recognizing the importance of spatial context of cells and molecules in situ. Furthermore, recent advances in the utility of hydrogels are no longer restricted to transcripts and proteins, but also incorporate functionalized lipid membrane (Götz et al., 2020). As the technologies continue to improve, the scientific community will be able to extract spatial multi-omic information from large-scale tissues with single-cell precision. Integrative organ-scale multi-omic analysis will play a key role in discovering the basic principles of biological systems and understanding disease mechanisms. A major challenge will be in making sense of the unprecedented amount of image-based data. Thus, development of robust and high-throughput algorithms that can extract quantitative information from multi-scale 3D images will be crucial. Also, interpreting the extracted spatial multi-omic information will likely require new mathematical and computational approaches. In the end, however, continuous progress in tissue transformation technologies and synergizing methods will unlock the untapped potential of holistic and integrative system-level biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors have no competing interests to declare
References
- Alon S, Goodwin DR, Sinha A, Wassie AT, Chen F, Daugharthy ER, Bando Y, Kajita A, Xue AG, Marrett K, Prior R, Cui Y, Payne AC, Yao CC, Suk HJ, Wang R, Yu CC, Tillberg P, Reginato P, … Boyden ES (2021). Expansion sequencing: Spatially precise in situ transcriptomics in intact biological systems. Science, 371(6528). 10.1126/science.aax2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y, & Nolan GP (2014). Multiplexed ion beam imaging of human breast tumors. Nature Medicine, 20(4), 436–442. 10.1038/nm.3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle M, Godefroy D, Couly G, Malone SA, Collier F, Giacobini P, & Chédotal A (2017). Tridimensional Visualization and Analysis of Early Human Development. Cell, 169(1), 161–173.e12. 10.1016/j.cell.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Bolin FP, Preuss LE, Taylor RC, & Ference RJ (1989). Refractive index of some mammalian tissues using a fiber optic cladding method. 28(12), 2297–2303. [DOI] [PubMed] [Google Scholar]
- Chang JB, Chen F, Yoon YG, Jung EE, Babcock H, Kang JS, Asano S, Suk HJ, Pak N, Tillberg PW, Wassie AT, Cai D, & Boyden ES (2017). Iterative expansion microscopy. Nature Methods, 14(6), 593–599. 10.1038/nmeth.4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, & Boyden ES (2015). Expansion Microscopy. Science, 347(6221), 543–548. 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang JB, Marblestone A, Church GM, Raj A, & Boyden ES (2016). Nanoscale imaging of RNA with expansion microscopy. Nature Methods, 13(8), 679–684. 10.1038/nmeth.3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, & Zhuang X (2015). Spatially resolved, highly multiplexed RNA profiling in single cells. Science, 348(6233), 1360–1363. 10.1126/science.aaa6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AS, Liu YC, Chiu SL, Hu SH, Huang CY, & Hsieh CH (2001). Three-dimensional mapping of brain neuropils in the cockroach, Diploptera punctata. Journal of Comparative Neurology, 440(1), 1–11. 10.1002/cne.1365 [DOI] [PubMed] [Google Scholar]
- Chozinski TJ, Halpern AR, Okawa H, Kim H, Tremel GJ, Wong ROL, & Vaughan JC (2016). Expansion microscopy with conventional antibodies and fluorescent proteins. Nature Methods, April, 1–7. 10.1038/nmeth.3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, & Deisseroth K (2013). Structural and molecular interrogation of intact biological systems. Nature, 497(7449), 332–337. 10.1038/nature12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close JL, Long BR, & Zeng H (2021). Spatially resolved transcriptomics in neuroscience. Nature Methods, 18(1), 23–25. 10.1038/s41592-020-01040-z [DOI] [PubMed] [Google Scholar]
- Codeluppi S, Borm LE, Zeisel A, La Manno G, van Lunteren JA, Svensson CI, & Linnarsson S (2018). Spatial organization of the somatosensory cortex revealed by osmFISH. Nature Methods, 15(11), 932–935. 10.1038/s41592-018-0175-z [DOI] [PubMed] [Google Scholar]
- Corsortium H (2019). The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature, 574(7777), 187–192. 10.1038/s41586-019-1629-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Gephart MGH, Barres BA, & Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. Proceedings of the National Academy of Sciences of the United States of America, 112(23), 7285–7290. 10.1073/pnas.1507125112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePas WH, Swarwalt-Lee R, Van Sambeek L, Kumar SR, Gradinaru V, & Newman DK (2016). Exposing the Three-Dimensional Biogeography and Metabolic States of Pathogens in Cystic Fibrosis Sputum via Hydrogel Embedding , Clearing , and rRNA Labeling. American Society For Microbiology, 7(5), 1–11. 10.1128/mBio.00796-16.Editor [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirckx JJJ, Kuypers LC, & Decraemer WF (2005). Refractive index of tissue measured with confocal microscopy. Journal of Biomedical Optics, 10(4), 044014. 10.1117/1.1993487 [DOI] [PubMed] [Google Scholar]
- Dodt HU, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgänsberger W, & Becker K (2007). Ultramicroscopy: Three-dimensional visualization of neuronal networks in the whole mouse brain. Nature Methods, 4(4), 331–336. 10.1038/nmeth1036 [DOI] [PubMed] [Google Scholar]
- Economo MN, Clack NG, Lavis LD, Gerfen CR, Svoboda K, Myers EW, & Chandrashekar J (2016). A platform for brain-wide imaging and reconstruction of individual neurons. ELife, 5(JANUARY2016), 1–22. 10.7554/eLife.10566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng CHL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, & Cai L (2019). Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature, 10.1038/s41586-019-1049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, & Dodt HU (2012). Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nature Protocols, 7(11), 1983–1995. 10.1038/nprot.2012.119 [DOI] [PubMed] [Google Scholar]
- Ertürk A, Mauch CP, Hellal F, Förstner F, Keck T, Becker K, Jährling N, Steffens H, Richter M, Hübener M, Kramer E, Kirchhoff F, Dodt HU, & Bradke F (2012). Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nature Medicine, 18(1), 166–171. 10.1038/nm.2600 [DOI] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, & Singer RH (1998). Visualization of single RNA transcripts in situ. Science, 280(5363), 585–590. 10.1126/science.280.5363.585 [DOI] [PubMed] [Google Scholar]
- Frei AP, Bava FA, Zunder ER, Hsieh EWY, Chen SY, Nolan GP, & Gherardini PF (2016). Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nature Methods, 13(3), 269–275. 10.1038/nmeth.3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambarotto D, Zwettler FU, Le Guennec M, Schmidt-Cernohorska M, Fortun D, Borgers S, Heine J, Schloetel JG, Reuss M, Unser M, Boyden ES, Sauer M, Hamel V, & Guichard P (2019). Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nature Methods, 16(1), 71–74. 10.1038/s41592-018-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Asano SM, Upadhyayula S, Pisarev I, Milkie DE, Liu TL, Singh V, Graves A, Huynh GH, Zhao Y, Bogovic J, Colonell J, Ott CM, Zugates C, Tappan S, Rodriguez A, Mosaliganti KR, Sheu SH, Pasolli HA, … Betzig E (2019). Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science, 363(6424). 10.1126/science.aau8302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S, & Nolan GP (2018). Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell, 174(4), 968–981.e15. 10.1016/j.cell.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz R, Kunz TC, Fink J, Solger F, Schlegel J, Seibel J, Kozjak-Pavlovic V, Rudel T, & Sauer M (2020). Nanoscale imaging of bacterial infections by sphingolipid expansion microscopy. Nature Communications, 11(1), 1–9. 10.1038/s41467-020-19897-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A, Chan KY, Dobreva T, Brown D, Balani DH, Boyce R, Kronenberg HM, Mcbride HJ, & Gradinaru V (2017). Bone CLARITY: Clearing, imaging, and computational analysis of osteoprogenitors within intact bone marrow. Science Translational Medicine, 9(387). 10.1126/scitranslmed.aah6518 [DOI] [PubMed] [Google Scholar]
- Guo SM, Veneziano R, Gordonov S, Li L, Danielson E, Perez de Arce K, Park D, Kulesa AB, Wamhoff EC, Blainey PC, Boyden ES, Cottrell JR, & Bathe M (2019). Multiplexed and high-throughput neuronal fluorescence imaging with diffusible probes. Nature Communications, 10(1). 10.1038/s41467-019-12372-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut G, Herrmann MD, & Pelkmans L (2018). Multiplexed protein maps link subcellular organization to cellular states. Science, 361(6401). 10.1126/science.aar7042 [DOI] [PubMed] [Google Scholar]
- Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, Kaneko T, Akagi T, Saito T, Saido T, & Miyawaki A (2015). ScaleS: An optical clearing palette for biological imaging. Nature Neuroscience, 18(10), 1518–1529. 10.1038/nn.4107 [DOI] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, & Miyawaki A (2011). Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nature Neuroscience, 14(11), 1481–1488. 10.1038/nn.2928 [DOI] [PubMed] [Google Scholar]
- Han Y, Kebschull JM, Campbell RAA, Cowan D, Imhof F, Zador AM, & Mrsic-Flogel TD (2018). The logic of single-cell projections from visual cortex. Nature, 556(7699), 51–56. 10.1038/nature26159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D (1969). A comparison of the crosslinking abilities of glutaraldehyde, formaldehyde and α-hydroxyadipaldehyde with bovine serum albumin and casein. Histochemie, 17(2), 151–161. 10.1007/BF00277781 [DOI] [PubMed] [Google Scholar]
- Hopwood D (1970). The reactions between formaldehyde, glutaraldehyde and osmium tetroxide, and their fixation effects on bovine serum albumin and on tissue blocks. Histochemie, 24(1), 50–64. 10.1007/BF00310003 [DOI] [PubMed] [Google Scholar]
- Horkay F, Tasaki I, & Basser PJ (2000). Osmotic Swelling of Polyacrylate Hydrogels in Physiological Salt Solutions. 84–90. 10.1021/bm9905031 [DOI] [PubMed] [Google Scholar]
- Insel TR, Landis SC, & Collins FS (2013). The NIH BRAIN Initiative. Science, 340(6133), 687–688. 10.1126/science.1239276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke MT, Fujimoto S, & Imai T (2013). SeeDB: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nature Neuroscience, 16(8), 1154–1161. 10.1038/nn.3447 [DOI] [PubMed] [Google Scholar]
- Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, & Nilsson M (2013). In situ sequencing for RNA analysis in preserved tissue and cells. Nature Methods, 10(9), 857–860. 10.1038/nmeth.2563 [DOI] [PubMed] [Google Scholar]
- Kebschull JM, Garcia da Silva P, Reid AP, Peikon ID, Albeanu DF, & Zador AM (2016). High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron, 91(5), 975–987. 10.1016/j.neuron.2016.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Cho JH, Murray E, Bakh N, Choi H, Ohn K, Ruelas L, Hubbert A, McCue M, Vassallo SL, Keller PJ, & Chung K (2015). Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proceedings of the National Academy of Sciences of the United States of America, 772(46), E6274–E6283. 10.1073/pnas.1510133112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku T, Guan W, Evans NB, Sohn CH, Albanese A, Kim J, Frosch MP, & Chung K (2020). Elasticizing tissues for reversible shape transformation and accelerated molecular labeling. Nature Methods, 10.1038/s41592-020-0823-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku T, Swaney J, Park JY, Albanese A, Murray E, Hun Cho J, Park YG, Mangena V, Chen J, & Chung K (2016). Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nature Biotechnology, 34(9), 973–981. 10.1038/nbt.3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, & Mason C (2013). ClearT: A detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development (Cambridge), 140(6), 1364–1368. 10.1242/dev.091844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson L, Frisén J, & Lundeberg J (2021). Spatially resolved transcriptomics adds a new dimension to genomics. Nature Methods, 18(1), 15–18. 10.1038/s41592-020-01038-7 [DOI] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Amamoto R, Peters DT, Turczyk BM, & Marblestone AH (2014). Highly Multiplexed Subcellular RNA Sequencing in Situ. Science, 343(March), 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenberg D, Ahyayauch H, & Goñi FM (2013). The mechanism of detergent solubilization of lipid bilayers. Biophysical Journal, 105(2), 289–299. 10.1016/j.bpj.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Fallahi-Sichani M, & Sorger PK (2015). Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nature Communications, 6, 1–7. 10.1038/ncomms9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, & Lichtman JW (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature, 450(7166), 56–62. 10.1038/nature06293 [DOI] [PubMed] [Google Scholar]
- Lubeck E, & Cai L (2012). Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nature Methods, 9(7), 743–748. 10.1038/nmeth.2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, & Cai L (2014). Single-cell in situ RNA profiling by sequential hybridization. Nature Methods, 11(4), 360–361. 10.1038/nmeth.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- M’Saad O, & Bewersdorf J (2020). Light microscopy of proteins in their ultrastructural context. Nature Communications, 11(1), 1–15. 10.1038/s41467-020-17523-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas W, Babayan BM, Uchida N, & Watabe-Uchida M (2017). Opposite initialization to novel cues in dopamine signaling in ventral and posterior striatum in mice. ELife, 6, 1–26. 10.7554/eLife.21886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas W, Bergan JF, Ogawa SK, Isogai Y, Venkataraju KU, Osten P, Uchida N, & Watabe-Uchida M (2015). Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. ELife, 4(August2015), 1–30. 10.7554/eLife.10032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Method of the Year 2020: spatially resolved transcriptomics. (2021). Nature Methods, 18(1), 1. 10.1038/s41592-020-01042-x [DOI] [PubMed] [Google Scholar]
- Micheva KD, & Smith SJ (2007). Array Tomography: A New Tool for Imaging the Molecular Architecture and Ultrastructure of Neural Circuits. Neuron, 55(1), 25–36. 10.1016/j.neuron.2007.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, & Zhuang X (2018). Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science, 362(6416). 10.1126/science.aau5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C, & Zhuang X (2016). High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proceedings of the National Academy of Sciences of the United States of America, 113(50), 14456–14461. 10.1073/pnas.1617699113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, Choi H, Park YG, Park JY, Hubbert A, McCue M, Vassallo S, Bakh N, Frosch MP, Wedeen VJ, Seung HS, & Chung K (2015). Simple, Scalable Proteomic Imaging for High-Dimensional Profiling of Intact Systems. Cell, 163(6), 1500–1514. 10.1016/j.cell.2015.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer WM, Martin AP, Flynn JR, Reed SL, White RG, Furbank RT, & Grof CPL (2015). PEA-CLARITY: 3D molecular imaging of whole plant organs. Scientific Reports, 5, 1–6. 10.1038/srep13492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Cai R, Quacquarelli FP, Ghasemigharagoz A, Lourbopoulos A, Matryba P, Plesnila N, Dichgans M, Hellal F, & Ertürk A (2016). Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nature Methods, 13(10), 859–867. 10.1038/nmeth.3964 [DOI] [PubMed] [Google Scholar]
- Park HE, Choi D, Park JS, Sim C, Park S, Kang S, Yim H, Lee M, Kim J, Pac J, Rhee K, Lee J, Lee Y, Lee Y, & Kim SY (2019). Scalable and Isotropic Expansion of Tissues with Simply Tunable Expansion Ratio. Advanced Science, 6(22). 10.1002/advs.201901673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Sohn CH, Chen R, Mccue M, Yun DH, Drummond GT, Ku T, Evans NB, Oak HC, Trieu W, Choi H, Jin X, Lilascharoen V, Wang J, Truttmann MC, Qi HW, Ploegh HL, Golub TR, Chen S, … Chung K. (2018). Protection of tissue physicochemical properties using polyfunctional. December. 10.1038/nbt.4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qidwai K, Afkhami M, & Day CE (2014). The Pathologist’s Guide to Fixatives. In Day CE (Ed.), Histopathology: Methods and Protocols (pp. 21–30). Springer New York. 10.1007/978-1-4939-1050-2_2 [DOI] [PubMed] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, & Osten P (2012). Serial two-photon tomography for automated ex vivo mouse brain imaging. Nature Methods, 9(3), 255–258. 10.1038/nmeth.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, C;atwprtjy M, Clevers H, Deplancke B, Dunham I, Eberwine J, Eils R, Enard W, Farmer A, Fugger L, Göttgens B, … Participants, H. C. A. M. (2013). Science Forum: The Human Cell Atlas. Science, 340(6133), 687–688.23661744 [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, & Tessier-Lavigne M (2014). IDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell, 159(4), 896–910. 10.1016/j.cell.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Renner M, Lancaster MA, Bian S, Choi H, Ku T, Peer A, Chung K, & Knoblich JA (2017). Self-organized developmental patterning and differentiation in cerebral organoids. The EMBO Journal, 36(10), 1316–1329. 10.15252/embj.201694700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DS, & Lichtman JW (2015). Clarifying Tissue Clearing. Cell, 162(2), 246–257. 10.1016/j.cell.2017.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka SK, Wang Y, Kishi JY, Zhu A, Zeng Y, Xie W, Kirli K, Yapp C, Cicconet M, Beliveau BJ, Lapan SW, Yin S, Lin M, Boyden ES, Kaeser PS, Pihan G, Church GM, & Yin P (2019). Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nature Biotechnology, 37(9), 1080–1090. 10.1038/s41587-019-0207-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Böckelmann R, Malykh Y, Gollnick H, Friedenberger M, Bode M, & Dress AWM (2006). Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nature Biotechnology, 24(10), 1270–1278. 10.1038/nbt1250 [DOI] [PubMed] [Google Scholar]
- Schwarz MK, Scherbarth A, Sprengel R, Engelhardt J, Theer P, & Giese G (2015). Fluorescent-protein stabilization and high-resolution imaging of cleared, intact mouse brains. PLoS ONE, 10(5), 1–26. 10.1371/journal.pone.0124650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Lubeck E, Schwarzkopf M, He T, Greenbaum A, & Sohn CH (2016). Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development, 143(15), 2862–2867. 10.1242/dev.138560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, Kongi K, Cantuti L, Hanisch UK, Philips MA, Rossner MJ, Mann M, & Simons M (2015). Cell type- and brain region-resolved mouse brain proteome. Nature Neuroscience, 18(12), 1819–1831. 10.1038/nn.4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva Santisteban T, Rabajania O, Kalinina I, Robinson S, & Meier M (2018). Rapid spheroid clearing on a microfluidic chip. Lab on a Chip, 18 (1), 153–161. 10.1039/c7lc01114h [DOI] [PubMed] [Google Scholar]
- Spalteholz W (1914). Über das Durchsichtigmachen von Über das Durchsichtigmachen von menschlichen und tierischen Präparaten (Leipzig: S. Hierzel; ). [Google Scholar]
- Stevens KR, Scull MA, Ramanan V, Fortin CL, Chaturvedi RR, Knouse KA, Xiao JW, Fung C, Mirabella T, Chen AX, McCue MG, Yang MT, Fleming HE, Chung K, De Jong YP, Chen CS, Rice CM, & Bhatia SN (2017). In situ expansion of engineered human liver tissue in a mouse model of chronic liver disease. Science Translational Medicine, 9(399). 10.1126/scitranslmed.aah5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D. en, Fan X, Shi Y, Zhang H, Huang Z, Cheng B, Tang Q, Li W, Zhu Y, Bai J, Liu W, Li Y, Wang X, Lei X, & Chen X (2021). Click-ExM enables expansion microscopy for all biomolecules. Nature Methods, 18(1), 107–113. 10.1038/s41592-020-01005-2 [DOI] [PubMed] [Google Scholar]
- Susaki EA, Shimizu C, Kuno A, Tainaka K, Li X, Nishi K, Morishima K, Ono H, Ode KL, Saeki Y, Miyamichi K, Isa K, Yokoyama C, Kitaura H, Ikemura M, Ushiku T, Shimizu Y, Saito T, Saido TC, … Ueda HR (2020). Versatile whole-organ/body staining and imaging based on electrolyte-gel properties of biological tissues. Nature Communications, 11(1). 10.1038/s41467-020-15906-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, Abe T, Kiyonari H, Shimizu Y, Miyawaki A, Yokota H, & Ueda HR (2014). Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell, 157(3), 726–739. 10.1016/j.cell.2014.03.042 [DOI] [PubMed] [Google Scholar]
- Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, & Deisseroth K (2016). Multiplexed Intact-Tissue Transcriptional Analysis at Cellular Resolution. Cell, 164(4), 792–804. 10.1016/j.cell.2016.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M, Ukai H, & Ueda HR (2014). Whole-body imaging with single-cell resolution by tissue decolorization. Cell, 159(4), 911–924. 10.1016/j.cell.2014.10.034 [DOI] [PubMed] [Google Scholar]
- Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, Bäckström A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, … Lundberg E (2017). A subcellular map of the human proteome. Science, 356(6340). 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk HJ, Yoshida F, Degennaro EM, Roossien DH, Gong G, Seneviratne U, Tannenbaum SR, Desimone R, Cai D, & Boyden ES (2016). Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nature Biotechnology, 34(9), 987–992. 10.1038/nbt.3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Ertürk A, Chung K, Gradinaru V, Chédotal A, Tomancak P, & Keller PJ (2020). Tissue clearing and its applications in neuroscience. Nature Reviews. Neuroscience 10.1038/s41583-019-0250-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson IM, Edlund K, Lundberg E, Navani S, Szigyarto CAK, Odeberg J, Djureinovic D, Takanen JO, Hober S, … Pontén F (2015). Tissue-based map of the human proteome. Science, 347(6220). 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, & Ponten F (2010). Towards a knowledge-based Human Protein Atlas. Nature Biotechnology, 28(12), 1248–1250. 10.1038/nbt1210-1248 [DOI] [PubMed] [Google Scholar]
- Wang G, Moffitt JR, & Zhuang X (2018). Multiplexed imaging of high-density libraries of RNAs with MERFISH and expansion microscopy. Scientific Reports, 8(1), 1–13. 10.1038/s41598-018-22297-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, Nolan GP, Bava FA, & Deisseroth K (2018). Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science, 361(6400), 1–18. 10.1126/science.aat5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnubst J, Bas E, Ferreira TA, Wu Z, Economo MN, Edson P, Arthur BJ, Bruns C, Rokicki K, Schauder D, Olbris DJ, Murphy SD, Ackerman DG, Arshadi C, Baldwin P, Blake R, Elsayed A, Hasan M, Ramirez D, … Chandrashekar J (2019). Reconstruction of 1,000 Projection Neurons Reveals New Cell Types and Organization of Long-Range Connectivity in the Mouse Brain. Cell, 179 (1), 268–281.e13. 10.1016/j.cell.2019.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, & Gradinaru V (2014). Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell, 158(4), 945–958. 10.1016/j.cell.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J, & Linnarsson S (2015). Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science, 347(6226), 1138–1141. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Chan KKH, Lai T, & Tang S (2013). Characterizing refractive index and thickness of biological tissues using combined multiphoton microscopy and optical coherence tomography. Biomedical Optics Express, 4(1), 38. 10.1364/boe.4.000038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X (2021). Spatially resolved single-cell genomics and transcriptomics by imaging. Nature Methods, 18(1), 18–22. 10.1038/s41592-020-01037-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwettler FU, Reinhard S, Gambarotto D, Bell TDM, Hamel V, Guichard P, & Sauer M (2020). Molecular resolution imaging by post-labeling expansion single-molecule localization microscopy (Ex-SMLM). Nature Communications, 11(1), 1–11. 10.1038/s41467-020-17086-8 [DOI] [PMC free article] [PubMed] [Google Scholar]


