Abstract
Animal models and traditional cell cultures are essential tools for drug development. However, these platforms can show striking discrepancies in efficacy and side effects when compared to human trials. These differences can lengthen the drug development process and even lead to drug withdrawal from the market. The establishment of preclinical drug screening platforms that have higher relevancy to physiological conditions is desirable to facilitate drug development. Here, a heart-on-a-chip platform, incorporating microgrooves and electrical pulse stimulations to recapitulate the well-aligned structure and synchronous beating of cardiomyocytes (CMs) for drug screening, is reported. Each chip is made with facile lithographic and laser-cutting processes that can be easily scaled up to high-throughput format. The maturation and phenotypic changes of CMs cultured on the heart-on-a-chip is validated and it can be treated with various drugs to evaluate cardiotoxicity and cardioprotective efficacy. The heart-on-a-chip can provide a high-throughput drug screening platform in preclinical drug development.
Keywords: cardiotoxicity, drug screening, electric stimulation, heart-on-a-chip, microgrooves
1. Introduction
The path from drug discovery to commercialization is challenging and requires a massive investment of time and money. A significant portion of this investment is lost in the failure to recognize severe drug-induced toxicities or poor efficacies in preclinical models.[1] Organ-on-a-chip systems have recently been developed for a variety of tissues and have gained extensive attention for improving the drug discovery process. These systems promise to be more accurate reflections of human physiology by recreating the tissue structure and environment in a miniaturized chip.[2,3] The development of these micro-physiological systems has leveraged advances in microfabrication to create advanced in vitro systems.[4] These systems have been developed for an array of organs, including the liver,[5] heart,[6] lung,[7] kidney,[8] and immune systems,[9] which could play crucial roles in the drug development process.[10,11]
The human heart continuously circulates blood through the vascular network to deliver nutrients and oxygen throughout the body. However, its central function also leaves the organ vulnerable to dangers from a variety of medical treatments. For instance, various Food and Drug Administration (FDA)-approved drugs have been withdrawn due to cardiotoxicity, such as dexfenfluramine for antiobesity treatment and dextro-propoxyphene for pain relief.[12] In addition, some anticancer treatments such as chemotherapy[13] and immunotherapy[14] can cause life-threatening heart damage or dysfunction that end their further administrations among certain patients.[15,16] Therefore, accurate preclinical evaluation of drug-associated cardiotoxicity or cardioprotective efficacy can expedite the drug development process and minimize the misallocation of resources for a wide range of drug candidates.[17]
Native cardiac tissue is mainly composed of highly organized cardiomyocytes (CMs) that beat synchronously.[18,19] The macroscale tissue-level behavior is crucial in differentiating them from cells cultured for conventional in vitro assays.[20] To close the gap between in vivo and in vitro models, various strategies have been implemented to recreate physiologically more relevant tissues in vitro.[21–23] For instance, bioinspired microstructures have been generated from extracellular matrix-mimicking hydrogel to guide the alignment of CMs.[21] To recapitulate the electrophysiological behavior of the CMs, conductive hydrogel-based scaffolds have been developed to provide an electroconductive microenvironment through the cells.[22] However, the conductive additives have been added in low concentrations and can generally lead to weak and asynchronous beating of CMs. Electrical field-based stimulation can also lead to enhanced CMs maturation and beating performance while dimensions of the in vitro model can be limited by the design of electrode.[23] To meet the need of screening cardiotoxicity of drugs, it is desirable to build in vitro cardiac tissue models that can enable high-throughput testing of “cardiac safety index” associated with cardiotoxic and cardioprotective drugs.[13]
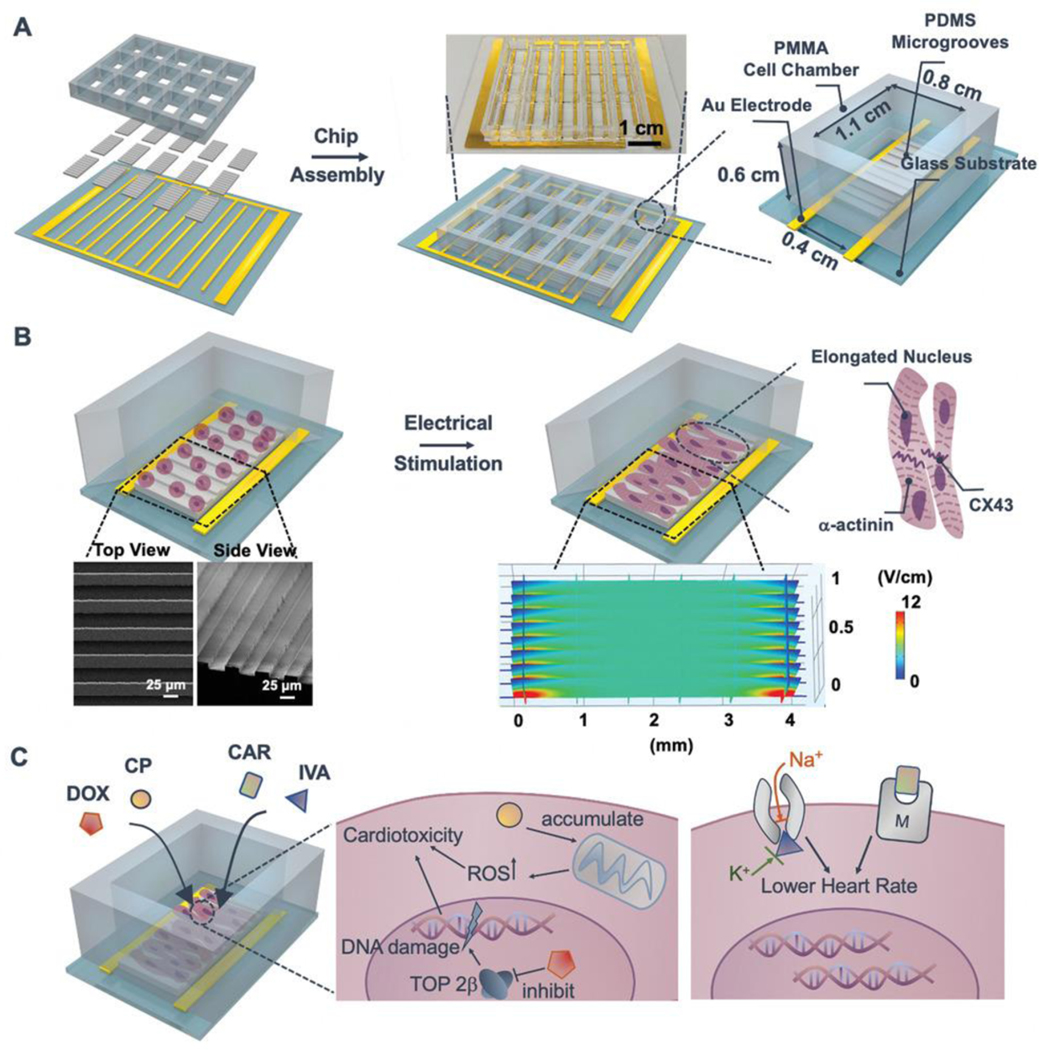
With the goal of creating a heart-mimicking high-throughput drug screening platform for cardiotoxicity, we aimed to design and develop a heart-on-a-chip platform with the following features: 1) facile fabrication, 2) controllable formation and maturation of cardiac tissues, and 3) scalable capacity for screening multiple drug dosages and candidates. The heart-on-a-chip is designed with polydimethylsiloxane (PDMS)-based micro-grooved structures (MGs) in each polymethyl methacrylate (PMMA)-based cell culture chamber, and assembled atop a comb-patterned gold electrode array for electrical stimulation (Figure 1A). The heart-on-a-chip promotes CM maturation by creating a biomimetic microenvironment in each chamber (Figure 1B) and it can be further used for drug screening (Figure 1C). To demonstrate the versatility of the heart-on-a-chip in studying cardiotoxic and cardioprotective efficacies, we choose clinically approved doxorubicin (DOX) and cyclophosphamide (CP) as model drugs to examine dose-dependent cardiotoxicity, and ivabradine (IVA) and carbachol (CAR) as candidates for ameliorating cardiotoxicity.
Figure 1.
Schematic for drug screening enabled by the heart-on-a-chip platform. A) The chip is composed of a 3 × 5 PMMA cell culture chamber, PDMS-molded MGs substrate, and comb-structure gold electrode array. Inset: the image of 15-chamber assembled heart-on-a-chip. B) CMs are seeded into MGs embedded chambers on Day 1; electrical stimulation is applied on Day 3 and lasts for 1 day. Enhanced maturation and beating behaviors of CMs are evidenced by increased alignment of α-actinin and CX43. Inset: SEM images of PDMS-based substrates with MGs structure. Inset: COMSOL simulation of the generated electrical field (≈5 V cm−1) within the MG embedded cell culture chamber by electrical pulse stimulation (biphasic, rectangular, 1 ms duration, 3.3 Hz, ±2 V). C) Drug-induced cardiotoxicity (anticancer drugs: DOX and CP) and cardioprotective efficacy (cardioprotective drugs: CAR and IVA) can be screened and evaluated in the heart-on-a-chip. DOX could inhibit topoisomerase II-β (TOP 2β) and cause damage to DNA, while CP could induce the oxidative stress (ROS accumulation) in mitochondria. IVA can block the Na+ and K+ channels to lower the beating rate; CAR is proposed to be the muscarinic (M) receptor agonist with efficacy in reducing heart beating rate.
2. Results and Discussions
2.1. Design and Fabrication of Heart-on-a-Chip
The heart-on-a-chip was fabricated using standard microfabrication strategies that enable facile scaling with minimal batch-to-batch variability. The heart-on-a-chip cell culture chamber was produced by laser cutting of PMMA to have 15 chambers and each chamber is ≈6 mm in height, ≈8 mm in width, and ≈11 mm in length. The compartmentalized design enabled testing of various drugs simultaneously on a single chip (Figure 1A). Laser cutting can easily generate diverse designs, which provide high flexibility in the number of samples to be tested in one run. Such versatility can be compatible with the commercial well-plate settings, which could be potentially compatible with current plate-reading equipment. The comb-patterned electrode was fabricated by an Au deposition method described previously[24] and integrated together with the PMMA chamber and PDMS MGs to establish an all-in-one heart-on-a-chip platform. Au was selected as the electrode material based on its excellent biocompatibility and conductivity. CMs were seeded into MGs embedded cell chambers for two days to adhere. Based on the reported electrical stimulation parameters,[23,25] 5 V cm−1 is sufficient for the stimulation of CMs and thus we applied pulsatile electrical field (biphasic, rectangular, 1 ms duration, 3.3 Hz, ±2 V) based on the distance between two parallel electrodes (0.4 cm). We further computationally simulated pulsatile electrical field in the device and electric field over the MG was found to be ≈5 V cm−1 (Figure 1B). We selected 3.3 Hz as the electrical stimulation frequency based on the report that stimulation at the natural heart beating rate of the neonatal rat (276 ± 74 beats per minute) can enhance CMs maturation.[26] The electrical stimulation started on Day 3 and lasted for one day to form the cardiac tissue samples for further characterizations or screening of drug candidates (Day 4). PDMS, a robust and biocompatible material, was used to generate MGs-patterned substrates through molding to replicate the microscale grooves to encourage CM alignment.[27] It is also an optically transparent material, facilitating microscope imaging.[28] In addition, PDMS was selected instead of a hydrogel because PDMS can achieve microscale patterns with high resolution. In addition, compared to a hydrogel, PDMS has negligible swelling and low absorption of hydrophilic drugs, which are desirable features for drug screening.[28,29] The height of each MG is ≈8 μm and different widths and spacings (25 μm width and 25 μm spacing, 35 μm width and 35 μm spacing, 25 μm width and 35 μm spacing, and 35 μm width and 25 μm spacing) of MGs were produced (Figure 1B; Figure S1A, Supporting Information) for optimization. 25 μm width and 25 μm spacing was selected for further study due to its enhanced lamella cardiac tissue formation (Figure 2A; Figure S1B, Supporting Information).
Figure 2.
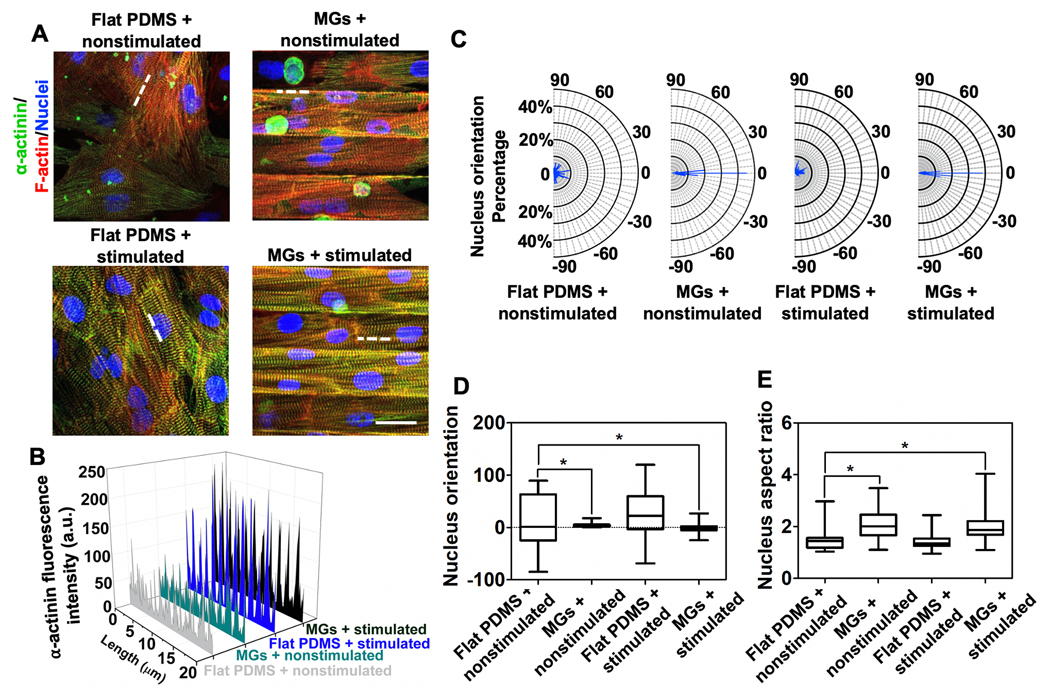
Characterization of on-chip CMs phenotype. A) α-actinin and F-actin staining of CMs cultured under Flat PDMS + nonstimulated (top, left), MGs + nonstimulated (top, right), PDMS + nonstimulated (bottom, left), and MGs + stimulated (bottom, right) conditions (Scale bar = 25 μm, CMs were seeded on Day 1, stimulated on Day 3, and imaged on Day 4.) B) Quantitative fluorescence analysis of α-actinin expression along the white dashed lines in (A). C) Frequency of CMs with different nucleus orientation under four different conditions. D) Nucleus orientation analysis of cultured CMs. E) Nucleus aspect ratios analysis of cultured CMs. (≈80 nuclei were analyzed for each group).
2.2. Characterization of CM Phenotype and Beating Behavior
We further characterized the phenotypic features of CMs cultured on the heart-on-a-chip. The MGs structure controlled the directionality of seeded CMs and improved the organization of contractile protein α-actinin (Figure 2A),[27] while stimulated CMs showed significantly enhanced α-actinin expression. Quantification of green fluorescence intensity (α-actinin) along the white dashed lines in Figure 2A confirmed the increased expression of α-actinin under the stimulated conditions (Figure 2B).[30] Additionally, the MGs enhanced CMs nucleus alignment as nearly 40% of CMs nuclei aligned parallel (0°) to the MGs while CMs cultured on flat PDMS mostly pointed to random directions (Figure 2C,D).[23] The width and spacing of MGs are optimized in the range of 25 to 35 μm based on the diameter of CMs (≈25 μm).[27] We examined various width and spacing combinations (25 μm width and 25 μm spacing, 35 μm width and 35 μm spacing, 25 μm width and 35 μm spacing, and 35 μm width and 25 μm spacing) based on their confinement effect to CMs (Figure S1, Supporting Information). We found that the one with 25 μm width and 25 μm spacing had the most significant effect in orienting the CMs (Figure 2C,D). The enhanced directionlity was mainly resulting from the MGs, while electrical stimulation slightly aligned the CMs in a cluster like manner.[24,27] Therefore, MGs could facilitate the formation of lamellar structured cardiac tissue. Furthermore, as an indicator of CMs maturation, we characterized the nucleus elongation by analyzing the aspect ratio of the nucleus. Notably, the MGs led to the appearance of nuclei with larger aspect ratios (ranging from 1.1 to 3.5), while electrical stimulation did not significantly elongate the nucleus (ranging from 1 to 2.4) (Figure 2E). This could be the influence from the confinement of MGs structure. Together, the MGs and stimulated condition led to more mature and highly aligned CMs within each cell culture chamber.
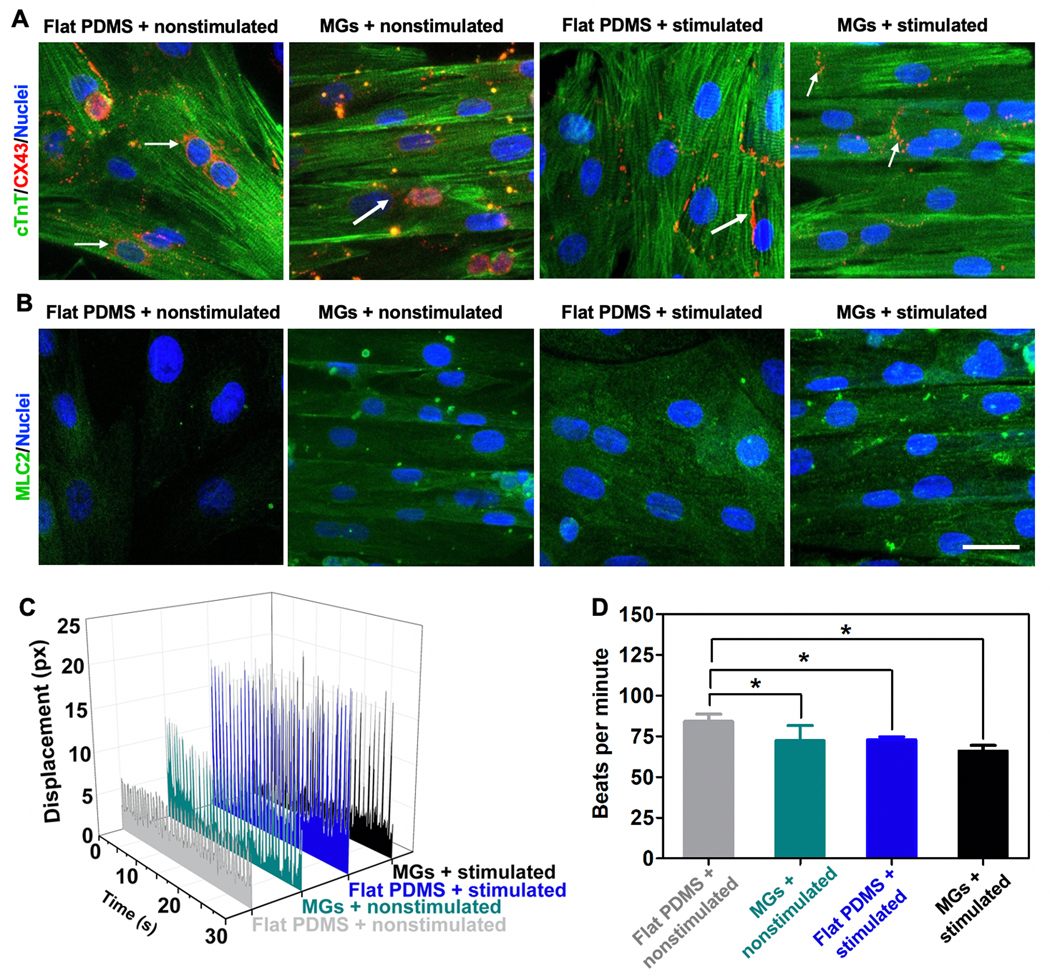
Apart from lamellar alignment of CMs, beating behavior is another crucial factor. Compared to the control (flat PDMS + nonstimulated) (Video S1, Supporting Information), MGs contributed to the lamellar alignment (Video S2, Supporting Information) and electrical stimulation contributed to the synchronous beating (Video S3, Supporting Information). Lamellar structured CMs appeared to beat synchronously in the presence of both MGs and electrical stimulation (Video S4, Supporting Information). As shown in Figure S2 of the Supporting Information, electrical stimulation (with or without MGs) could induce the rhythmic change in displacement over the monitoring period. Combined with MGs, CMs could beat synchronously while the flat PDMS + nonstimulated group exhibited nonsynchronous beating. Intrinsically, phenotypic changes in the CMs paved the basis for the synchronous beating behavior with Connexin 43 (CX43) expression localized to the cell surface instead of surrounding the nucleus as indicated by white arrows in Figure 3A, which was mostly due to direct electrical stimulation. Notably, with the help of MGs, CX43 expression was perpendicular to the direction of MGs and indicated isotropic synchronous beating behavior.[31] Furthermore, the maturity of the CMs was verified by Myosin Light Chain 2 (MLC2) staining (Figure 3B), where both MGs and stimulated conditions enhanced the maturation of CMs showing significantly increased MLC2 expression.[31] Most CMs attached to the substrate on Day 3 and then the electrical stimulation was initiated. One day of electrical stimulation can increase the maturation of CMs as evidenced by enhanced MLC2 expression (Figure 3B) and as shown in other works.[6,27] The electrical stimulation was maintained during the tissue exposure to the drugs (Days 4–5). Both MLC2 fluorescence intensity analysis and quantitative reverse transcription PCR (RT-qPCR) test (Figure S3A,B, Supporting Information) validated that the combined effects of MGs and electrical stimulation lead to enhanced expression of MLC2, indicating the enhanced maturation of CMs. In addition, benefiting from the combined effects, CX43 expressions migrated from the nuclei to cell membranes of CMs and it also exhibited increased expression as evidenced by RT-qPCR (Figure S3C, Supporting Information).[20] Furthermore, the beating behavior of the CMs under the presence of stimulated conditions was stronger as evidenced by larger displacement change of the cells compared to nonstimulated group (Figure 3C). In addition, the beating rate was lowered probably due to the stronger beating behavior (Figure 3D). In summary, the heart-on-a-chip facilitated stronger and unidirectional beating of CMs that more closely represented in vivo physiology than standard 2D culture.
Figure 3.
Characterization of on-chip beating behaviors of CMs. A) cTnT and CX43 and B) MLC2 staining of CMs cultured under four different conditions (from left to right): flat PDMS + nonstimulated, MGs + nonstimulated, PDMS + nonstimulated, and MGs + stimulated conditions. (Scale bar = 25 μm, CMs were seeded on Day 1, stimulated on Day 3, and imaged on Day 4.) C) Beating behavior of CMs was monitored over 30 s and represented as displacement change of cell morphology under four different conditions. D) Quantification of beating behavior of CMs represented as beats per minute. (Data represent mean ± SD, n = 3; *p < 0.05, **p < 0.01, ***p < 0.001).
2.3. Cardiotoxicity of Anticancer Drugs on the Heart-on-a-Chip
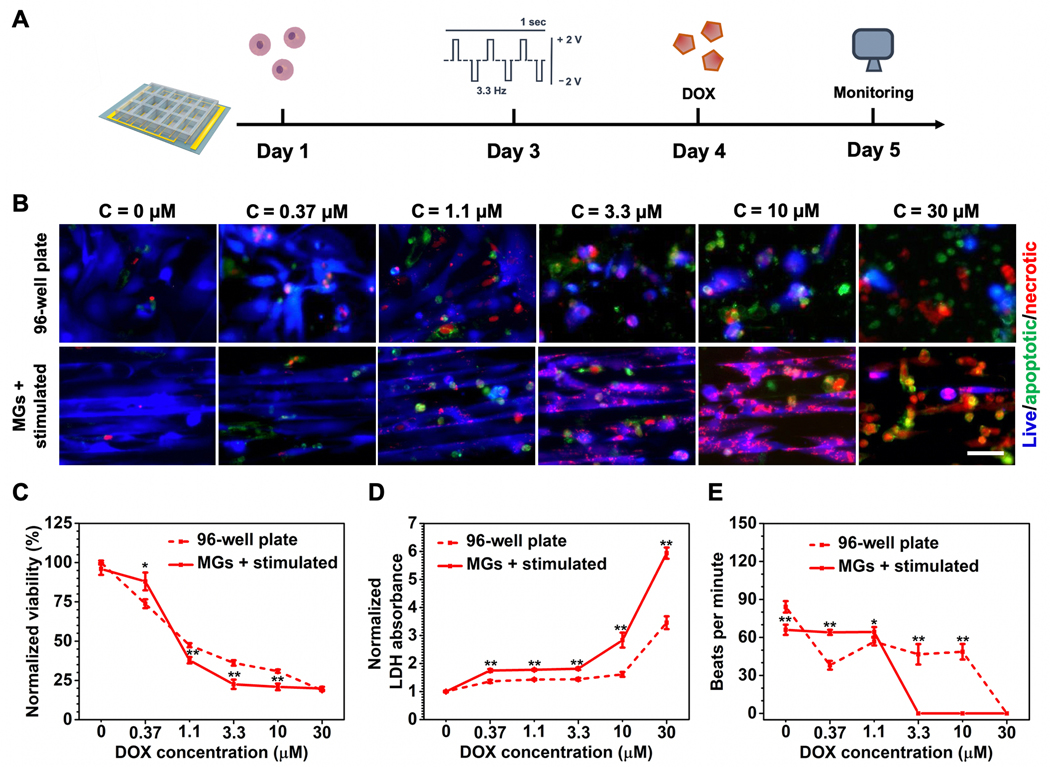
To demonstrate the applicability of the developed heart-on-a-chip for drug screening, we first selected two frequently used anticancer drugs (DOX and CP) to examine their cardiotoxicity on the heart-on-a-chip. Cardiac dysfunction is a common adverse effect of clinically approved chemotherapy and immunotherapy regimens.[32] It has been demonstrated that both DOX and CP can induce severe heart damage in which patients with normal cardiac function can suffer from lethal cardiac complications in a dose-dependent manner.[33] Failure to mimic the basic physiological features of cardiac tissue can lead to inaccurate interpretations of dose-induced cardiotoxicity in vitro. We first screened the potent chemotherapeutic drug DOX, which has documented severe cardiotoxicity among patients.[33] The timeline of cardiotoxicity evaluations enabled by heart-on-a-chip was shown in Figure 4A, where predetermined drug regimens were added into cell culture chamber on Day 4 after the formation of more matured cardiac testing samples. Compared to traditional 2D well plate-based in vitro assays, mature cardiac tissues in the heart-on-a-chip demonstrated an increased sensitivity to DOX-induced toxicity (Figure 4B). Quantitative analysis of cell viability by Cell Counting Kit-8 (CCK-8) (Figure 4C) showed consistent results. More significantly, a clinically relevant indicator, secretion of lactate dehydrogenase (LDH),[34] indicated higher percentage of CMs dysfunction in the heart-on-a-chip compared to the control well plate group under increasing DOX doses (Figure 4D). Similar discrepancies between the heart-on-a-chip and well plate group were also observed in the CP screening, in which increased damage was found in the heart-on-a-chip with lower viability and increased release of LDH (Figure S4, Supporting Information).
Figure 4.
Cardiotoxicity assessment induced by DOX. A) Schematic timeline of cardiotoxicity evaluations enabled by heart-on-a-chip. B) Representative fluorescent images of CMs viability treated by different dosages of DOX. (Scale bar = 50 μm, CMs were seeded on Day 1, stimulated on Day 3, followed by the addition of DOX on Day 4, and imaged on Day 5.) C) CCK-8 assay for quantitative analysis of CM viability treated by increasing doses of DOX. D) LDH assay for quantitative analysis of CMs viability treated by increasing dosage of DOX. E) Quantification of beating behavior of CMs treated by increasing dosages of DOX represented as beats per minute. (4C and 4D data are mean ± SEM, n ≥ 3; 4E data are mean ± SD, n = 3; *p < 0.05, **p < 0.01, ***p < 0.001).
Furthermore, one common side effect of DOX treatment is increased heart rate.[35] As shown in Figure 4E, this aspect was not observed in the control group, where beating frequency severely diminished starting from even the lowest dosage. Most cells died in the control group and the remaining live cells were beating at random asynchronous rates. However, more rhythmic beating behavior was observed in CMs cultured on the heart-on-a-chip and this behavior was maintained with the increasing DOX dosage up to 1.1 × 10−6 m (≈60 beats min−1) even though cell viability was only around 40%. The beating halted with a further increase in DOX concentration due to complete dysfunction and/or death of the CMs. Similarly, clinical manifestations of CP-induced cardiotoxicity often include tachyarrhythmias with changes in the rate of heart beat.[36] As shown in Figure S4E of the Supporting Information, the increasing frequency of heart beats was notably recaptured at lower dosage range (from 3.7 × 10−6 to 11 × 10−6 m) on the heart-on-a-chip while heart beats in the control group exhibited final increase at 300 × 10−6 m. The clinically relevant manifestations were demonstrated in the heart-on-a-chip group and they could be the result of the enhanced maturation of CMs in the heart-on-a-chip. The dose-specific balance between anticancer efficacy and severe heart damage should be studied prior to drug administration and, with platforms like ours, promisingly on an individual patient basis. The traditional strategy for determining the safety range of an anticancer agent might give inaccurate information because of the distinct cell behavior of mature tissue that cannot be recreated by traditional in vitro models. The ability to recapture several clinical cardiotoxic manifestations in the heart-on-a-chip also makes it suitable for drug response predictions and provides a platform to study clinically applicable combination therapies for ameliorating cardiotoxicity by coadministering cardioprotective therapeutics.
2.4. Cardioprotective Efficacy Study on the Heart-on-a-Chip
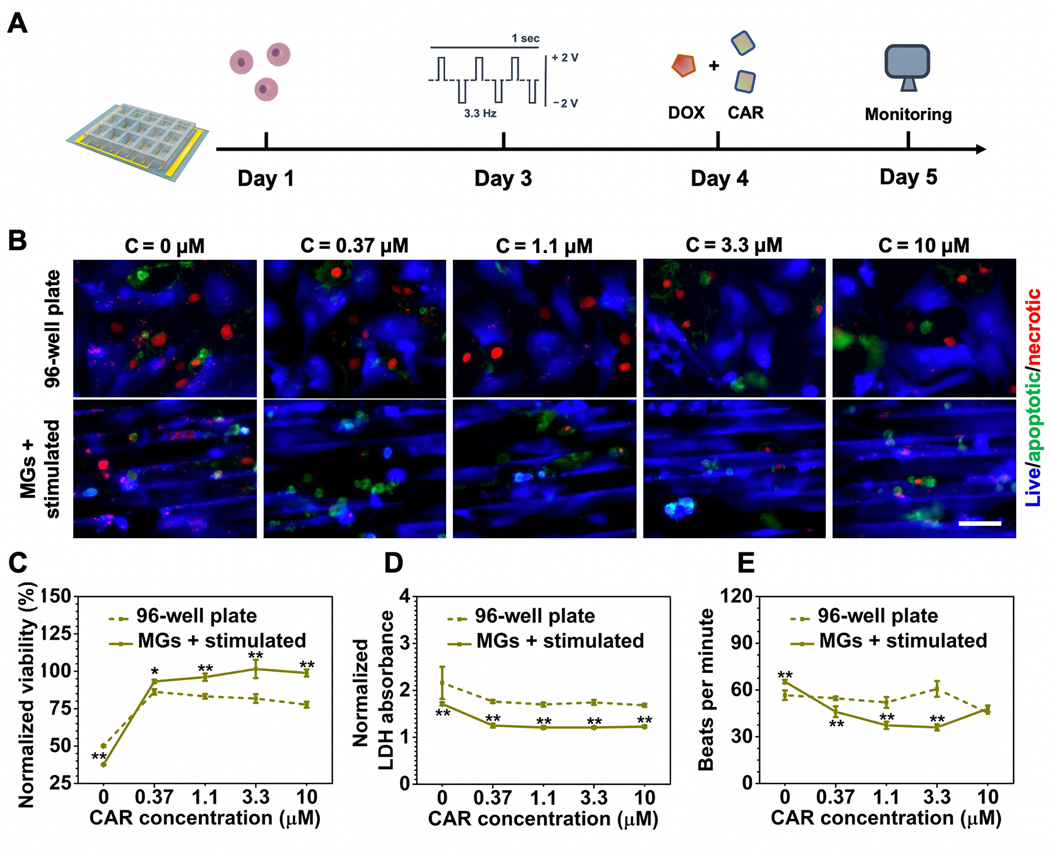
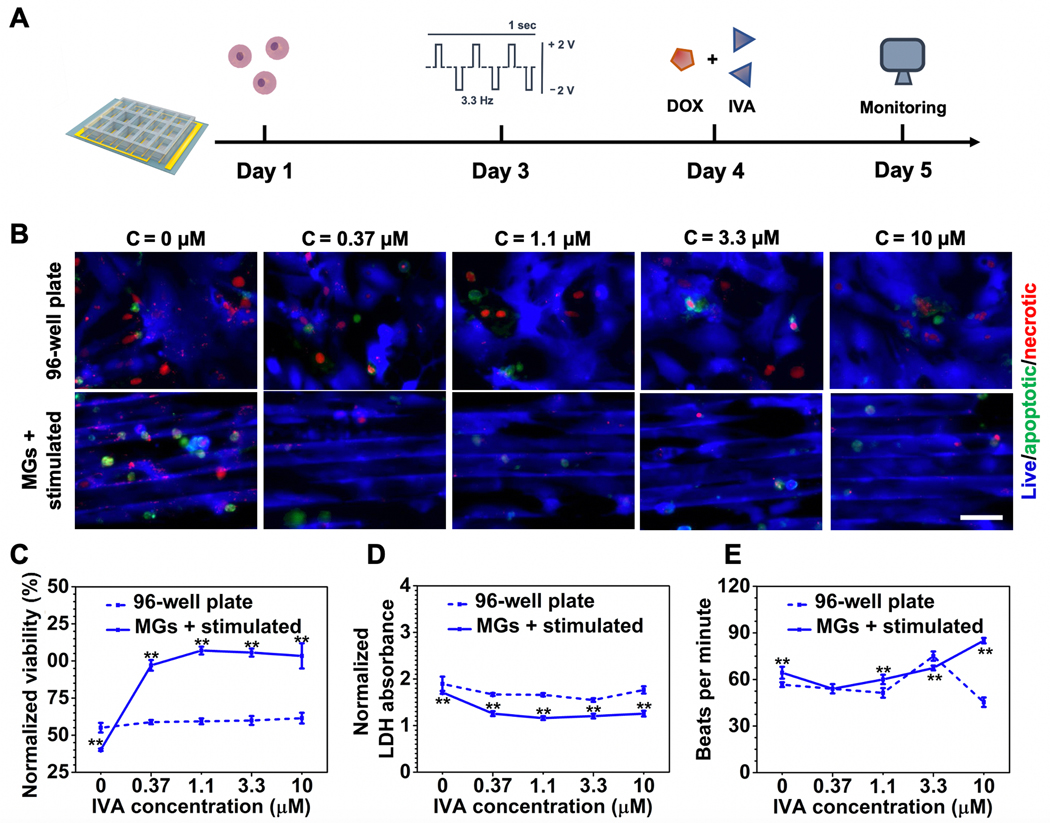
To repurpose cardioprotective agents as potential combination therapies for anticancer treatment, we examined CAR and IVA on the heart-on-a-chip. IVA is an FDA-approved therapy for heart failure, while CAR has been developed as an intraocular solution for glaucoma.[6,37] Both drugs have been studied for their potential to be repurposed as ameliorative agents for anticancer drugs but have not gained FDA approval for such indications.[36] Both CAR and IVA exhibited no cardiotoxicity and can efficiently decrease heart beating rates in a dose-dependent manner (Figure S5, Supporting Information). Here, we tested their cardioprotective efficacy in anticancer combination therapies on the heart-on-a-chip. As a broad-spectrum anthracycline anticancer drug, cardiomyopathy is one of the most severe side effects of DOX treatment and has a limited number of options to remediate its effects. Based on our previous data, DOX dosage was applied at a concentration of 1.1 × 10−6 m because the cells showed reduced viability (≈40%) while still maintaining beating behavior (≈60 beats min−1) on the heart-on-a-chip. We first evaluated CAR for their efficacy at ameliorating the cytotoxicity of DOX (Figure 5A). By increasing the dosage of CAR from 0 to 10 × 10−6 m, CMs on the heart-on-a-chip showed a significantly greater response to the protective agents compared with the control groups as shown in Figure 5B. In addition, quantitative analysis by CCK-8 and LDH assay also corresponded to this trend (Figure 5C,D). Notably, CAR exhibited effective decrease of heat beating rate (Figure 5E). Although the protective effect was observed in both groups, the drugs rendered a greater impact in the heart-on-a-chip, indicating that the maturity of the microtissue may enhance the drug efficacy. Next, we screened the efficacy of IVA in the similar regimens combined with DOX (Figure 6A). In terms of CM viability, IVA exhibited a similar trend as CAR where cardioprotective effect was more drastically demonstrated on the heart-on-a-chip group but not the well plate group (Figure 6B–D). However, CAR treatment on the heart-on-a-chip demonstrated greater potency in decreasing beating rate compared to IVA (Figure 6E). Drug repurposing is currently under extensive research and our results indicated that CAR outperformed IVA in ameliorating DOX-induced elevation in beating rates, which may give insight on screening existing compounds for new treatments.
Figure 5.
Cardioprotective efficacy assessment of CAR in combination therapy with DOX. A) Schematic timeline of cardioprotective efficacy evaluations enabled by the heart-on-a-chip. B) Representative fluorescent images of CM viability treated by increasing the dosage of CAR under the same DOX conentration (1.1 × 10−6 M). (Scale bar = 50 μm, CMs were seeded on Day 1, stimulated on Day 3, followed by the addition of DOX and CAR on Day 4, and imaged on Day 5.) C) CCK-8 assay for quantitative analysis of CM viability treated with increasing dosage of CAR under the same DOX concentration (1.1 × 10−6 M). D) LDH assay for quantitative analysis of CM viability treated by increasing dosage of CAR under the same DOX concentration (1.1 × 10−6 M). E) Quantification of beating behavior of CMs cultured under increasing dosages of CAR under the same DOX concentration (1.1 × 10−6 M) represented as beats per minute. (5C and 5D data are mean ± SEM, n ≥ 3; 5E data are mean ± SD, n = 3; *p < 0.05, **p < 0.01, ***p < 0.001).
Figure 6.
Cardioprotective efficacy assessment of IVA in combination therapy with DOX. A) Schematic timeline of cardioprotective efficacy evaluations enabled by the heart-on-a-chip. B) Representative fluorescent images of CM viability treated by increasing dosages of IVA under the same DOX concentration (1.1 × 10−6 M). (Scale bar = 50 μm, CMs were seeded on Day 1, stimulated on Day 3, following the addition of DOX and IVA on Day 4, and imaged on Day 5.) C) CCK-8 assay for quantitative analysis of CM viability treated with increasing dosage of IVA under the same DOX concentration (1.1 × 10−6 M). D) LDH assay for quantitative analysis of CM viability treated by increasing dosage of IVA under the same DOX concentration (1.1 × 10−6 M). E) Quantification of beating behavior of CMs cultured under increasing dosages of IVA under the same DOX concentration (1.1 × 10−6 M) represented as beats per minute. (6C and 6D data are mean ± SEM, n ≥ 3; 6E data are mean ± SD, n = 3; *p < 0.05, **p < 0.01, ***p < 0.001).
Combinations of anticancer and cardioprotective agents are commonly coadministrated in the clinical setting to reduce life-threatening adverse effects associated with heart damage while maintaining antitumor efficacy.[36,38] Our heart-on-a-chip platform corroborated the efficacy of potential cardioprotective drug candidates and demonstrated a differential drug response compared to traditional in vitro assays in terms of cell viability and beating rate. Based on the greater maturity of the cardiac tissue-like constructs, we believe that the results from the heart-on-a-chip more closely resemble the clinical efficacy. Other studies have designed heart-on-a-chip systems for screening a variety of drugs. However, they generally fail to recapture either the alignment or synchronous beating of CMs.[39] Integration of both characteristics of cardiac microtissues would be desirable for drug screening.[23] High-throughput screening is an important feature for drug development platforms, which is mostly limited to traditional 2D cultures.[13] Multiple clinical trials are ongoing to test a variety of drug candidates to ameliorate cardiotoxicity. Our platform is suitable to meet the increasing demand for testing a wide range of candidates by housing a physiologically relevant tissue to yield more accurate drug response evaluations. In addition, the whole drug screening process from CMs seeding and cardiac-tissue formation to final drug response monitoring takes 5 days, which could be suitable for a rapid drug screening timeframe in industrial and clinical settings.
In this study, we developed a heart-on-a-chip device that is integrated with both MG structures and gold electrodes. The synergistic effects of MGs and electrical stimulation could induce the formation of well-aligned cardiac microtissues that beat synchronously. Specifically, our culturing process only requires the direct seeding of CMs without further manipulations, such as mixing with a hydrogel-based matrix.[40,41] In addition, the process of generating CMs with synchronized beating takes only 3 days, and the entire drug screening process takes 5 days. The process fits rapid drug screening for preclinical applications.[42] Furthermore, our platform is scalable because of using comb-structured electrodes and cell culture chambers with tunable sizes and shapes. The comb-structured electrode array only requires one connection per chip,[43] and facile laser cutting could produce cell culture chambers with various sizes and designs.[44]
3. Conclusions
In summary, we have developed a heart-on-a-chip device that is designed with both MG structures and embedded electrodes to culture mature cardiac microtissues. We manufactured the system with facile microfabrication processes that can be easily scaled up. These advantages are desirable for single drug or combinational therapy development in the preclinical settings as they can effectively screen large drug panels in a relatively short timeframe. Although we optimized the device specifically for rat CMs, the physical features (MG parameters) and electrical stimulation parameters of the device can be easily adjusted to meet the requirements of different cell sources or cell types. Patient-derived cells and myocytes from other tissues, such as skeletal muscle cells, can also be incorporated. The ability to generate mature phenotypes of cardiac tissues and the modularity of the heart-on-a-chip are its major advantages that make it applicable as a preclinical drug screening tool. Its ease of use and capability to yield more relevant information on drug response make it a powerful tool in rapid drug development.
4. Experimental Section
Materials:
All reagents and chemicals were obtained from Sigma-Aldrich unless stated otherwise. Tissue culture plastics were obtained from Fisher Scientific. Cell culture reagents and media were bought from Gibco. DOX and CP were obtained from Oakwood Products, Anti-CX43 antibody (ab11370), anti-α-Actinin antibody (ab210557), anti-Cardiac Troponin T (cTnT) antibody (ab8295), anti-MLC2 antibody (ab79935), Alexa Fluor 488 antibody (ab150081), Apoptosis/Necrosis Assay Kit (blue, green, red) (ab176749), and LDH Assay Kit (Cytotoxicity) (ab65393) were obtained from Abcam. Alexa Fluor 488 antibody (A11059), Alexa Fluor 555 antibody (A21428), Texas Red phalloidin (T7471), CCK-8, Fibronectin, and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Invitrogen.
Fabrication of Microgroove Array:
A master mold with 8 μm thick was made via photolithography process using SU-8 (SU-8 2050, negative photoresist, Micro-Chem Corp.).[38] Briefly, a photomask was used to block photoresist exposure to ultraviolet (UV) in predesigned areas spun on a cleaned silicon wafer. The photoresist was developed in SU-8 developer and dried with nitrogen gas to yield the MG structures. The resulting MG structures were treated with silane (trimethyl chlorosilane) at room temperature for 3 min in a vacuum for easy removal of the PDMS from the master mold.[45] PDMS was produced by mixing prepolymer and curing agent (10:1 ratio (w/w)) (Sylgard 184, Dow Corning Co.). The mixture of the prepolymer and curing agent was gently deposited onto the SU-8 master mold, degassed in a desiccator, and baked at 80 °C for 2 h. Four types of MGs with different widths and spacings were generated: 25 μm width with 25 μm spacing, 35 μm width with 35 μm spacing, 25 μm width with 35 μm spacing, and 35 μm width with 25 μm spacing.
Fabrication of Gold Electrode Array:
The area between electrodes was covered with PMMA pieces to generate the desired pattern. The surface was then etched by O2 plasma at 100 W, 20 sccm O2, and 100 mTorr (etching rate 100 nm min−1) for 10 min. The sidewall of the grating ridges was inclined after etching, which ensures the deposition of a continuous metal film. Subsequently, the electrode area was coated with 30 nm Cr adhesion layer and 70 nm Au conducting layer. Au electrolyte (Tecknic Inc.) was used for electroplating to deposit ≈800 nm Au film on the electrode area.[24]
Assembly of the Heart-on-a-Chip:
The chambers of the heart-on-a-chip are made of PMMA by laser cutting (Universal Laser System VLS 2.3 say model). The 3 × 5 array of chambers, each 8 mm in width, 11 mm in length, and 6 mm in height, was designed to match the size of the Au electrode array. The chamber array was sealed to the gold electrode substrate using uncured PDMS. A thin layer of PDMS mixture of prepolymer and curing agent (10:1 ratio (w/w)) was applied to the bottom of PMMA chamber array. The assembled chamber array and gold electrode substrate were baked at 80 °C for 1 h. Previously fabricated PDMS MG slides were put inside each chamber and oriented perpendicular to the underlying electrodes. As a control, smooth PDMS slides were used between the electrodes.
Isolation of CMs:
Neonatal rat ventricular myocytes, provided by David Geffen School of Medicine at University of California, Los Angeles, were isolated from 1 to 3-day-old Sprague Dawley rats. After harvesting the tissue, the samples were digested with trypsin and collagenase type II digestion under the approval of the Institute’s Committee on Animal Care. The CMs were suspended at a density of 1.0 × 106 mL−1 in 10% fetal calf serum (FBS) DMEM with 0.1 × 10−3 m 5-bromo-2-deoxyuridine for future cell seeding and culture.
Device Cell Seeding and Culture:
Prior to cell seeding, devices were sterilized by UV-ozone exposure for 10 min. Subsequently, chambers were incubated with a 1 mg mL−1 solution of fibronectin in PBS for 2 h. Fibronectin solution was aspirated, and chambers were seeded with primary CMs at a density of 1.8 × 105 cm−2 in 10% FBS DMEM media. The cell media was changed every other day.
Electrical Stimulation:
After cells were incubated for 48 h without electrical stimulation, the electrode was connected to AFG1000 Arbitrary Function Generator (Tektronix Inc., OR, USA). Electrical stimulation was set in the form of symmetric biphasic pulses (3.3 Hz, amplitude of 5 V cm−1 per phase, and duration of 1 ms per phase). The desired stimulation regime was verified with an oscilloscope. For enhancing CMs maturation, electrical stimulation lasted from day 3 to day 4. For drug screening, electrical stimulation was maintained from day 4 to day 5.
Immunostaining and Fluorescent Microscopy:
To visualize morphology and phenotype of CMs cultured under different conditions, immunofluorescent staining was performed according to the vendor’s protocol. At day 4, cell-seeded chambers were rinsed with DPBS, fixed and stained with Texas Red phalloidin and DAPI to visualize the F-actin and nuclei, respectively. For immunofluorescent analysis, samples were blocked and primary antibodies (anti-α-actinin, 1:100; anti-Cx43, 1:100; anti-cTnT, 1:100; anti-MLC2, 1:100) were added for 1 h at room temperature. The samples were washed three times in PBS for 3–5 min. Secondary antibodies were then added (Alexa Fluor 488 antibody, Alexa Fluor 488 antibody, Alexa Fluor 555 antibody, 1:800) and incubated at room temperature for 1 h. The samples were washed three times in PBS for 3–5 min. Finally, samples were washed twice with 300 × 10−9 m DAPI containing PBS for 3–5 min each. Fluorescence imaging was performed by confocal microscopy (LSM880, Zeiss).
Quantification of Nuclei Elongation and Alignment:
Following a previously reported method,[46] fluorescent images of DAPI-stained nuclei acquired by confocal microscopy were processed using ImageJ to convert to a binary image and identify individual nucleus as an ellipse. For alignment analysis, the angle between the major elliptical axis of the elongated nuclei and the MGs direction was quantified as the angle of deviation. For cells cultured on the control surfaces without MGs, the angle was taken to be the angle between the long axis of the nucleus and the electric field directions or a fixed horizontal line in nonstimulated samples.
Video Imaging and Beating Behavior Quantification:
To quantify beating behavior, CMs were imaged with movies taken by camera attached fluorescence microscope (Zeiss Axio Observer, Zeiss) at 400 × magnification. Video sequences were digitized at 30 fps and CM beating behavior was analyzed as previously described.[47] Video was exported by Adobe Premiere CC software for the change of video format to AVI format. Video was rendered with VirtualDub software to make it compatible with ImageJ software. Region of interest (ROI) was selected by using a circle tool. A time series analyzer V3 plugin was used directly to analyze the dynamic pixel changes in the selected ROI. The beats per minute across different conditions were quantified for analysis of beating frequency.
RT-qPCR:
RNA was extracted from CMs using the TRIZOL (TAKARA) according to the manufacturer’s protocol. Reverse transcription was performed using the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (TAKARA). Quantitative PCR was performed with the SYBR Premix Ex Taq II Kit (Tli RNaseH Plus) (TAKARA). The cycling conditions were 95 °C for 10 min and then 40 cycles of (95 °C for 10 s, 57 °C for 30 s, and 72 °C 32 s). The relative expression of target genes was calculated using GADPH as a reference gene. The following primer sets were used for amplifications: MLC2-Fw, 5′-TCTCCATGTTTGAGCAGACC-3′; MLC2-Rv, 5′- TTTTCACGTTCACTCGTCCG-3′; CX43-Fw, 5′- AACATGCACCTAGGGTGTTC-3′; Cx43-Rv, 5′-TGTACCTCCCTTATCCCCAC-3′; GADPH-Fw, 5′-TGTGTCCGTCGTGGATCTGA-3′; GADPH-Rv, 5′-TTGCTGTTGAAGTCGCAGGAG-3′.
Drug Treatment:
Drug treatment occurs on day 4. For cardiotoxicity analysis, DOX or CP were applied for 24 h. For studies of cardioprotective efficacy against anticancer drugs, CAR and DOX or IVA and DOX were coadministered for 24 h. At the end of cultivation, cell viability and beating behavior were detected as described previously.
Cell Viability Assays:
CCK-8 was used to assess cell viability. 10 μL CCK-8 was added in every chamber and cultivated at 37 °C for 2 h. Absorbance was measured at 450 nm and the experiment was repeated three times. The Apoptosis/Necrosis Assay Kit was employed to assess cell viability after drug treatments for 24 h. After washing with PBS twice, cells were imaged using an inverted fluorescence microscope (Zeiss Axio Observer, Zeiss). The release of LDH by CMs was determined using the LDH Assay kit. Briefly, following the drug treatment, the medium in each chamber was collected and centrifuged at 3000 × g for 1 min. The supernatant was collected and analyzed based on the manufacturer’s instructions. Absorbance readings of LDH reaction products were measured at 450 nm using a SpectraMax M5 plate reader (Molecular Devices).
Statistical Analysis:
Statistical analysis was performed using GraphPad Prism 6 software package. Unpaired two-tailed Student’s t-test was used to characterize the difference between each group. Data were presented as mean ± SEM or mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 were considered to be statistically significant.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge funding from the National Institutes of Health (GM126571).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Li Ren, Prof., Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA; Key Laboratory for Space Bioscience and Biotechnology, School of Life Science, Northwestern Polytechnical University, Xi’an, Shanxi 710072, China; Research Centre of Microfluidic Chip for Health Care and Environmental Monitoring, Yangtze River Delta Research Institute of Northwestern Polytechnical University in Taicang, Suzhou, Jiangsu 215400, China.
Xingwu Zhou, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA; Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Rohollah Nasiri, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA; Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA; Department of Mechanical Engineering, Sharif University of Technology, Tehran 11365, Iran.
Dr. Jun Fang, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Prof. Xing Jiang, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA; School of Nursing, Nanjing University of Chinese Medicine, Nanjing 210023, China.
Canran Wang, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA; Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Dr. Moyuan Qu, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA; State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China.
Haonan Ling, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA; Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Yihang Chen, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA; Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Yumeng Xue, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA; Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Martin C. Hartel, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Peyton Tebon, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA; Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Dr. Shiming Zhang, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Dr. Han-Jun Kim, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA; Terasaki Institute for Biomedical Innovation, Los Angeles, CA 90024, USA.
Prof. Xichen Yuan, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA; Key Laboratory for Space Bioscience and Biotechnology School of Life Science, Northwestern Polytechnical University, Xi’an, Shanxi 710072, China; Research Centre of Microfluidic Chip for Health Care and Environmental Monitoring, Yangtze River Delta Research Institute of Northwestern Polytechnical University in Taicang, Suzhou, Jiangsu 215400, China.
Prof. Amir Shamloo, Department of Mechanical Engineering, Sharif University of Technology, Tehran 11365, Iran
Prof. Mehmet Remzi Dokmeci, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA; Department of Radiology, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA 90095, USA; Terasaki Institute for Biomedical Innovation, Los Angeles, CA 90024, USA.
Prof. Song Li, Department of Medicine, University of California, Los Angeles, Los Angeles, CA 90095, USA
Prof. Ali Khademhosseini, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA; Department of Radiology, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA 90095, USA; Terasaki Institute for Biomedical Innovation, Los Angeles, CA 90024, USA; Jonsson Comprehensive Cancer Center, University of California, Los Angeles, Los Angeles, CA 90095, USA; Department of Chemical and Biomolecular Engineering, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Prof. Samad Ahadian, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA; Terasaki Institute for Biomedical Innovation, Los Angeles, CA 90024, USA.
Dr. Wujin Sun, Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA 90095, USA Center for Minimally Invasive Therapeutics, California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA; Terasaki Institute for Biomedical Innovation, Los Angeles, CA 90024, USA.
References
- [1].Esch EW, Bahinski A, Huh D, Nat. Rev. Drug Discovery 2015, 14, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ronaldson-Bouchard K, Vunjak-Novakovic G, Cell Stem Cell 2018, 22, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sun W, Lee J, Zhang S, Benyshek C, Dokmeci MR, Khademhosseini A, Adv. Sci. 2019, 6, 1801039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA, Massa S, Riahi R, Chae S, Hu N, Avci H, Zhang W, Silvestri A, Sanati Nezhad A, Manbohi A, De Ferrari F, Polini A, Calzone G, Shaikh N, Alerasool P, Budina E, Kang J, Bhise N, Ribas J, Pourmand A, Skardal A, Shupe T, Bishop CE, Dokmeci MR, Atala A, Khademhosseini A, Proc. Natl. Acad. Sci. USA 2017, 114, E2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jang K-J, Otieno MA, Ronxhi J, Lim H-K, Ewart L, Kodella KR, Petropolis DB, Kulkarni G, Rubins JE, Conegliano D, Nawroth J, Simic D, Lam W, Singer M, Barale E, Singh B, Sonee M, Streeter AJ, Manthey C, Jones B, Srivastava A, Andersson LC, Williams D, Park H, Barrile R, Sliz J, Herland A, Haney S, Karalis K, Ingber DE, Hamilton GA, Sci. Trans. Med. 2019, 11, 5516. [DOI] [PubMed] [Google Scholar]
- [6].Zhao Y, Rafatian N, Feric NT, Cox BJ, Aschar-Sobbi R, Wang EY, Aggarwal P, Zhang B, Conant G, Ronaldson-Bouchard K, Pahnke A, Protze S, Lee JH, Davenport Huyer L, Jekic D, Wickeler A, Naguib HE, Keller GM, Vunjak-Novakovic G, Broeckel U, Backx PH, Radisic M, Cell 2019, 176, 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE, Science 2010, 328, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wilmer MJ, Ng CP, Lanz HL, Vulto P, Suter-Dick L, Masereeuw R, Trends Biotechnol. 2016, 34, 156. [DOI] [PubMed] [Google Scholar]
- [9].Jones CN, Dalli J, Dimisko L, Wong E, Serhan CN, Irimia D, Proc. Natl. Acad. Sci. USA 2012, 109, 20560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Herland A, Maoz BM, Das D, Somayaji MR, Prantil-Baun R, Cronce Novak, M., Huffstater T, Jeanty SSF, Ingram M, Chalkiadaki A, Chou D. Benson, Marquez S, Delahanty A, Jalili-Firoozinezhad, Milton Y, Sontheimer-Phelps A, Swenor B, Levy O, Parker KK, Przekwas A, Ingber DE, Nat. Biomed. Eng 2020, 4, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Novak R, Ingram M, Marquez S, Das D, Delahanty A, Herland A, Maoz BM, Jeanty SSF, Somayaji MR, Burt M, Calamari E, Chalkiadaki A, Cho A, Choe Y, Chou DB, Cronce M, Dauth S, Divic T, Fernandez-Alcon J, Ferrante T, Ferrier J, FitzGerald EA, Fleming R, Jalili-Firoozinezhad S, Grevesse T, Goss JA, Hamkins-Indik T, Henry O, Hinojosa C, Huffstater T, Jang K-J, Kujala V, Leng L, Mannix R, Milton Y, Nawroth J, Nestor BA, Ng CF, O’Connor B, Park T-E, Sanchez H, Sliz J, Sontheimer-Phelps A, Swenor B, Thompson G, Touloumes GJ, Tranchemontagne Z, Wen N, Yadid M, Bahinski A, Hamilton GA, Levner D, Levy O, Przekwas A, Prantil-Baun R, Parker KK, Ingber DE, Nat. Biomed. Eng. 2020, 4, 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fung M, Thornton A, Mybeck K, J. H.-h. Wu, K. Hornbuckle, E. Muniz, Drug Inf. J. 2001, 35, 293. [Google Scholar]
- [13].Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, Churko JM, Kitani T, Wu H, Holmström A, Matsa E, Zhang Y, Kumar A, Fan AC, Del Álamo JC, Wu SM, Moslehi J, Mercola M, Wu JC, Sci. Trans. Med. 2017, 9, 2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Heinzerling L, Ott PA, Hodi FS, Husain AN, Tajmir-Riahi A, Tawbi H, Pauschinger M, Gajewski TF, Lipson EJ, Luke JJ, ImmunoTher. Cancer 2016, 4, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ferri N, Siegl P, Corsini A, Herrmann J, Lerman A, Benghozi R, Pharmacol. Therapeut. 2013, 138, 470. [DOI] [PubMed] [Google Scholar]
- [16].Cardoso R, Nasir K, Nat. Rev. Cardiol. 2019, 16, 387. [DOI] [PubMed] [Google Scholar]
- [17].Mathur A, Ma Z, Loskill P, Jeeawoody S, Healy KE, Adv. Drug Delivery Rev. 2016, 96, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang F, Zhang N, Meng H-X, Liu H-X, Lu Y-Q, Liu C-M, Zhang Z-M, Qu K-Y, Huang N-P, ACS Biomater. Sci. Eng. 2019, 5, 3022. [DOI] [PubMed] [Google Scholar]
- [19].KlÉBer AG, Rudy Y, Physiol. Rev. 2004, 84, 431. [DOI] [PubMed] [Google Scholar]
- [20].Marsano A, Conficconi C, Lemme M, Occhetta P, Gaudiello E, Votta E, Cerino G, Redaelli A, Rasponi M, Lab Chip 2016, 16, 599. [DOI] [PubMed] [Google Scholar]
- [21].Hosseini V, Ahadian S, Ostrovidov S, Camci-Unal G, Chen S, Kaji H, Ramalingam M, Khademhosseini A, Tissue Eng, Part A 2012, 18, 2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shin SR, Zihlmann C, Akbari M, Assawes P, Cheung L, Zhang K, Manoharan V, Zhang YS, Yüksekkaya M, Wan K.-t., Nikkhah M, Dokmeci MR, Tang X, Khademhosseini A, Small 2016, 12, 3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xiao Y, Zhang B, Liu H, Miklas JW, Gagliardi M, Pahnke A, Thavandiran N, Sun Y, Simmons C, Keller G, Radisic M, Lab Chip 2014, 14, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Au HTH, Cui B, Chu ZE, Veres T, Radisic M, Lab Chip 2009, 9, 564. [DOI] [PubMed] [Google Scholar]
- [25].Song B, Gu Y, Pu J, Reid B, Zhao Z, Zhao M, Nat. Protoc. 2007, 2, 1479. [DOI] [PubMed] [Google Scholar]
- [26].Godier-Furnémont AF, Tiburcy M, Wagner E, Dewenter M, Lämmle S, El-Armouche A, Lehnart SE, Vunjak-Novakovic G, Zimmermann W-H, Biomaterials 2015, 60, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan HY, Yadid M, Park SJ, Kotikian A, Nesmith AP, Campbell PH, Vlassak J, Lewis JA, Parker KK, Nat. Mater. 2017, 16, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Halldorsson S, Lucumi E, Gomez-Sjoberg R, Fleming RMT, Biosens. Bioelectron. 2015, 63, 218. [DOI] [PubMed] [Google Scholar]
- [29].Zhu JX, Zhou XW, Kim HJ, Qu MY, Jiang X, Lee K, Ren L, Wu QZ, Wang CR, Zhu XM, Tebon P, Zhang SM, Lee J, Ashammakhi N, Ahadian S, Dokmeci MR, Gu Z, Sun WJ, Khademhosseini A, Small 2020, 16, 9. [Google Scholar]
- [30].Pasqualini FS, Sheehy SP, Agarwal A, Aratyn-Schaus Y, Parker KK, Stem Cell Rep. 2015, 4, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schulz R, Gorge PM, Gorbe A, Ferdinandy P, Lampe PD, Leybaert L, Pharmacol. Therapeut. 2015, 153, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yeh ETH, Bickford CL, J. Am. Coll. Cardiol. 2009, 53, 2231. [DOI] [PubMed] [Google Scholar]
- [33].Octavia Y, Tocchetti CG, Gabrielson KL, Janssens S, Crijns HJ, Moens AL, J. Mol. Cell. Cardiol. 2012, 52, 1213. [DOI] [PubMed] [Google Scholar]
- [34].Zhao LS, Qi Y, Xu LN, Tao XF, Han X, Yin LH, Peng JY, Redox Biol. 2018, 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].El-Naggar AE, El-Gowilly SM, Sharabi FM, J. Cardiovasc. Pharm. 2018, 72, 22. [DOI] [PubMed] [Google Scholar]
- [36].Dhesi S, Chu MP, Blevins G, Paterson I, Larratt L, Oudit GY, Kim DH, J. Investig. Med. High Impact Case Rep. 2013, 1, 2324709613480346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE, Colvin MM, Drazner MH, Filippatos G, Fonarow GC, Givertz MM, Hollenberg SM, Lindenfeld J, Masoudi FA, McBride PE, Peterson PN, Stevenson LW, Westlake C, Circulation 2016, 134, 282. [DOI] [PubMed] [Google Scholar]
- [38].Zuo GF, Ren XM, Qian XS, Ye P, Luo J, Gao XF, Zhang JJ, Chen SL, J. Cell. Physiol. 2019, 234, 1925. [DOI] [PubMed] [Google Scholar]
- [39].Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks N, Mandegar M, Conklin BR, Lee LP, Healy KE, Sci. Rep. 2015, 5, 8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang S. Masse, Gagliardi M, A., Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, adisic M, Nat. Methods 2013, 10, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhao Y, Wang EY, Davenport LH, Liao Y, Yeager K, Vunjak-Novakovic G, Radisic M, Zhang B, Adv. Healthcare Mater. 2019, 8, 1801187. [DOI] [PubMed] [Google Scholar]
- [42].Rothbauer M, Rosser JM, Zirath H, Ertl P, Curr. Opin. Biotechnol. 2019, 55, 81. [DOI] [PubMed] [Google Scholar]
- [43].Arya SK, Pui TS, Wong CC, Kumar S, Rahman AR, Langmuir 2013, 29, 6770. [DOI] [PubMed] [Google Scholar]
- [44].Klank H, Kutter JP, Geschke O, Lab Chip 2002, 2, 242. [DOI] [PubMed] [Google Scholar]
- [45].Ren L, Liu W, Wang Y, Wang JC, Tu Q, Xu J, Liu R, Shen SF, Wang J, Anal. Chem. 2013, 85, 235. [DOI] [PubMed] [Google Scholar]
- [46].Tsang KM, Annabi N, Ercole F, Zhou K, Karst D, Li F, Haynes M, Evans RA, Thissen H, Khademhosseini A, Forsythe JS, Adv. Funct. Mater. 2015, 25, 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sampurna B, Audira G, Juniardi S, Lai Y-H, Hsiao C-D, Inventions 2018, 3, 21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.