Abstract
Objectives:
Many studies suggest a strong familial component to fibromyalgia (FM). However, these studies have nearly all been confined to individuals with “primary” FM, i.e. FM without any other accompanying disorder. The current 2011–16 criteria for diagnosing FM construct a score using a combination of the number of painful body sites and the severity of somatic symptoms (FM-score). We estimated the genetic heritability of FM-score across sex and age groups to identify subgroups of individuals with greater heritability, which may help in the design of future genetic studies.
Methods:
We collected data on 26,749 individuals of European ancestry undergoing elective surgery at the University of Michigan (Michigan Genomics Initiative study, MGI). We estimated the SNP-based heritability of FM-score by age and sex categories using genome-wide association study (GWAS) data and a linear mixed model.
Results:
Overall, FM-score had an estimated heritability of 13.9% (SE=2.9%). Estimated FM-score heritability was highest in individuals ≤ 50 years of age (23.5%; SE=7.9%) and lowest in individuals >60 years (7.3%; SE=8.1%). These patterns remained the same when we analyzed FM as a case-control phenotype. Even through women had approximately 30% higher average FM-score than males across age categories, FM-score heritability did not differ significantly by sex.
Conclusion:
Younger individuals appear to have a much stronger genetic component to the FM-score than older individuals. Older individuals may be more likely to have what previously had been called “secondary FM.” Regardless of the cause, these results have implications for future genetic studies of FM and associated conditions.
Background:
Fibromyalgia (FM) is a symptom complex characterized by widespread pain accompanied by somatic symptoms such as fatigue, sleep and memory problems. Nearly all recent research studies of FM have focused on what is termed “primary FM”, which is FM without any other identifiable autoimmune or structural causes of pain. However similar symptom complexes are observed in individuals with identifiable causes of pain such as autoimmune disorders and chronic diseases. This form of FM is thought to be more similar to animal and human studies of “central sensitization”, where ongoing nociceptive input is required to drive the processes of central sensitization, at both the level of the spinal cord and brain 1–6.
Individuals with primary FM typically begin developing pain in their childhood or teens and are often diagnosed with regional pain conditions early in their life before finally being diagnosed with FM. Primary FM occurs preferentially in females, is strongly familial, and co-aggregates with other regional pain conditions in both individuals and families 7–12. In contrast to primary FM, pain with identifiable causes can be caused by pain-related diseases, such as osteoarthritis, which often occur later in life and less is known about its heritability. Understanding the pathogenic differences between FM with or without identifiable causes of pain may lead to different treatments for the two forms of FM. For example, central nervous system drugs and primary FM therapies may be less effective than identification and treatment of the ongoing nociceptive input7,13.
Candidate gene and genome-wide association studies (GWAS) comparing variant allele frequencies in FM cases and controls have been performed but many of the results have been inconsistently noted or replicated 14. However, the studies to date have been small in size (N < 1000) and would only have been able to identify common genetic variants with very large effects 14,15.
Population16 or hospital-based17 cohorts provide an opportunity to study the genetics of a wider spectrum of pain in much larger samples, although it may not always be possible to differentiate patients with or without any identifiable causes of pain given available survey or medical records and the high prevalence of individuals with peripheral sources of ongoing nociceptive input. Large studies of quantitative pain phenotypes can potentially contribute to the understanding of disease processes. A multi-site chronic pain (MCP) GWAS 18 performed using the large-scale UK Biobank data, found 76 independent MCP-associated variants at 39 loci and estimated MCP heritability of 10.2%.
Estimates of disease or trait heritability are of interest because they give a sense of the genetic contribution to the measured trait. Heritability has traditionally been estimated from family and twin studies, which require intensive participant recruitment. But narrow sense heritability (additive components) can also be estimated from cohort or case-control based study GWAS data 19,20. The estimation of heritability from GWAS data is based on the idea that if genetics underlie the predisposition to disease, individuals with more similar levels of a trait or with a disease will tend to share more alleles than individuals with less similar trait levels or without the disorder21. For example, estimates of heritability based on GWAS data in non-familial data range from 55% to 81% for height 19,22, 23% to 51% for BMI 20,23 and 37% to 50% for depression 24,25. The GWAS-based estimates are usually smaller than those estimated from familial data or twin studies, likely because they only capture additive effects from the variant classes included in the estimation (narrow sense heritability) 26.
The genetic contributions to trait level or disease risk can vary by age or sex 27, and these differences can be assessed by estimating heritability in subgroups of samples. For example, many physical measures including basal metabolic rate, systolic blood pressure, body mass index and neck pain appear to be more heritable in younger individuals 28, potentially because trait differences at younger ages are less driven by environmental factors or because the process that causes pain at older ages are different or more diverse from those at younger ages.
Our goal is to estimate the heritability of FM and of a continuous measure of FM severity and to ask if heritability differs by patient sex and/or age at assessment. To do this, we measured FM severity using patient completed 2011 Survey Criteria for FM 29,30 from 26,749 European-ancestry individuals undergoing elective surgery from the Michigan Genomics Initiative. Using GWAS data we estimated the heritability of a continuous phenotype, FM severity (FM-score), across age categories. We also dichotomized FM scores as a case-control phenotype according to two different definitions and estimated their heritability across age categories. Further, we estimated the genetic correlation of FM score with several psychiatric, personality and autoimmune traits using publicly available GWAS summary statistics.
Methods.
Patients.
Participants were prospectively recruited into the Michigan Genomics Initiative (MGI), an institutional biorepository at the University of Michigan. All patients were ≥ 18 years of age and were scheduled to have an elective surgery on the day of their recruitment. We excluded patients who did not speak English, were unable to provide written informed consent, or were currently imprisoned. We obtained written informed consent from all patients for use of their clinical data and DNA for research purposes. This study was approved by the University of Michigan IRB (IRB ID HUM00099605).
Genotyping.
We genotyped DNA from blood samples using customized versions of Illumina HumanCoreExome v12.1 array and applied quality control filters (See Supplementary Section A and B). After sample quality control, 37,412 samples remained with 462,868 polymorphic variants. Average genotyping rate of the included samples and variants was 99.96%.
We projected the genotype data for the 37,412 samples on the principal components of Human Genome diversity project (HGDP) data using TRACE 31 to infer the genetic ancestry of the samples. Of these samples, 31,730 (84.8%) individuals were inferred to be of European ancestry. We estimated the sample kinship 32 and retained 30,431 samples who had less than a second-degree relationship. We next performed principal component analysis on their genotype data. We excluded 33 samples who were outliers based on the first and second principal components resulting in 30,398 samples with genotype data. (See Supplementary Section C and Supplementary Figure 1).
Phenotyping.
We phenotyped patients preoperatively using a self-report questionnaire of widespread pain and psychological status, based on the American College of Rheumatology (ACR) survey criteria for FM. These criteria for FM, conceptualized in 2011, represent a validated self-report measure based on presence of widespread pain and comorbid symptoms 29,30. To calculate the Widespread Pain Index (WPI), we assessed 19 specific body areas using the Michigan Body Map (score 0–19) as described in the ACR survey criteria 30. To calculate Symptoms Severity Index (SSI) we used the comorbid Symptom Severity scale with questions on fatigue, trouble thinking or remembering, waking up tired, pain or cramps in the lower abdomen, depression, and headache (score 0–12). Following Wolfe et al 30 we summed the WPI and SSI to create FM scores (potential range 0 to 31).
To dichotomize patients as FM-cases or controls based on FM-score and its components, we used two definitions,
FM-2011: any individual with FM-score ≥ 13 was defined as a FM-Criteria2011 case
FM-2016-modified: any individual with 4 of the 5 main body regions having pain and WPI-score ≥ 7 and SSI-score ≥ 5 or WPI-score 4–6 and SSI-score ≥ 9.
FM-2016-modified is a modified version of the criteria outlined by Wolfe et al 33 in 2016, adapted to our study according to the availability of data (See Supplementary Section D for details). All individuals not defined as a case were defined as controls.
Of 30,398 European ancestry individuals with genotype data, 3,649 (12.0%) did not have an FM score (WPI was missing for 2,708 (8.9%) and SSI was missing for 3,494 (11.5%) individuals), leaving 26,749 individuals for analysis.
Log transformation of FM, WPI and SSI.
Since the distribution of FM was highly skewed, we added a small constant (0.1) to the FM score to retain individuals with a value of zero in the analysis, and log transformed the adjusted score. We regressed the log-transformed FM (Log-FM) score on the age, age2 and sex using a linear regression model and inverse normalized the residuals (inverse normalized FM score). The same transformations were applied to WPI and SSI.
Estimation of heritability and genetic correlation.
The genetic contribution to a phenotype can be measured by heritability, the fraction of trait variation explained by genetic variation. We estimated heritability of FM-score, WPI and SSI using a linear mixed effect model (LMM) 34.
Where y is the vector of phenotype values for n individuals, in this case the inverse normalized FM-score (or inverse normalized WPI/SSI score); x is a matrix of non-genetic covariates containing the top 10 genetic principal components and a binary variable with levels 0 and 1 indicating the genotype array (UM_HUNT_Biobank or UM_HUNT_Biobank_v1–1); is the vector of corresponding fixed effects; is a random vector of genetic contributions to the phenotypes, random effects, with being the genetic relatedness matrix (GRM) between the pairs of individuals (See Supplementary Section E for the construction of GRM); is the genetic variability contributed by the genetic relatedness of the samples; is the residual variance of the model. The heritability of the phenotype y is estimated as . We used Genome-wide Complex Trait Analysis (GCTA) 35 to fit this model and estimate the heritabilities. We further used a likelihood-ratio-test (LRT) to evaluate the significance of the estimated heritability. We note here that ℎ2 used here is a narrow sense measure of heritability. Thus ℎ2 only measures the fraction of variability in trait explained by the additive effects of the variants in the array which is lower than the total trait heritability. For a case-control phenotype (binary), we use the same linear mixed model but with liability scale adjusting for ascertainment probabilities of the cases by the population prevalence 36. Here, we have used two different ways to dichotomize the individuals with FM-score into cases and controls. For both the case-control definitions to estimate the heritability of FM as a case-control phenotype we used a population prevalence of 2% 37.
The genetic overlap between two phenotypes y and w can be measured by the genetic correlation 20. We estimated the genetic correlation among FM-score, SSI, WPI using multiple linear mixed model. For phenotype, w, we used the same mixed model (LMM) as for y
with the additional assumption is defined as the co-heritability between the phenotypes y and w. The genetic correlation between the phenotypes is then defined as We used Phenix 38 to fit the model and calculate the co-heritability and subsequently estimated the genetic correlations. Further, we estimated the genetic correlation of FM score with selected traits using publicly available summary statistics from existing genetic association studies using LD score regression39–41.
Results
We included in our analysis 26,749 European ancestry patients who were scheduled for elective surgery. These patients had an average age of 54.2 years (SD = 15.9) and 53.2% were female. The distribution of the FM scores for 26,749 individuals is shown in Figure 1. 10.8% of the sample reported FM score of 0 (both the WPI and SSI scores equal 0) (see Supplementary Figure 2 for WPI and SSI distributions).
Figure 1:
Distribution of FM Scores in the sample included for analysis. The range of FM scores is from 0 to 31. A blue vertical line corresponding to the FM-score of 13 shows the cut-off for being treated as FM. Any sample having a FM-score above or equal to 13 is clinically considered to be an FM patient.
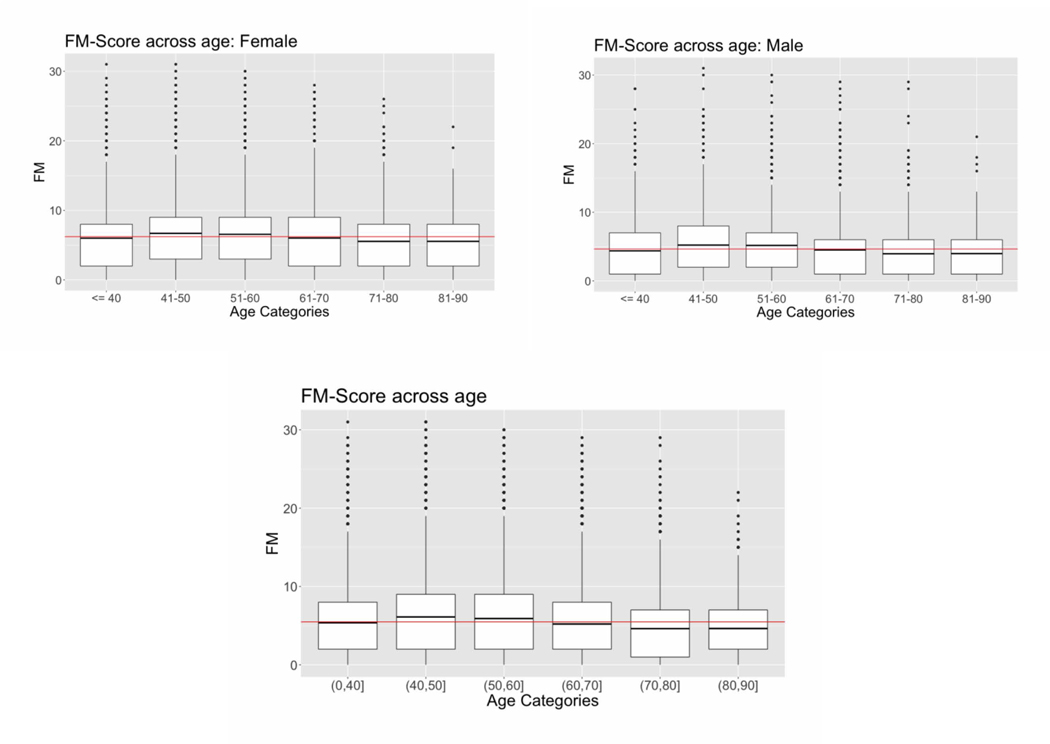
FM scores by age categories and by sex are shown in Figure 2. When divided into age subcategories of 10 years, the age categories 40–50 and 50–60 had significantly higher FM-scores compared to the age category ≤ 40 and the age categories 60–70, 70–80 and 80–90 had lower FM-score compared to the age category ≤ 40 (Figure 2). This pattern was consistent for females and males (Figure 2) (for regression estimates see Supplementary Table 1). In the total sample, the average FM score (mean ± SD) was higher in females than in males (6.2 ± 4.9 versus 4.6 ± 4.2 respectively, mean female FM score / mean male FM score = 1.34) (see Supplementary Table 2 for the FM scores for patients dichotomized at 50 years).
Figure 2:
Distribution of Fibromyalgia (FM) scores in women, men, and the entire sample, divided into 10- year age categories. Data are shown as box plots, where the boxes represent the 25th to 75th percentiles, the lines within the boxes represent the median, and the lines outside the boxes represent the 10th and 90th percentiles. Circles indicate outliers. Red lines indicate the mean FM score across all age categories.
We asked if the components of FM score, measurements of pain at different sites of the body (WPI) and symptoms of pain (SSI) had consistent trends across age and sex (Supplementary Table 3). Younger (age ≤ 50) individuals had a lower WPI-score than the older (age > 50) individuals (1.8 ± 2.6 versus 2.0 ± 2.6 respectively). In contrast, younger individuals had a higher SSI-score than the older individuals (3.8 ± 2.9 versus 3.4 ± 2.8) (Supplementary Table 3). Females had higher WPI and SSI scores than males (2.1 ± 2.8 versus 1.6 ± 2.3 respectively for WPI and 4.1 ± 2.9 versus 3.0 ± 2.6 for SI).
To evaluate the significance of age and sex on FM-score related measures, we used a multiple linear regression model simultaneously adjusting for age, age2 and sex. We found that age, age2 and sex were significantly associated with FM, WPI and SSI (p-value<0.05) (Supplementary Table 4). To determine if the effect of age (younger or older) on FM score varied by sex or the effect of sex on FM score varied by age category (younger or older), we tested for an interaction between age category and sex. We did not find significant evidence that the effect of sex varied between older and younger individuals or that the effect of age varied between sex.
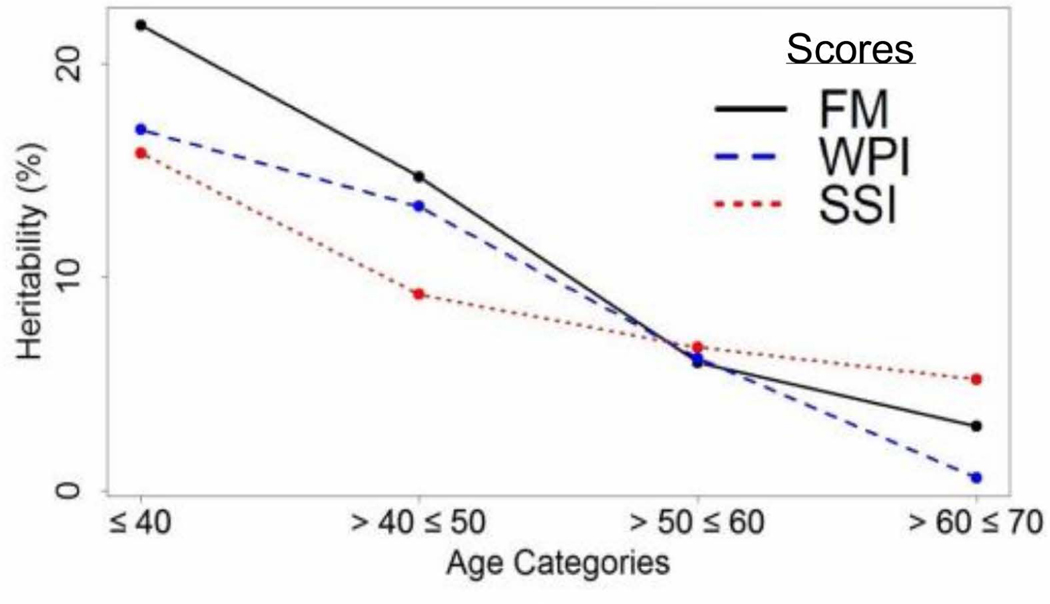
To estimate the genetic contribution to FM score, we calculated the genotype-based heritability of the inverse normalized FM score. We constructed the GRM using the common variants (minor allele frequency > 5%) and fit a linear mixed model (See Methods). Estimated FM score heritability was 13.9% (SD=2.9%; p-value= 1.59 × 10−7). To examine if the heritability differed by age, we divided the sample into 10-year age categories. Patients with age ≤ 40 years had the highest heritability, estimated at 22.8% (SD=13.4%) and those aged 60–70 years had the lowest heritability, estimated at 3%, although no age category was significantly heritable on its own. We saw similar trends for WPI and SSI (Figure 3).
Figure 3:
Heritability of FM, WPI and SSI scores by age categories. The vertical axis represents the estimated heritability (in percentage) while the horizontal axis represents the different age categories from younger to older (left to right) in groups of 10 years.
To obtain larger age subgroups, we dichotomized patients by age cut-offs and estimated the heritability in the corresponding age groups (Table 1). For each age cut-off we observed that younger individuals had consistently higher heritability of FM score than older individuals. For example, individuals with age ≤ 50 years had an estimated heritability of 23.5% (SD=7.9%; p-value =3.0 × 10−4) and those with age > 50 had an estimated heritability of 8.6% (SD=5.6%; p-value = 0.12). This means that the estimated heritability of FM for individuals ≤ 50 years is significantly higher than 0. Conversely, the heritability for individuals > 50 years is low and could not be distinguished from 0 in this sample.
Table 1:
Heritability of inverse normalized FM score (see Methods) in different age categories with the corresponding p-values.
| Age Category (years) |
N | Heritability |
||
|---|---|---|---|---|
| Estimate (%) | SE (%) | P-value | ||
|
| ||||
| ≤40 | 5,693 | 22.8 | 13.4 | 0.09 |
| >40 | 21,056 | 8.1 | 4.9 | 0.06 |
|
| ||||
| ≤50 | 10,201 | 23.5 | 7.9 | 3.0 ×10−4 |
| >50 | 16,548 | 8.6 | 5.6 | 0.12 |
|
| ||||
| ≤60 | 16,687 | 12.4 | 5.5 | 0.01 |
| >60 | 10,062 | 7.5 | 8.1 | 0.41 |
|
| ||||
| All | 26,749 | 13.9 | 2.9 | 1.6 × 10−7 |
When we repeated the analysis by age category separately for males and females, we found that females had slightly higher estimated heritabilities than males in almost all age categories. However, we found no evidence of a significant difference in the estimated heritabilities between males and females (Supplementary Section F, Supplementary Table 5).
To assess the heritability in individuals more likely to have FM we used the two definitions of FM cases, 1) any individual with FM-score ≥ 13 was defined as a FM-2011 case22 (Number of cases= 2,304; sample prevalence = 8.6%) and 2) any individual with 4 of the 5 main body regions having pain and WPI-score ≥ 7 and SSI-score ≥ 5 or WPI-score 4–6 and SSI-score ≥ 9 defined as a FM-2016-modified case (Number of cases= 1,319; sample prevalence = 4.9%) (adapted from Wolfe et al 2016). All individuals not defined as a case were defined as controls. All but 48 of the FM-2016-modified cases were also defined as FM-2011 cases. We estimated heritabilities as 8.6% (p-value=0.005) for FM-2011 and 7.9% (p-value =0.41) for FM-2016-modified. When divided into age categories, we observed higher estimated heritability for younger individuals compared to older individuals, suggesting that the overall trends by age held true in the data across FMness measured by different criteria (Supplementary Table 6A and 6B).
To understand the contributions of the WPI and SSI to the age-based FM score heritability trends, we estimated the genetic correlations of FM score with WPI and SSI by 10-year age categories (Figure 4). The estimated FM score-WPI genetic correlation varied from 38% for the younger individuals (age ≤ 40), to 56% for the individuals with age > 40 ≤ 50 and age > 50 ≤ 60 and 30% for the older group of individuals (age > 60 and ≤ 70). The FM score-SSI genetic correlation varied from 57% in individuals with age ≤ 40 to 88% in individuals with age > 60 and ≤ 70. The estimated genetic correlation between WPI and SSI varied between 55% for the individuals with age > 40 and ≤ 50 to 6% for the older individuals (age > 60 and ≤ 70). These genetic correlation estimates show that for younger individuals, both WPI and SSI contribute substantially towards the genetic components of FM, while in the older individuals SSI appears to be the dominant component. Further, our estimates show that, for younger individuals, WPI and SSI have a substantial shared genetic component, while in older individuals, they have a low genetic correlation. Using FM-2011 and FM-2016-modified definitions to dichotomize individuals to cases and controls, we found similar patterns of genetic correlations across age categories (Supplementary Figure 3).
Figure 4:
Estimated pairwise genetic correlation matrices between FM, WPI and SSI by age categories. Different colors represent different combinations of traits used to estimate the genetic correlation.
We next tested for co-heritability of FM score with traits that might a priori be expected to be correlated with FM score or were found to be significant in the UK Biobank study of multi-site chronic pain (MCP). FM score had a significant genetic correlation with psychiatric disorders such as ADHD, neuroticism, major depressive symptoms, subjective well-being and depressive symptoms (−0.26 to 0.78; Table 2). We also found significant genetic correlation of FM score with immune and autoimmune diseases like asthma and rheumatoid arthritis (0.31 to 0.35; Table 2). To understand the contribution of SSI and WPI to the genetic correlations of FM score, we separately estimated their genetic correlations with the same traits. For almost all tested traits, SSI had similar estimated genetic correlations as FM score. WPI had similar estimated genetic correlations as FM score for asthma and rheumatoid arthritis, and of the tested traits, these two were the most strongly genetically correlated with WPI. In contrast, WPI showed much lower genetic correlation with psychiatric traits (−0.12 and 0.19) than we found for FM score or SSI, although nominal significance for genetic correlation of WPI with ADHD and neuroticism in younger individuals. Multiple phenotypes that were reported to have significant genetic correlation with MCP in the UK Biobank study did not have significant genetic correlation with FM score, WPI or SSI. However, the directions of genetic correlations for these traits were highly consistent between the UK Biobank and our sample (9 out of the 10 reported traits in Table 2 had the same direction of effects). Further, the estimated genetic correlations for these traits were highly correlated (r > 0.9) to those estimated in UK Biobank sample for MCP.
Table 2:
Genetic correlation of FM score, WPI score and SSI score with selected traits for overall sample as well as across age categories.
| Trait | PMID | Genetic Correlation (All individuals) |
Genetic Correlation (individuals with age ≤ 50) |
Genetic Correlation (individuals with > 50) |
Genetic correlation in UK Biobank |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FM | WPI | SSI | FM | WPI | SSI | FM | WPI | SSI | MCP | ||
| Depressive Symptoms P | 27089181 | 0.53* | 0.11 | 0.49* | 0.63* | 0.12 | 0.59* | 0.49* | 0.07 | 0.44* | 0.59* |
| Major Depressive disorder P | 29700475 | 0.40* | 0.08 | 0.43* | 0.48* | 0.07 | 0.50* | 0.36* | 0.09 | 0.37* | 0.53* |
| Bipolar disorder P | 21926972 | 0.08 | 0.03 | 0.09 | 0.13 | 0.04 | 0.14* | 0.06 | 0.04 | 0.07 | 0.02 |
| Attention deficit hyperactivity disorder P | 27663945 | 0.78* | 0.19* | 0.66* | 0.81* | 0.20* | 0.80* | 0.53* | 0.15 | 0.54* | Not reported |
| Subjective Well Being P | 27089181 | −0.26* | −0.12 | −0.28* | −0.25* | −0.11 | −0.23* | −0.21* | −0.14 | −0.22* | Not reported |
| PGC Cross disorder analysis P | 24353885 | 0.28* | 0.15 | 0.33* | 0.32* | 0.13 | 0.34* | 0.25* | 0.11 | 0.30* | 0.13* |
| Autism Spectrum disorder P | 30804558 | 0.04 | −0.01 | 0.05 | −0.01 | −0.02 | 0.04 | 0.05 | 0.01 | 0.07 | −0.10* |
| Anorexia Nervosa P | 24514567 | −0.09 | −0.06 | −0.12 | −0.13 | −0.07 | −0.17* | −0.07 | −0.06 | −0.10 | −0.06* |
| Rheumatoid Arthritis A | 24390342 | 0.35* | 0.38* | 0.26* | 0.38* | 0.39* | 0.25* | 0.31* | 0.32* | 0.19* | 0.16* |
| Asthma A | 17611496 | 0.31* | 0.33* | 0.30* | 0.40* | 0.45* | 0.39* | 0.28* | 0.30* | 0.22 | 0.22* |
| Primary biliary cirrhosis A | 26394269 | 0.13 | 0.16 | 0.12 | 0.15 | 0.16 | 0.09 | 0.11 | 0.13 | 0.06 | 0.10* |
| Neuroticism R | 27089181 | 0.39* | 0.16 | 0.35* | 0.45* | 0.17* | 0.41* | 0.37* | 0.13 | 0.38* | 0.40* |
| Sleep duration S | 27494321 | −0.03 | 0.05 | −0.11 | −0.02 | 0.01 | −0.08 | −0.03 | 0.02 | −0.08 | Not reported |
| Multi-site Chronic Pain | 31194737 | 0.46* | 0.38* | 0.29* | 0.49* | 0.43* | 0.31* | 0.41* | 0.37* | 0.21* | – |
Significant (p-value < 0.05) estimates are marked as
Superscript index:
= Psychiatric trait;
= immune/autoimmune trait;
= Personality trait;
= Sleeping trait.
We also estimated the genetic correlation of FM score, WPI and SSI with the UK Biobank MCP. We found significant genetic correlations of MCP with FM-score (0.46), WPI (0.38) and SSI (0.29). For younger individuals the genetic correlations were slightly higher than those estimated for older individuals (Table 2).
Conclusions
The present study is the largest to date to examine the genetic contributions to the FM-score (a composite measure of WPI and SSI). We found that FM score is more heritable in younger than in older individuals within this hospital-based sample. Thus, the variability in FM score for younger individuals, which is potentially more likely to be primary FM, appears to be more driven by genetic factors shared across individuals than in older individuals. Older individuals may have a greater contribution of environmental factors to pain, a greater diversity of conditions that increase pain, and/or more susceptibility towards nociceptive pain.
We found that there is a substantial genetic correlation of FM with both WPI and SSI for younger individuals, indicating that both the pain component (WPI) and the comorbid symptoms component (SSI) jointly contribute to the genetic architecture of FM for the younger individuals. In contrast, for the older individuals, the heritability of FM is more highly correlated with comorbid (SSI) component than with the pain (WPI) component. Overall, our results suggest that genetic studies of FM might have differing results dependent on the age of the participants.
If FM in younger individuals stayed constant throughout their lives one would expect FM measures to slowly increase with age as pain from chronic diseases increases, but in this study, we see that the mean FM score (in individuals undergoing elective surgery) is slightly lower in older individuals with age. Other studies have shown that the incidence and prevalence of FM wanes over time. Wolfe et. al. 37 showed that the prevalence of chronic widespread pain in Kansas peaked at age 60–69 and then decreased in older individuals. Vincent et. al. 42 used the 2011 FM Survey criteria to show a prevalence of FM in the general Minnesota population at 8.4% for ages 21–39, 6.0% for ages 40–59, and 3.8% for individuals over age 6027.
Given an individual’s genetic information is constant from birth until death (notwithstanding epigenetic modifications), our results suggest that, for a set number of FM cases, inclusion of younger individuals might increase the power to detect primary FM. All previous GWAS or large candidate gene studies in FM were composed of many individuals over 50 years of age or have not reported age 15,42,43. Thus, all of the large-scale genetic studies performed to date in FM have included sizable numbers of older individuals, where the genetic contributions to FM or FM symptoms might have been lower or different than in younger cohorts.
Differentiating individuals with primary FM from those with pain from an identifiable source, in a hospital or electronic health record (EHR)-based cohort is difficult even with the potentially large arrays of phenotype data. Although EHR-based studies can reduce diagnosis/reporting misclassifications and recall bias when compared to cohort studies, ongoing sources of nociceptive pain might still be present without a diagnosis and hence not identified in an EHR-based study. In particular, nociceptive pain might be relatively more common in the population we have considered here, which is based on elective surgery. How to distinguish between pain with or without identifiable causes in such an EHR-based study remains a largely unanswered question. This in turn impedes our and other EHR-based studies abilities to isolate the patients with primary FM. Johnston et al. 18 in their UK Biobank based study of multi-site chronic pain did not report if individuals have identifiable or non-identifiable sources of pain. Given our current measures of FM we cannot definitively say if the older individuals we have classified as being FM cases have primary FM or have pain from an identifiable source. Thus, we do not have data to speak to if primary FM has a smaller genetic component in older individuals than it does in younger individuals.
Although across age categories females had higher FM scores than males, we do not find evidence that heritability varies by sex. This is possible because women can have a higher mean FM score value than men, for example because sex-related factors cause higher FM score values in females, but still have the same amount of FM score variability explained by genetic variation. We can interpret this as follows: although the average FM-scores in females are higher than in men, the genetic contributions to FM score variability do not differ significantly by sex in this sample size.
FM co-occurs with multiple diseases, suggesting there are shared genetic factors underlying these diseases. We found that FM score and SSI have a strong genetic correlation with several psychiatric and personality syndromes indicating a substantial genetic overlap between them, potentially because psychological measures are part of the FM score. However, our genetic correlation findings for FM score are in agreement with results from a UK Biobank study (n=387,649) by Johnston et al., that found a genetic overlap between MCP and several psychiatric conditions. FM score and WPI were more strongly positively genetically correlated with asthma and rheumatoid arthritis than was MCP from the UK Biobank. For rheumatoid arthritis this may be due to an enrichment of individuals with rheumatoid arthritis in our surgical patient population compared to the more general UK Biobank population.
One limitation of our study is that our sample is not population-based. Individuals who were scheduled to have surgery were eligible for recruitment in the study and are more likely to suffer from pain and to be enriched for particular FM-related disorders. Additionally, we have used quantile-based inverse normalization of FM score which can affect the power to detect heritability.
Overall, this study highlights the importance of considering the age distribution of individuals when designing a genetic association study of FM. These data support the distinction that there are (at least) two different forms of FM: one that occurs in younger individuals and is strongly genetically driven – and the other that occurs in older individuals and can be driven by a variety of non-genetic factors and other conditions that cause pain.
Supplementary Material
Acknowledgments
This work was supported by P50 AR070600 and R01 DA038261 grants from the National Institute of Health. The authors acknowledge the University of Michigan Medical School Central Biorepository for providing bio-specimen storage, management, and distribution services in support of the research reported in this publication.
References
- 1.Schmidt-Wilcke T, Clauw DJ. Fibromyalgia: From pathophysiology to therapy. Nat Rev Rheumatol. 2011. doi: 10.1038/nrrheum.2011.98 [DOI] [PubMed] [Google Scholar]
- 2.Clauw DJ. Fibromyalgia: A clinical review. JAMA - J Am Med Assoc. 2014. doi: 10.1001/jama.2014.3266 [DOI] [PubMed] [Google Scholar]
- 3.Arnold LM, Hudson JI, Hess E V., et al. Family Study of Fibromyalgia. Arthritis Rheum. 2004. doi: 10.1002/art.20042 [DOI] [PubMed] [Google Scholar]
- 4.Hudson JI, Goldenberg DL, Pope HG, Keck PE, Schlesinger L. Comorbidity of fibromyalgia with medical and psychiatric disorders. Am J Med. 1992. doi: 10.1016/0002-9343(92)90265-D [DOI] [PubMed] [Google Scholar]
- 5.Aaron LA, Buchwald D. A review of the evidence for overlap among unexplained clinical conditions. Ann Intern Med. 2001. [DOI] [PubMed] [Google Scholar]
- 6.Kato K, Sullivan PF, Evengård B, Pedersen NL. A population-based twin study of functional somatic syndromes. Psychol Med. 2009. doi: 10.1017/S0033291708003784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buskila D, Neumann L, Hazanov I, Carmi R. Familial aggregation in the fibromyalgia syndrome. Semin Arthritis Rheum. 1996. doi: 10.1016/S0049-0172(96)80011-4 [DOI] [PubMed] [Google Scholar]
- 8.Woolf CJ. The pathophysiology of peripheral neuropathic pain--abnormal peripheral input and abnormal central processing. Acta Neurochir Suppl (Wien). 1993. [DOI] [PubMed] [Google Scholar]
- 9.Woolf CJ, Thompson SWN. The induction and maintenance of central sensitization is dependent on N-methyl-d-aspartic acid receptor activation; implications for the treatment of post-injury pain hypersensitivity states. Pain. 1991. doi: 10.1016/0304-3959(91)90100-C [DOI] [PubMed] [Google Scholar]
- 10.Sluka KA. Pain mechanisms involved in musculoskeletal disorders. J Orthop Sports Phys Ther. 1996. doi: 10.2519/jospt.1996.24.4.240 [DOI] [PubMed] [Google Scholar]
- 11.Sluka KA, Clauw DJ. Neurobiology of fibromyalgia and chronic widespread pain. Neuroscience. 2016. doi: 10.1016/j.neuroscience.2016.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woolf CJ. Central sensitization: Implications for the diagnosis and treatment of pain. Pain. 2011;152(Supplement):S2–S15. doi: 10.1016/j.pain.2010.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ablin JN, Buskila D. Update on the genetics of the fibromyalgia syndrome. Best Pract Res Clin Rheumatol. 2015. doi: 10.1016/j.berh.2015.04.018 [DOI] [PubMed] [Google Scholar]
- 14.Lee YH, Choi SJ, Ji JD, Song GG. Candidate gene studies of fibromyalgia: a systematic review and meta-analysis. Rheumatol Int. 2012;32(2):417–426. doi: 10.1007/s00296-010-1678-9 [DOI] [PubMed] [Google Scholar]
- 15.Docampo E, Escaramís G, Gratacòs M, et al. Genome-wide analysis of single nucleotide polymorphisms and copy number variants in fibromyalgia suggest a role for the central nervous system. Pain. 2014;155(6):1102–1109. doi: 10.1016/j.pain.2014.02.016 [DOI] [PubMed] [Google Scholar]
- 16.Bahcall OG. UK Biobank — a new era in genomic medicine. Nat Rev Genet. 2018. doi: 10.1038/s41576-018-0065-3 [DOI] [PubMed] [Google Scholar]
- 17.Wolford BN, Willer CJ, Surakka I. Electronic health records: The next wave of complex disease genetics. Hum Mol Genet. 2018. doi: 10.1093/hmg/ddy081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston KJA, Adams MJ, Nicholl BI, et al. Genome-wide association study of multisite chronic pain in UK Biobank. PLoS Genet. 2019. doi: 10.1371/journal.pgen.1008164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaitlen N, Pasaniuc B, Sankararaman S, et al. Leveraging population admixture to characterize the heritability of complex traits. Nat Genet. 2014. doi: 10.1038/ng.3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, Stephens M. Efficient Algorithms for Multivariate Linear Mixed Models in Genome-wide Association Studies. Nat Genet. 2014;11(4):407–409. doi: 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen H, Buskila D, Neumann L, Ebstein RP. Confirmation of an association between fibromyalgia and serotonin transporter promoter region (5-HTTLPR) polymorphism, and relationship to anxiety-related personality traits [5]. Arthritis Rheum. 2002. doi: 10.1002/art.10103 [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silventoinen K, Magnusson PKE, Tynelius P, Kaprio J, Rasmussen F. Heritability of body size and muscle strength in young adulthood: A study of one million Swedish men. Genet Epidemiol. 2008. doi: 10.1002/gepi.20308 [DOI] [PubMed] [Google Scholar]
- 24.Lohoff FW. Overview of the genetics of major depressive disorder. Curr Psychiatry Rep. 2010. doi: 10.1007/s11920-010-0150-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan PF, Neale MC, Kendler KS. Genetic Epidemiology of Major Depression: Review and Meta-Analysis. Am J Psychiatry. 2000;157(10):1552–1562. doi: 10.1176/appi.ajp.157.10.1552 [DOI] [PubMed] [Google Scholar]
- 26.Mayhew AJ, Meyre D. Assessing the Heritability of Complex Traits in Humans: Methodological Challenges and Opportunities. Curr Genomics. 2017. doi: 10.2174/1389202918666170307161450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan L, Ober C, Abney M. Heritability estimation of sex-specific effects on human quantitative traits. Genet Epidemiol. 2007. doi: 10.1002/gepi.20214 [DOI] [PubMed] [Google Scholar]
- 28.Ge T, Chen CY, Neale BM, Sabuncu MR, Smoller JW. Phenome-wide heritability analysis of the UK Biobank. PLoS Genet. 2017. doi: 10.1371/journal.pgen.1006711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfe F, Clauw DJ, Fitzcharles MA, et al. The American College of Rheumatology preliminary diagnostic criteria for fibromyalgia and measurement of symptom severity. Arthritis Care Res. 2010. doi: 10.1002/acr.20140 [DOI] [PubMed] [Google Scholar]
- 30.Wolfe F, Clauw DJ, Fitzcharles MA, et al. Fibromyalgia criteria and severity scales for clinical and epidemiological studies: A modification of the ACR preliminary diagnostic criteria for fibromyalgia. J Rheumatol. 2011. doi: 10.3899/jrheum.100594 [DOI] [PubMed] [Google Scholar]
- 31.Wang C, Zhan X, Liang L, Abecasis GR, Lin X. Improved Ancestry Estimation for both Genotyping and Sequencing Data using Projection Procrustes Analysis and Genotype Imputation. Am J Hum Genet. 2015. doi: 10.1016/j.ajhg.2015.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfe F, Clauw DJ, Fitzcharles MA, et al. 2016 Revisions to the 2010/2011 fibromyalgia diagnostic criteria. Semin Arthritis Rheum. 2016. doi: 10.1016/j.semarthrit.2016.08.012 [DOI] [PubMed] [Google Scholar]
- 34.Kang HM, Sul JH, Service SK, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010. doi: 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011. doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011. doi: 10.1016/j.ajhg.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe F, Ross K, Anderson J, Russell IJ, Hebert L. The prevalence and characteristics of fibromyalgia in the general population. Arthritis Rheum. 1995. doi: 10.1002/art.1780380104 [DOI] [PubMed] [Google Scholar]
- 38.Dahl A, Iotchkova V, Baud A, et al. A multiple-phenotype imputation method for genetic studies. Nat Genet. 2016;48(4):466–472. doi: 10.1038/ng.3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan B, Loh PR, Finucane HK, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng J, Erzurumluoglu AM, Elsworth BL, et al. LD Hub: A centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017. doi: 10.1093/bioinformatics/btw613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vincent A, Lahr BD, Wolfe F, et al. Prevalence of Fibromyalgia: A Population-Based Study in Olmsted County, Minnesota, Utilizing the Rochester Epidemiology Project. Arthritis Care Res (Hoboken). 2013;65(5):786–792. doi: 10.1002/acr.21896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Offenbaecher M, Bondy B, De Jonge S, et al. Possible association of fibromyalgia with a polymorphism in the serotonin transporter gene regulatory region. Arthritis Rheum. 1999. doi: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.