ABSTRACT
Rapid and accurate diagnostic testing is essential to bring the ongoing COVID-19 pandemic to an end. As the demand for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) testing continues to increase amid supply shortages, many laboratories have investigated the use of sources other than nasopharyngeal (NP) swabs. Saliva and midturbinate (MT) nasal swabs are attractive alternatives, as they allow for self-collection and are well accepted by patients. Saliva also requires limited consumables. We compared the performance of health care provider-collected NP swabs, patient-collected MT swabs, and patient-collected saliva specimens for SARS-CoV-2 detection using a laboratory-developed PCR assay that had received Emergency Use Authorization by the FDA. Of 281 total evaluable samples, 33 (11.7%) NP swabs, 33 (11.7%) MT swabs, and 32 (11.4%) saliva specimens were positive for SARS-CoV-2 following resolution of discordant results. Compared to NP swabs, saliva exhibited a sensitivity of 90.9% (30/33) and specificity of 99.2% (246/248), while patient-collected MT swabs exhibited a sensitivity of 93.9% (31/33) and specificity of 99.2% (246/248). When comparing to the consensus standard, the sensitivity was found to be 100% (31/31) for both NP and MT swabs and 96.8% (30/31) for saliva specimens, while specificity was the same in both NP swabs and saliva specimens (98.8% [247/250]) and 99.2% (248/250) for MT swabs. Pretreatment of saliva with proteinase K and heating for 15 min prior to extraction reduced the invalid rate from 26.7% (52/195) to 0% (0/195). These data show that midturbinate nasal swabs and saliva are suitable sources for self-collection in individuals who require routine monitoring for SARS-CoV-2 infection.
KEYWORDS: COVID-19, NAAT, midturbinate, saliva
INTRODUCTION
Control of coronavirus disease 2019 (COVID-19) relies on rapid diagnosis and isolation of positive cases. Currently, nucleic acid amplification tests (NAATs), such as PCR, are the most sensitive and specific means for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Testing demand has increased dramatically over the last year to include not only symptomatic patients, but also screening of health care providers (HCP), asymptomatic individuals prior to medical procedures, and serial testing of persons residing in living situations with a high risk of transmission and/or poor outcomes (e.g., long-term care residents). At-home collection has been considered a screening strategy for patient populations that require routine monitoring.
While the nasopharyngeal (NP) swab is considered the “gold standard” specimen type for SARS-CoV-2 detection by NAAT, it is a relatively invasive and uncomfortable sample type for patients. Further, NP swabs must be collected by a trained HCP wearing personal protective equipment (PPE) and is not a suitable option for self-collection (1). In recent years, midturbinate (MT) nasal swabs, sometimes referred as “deep nasal swabs,” have gained recognition as an acceptable alternative collection method for the detection of respiratory viruses, including SARS-CoV-2 (1–3). These studies have shown that MT swabs exhibit sensitivity similar to that of NP swabs, with the potential application for self-collection (4). Saliva has also been investigated as a potential specimen type for self-collection, as it is easily obtained, more tolerated by patients, and shows sensitivity similar to that of NP swabs (1, 5–17). The Infectious Diseases Society of America (IDSA) “Guidelines on the Diagnosis of COVID-19: Molecular Diagnostic Testing” (1) provides a thorough comparison of SARS-CoV-2 detection by NAAT using MT swabs, saliva, oropharyngeal swabs, and NP swabs. Saliva is also an appealing specimen type for SARS-CoV-2 detection, as its collection does not require a swab, and some methods do not require the use of transport media (18).
Although an attractive option for self-collection, saliva may contain PCR inhibitors and is often viscous in nature, which can lead to decreased extraction efficiency and an increased number of invalid results. Such results can lead to increased costs associated with specimen recollection and repeat testing, as well as delayed clinical management decisions. The objective of this study was to evaluate self-collected saliva and MT swabs as alternative specimen types for detection of SARS-CoV-2, as well as to optimize the processing of saliva specimens prior to testing.
MATERIALS AND METHODS
Clinical specimens and study design.
This study was approved by the Institutional Review Board at Mayo Clinic. Adults, age 18 or older, who presented at an outpatient collection site for SARS-CoV-2 molecular testing were invited to participate. Patients who had provided consent first collected a saliva specimen using the OMNIgene-ORAL (OM-505) collection kit (DNA Genotek, Ottawa, Ontario, Canada) and then collected a flocked MT swab (Copan Diagnostics, Murrieta, CA) specimen from both nostrils. Patients followed written instructions for self-collection of saliva and MT specimens. Collection was observed by an HCP, but no oral instructions were provided to study participants. Following self-collection of saliva and MT swab specimens, an NP swab was collected by an HCP according to routine protocol. All three specimens were transported to the testing laboratory at 2 to 8°C for SARS-CoV-2 PCR. The majority of specimens were tested within 3 days of collection. In the event that initial testing was delayed by >3 days postcollection, specimens were frozen at −80°C for up to 2 weeks prior to testing. In addition, all enrolled participants were surveyed regarding their preferred collection type.
Specimen extraction and testing.
NP and MT swab specimens were extracted on either the NucliSENS EMAG (bioMérieux; Durham, NC) or the Hamilton Microlab STAR (Hamilton; Reno, NV), while saliva specimens were extracted on the EMAG, the STAR, or the MagNA Pure 96 (Roche; Indianapolis, IN). The EMAG utilizes NucliSENS extraction reagents, the STAR utilizes the Maxwell HT viral total nucleic acid (TNA) kit (Promega; Madison, WI) for extraction, and the MagNA Pure 96 utilizes the MagNA Pure 96 DNA and viral NA large-volume kit. In a biosafety cabinet, raw specimen was first combined with lysis buffer corresponding with the extraction system used (EMAG, 0.2 ml of specimen to 2.0 ml of lysis buffer; STAR, 0.3 ml of specimen to 0.3 ml of lysis buffer; and MagNA Pure 96, 0.2 ml of specimen to 0.3 ml of lysis buffer) and incubated for 10 min. Two hundred microliters of lysed specimen was extracted with an output volume of 100 μl. Extracts were tested by the Mayo Clinic laboratory-developed test (LDT) that was granted Emergency Use Authorization (EUA) for detection of SARS-CoV-2 RNA by PCR (19). The LDT is a TaqMan real-time reverse transcription-PCR assay that targets the nucleocapsid region of the SARS-CoV-2 genome, along with an RNase P internal-control (IC) target. Fifteen microliters of master mix was combined with 5 μl of nucleic acid eluate and amplified on the LightCycler 480 (Roche Applied Sciences; Indianapolis, IN) using a 2-step reverse transcription-PCR. Results were reported as “detected,” “undetected,” “indeterminate,” or “inconclusive” (i.e., internal-control failure). Indeterminate results exhibit an amplification curve with no software-reported crossing point (Cp) value, which is often indicative of a SARS-CoV-2 RNA concentration near the limit of detection of the assay. After increased inhibition rates were observed in saliva specimens, an upfront proteolytic digestion protocol was investigated: all previously collected saliva specimens with sufficient volume were digested prior to lysis by combining 500 μl of raw specimen with 100 μl of proteinase K (20 mg/ml) and then heating/shaking on a thermomixer at 55°C and 1,500 rpm for 15 min. This protocol is currently utilized in our laboratory for digestion of other mucoid respiratory specimens (i.e., bronchoalveolar lavage fluid and sputum) (20).
Resolution of discordant results.
The NP swab result was considered the reference method for calculating sensitivity and specificity. In the event that one or more specimens from a study participant yielded a result discordant with that of the NP swab (i.e., undetected in the saliva and/or MT swab specimens and detected in the NP swab), testing on all three specimens was repeated in triplicate. Because false-positive NAATs are infrequent, all initially positive results and any subsequent positive results obtained during repeat testing were counted as true positives. Results are presented both prior to and after resolution of discordant results. Samples were stored at −80°C for up to 70 days prior to repeat testing.
RESULTS
Enrolled participants and survey results.
A total of 332 participants consented to participation in the study. Of these, 300 participants were able to successfully provide all three specimen types for testing. When surveyed regarding their preferred collection type, 258 (77.7%) participants preferred self-collected saliva, 47 (14.2%) preferred the self-collected MT swab, 21 (6.3%) preferred the HCP-collected NP swab, and 6 (1.8%) of participants gave no response.
Initial comparison of NP swab, MT swab, and saliva specimens.
Clinician-collected NP swabs, self-collected MT swabs, and self-collected saliva specimens from 212 patients were initially extracted on the EMAG or STAR instrument and tested. Aside from being aliquoted into lysis buffer, saliva specimens were not digested prior to extraction. Of these, 27 (12.7%) NP swabs, 23 (10.8%) MT swabs, and 14 (6.6%) saliva specimens were positive for SARS-CoV-2. Average crossing point (Cp) values of positive specimens for each specimen type were 29.8 (NP), 30.2 (MT), and 32.5 (saliva). In addition, 32.1% (68/212) of saliva specimens yielded an invalid result during initial testing due to presumed inhibition of the RNase P internal-control target.
Comparison of saliva specimen results before and after processing.
After increased inhibition rates were observed in saliva specimens, all previously tested specimens were digested using the protocol outlined in Materials and Methods. Of the 212 specimens initially tested, 195 had sufficient volume remaining for digestion. All digested saliva specimens underwent upfront lysis followed by routine extraction on the MagNA Pure 96 (total nucleic acid kit), which had showed increased extraction efficiency for viscous specimen types in prior studies compared with other evaluated systems (21). In total, 195 saliva specimens were tested both prior to and following digestion. Without upfront digestion, 13 (6.7%) saliva specimens were positive for SARS-CoV-2, 2 (1.0%) were indeterminate, and 52 (26.7%) yielded invalid results during initial testing. Following digestion using the aforementioned protocol, SARS-CoV-2 was detected in 19 (9.7%) saliva specimens, 2 (2.0%) yielded indeterminate results, and all 195 (100%) saliva specimens yielded valid results during initial testing. For the 10 saliva specimens that were positive for SARS-CoV-2 both before and after digestion, the target Cp value decreased by an average of 2.0 cycles following digestion. In addition, the RNase P Cp value decreased by an average of 1.8 cycles in all saliva specimens following digestion. Given these improved results, the saliva digestion protocol was applied to all additional saliva specimens collected during this study, followed by extraction on the MagNA Pure 96. Extraction of the NP and MT swab specimens remained on the EMAG and Hamilton instruments.
Comparison of digested saliva, MT swab, and NP swab specimens.
A total of 281 digested saliva specimens were compared with concurrently collected NP and MT swab specimens.
(i) Initial results without discordant-result analysis.
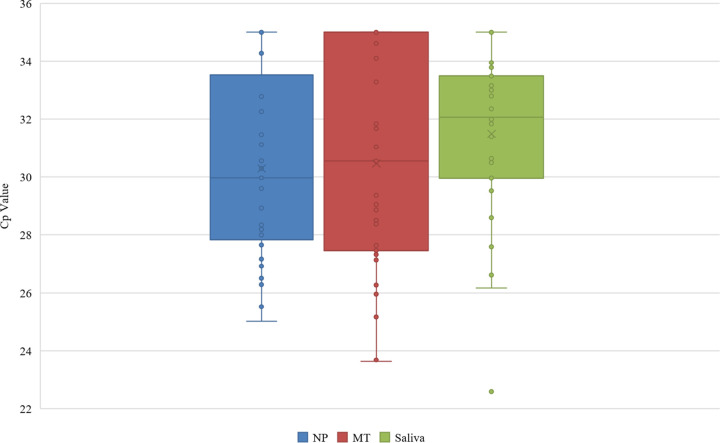
Of the 281 evaluable specimens, 32 (11.4%) NP swabs, 32 (11.4%) MT swabs, and 30 (10.7%) saliva specimens were initially positive for SARS-CoV-2. In addition, 1 (0.3%) NP swab and 4 (1.4%) saliva specimens yielded an indeterminate result (Fig. 1). Compared to NP swabs, MT swabs exhibited a sensitivity of 90.6% (29/32) and specificity of 98.8% (246/249), while saliva specimens exhibited a sensitivity of 87.5% (28/32) and specificity of 99.2% (247/249) (Table 1). In addition to comparing to NP swabs, results were also compared to the “consensus standard,” which was defined as the result obtained from ≥2 of the 3 specimen types. Compared to the consensus standard, NP swabs and MT swabs both exhibited a sensitivity of 96.8% (30/31), while sensitivity in saliva specimens was slightly lower (93.5% [29/31]) (Table 2). In addition, specificity was the same in both NP swabs and saliva specimens (98.8% [247/250]), while a specificity of 99.2% (248/250) was observed for MT swabs. Average SARS-CoV-2 Cp values of positive specimens for each specimen type were 30.1 (NP), 30.6 (MT), and 31.4 (saliva) (Fig. 2). For 33.3% (10/30) of positive saliva specimens, the Cp value was lower than that of the corresponding NP swab specimen, indicating that more SARS-CoV-2 RNA was present in the saliva specimens than in the NP swabs. Additionally, the average Cp values for the RNase P internal-control target were 28.6, 30.0, and 25.6 for NP swabs, MT swabs, and saliva, respectively.
FIG 1.
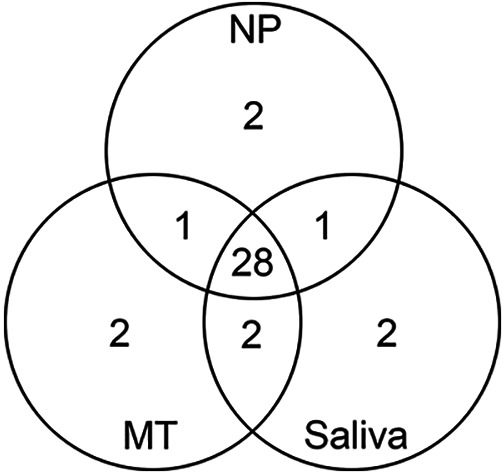
Correlation of positive SARS-CoV-2 PCR results between paired NP swabs, MT swabs, and saliva specimens prior to discordant result analysis. Twenty-eight patients were positive for SARS-CoV-2 by all three specimen types, while 3 patients were positive by 2 of 3 specimen types. Two patients were positive by NP or MT swab specimens only, while 1 patient was positive by saliva only.
TABLE 1.
Comparison of provider-collected NP swabs to patient-collected MT swabs and patient-collected saliva specimens for detection of SARS-CoV-2 RNA by real-time PCR
| Sample type and SARS-CoV-2 result | No. of NP swabs with a SARS-CoV-2 result of: |
||
|---|---|---|---|
| Detected | Indeterminate | Undetected | |
| MT swab | |||
| Detected | 29 | 0 | 3a |
| Indeterminate | 0 | 0 | 0 |
| Undetected | 3b | 1c | 245 |
| Saliva | |||
| Detected | 28 | 0 | 2d |
| Indeterminate | 2e | 0 | 2f |
| Undetected | 2g | 1h | 244 |
One of these 3 samples repeated as detected (i.e., positive) in both the NP and MT swabs. The remaining two samples repeated as undetected (i.e., negative) in both the NP and MT swabs. Both MT swabs had an initial Cp value of 35.0, suggesting that a low level of SARS-CoV-2 RNA was present in these specimens.
One of these 3 samples repeated as detected in both the NP and MT swabs. The remaining two samples repeated as detected in the NP swab and undetected in the MT swab. Both NP swabs had an initial Cp value of 35.0, suggesting that a low level of SARS-CoV-2 RNA was present in these specimens.
This MT swab sample repeated as undetected, while the NP swab was indeterminate upon repeat testing.
One of these samples repeated as detected in both the NP swab and saliva specimen, while the second sample repeated as undetected in the NP swab and indeterminate in the saliva specimen. The saliva specimen had an initial Cp value of 35.0, suggesting that a low level of SARS-CoV-2 RNA was present in this specimen.
One of these samples repeated as detected in both the NP swab and saliva specimen, while the second sample repeated as detected in the NP swab and indeterminate in the saliva specimen.
One sample repeated as undetected in both the NP swab and saliva specimen. The second sample repeated as undetected in the NP swab and detected in the saliva specimen.
Both saliva specimens repeated as undetected, while both NP swabs were detected upon repeat testing. Both NP swabs had an initial Cp value of 35.0, suggesting that a low level of SARS-CoV-2 RNA was present in these specimens.
The NP swab repeated as indeterminate, while the saliva specimen repeated as undetected upon repeat testing.
TABLE 2.
Comparison of provider-collected NP swabs, patient-collected MT swabs, and patient-collected saliva specimens to consensus results among three specimen types tested for SARS-CoV-2 RNA by real-time PCR
| Sample type and SARS-CoV-2 result | No. of specimens with a consensus SARS-CoV-2 result of: |
||
|---|---|---|---|
| Detected | Indeterminate | Undetected | |
| NP swab | |||
| Detected | 30 | 0 | 2a |
| Indeterminate | 0 | 0 | 1b |
| Undetected | 1c | 0 | 247 |
| MT swab | |||
| Detected | 30 | 0 | 2d |
| Indeterminate | 0 | 0 | 0 |
| Undetected | 1e | 0 | 248 |
| Saliva specimen | |||
| Detected | 29 | 0 | 1f |
| Indeterminate | 2g | 0 | 2h |
| Undetected | 0 | 0 | 247 |
Both samples repeated as detected (i.e., positive) in the NP swab and undetected (i.e., negative) in both the MT swab and saliva specimen. Both NP swabs had an initial Cp value of 35.0, suggesting that a low level of SARS-CoV-2 RNA was present in these specimens.
This sample repeated as indeterminate in the NP swab and undetected in the MT swab and saliva specimens.
This sample repeated as detected in all three specimen types.
Both of these samples repeated as undetected in all three specimen types. Both MT swabs had an initial Cp value of 35.0, suggesting that a low level of SARS-CoV-2 RNA was present in these specimens.
This sample repeated as detected in all three sample types.
This sample repeated as undetected in both the NP and MT swabs and indeterminate in the saliva specimen. The saliva specimen had an initial Cp value of 35.0, suggesting that a low level of SARS-CoV-2 RNA was present in this specimen.
One of these samples repeated as detected in all three specimen types, while the second sample repeated as detected in the NP and MT swabs and indeterminate in the saliva specimen.
One sample repeated as undetected in all three specimen types. The second sample repeated as undetected in the NP and MT swabs and detected in the saliva specimen.
FIG 2.
Comparison of crossing point (Cp) values for positive NP swab, MT swab, and saliva specimens. Note that the maximum Cp value of the assay is 35.
(ii) Results following discordant-result analysis.
Following discordant-result analysis, 33 (11.7%) NP swabs, 33 (11.7%) MT swabs, and 32 (11.4%) saliva specimens were positive for SARS-CoV-2, while 1 (0.3%) NP swab and 2 (0.7%) saliva specimens yielded indeterminate results. The sensitivity and specificity of MT swabs improved to 93.9% (31/33) and 99.2% (246/248), respectively. Similarly, saliva specimens exhibited a sensitivity of 90.9% (30/33) and specificity of 99.2% (246/248) following discordant-result analysis. Compared to the consensus standard, the sensitivity improved to 100% (31/31) for both NP and MT swabs and 96.8% (30/31) for saliva specimens following discordant-result resolution, while the specificity of all three specimen types remained the same.
DISCUSSION
In this prospective study, we used an optimized protocol for saliva digestion and demonstrated that patient-collected saliva and MT swab specimens provide sensitivity similar to but slightly lower than that of HCP-collected NP swabs for SARS-CoV-2 detection by PCR.
Although previous studies have investigated saliva as a potential source for SARS-CoV-2 testing, the Infectious Diseases Society of America (IDSA) guidelines were only recently (December 2020) updated to recommend it as a standalone source. Saliva is known to be a difficult specimen type to work with due to its inhibitory and viscous nature. As expected, other studies have observed higher invalid rates for saliva specimens than for NP swabs (1). By combining saliva specimens with proteinase K and performing a 15-min heating/shaking step, we observed the invalid rate decrease from 26.7% (52/195) before digestion to 0% (0/195) following digestion. Implementation of an upfront digestion protocol for saliva specimens can aid in preventing increased costs and delayed results due to repeat testing.
Because patient-collected MT swab and saliva specimens showed a slight decrease in sensitivity compared to that of NP swabs, the potential for false-negative results is a small but significant concern. However, it is important to note that none of the three specimen types detected all potential SARS-CoV-2 cases and that the NP swab is an imperfect reference method due to sampling and collection technique variability (1, 22). This was observed in our study, in which MT swab specimens provided the same number of positive results as NP swab specimens following discordant-result resolution (n = 33). After discordant-result analysis, patient-collected MT swabs showed slightly higher sensitivity (93.9% [31/33]) than that of saliva specimens (90.9% [30/33]) compared to NP swabs, while the specificities of both specimen types were the same (99.2% [246/248]). When comparing to the consensus standard, the sensitivity of both NP and MT swabs was found to be 100% (31/31), while that of saliva specimens was 96.8% (30/31) following discordant-result analysis. In addition, both NP swabs and saliva specimens exhibited a specificity of 98.8% [247/250]), while a specificity of 99.2% (248/250) was observed for MT swabs following discordant-result analysis. It should be noted that the majority of discrepant results yielded Cp values of 35.0 or indeterminate results, suggesting that a low level of SARS-CoV-2 RNA was present in these samples. It is well known that samples with RNA concentrations at or near the limit of detection of the assay may not yield reproducible results, which can impact agreement among sample types. Several meta-analyses evaluate alternative specimen types for SARS-CoV-2 testing in comparison to NP swabs, which synthesizes a larger data set and decreases bias generated by disagreement in low-titer specimens (16, 17, 22).
Although there was not a large degree of variability among the average Cp values observed in each specimen type, 33.3% (10/30) of positive saliva specimens yielded Cp values lower than that of the corresponding NP swab specimen. While this suggests that more SARS-CoV-2 RNA was present in the saliva specimens for these patients, caution should be taken when using Cp or cycle threshold (CT) values to assess patient infectivity or potential outcomes. Several preanalytical variables can impact Cp or CT values, including specimen type, quality of collection, illness progression, transport media type, and specimen storage. In addition, analytical variables such as sampling/aliquoting, extraction efficiency, input/output volumes, and amplification protocol further contribute to inconsistency of these values (23).
Validating multiple sources for self-collection is advantageous, as it allows laboratories to pivot when faced with various supply shortages. Further, MT swabs and saliva offer unique advantages for self-collection. When surveyed regarding their preferred collection type, the majority of study participants (77.7%) indicated that they preferred self-collected saliva over both the self-collected MT swab and the HCP-collected NP swab. Although a lower percentage of participants (14.2%) preferred self-collected MT swabs, the preference for this collection type was still more than twice that of HCP-collected NP swabs (6.3%). Both self-collected saliva and self-collected MT swabs provide a more comfortable collection option than NP swabs, which can have a significant impact on compliance in those patients who require routine monitoring. Considering the ongoing supply issues many laboratories continue to face, required consumables and reagents must also be considered for both specimen types. Although our study utilized a saliva collection device containing a proprietary stabilizing medium, saliva can also be collected into a sterile container and does not require the use of a swab. Postcollection, we found that saliva specimens required upfront digestion using proteinase K to ensure lower PCR inhibition rates, whereas MT swabs did not (8, 24, 25).
Our study has several limitations. First, all digested saliva specimens were extracted on the MagNA Pure 96, while NP and MT swab specimens were extracted on either the EMAG or STAR. As mentioned previously, interinstrument comparability studies showed equivalent extraction results for swab specimens, but the MagNA Pure 96 showed an improved extraction efficiency of viscous specimens like saliva. Our study also did not look at the time elapsed from symptom onset to specimen collection, which could impact the sensitivity of detecting SARS-CoV-2 RNA. Lastly, our study did not attempt to quantify the amount of SARS-CoV-2 RNA present for comparison between specimen types but rather utilized Cp values as an approximation of viral load.
In conclusion, both patient-collected MT nasal swabs and saliva are suitable specimen types for SARS-CoV-2 PCR, as they both exhibit sensitivity and specificity comparable to those of provider-collected NP swabs. Because saliva specimens may contain PCR inhibitors, it is important to establish an upfront digestion protocol to optimize extraction efficiency and assay performance. Saliva can be a useful tool for facilitating routine monitoring of individuals at increased risk for SARS-CoV-2 infection, as well as provide labs with an additional testing option amid ongoing supply challenges.
ACKNOWLEDGMENT
We thank the Clinical Virology Laboratory at Mayo Clinic for their technical assistance with this study.
Contributor Information
Bobbi S. Pritt, Email: pritt.bobbi@mayo.edu.
Michael J. Loeffelholz, Cepheid
REFERENCES
- 1.Hanson KE, Caliendo AM, Arias CA, Hayden MK, Englund JA, Lee MJ, Loeb M, Patel R, El Alayli A, Altayar O, Patel P, Falck-Ytter Y, Lavergne V, Morgan RL, Murad MH, Sultan S, Bhimraj A, Mustafa RA. 2020. The Infectious Diseases Society of America guidelines on the diagnosis of COVID-19: molecular diagnostic testing. Version 2.0.0. Available at https://www.idsociety.org/practice-guideline/covid-19-guideline-diagnostics/. Accessed 19 May 2021. [DOI] [PMC free article] [PubMed]
- 2.Tu YP, Jennings R, Hart B, Cangelosi GA, Wood RC, Wehber K, Verma P, Vojta D, Berke EM. 2020. Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med 383:494–496. 10.1056/NEJMc2016321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCulloch DJ, Kim AE, Wilcox NC, Logue JK, Greninger AL, Englund JA, Chu HY. 2020. Comparison of unsupervised home self-collected midnasal swabs with clinician-collected nasopharyngeal swabs for detection of SARS-CoV-2 infection. JAMA Netw Open 3:e2016382. 10.1001/jamanetworkopen.2020.16382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larios OE, Coleman BL, Drews SJ, Mazzulli T, Borgundvaag B, Green K, STOP-Flu Study Group, McGeer AJ. 2011. Self-collected mid-turbinate swabs for the detection of respiratory viruses in adults with acute respiratory illnesses. PLoS One 6:e21335. 10.1371/journal.pone.0021335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azzi L, Baj A, Alberio T, Lualdi M, Veronesi G, Carcano G, Ageno W, Gambarini C, Maffioli L, Saverio SD, Gasperina DD, Genoni AP, Premi E, Donati S, Azzolini C, Grandi AM, Dentali F, Tangianu F, Sessa F, Maurino V, Tettamanti L, Siracusa C, Vigezzi A, Monti E, Iori V, Iovino D, Ietto G, ASST dei Sette Laghi Rapid Salivary Test Nurse staff Research Group, Grossi PA, Tagliabue A, Fasano M. 2020. Rapid salivary test suitable for a mass screening program to detect SARS-CoV-2: a diagnostic accuracy study. J Infect 81:e75–e78. 10.1016/j.jinf.2020.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanson KE, Barker AP, Hillyard DR, Gilmore N, Barrett JW, Orlandi RR, Shakir SM. 2020. Self-collected anterior nasal and saliva specimens versus health care worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol 58:e01824-20. 10.1128/JCM.01824-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasaki S, Fujisawa S, Nakakubo S, Kamada K, Yamashita Y, Fukumoto T, Sato K, Oguri S, Taki K, Senjo H, Sugita J, Hayasaka K, Konno S, Nishida M, Teshima T. 2020. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect 81:e145–e147. 10.1016/j.jinf.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landry ML, Criscuolo J, Peaper DR. 2020. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol 130:104567. 10.1016/j.jcv.2020.104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung EC, Chow VC, Lee MK, Lai RW. 2021. Deep throat saliva as an alternative diagnostic specimen type for the detection of SARS-CoV-2. J Med Virol 93:533–536. 10.1002/jmv.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCormick-Baw C, Morgan K, Gaffney D, Cazares Y, Jaworski K, Byrd A, Molberg K, Cavuoti D. 2020. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2. J Clin Microbiol 58:e01109-20. 10.1128/JCM.01109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Migueres M, Mengelle C, Dimeglio C, Didier A, Alvarez M, Delobel P, Mansuy JM, Izopet J. 2020. Saliva sampling for diagnosing SARS-CoV-2 infections in symptomatic patients and asymptomatic carriers. J Clin Virol 130:104580. 10.1016/j.jcv.2020.104580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto MP, Darles C, Valero E, Benner P, Dutasta F, Janvier F. 8August2020. Posterior oropharyngeal saliva for the detection of SARS-CoV-2. Clin Infect Dis 10.1093/cid/ciaa1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams E, Bond K, Zhang B, Putland M, Williamson DA. 2020. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol 58:e00776-20. 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, Warren JL, Geng B, Muenker MC, Moore AJ, Vogels CBF, Petrone ME, Ott IM, Lu P, Venkataraman A, Lu-Culligan A, Klein J, Earnest R, Simonov M, Datta R, Handoko R, Naushad N, Sewanan LR, Valdez J, White EB, Lapidus S, Kalinich CC, Jiang X, Kim DJ, Kudo E, Linehan M, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Weizman OE, Wong P, Yang Y, Bermejo S, Odio CD, Omer SB, Dela Cruz CS, Farhadian S, Martinello RA, Iwasaki A, Grubaugh ND, Ko AI. 2020. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med 383:1283–1286. 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fakheran O, Dehghannejad M, Khademi A. 2020. Saliva as a diagnostic specimen for detection of SARS-CoV-2 in suspected patients: a scoping review. Infect Dis Poverty 9:100. 10.1186/s40249-020-00728-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. 20April2021. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: a systematic review and meta-analysis. J Clin Microbiol 10.1128/JCM.02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsang NNY, So HC, Ng KY, Cowling BJ, Leung GM, Ip DKM. 12April2021. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis. Lancet Infect Dis 10.1016/S1473-3099(21)00146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YG, Yun SG, Kim MY, Park K, Cho CH, Yoon SY, Nam MH, Lee CK, Cho YJ, Lim CS. 2017. Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription-PCR. J Clin Microbiol 55:226–233. 10.1128/JCM.01704-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodino KG, Espy MJ, Buckwalter SP, Walchak RC, Germer JJ, Fernholz E, Boerger A, Schuetz AN, Yao JD, Binnicker MJ. 2020. Evaluation of saline, phosphate-buffered saline, and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testing. J Clin Microbiol 58:e00590-20. 10.1128/JCM.00590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binnicker MJ, Buckwalter SP, Eisberner JJ, Stewart RA, McCullough AE, Wohlfiel SL, Wengenack NL. 2007. Detection of Coccidioides species in clinical specimens by real-time PCR. J Clin Microbiol 45:173–178. 10.1128/JCM.01776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bommersbach CE, Buckwalter SP, Espy MJ, Mandrekar J, Binnicker MJ, Schuetz AN. 2019. Comparison of five nucleic acid extraction platforms on a variety of clinical specimens. Microbe 2019.
- 22.Butler-Laporte G, Lawandi A, Schiller I, Yao M, Dendukuri N, McDonald EG, Lee TC. 2021. Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and meta-analysis. JAMA Intern Med 181:353–360. 10.1001/jamainternmed.2020.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhoads D, Peaper DR, She RC, Nolte FS, Wojewoda CM, Anderson NW, Pritt BS. 2021. College of American Pathologists (CAP) Microbiology Committee Perspective: caution must be used in interpreting the cycle threshold (Ct) value. Clin Infect Dis 72:e685–e686. 10.1093/cid/ciaa1199. [DOI] [PubMed] [Google Scholar]
- 24.Chu AW, Chan WM, Ip JD, Yip CC, Chan JF, Yuen KY, To KK. 2020. Evaluation of simple nucleic acid extraction methods for the detection of SARS-CoV-2 in nasopharyngeal and saliva specimens during global shortage of extraction kits. J Clin Virol 129:104519. 10.1016/j.jcv.2020.104519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barat B, Das S, De Giorgi V, Henderson DK, Kopka S, Lau AF, Miller T, Moriarty T, Palmore TN, Sawney S, Spalding C, Tanjutco P, Wortmann G, Zelazny AM, Frank KM. 2021. Pooled saliva specimens for SARS-CoV-2 testing. J Clin Microbiol 59:e02486-20. 10.1128/JCM.02486-20. [DOI] [PMC free article] [PubMed] [Google Scholar]