ABSTRACT
Longitudinal studies assessing durability of the anti-severe acute respiratory syndrome coronavirus 2 (anti-SARS-CoV-2) humoral immune response have generated conflicting results. This has been proposed to be due to differences in patient populations, the lack of standardized methodologies, and the use of assays that measure distinct aspects of the humoral response. SARS-CoV-2 antibodies were serially measured in sera from a cohort of 44 well-characterized convalescent plasma donors over 120 days post-COVID-19 symptom onset, utilizing eight assays, which varied according to antigen source, the detected antibody isotype, and the activity measured (i.e., binding, blocking, or neutralizing). While the majority of assays demonstrated a gradual decline in antibody titers over the course of 120 days, the two electrochemiluminescence immunoassay Roche assays (Roche Diagnostics Elecsys anti-SARS-CoV-2 [qualitative, nucleocapsid based] and Roche Diagnostics Elecsys anti-SARS-CoV-2 S [semiquantitative, spike based]), which utilize dual-antigen binding for antibody detection, demonstrated stable and/or increasing antibody titers over the study period. This study is among the first to assess longitudinal, rather than cross-sectional, SARS-CoV-2 antibody profiles among convalescent COVID-19 patients, primarily using commercially available serologic assays with Food and Drug Administration emergency use authorization. We show that SARS-CoV-2 antibody detection is dependent on the serologic method used, which has implications for future assay utilization and clinical value.
KEYWORDS: COVID-19, SARS-CoV-2, serology, antibody, dual-antigen binding assays, antibodies, immunoassays
INTRODUCTION
With the deployment of vaccines and therapeutic antibody products targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), there remains a need to correlate antibody titers and/or neutralizing activity with protective immunity. Additionally, understanding the durability of the SARS-CoV-2 antibody responses elicited following natural infection, vaccination, or administration of immunoglobulin therapy will be essential to guide the ongoing COVID-19 public health response and to optimize individualized patient care. Based on previous experience with human coronaviruses, it is anticipated that the humoral immune response to SARS-CoV-2 infection or vaccination will wane; however, the time frame, extent, and clinical implications of this remain unclear (1–3).
Prior reports have generated conflicting findings with respect to persistence of measurable SARS-CoV-2 antibodies following natural infection. In a cohort of 188 patients with prior mild COVID-19, cross-sectional analysis of antibody endpoint titers using laboratory-developed sandwich enzyme-linked immunosorbent assays (ELISAs) measuring IgG antibodies against different recombinant SARS-CoV-2 antigens, including the nucleocapsid (NC), spike (S), and receptor binding domain (RBD) of the S1 subunit of the S protein, reported only modest declines in antibodies over a 6-month period post-symptom onset (4). The authors also replicated these data using a pseudovirus neutralizing antibody (nAb) assay. These results corroborate earlier findings in a large population of Icelanders where it was demonstrated that SARS-CoV-2 IgG seropositivity was maintained without any appreciable decline in antibody titer for at least 4 months after initial diagnosis of COVID-19 (5).
In stark contrast to these findings, however, multiple studies have demonstrated significant declines in the SARS-CoV-2 antibody response within 2 to 4 months after symptom onset (6, 7). Studies utilizing nAb assays to specifically measure the functional portion of the humoral response have also reported conflicting results, with one study indicating that nAb titers are stable out to 5 months post-symptom onset, while others report dramatic decreases in nAb titers within 3 months of symptom onset (7, 8). Collectively, the clinical application of these findings is challenging given that the majority of these studies used nonstandardized, site-specific laboratory-developed tests (LDTs) and not more widely available assays with Food and Drug Administration emergency use authorization (FDA EUA). Furthermore, most of these previous reports used numerical data from qualitative assays, which limits the communicability of data across studies. Few studies have been performed using the more widely available testing methods to measure SARS-CoV-2 antibodies longitudinally in a single, well-characterized patient cohort.
Here, using serially collected samples from 44 COVID-19-confirmed convalescent plasma (CCP) donors, we investigated antibody persistence using eight different SARS-CoV-2 antibody assays, including five binding and three nAb assays, six with FDA EUA, and show assay-dependent differential antibody trending over a 4-month period post-SARS-CoV-2 infection.
MATERIALS AND METHODS
Patient cohort.
Patients (N = 44) selected for inclusion in this cohort had provided at least two serum samples between April and July 2020 and were all characterized as having mild COVID-19 due to recovery in the outpatient setting. Serum samples were collected from these COVID-19 convalescent plasma donors in serum separator tubes (SSTs) at the time of plasma donation at Mayo Clinic (Rochester, MN). Donor eligibility required a PCR-confirmed diagnosis of SARS-CoV-2 infection and donation at least 14 days after resolution of all symptoms (excluding anosmia) as determined by a medical professional. At the time of collection, serum samples were tested on the Euroimmun anti-SARS-CoV-2 IgG ELISA and the Ortho-Clinical Diagnostics anti-SARS-CoV-2 IgG assay and were subsequently frozen at −20°C. Aliquots were stored frozen at −20°C until testing was performed on additional platforms. The length of time samples were stored at −20°C prior to completion of testing varied between 1 and 5 months. Samples went through up to three freeze-thaw cycles between testing. Testing of these serum samples was performed at Mayo Clinic for all assays, with exception of the NeuCovix-HT assay which was performed at Arizona State University. This study was approved by the institutional review board of Mayo Clinic in Rochester, MN.
SARS-CoV-2 serologic assay methods.
All serologic assays with FDA EUA were performed and resulted per manufacturers’ instructions. These assays include the following.
(i) Abbott Laboratories SARS-CoV-2 IgG (Abbott Park, IL).
The Abbott SARS-CoV-2 IgG assay is a qualitative, paramagnetic microparticle-based chemiluminescent immunoassay (CIA) which detects IgG-class antibodies against the SARS-CoV-2 nucleocapsid (NC) protein. Testing was performed on the Abbott ARCHITECT i2000SR system. Results were calculated by dividing the sample relative light unit (RLU) by the mean calibrator RLU to generate a signal-to-cutoff (S/Co) value. S/Co values of ≥1.40 or <1.40 were considered positive or negative for the presence of anti-NC IgG, respectively (https://www.fda.gov/media/137383/download).
(ii) Ortho-Clinical Diagnostics anti-SARS-CoV-2 IgG (Rochester, NY).
The Ortho-Clinical anti-SARS-CoV-2 IgG assay is a qualitative CIA which detects IgG-class antibodies against the SARS-CoV-2 spike (S) protein. Testing was performed on the Ortho-Clinical Diagnostics VITROS 3600 platform. Results were calculated by dividing the sample chemiluminescence signal by the assay calibrator signal to generate a S/Co value. S/Co values of ≥1.00 and <1.00 were considered positive or negative for the presence of anti-S IgG, respectively (https://www.fda.gov/media/137363/download).
(iii) Euroimmun anti-SARS-CoV-2 IgG ELISA (Lubeck, Germany).
The Euroimmun anti-SARS-CoV-2 IgG assay is a qualitative enzyme-linked immunosorbent assay (ELISA) which detects IgG-class antibodies against subunit 1 (S1) of the SARS-CoV-2 S protein. Testing was performed on the Agility automated ELISA processor (Dynex Technologies, Inc., Chantilly, VA). Results were calculated by dividing the sample optical density (OD) by the mean of the duplicate calibrator OD value to determine an index value. Index values of ≥1.1, 0.8 to <1.1, and <0.8 were considered positive, indeterminate, and negative for the presence of anti-S1 IgG, respectively (https://www.fda.gov/media/137609/download).
(iv) Roche Diagnostics Elecsys anti-SARS-CoV-2 (Indianapolis, IN).
The Roche Elecsys anti-SARS-CoV-2 assay is a qualitative electrochemiluminescence immunoassay (ECLIA) which detects total antibodies (predominantly IgG and may also capture IgM and IgA) against the SARS-CoV-2 NC protein. Testing was performed on the Roche Diagnostics cobas e 801 platform. Results were determined by calculating the cutoff index (COI) for the sample by dividing the specimen electrochemiluminescence signal by that of the assay calibrator. COI values of ≥1.1 or <1.1 were considered positive or negative for the presence of total antibodies against NC, respectively (https://www.fda.gov/media/137605/download).
(v) Roche Diagnostic Elecsys anti-SARS-CoV-2 S (Indianapolis, IN).
The Roche Elecsys anti-SARS-CoV-2 S assay is a semiquantitative ECLIA which detects total antibodies (predominantly IgG and may also capture IgM and IgA) against the SARS-CoV-2 receptor binding domain (RBD). Testing was performed on the Roche Diagnostics cobas e 801 analyzer. The assay is standardized against an internal Roche Diagnostics anti-RBD monoclonal antibody mixture, with a 1 nM concentration corresponding to 20 units/ml (U/ml) by the assay. Results in units per milliliter were determined by comparing the sample electrochemiluminescence signal to that of the standard calibration curve. Values of ≥0.8 or <0.8 were considered positive or negative for the presence of total antibodies against RBD, respectively (https://www.fda.gov/media/144037/download).
(vi) GenScript cPASS SARS-CoV-2 neutralization antibody detection kit (Piscataway, NJ).
The GenScript cPASS assay is a qualitative blocking ELISA for detection of total nAb to SARS-CoV-2. Briefly, recombinant horseradish peroxidase (HRP)-conjugated RBD and solid-phase bound human angiotensin converting enzyme 2 (ACE2) are mixed with patient samples. In the presence of nAb, the RBD-ACE2 interaction is disrupted, leading to a loss of HRP signal; in contrast, in the absence of nAb, HRP-conjugated RBD will bind to ACE2 and a colorimetric signal will be generated following addition of substrate. The percent signal inhibition (PSI) was determined by dividing the sample OD by the negative-control OD, with PSI values of ≥30% or <30% considered positive or negative for the presence of nAb to SARS-CoV-2, respectively (https://www.fda.gov/media/143583/download).
Two assays without FDA EUA were also used to evaluate this cohort of samples. These included the NeuCovix-HT neutralizing antibody assay, which was performed for research purposes only, and the Mayo Clinic neutralizing antibody assay, which was submitted for FDA EUA. As a result of changing EUA requirements for SARS-CoV-2 laboratory developed tests, EUA for the Mayo Clinic neutralizing antibody assay was not further pursued. These assays were performed as follows.
(vii) Mayo Clinic neutralizing antibody assay.
The Mayo Clinic nAb test is a modification of the quantitative IMMUNO-COV assay (Imanis Life Sciences, Rochester, MN), which is an in vitro pseudovirus neutralizing assay that measures the ability of patient sera to prevent replication-competent vesicular stomatitis virus (VSV) expressing the SARS-CoV-2 spike protein from infecting engineered cells in a 96-well plate format using a luciferase reporter protein (9). Briefly, patient serum samples were diluted 1:40 in Opti-MEM and mixed 1:1 with 40 μl of recombinant virus in Opti-MEM, for a final serum dilution of 1:80. This mixture was incubated at room temperature for 30 min. Subsequently, 100 μl of the specimen-virus mixture was added to each well of a 96-well plate containing a 1:1 mixture of two distinct subclones of Vero cells that express complementary components of a dual split protein (DSP) luciferase reporter, variant 1 or variant 2. To generate the 96-well plates, 30,000 Vero DSP1 and −2 cells were premixed at a ratio 1:1 in 50 μl of culture medium and plated into each well 24 h prior to usage of the plate. The cells were incubated with the specimen-virus mixture for 18 h to allow for infection. A stable live-cell substrate, EnduRen (Promega, Fitchburg, WI), was then added to the culture medium for an additional 6 h. The EnduRen is taken up by the Vero cells and converted into coelenterazine, the substrate for Renilla luciferase. Functional Renilla luciferase converts coelenterazine into coelenteramide, emitting luminescence. A TECAN Infinite M200 Pro plate reader equipped with a luminescence photomultiplier tube (PMT) was used to measure the luminescence (relative light units [RLU]).
In the absence of nAb, the VSV-SARS-CoV-2 recombinant virus infects the Vero cells, resulting in membrane expression of the SARS-CoV-2 spike protein, which drives intercellular fusion between the Vero-DSP1 and Vero-DSP2, bringing together the split luciferase protein to form functional luciferase enzyme. The presence of nAb in a sample blocks infection by VSV-SARS-CoV-2, which prevents virus-induced intercellular fusion of the Vero-DSP1 and Vero-DSP2 cell lines. In the absence of intercellular fusion, the luciferase split products are not brought into close proximity; therefore, functional luciferase is not formed, leading to a reduction in RLUs.
The percentage of sample well signal (RLUsample) relative to the average single-point calibrator signal (RLUCal) was calculated. This percentage is referred to as residual activity (e.g., the amount of residual activity of VSV-SARS-CoV-2 virus after neutralization by patient antibodies). The calibrator was generated using a humanized monoclonal antibody (MAb) with neutralizing activity against the SARS-CoV-2 spike protein, diluted into pooled donor sera collected pre-November 2019. The MAb concentration used in the calibrator corresponded to a 50% reduction in infectivity of the recombinant VSV-SARS-CoV-2. Patient wells with RLUs of less than that of the calibrator were considered positive, and a titration was performed. This corresponds to a percentage of <100% ([RLUsample/RLUCal] × 100).
Serum samples were tested initially at a 1:80 dilution in duplicates. Samples with an average RLU higher that that of the calibrator, indicative of the absence of nAb, were reported as “negative,” and no further testing was performed. Samples with an RLU lower than that of the calibrator were serially diluted 1:80 to 1:2,560. The last dilution for which the average RLU was lower than that of the calibrator was considered the endpoint titer.
(viii) NeuCovix-HT assay.
The NeuCovix-HT neutralizing antibody assay is a commercialized kit assay (AXIM Biotechnologies, Inc., San Diego, CA) that utilizes Perkin Elmer’s AlphaLISA technology (Perkin Elmer, Waltham, MA) to detect antibodies that block the interaction of the SARS-CoV-2 RBD and the ACE2 receptor. When antibodies that block RBD from binding to ACE2 are absent, RBD-Acceptor beads bind to ACE2-biotin, and the complex binds to donor beads coated with streptavidin, bringing the beads into close proximity (<200 nm). Excitation of the donor beads by a 680-nm red laser provokes the release of singlet oxygen molecules that triggers a cascade of energy transfer in the acceptor beads, resulting in a sharp peak of light emission at 615 nm. If nAbs are present, the RBD acceptor beads cannot maintain close proximity to the ACE2-biotin molecules; therefore, singlet oxygen excitation cannot occur with the acceptor beads.
Biotinylated ACE2, RBD protein, calibrators, and controls were produced and provided by AXIM Biotechnologies, Inc., San Diego, CA. The calibrator is a human SARS-CoV-2 nAb that was recombinantly produced in HEK-293 cells. This recombinant antibody recognizes the SARS-CoV-2 spike protein RBD domain and inhibits interaction between SARS-CoV-2 RBD and ACE2 with a 50% inhibition concentration (IC50) of 1.5 μg/ml. Calibrators for the standard curve were prepared by diluting the nAb in AlphaLISA assay buffer to known concentrations. Streptavidin donor beads (catalog number [cat. no.] 6760002), functional acceptor beads (cat. no. 5772002B), and AlphaLISA assay buffer (cat. no. AL000F) were acquired from PerkinElmer (Waltham, MA). RBD protein was conjugated to the acceptor beads according to the manufacturer’s instructions. The assay was performed in white ProxiPlate-384 Plus plates (Perkin Elmer, cat. no. 6008280).
A working solution of biotinylated ACE2 at 0.125 μg/ml was prepared in 1× AlphaLISA assay buffer immediately before running the assay. A working solution of AlphaLISA RBD acceptor beads was prepared by diluting the stock 5-mg/ml solution to 40 μg/ml in 1× AlphaLISA buffer. Eight microliters of working ACE2-biotin solution was dispensed into the wells of a ProxiPlate-384 Plus plate followed by 2 μl of serum sample or controls/calibrators. Subsequently, 5 μl of RBD acceptor bead working solution was added to the wells containing the ACE2-biotin–sample mixture. The reaction mixture was then incubated without mixing for 1 h at room temperature. Within 30 min, a working solution of streptavidin (SA) donor beads was diluted to 100 μg/ml under subdued lighting.
Once incubation of the ACE2-biotin–sample–RBD-bead mixture described above was complete, 5 μl of working streptavidin donor-bead solution was dispensed to each reaction well. The plate was incubated for an additional 30 min at room temperature, and then the AlphaLISA signal was measured with a Perkin-Elmer Fusion α-FP-HT multilabel reader equipped with the ALPHA module using the AlphaScreen standard settings.
A standard curve was generated using eight 2-fold serially diluted calibrators run in parallel with the patient samples. Background signal was subtracted from the raw signal of each well, including calibrators, controls, and patient samples. The standard curve was generated by plotting the AlphaLISA counts versus the concentration of the nAb calibrator. Calculated relative concentration units (RCUs) were derived using a nonlinear 4-parameter logistic equation in GraphPad Prism (GraphPad Software Inc., La Jolla, CA). A positive cutoff of ≥0.2 RCU was utilized.
Data analysis.
Antibody titers from serially collected samples were analyzed for trends over time using separate linear mixed-effects models for each assay under study. The models included a fixed effect of “time,” which was defined as the number of days from symptom onset to serum collection. In the course of modeling, both random intercepts and slopes and random intercept only were considered. With only up to three time points per patient, the random intercept-only model was selected for the final analysis.
To graphically present trends in antibody titers over time succinctly, two methods were used. First, days from symptom onset was categorized into time epochs, and boxplots of observed values were plotted. This approach does not account for the clustering of observations within patients and is intended as a representation of the general trends over time (Fig. 1). Second, patient-specific longitudinal trajectory plots, overlaid with model-based estimate of the trend, were constructed (Fig. 2). For the Euroimmun IgG ELISA and the Mayo Clinic nAb assays, measurements below the lower limit of detection were imputed to equal half the lower limit of detection. For NeuCovix-HT, a censored regression was performed using Gauss-Hermite quadrature. Assay result correlation was assessed using the Kendall rank correlation τp. Statistical analyses were conducted using R version 3.6.2 or JMP Pro version 14.1.0.
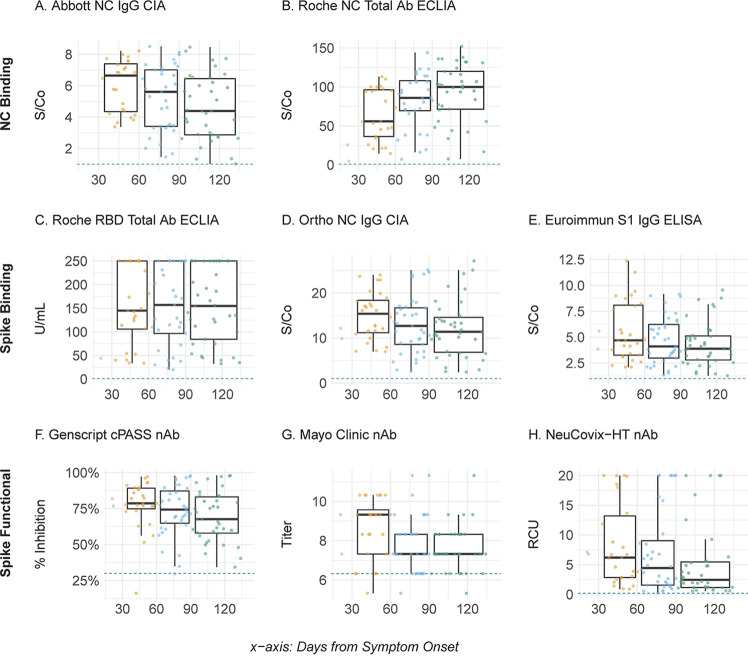
FIG 1.
SARS-CoV-2 antibody titers relative to time post-symptom onset. (A to H) Assay-specific antibody levels for each donor sample (individual filled circles) were plotted as a function of time post-symptom onset. Results were binned at 30 to 60 days (orange), 60 to 90 days (blue), and 91+ days (green). Data collected prior to day 30 are shown in gray due to limited sample size (n = 12). The horizontal boxplot lines indicate the median values, boxes indicate the 25th and 75th percentiles, and vertical lines indicate the largest values within 1.5 times the interquartile range (IQR) greater than the 75th percentile and the smallest values within 1.5 times the IQR lower than the 25th percentile. The horizontal dotted lines indicate the positive cutoff threshold of each assay. Mayo Clinic nAb is shown on the log2 scale. Abbreviations: S/Co, signal to cutoff; NC, nucleocapsid protein; ECLIA, electrochemiluminescent immunoassay; CIA, chemiluminescent immunoassay; RBD, receptor binding domain of spike glycoprotein; nAb, neutralizing antibody; NC, nucleocapsid protein; S, spike glycoprotein; S1, subunit 1 of spike glycoprotein; RCU, relative concentration unit.
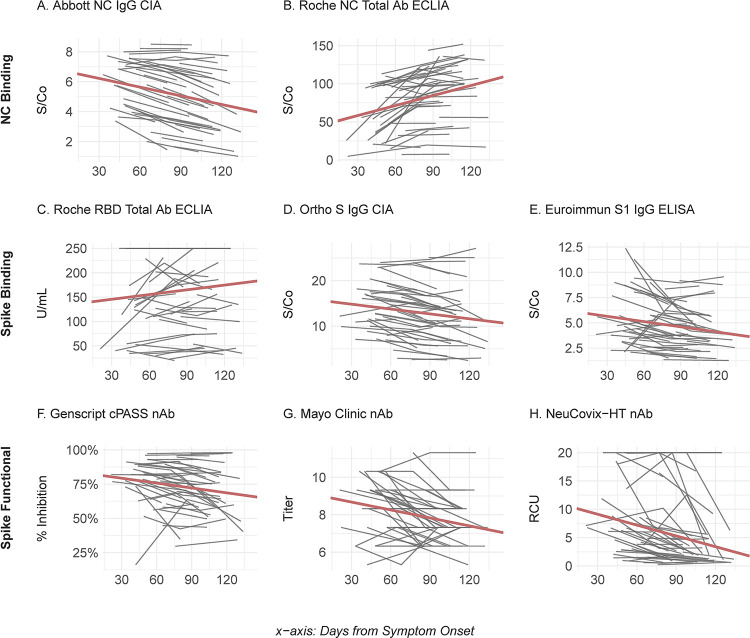
FIG 2.
Longitudinal SARS-CoV-2 antibody levels in COVID-19 convalescent plasma donors. (A to H) Data plotted are the individual patient (n = 44) linear trajectories (in gray) for specimens collected after onset of COVID-19. Table S2 in the supplemental material presents the slope estimates and P values associated with the trends. The bold red lines represent the model-based reference lines for the fixed effects. See Fig. 1 legend for abbreviations.
RESULTS
The mean CCP donor age was 47 years (range, 23 to 73 years), sexes were equally represented, and all exhibited mild to moderate COVID-19 but recovered in the outpatient setting with a 15-day mean symptom duration (range, 2 to 46 days) (see Table S1 in the supplemental material). CCP and serum were provided twice by 26 donors and on three occasions by 18 donors. The first CCP donation was provided a mean of 54 days (range, 21 to 95 days) and the last at 105 days (range, 64 to 134 days) post-symptom onset. From this cohort, a total of 106 serum samples were evaluated, with 99 samples tested by all eight assays, including two for qualitative IgG detection against subunit 1 (S1) of the S glycoprotein (Ortho-Clinical Diagnostics CIA and Euroimmun Inc. ELISA), one for qualitative detection of total antibodies against the NC antigen (Roche Diagnostics NC ECLIA), one that semiquantitatively detects total antibodies against RBD (Roche Diagnostics RBD ECLIA), one for qualitative detection of IgG against the NC antigen (Abbott CIA), and one surrogate, qualitative neutralization assay measuring inhibition of the RBD-ACE2 interaction (GenScript cPASS). The two other nAb assays, both without FDA EUA, include the semiquantitative NeuCovix-HT assay, also a surrogate RBD-ACE2 inhibition assay, and a semiquantitative live-cell neutralization assay expressing the SARS-CoV-2 spike protein in VSV (Mayo Clinic nAb). Seven of the 106 serum samples had insufficient volume for testing by the Abbott NC IgG CIA and were only tested by seven of the eight methods.
We found high clinical sensitivity (97.2% to 100%) among all eight assays, at each time point, which is consistent with previously reported performance characteristics (9–11). Only four CCP donors reverted to seronegative status by a single assay at the final time point measured, including three donors tested by the Abbott NC IgG CIA at days 103, 129, and 132 and one donor tested by the Mayo Clinic nAb assay at day 119 post-symptom onset (Fig. 1). Longitudinal analysis assessing the relative change in antibody titers revealed a statistically significant (P < 0.05) downward trend for six of the assays, including all three nAb assays and three binding antibody assays, including the Abbott NC IgG CIA, the Euroimmun Inc. S IgG ELISA and the Ortho-Clinical Diagnostics S1 CIA (Fig. 2; see also Table S2). In contrast, both the Roche NC and RBD Total Ab ECLIAs showed a statistically significant increase in antibody titers over time (Fig. 1). Finally, quantitative SARS-CoV-2 serologic assay results were correlated and found to be highly variable, with Kendall’s τp ranging from 0.19 for the Roche NC Total Ab ECLIA and Mayo Clinic nAb to 0.76 between the Ortho-Clinical Diagnostics NC IgG CIA and GenScript cPASS nAb assay (Fig. 3). Spike-based binding assays had the best correlation as a group (τp, 0.74, 0.73, and 0.64) compared to those for the spike based functional assays (τp, 0.68, 0.60, and 0.53) and NC-based binding assays (τp, 0.26). The NC-based assays had poor correlation with spike-based binding assays and spike-based functional assays (τp, 0.19 to 0.57).
FIG 3.
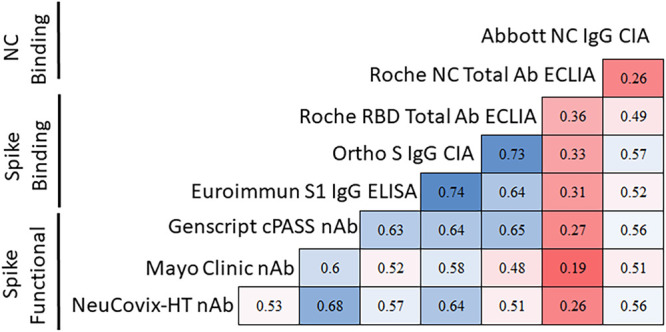
SARS-CoV-2 antibody titer correlation across assays. The correlation of titers between different assays is shown in a matrix format using the Kendall rank correlation τp. Blue indicates a higher degree of correlation and red indicates a lower degree of correlation.
DISCUSSION
Using a well-characterized serial sample set from COVID-19-confirmed CCP donors, we show differential antibody persistence among eight unique SARS-CoV-2 serologic methods, including both binding and nAb assays with and without FDA EUA. Our findings suggest that there is prolonged antibody detection using two ECLIA-based methods targeting total antibodies against the NC or RBD antigens. Although the precise cause for this remains undefined, differences related to individual assay detection methods and the targeted antibody type likely play a significant role.
Maturation of the humoral immune response in immunocompetent individuals progresses through the initial secretion of typically low-affinity antibodies from short-lived plasmablasts, which peak 2 to 3 weeks postinfection, to affinity maturation and secretion of high-affinity antibodies by long-lived plasma cells (12, 13). This maturation process may explain the observed increase in relative antibody levels by the Roche ECLIAs, which unlike other methods, are based on antibody detection via a dual-antigen bridging method to capture bivalent binding SARS-CoV-2-specific immunoglobulins. This dual-binding requirement may reflect detection of high-affinity and -avidity antibodies (Roche Diagnostics, personal communication), although further studies are warranted to better characterize this possibility. This alternative antibody capture method may also clarify the difference in documented antibody durability between the Roche anti-NC ECLIA and the Abbot CIA, which also detects anti-NC antibodies but uses a standard, single-antigen-binding sandwich immunoassay format. Our findings are consistent with recent studies showing increasing anti-NC antibody levels over time by the Roche ECLIA and also support prior findings suggesting that antibody levels are not solely related to the SARS-CoV-2 antigen used in serologic LDTs (4, 14). Our data further build on the likelihood that the difference in antibody persistence may be method dependent, by demonstrating that a similar increase in antibody levels post-symptom onset occurs when using the more recently released semiquantitative Roche ECLIA, which detects total antibodies against the RBD.
Differences in the relative antibody durability in our study is unlikely to be strictly isotype related. Anti-SARS-CoV-2 IgM-class antibodies decline rapidly prior to IgG, with minimal detection by 12 weeks postinfection (15). Yet, we observed increasing antibody levels using total immunoglobulin targeted assays, whereas all methods specific for anti-SARS-CoV-2 IgG showed declining levels over time. Notably, all three functional nAb assays also showed declining serum potency to block the RBD-ACE interaction. One likely explanation for this is that these functional assays are concentration sensitive but less dependent on antibody avidity or affinity. Prior LDT neutralization assays for SARS-CoV-2 have shown modest but slowly declining nAb levels over time (8). Live pseudovirus neutralizing assays often involve longer periods of coincubation between the virus and patient sera, which may provide ample time for lower-affinity/avidity antibodies to engage their target (16). The findings in our live recombinant SARS-CoV-2-neutralizing assay were confirmed here using two additional LDT neutralization assays, one being the only SARS-CoV-2 nAb assay with FDA EUA (GenScript cPASS nAb).
While qualitative interpretations from all eight assays demonstrated strong agreement, the association of numerical results between assays were highly variable, with Kendall’s τp ranging from 0.19 (Roche NC Total Ab ECLIA and Mayo Clinic nAb) to 0.76 (Ortho-Clinical Diagnostics S IgG CIA and GenScript cPASS nAb) when utilizing the baseline sample for each donor. Numerical results generated by each of these assays are referenced against either a single or a multipoint calibration curve. However, as of the writing of the manuscript, these assays are not calibrated against a common reference standard, such as the World Health Organization (WHO) international SARS-CoV-2 antibody standard (first released in December 2020), which partially explains the low correlation of numerical results (17). Standardization of SARS-CoV-2 serologic assays using reference material, such as that available through the WHO, would help to minimize interassay variability among both current and future serologic methods. Such standardization will also be critical for SARS-CoV-2 quantitative and semiquantitative serologic methods, should an antibody-based correlate or surrogate of protective immunity be identified, similar to what has been established for other vaccine-preventable diseases (17).
In conclusion, our study is among the first to assess longitudinal, rather than cross-sectional, SARS-CoV-2 antibody profiles among convalescent COVID-19 patients, primarily using commercially available serologic assays with FDA EUA instead of focusing on more-restricted-use LDTs. Based on our findings, it will be important to determine if the specific antibodies targeted by the Roche ECLIA assays are more reflective of protective or long-term immunity. Our study has a number of limitations, including the assessment of numerical output values of assays that are designed to be strictly qualitative in nature. Additionally, a limited number of CCP donors were available with serial samples, and we were only able to assess antibody longevity up to approximately 4 months following symptom onset. Further studies evaluating serial samples collected beyond this time point are necessary to continue to define the durability of the antibody response both following natural infection, including with the newly circulating and emerging SARS-CoV-2 variants, and following vaccination. The implications of better defining the duration of detectable antibody responses over time may become more relevant in the future should a correlate of protective immunity be identified and subsequent serologic testing, using calibrated assays, come to play a more prominent role in clinical management.
Footnotes
Supplemental material is available online only.
Contributor Information
John R. Mills, Email: mills.john2@mayo.edu.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.Tang F, Quan Y, Xin ZT, Wrammert J, Ma MJ, Lv H, Wang TB, Yang H, Richardus JH, Liu W, Cao WC. 2011. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol 186:7264–7268. 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 2.Callow KA, Parry HF, Sergeant M, Tyrrell DA. 1990. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect 105:435–446. 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu LP, Wang NC, Chang YH, Tian XY, Na DY, Zhang LY, Zheng L, Lan T, Wang LF, Liang GD. 2007. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis 13:1562–1564. 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, Nakao C, Rayaprolu V, Rawlings SA, Peters B, Krammer F, Simon V, Saphire EO, Smith DM, Weiskopf D, Sette A, Crotty S. 2021. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371:eabf4063. 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, Arnthorsson AO, Helgason D, Bjarnadottir K, Ingvarsson RF, Thorsteinsdottir B, Kristjansdottir S, Birgisdottir K, Kristinsdottir AM, Sigurdsson MI, Arnadottir GA, Ivarsdottir EV, Andresdottir M, Jonsson F, Agustsdottir AB, Berglund J, Eiriksdottir B, Fridriksdottir R, Gardarsdottir EE, Gottfredsson M, Gretarsdottir OS, Gudmundsdottir S, Gudmundsson KR, Gunnarsdottir TR, Gylfason A, Helgason A, Jensson BO, Jonasdottir A, Jonsson H, Kristjansson T, Kristinsson KG, Magnusdottir DN, Magnusson OT, Olafsdottir LB, Rognvaldsson S, Le Roux L, Sigmundsdottir G, Sigurdsson A, Sveinbjornsson G, Sveinsdottir KE, Sveinsdottir M, Thorarensen EA, Thorbjornsson B, Thordardottir M, Saemundsdottir J, et al. 2020. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 383:1724–1734. 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF, Wang DQ, Hu Y, Ren JH, Tang N, Xu YY, Yu LH, Mo Z, Gong F, Zhang XL, Tian WG, Hu L, Zhang XX, Xiang JL, Du HX, Liu HW, Lang CH, Luo XH, Wu SB, Cui XP, Zhou Z, Zhu MM, Wang J, Xue CJ, Li XF, Wang L, Li ZJ, Wang K, Niu CC, Yang QJ, Tang XJ, Zhang Y, Liu XM, Li JJ, Zhang DC, Zhang F, Liu P, Yuan J, Li Q, Hu JL, Chen J, Huang AL. 2020. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 26:845–848. 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 7.Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, Hemmings O, O'Byrne A, Kouphou N, Galao RP, Betancor G, Wilson HD, Signell AW, Winstone H, Kerridge C, Huettner I, Jimenez-Guardeno JM, Lista MJ, Temperton N, Snell LB, Bisnauthsing K, Moore A, Green A, Martinez L, Stokes B, Honey J, Izquierdo-Barras A, Arbane G, Patel A, Tan MKI, O'Connell L, O'Hara G, MacMahon E, Douthwaite S, Nebbia G, Batra R, Martinez-Nunez R, Shankar-Hari M, Edgeworth JD, Neil SJD, Malim MH, Doores KJ. 2020. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol 5:1598–1607. 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, McMahon M, Meade P, Mendu DR, Muellers K, Stadlbauer D, Stone K, Strohmeier S, Simon V, Aberg J, Reich DL, Krammer F, Cordon-Cardo C. 2020. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 370:1227–1230. 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnurra C, Reiners N, Biemann R, Kaiser T, Trawinski H, Jassoy C. 2020. Comparison of the diagnostic sensitivity of SARS-CoV-2 nucleoprotein and glycoprotein-based antibody tests. J Clin Virol 129:104544. 10.1016/j.jcv.2020.104544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merrill AE, Jackson JB, Ehlers A, Voss D, Krasowski MD. 2020. Head-to-head comparison of two SARS-CoV-2 serology assays. J Appl Lab Med 5:1351–1357. 10.1093/jalm/jfaa125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theel ES, Harring J, Hilgart H, Granger D. 2020. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin Microbiol 58:e01243-20. 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen HN. 2014. Affinity enhancement of antibodies: how low-affinity antibodies produced early in immune responses are followed by high-affinity antibodies later and in memory B-cell responses. Cancer Immunol Res 2:381–392. 10.1158/2326-6066.CIR-14-0029. [DOI] [PubMed] [Google Scholar]
- 13.McCarthy KR, Raymond DD, Do KT, Schmidt AG, Harrison SC. 2019. Affinity maturation in a human humoral response to influenza hemagglutinin. Proc Natl Acad Sci U S A 116:26745–26751. 10.1073/pnas.1915620116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muecksch F, Wise H, Batchelor B, Squires M, Semple E, RIchardson C, McGuire J, Clearly S, Furrie E, Neil G, Hay G, Templeton K, Lorenzi JCC, Hatziioannou T, Jenks S, Bieniasz PD. 6August2020. Longitudinal analysis of clinical serology assay performance and neutralising antibody levels in COVID19 convalescents. medRxiv 10.1101/2020.08.05.20169128. [DOI] [Google Scholar]
- 15.Roltgen K, Powell AE, Wirz OF, Stevens BA, Hogan CA, Najeeb J, Hunter M, Wang H, Sahoo MK, Huang C, Yamamoto F, Manohar M, Manalac J, Otrelo-Cardoso AR, Pham TD, Rustagi A, Rogers AJ, Shah NH, Blish CA, Cochran JR, Jardetzky TS, Zehnder JL, Wang TT, Narasimhan B, Gombar S, Tibshirani R, Nadeau KC, Kim PS, Pinsky BA, Boyd SD. 2020. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol 5:eabe0240. 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoury DS, Wheatley AK, Ramuta MD, Reynaldi A, Cromer D, Subbarao K, O'Connor DH, Kent SJ, Davenport MP. 2020. Measuring immunity to SARS-CoV-2 infection: comparing assays and animal models. Nat Rev Immunol 20:727–738. 10.1038/s41577-020-00471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattiuzzo G, Bentley EM, Hassall M, Routley S, Richardson S, Bernasconi V, Kristiansen P, Harvala H, Roberts D, Semple MG, Turtle LCW, Openshaw PJM, Balillie K, Nissen-Meyer LSH, Brantsaeter ABBH, Atkinson E, Rigsby P, Padley D, Almond N, Rose NJ, Page M. 2020. Establishment of the WHO International Standard and Reference Panel for anti-SARS-CoV-2 antibody. https://cdn.who.int/media/docs/default-source/biologicals/ecbs/bs-2020-2403-sars-cov-2-ab-ik-17-nov-2020_4ef4fdae-e1ce-4ba7-b21a-d725c68b152b.pdf?sfvrsn=662b46ae_8&download=true. Accessed 2 February 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2. Download JCM.01231-21-s0001.pdf, PDF file, 0.2 MB (215.8KB, pdf)