ABSTRACT
Antigen-based rapid diagnostic tests (RDTs) are used in children despite the lack of data. We evaluated the diagnostic performance of the Panbio-COVID-19 Ag Rapid Test Device (P-RDT) in children. Symptomatic and asymptomatic participants 0 to 16 years old had two nasopharyngeal swabs (NPS) for both reverse transcription-PCR (RT-PCR) and P-RDT. A total of 822 participants completed the study, of which 533 (64.9%) were symptomatic. Among the 119 (14.5%) RT-PCR-positive patients, the P-RDT sensitivity was 0.66 (95% confidence interval [CI] 0.57 to 0.74). Mean viral load (VL) was higher among P-RDT-positive patients than negative ones (P < 0.001). Sensitivity was 0.91 in specimens with VL of >1.0E6 IU/ml (95% CI 0.83 to 0.99) and decreased to 0.75 (95% CI 0.66 to 0.83) for specimens >1.0E3 IU/ml. Among symptomatic participants, the P-RDT displayed a sensitivity of 0.73 (95% CI 0.64 to 0.82), which peaked at 1.00 at 2 days post-onset of symptoms (DPOS) (95% CI 1.00 to 1.00), then decreased to 0.56 (95% CI 0.23 to 0.88) at 5 DPOS. There was a trend toward lower P-RDT sensitivity in symptomatic children <12 years (0.62 [95% CI 0.45 to 0.78]) versus ≥12 years (0.80 [95% CI 0.69 to 0.91]; P = 0.09). In asymptomatic participants, the P-RDT displayed a sensitivity of 0.43 (95% CI 0.26 to 0.61). Specificity was 1.00 in symptomatic and asymptomatic children (95% CI 0.99 to 1.00). The overall 73% and 43% sensitivities of P-RDT in symptomatic and asymptomatic children, respectively, was below the 80% cutoff recommended by the World Health Organization. We observed a correlation between VL and P-RDT sensitivity, as well as variation of sensitivity according to DPOS, a major determinant of VL. These data highlight the limitations of RDTs in children, with the potential exception in early symptomatic children ≥12yrs.
KEYWORDS: COVID-19, children, SARS-CoV-2, antigen-based rapid diagnostic tests, diagnostics, pediatric infectious disease, rapid diagnostic tests
INTRODUCTION
The current coronavirus disease 19 (COVID-19) pandemic induces the need for widespread SARS-CoV-2 testing to control virus circulation. The rapid identification of SARS-CoV-2-infected individuals is important whether persons are symptomatic or not, as the role of asymptomatic persons in SARS-CoV-2 transmission is still unclear. Therefore, easy to use, affordable, and rapid diagnostic methods are required in addition to the gold standard of reverse transcription-PCR (RT-PCR) (1). These devices are increasingly helpful in settings where results are immediately needed, access to a testing facility is limited, or in case of shortage of RT-PCR reagents. Several antigen-based rapid diagnostic tests (RDTs) have been marketed to fill this gap. Among them, the Panbio-COVID-19 Ag Rapid Test Device (referred to here as P-RDT) has displayed an overall sensitivity ranging between 61% and 92% (2–7) in adults compared to nasopharyngeal RT-PCR. Sensitivity was improved for higher viral loads (VLs), shorter duration of symptoms and/or in symptomatic patients (2–6). In symptomatic children, the overall sensitivity of the P-RDT was 45 to 78% (5, 8, 9), but published data do not take into account the effect of VL nor the duration of symptoms. Moreover, to our knowledge, no study has evaluated this assay in asymptomatic children at the time of sampling. The aim of the present study was to provide an independent evaluation of the diagnostic performance of the P-RDT in a large cohort of symptomatic and asymptomatic children. We also aimed to identify situations with optimal P-RDT sensitivity by accounting for VL, day post onset of symptoms (DPOS), type and number of symptoms.
MATERIALS AND METHODS
Setting.
This single-center prospective diagnostic study was performed in Geneva University Hospitals’ (HUG) pediatric testing center, from 10 November 2020 to 26 March 2021, with a maximum 14-day incidence of 1,199/100,000 at the time of study onset (10). Participants 0 to 16 years old who presented with the need for SARS-CoV-2 RT-PCR testing were approached. The indication for RT-PCR testing in symptomatic participants was symptoms suggestive of SARS-CoV-2 infection according to local governmental testing criteria. The indications for RT-PCR testing in asymptomatic participants were notification by local health authorities after contact with a laboratory confirmed a SARS-CoV-2-infected person and pretravel testing. The presence or absence of exposure to a SARS-CoV-2 infected person was not documented.
Study procedures.
For each enrolled participant, two nasopharyngeal swabs (NPS) were collected. Nurses were trained to perform NPS testing through a standardized video-documented procedure. First, a standard flocked swab placed in viral transport media (VTM) was used for viral genome detection by RT-PCR. The second swab, provided in the P-RDT kit, was obtained from the contralateral or ipsilateral nostril and the antigen test was performed immediately at the testing center as per the manufacturer’s instructions. All study participants and/or caregivers provided written informed consent prior to specimen collection. The study was approved by the local research ethics board (Commission cantonale d'éthique de la recherche number 2020-02323).
P-RDT testing.
The Panbio-COVID-19 Ag Rapid Test Device for nasopharyngeal use (Abbott Rapid Diagnostics, USA; ref 41FK10) was chosen for the current study based on adult data from our institution showing optimal sensitivity and ease of use. The P-RDT was used as recommended by the manufacturers, using materials provided in the kit only. P-RDT results were read independently by two members of the study team, both being blinded to the result assigned by their pair as well as to the clinical presentation of the participant. Any discrepant result was considered positive when any of the above-mentioned readers set a positive diagnosis.
RT-PCR testing.
RT-PCR testing was performed either on the Cobas SARS-CoV-2 assay (cobas SARS-CoV-2 Test, Cobas 6800, Roche, Switzerland) or on the TaqPath (Applied Biosystems, Thermo Fisher Scientific, Waltham, USA) RT-PCR assay, using NPS in 3 ml VTM. In order to transform cycle threshold (CT) values into IU/ml, serial dilutions of the first World Health Organization (WHO) international standard for SARS-CoV-2 RNA (11) (National Institute for Biological Standards and Control [NIBSC], Potters Bar, United Kingdom; product code: 20/146) were performed to calibrate both RT-PCR assays, as per the manufacturer’s instructions. All VLs were calculated for originals specimens in log IU/ml of VTM from the CT values using the following formulas: for Cobas (E gene target), VL (log IU/ml) = (CT value − 42.59)/−3.096 and for TaqPath (N gene target), VL (log IU/ml) = (CT value − 44.333)/−3.08.
Data collection.
The following data collected at enrollment were managed using RedCap electronic data capture tools hosted at HUG: date of enrollment, number of days post onset of symptoms (DPOS), gender, age, type of symptoms (nasal discharge, cough, dyspnea, dysphagia, dysgueusia, anosmia, vomiting, diarrhea, fever, chills, decreased intake, headache, myalgia, fatigue and irritability, abdominal pain, and/or nausea), and comorbidities if present (chronic respiratory disease, cardiopathy, immunosuppression, cancer, diabetes, obesity, hypertension, and organ failure). P-RDT results, RT-PCR and VL/CT values were included subsequently.
Statistics.
Before study onset, a sample size was calculated to have sufficient power to generate a 95% confidence interval (CI) with a lower bound above the WHO target of 80% if the prevalence was 25% (corresponding to the pediatric positivity rate at study onset) and the measured sensitivity of 85% (corresponding to the sensitivity reported in adults in our institution) (6). The sample size was therefore estimated at 654 participants. Continuous variables were expressed by their mean ± standard deviation (SD) and median (interquartile range [IQR]) upon variable distribution. Categorical variables were presented by their frequencies and relative proportions. For comparisons of continuous variables, parametric Student t tests and nonparametric Mann-Whitney tests upon variable distribution were used. For categorical variables, either Chi-square or Fischer’s exact tests were performed, depending on applicability. Statistical analyses were processed using SPSS software v23.0 (IBM Corp., Armonk, NY). Statistical significance was defined as P < 0.05 (two-sided).
Data availability.
Study protocols, statistical analysis plan, and individual participant data that underlie the results reported in this article can be made available (after deidentification) to researchers who make a methodologically sound proposal. Proposals should be directed to arnaud.lhuillier@hcuge.ch. To gain access, data requestors will need to sign a data access agreement.
RESULTS
A total of 885 pediatric participants were enrolled. Among them, 63 were subsequently excluded (Fig. S1 in the supplemental material). A total of 822 participants completed the study and had both RT-PCR and P-RDT performed. The demographics of symptomatic and asymptomatic study participants are detailed in Table 1.
TABLE 1.
Study participants’ demographics
| Characteristicsa | Symptomatic (n = 533) | Asymptomatic (n = 289) | Combined (n = 822) | P valueb |
|---|---|---|---|---|
| Median age (±IQR) | 12.1 (9.4–14.5) | 10.9 (8.5–13.7) | 11.8 (9.0–14.3) | 0.002 |
| Female sex, n (%) | 266 (49.9) | 138 (47.8) | 404 (49.1) | 0.555 |
| Comorbidities, n (%) | ||||
| Chronic respiratory disease | 33 (6.2) | 13 (4.5) | 46 (5.6) | |
| Obesity | 5 (2.8) | 10 (3.5) | 25 (3.0) | |
| Diabetes | 1 (0.2) | 3 (1.0) | 4 (0.5) | |
| Hypertension | 2 (0.4) | 0 | 2 (0.2) | |
| Cancer | 0 | 2 (0.7) | 2 (0.2) | |
| Cardiopathy | 1 (0.2) | 0 | 1 (0.1) | |
| Other immunosuppression | 1 (0.2) | 0 | 1 (0.1) | |
| Chronic liver failure | 1 (0.2) | 0 | 1 (0.1) | |
| Result of RT-PCR, n (%) | 0.014 | |||
| Negative | 444 (83.3) | 259 (89.6) | 703 (85.5) | |
| Positive | 89 (16.7) | 30 (10.4) | 119 (14.5) | |
| Mean log RNA IU/ml (± SD) | 5.9 (±1.8) | 4.1 (±1.9) | 5.5 (±2.0) | <0.001 |
RT-PCR, reverse transcription-PCR; IQR, interquartile range; SD, standard deviation.
Symptomatic versus asymptomatic participants.
Overall, 14.5% (119/822) were positive by RT-PCR with a mean RNA VL of 5.5 log IU/ml (SD 2.0) (Table 1). Among the 822 P-RDT performed, only one P-RDT result displayed a discrepant interpretation between the two observers (κ = 0.999). The corresponding patient was subsequently considered positive for the purpose of the analysis, leaving an overall positivity rate of 9.6% (79/822). The P-RDT’s sensitivity and specificity when challenged against RT-PCR were 0.66 (95% CI 0.57 to 0.74) and 1.00 (95% CI 1.00 to 1.00), respectively (Table 2, Table S1). Mean VL was higher among positive P-RDT specimens than negative ones (6.4 log IU/ml [SD 1.4] versus 3.7 [SD 1.6]; P < 0.001) (Fig. 1).
TABLE 2.
Diagnostic accuracy of the Panbio RDT
| Characteristicsa | Symptomatic | Asymptomatic | Combined |
|---|---|---|---|
| Sensitivity (95% CI) | 0.73 (0.64–0.82) | 0.43 (0.26–0.61) | 0.66 (0.57–0.74) |
| Specificity (95% CI) | 1.00 (0.99–1.00) | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) |
| Positive predictive value (95% CI) | 0.98 (0.92–1.00) | 1.00 (1.00–1.00) | 0.99 (0.96–1.01) |
| Negative predictive value (95% CI) | 0.95 (0.92–0.97) | 0.94 (0.91–0.97) | 0.95 (0.93–0.96) |
CI, confidence interval.
FIG 1.
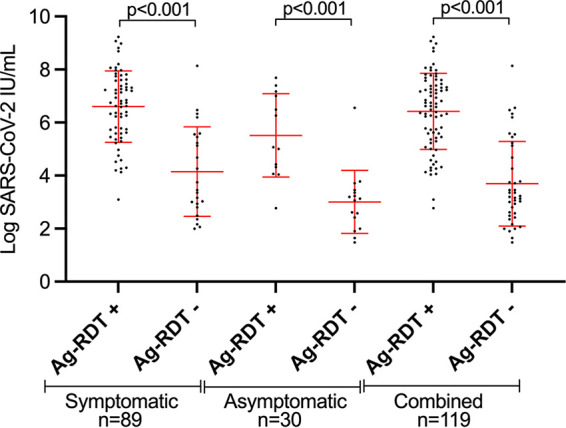
Mean (standard deviation) SARS-CoV-2 viral load expressed in log IU/ml among RT-PCR-positive individuals according to Panbio RDT results. RDT, antigen-based rapid diagnostic test; RT-PCR, reverse transcription-PCR; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Sensitivity varied according to RT-PCR VL, even though false-negative results occurred throughout all VL values. Sensitivity was highest at 0.97 in specimens with VL of >1.0E7 IU/ml (95% CI 0.91 to 1.00), decreased slightly to 0.91 (95% CI 0.83 to 0.99) for specimens of >1.0E6 IU/ml, and to 0.87 (95% CI 0.79 to 0.94) and 0.86 (95% CI 0.79 to 0.94) for specimens of >1.0E5 IU/ml and >1.0E4 IU/ml, respectively. Sensitivity then dropped to 0.75 (95% CI 0.66 to 0.83) and 0.69 (95% CI 0.61 to 0.78) for specimens of >1.0E3 IU/ml and >1.0E2 IU/ml, respectively (Fig. 2). Of note, nine P-RDT-negative specimens that showed a high viral load by RT-PCR were retested using another RT-PCR assay which confirmed high viral load in every specimen.
FIG 2.
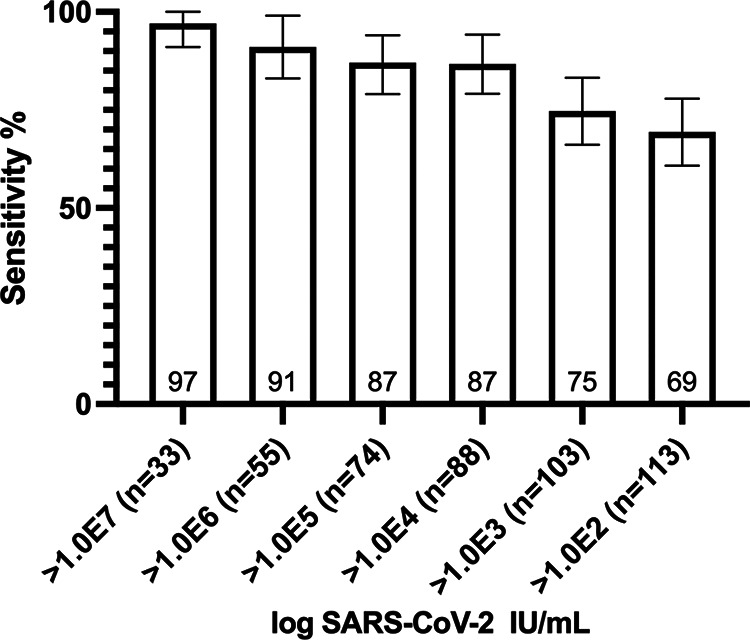
Sensitivity (95% confidence interval) of Panbio RDT according to SARS-CoV-2 viral load expressed in log IU/ml. RDT, antigen-based rapid diagnostic test; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Mean viral load was lower in children <12 years old than in older children (5.0 log IU/ml [SD 2.0] versus 5.9 [SD 1.9]; P = 0.011) and there was a trend toward lower P-RDT sensitivity in children <12 years old (0.57 [95% CI 0.44 to 0.70]) than in older children (0.74 [95% CI 0.63 to 0.85]; P = 0.057). Additional demographics stratified by age group are detailed in Table S2.
Symptomatic participants.
Among the 533 (64.9%) symptomatic participants, median duration of symptoms at time of testing was 2 days (IQR 1 to 3) (Table S3). The most frequently reported symptoms were headache (56%), nasal discharge (56%), cough (45%), and fatigue (44%) (Table S3). Eighty-nine symptomatic patients (16.7%) were positive by RT-PCR with a mean RNA VL of 5.9 log IU/ml (SD 1.8) (Table 1). The P-RDT displayed an overall sensitivity and specificity of 0.73 (95% CI 0.64 to 0.82) and 1.00 (95% CI 0.99 to 1.00), respectively (Table 2, Table S1). For specimens with VL of >1.0E6 IU/ml, sensitivity was 0.92 (95% CI 0.84 to 1.00). Mean VL was higher among positive P-RDT specimens than negative ones (6.6 log IU/ml [SD 1.3] versus 4.1 [SD 1.7]; P < 0.001) (Fig. 1).
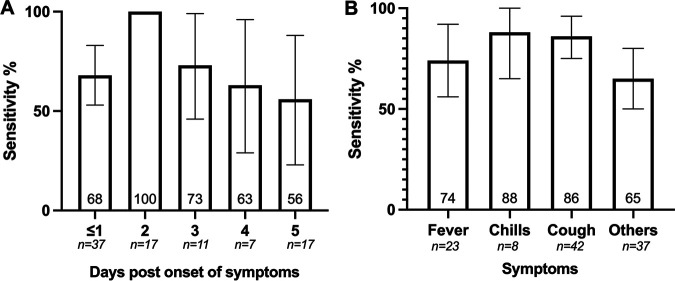
Sensitivity was 0.68 at 0 to 1 DPOS (95% CI 0.53 to 0.83), peaked at 1.00 at 2 DPOS (95% CI 1.00 to 1.00), then gradually decreased to 0.73 (95% CI 0.46 to 0.99), 0.63 (95% CI 0.29 to 0.96), and 0.56 (95% CI 0.23 to 0.88) at 3, 4, and 5 DPOS, respectively (Fig. 3A). False-negative results occurred across all DPOS except at 2 DPOS.
FIG 3.
Sensitivity (95% confidence interval) of Panbio RDT according to days post onset of symptoms (A) and clinical symptoms (B). RDT, antigen-based rapid diagnostic test.
Additionally, we analyzed sensitivity according to typical acute COVID-19 symptoms. For this analysis, only objective symptoms were reported because of the low ability of children to report more subjective symptoms, such as anosmia, even though very suggestive of COVID-19. Sensitivity was highest in the presence of chills (0.88 [95% CI 0.65 to 1.00]) and cough (0.86 [95% CI 0.75 to 0.96]), followed by fever (0.74 [95% CI 0.56 to 0.92]) and then nonspecific symptoms (0.65 [95% CI 0.50 to 0.80]) (Fig. 3B). Interestingly, sensitivity was significantly better in participants reporting >2 symptoms (0.78 [95% CI 0.68 to 0.87]) than in those reporting only 1 to 2 symptoms (0.53 [95% CI 0.29 to 0.77]; P = 0.038).
Among symptomatic participants, mean viral load did not significantly differ between children <12 years old and older children (5.5 log IU/ml [SD 1.8] versus 6.2 [SD 1.8]; P = 0.085). Moreover, median duration of symptoms did not differ between children <12 years old and older children (2 days [IQR 1 to 4] for both groups; P = 0.790). There was a trend toward lower P-RDT sensitivity in children <12 years old (0.62 [95% CI 0.45 to 0.78]) than in older ones (0.80 [95% CI 0.69 to 0.91]; P = 0.09).
Asymptomatic participants.
Among the 289 (35.1%) asymptomatic participants at the time of sampling, 10.4% (30/289) were positive by RT-PCR with a mean RNA VL of 4.1 log IU/ml (SD 1.9), which was significantly lower than found in symptomatic participants (P < 0.001) (Table 1). The P-RDT displayed an overall sensitivity and specificity of 0.43 (95% CI 0.26 to 0.61) and 1.00 (95% CI 1.00 to 1.00), respectively (Table 2, Table S1). For specimens with VL >1.0E6 IU/ml, sensitivity was 0.86 (95% CI 0.60 to 1.00). Mean VL was higher among positive P-RDT specimens than in negative ones (5.5 log IU/ml [SD 1.6] versus 3.0 [SD 1.2]; P < 0.001) (Fig. 1). Mean viral load did not significantly differ between children <12 years old and older children (4.3 [SD 2.0] versus 3.5 [SD 0.8]; P = 0.158).
DISCUSSION
This study prospectively evaluated the diagnostic accuracy of the Panbio-COVID-19 RDT in the clinical setting in more than 800 symptomatic and asymptomatic children, taking into account VL, DPOS, and clinical parameters such as number and type of symptoms. The study was performed during a period of sustained virus circulation, with an overall positivity RT-PCR rate of 17% among symptomatic participants. The major finding of our work is an overall suboptimal 66% sensitivity of the assay, ranging between 43% and 73% in asymptomatic and symptomatic children, respectively. On the other hand, specificity was 100% regardless of the presence or absence of symptoms. It would therefore seem very unlikely that children are unnecessarily sent into quarantine, which is important from a public health perspective. The WHO RDT target product profile cutoffs of ≥80% for sensitivity and ≥97% for specificity were achieved for specificity but not for sensitivity (12). The relatively low sensitivity of the P-RDT is in line with previous data showing an assay sensitivity of 45 to 78% among symptomatic children (5, 8, 9), and confirms that the assay sensitivity is lower than that in symptomatic adults in whom the largest studies report sensitivity between 67 and 92% (2, 3, 5–7). However, among symptomatic children with high VL, the assay’s sensitivity seemed only marginally lower than symptomatic adults with high VL (6). The suboptimal sensitivity of the assay in children is most likely explained by the increasingly recognized evidence that children have lower SARS-CoV-2 VLs than adults. Indeed, although initial studies suggested similar VLs in adults and children, they were limited in sample size and did not take into account DPOS (13–15), which is a major determinant of VL (16). Recently, studies on larger data sets and/or taking into account DPOS have shown that SARS-CoV-2-infected children have significantly lower VLs than adults (17–19). Another possible explanation for the lower sensitivity could be sampling bias related to the technical challenge of the NPS procedure in children, given that the swab for the P-RDT testing was the second one to be performed. False-negative RDT results have also been observed in adult RDT studies, even though less frequently (6), and are unlikely to be caused by SARS-CoV-2 variants. Indeed, no mutation in the N gene possibly causing false-negative RDTs in circulating SARS-CoV-2 variants have been identified and, so far, all variants are detected with RDTs with comparable sensitivity to earlier circulating variants (20).
Novel findings in our study relate to the evaluation of P-RDT sensitivity in the light of several factors such as VL, DPOS, type and number of symptoms, which has not been reported so far in the pediatric population. As expected, VL was higher among those with positive P-RDT (true positives) than among those with negative P-RDT (false negatives), as already shown in the adult setting (6). Similarly, and as previously shown in adults (2, 3, 6), sensitivity was correlated with VL, peaking at 97% in specimens with >1.0E7 IU/ml and dropping to 75% in specimens with >1.0E3 IU/ml. Sensitivity remained at >80% in specimens with >1.0E4 IU/ml. However, the presence of false-negative P-RDT results in participants with VLs compatible with shedding of infectious virus is important from a public health perspective, as these would not be identified by P-RDT despite being likely contagious. Another interesting finding was the impact of DPOS on the assay sensitivity. Sensitivity was optimal at 2 DPOS, when VL is expected to peak (21, 22). These findings somehow differ from adult data, where sensitivity remained high throughout the first five DPOS (despite being lower at 0 DPOS) (6), likely here again reflecting the impact of higher VLs in adults on P-RDT sensitivity. Similarly, the trend toward lower sensitivity in children <12 years old is likely explained by lower VLs in younger children, as seen in this study and other publications (17–19). Among symptomatic participants, sensitivity was better in those with COVID-19-typical symptoms, as previously shown in adults (6), but also in those with >2 symptoms. Interestingly, the sensitivity of P-RDT in children asymptomatic at time of sampling was low at 43%. This is probably related to the fact that asymptomatic children in our data set had significantly lower VL than symptomatic children.
The strength of our study is related to the large size of a purely pediatric data set. The large subset of asymptomatic children at the time of sampling, and the analysis of diagnostic accuracy based on VL, DPOS, number and specific symptoms, represent additional strengths and novelties. With a majority of mild clinical presentation and 25% being asymptomatic among RT-PCR-positive cases, our data set is representative of the majority of SARS-CoV-2-infected children, even though the extent of the pediatric contribution to community transmission is still debated.
Our study has several limitations. First, the evaluation was based on one RDT only. Comparative studies have shown similar or reduced performance of other RDTs compared to the P-RDT (6, 23). It is therefore highly unlikely that another RDT would perform significantly better in children than the P-RDT. Second, the study was performed using two different validated RT-PCR assays as gold standards, although 90% of the specimens were tested on the Cobas assay, and VLs were reported in IU/ml, allowing comparison between assays. Third, the study was conducted in a high-prevalence setting. Extrapolating the findings to low-prevalence settings must be done with caution. Then, providing that the NPS for P-RDT was always performed after the NPS for RT-PCR, one cannot exclude that the second procedure was more challenging to perform. Finally, we did not evaluate the performance of P-RDT on oropharyngeal, nasal, or saliva specimens. However, given the fact that VL is lower in these anatomical compartments compared to NPS (24–27), one can expect even lower sensitivity of P-RDT if used on oropharyngeal, nasal, or saliva specimens.
In conclusion, this independent study confirms the respective suboptimal sensitivity of P-RDT in symptomatic children and its poor sensitivity in children asymptomatic at the time of sampling, providing additional evidence for cautious routine use of these tests for the detection of SARS-CoV-2, both in symptomatic and asymptomatic children. This study also highlights the impact of VL, DPOS, and clinical presentation on the assay’s sensitivity and shows that the sensitivity was ≥80% in participants with medium and high VLs, suggesting reliable identification of contagious individuals (16, 28). However, it should be discussed whether missing individuals with lower VLs is acceptable, since they might subsequently have an increase in their VL and become contagious. Therefore, public health benefits of rapidly identifying infected children should be balanced with the disadvantages of missed diagnoses (29). For individual diagnosis, P-RDT seems a decent alternative to RT-PCR in symptomatic children ≥12 years, especially if tested at <5 DPOS, based on our findings. For mass pediatric screening, however, such as in school settings or institutions, providing there is no vulnerable contact person, the suboptimal sensitivity of P-RDT is likely outweighed by the advantages of P-RDT, allowing rapid identification of most infected individuals without the need of a laboratory facility.
ACKNOWLEDGMENTS
We thank our medical students (Mélina Ben Allel, Emmanuelle Mohbat, Liv Mahler, James Nef, Mia Lidén, Eloïse Sattonnay, Estelle Delamare, Eva Dufeil, Mariella Baccaro, Micaela Ruef, Natacha Pougnier, Sami Uslu, Sara Bekiri, Sara Moreira, Sophie Margot, Thomas Demaurex, and Vanesa Redzepi) for their help in the recruitment of patients, nurses and staff of the Pediatric Emergency Department of our institution for their work, as well as participants for their willingness to participate in the study. We also thank Sabine Yerly and Aline Mamin for performing the calibration of the WHO international standard for SARS-CoV-2 RNA.
The study was supported by the Geneva Centre for Emerging Viral Diseases. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Arnaud G. L’Huillier, Email: arnaud.lhuillier@hcuge.ch.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masia M, Fernandez-Gonzalez M, Sanchez M, Carvajal M, Garcia JA, Gonzalo-Jimenez N. 2021. Nasopharyngeal Panbio COVID-19 antigen performed at point-of-care has a high sensitivity in symptomatic and asymptomatic patients with higher risk for transmission and older age. Open Forum Infect Dis 8:ofab059. 10.1093/ofid/ofab059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drevinek P, Hurych J, Kepka Z, Briksi A, Kulich M, Zajac M. 2020. The sensitivity of SARS-CoV-2 antigen tests in the view of large-scale testing. MedRxiv 10.1101/2020.11.23.20237198. [DOI] [PubMed] [Google Scholar]
- 4.Linares M, Perez-Tanoira R, Carrero A, Romanyk J, Perez-Garcia F, Gomez-Herruz P, Arroyo T, Cuadros J. 2020. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol 133:104659. 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albert E, Torres I, Bueno F, Huntley D, Molla E, Fernandez-Fuentes MA, Martinez M, Poujois S, Forque L, Valdivia A, Solano de la Asuncion C, Ferrer J, Colomina J, Navarro D. 2021. Field evaluation of a rapid antigen test (Panbio COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect 27:472. 10.1016/j.cmi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger A, Nsoga MTN, Perez-Rodriguez FJ, Aad YA, Sattonnet-Roche P, Gayet-Ageron A, Jaksic C, Torriani G, Boehm E, Kronig I, Sacks JA, de Vos M, Bausch FJ, Chappuis F, Renzoni A, Kaiser L, Schibler M, Eckerle I. 2021. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS One 16:e0248921. 10.1371/journal.pone.0248921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alemany A, Baro B, Ouchi D, Rodo P, Ubals M, Corbacho-Monne M, Vergara-Alert J, Rodon J, Segales J, Esteban C, Fernandez G, Ruiz L, Bassat Q, Clotet B, Ara J, Vall-Mayans M, C GB, Blanco I, Mitja O. 2021. Analytical and clinical performance of the panbio COVID-19 antigen-detecting rapid diagnostic test. J Infect 82:186–230. 10.1016/j.jinf.2020.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Donapetry P, Garcia-Clemente P, Bloise I, Garcia-Sanchez C, Sanchez Castellano MA, Romero MP, Gutierrez Arroyo A, Mingorance J, de Ceano-Vivas La Calle M, Garcia-Rodriguez J, SARS-CoV-2 Working Group . 2021. Think of the children: evaluation of SARS-CoV-2 rapid antigen test in pediatric population. Pediatr Infect Dis J 40:385–388. 10.1097/INF.0000000000003101. [DOI] [PubMed] [Google Scholar]
- 9.Villaverde S, Dominguez-Rodriguez S, Sabrido G, Perez-Jorge C, Plata M, Romero MP, Grasa CD, Jimenez AB, Heras E, Broncano A, Nunez MDM, Illan M, Merino P, Soto B, Molina-Arana D, Bermejo A, Mendoza P, Gijon M, Perez-Moneo B, Moraleda C, Tagarro A, Epidemiological Study of COVID-19 in Children of the Spanish Society of Pediatric (EPICO-AEP) Working Group . 2021. Diagnostic accuracy of the Panbio severe acute respiratory syndrome coronavirus 2 antigen rapid test compared with reverse-transcriptase polymerase chain reaction testing of nasopharyngeal samples in the pediatric population. J Pediatr 232:287–289. 10.1016/j.jpeds.2021.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swiss Federal Office of Public Health. SARS-CoV-2 epidemiological monitoring. https://www.covid19.admin.ch/fr/epidemiologic/case?detTime=phase2b&detSum=14d_sum#showDetail2021. Accessed 13 July 2021.
- 11.Bentley E, Mee ET, Routley S, Mate R, Fritzsche M, Hurley M, Le Duff Y, Anderson R, Hockley J, Rigsby P, Page M, Rose N, Mattiuzzo G, Collaborative Study Group . 2020. Collaborative study for the establishment of a WHO international standard for SARS-CoV-2 RNA. Expert Committee on Biological Standardization, World Health Organization, Geneva, Switzerland. [Google Scholar]
- 12.Word Health Organization. COVID-19 target product profiles for priority diagnostics to support response to the COVID-19 pandemic. https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1. Accessed 8 April 2021.
- 13.Heald-Sargent T, Muller WJ, Zheng X, Rippe J, Patel AB, Kociolek LK. 2020. Age-related differences in nasopharyngeal severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) levels in patients with mild to moderate coronavirus disease 2019 (COVID-19). JAMA Pediatr 174:902–903. 10.1001/jamapediatrics.2020.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baggio S, L’Huillier AG, Yerly S, Bellon M, Wagner N, Rohr M, Huttner A, Blanchard-Rohner G, Loevy N, Kaiser L, Jacquerioz F, Eckerle I. 2020. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral load in the upper respiratory tract of children and adults with early acute coronavirus disease 2019 (COVID-19). Clin Infect Dis 73:148–150. 10.1093/cid/ciaa1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yonker LM, Neilan AM, Bartsch Y, Patel AB, Regan J, Arya P, Gootkind E, Park G, Hardcastle M, St John A, Appleman L, Chiu ML, Fialkowski A, De la Flor D, Lima R, Bordt EA, Yockey LJ, D'Avino P, Fischinger S, Shui JE, Lerou PH, Bonventre JV, Yu XG, Ryan ET, Bassett IV, Irimia D, Edlow AG, Alter G, Li JZ, Fasano A. 2020. Pediatric severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): clinical presentation, infectivity, and immune responses. J Pediatr 227:45–52. 10.1016/j.jpeds.2020.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brunink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 17.Euser S, Aronson S, Manders I, Van Lelyveld S, Herpers B, Sinnige J, et al. 2021. SARS-CoV-2 viral load distribution reveals that viral loads increase with age: a retrospective cross-sectional cohort study. medRxiv 10.1101/2021.01.15.21249691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellon M, Baggio S, Bausch FJ, Spechbach H, Salamun J, Genecand C, Tardin A, Kaiser L, L'Huillier AG, Eckerle I. 2021. SARS-CoV-2 viral load kinetics in symptomatic children, adolescents and adults. Clin Infect Dis ciab396. 10.1093/cid/ciab396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones TC, Biele G, Muhlemann B, Veith T, Schneider J, Beheim-Schwarzbach J, Bleicker T, Tesch J, Schmidt ML, Sander LE, Kurth F, Menzel P, Schwarzer R, Zuchowski M, Hofmann J, Krumbholz A, Stein A, Edelmann A, Corman VM, Drosten C. 2021. Estimating infectiousness throughout SARS-CoV-2 infection course. Science eabi5273. 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bekliz M, Adea K, Essaidi-Laziosi M, Sacks JA, Escadafal C, Kaiser L, Eckerle I. 2021. Analytical comparison of nine SARS-CoV-2 antigen-detecting rapid diagnostic tests for emerging SARS-CoV-2 variants. medRxiv 10.1101/2021.05.31.21258111. [DOI] [Google Scholar]
- 21.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, Guo Q, Song T, He J, Yen HL, Peiris M, Wu J. 2020. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 382:1177–1179. 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh KA, Jordan K, Clyne B, Rohde D, Drummond L, Byrne P, Ahern S, Carty PG, O'Brien KK, O'Murchu E, O'Neill M, Smith SM, Ryan M, Harrington P. 2020. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect 81:357–371. 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corman VM, Haage VC, Bleicker T, Schmidt ML, Muhlemann B, Zuchowski M, Jo WK, Tscheak P, Moncke-Buchner E, Muller MA, Krumbholz A, Drexler JF, Drosten C. 2021. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study. Lancet Microbe 2:e311–e319. 10.1016/S2666-5247(21)00056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calame A, Mazza L, Renzoni A, Kaiser L, Schibler M. 2021. Sensitivity of nasopharyngeal, oropharyngeal, and nasal wash specimens for SARS-CoV-2 detection in the setting of sampling device shortage. Eur J Clin Microbiol Infect Dis 40:441–445. 10.1007/s10096-020-04039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JH, Yip CC, Poon RW, Chan KH, Cheng VC, Hung IF, Chan JF, Yuen KY, To KK. 2020. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg Microbes Infect 9:1356–1359. 10.1080/22221751.2020.1775133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber M, Schreiber PW, Scheier T, Audigé A, Buonomano R, Rudiger A, et al. 2021. High efficacy of saliva in detecting SARS-CoV-2 by RT-PCR in adults and children. medRxiv 10.1101/2020.12.01.20241778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fougère Y, Schwob JM, Miauton A, Hoegger F, Opota O, Jaton K, et al. 2021. Performance of RT-PCR on saliva specimens compared to nasopharyngeal swabs for the detection of SARS-CoV-2 in children: a prospective comparative clinical trial. medRxiv 10.1101/2021.02.27.21252571. [DOI] [PubMed] [Google Scholar]
- 28.L'Huillier AG, Torriani G, Pigny F, Kaiser L, Eckerle I. 2020. Culture-competent SARS-CoV-2 in nasopharynx of symptomatic neonates, children, and adolescents. Emerg Infect Dis 26:2494–2497. 10.3201/eid2610.202403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, Hay JA, Tambe M, Mina MJ, Parker R. 2021. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv 7:eabd5393. 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and Tables S1 to S3. Download JCM.00991-21-s0001.pdf, PDF file, 0.07 MB (69.2KB, pdf)
Data Availability Statement
Study protocols, statistical analysis plan, and individual participant data that underlie the results reported in this article can be made available (after deidentification) to researchers who make a methodologically sound proposal. Proposals should be directed to arnaud.lhuillier@hcuge.ch. To gain access, data requestors will need to sign a data access agreement.