Abstract
Background:
Although pathogenic 22q11.2 deletions are an important cause of developmental delays and lifelong disease burden, their variable and complex clinical expression contributes to under-recognition, delayed molecular diagnosis and uncertainty about prevalence. We sought to estimate the contemporary live-birth prevalence of typical 22q11.2 deletions using a population-based newborn screening sample and to examine data available for associated clinical features.
Methods:
Using DNA available from an unbiased sample of about 12% of all dried blood spots collected for newborn screening in Ontario between January 2017 and September 2018, we prospectively screened for 22q11.2 deletions using multiplex quantitative polymerase chain reaction assays and conducted independent confirmatory studies. We used cross-sectional analyses to compare available clinical and T-cell receptor excision circle (TREC, used in newborn screening for severe combined immunodeficiency) data between samples with and without 22q11.2 deletions.
Results:
The estimated minimum prevalence of 22q11.2 deletions was 1 in 2148 (4.7 per 10 000) live births (95% confidence interval [CI] 2.5 to 7.8 per 10 000), based on a total of 30 074 samples screened, with 14 having confirmed 22q11.2 deletions. Of term singletons, samples with 22q11.2 deletions had significantly younger median maternal age (25.5 v. 32.0 yr, difference −6.5 yr, 95% CI −7 to −2 yr), a greater proportion with small birth weight for gestational age (odds ratio 7.00, 95% CI 2.36 to 23.18) and lower median TREC levels (108.9 v. 602.5 copies/3 μL, p < 0.001).
Interpretation:
These results indicate that the 22q11.2 deletion syndrome is one of the most common of rare genetic conditions and may be associated with relatively younger maternal ages and with prenatal growth abnormalities. The findings support the public health importance of early — prenatal and neonatal — diagnosis that would enable prompt screening for and management of well-known actionable features associated with 22q11.2 deletions.
The 22q11.2 deletion syndrome (22q11.2DS; Online Mendelian Inheritance in Man [OMIM] 188400/192430), previously called DiGeorge or velocardiofacial syndrome, is an important genetic condition associated with recurrent 22q11.2 microdeletions and highly penetrant expression.1 Features include developmental delay, intellectual disability, congenital cardiac or palatal anomalies (or both), pediatric immunodeficiency, and treatable endocrinologic and neuropsychiatric conditions. Variable presentation, often without major anatomic anomalies, contributes to clinical under-recognition and diagnostic delay, often with many years before molecular diagnosis.1,2
We are unaware of any contemporary population-based live-birth prevalence estimates for 22q11.2 deletions based on newborn screening data. Prevalence estimates vary widely, most commonly reported as 1.7 to 3.3 per 10 000 live births.1 Dating back to 1996,3 previous estimates have used multiple strategies, including ascertainment from birth defects registries,3,4 infants with congenital cardiac disease4,5 or clinically indicated genetic testing results.6,7 One study used 25 704 newborn screening samples selected from individuals born between 1981 and 2005 to retrospectively identify 22q11.2 deletions, but that study excluded neonatal and early infant deaths.8 Newborn screening programs using T-cell receptor excision circles (TRECs) for identification of severe combined immunodeficiency can detect some individuals with 22q11.2DS (those with neonatal immunodeficiency9–12); however, phenotypically based methods are unlikely to be sufficient for population-wide detection of pathogenic deletions with such variable expression.10,11
Given the morbidity and mortality associated with 22q11.2DS, which extend throughout the lifespan,1,13–16 it has been proposed that 22q11.2 deletions be added to newborn screening panels, a plan endorsed by families of affected individuals. 14 In addition to newborn screening considerations, estimates from prenatal studies and technologic advances in prenatal screening for 22q11.2 deletions (e.g., noninvasive prenatal testing) have increased the urgency of determining the current live-birth prevalence.15,16 We sought to estimate the minimum live-birth prevalence of typical 22q11.2 deletions using contemporary population-based newborn screening data. We also examined available clinical data, including TREC results.
Methods
Study design and setting
We employed a cross-sectional study design to estimate the prevalence of 22q11.2 deletions by systematically screening for the most common pathogenic 22q11.2 deletions (Figure 1)1 within a subset of prospectively collected Ontario newborn screening samples. We also aimed to compare newborn screening results for TREC and clinical variables between those with and without 22q11.2 deletions.
Figure 1:
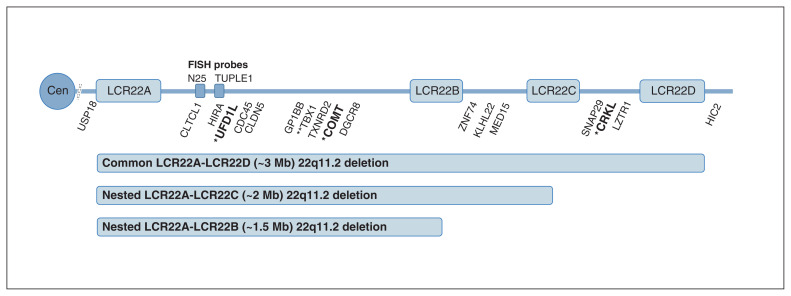
Illustration of the common 22q11.2 deletion (about 3 megabases [Mb]) and the rarer proximal nested 22q11.2 deletions (about 2 Mb and about 1.5 Mb), as well as the approximate positions of genes for probes used to detect these deletions: 3 primary screening quantitative real-time polymerase chain reaction (qPCR) probes (UFD1L, COMT, CRKL, bold font, single asterisk), 15 genes for confirmatory multiplex ligation-dependent probe amplification studies (16 probes, including 2 at TBX1, and flanking probes at USP18 and HIC2), and 1 secondary screening qPCR probe (TBX1, 2 asterisks). Also shown are the relative positions of the low copy repeat (LCR) sequences (segmental duplications) that predispose this complex genomic region to de novo 22q11.2 deletion events at gametogenesis, and probes (N25 and TUPLE1) commonly used for targeted fluorescence in situ hybridization (FISH) studies that cannot determine the length of deletions. Clinical genome-wide microarray, the current standard for detecting pathogenic copy number variation,1,17 provides information on deletion length and extent. Note: Cen = centromere.
We used anonymized dried blood spot samples collected by Newborn Screening Ontario between January 2017 and September 2018.1 The total number of samples studied was determined by power analysis for this rare disease and by funding availability (Appendix 1, Supplemental methods, available at www.cmajopen.ca/content/9/3/E802/suppl/DC1).
Data sources
We analyzed neonatal dried blood spot–derived samples collected as part of Newborn Screening Ontario’s newborn screening program, as per the organization’s policy on storage and secondary use of samples. Residual DNA from the TREC quantitative real-time polymerase chain reaction (qPCR) assay, a part of Ontario’s newborn screening program for severe combined immunodeficiency,18 was saved for use in this study and was available for all collected samples (i.e., there were no exclusions). Clinical data for the neonates screened were entered at the point of care.
Laboratory investigations and clinical variables
The primary qPCR screening assay for 22q11.2 deletions comprised primers and probes for three 22q11.2 deletion region genes (Figure 1): UFD1L and COMT (located in the low copy repeat LCR22A–LCR22B region) and CRKL (located in the LCR22C–LCR22D region), with RPPH1 used as a reference gene for appropriate DNA extraction and relative quantification (Appendix 1, Supplemental methods, Table S1 and Table S2). For each probe, the relative quantification value was calculated and a cut-off value defined using the area under the receiver operating characteristic curve (Appendix 1, Supplemental methods, Table S3). A screen-positive sample was defined as having a putative deletion of all three 22q11.2 region probes (i.e., a suspected LCR22A–LCR22D deletion) or deletion of both the UFD1L and COMT probes but not the CRKL probe (i.e., a suspected LCR22A–LCR22B or LCR22A–LCR22C deletion; see Figure 1).1 Screen-positive samples with sufficient DNA for an additional dried blood spot punch were then subjected to a secondary qPCR screening assay using the same reference probe but a different 22q11.2 probe (TBX1; Appendix 1, Supplemental methods, Table S3).
Initially, the primary screen-positive samples that also had a TBX1 relative quantification value below (or near) an established cut-off (Appendix 1, Supplemental methods, Table S3)19 were prioritized for standard multiplex ligation-dependent probe amplification (MLPA) assays (MRC Holland, Amsterdam, Netherlands); 6 screen-negative samples served as controls for MLPA normalization (Appendix 1, Supplemental methods). Subsequently, the remainder of the screen-positive samples were subjected to MLPA confirmatory testing (Appendix 1, Supplemental methods). Samples were deemed to have a confirmed 22q11.2 deletion if they screened positive on the initial 3-probe qPCR assay and if MLPA determined the presence of a common or proximal nested 22q11.2 deletion (Figure 1). Methodologic details, including DNA extraction and creation of dried blood spot quality control material for all qPCR and MLPA assays, are provided in Appendix 1, Supplemental methods.
Available clinical variables were confined to maternal age, newborn sex, birth weight, gestational age, neonatal transfusion status and neonatal feeding type. Sample size varied for each variable because of missing data. We inspected the data for outliers and excluded any data points that appeared to be possible data entry errors.
Statistical analysis
We calculated a live-birth minimum prevalence estimate of the 22q11.2 deletion by dividing the number of newborn screening samples with MLPA-confirmed 22q11.2 deletion by the total number of newborn samples assessed. We calculated 95% confidence intervals (CIs) for prevalence using the binomial distribution. For the subgroup of singleton newborns born at term (defined as ≥ 37 wk and < 42 wk gestational age20), we compared TREC values and other available clinical variables between those with a confirmed 22q11.2 deletion and the remaining population-based sample using the χ2 or Fisher exact test for categorical variables and the Mann–Whitney U test for non-normally distributed continuous data. For the clinical variables, we analyzed only data for the singleton term newborns because multiple-gestation and preterm births are likely to affect the variables studied, such as TREC levels.12 We also calculated 95% CIs for differences between medians, and we calculated odds ratios with 95% CIs for proportions.21
We performed statistical analyses using SAS software, version 9.4 (SAS Institute) and R statistical software (version 4.0.2). We defined statistical significance as p < 0.05, 2-tailed.
Ethics approval
This study was approved by the Children’s Hospital of Eastern Ontario Research Ethics Board and the Centre for Addiction and Mental Health Research Ethics Board. In accordance with guidelines from these bodies regarding storage and secondary use of dried blood spot samples that are designed to ensure that the samples remain unidentifiable, minimal clinical data for the newborns were available.
Results
A total of 30 074 anonymized dried blood spot samples collected by Newborn Screening Ontario were available for study, corresponding to 12% of all infants born in Ontario, Canada, during the 21-month period of data collection. Because of intermittent sampling, nearly all of these samples (30 017/30 074 [> 99%]) were from infants born in 11 of the 21 months. For the overall sample, available clinical data showed that 14 662 (49%) were known females, 29 087 (97%) were known singletons, and 987 (3%) were known multiple births. The infants were born at mean gestational age 39 (standard deviation 2) weeks, with a total of 2595 (9%) being preterm. For the subsample of 29 087 singletons, 2002 were preterm births, 64 were post-term births, and 573 had no gestational age data available; thus, there were a total of 26 448 singleton term births.
Estimated live-birth prevalence of the 22q11.2 deletion
Screening and confirmatory assay results provided a minimum estimate of live-birth prevalence of the pathogenic 22q11.2 deletion of 1 in 2148 (4.7 per 10 000, 95% CI 2.5 to 7.8 per 10 000), based on 30 074 Ontario newborn screening samples. There were 14 samples that had screen-positive results on the primary qPCR assay and that had MLPA confirmation (Appendix 1, Supplemental results): 11 (79%) with the common LCR22A–LCR22D 22q11.2 deletion and 3 (21%) with proximal nested deletions (2 LCR22A–LCR22B and 1 LCR22A–LCR22C) (Figure 1).
Clinical variables
All 14 newborn screening samples with a 22q11.2 deletion were singleton births. There were 13 term births and 1 preterm birth. Of the total 26 448 singleton term births, we compared the 13 with a 22q11.2 deletion to the remaining population sample who had available data for sex, birth weight and gestational age (n = 26 305). Table 1 summarizes the results (see footnotes for details of sample sizes for each variable).
Table 1:
Demographic and clinical characteristics for the subset of term singleton newborns studied,* comparing those having a 22q11.2 deletion with the remainder of the population sampled
| Demographic or clinical variable | Newborn group;* no. (%) of newborns† | OR (95% CI)‡ | |
|---|---|---|---|
| With 22q11.2 deletion (maximum n = 13) | Remaining population (maximum n = 26 305) | ||
| Sex, male | 4 (31) | 13 467 (51) | 0.42 (0.12 to 1.35) |
| Birth weight for gestational age§ | |||
| < 10th percentile | 6 (46) | 2869 (11) | 7.00 (2.36 to 23.18) |
| < 3rd percentile¶ | 2 (15) | 819 (3) | 5.66 (0.90 to 23.80) |
| Neonatal transfusion** | 0 (0) | 12 (0.06) | 0.00 (0.00 to 834.95) |
| Complex neonatal feeding**†† | 2 (18) | 183 (0.8) | 29.41 (4.53 to 134.65) |
| Gestational age, wk, median (IQR) | 39.0 (38.0 to 39.6) | 39.3 (38.5 to 40.2) | −0.3 (−1.1 to 0.2)‡‡ |
| Maternal age,** yr, median (IQR) | 25.5 (24.0 to 29.5) | 32.0 (28.0 to 35.0) | −6.5 (−7 to −2)‡‡ |
Note: CI = confidence interval, IQR = interquartile range, OR = odds ratio.
Only singletons born at term with available data for sex, birth weight and gestational age are included. “Term” was defined as 37 weeks ≤ gestational age < 42 weeks; all multiple births were excluded. Some data were missing for other variables (see details below). Also, for 1 individual of the 13 in the 22q11.2 deletion group and 109 of the 26 305 individuals in the remaining population group, the DNA was obtained before 24 hours of age, which might be considered a less-than-satisfactory sample, given that certain newborn screening tests may be less sensitive with DNA sampled in this period.
Except where indicated otherwise (i.e., for the continuous variables gestational age and maternal age).
Except where indicated otherwise (i.e., for the continuous variables gestational age and maternal age), the entries in this column are ORs quantifying the association between each variable of interest and the 22q11.2 deletion, with 95% CI. For ease of interpretation, the following variables were statistically significant with a p value < 0.05: birth weight for gestational age < 10th percentile, complex neonatal feeding and maternal age.
Percentiles were calculated based on a Canadian reference set of all singletons born in Canada between 1994 and 1996 (with the exception of Ontario).22
Among female infants, the proportion with birth weight for gestational age below the third percentile was significantly greater in the 22q11.2 deletion group (n = 2/9 [22%] v. n = 401/12 838 [3%], p = 0.03).
Some data were missing for each of the following variables: neonatal transfusion data were available for n = 11 from the 22q11.2 deletion group and n = 21 699 from the remaining population; neonatal feeding data were available for n = 11 from the 22q11.2 deletion group and n = 24 414 from the remaining population; maternal age data were available for n = 12 from the 22q11.2 deletion group and n = 25 999 from the population group (the latter excluding 1 individual with maternal age recorded as > 60 yr, presumed to be a data entry error).
“Complex neonatal feeding” refers to neonates who required total parenteral nutrition and those who were designated to receive nothing by mouth (“nil per os” or NPO) as either their sole method of feeding or in combination with another feeding type. The 2 individuals with 22q11.2 deletion and a complex feeding type were both recorded as NPO at birth.
Entry shown is the difference between medians (22q11.2 deletion group minus population group), with 95% CI.
Median maternal age (with interquartile range [IQR]) was significantly younger for those with a 22q11.2 deletion (25.5 [IQR 24.0–29.5] yr v. 32.0 [IQR 28.0–35.0] yr; difference −6.5 yr, 95% CI −7 to −2 yr; Table 1). Those with a 22q11.2 deletion also had a higher prevalence of low (< 10th percentile) birth weight for gestational age (odds ratio [OR] 7.00, 95% CI 2.36 to 23.18; Table 1). A complex neonatal feeding type was more likely in the 22q11.2 deletion subgroup (OR 29.41, 95% CI 4.53 to 134.65; Table 1). There were no significant differences for the other variables examined (Table 1).
TREC values
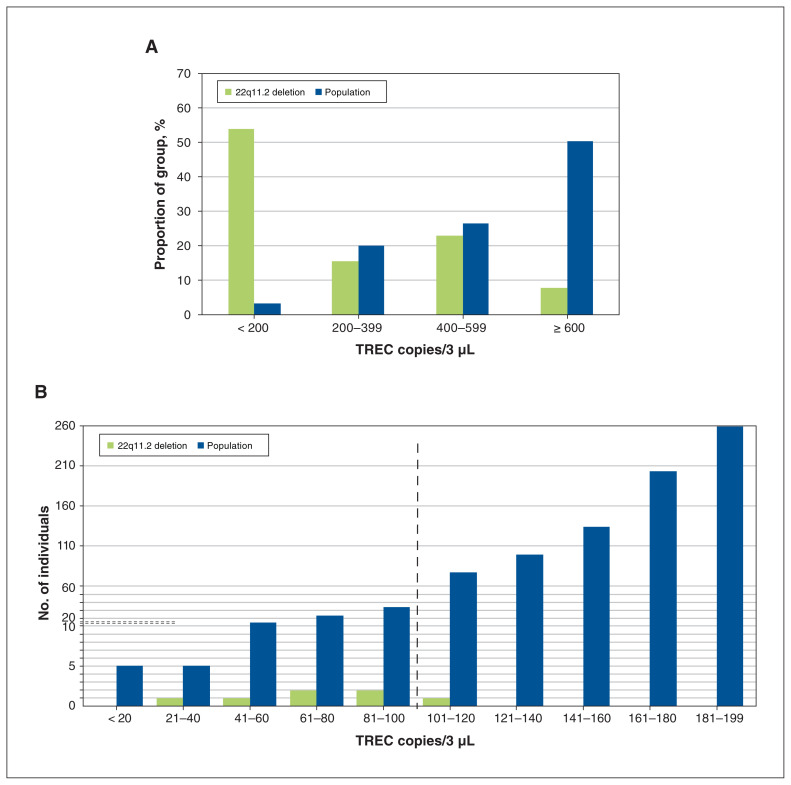
Term singleton samples with a 22q11.2 deletion had significantly lower median TREC values than the remaining population-based samples (108.9 v. 602.5 copies/3 μL, p < 0.001). The proportion with fewer than 200 TREC copies/3 μL was greater in the 22q11.2 deletion subgroup (Figure 2A). Six (46%) of this 22q11.2 deletion subgroup met the initial clinical newborn screening cut-off value for severe combined immunodeficiency of 100 or fewer TREC copies/3 μL (Figure 2B), compared with 81 (0.3%) of the population-based samples (p < 0.001).
Figure 2:
(A) Severe combined immunodeficiency screening results showing distribution of T-cell receptor excision circle (TREC) copies/3 μL for term singleton newborn screening samples, by 22q11.2 deletion status. For the term singleton subsample, the majority (7/13, 54%) of the 22q11.2 deletion group had fewer than 200 TREC copies/3 μL (about the 3rd percentile), whereas the greatest proportion of the remaining population-based group (13 297 [50%] of 26 435) had at least 600 TREC copies/3 μL. (B) Subset of term singleton newborn screening samples with lowest TREC values (< 200 copies/3 μL). Shown here are detailed distribution results for samples from the term singleton subsample. Overall, there were 859 with fewer than 200 TREC copies/3 μL, 7 with confirmed 22q11.2 deletion and 852 from the remaining population-based group. The dashed horizontal lines indicate where the scale changes for fine gradations: below 10, each mark on the y axis indicates 1 individual, and above 10, each mark on the y axis indicates 10 individuals. Six (46%) of the overall 13 term singleton samples with a confirmed 22q11.2 deletion had no more than 100 TREC copies/3 μL (to the left of the vertical dashed line), compared with 81 (0.3%) of the remaining population-based group (n = 26 435, p < 0.001). Currently, 100 TREC copies/3 μL is the cut-off in Ontario to undergo a secondary, more accurate TREC assay for final reporting of severe combined immunodeficiency identified by newborn screening.
For newborn screening in Ontario, all samples meeting this initial cut-off proceed to a second, confirmatory TREC assay, run in duplicate, with a cut-off of no more than 75 copies/3 μL. Of the total 87 samples proceeding to this secondary assay, 11 met this clinical cut-off: 1 (8%) from the 22q11.2 deletion group and 10 (0.04%) from the remaining population-based group (p = 0.005).
Interpretation
The estimated minimum prevalence of the 22q11.2 deletion in the Ontario newborn screening sample that we studied was 1 in 2148 (4.7 per 10 000), with the majority of confirmed deletions spanning the full LCR22A–LCR22D region. Among singletons born at term, those with a 22q11.2 deletion had significantly younger maternal age, lower TREC levels and a higher proportion with small birth weight for gestational age compared with the remaining population sample.
Extrapolating our results to the approximately 140 000 live births annually in Ontario,23 we could expect about 66 births with a 22q11.2 deletion each year. For context, comparable prevalence estimates using data from Canada and the United States for other genetic disorders (e.g., cystic fibrosis) are provided in Table 2; of these, only Down syndrome is more common than 22q11.2DS.18,24,25
Table 2:
Live-birth prevalence of familiar chromosomal abnormalities and selected conditions included in newborn screening programs
| Condition | Population-based live-birth prevalence | |
|---|---|---|
| Estimate | Per 10 000 | |
| Familiar chromosomal abnormalities | ||
| Trisomy 21* | 1 in 750 | 13.33 |
| Trisomy 18† | 1 in 5000 | 2.00 |
| Trisomy 13† | 1 in 16 000 | 0.63 |
| Selected conditions included in newborn screening‡ | ||
| Congenital hypothyroidism | 1 in 3000 | 3.33 |
| Cystic fibrosis | 1 in 3600 | 2.78 |
| Phenylketonuria | 1 in 12 000 | 0.83 |
| SCID | 1 in 50 000 to 1 in 100 000 | 0.1 to 0.2 |
The prevalence of 22q11.2 deletions estimated in this study is higher than previous prevalence estimates based on different sampling methods, but remains in line with the description of 22q11.2DS as a rare disease (defined as < 1 in 2000 or < 5 per 10 000).1 22q11.2DS has historically presented a substantial diagnostic challenge for clinicians, with clinical diagnosis based on obvious congenital anomalies that do not predict the intellectual or neuropsychiatric outcomes of most concern to parents.1,2 Most affected newborns would be expected to have unaffected parents.1 However, improved pediatric care over many decades, as well as the limited effects of a 22q11.2 deletion on reproductive fitness when major neuropsychiatric phenotypes are absent, could lead to increasing numbers of affected parents, many of whom would be expected to be undiagnosed.26
In contrast to previous studies providing prevalence estimates for 22q11.2DS,3–8,27 the main strength of this study was the use of an unselected contemporary newborn screening sample that was uniformly assessed using a standard method (multiplex qPCR), which is often used for newborn screening.
Several previous studies used clinically ascertained samples with the 22q11.2 deletion3–7 and may have underestimated the prevalence of 22q11.2DS, given its variable clinical phenotype, which may not include typically associated congenital anomalies.1 Only 2 previous population-based studies based their prevalence estimates solely on molecular genetic data.8,27 One was a Danish study restricted to residents 1 year of age or older, which retrospectively examined dried blood spots banked over a 24.5-year period to 2005;8 that study could not account for infant mortality.13 The other was a Norwegian study restricted to newborns that required both parents to consent to participation in a genetic research study.27 The 22q11.2 deletions identified in these 2 studies represent bookends of lowest (n = 1 in 12 252, or 0.82 per 10 000)27 and highest (n = 7 in 25 704, or 2.7 per 10 000)8 estimated live-birth prevalence before the current study.
In the current study, as expected, the majority of 22q11.2 deletions spanned the full LCR22A–LCR22D region (Figure 1).1 However, rarer proximal nested 22q11.2 deletions comprised 3 (21%) of the 14 confirmed 22q11.2 deletions, a higher prevalence than reported in large clinically ascertained samples.1,28 This raises the possibility that nested 22q11.2 deletions may have a somewhat lower penetrance for typical anatomic features leading to clinical detection2 compared with the common LCR22A–LCR22D deletion.28
In contrast to more familiar chromosomal abnormalities, such as trisomy 21, the limited clinical data available for the samples in the current study show that pathogenic 22q11.2 deletions may be associated with earlier, not late, maternal age.29 Also, we found that the 22q11.2 deletion may be associated with mild abnormalities of fetal growth, consistent with results from a retrospective study of adults with 22q11.2DS.30
Notably, the TREC results, while consistent with previous TREC-based newborn screening studies showing a higher prevalence of the 22q11.2 deletion with low values,9,10 indicated that only a minority of all confirmed 22q11.2 deletion samples would be detected using a newborn screening strategy based on severe combined immunodeficiency. This finding was foreshadowed by the results of a previous retrospective study11 and provides further support for developing genetically based newborn screening for 22q11.2 deletions. For any such proposal, screening costs per newborn must be low (e.g., < US$7 per sample31), methods must be scalable within current clinical newborn screening laboratories, and feasible plans for confirmatory studies and clinical referral for infants with confirmed positive screening results are needed.32
Our findings set the stage for future prospective studies to further refine prevalence estimates of high-impact 22q11.2 deletions, including the rarer proximal nested deletions. Large, multicentre newborn screening studies involving diverse jurisdictions could allow determination of factors that may affect 22q11.2DS prevalence, such as ethnicity and cultural factors, availability of prenatal screening33 and reproductive technologies. If ethics approval could be obtained, assessment of phenotypic prenatal and postnatal data, details about newborns receiving an early clinical diagnosis of 22q11.2DS, determination of inherited and de novo 22q11.2 deletion status and ability to provide parents of newborns with the 22q11.2DS diagnosis after clinical laboratory confirmation would offer substantially improved understanding of this important condition. A recent study of prospective mothers and previous reports would support such a study design.14,33
Limitations
The main limitation of this and other 22q11.2DS prevalence studies is the sample size. Larger, comparably ascertained samples are needed to refine live-birth prevalence estimates and improve knowledge about associated clinical features. Nonetheless, the results complement previous estimates using other designs (e.g., those based on congenital physical features and clinical recognition3–7) and add to studies showing high prenatal prevalence of the 22q11.2 deletion15 and strong association with fetal loss (stillbirths and miscarriages).34,35
In addition, because of restrictions related to de-identified dried blood spot samples set by the research ethics boards, minimal clinical data were available, which prevented us from learning about factors that could affect the prevalence of the 22q11.2 deletion or about the outcomes of these newborns, including whether and when any received a clinical diagnosis of 22q11.2DS.
This study used unbiased but intermittent sampling with a non-uniform distribution over a 21-month period and did not include confirmatory assays for all 30 074 samples. We did not have the ability to calculate true positive and true negative rates or to evaluate the specific qPCR-based assays used, although we note that qPCR is already a standard method used in existing newborn screening programs. MLPA is also used in some clinical laboratories and has a sensitivity and specificity of 99% and 97%, respectively, for the detection of 22q11.2 deletions.36
The estimate of live-birth prevalence determined in this study is referred to as a “minimum” prevalence because it could not be less but could possibly be higher. However, the results for the samples with screening results in the range of the screen-positive samples but where MLPA confirmed absence of the 22q11.2 deletion suggest that this would be unlikely. A well-designed and adequately powered study of newborn screening methods for 22q11.2 deletions is needed.
Conclusion
The results of this study provide a contemporary live-birth prevalence estimate for pathogenic 22q11.2 deletions that indicates 22q11.2DS to be one of the most common of rare genetic conditions. The clinical findings of this study, including increased prevalence of small-for-gestational-age infants with 22q11.2 deletions and young maternal age, together with what is well known about the 22q11.2 deletion, support the public health importance of early (i.e., prenatal and neonatal) diagnosis. Early diagnosis would enable prompt screening, detection and treatment initiation for actionable features associated with 22q11.2 deletions.
Appendix
Acknowledgements
The authors thank personnel of the Newborn Screening Ontario program at the Children’s Hospital of Eastern Ontario for sample collection and laboratory investigations, as well as personnel affiliated with the Clinical Genetics Research Program at the Centre for Addiction and Mental Health (CAMH) and the Dalglish Family 22q Clinic at the Toronto General Hospital for their assistance with data checking and clinical context. The authors thank Marcos Sanches, biostatistician at CAMH, for assistance with the statistical analyses. They also thank individuals involved in the original conceptualization and probe testing, as well as colleagues, patients and family members whose initiative and motivation have inspired efforts to develop newborn screening for 22q11.2 deletion syndrome. The authors also acknowledge those who attended and supported the initiative to introduce newborn screening for 22q11.2 deletion syndrome to the Secretary’s Advisory Committee on Heritable Disorders in Newborns and Children in Washington, DC, in 2012.
Footnotes
Competing interests: None declared.
This article has been peer reviewed.
Contributors: Anne Bassett, Dennis Bulman and Aoy Tomita-Mitchell were involved in conceptualization and design of the study. Mylene Theriault, Kristin Kernohan and Pranesh Chakraborty were involved in data acquisition. Mylene Theriault, Christina Blagojevic and Tracy Heung were involved in data analysis. Christina Blagojevic, Tracy Heung and Anne Bassett were involved in interpretation of the data. Christina Blagojevic drafted the manuscript, which was critically revised primarily by Anne Bassett with contributions from all other authors. All of the authors provided approval of the manuscript for publication and agreed to be accountable for the work.
Funding: This work was funded by a McLaughlin Centre Accelerator Grant (Anne Bassett, Dennis Bulman), with support from Canadian Institutes of Health Research grant PJT-148924 (Anne Bassett) and, for prior work relevant to probe development, a National Institute of Child Health and Human Development grant 1R21-HD-060309-01 and funding from the Audrey Hohenwalter VCFS Foundation (Aoy Tomita-Mitchell). Anne Bassett holds the Dalglish Chair in 22q11.2 Deletion Syndrome at the University Health Network and the University of Toronto.
Data sharing: Data sharing is precluded as per stipulations of the research ethics boards that approved the study.
Supplemental information: For reviewer comments and the original submission of this manuscript, please see www.cmajopen.ca/content/9/3/E802/suppl/DC1.
References
- 1.McDonald-McGinn DM, Sullivan KE, Marino B, et al. 22q11.2 deletion syndrome. Nat Rev Dis Primers. 2015;1:15071. doi: 10.1038/nrdp.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer LD, Butcher NJ, Boot E, et al. Elucidating the diagnostic odyssey of 22q11.2 deletion syndrome. Am J Med Genet A. 2018;176:936–44. doi: 10.1002/ajmg.a.38645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tézenas Du Montcel S, Mendizabai H, Aymé S, et al. Prevalence of 22q11 microdeletion. J Med Genet. 1996;33:719. doi: 10.1136/jmg.33.8.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botto LD, May K, Fernhoff PM, et al. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics. 2003;112:101–7. doi: 10.1542/peds.112.1.101. [DOI] [PubMed] [Google Scholar]
- 5.Goodship J, Cross I, LiLing J, et al. A population study of chromosome 22q11 deletions in infancy. Arch Dis Child. 1998;79:348–51. doi: 10.1136/adc.79.4.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devriendt K, Fryns JP, Mortier G, et al. The annual incidence of DiGeorge/velocardiofacial syndrome. J Med Genet. 1998;35:789–90. doi: 10.1136/jmg.35.9.789-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oskarsdóttir S, Vujic M, Fasth A. Incidence and prevalence of the 22q11 deletion syndrome: a population-based study in Western Sweden. Arch Dis Child. 2004;89:148–51. doi: 10.1136/adc.2003.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen L, Sparsø T, Weinsheimer SM, et al. Prevalence of rearrangements in the 22q11.2 region and population-based risk of neuropsychiatric and developmental disorders in a Danish population: a case-cohort study. Lancet Psychiatry. 2018;5:573–80. doi: 10.1016/S2215-0366(18)30168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin-Nalda A, Cueto-González AM, Argudo-Ramírez A, et al. Identification of 22q11.2 deletion syndrome via newborn screening for severe combined immunodeficiency. Two years’ experience in Catalonia (Spain) Mol Genet Genomic Med. 2019;7:e1016. doi: 10.1002/mgg3.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barry JC, Blaine Crowley T, Jyonouchi S, et al. Identification of 22q11.2 deletion syndrome via newborn screening for severe combined immunodeficiency. J Clin Immunol. 2017;37:476–85. doi: 10.1007/s10875-017-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingman Framme J, Borte S, von Döbeln U, et al. Retrospective analysis of TREC based newborn screening results and clinical phenotypes in infants with the 22q11 deletion syndrome. J Clin Immunol. 2014;34:514–9. doi: 10.1007/s10875-014-0002-y. [DOI] [PubMed] [Google Scholar]
- 12.Routes JM, Grossman WJ, Verbsky J, et al. Statewide newborn screening for severe T-cell lymphopenia. JAMA. 2009;302:2465–70. doi: 10.1001/jama.2009.1806. [DOI] [PubMed] [Google Scholar]
- 13.Repetto GM, Guzmán ML, Delgado I, et al. Case fatality rate and associated factors in patients with 22q11 microdeletion syndrome: a retrospective cohort study. BMJ Open. 2014;4:e005041. doi: 10.1136/bmjopen-2014-005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costain G, Chow EWC, Ray PN, et al. Caregiver and adult patient perspectives on the importance of a diagnosis of 22q11.2 deletion syndrome. J Intellect Disabil Res. 2012;56:641–51. doi: 10.1111/j.1365-2788.2011.01510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grati FR, Molina Gomes D, Ferreira JCPB, et al. Prevalence of recurrent pathogenic microdeletions and microduplications in over 9500 pregnancies. Prenat Diagn. 2015;35:801–9. doi: 10.1002/pd.4613. [DOI] [PubMed] [Google Scholar]
- 16.Grati FR, Gross SJ. Noninvasive screening by cell-free DNA for 22q11.2 deletion: benefits, limitations, and challenges. Prenat Diagn. 2019;39:70–80. doi: 10.1002/pd.5391. [DOI] [PubMed] [Google Scholar]
- 17.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newborn Screening Ontario. Newborn screening manual: a guide for newborn care providers. Edition 2.1. Ottawa: Children’s Hospital of Eastern Ontario; 2018. [Google Scholar]
- 19.Tomita-Mitchell A, Mahnke DK, Larson JM, et al. Multiplexed quantitative real-time PCR to detect 22q11.2 deletion in patients with congenital heart disease. Physiol Genomics. 2010;42A:52–60. doi: 10.1152/physiolgenomics.00073.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spong CY. Defining ‘term’ pregnancy: recommendations from the Defining ‘Term’ Pregnancy Workgroup. JAMA. 2013;309:2445–6. doi: 10.1001/jama.2013.6235. [DOI] [PubMed] [Google Scholar]
- 21.Fay MP. Confidence intervals that match Fisher’s exact or Blaker’s exact tests. Biostatistics. 2010;11:373–4. doi: 10.1093/biostatistics/kxp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer MS, Platt RW, Wen SW, et al. Fetal/Infant Health Study Group of the Canadian Perinatal Surveillance System. A new and improved population-based Canadian reference for birth weight for gestational age. Pediatrics. 2001;108:E35. doi: 10.1542/peds.108.2.e35. [DOI] [PubMed] [Google Scholar]
- 23.Table: 13-10-0414-01: Live births, by place of residence of mother. Ottawa: Statistics Canada; [accessed 2020 Oct. 24]. Available https://www150.statcan.gc.ca/t1/tbl1/en/tv.action?pid=1310041401. [Google Scholar]
- 24.Down syndrome surveillance in Canada 2005–2013. Ottawa: Public Health Agency of Canada; [accessed 2020 Oct. 24]. modified 2017 Mar 2. Available: https://www.canada.ca/en/public-health/services/publications/healthy-living/down-syndrome-surveillance-2005-2013.html. [Google Scholar]
- 25.Genetics [website] Bethesda (MD): US National Library of Medicine; [accessed 2020 Oct. 24]. Available https://medlineplus.gov/genetics/ [Google Scholar]
- 26.Costain G, Chow EWC, Silversides CK, et al. Sex differences in reproductive fitness contribute to preferential maternal transmission of 22q11.2 deletions. J Med Genet. 2011;48:819–24. doi: 10.1136/jmedgenet-2011-100440. [DOI] [PubMed] [Google Scholar]
- 27.Smajlagić D, Lavrichenko K, Berland S, et al. Population prevalence and inheritance pattern of recurrent CNVs associated with neurodevelopmental disorders in 12,252 newborns and their parents. Eur J Hum Genet. 2021;29:205–15. doi: 10.1038/s41431-020-00707-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo T, Diacou A, Nomaru H, et al. International Chromosome 22q11.2; International 22q11.2 Brain and Behavior Consortia. Deletion size analysis of 1680 22q11.2DS subjects identifies a new recombination hotspot on chromosome 22q11.2. Hum Mol Genet. 2018;27:1150–63. doi: 10.1093/hmg/ddy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delio M, Guo T, McDonald-McGinn DM, et al. Enhanced maternal origin of the 22q11.2 deletion in velocardiofacial and DiGeorge syndromes. Am J Hum Genet. 2013;92:439–47. doi: 10.1016/j.ajhg.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van L, Butcher NJ, Costain G, et al. Fetal growth and gestational factors as predictors of schizophrenia in 22q11.2 deletion syndrome. Genet Med. 2016;18:350–5. doi: 10.1038/gim.2015.84. [DOI] [PubMed] [Google Scholar]
- 31.Wells J, Rosenberg M, Hoffman G, et al. A decision-tree approach to cost comparison of newborn screening strategies for cystic fibrosis. Pediatrics. 2012;129:e339–47. doi: 10.1542/peds.2011-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson JM, Jungner G. Principles and practice of screening for disease [article in Spanish] Bol Oficina Sanit Panam. 1968;65:281–393. [PubMed] [Google Scholar]
- 33.Kagan KO, Hoopman M, Pfaff T, et al. First trimester screening for common trisomies and microdeletion 22q11.2 syndrome using cell-free DNA: a prospective clinical study. Fetal Diagn Ther. 2020;47:841–52. doi: 10.1159/000510069. [DOI] [PubMed] [Google Scholar]
- 34.Maisenbacher MK, Merrion K, Pettersen B, et al. Incidence of the 22q11.2 deletion in a large cohort of miscarriage samples. Mol Cytogenet. 2017;10:6. doi: 10.1186/s13039-017-0308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy UM, Page GP, Saade GR, et al. NICHD Stillbirth Collaborative Research Network. Karyotype versus microarray testing for genetic abnormalities after stillbirth. N Engl J Med. 2012;367:2185–93. doi: 10.1056/NEJMoa1201569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vorstman JAS, Jalali GR, Rappaport EF, et al. MLPA: a rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum Mutat. 2006;27:814–21. doi: 10.1002/humu.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.