Abstract
Transfer RNAs (tRNAs) are key players in protein synthesis. To be fully active, tRNAs undergo extensive post-transcriptional modifications, including queuosine (Q), a hypermodified 7-deaza-guanosine present in the anticodon of several tRNAs in bacteria and eukarya. Here, molecular and biochemical approaches revealed that in the protozoan parasite Trypanosoma brucei, Q-containing tRNAs have a preference for the U-ending codons for asparagine, aspartate, tyrosine and histidine, analogous to what has been described in other systems. However, since a lack of tRNA genes in T. brucei mitochondria makes it essential to import a complete set from the cytoplasm, we surprisingly found that Q-modified tRNAs are preferentially imported over their unmodified counterparts. In turn, their absence from mitochondria has a pronounced effect on organellar translation and affects function. Although Q modification in T. brucei is globally important for codon selection, it is more so for mitochondrial protein synthesis. These results provide a unique example of the combined regulatory effect of codon usage and wobble modifications on protein synthesis; all driven by tRNA intracellular transport dynamics.
INTRODUCTION
Transfer RNAs (tRNAs) bear the most extensive diversity of post-transcriptional modifications among all nucleic acids in cells (1). Several complex modifications exist in the anticodon stem-loop region of tRNAs, which may act to ensure accuracy and fidelity during translation (2). Queuosine (Q), a hypermodified 7-deaza-guanosine, is one of the most chemically intricate modifications described to date. Q is found at the wobble position 34 of tRNAs that contain a 5′-GUN-3′ anticodon sequence, where N represents any of the canonical nucleotides, which includes tRNAHisGUG, tRNAAspGUC, tRNAAsnGUU, tRNATyrGUA (3). Q is found in both bacteria and eukarya, and catalyzed by tRNA guanine transglycosylase (TGT) (4). In bacteria, Q is synthesized de novo, wherein guanosine is modified by five sequential enzymatic reactions to form preQ1, which is then incorporated into tRNAs by the bacterial TGT homodimer (bTGT). PreQ1 is further modified to Q by two enzymatic steps on the tRNA itself (4). A related 7-deaza-guanosine derivative called archaeosine (G+) is present at position 15 on the D-loop of several tRNAs in archaea. Archaeal TGT is a homolog of bacterial TGT and catalyzes the exchange of guanine with preQ0, a precursor common to the biosynthesis of both G+ and Q (5,6). Eukaryotes lack the enzymes required for de novo synthesis of queuosine and hence rely on nutrient sources and additionally, in the case of mammals, their gut microbiome. Queuine (q, free base of queuosine) is recognized by eukaryotic TGT (eTGT), and exchanged with guanine on the tRNA (7). In mice and humans, TGT exists as a heterodimer of queuine tRNA ribosyltransferase 1 (QTRT1), and queuine tRNA ribosyltransferase domain containing 1 (QTRTD1) (8).
Because of its position at the wobble base in the anticodon, Q may play a role in codon recognition. Morris and co-workers proposed a model where the Q modification may influence cellular growth and differentiation by codon-biased regulation of protein synthesis (9). They carried out an in silico analysis which showed that the NAU/NAC ratio of usage of cognate codons of Q-containing tRNAs, is much higher in housekeeping genes (>0.81) than in onco-developmental gene transcripts (<0.35) (9), which could lead to differential protein synthesis in healthy v/s cancer cells. In Xenopus oocytes, Q-containing tRNAs eliminate the proposed codon preference by efficiently recognizing either NAU or NAC codons, whereas the unmodified tRNAs preferably recognize NAC (10). In drosophilid lineage, Q mediates a shift in codon preference within a genome, during various developmental stages (11,12). Recently, Tuorto et al. showed that Q‐containing tRNA levels control translational speed at their cognate and near‐cognate codons in humans (13). They also reported that the loss of Q resulted in the induction of the unfolded protein response, in humans and mice (13). Thus, there is evidence supporting the role of queuosine tRNA modification in regulating codon-biased translation, although its impact appears to vary between organisms.
The role of tRNA modifications is particularly relevant in medically and economically important trypanosomatid parasites. Trypanosoma brucei is a unicellular dixenous parasite, belonging to the order Kinetoplastida. Its nuclear genome is organized in a unique way, with several unrelated genes clustered together and transcribed as a polycistronic unit (14,15). As a result, these parasites lack most of the transcriptional control found in other organisms, with the bulk of gene expression regulation occurring post-transcriptionally. Additionally, they undergo extensive morphological and metabolic remodeling, during their life cycle, which includes switching from glycolysis in the mammalian host to oxidative phosphorylation inside the insect vector, continually changing their surface glycoprotein composition to evade the immune system, etc. (16). These events demand rapid changes in translation of developmentally regulated proteins. In such situations, changes in post-transcriptional modifications of tRNAs may be essential for the survival or virulence of the parasite, which in the case of Q implies two different environments to supply queuine: the gut of the insect and the bloodstream of mammals.
Complicating things further is the fact that the mitochondrial genome of T. brucei does not encode any tRNAs. Consequently, they must import the whole set of tRNAs from the cytoplasm to be utilized for organellar translation (17). tRNA modifications present a possible way to regulate import into the mitochondria or to adapt to the mitochondrial translation machinery, which due to its ancestry may be more bacterial in nature. However, there need not be a universal function of tRNA modifications in mitochondrial import and these may likely be case specific, depending on the type of tRNA, modification or the organism. For example, cytosol-specific 2-thiolation of tRNAGlu is thought to act as a negative determinant of import in Leishmania tarentolae, representing the only known example of a modification affecting tRNA import (18). However, in T. brucei, tRNAGlu is imported independent of thiolation (19). Likewise, thiolation is not an import determinant in the apicomplexan parasite Toxoplasma gondii (20). Thus, once again, the extent to which a modification may impact tRNA mitochondria-import also seems to differ between different protozoans.
In the current study, we have explored the function of queuosine tRNA modification in T. brucei. Using bioinformatics and molecular biology approaches, we have identified two subunits of the TbTGT complex, and established their function, localization and physical interaction. Further, we aimed to study the mechanism by which this modification differentially influences translation. We analyzed the combined effects of differential codon usage, and Q-tRNA levels, on cytosolic translation. Furthermore, as the mitochondrion is one of the organelles, which undergoes major transformation during the life cycle of Trypanosoma, we studied the role of Q in mitochondrial translation. As expected, queuosine functions in codon-biased translation in the cytoplasm, but surprisingly Q-tRNAs are preferentially imported into the mitochondria to play a role in translation of U-rich mitochondrial mRNAs. The combined effects of its role in transport and organellar protein synthesis is critical for mitochondrial function as shown here.
MATERIALS AND METHODS
Cell culture, generation of cell lines and q depletion experiments
Wild type (WT), procyclic T. brucei 29–13 cells (21) were grown in SDM-79 medium (22) supplemented with 10% fetal bovine serum. RNAi construct was generated by cloning region of the coding sequence of TbTGT1 into the plasmid vector p2T7-177 using following oligonucleotides: TbTGT1-F: CCCAAGCTTCTAAGACCAGGGGAAGATATACTTAAT; TbTGT1-R: CGCGGATCCCGATAGGAGTGTAGCCCCAA. Cloning sites for HindIII and BamHI are underlined in the sequence. RNAi was induced by the addition of 1 μg/ml of tetracycline to the media, generating dsRNA from head-to-head orientated T7 promoters (21).
For tagging of the endogenous TbTGT proteins: TbTGT1 was tagged at the c-terminus, with myc tag using pMOTag 4M vector (with phleoR), as described (23). TbTGT2 was tagged at the c-terminus, with V5 tag, using a modified pPOTv4 vector (with puroR) (24). Primers used for PCR were as following: TGT1myc-F: GAATACGTACAGAAGTTTTTTTTAGGATATTACCCGGCACGGGATTACCCAAAGTGGATTGTTGATGCACTTGCTTCTGTCAGTGTTGAGCTCCCCCGCCCGCTCGAGATGGAGCAGAAG, TGT1myc-R: GAGAATACCAAAAGAACACTAAAAAAGACACCGATTCGTGTTATTGAAACCTTATTCCTTATTCAGCCCGATGCGTTACCTTTCCCCTAATATGGCGGCCGCTCTAGAACTAGTGGAT; TGT2V5-F: TTCATAATTTAGCCTTCCTGGTGCAGCTAATAGGGCTTTACAGAAGGTCAACTGCGGAGGCTCGAGAAGTCTGCTAACTTGGGTGCTGGCACAGTTGCCGCTCGAGATGTACCCTTACG, TGT2V5-R: AGACAGAGCAGGAGTTTCCATAGAAAGGAGAACGTGAAAGAAAAAAAATTATGCGATCATCATTATCATATTCGTTCATTAAGTTGAATGTTCTGGCGGCCGCTCTAGAACTAGTGGAT.
Resultant linear PCR products containing the tag and a resistance marker, flanked by the gene CDS and 3′UTR on either side, were transfected into procyclic stage of T. brucei by electroporation. Positive clones were selected by appropriate antibiotic and confirmed by genomic DNA PCR and western blotting.
For queuine depletion experiments, procyclic form 29-13 cells were washed with phosphate-buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) and resuspended in fresh SDM79 medium supplemented with 10% dialyzed fetal bovine serum (DFBS, 10,000 mw cut-off) to remove queuine. The cells were either subsequently supplemented with 20 nM synthetic queuine (+ q) or vehicle control dimethyl sulfoxide (− q). For E. coli RNA add-back experiments, cells depleted for queuine for 48 hrs were supplemented with total RNA isolated from either WT or TGT−/− E. coli K12 strains. (100 μg total RNA/ml of T. brucei culture)
Northern blot analysis
Total RNA was isolated as previously described (25). For APB-affinity electrophoresis, samples were deacylated by incubation in 100 mM Tris pH 9.0 for 30 min. For oxidation control, RNA was deacylated and added to a solution containing 50 mM Tris pH 5.0 and 2 mM NaIO4 for 2 h at 37°C in the dark. The oxidation reaction was quenched with 2.5 mM glucose before use. 25 mg of 3-aminophenylboronic acid (APB) (Sigma) was added to 10 ml of 8M-urea 8% polyacrylamide mix before solidifying (26). Electrophoresis of APB gels was carried out for approximately 5 hrs at 75 V, at 4°C and electroblotted to Zeta-probe membranes. The membrane was then UV cross-linked for 1 min. Northern hybridization was performed according to the manufacturer (Bio-Rad) using γ-ATP 32P-labeled oligonucleotides. Following hybridization, membranes were exposed overnight on a phosphoimager screen. Blots were analyzed using a Typhoon FLA 9000 scanner and the ImageQuant TL software (GE Healthcare). Probes used for Northern hybridization were as follows: tRNATyr: GTGGTCCTTCCGGCCGGAATCGAA; tRNAGlu: TTCCGGTACCGGGAATCGAAC; tRNAHis: CCACTCAACTATCTTCCC; tRNAAsp: CGGGTCACCCGCGTGACAGG; tRNAAsn: CTCCTCCCGTTGGATTCG; 7SL: GCTGCTACTGGGAGCTTCTCATAC; 12S rRNA: AGGAGAGTAGGACTTGCCCT; E. coli tRNAAsp: AACGGACGGGACTCGAACCCGCGAC; E. coli tRNAIle: CCTGAGTGGACTTGAACCACCGACC.
Immunofluorescence
Procyclic T. brucei 29–13 cells co-expressing TbTGT1-myc and TbTGT2-V5 were stained with 200 nM Mitotracker red for 30 min, at 27°C. 1 × 107 cells were centrifuged and washed once with PBS. Pellet was resuspended in 4% paraformaldehyde and the cells were fixed on Superfrost Plus slides (Thermo Scientific) for 20 min. Cells were permeabilized in ice-cold methanol for 20 min and washed with PBS. All subsequent steps were carried out in humid chamber. Cells were blocked with 5% milk in PBS-Tween, and incubated for 2 hrs, with polyclonal rabbit antibodies specific to c-myc (Sigma) or V5 epitope tag (Sigma), diluted 1:200 in 5% milk in PBS-T. The cells were washed thrice with PBS and incubated with goat anti-rabbit IgG secondary antibodies labeled with Alexa488™. Finally, the slides were washed thrice with PBS and counter stained with DAPI (Invitrogen) to stain DNA and visualized on Zeiss fluorescent microscope.
Co-immunoprecipitations
For co-immunoprecipitation (co-IP) analysis, cleared lysate was prepared by lysis of 2 × 108 cells in 1 ml of IPP150 (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40) with 1% Triton X-100 for 30 min, followed by centrifugation at 10 000 g at 4°C. For each immunoprecipitation, 0.5 ml of cleared lysate (1 × 108cells) was incubated overnight with 1 μl of rabbit antibody specific for either myc or V5 epitope tags. 100 mg of Protein A Sepharose™ Cl-4B beads (GE Healthcare) were swollen in 0.1 M PBS for 1 h and washed twice with 1 ml of PBS with 0.1% BSA and once with 1 ml of IPP150. Beads were then incubated for 4 h with rotation at 4°C with cleared lysate/antibody. After incubation, the supernatant was removed, and beads were washed four times with 1 ml of IPP150. Complexes bound to beads were eluted with 100 μl of 2× SDS sample buffer and heated for 5 min at 95°C. co-IPs were analyzed by SDS-PAGE and western blotting. To rule out non-specific precipitations, IPs were carried out in cell lines where only one of the subunits was tagged. An IP with V5 antibody was carried out in TGT1-myc cell line, lacking V5 tag for TGT2 and IP with myc tag was carried out in TGT2-V5 cell line where TGT1 was untagged.
qRT-PCR
cDNA was prepared from 1 μg of total RNA, as per manufacturer's instruction (QuantiTect™ Reverse Transcription kit, Qiagen), using oligo dT primer. Quantitative reverse-transcription (qRT-PCR) was carried out using Power SYBR™ green master mix according to manufacturer's instructions (Applied Biosystems™) with following primers: TbTGT1-F: AACTTGGCGTACCTCATCAA, TbTGT1-R: GGGAGCTCAACACTGACAGA.
Dual luciferase reporter construct design and assay
Renilla luciferase sequence was kept unaltered, while the cognate codons of Q-tRNAs in the firefly luciferase were either changed to all C-ending (NAC) or all U-ending (NAU) codons. These two recoded constructs were synthesized by Eurofins Genomics. All three constructs were then cloned (separately) into pT7V5 plasmid (with puroR), linearized with NotI and electroporated into procyclic 29–13 cells. Positive clones were selected with puromycin, luciferase expression was confirmed by western blotting. The cells were grown in presence or absence of queuine as described earlier. The reporter assay was performed using Promega Dual Luciferase kit, as per manufacturer's protocol. Briefly, cells induced O/N with tetracycline, were washed once with PBS and resuspended in PLB buffer in 96 well transparent flat bottom plates. The plate was incubated at room temperature for 15 min with constant shaking. The substrate for Firefly luciferase LARII was added and luminescence (FLuc) was measured in Tecan Spark Luminometer. Subsequently, the Stop & Glo reagent (containing quencher for Firefly luminescence as well as substrate for Renilla) was added and Renilla luminescence (RLuc) was measured. The ratio of FLuc/RLuc was calculated and the values were further normalized to the WT construct.
Organellar fractionation
4 × 108 procyclic T. brucei 29–13 cells were harvested by centrifugation, washed in PBS, resuspended in 500 μl SoTE (0.6 M Sorbitol, 2 mM EDTA, 20 mM Tris–HCl pH 7.5) and lysed with 500 μl SoTE containing 0.1% digitonin. The cells were mixed thoroughly and incubated on ice for 5 min before centrifugation (4°C, 8000 g, 5 min). The supernatant was treated as the cytosolic fraction. The pellet was resuspended in 500 μl SoTE supplemented with 2 μg/ml RNaseA, 1μl DNase I (2 units, New England Biolabs), 3 mM MgCl2 and incubated on ice for 15 min. The resultant pellet after centrifugation (4°C, 8000 g, 5 min), was treated as the organellar fraction. RNA was isolated from these enriched fractions along with total RNA from whole cells and resolved by APB-affinity PAGE and northern blotting.
Mitochondrial codon usage analysis
Codon usage analysis for mitochondrial genes was performed using codon usage calculator at the sequence manipulation site available online (27). Frequencies for each codon were calculated as the number of NAC or NAU codons divided by the total number of (NAC + NAU) in a transcript. Codon frequencies per thousand were calculated by dividing the number of occurrences of each type of codon by the total number of codons analyzed and normalized to 1000. Data about nuclear codon usage was extracted from Codon and Codon Pair Usage Tables (CoCoPUTs) (28), HIVE codon usage tables (CUTs) database (29), for NCBI:txid5702 (Trypanosoma brucei brucei).
In vitro mitochondrial import assay for tRNAs
Mitochondria were isolated from procyclic T. brucei 29–13 cells by hypotonic lysis following protocols previously described (30). For import assays, 1 mg of purified mitochondria were incubated with 50–100 000 cpm of radioactively labeled tRNA, 0.25 M sucrose, 20 mM Tris–HCl (pH 8.0), 1 mM ATP, 2 mM DTT, 10 mM MgCl2 and 2 mM EDTA; for 10, 20, 30, 40 or 60 min, 27°C. After each time point, 100 U of micrococcal nuclease and 5 mM CaCl2 were added, and the reaction was incubated an additional 30 min to digest the tRNAs that were not imported into the mitochondria. Reactions were then stopped by the addition of 10 mM EGTA (pH 8.0). To purify the protected tRNAs, the mitochondria were washed with 0.25 M sucrose/20 mM Tris–HCl (pH 8.0), pelleted, suspended in 90 μl of 10 mM Tris–HCl (pH 8.0), 1 mM EDTA, and phenol extracted, followed by ethanol precipitation. The radioactively labeled tRNAs were separated by electrophoresis through 7 M urea/ 6% polyacrylamide gels. Gels were dried and exposed to a phosphoimager screen and analyzed using a Typhoon FLA 9000 scanner and the ImageQuant TL software (GE Healthcare). Whenever native substrates were used, they were hybridized with an antisense biotinylated oligonucleotide specific for each species and purified by affinity chromatography through streptavidin beads as described previously (31). Native tRNAs were 5′-end-labeled with γ-32P ATP. An aliquot of RNA isolated from each time-point of the assay was radiolabeled with γ-32P ATP and resolved on 7 M urea/ 6% polyacrylamide gels. The gels were dried and exposed to the phosphoimager screen for autoradiography. A parallel assay was performed using the same materials (technical replicate) and the isolated RNA was used for northern blotting. These northerns were probed with radiolabeled probe against tRNAGlu.
In vivo mitochondrial translation assay and two-dimensional gel electrophoresis
In vivo translation and 2D gel electrophoresis was carried out as described previously (32). Briefly, 1 × 107 cells were harvested, washed twice with sterile SoTE (0.6 M sorbitol, 20 mM Tris–HCl, pH 7.5, 2 mM EDTA). Pelleted cells were resuspended in 90 μl SoTE, with 1 μl of 100 mM DTT and the cytosolic translation was inhibited with 1 μl of 100 mg/ml cycloheximide for 10 min at 27°C. 10 μl of EasyTag™ EXPRE35S protein labeling mix was added to the cells and incubated for 1 hr, at 27°C. Cells were recovered by a 5 min centrifugation at 2000 g. The pellet was resuspended in 200 μl of SoTE and stored at –80°C until further use. Before loading on the electrophoretic gel, 50 μl of the labeled cell lysate was centrifuged and the pellet was resuspended in 100 μl of loading buffer containing 250 mM Tris–HCl, pH 6.8, 2% SDS, 2% β-mercaptoethanol, 30% glycerol and 0.01% bromophenol blue, and incubated at 37 °C for 30 min. Samples were cleared by centrifugation at 14,000 g for 15 min and separated on 1 mm-thick 9.5% Tris-Glycine SDS gel (stacking gel: ∼3 cm; resolving gel: ∼15 cm) at a constant current not exceeding 20 mA. The gel slice (∼5 mm wide) was incubated for 30 min at 37°C in 125 mM Tris–HCl, pH 6.8, 1% β-mercaptoethanol, 1% SDS and transferred onto 1.5 mm-thick 14% gel (same dimensions as 9.5% gel), cast with a single broad-well comb. The second dimension gel was run overnight, at a constant current of 6 mA. Gels were fixed for 40 min in 10% Acetic Acid, 50% MeOH, stained in Coomassie blue R250, de-stained in 10% acetic acid, 10% methanol, and scanned. For autoradiography, gels were further washed three times in water for 20 min, incubated in 1M salicylate for 60 min, dried and exposed to a phosphoimager screen and analyzed using a Typhoon FLA 9000 scanner and the ImageQuant TL software (GE Healthcare) (32).
Cytochrome c oxidase activity assay
Mitochondrial vesicles were isolated from 2.5 × 108 cells by hypotonic lysis as described previously (33) and stored at -80°C. For preparation of mitochondrial lysate, the pellets were resuspended in 40 μl 1M aminocaproic acid (ACA) and lysed with 10 μl 10% dodecyl maltoside. The samples were incubated on ice for 1 h and centrifuged at 15 000 g for 30 min. The supernatant was used as mitochondrial lysate. Protein concentrations were determined by BCA kit, according to manufacturer's instructions (Pierce, Thermo Fischer).
Cytochrome c oxidase (complex IV) activity was measured as described previously (33). Briefly, in a 1 ml cuvette containing COX buffer (40 mM sodium phosphate buffer, pH 7.4; 0.5 mM EDTA; 20 μM horse heart cytochrome c; 30 μM sodium ascorbate; 0.005% dodecyl maltoside) 20 μl of mitochondrial lysate was added. Antimycin A (Sigma) at final concentration 300 ng/ml was used to inhibit the interfering reductase activity selectively. The change in absorbance at 550 nm was measured every 20 s for 10 min. A unit of activity was defined as the amount of enzyme that catalyzes the oxidation of 1 mmol of cytochrome c per minute, assuming an extinction coefficient of 21.1 mM–1 cm–1 (33).
Digestion of LC–MS/MS analysis
Total RNA from procyclic T. brucei 29–13 cells was isolated as described previously (25). Digestionof RNA was performed as previously described (34). Separation was accomplished by reverse-phaseliquid chromatography using an Acquity UPLC HSS T3, 1.8 µm, 1 mm X 100 mm column, (Waters, Milford, MA) on a Ultimate 3000 UHPLC system (Thermo Fisher Scientific, San Jose, CA). Mobile phase A consisted of 5.3 mM ammonium acetate in LC-MS grade water, pH 5.3. Mobile phase B consisted of a 60:40 mixture of 5.3 mM ammonium acetate and acetonitrile with a gradient of 0% B (from 0 to 1.8 min), 2% B at 3 to 3.5 min, 3% B at 4.1 min, 5% B at 7 min, 25% B at 9 min, 35% B at 15 min, 99% B at 15.5 min (hold for 4.5 min), 99% B at 20 min then returning to 0% B at 25.5 min at a flow rate of 100 µL min-1. The column temperature was set at 30 °C. Data was acquired on an AB Sciex TripleTOF 5600+ mass spectrometer having a DuoSpray Ion Source utilizing Analyst 1.7. The method consisted of two experiments, a Positive TOF MS scan (accumulation time 250 ms) followed by a Positive Product Ion scan set at 410.00 Da: unit resolution (accumulation time 100 ms). Intensity threshold was set to 1 cps. Source settings were GS1, GS2 and CUR gases at 20, 10, and 10, respectively. Source temperature was set at 350.00 with an Ion spray voltage (Floating) of 5000.00. Mass range for Positive TOF MS scan 200 to 900 Da and 100 to 600 for the Positive Product Scan. Data was processed using AB Sciex Peak View 2.2.
RESULTS
T. brucei genome encodes two paralogs of TGT
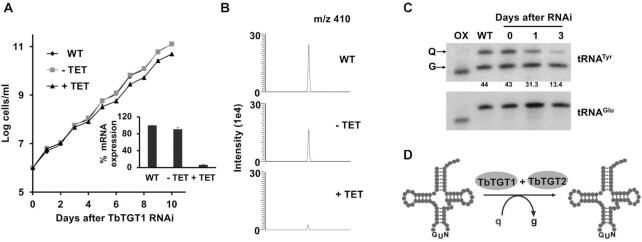
Previously, we have reported the presence of queuosine modification in all four tRNAs (Tyr, His, Asp, Asn) with GUN anticodons in T. brucei (25). To identify tRNA guanine transglycosylase (TGT) paralogs in T. brucei, we performed a database search using human TGT proteins (35) (QTRT1 and QTRTD1) as a BLAST query for the kinetoplastid genome database (TriTrypDB (36)). We identified two putative paralogs TbTGT1 (Tb927.6.3130) and TbTGT2 (Tb927.5.3520) with a calculated molecular mass of 44.74 and 33.17 kDa, respectively. TbTGT1 is homologous to human QTRT1 subunit (54% similarity, E value: 1e–126, NCBI Blast) and contains all the conserved catalytic Asp residues required for the transglycosylase activity (37,38) (Supplementary Figure S1). Meanwhile, TbTGT2 is more divergent (29% similarity with QTRTD1, E value: 4e–17, NCBI Blast) with no active Asp residues. The three cysteine and one histidine residues (CCCH) responsible for Zn2+ binding, are present in both (Supplementary Figure S1), suggesting a similar subunit composition to the human TGT (8). We have already described one of the TbTGT proteins (Tb927.5.3520), referred to, hereafter as TbTGT2 and showed that while the down-regulation of TbTGT2 does not affect cell viability, it is necessary for the formation of Q-modified tRNAs (25). To evaluate the physiological role of TbTGT1, we downregulated its expression by RNAi, in the procyclic (insect) stage of the parasite. Briefly, a portion of the open reading frame of TbTGT1 was cloned into tetracycline-inducible p2T7-177 RNAi vector, the construct was linearized, used to transform T. brucei cells and establish clonal cell lines as described in the methods section. TbTGT1 RNAi cell lines were grown in the presence (RNAi induced, +TET) or absence of (RNAi uninduced, –TET) tetracycline. Down-regulation of the mRNA levels was confirmed by qRT-PCR (Figure 1A, inset). The knock-down of TbTGT1 resulted in a slight growth defect (Figure 1A). Next, we investigated the involvement of TbTGT1, in Q modification of tRNAs. Total RNA from TbTGT1 RNAi cells was analyzed using aminoacryloylphenyl boronic acid (APB)-affinity electrophoresis, followed by northern blotting (26). RNA isolated following three days of RNAi induction showed 87% reduction in the intensity of the slower migrating band corresponding to Q modified tRNATyr (Figure 1C). The knock-down of TbTGT1 had no effect on the levels of tRNAGlu, which is not a substrate for TGT. The decrease mentioned above in queuosine modification levels was also verified by mass spectrometry of total RNA isolated from RNAi induced cells and compared to RNA isolated from either wild-type or uninduced cells (Figure 1B). Combined with our previous analysis of TbTGT2 (25), these results suggest that both TbTGT1 and TbTGT2 are necessary for queuosine modification in tRNAs in T. brucei (Figure 1D).
Figure 1.
TbTGT1 is necessary for Q formation in the tRNAs. (A) Growth curve of T. brucei procyclic form (PF) cells where TbTGT1 expression has been silenced by RNAi (+TET) compared to WT cells or an uninduced control (−TET). The data represents mean ± SD from three independent experiments. RNAi knock-downs were confirmed by qRT PCR (inset) and presented as % relative TGT1 mRNA expression, normalized to WT; (B) Extracted ion chromatogram for Q 410 m/z comparing samples from WT, TbTGT1 RNAi induced (+TET) and uninduced (−TET). Peak corresponding to m/z of 410 is 7 times reduced in +TET sample than –TET control. (C) APB gel - Northern hybridization showing the effect of RNAi knock-down of TbTGT1 in T. brucei on the levels of queuosine of tRNATyr (Numbers under the blot indicate %Q levels, calculated by dividing the intensity of the Q band by the sum of the Q and G bands, and multiplied by 100). tRNAGlu was used as a loading control; Oxidized RNA was used as a negative control (OX). 3-Aminophenylboronic acid (APB) has affinity for the cis-diol groups present in queuosine. As a result, Q-tRNAs migrate slower on the gel, leading to separation of modified (Q) and unmodified (G) tRNAs. In oxidized control (OX), the tRNAs are treated with periodate, to oxidize all cis-diol groups (queuosine as well as terminal 3′ ribose). (D) Schematic representation of enzymatic substitution of guanine (g) with queuine (q) by TbTGT heteromer.
TbTGT1 and TbTGT2 localize to the nucleus and form a heteromeric complex
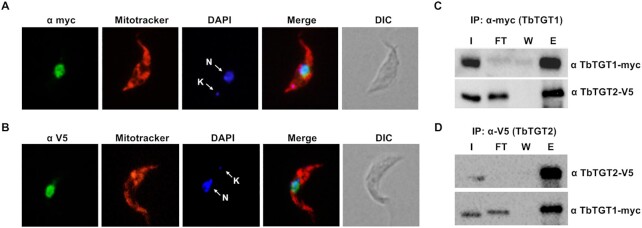
We previously reported that Q formation is a nuclear event (25). To further corroborate this, we determined the localization of both TGT proteins using endogenously tagged TbTGT1-myc and TbTGT2-V5. In addition, Mitotracker red was used to stain the mitochondria while DAPI was used to mark the DNA. The cells were visualized by fluorescence microscopy. Both TbTGT1 and TbTGT2 localized to the nucleus in T. brucei (Figure 2A and B); contrary to the report in humans and mice, where these proteins localize to the outer surface of the mitochondria (35). We did not observe any significant localization to the mitochondria or the cytosol. In a previous study, we demonstrated the existence of a retrograde nuclear import pathway in T. brucei (25). The localization of the endogenously tagged TbTGTs, further supports the finding that the cytosolically spliced tRNATyr is imported back to the nucleus in order to obtain the queuosine modification. Unlike its bacterial counterpart where a single TGT enzyme forms a functional homodimer, eukaryotic TGTs may exist as a heterodimer. So far, this has been experimentally supported in metazoans: rabbit erythrocytes (39); bovine liver (40); mice (35) and humans (8). In order to investigate the interactions between the two TGT proteins of T. brucei, a cell line co-expressing endogenously tagged TbTGT1 and TbTGT2 was used for co-immunoprecipitation (co-IP) analysis. Briefly, the cleared lysate was incubated with antibodies specific for either of the epitope tags, bound to protein A sepharose beads and eluted in SDS buffer. When TbTGT1 was used as bait for IP, TbTGT2 was detected in the eluate (Figure 2C). A reverse IP was performed using TbTGT2 as bait, to confirm this result, and as expected, TbTGT1 was detected in the eluate (Figure 2D). To rule out non-specific interactions, we performed IPs in cell lines where only one of the subunits was tagged. An IP with V5 antibody was performed in TGT1-myc tagged cell line, where it failed to pull down the myc-tagged subunit, and vice-versa (Supplementary Figure S2). These findings confirm specific interactions between two TGT subunits of T. brucei, in vivo.
Figure 2.
TbTGT1 and TbTGT2 form a complex and localize to the nucleus. Immunofluorescence assay performed with cells expressing endogenously epitope-tagged (A) TbTGT1 (myc) and (B) TbTGT2 (V5) (in green in both cases). Mitotracker was used to stain the mitochondria (red) while DAPI stained the nuclear (white arrow, N) and kinetoplast (mitochondrial) (white arrow, K) DNA (blue). DIC refers to a phase-contrast image. (C) Immunoprecipitation of TbTGT1-myc with anti-myc tag antibody. The lysates were analyzed by western blots against anti-myc and anti-V5 antibodies. (D) Immunoprecipitation of TbTGT2-V5 with anti-V5 tag antibody. (I: input, FT: flow-through, W: wash, E: eluate) The lysates were analyzed by western blots against anti-myc and anti-V5 antibodies; results are representative of three independent experiments.
T. brucei obtain queuine from their environment
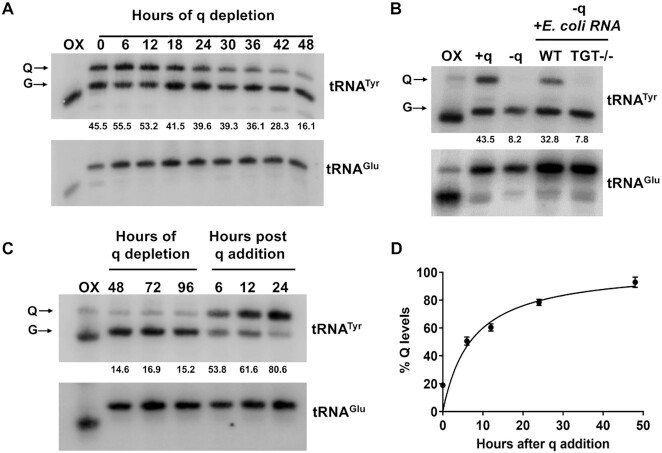
As eukaryotes cannot synthesize queuine, the free base of queuosine, they either rely on their diet/environment to obtain free queuine (7). For T. brucei grown in culture, fetal bovine serum (FBS) is the only source of queuine. Thus, when these cells are grown using dialyzed FBS (dialyzed for small molecules including queuine), their tRNAs lack the queuosine modification. We carried out time course experiments to determine the dynamics of disappearance of Q from tRNAs and noticed a reduction of Q levels in tRNATyr, beginning from 18–24 hrs of growth in q-deficient medium (Figure 3A). This experiment confirmed that T. brucei is not able to synthesize queuine. Further, we checked the ability of T. brucei cells to incorporate queuosine into tRNA using two different sources of queuine. First, we provided it in the form of total RNA isolated from bacteria or their degradation products and observed that the cells were able to use it to modify their own tRNAs, indicating that T. brucei is able to salvage queuine from bacterial RNA. As a control, we used a TGT deletion mutant (TGT −/−) E. coli, which does not contain the queuosine modification (Figure 3B, Supplementary Figure S3), which expectedly failed to complement Q formation in T. brucei tRNA. Secondly, we obtained similar results using synthetic queuine, where significant levels of queuosine modification were seen as early as 6 hrs after queuine addition (Figure 3C). Densitometric analysis showed that the levels of Q modification saturate by 48 h (Figure 3D). These experiments confirmed, that similar to other eukaryotes, in T. brucei, levels of Q-tRNA modification are influenced by the availability of queuine in their micro-environment. To determine if absence of q in this experimental setup leads to any growth defect, the cells were grown in SDM-79 using dialyzed FBS, supplemented with either synthetic queuine (+q) or vehicle control (−q). We did not observe any pronounced growth defect over 10 days (Supplementary Figure S6F).
Figure 3.
T. brucei cells obtain queuine from their environment. (A) APB gel—northern hybridization of total RNA isolated at indicated time points, from T. brucei cells depleted for queuine. (B) Northern blot of cells depleted for queuine (−q) and then fed with total RNA isolated from either WT or TGT−/− E. coli. +q lane indicates Q levels in cells prior to queuine depletion. (C) Time-course analysis of cells depleted for queuine and then fed with 20 nM synthetic queuine. All membranes were probed with a 32P radiolabeled probe against tRNATyr. tRNAGlu was used as a negative control (numbers under the blots indicate % Q levels). For panels A–C, oxidized RNA was used as a negative control (OX). (D) Densitometric analysis of percent of tRNATyr modified over time after addition of queuine, over three independent experiments.
Queuosine is important for codon-biased translation
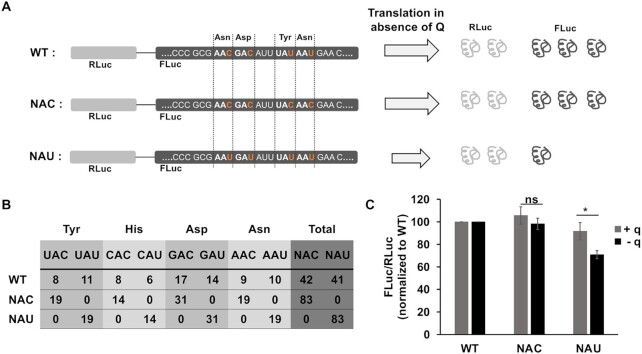
To investigate the role of Q in cytosolic codon-biased translation, we employed the dual luciferase reporter system, where the Renilla fly luciferase (RLuc) is fused to Firefly luciferase (FLuc) with a linker (41). Since these enzymes use different substrates, it is possible to quantify differential expression of these enzymes simultaneously, with luminescence assays. We constructed two codon re-engineered constructs, where the RLuc sequence was kept unaltered, but in the FLuc sequence, the cognate codons for all 4 Q-containing tRNAs (tRNAHisGUG, tRNAAspGUC, tRNAAsnGUU, tRNATyrGUA) were changed either to all NAC or all NAU codons, as opposed to the WT FLuc, which contained approximately equal number of NAC and NAU codons. The total number of NAC and NAU codons are enumerated in (Figure 4B). We hypothesized based on available literature, that Q might be more efficient than G, in decoding U-ending codons (10,13). Thus, in the absence of Q we might observe a decreased translation of NAU luciferase, and subsequently, a decrease in the luminescence (Figure 4A). We transfected these three constructs into T. brucei 29–13 cells. Transcription of the dual luciferase constructs was induced with tetracycline, and the cell lines were grown in presence or absence of queuine. When we measured the luminescence of FLuc and normalized it to RLuc, we observed that the signal of FLuc was marginally upregulated for NAC construct, with or without queuine, as compared to WT (Figure 4C). However, the NAU construct showed significant decrease (∼30%) in the levels of FLuc luminescence in the absence of queuine (Figure 4C), suggesting that U-rich codons might be translated less efficiently in the absence of queuine.
Figure 4.
Queuosine is important for cytosolic codon-biased translation. (A) Schematic representation of dual luciferase construct in which Renilla fly luciferase (RLuc) is expressed in-frame with a codon-reengineered Firefly luciferase (FLuc), where the original sequence was left intact (WT), or all the Asn, Asp, Tyr and His codons were changed to NAC or NAU as indicated schematically in panel A. If Q has a role in translation of U-rich mRNAs, in absence of Q-tRNAs, NAU luciferase would be translated inefficiently, as depicted by the shortened grey arrow, leading to reduction in the levels of Firefly luciferase. Renilla luciferase levels, however, should remain comparable across different cell lines, as they are not subject to codon changes. (B) Number of cognate codons for Q-tRNAs in the three constructs. (C) Ratio of luminescence of FLuc to RLuc, further normalized to the WT construct. For this experiment, the cells were depleted for queuine for 48 h, and subsequently supplemented with 20 nM synthetic queuine (+ q) or vehicle control (− q) for 24 h. The graphs are representative of three independent experiments (* P <0.01, ns: not significant).
The connection between Q modification, codon usage and mitochondrial import
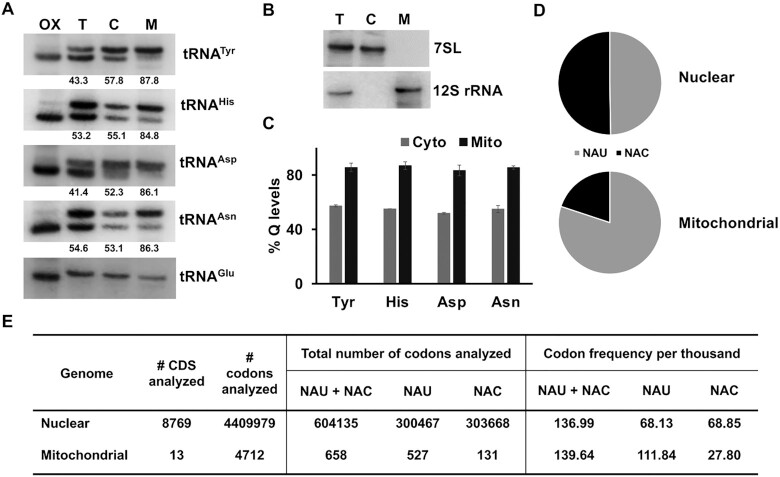
As tRNA modifications have been implicated in regulating mitochondrial functions (42) (possibly through translation regulation) and in several cases their absence in tRNAs is associated with disease (42), we explored the role of queuosine in the mitochondrion of T. brucei. We obtained organellar fractions, using digitonin permeabilization, and then performed northern blots to assess the levels of Q-containing tRNAs while comparing cytosolic and mitochondrial fractions. We observed, that 85% of the mitochondrial fraction of tRNATyr, is modified with Q, while only 55% of the cytosolic fraction was modified (Figure 5A and C). A similar result was observed for tRNAHis, tRNAAsp and tRNAAsn (Figure 5A and C). The purity of these fractions was verified using 7SL RNA and 12S rRNA as cytosolic and mitochondrial markers respectively (Figure 5B). This observation opens several questions, regarding the import of tRNAs into the mitochondria. It is important to emphasize here, that in T. brucei, the mitochondrial genome does not encode any tRNAs, and the whole set is imported from the cytosol (43). The mitochondrial genome of T. brucei encodes 18 protein coding ORFs, two ribosomal RNAs, and hundreds of guide RNAs required for RNA editing (44). The protein-coding genes mostly include subunits of respiratory chain complexes I, III, IV and V, as well as two ribosomal proteins (45). As stated before, Q-containing tRNAs can decode their cognate codons, (NAC or NAU) with different efficiency (10). When we carried out codon usage analysis of several mitochondrially encoded mRNAs, we found that almost all the mRNAs analyzed favored U-ending codons (Figure 5D and E, Supplementary Figure S4). In the nuclear genome on the other hand, the overall frequency of NAC and NAU codons is approximately equal (Figure 5D and E). This reversal of codon usage is typical of most eukaryotes and combined with the fact that the levels of Q modification are significantly different in the two compartments, raises the question whether mitochondria preferentially import Q-modified tRNAs.
Figure 5.
Levels of queuosine tRNA modification and codon usage in the mitochondria of T. brucei. (A) Northern blot analysis of total RNA isolated from whole cells (T), cytosolic (C), and mitochondrial (M) fractions. Membranes were probed with radioactively labeled probe specific for the tRNA as indicated to the right of each blot (numbers under the blots indicate % Q levels). Oxidized RNA was used as a negative control (OX). (B) Control Northern blots to assess the purity of organellar fractions, probed for 7SL (cytosolic control) and 12S rRNA (mitochondrial control). The blots in panels A and B are representative of three independent experiments. (C) Densitometric analysis of Queuosine levels in the cytosolic (Cyto) and mitochondrial (Mito) fractions of tRNATyr, tRNAHis, tRNAAsp and tRNAAsn across three independent experiments. (D) Fraction of NAU and NAC synonymous codons in 8769 nuclearly encoded mRNAs and 13 mitochondrially encoded mRNAs. (E) codon frequencies per thousand for NAU and NAC codons in nuclear v/s mitochondrial genomes of T. brucei. Codon usage data for nuclearly encoded transcripts was extracted from Codon and Codon Pair Usage Tables (CoCoPUTs) (28), HIVE CUTs database (29), for NCBI:txid5702 (Trypanosoma brucei brucei). Mitochondrial codon usage was calculated as described in the methods section.
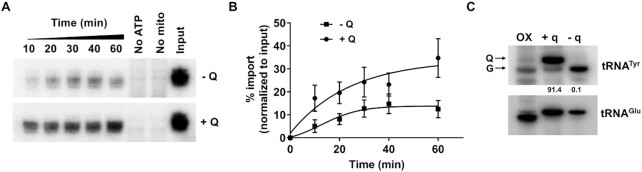
To determine the effect of Q on the import of tRNAs into the mitochondria we employed in vitro import assay (30). Briefly, we purified mitochondria from T. brucei cells by hypotonic lysis followed by percol gradient centrifugation. In parallel, total tRNA was isolated from cells grown with or without queuine, native tRNATyr was isolated from these samples by biotin-streptavidin oligonucleotide affinity purification (31) and radiolabeled with γ-ATP 32P. The amount of radiolabeled tRNA protected from micrococcal nuclease inside the mitochondria over different time intervals was evaluated as a measure of tRNA import as previously described (30). We observed that Q-modified tRNATyr was imported into isolated mitochondria, 3–4-fold more efficiently as compared to unmodified tRNATyr (Figure 6A–C). In addition, an aliquot of RNA isolated from each time-point of the assay was radioactively labeled and separated in a denaturing acrylamide gel. We observed comparable recovery of tRNAs as well as guide RNAs from the mitochondria following the assay in both the –Q (Supplementary Figure S5A) and +Q (Supplementary Figure S5B) reactions. Additionally, RNA isolated from a technical replicate of the above-mentioned experiment was used for northern blots using a radioactive oligonucleotide prove specific for tRNAGlu; an unrelated non-Q containing tRNA. Again no differences in the levels of mitochondrial tRNAGlu were observed; similar amounts of this tRNA were recovered after the assay in all time points, in both –Q (Supplementary Figure S5A, lower panel) and +Q reactions (Supplementary Figure S5B, lower panel). In a parallel experiment, the total RNA used to purify native tRNATyr was also analyzed by northern hybridization with a radiolabeled probe specific for tRNATyr to confirm the presence or absence of Q, even in this assay no significant differences in the total amount of this tRNA were observed (Figure 6C). Taken together, these experiments show that mitochondria preferentially import Q-modified tRNAs from the cytoplasm and the differences between the partitioning of the Q-modified and unmodified tRNAs observed in vitro faithfully represent the in vivo partitioning. Furthermore, neither the results from our in vitro assay nor those from the in vivo experiments can be ascribed to either technical difficulty with tRNA recovery or differential stability between the Q-modified and unmodified tRNAs.
Figure 6.
Q-modified tRNATyr is preferentially imported into the mitochondria. (A) Autoradiographs showing the in vitro import of native 32P labeled tRNATyr purified from T. brucei cells grown with or without queuine. ‘No ATP’ refers to the control where ATP was not added to the reaction. ‘No mito’ refers to the negative control where nitochondria were not added. (B) Densitometric analysis of in vitro tRNA import assays from four independent experiments (P value: 0.007, Student's t test for 60 min timepoint). (C) Total RNA used to purify the native tRNAs used in the in vitro import assays, probed for tRNATyr and tRNAGlu (numbers under the blot indicate %Q levels).
Lack of Q in the tRNAs leads to reduction in mitochondrial protein synthesis and affects mitochondrial function
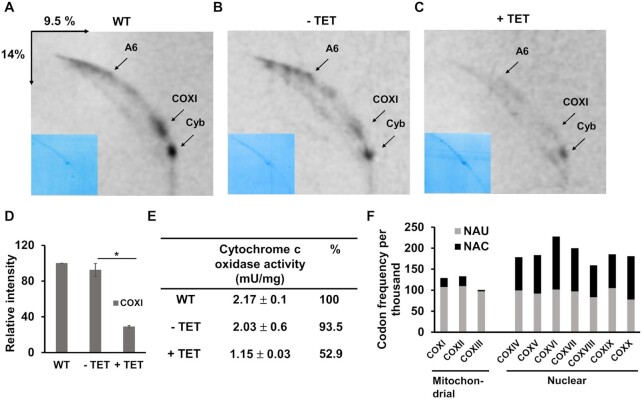
In order to quantify mitochondrial translation in the presence and absence of queuosine modification, we inhibited cytosolic translation with cycloheximide in the TbTGT1 RNAi cell line, and pulse-labeled the cells with 35S methionine and cysteine. This technique allowed us to monitor de novo protein synthesis in the mitochondria. When we resolved the proteins by 2-dimensional gel electrophoresis, we observed that, while the signal for WT and uninduced (−TET) samples was comparable (Figure 7A and B), a significant reduction in mitochondrial protein synthesis was observed in the RNAi induced (+TET) cell line (Figure 7C). These gels were also stained with Coomassie brilliant blue as loading control (insets, Figure 7A–C). The three most abundant mitochondrial translation products ATP synthase subunit 6 (A6), Apocytochrome b (Cyb) and Cytochrome C oxidase subunit I (COXI), can be easily identified using this approach (32,46,47). Semi quantitative densitometric analysis over three independent experiments, showed a 3-fold decrease in the synthesis of COXI, in the absence of queuosine (Figure 7D). To determine if this reduction in protein synthesis impacts the function of respiratory complexes, we measured the activity of complex IV (cIV) in isolated mitochondria, by spectrophotometric assay. We observed ∼47% reduction in cIV activity after TbTGT1 knock-down (Figure 7E). Comparing the codon usage of various subunits of cIV, we noticed that the mitochondrially encoded subunits (COXI-III), predominantly use the NAU codons while no such bias was observed in the nuclearly encoded subunits (Figure 7F, Supplementary Table S3). Indeed, this phenomenon is not restricted to cIV, but a similar codon bias was observed in mitochondrially v/s nuclearly encoded subunits of respiratory complexes I, III and V (Supplementary Figure S4, Supplementary Tables S1–S2 and S4).
Figure 7.
Lack of Q in the tRNAs leads to reduction in mitochondrial protein synthesis. Autoradiographs of 2D gel electrophoresis of total mitochondrial 35S methionine–cysteine labeled proteins used to assess de novo mitochondrial translation. For this experiment, WT (A), TbTGT1 RNAi uninduced (−TET) (B) and induced (+TET) (C) cells were labeled with 35S for mitochondrial translation and resolved on 2D gels (insets: Coomassie staining for loading control). (D) Semi-quantitative densitometric analysis of de novo synthesis of COXI from three independent experiments. Values were normalized to WT signals. (* P <0.05). (E) Spectrophotometric assay for cIV for WT, TbTGT1 RNAi uninduced (−TET) and induced (+TET) cells. Activity was measured in mitochondrial lysates prepared from three independent experiments. One unit (U) of activity catalyzes the oxidation of 1 mmol of cytochrome c per min. Specific activity is calculated as U per mg of mitochondrial protein. Mean ± SD values are shown. (F) Codon usage analysis for NAU and NAC codons in mitochondrially v/s nuclearly encoded subunits of respiratory complex IV. COX: Cytochrome c oxidase (The roman numeral after COX indicates the number of subunit).
A similar analysis of mitochondrial protein synthesis was carried out in cells depleted for or supplemented with synthetic queuine. As expected, we observed a significant reduction in protein synthesis in absence of queuine (Supplementary Figure S6A and B). Semi quantitative densitometric analysis over three independent experiments, showed a 2–3-fold decrease in the synthesis of A6, Cyb and COXI, in the absence of queuine (Supplementary Figure S6C). Total RNA from this experiment was also separated on APB-affinity gel, followed by northern blotting, to confirm the presence and absence of Q-tRNAs (Supplementary Figure S6D). Mitochondria isolated from queuine depleted cells showed a 45% decrease in the activity of cIV, as measured spectrophotometrically (Supplementary Figure S6E). To determine if absence of q in this experimental setup leads to any growth defect, the cells were grown in SDM-79 using dialyzed FBS, supplemented with either synthetic queuine (+q) or vehicle control (−q). We did not observe any pronounced growth defect over 10 days (Supplementary Figure S6F).
DISCUSSION
In this study, we describe the presence of two paralogs (TbTGT1 and TbTGT2) of the TGT enzyme responsible for queuosine modification in Trypanosoma brucei. It was already reported in metazoans by in vitro reconstitution experiments, that the two TGT subunits form a heterodimer (8,35). In T. brucei, we found that TGT1 and TGT2 belong to the same complex in vivo, although whether they interact directly or are part of a larger complex through indirect interaction is yet to be determined. However, the TbTGT complex differs significantly from that of other organisms studied so far in being exclusively localized to the nucleus, while other eukaryotic TGTs associate with the outer membrane of the mitochondria. Nuclear localization of TbTGT is in line with our recent discovery of a retrograde nuclear import pathway (25), whereby the only intron-containing tRNA of T. brucei, tRNATyr, is spliced in the cytoplasm and then imported back to the nucleus to be modified with queuosine (25). This type of transport dynamics led us to explore the function of queuosine in T. brucei.
We show here, that like other eukaryotes studied so far, T. brucei cannot synthesize queuine, but obtain it from their environment. They are able to import queuine from environment, as well as salvage it from recycled nucleotides. The observation that T. brucei are able to obtain queuine from total RNA of E. coli, leads us to speculate the existence of a queuine salvage pathway analogous to that described by Zallot et al. with yeast complementation experiments showing that members of the DUF2419 protein family from several eukaryotic species are able to rescue queuosine modification by queuine salvage (7).
The codons, for all four amino acids, decoded by Q-containing tRNAs belong to two-fold degenerate codon boxes, with only one tRNA isoacceptor per box. Q is therefore proposed to affect codon recognition, as suggested by various earlier studies in other systems. Q-containing tRNAs may be involved in equilibrating codon bias between their cognate codons, while unmodified tRNAs preferably recognize NAC over NAU codons (10–13). Using a dual luciferase reporter system, we analyzed the correlation of codon usage and the queuosine modification. In the absence of Q, cells were able to synthesize significantly less amount of firefly luciferase, when its mRNA had U as opposed to C in third position of codons for tyrosine, histidine, aspartic acid and asparagine. Dedon and Begley coined the term modification tunable transcripts (MoTTs), to describe transcripts, which exhibit codon bias differentially translated by wobble tRNA modifications (48). On similar lines, we observe that Q-modified tRNAs might differentially translate the luciferase mRNA based on its codon usage. This suggest that more global changes in the relative availability of queuine, in different hosts, may affect the levels of Q-tRNA; such changes may in turn modulate the levels of a subset of proteins throughout the life cycle of T. brucei, adding another layer to the developmental-stage specific translation regulation. Testing this hypothesis will require further experimentation.
Q depletion experiments have been carried in various systems. In C. elegans (49) and C. reinhardtii (50), there were no obvious effects of Q depletion, whereas D. melanogaster (51) and D. discoidium (52) found difficulties in responding to certain stress stimuli, although no effect was seen in stress-free conditions. Therefore, it is safe to assume, that Q might be more relevant during processes that require rapid changes in translation rates. During the life cycle of T. brucei, the parasite has to adjust to several different environments (53) and undergo extensive metabolic remodeling. In the bloodstream of the mammalian host, due to an abundance of glucose, the parasites rely on glycolysis and substrate-level phosphorylation to generate ATP (54). However, in the relatively glucose-poor environment of the insect midgut, they have to utilize the amino acid proline as a substrate. Proline is metabolized in the mitochondrion by a subset of enzymes of the tricarboxylic acid cycle (where it is converted to 2-ketoglutarate and subsequently to succinate), coupled with oxidative phosphorylation (55). Consequently, the mitochondrion is one of the most developmentally affected organelles during this transition. In turn, the procyclic (insect midgut) form parasites have a highly branched and metabolically active mitochondrion, whereas in the bloodstream (mammalian) form, the mitochondrion is significantly reduced and inactive in terms of oxidative phosphorylation (56).
We found the mitochondrial mRNAs of T. brucei to be abundant in U-rich cognate codons of Q-tRNAs. This finding in itself is not surprising, given that most mitochondrial genomes display AT bias at the third position of the codons. Added to this is the unique RNA editing mechanism present in the mitochondria of kinetoplastids, which involves extensive U insertions or deletions, resulting mostly in U-rich mRNAs (57,58). Perhaps the more intriguing consequence of these observations is the need to efficiently and rapidly decode the U-rich codons to meet the translational demands. However, the situation in T. brucei is even more complicated, the mitochondrial genome of T. brucei does not encode any tRNAs, hence they must import the whole set from the cytoplasm (43). We observed that the mitochondrial fraction of GUN-tRNAs is almost completely modified with queuosine. There could be various underlying mechanisms causing this. We found that in vitro, the mitochondria import Q-modified tRNAs more readily, than the unmodified ones. These results suggest that the mitochondria have a mechanism to either preferentially import Q-modified tRNAs and/or exclude the unmodified ones. Significantly, the steady-state levels of Q-modified and unmodified tRNAs in the cytoplasm is nearly equal (∼55% modified and 45% unmodified, see Figure 5A and C), thus the preferential import of Q-modified tRNA is not merely a reflection of the relative concentrations of the modified tRNA in the cytoplasm. Alternatively, it has been reported that the elongation factor 1a (EF-1a) is a necessary cofactor for the import, selectively binding aminoacylated tRNAs and delivering them to the mitochondrial outer membrane ready to be imported (59). However, our results with the in vitro import assay suggest that preferential import of Q-containing tRNAs is independent of any cytosolic factors. EF-1a independent import of unspliced tRNATyr has also been reported in Leishmania tarentolae (60). It is also possible that the determinant for the selective import is not queuosine itself, but another modification dependent on Q. It is now becoming increasingly clear that not only the presence of a set of certain modifications on the tRNA is important, but their sequence of appearance is also most critical. For example, the presence of Q in tRNAAsp increases the methylation of C38 (m5C) adjacent to the anticodon sequence in Schizosaccharomyces pombe (61). Although this methylation is absent in T. brucei (62), one cannot rule out any other modification being subject to change depending on queuosinylation. Alternatively, or simultaneously, the Q modified tRNAs could have differential turn over in the mitochondria, leading to their accumulation. A recent study has shown that presence of Q in the anticodon of tRNAs protects them against the anticodon nuclease angiogenin in human cells (63). It would require extensive further experimentation to verify either theory.
We see significant impairment of mitochondrial protein synthesis in absence of Q. Respiratory complex IV (cIV) in trypanosomatids, is composed of three large mitochondrially encoded subunits (COX I-III) and 11–15 smaller nuclearly encoded subunits (64). Thus, impairment of mitochondrial translation would adversely affect the assembly of the complex, and result in fewer functionally active complexes. Indeed, we observe a significant decrease in the activity of cIV, in cells lacking Q modification. These effects might become critical in stress conditions, inside the challenging environment of the hosts of T. brucei.
The combined effect of codon usage and modification levels on mitochondrial protein synthesis, further strengthens our hypothesis that Q plays a critical role in decoding U-rich codons. Although mechanisms of cytosolic and mitochondrial translation differ, parallels could be drawn to certain extent. In the absence of Q, translation might be stalled at NAU codons, leading to premature terminations and the induction of unfolded protein response. Further studies inside the mammalian host of T. brucei would illuminate the physiological relevance of queuosine modification in the face of unique translational demands. Given its parasitic lifestyle, between different hosts that could lead to differential availability of queuine, a complex picture of translation regulation by environmental as well as intrinsic factors begins to form.
In conclusion, we show here that the unique nuclear localization of the TbTGT enzyme may provide for more efficient crosstalk between tRNA trafficking and modification status, while ensuring that fully processed and modified tRNAs are supplied to the mitochondria. Moreover, the mitochondria of T. brucei preferentially import Q-modified tRNAs, and also use them most advantageously, to translate the U-rich mRNAs. Taken together, our results provide a unique example of the interdependency of codon usage and wobble modifications, and their combined regulatory effect on translation in T. brucei; all dictated by tRNA intracellular transport dynamics.
Supplementary Material
ACKNOWLEDGEMENTS
We thank G. Klebe, for the gift of synthetic queuine and V. de Crécy-Lagard for the TGT−/− E. coli K12 strain. We thank A. Zíková for useful suggestions concerning mitochondrial physiology of trypanosomes. We would like to thank all members of the Alfonzo and Paris laboratories for their comments and suggestions.
Contributor Information
Sneha Kulkarni, Institute of Parasitology, Biology Centre, Czech Academy of Sciences, České Budějovice, Czech Republic; Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic.
Mary Anne T Rubio, Institute of Parasitology, Biology Centre, Czech Academy of Sciences, České Budějovice, Czech Republic; Department of Microbiology and The Center for RNA Biology, The Ohio State University, Columbus, OH, USA.
Eva Hegedűsová, Institute of Parasitology, Biology Centre, Czech Academy of Sciences, České Budějovice, Czech Republic.
Robert L Ross, Metabolomics Mass Spectrometry Core, Department of Cancer Biology, University of Cincinnati, Cincinnati, OH, USA.
Patrick A Limbach, Rieveschl Laboratories for Mass Spectrometry, Department of Chemistry, University of Cincinnati, Cincinnati, OH, USA.
Juan D Alfonzo, Department of Microbiology and The Center for RNA Biology, The Ohio State University, Columbus, OH, USA.
Zdeněk Paris, Institute of Parasitology, Biology Centre, Czech Academy of Sciences, České Budějovice, Czech Republic; Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM058843 to P.A.L. and GM132254, GM084065-11 to J.D.A.]; Czech Science Foundation [15-21450Y, 20-11585S to Z.P.]; ERDF/ESF project Centre for Research of Pathogenicity and Virulence of Parasites [CZ.02.1.01/0.0/0.0/16 019/0000759 to Z.P.]; University of South Bohemia [036/2017/P to S.K.]. Funding for open access charge: Biology Centre, Czech Academy of Sciences, České Budějovice, Czech Republic.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grosjean H., Sprinzl M., Steinberg S.. Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie. 1995; 77:139–141. [DOI] [PubMed] [Google Scholar]
- 2.Phizicky E.M., Alfonzo J.D.. Do all modifications benefit all tRNAs. FEBS Lett. 2010; 584:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinayak M., Pathak C.. Queuosine modification of tRNA: its divergent role in cellular machinery. Biosci. Rep. 2010; 30:135–148. [DOI] [PubMed] [Google Scholar]
- 4.Curnow A.W., Garcia G.A.. tRNA-guanine transglycosylase from Escherichia coli. J. Biol. Chem. 1995; 270:17264–17267. [DOI] [PubMed] [Google Scholar]
- 5.Gregson J.M., Crain P.F., Edmonds C.G., Gupta R., Hashizume T., Phillipson D.W., Mccloskey J.A.. Structure of the archaeal transfer RNA nucleoside G*-15 (2-amino-4, 7-dihydro-4-oxo-7-ß-D-ribofuranosyl-1H-pyrrolo[2, 3-d]pyrimidine-5-carboximidamide (Archaeosine)). J. Biol. Chem. 1993; 268:10076–10086. [PubMed] [Google Scholar]
- 6.Iwata-Reuyl D.Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorg. Chem. 2003; 31:24–43. [DOI] [PubMed] [Google Scholar]
- 7.Zallot R., Brochier-Armanet C., Gaston K.W., Forouhar F., Limbach P.A., Hunt J.F., de Crécy-Lagard V.. Plant, animal, and fungal micronutrient queuosine is salvaged by members of the DUF2419 protein family. ACS Chem. Biol. 2014; 9:1812–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y.C., Kelly V.P., Stachura S.V, Garcia G.A.. Characterization of the human tRNA-guanine transglycosylase: Confirmation of the heterodimeric subunit structure. RNA. 2010; 16:958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris R.C., Brown K.G., Elliott M.S.. The effect of queuosine on tRNA structure and function. J. Biomol. Struct. Dyn. 1999; 16:757–774. [DOI] [PubMed] [Google Scholar]
- 10.Meier F., Suter B., Grosjean H., Keith G., Kubli E.. Queuosine modification of the wobble base in tRNA-His influences ‘in vivo’ decoding properties. EMBO J. 1985; 4:823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaborske J.M., Bauer DuMont V.L., Wallace E.W.J., Pan T., Aquadro C.F., Drummond D.A.. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol. 2014; 12:e1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiari Y., Dion K., Colborn J., Parmakelis A., Powel J.R.. On the possible role of tRNA base modifications in the evolution of codon usage: queuosine and Drosophila. J. Mol. Evol. 2010; 70:339–345. [DOI] [PubMed] [Google Scholar]
- 13.Tuorto F., Legrand C., Cirzi C., Federico G., Liebers R., Müller M., Ehrenhofer-Murray A.E., Dittmar G., Gröne H.-J., Lyko F.. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 2018; 37:e99777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer S.Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol. Biochem. Parasitol. 2012; 181:61–72. [DOI] [PubMed] [Google Scholar]
- 15.Kelly S., Kramer S., Schwede A., Maini P.K., Gull K., Carrington M.. Genome organization is a major component of gene expression control in response to stress and during the cell division cycle in trypanosomes. Open Biol. 2012; 2:120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenn K., Matthews K.R.. The cell biology of Trypanosoma brucei differentiation. Curr. Opin. Microbiol. 2007; 10:539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubio M.A.T., Alfonzo J.D.. Editing and modification in trypanosomatids: the reshaping of non-coding RNAs. Top. Curr. Gen. 2005; 12:71–86. [Google Scholar]
- 18.Kaneko T., Suzuki T., Kapushoc S.T., Rubio M.A., Ghazvini J., Watanabe K., Simpson L., Suzuki T.. Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: Implication for tRNA sorting mechanism. EMBO J. 2003; 22:657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paris Z., Rubio M.A.T., Lukes J., Alfonzo J.D.. Mitochondrial tRNA import in Trypanosoma brucei is independent of thiolation and the Rieske protein. RNA. 2009; 15:1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esseiva A.C., Naguleswaran A., Hemphill A., Schneider A.. Mitochondrial tRNA import in Toxoplasma gondii. J. Biol. Chem. 2004; 279:42363–42368. [DOI] [PubMed] [Google Scholar]
- 21.Wirtz E., Leal S., Ochatt C., Cross G.A.M.. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999; 99:89–101. [DOI] [PubMed] [Google Scholar]
- 22.Brun R., Schonenberger M.. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979; 36:289–292. [PubMed] [Google Scholar]
- 23.Oberholzer M., Morand S., Kunz S., Seebeck T.. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 2006; 145:117–120. [DOI] [PubMed] [Google Scholar]
- 24.Dean S., Sunter J., Wheeler R.J., Hodkinson I., Gluenz E., Gull K.. A toolkit enabling efficient, scalable and reproducible gene tagging in trypanosomatids. Open Biol. 2015; 5:140197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kessler A.C., Kulkarni S.S., Paulines M.J., Rubio M.A.T., Limbach P.A., Paris Z., Alfonzo J.D.. Retrograde nuclear transport from the cytoplasm is required for tRNATyr maturation in T. brucei. RNA Biol. 2017; 6286:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igloi G.L., Kössel H.. Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Application of polyacrylamide containing a covalently bound boronic acid. Nucleic. Acids. Res. 1985; 13:6881–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stothard P.The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques. 2000; 28:1102–1104. [DOI] [PubMed] [Google Scholar]
- 28.Alexaki A., Kames J., Holcomb D.D., Athey J., Santana-Quintero L.V., Lam P.V.N., Hamasaki-Katagiri N., Osipova E., Simonyan V., Bar H.et al.. Codon and codon-pair usage tables (CoCoPUTs): facilitating genetic variation analyses and recombinant gene design. J. Mol. Biol. 2019; 431:2434–2441. [DOI] [PubMed] [Google Scholar]
- 29.Athey J., Alexaki A., Osipova E., Rostovtsev A., Santana-Quintero L.V., Katneni U., Simonyan V., Kimchi-Sarfaty C.. A new and updated resource for codon usage tables. BMC Bioinformatics. 2017; 18:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapushoc S.T., Alfonzo J.D., Rubio M.A.T., Simpson L.. End processing precedes mitochondrial importation and editing of tRNAs in Leishmania tarentolae. J. Biol. Chem. 2000; 275:37907–37914. [DOI] [PubMed] [Google Scholar]
- 31.Crain P.F., Alfonzo J.D., Rozenski J., Kapushoc S.T., McCloskey J.A., Simpson L.. Modification of the universally unmodified uridine-33 in a mitochondria-imported edited tRNA and the role of the anticodon arm structure on editing efficiency. RNA. 2002; 8:752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horváth A., Berry E., Maslov D.A.. Translation of the edited mRNA for cytochrome b in trypanosome mitochondria. Science. 2000; 287:1639–1640. [DOI] [PubMed] [Google Scholar]
- 33.Horváth A., Horáková E., Dunajčíková P., Verner Z., Pravdová E., lapetová I., Cuninková L., Lukeš J.. Downregulation of the nuclear-encoded subunits of the complexes III and IV disrupts their respective complexes but not complex I in procyclic Trypanosoma brucei. Mol. Microbiol. 2005; 58:116–130. [DOI] [PubMed] [Google Scholar]
- 34.Ross R., Cao X., Yu N., Limbach P.A.. Sequencemapping of transfer RNA chemical modifications by liquid chromatography tandemmass spectrometry. Methods. 2016; 107:73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boland C., Hayes P., Santa-Maria I., Nishimura S., Kelly V.P.. Queuosine formation in eukaryotic tRNA occurs via a mitochondria-localized heteromeric transglycosylase. J. Biol. Chem. 2009; 284:18218–18227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B.P., Carrington M., Depledge D.P., Fischer S., Gajria B., Gao X.et al.. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic. Acids. Res. 2010; 38:D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romier C., Reuter K., Suck D., Ficner R.. Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange. EMBO J. 1996; 15:2850–2857. [PMC free article] [PubMed] [Google Scholar]
- 38.Xie W., Liu X., Huang R.H.. Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate. Nat. Struct. Biol. 2003; 10:781–788. [DOI] [PubMed] [Google Scholar]
- 39.Howes N.K., Farkas W.R.. Studies with a homogeneous enzyme from rabbit erythrocytes catalyzing the insertion of guanine into tRNA. J. Biol. Chem. 1978; 253:9082–9087. [PubMed] [Google Scholar]
- 40.Slany R.K., Müller S.O.. tRNA-guanine transglycosylase from bovine liver. Eur. J. Biochem. 1995; 230:221–228. [DOI] [PubMed] [Google Scholar]
- 41.Hannah R., Sherf B.A., Navarro S.L., Hannah R.R., Wood K.V.. Dual-luciferase TM reporter assay: an advanced co-reporter technology integrating firefly and renilla luciferase assays. Promega notes. 1996; 57:2–8. [Google Scholar]
- 42.Bohnsack M.T., Sloan K.E.. The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. 2018; 75:241–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mottram J.C., Bell S.D., Nelson R.G., Barry J.D.. tRNAs of Trypanosoma brucei. J. Biol. Chem. 1991; 266:18313–18317. [PubMed] [Google Scholar]
- 44.Aphasizheva I., Alfonzo J., Carnes J., Cestari I., Cruz-Reyes J., Göringer H.U., Hajduk S., Lukeš J., Madison-Antenucci S., Maslov D.A.et al.. Lexis and grammar of mitochondrial RNA processing in trypanosomes. Trends Parasitol. 2020; 36:337–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramrath D.J.F., Niemann M., Leibundgut M., Bieri P., Prange C., Horn E.K., Leitner A., Boehringer D., Schneider A., Ban N.. Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes. Science. 2018; 362:eaau7735. [DOI] [PubMed] [Google Scholar]
- 46.Škodová-Sveráková I., Horváth A., Maslov D.A.. Identification of the mitochondrially encoded subunit 6 of F1FO ATPase in Trypanosoma brucei. Mol. Biochem. Parasitol. 2015; 201:135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aphasizheva I., Maslov D., Wang X., Huang L., Aphasizhev R.. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol. Cell. 2011; 42:106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dedon P.C., Begley T.J.. A system of RNA modifications and biased codon use controls cellular stress response at the level of translation. Chem. Res. Toxicol. 2014; 27:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaur R., Björk G.R., Tuck S., Varshney U.. Diet-dependent depletion of queuosine in tRNAs in Caenorhabditis elegans does not lead to a developmental block. J. Biosci. 2007; 32:747–754. [DOI] [PubMed] [Google Scholar]
- 50.Kirtland G.M., Morris T.D., Moore P.H., O’Brian J.J., Edmonds C.G., McCloskey J.A., Katze J.R.. Novel salvage of queuine from queuosine and absence of queuine synthesis in Chlorella pyrenoidosa and Chlamydomonas reinhardtii. J. Bacteriol. 1988; 170:5633–5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siard T.J., Jacobson K.B., Farkas W.R.. Queuine metabolism and cadmium toxicity in Drosophila melanogaster. Biofactors. 1991; 3:41–47. [PubMed] [Google Scholar]
- 52.Schachner E., Aschhoff H.J., Kersten H.. Specific changes in lactate levels, lactate dehydrogenase patterns and cytochrome b559 in Dictyostelium discoideum caused by queuine. Eur. J. Biochem. 1984; 139:481–487. [DOI] [PubMed] [Google Scholar]
- 53.Matthews K.R.The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 2005; 118:283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bringaud F., Rivière L., Coustou V.. Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol. Biochem. Parasitol. 2006; 149:1–9. [DOI] [PubMed] [Google Scholar]
- 55.Coustou V., Biran M., Breton M., Guegan F., Rivière L., Plazolles N., Nolan D., Barrett M.P., Franconi J.M., Bringaud F.. Glucose-induced remodeling of intermediary and energy metabolism in procyclic Trypanosoma brucei. J. Biol. Chem. 2008; 283:16343–16354. [DOI] [PubMed] [Google Scholar]
- 56.Verner Z., Basu S., Benz C., Dixit S., Dobáková E., Faktorová D., Hashimi H., Horáková E., Huang Z., Paris Z.et al.. Malleable mitochondrion of Trypanosoma brucei. Int. Rev. Cell Mol. Biol. 2015; 315:73–151. [DOI] [PubMed] [Google Scholar]
- 57.Benne R.RNA-editing in trypanosome mitochondria. BBA - Gene Struct. Expr. 1989; 1007:131–139. [DOI] [PubMed] [Google Scholar]
- 58.Simpson L., Shaw J.. RNA editing and the mitochondrial cryptogenes of kinetoplastid protozoa. Cell. 1989; 57:355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bouzaidi-Tiali N., Aeby E., Charrière F., Pusnik M., Schneider A.. Elongation factor 1a mediates the specificity of mitochondrial tRNA import in T. brucei. EMBO J. 2007; 26:4302–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sbicego S., Nabholz C.E., Hauser R., Blum B., Schneider A.. In vivo import of unspliced tRNA(Tyr) containing synthetic introns of variable length into mitochondria of Leishmania tarentolae. Nucleic Acids Res. 1998; 26:5251–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ehrenhofer-Murray A.E.Cross-talk between Dnmt2-dependent tRNA methylation and queuosine modification. Biomolecules. 2017; 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Militello K.T., Chen L.M., Ackerman S.E., Mandarano A.H., Valentine E.L.. A map of 5-methylcytosine residues in Trypanosoma brucei tRNA revealed by sodium bisulfite sequencing. Mol. Biochem. Parasitol. 2014; 193:122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X., Matuszek Z., Huang Y., Parisien M., Dai Q., Clark W., Schwartz M.H., Pan T.. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA. 2018; 24:1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zíková A., Panigrahi A.K., Uboldi A.D., Dalley R.A., Handman E., Stuart K.. Structural and functional association of Trypanosoma brucei MIX protein with cytochrome c oxidase complex. Eukaryot. Cell. 2008; 7:1994–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.