Abstract
Long-read whole-genome sequencing analysis of DNA methylation would provide useful information on the chromosomal context of gene expression regulation. Here we describe the development of a method that improves the read length generated by using the bisulfite-sequencing-based approach. In this method, we combined recently developed enzymatic base conversion, where an unmethylated cytosine (C) should be converted to thymine (T), with nanopore sequencing. After methylation-sensitive base conversion, the sequencing library was constructed using long-range polymerase chain reaction. This type of analysis is possible using a minimum of 1 ng genomic DNA, and an N50 read length of 3.4–7.6 kb is achieved. To analyze the produced data, which contained a substantial number of base mismatches due to sequence conversion and an inaccurate base read of the nanopore sequencing, a new analytical pipeline was constructed. To demonstrate the performance of long-read methylation sequencing, breast cancer cell lines and clinical specimens were subjected to analysis, which revealed the chromosomal methylation context of key cancer-related genes, allele-specific methylated genes, and repetitive or deletion regions. This method should convert the intractable specimens for which the amount of available genomic DNA is limited to the tractable targets.
INTRODUCTION
In mammals, cytosine (C) in CpG dinucleotides is predominantly modified by DNA methyltransferases to 5-methylcytosine (mC) (1). C methylation plays the most pivotal role in epigenetic regulation of gene expressions. Active promoters are supposed to be mostly unmethylated, but gene expression repression is often mediated by mC. DNA-methylation-mediated gene expression regulation plays various roles in many aspects of biological processes, such as normal development and disease progression. A well-studied case involves the imprinting of the maternal or paternal genes in normal cells, where the gene locus, an array of gene loci, or even the whole X chromosome is collectively methylated and thus silenced. In cancers, genomic DNA (gDNA) is supposed to be generally hypomethylated, whereas some specific regions harboring tumor suppressor gene loci are often hyper-methylated (2). In either normal or disease circumstances, DNA methylation is supposed to occur in a relatively wide genomic region. However, our current knowledge of DNA methylation remains a patchwork of fragmented information obtained from short-read sequencing. Comprehensive elucidation of DNA methylation on chromosomal-level allelic backgrounds has yet to be attained.
This is partly due to the limitations of commonly employed analytical methods for DNA methylation. Detection of mC from unmethylated C is commonly achieved through bisulfite sequencing, which involves converting unmethylated C to uracil (U) via a chemical reaction of the bisulfite treatment (3). As this reaction is conducted in a chemically harsh condition, a substantial portion of DNA is fragmented and degraded. Therefore, it is difficult to obtain and analyze intact DNA fragments that are sufficiently long for subsequent long-read sequencing (4). It is also suggested that a severe bias is introduced in the representation of gDNA during the bisulfite reaction process, which skews the equal distribution of the generated sequence information (5). To address these problems, relatively non-destructive methods for base conversion have recently been developed (6,7). The novel enzymatic methyl sequencing method, named ‘EMseq,’ utilizes oxidation of the TET enzyme from methylcytosine (mC) and hydroxymethylcytosine (hmC) to carboxyl cytosine (caC) for protection, and deamination by the APOBEC enzyme for conversion from unmethylated C to U (6). Subsequently, unmethylated C is sequenced as thymine (T), in a process that is similar to bisulfite sequencing. Since this is an enzymatic reaction, which is conducted in a much milder condition, fragmentation, and representation bias of material DNA should be less severe. In fact, enzymatic conversion of unmethylated cytosines allows for the production of long DNA fragments that are unattainable through bisulfite conversion (6,8). While the standard methods of WGBS and EMseq are unable to distinguish between mC and hmC, the modified methods can detect either mC or hmC (8,9).
We wondered if the EMseq approach may enable long-read whole-genome analysis of DNA methylation. For the purpose of long-read sequencing, we employed a nanopore sequencer. A nanopore sequencer can potentially read DNA molecules that are longer than 100 kb (10). It has also been reported that modifications in DNA and RNA can be detected from the electric signal patterns observed for mC of DNA and RNA, hmC of DNA, and unmethylated C (11,12). In fact, several tools based on machine learning have been developed (13–15). However, detection accuracy of mC is usually less than 90%, which hinders precise interpretation of the obtained data (14). Moreover, this direct detection method requires the gDNA of 0.5–1 μg as a starting material, which is not practically available for many biological targets. Recently, the efficacy of lrTAPS, which combines a base-conversion method of mC or hmC and long-read sequencing, and LR-EMseq, which combines EMseq and long-read sequencing, was studied (8,16). LrTAPS and LR-EMseq employed a locus-specific polymerase chain reaction (PCR) and did not apply whole-genome analysis.
Here, we describe the development of a new method for whole-genome methylome analysis using long-read sequencing, which could be performed from 1 to 100 ng of input DNA. We combined the nanopore sequencing and EMseq methods, in which base-converted DNA through an enzymatic reaction, EM, was sequenced using the nanopore sequencer (designated as nanoEM hereafter). To analyze the nanoEM data, a bioinformatics pipeline was required and, thus, was developed, starting from long-read mapping of the mostly C–T converted bases from generally error-prone data. For validation and comparison purposes, we also conducted whole-genome sequencing (WGS) using PromethION, from which methylation was called by directly resolving electric signals, whole-genome bisulfite sequencing (WGBS) and EMseq using a short-read Illumina sequencer. To demonstrate the performance of the developed method, we applied nanoEM analysis for methylation analysis of breast cancers. We first analyzed two cell lines, MDA-MB-231 (MB231) and BT474. The MB231 cell line is derived from a so-called triple-negative-type breast cancer, in which none of the estrogen receptor (ER), progesterone receptor (PGR), or human epidermal growth factor receptor 2 (HER2) was expressed. The BT-474 cell line is derived from a luminal B breast cancer, in which PGR, HER2, and ER are expressed. These cell lines were selected to represent cells of relatively high and low methylation statuses for MB231 and BT474 cells. Later, we further demonstrate its performance using two clinical breast cancer specimens.
MATERIALS AND METHODS
Cultivation of breast cancer cell lines
MDA-MD-231 (ATCC, HTB-20) (17) and BT-474 (ATCC, HTB-20) (18) were cultured in L15 medium and RPMI medium (FUJIFILM Wako Pure Chemical) containing l-glutamine supplemented with 10% FBS (Corning) and 1× Antibiotic-Antimycotic (Thermo Fisher Scientific), respectively.
Clinical specimens
Informed consent was obtained from all patients. This study was approved by the Clinical Ethics Committee of St. Marianna University School of Medicine (IRB#: 2297-i103) and the Research Ethics Committee of the University of Tokyo (IRB#: 18-235). The fresh frozen clinical specimens of breast cancer and matched normal tissues that were used in this study were obtained from St. Marianna University School of Medicine Hospital. Cases 7 and 8 were ‘ER-negative, PGR-negative, and HER2-positive,’ and ‘ER-positive, PGR-positive, and HER2-negative’ breast cancer, respectively, as diagnosed from a histopathological viewpoint. The specimen for Case 7 was identical with the specimen that was used in our previous study (19).
Extraction of gDNA
We extracted gDNA from the cultured cells and the clinical specimens using the MagAttract HMW DNA Kit (Qiagen) or smart DNA prep (a) (Analytik Jena) according to the manufacturer's instructions. Extracted gDNA was quantified using Genomic DNA ScreenTape assay and a 2200 TapeStation system (Agilent Technologies). We only used gDNA for which the DNA integrity number (DIN) was ∼9 except for the tumor tissue of Case 8, for which the DIN was 6.6.
Enzymatic methyl sequencing using a short-read sequencer
gDNA of the cell lines was fragmented using an M220 Focused-ultrasonicator (Covaris) with the following settings: 50 W peak incident power, 20% duty factor, 200 cycles per burst, and 60 s treatment time. Then, 200 ng of fragmented DNA was applied in the preparation of a library for EMseq. The EMseq libraries were prepared using the EMseq Kit (NEBNext Enzymatic Methyl-seq Kit; New England BioLabs) according to the manufacturer's instructions. Briefly, the end of the fragmented DNA was repaired and dA-tailed. An EMseq adapter was ligated to the end-prepped DNA. Then, mC and hmC of the ligated DNA were oxidized using the TET2 enzyme to protect against the deamination reaction. To convert umC to U, umC of the oxidized DNA was deaminated by the APOBEC enzyme. The deaminated DNA was amplified by four cycles of PCR using Q5U Master Mix and primers for the Illumina sequencer. The amplified libraries were purified using sample purification beads according to the purification protocol for longer insert sizes. The libraries were quantified using a High Sensitivity DNA Kit and Bioanalyzer (Agilent Technologies). Pair-end sequencing of 150 bp was performed using the NovaSeq 6000 system (Illumina).
WGBS using a short-read sequencer
gDNA of the cell lines was fragmented using an M220 Focused-ultrasonicator (Covaris) with the following settings: 50 W peak incident power, 200% duty factor, 200 cycles per burst, and 75 s treatment time. The WGBS library was prepared from 200 ng of fragmented DNA, utilizing the EMseq Kit for end prep, adapter ligation, and PCR as well as the EZ DNA Methylation-Gold Kit (Zymo Research) for bisulfite conversion. Briefly, end prep and adapter ligation were performed according to the protocol of the EMseq Kit. The adapter-ligated DNA was treated by bisulfite and deaminated according to the protocol of the EZ DNA Methylation-Gold Kit. The bisulfite-converted DNA was amplified in four cycles of PCR using Q5U Master Mix and primers for the Illumina sequencing. The amplified libraries were purified using sample purification beads according to the purification protocol for standard insert sizes. The libraries were quantified using a High Sensitivity DNA Kit and Bioanalyzer (Agilent Technologies). Pair-end sequencing of 150 bp were performed using the NovaSeq 6000 system (Illumina).
WGS using a short-read sequencer
gDNA of clinical samples was fragmented by focused-ultrasonicator M220 (Covaris) with the following settings: 50 W peak incident power, 20% duty factor, 200 cycle per burst, and 75 s treatment time. The WGS library was prepared from 100 ng of the fragmented DNA, using TruSeq Nano DNA Library Prep Kit (Illumina) according to the manufacturer's instructions. The libraries were quantified using High Sensitivity DNA Kit and Bioanalyzer (Agilent Technologies). Pair-end sequencing of 150 bp were performed using NovaSeq6000 system (Illumina).
WGS using a nanopore sequencer
WGS of the two cell lines and clinical samples using PromethION was performed as described previously (10). Then, 1–1.5 μg of the high-molecular-weight gDNA was used to prepare the PromethION library. The libraries were prepared using a Ligation Sequencing Kit (SQK-LSK109, Oxford Nanopore Technologies) according to the manufacturer's protocol. The prepared libraries were sequenced using PromethION flow cells (FLO-PRO002, Oxford Nanopore Technologies). The sequencing data were base called using Guppy.
Nanopore enzymatic methyl sequencing (nanoEM)
gDNA was fragmentated using g-TUBE (Covaris) by double centrifugation at 4700 g for 1 min. Then, 1–100 ng of fragmented DNA was oxidized and deaminated in four reaction tubes using the NEBNext Enzymatic Methyl-seq Kit (New England BioLabs) following the manufacturer's instructions. The ends of the fragmented DNA were repaired and dA-tailed. An EMseq adapter was ligated to the end-prepped DNA. Then, the mC and hmC of the ligated DNA were oxidized using the TET2 enzyme to protect against the deamination reaction. To convert umC to U, the umC of the oxidized DNA was deaminated by the APOBEC enzyme. For beads purification after APOBEC conversion, the deaminated DNA was eluted by nuclease-free water. The eluted DNA was amplified using EMseq Index primers of the NEBNext Enzymatic Methyl-seq Kit and KOD Multi & Epi (TOYOBO) (1 cycle of 2 min at 94°C; 16 cycles of 10 s at 98°C, 30 s at 62°C, and 10 min at 68°C; 1 cycle of 10 min at 68°C, held at 4°C) or KOD One PCR Master Mix (TOYOBO) (12–21 cycles of 10 s at 98°C, 5 s at 57°C, and 15 min at 68°C, held at 4°C). The amplified DNA was purified using 1× volume of AMPureXP beads (Beckman Coulter) or a DNA Clean & Concentrator-5 Kit (Zymo). Size selection of the purified DNA was performed using 0.82–0.9× volume of ProNex Size-Selective Chemistry using the ProNex Size-Selective DNA Purification System (Promega).
From 200 to 1000 ng of the size-selected DNA, the library for PromethION (Oxford Nanopore Technologies) was prepared with a 1D Ligation Sequencing Kit (SQK-LSK109, Oxford Nanopore Technologies) following the manufacturer's instructions. Then, 100–300 ng of the prepared libraries was inputted to PromethION. The sequencing data was base called using Guppy.
SNP calling
For single nucleotide polymorphism (SNP) calling, we utilized public WGS data from the Cancer Cell Line Encyclopedia (CCLE) (20). The raw sequence data were obtained from the Sequence Read Archive with the following accession numbers: SRR8652107 for MDA-MB-231 and SRR8639205 for BT-474. The adapter sequence and bases with low quality were trimmed using Trim Galore v 0.6.0 (https://github.com/FelixKrueger/TrimGalore) with the following parameters: ‘–quality 20 –phred33 –stringency 3 –length 20 –illumina –paired –trim1.’ The trimmed reads were aligned to a human reference genome GRCh38.p12 using BWA v0.7.17-r1188 with the default parameters (21). The reads derived from PCR duplicates were removed using MarkDuplicates of Picard tools v2.0.1 and samtools v1.9. SNPs were called using HaplotypeCaller of GATK v4.0.12.0. We conducted base quality score recalibration using BaseRecalibrator of GATK with dbSNP, mills indel, and 1000 Genomes Project Phase I indel calls as known sites. We also performed base quality score recalibration using ApplyBQSR of GATK.
Data analysis of WGBS and EMseq using a short-read sequencer
The sequence reads of WGBS and EMseq were adapter-trimmed using Trim Galore v0.5.0 (https://github.com/FelixKrueger/TrimGalore) with the default parameters. The trimmed reads were aligned to GRCh38.p12 using Bismark v0.22.1 with the following parameters: ‘–multicore 10 -X 1000.’ The reads that originated from PCR duplicates were removed using deduplicate_bismark with the default parameter (22). The methylation information was extracted with a bismark_methylation_extractor using the following parameters: ‘–multicore 10 –ignore 11 –ignore_3prime 1 –ignore_r2 5 –ignore_3prime_r2 2 –gzip –bedGraph –buffer_size 90G.’
Methylation calling from the nanoEM dataset
We mapped nanoEM data to reference the genome by using the bismark-like method (22). First, we converted all C/Gs in the human reference genome to T/As. Second, we converted all C/Gs in the sequencing data to G/As. Third, we mapped the converted sequencing data to the converted reference genome by using minimap2 (version 2.16-r922) with the ‘map-ont’ option (23). Finally, we chose the best unique alignment by mapping the quality. We detected methylated or unmethylated C by using the sambamba mpileup command (version 0.7.1, default parameters) with CpG sites extracted from the reference genome (24). The scripts and the detailed explanations of our pipeline used for data analyses of nanoEM is available in a GitHub repository at https://github.com/yos-sk/nanoEM.
Methylation calling from the PromethION WGS dataset
We mapped PromethION WGS data to a human reference genome, GRCh38.p12, by using minimap2 (version 2.16-r922). Then, we extracted the mapping results using a threshold of mapping quality greater than 20. We indexed fastq data with fast5 data and called CpG methylation by using the call-methylation function of nanopolish version 0.11.1 (https://github.com/jts/nanopolish) with a pretrained 6-mer DNA model for R9.4 chemistry. Then, we calculated the frequency of methylated C.
RESULTS AND DISCUSSION
Comparison between EMseq and WGBS
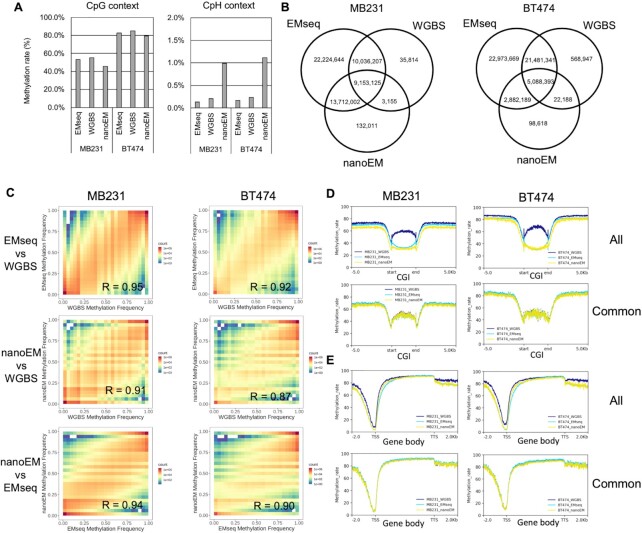
First, to evaluate the EM conversion, we conducted whole-genome EM Sequencing using an Illumina sequencer. For the purpose of comparison, we also performed WGBS using the same material. For this analysis, we used the two breast cancer cell lines, MDA-MB-231 (MB231) and BT-474 (BT474). An average of 473 741 078 read pairs, which is at ×47 coverage of the human genome, was obtained (Supplementary Table S1). Overall, the methylation rates in the CpG context as measured by EMseq and WGBS were 53.3% and 55.1% in MB231 and 82.5% and 84.9% in BT474, respectively (Figure 1A). The observed methylation rates were consistent between EMseq and WGBS. The methylation rates (0.13–0.23%) at the CpH sites (H = A/C/T), which are believed to be rare in most mammalian cells except for neuronal, embryonic, and germ cells (25), were detected in both analyses, validating the accuracy of conversion in both methods (Figure 1A). Then, we compared the number of CpGs that were covered by five reads or more. Out of the total of 58,351,766 CpG sites in the human genome, except for chromosome Y, 19 228 301–27 160 869 (33.0–46.5%) of the CpGs were covered by WGBS, but 52 425 592–55 125 978 (89.8–94.5%) of CpGs were detected using EMseq (Figure 1B and Supplementary Figure S1). EMseq covered more CpH sites than WGBS (Supplementary Figure S2A). To consider the dependency of coverage on the sequencing depth, we serially diluted the reads and compared the number of covered CpG sites, depending on the given sequencing depth between methods. We confirmed that, generally, EMseq showed higher CpG coverage than did WGBS (Supplementary Figure S3A). In particular, whereas WGBS showed significantly low coverage on CpG islands (CGIs) because of the possible representation bias against the GC-rich regions during library construction, EMseq showed less bias at those particularly biologically relevant sites (Supplementary Figure S3B). In a comparison between coverage and GC content, WGBS had a poorer coverage in high GC content than EMseq (Supplementary Figure S4).
Figure 1.
Comparison among EMseq, WGBS, and nanoEM. (A) The average cytosine methylation rate by CpG and CpH contexts (H = A/C/T), which were measured by EMseq, WGBS, and nanoEM. Cytosines covered by at least five reads were used for calculation. (B) Venn diagrams of the CpG sites, regardless of their modification status, that were covered by at least five reads using EMseq, WGBS, and nanoEM. (C) The correlation of the methylation rate of CpG with EMseq, WGBS, and nanoEM. Pearson's correlations are shown in the graphs. (D, E) Average methylation rate of the CpG sites covered by at least five reads around CGIs and gene bodies of active transcribed genes expressed >10 transcript per million (TPM) using each method (top). Average methylation rate of CpG sites commonly covered in EMseq, WGBS, and nanoEM (bottom). The plots were prepared using deeptools v 3.4.2 (49). CGI and gene body were scaled to 3000 and 5000 bp by deeptools, respectively. The coordinates of CGI and transcript model of Gencode v33 were obtained from the University of California–Santa Cruz (UCSC) Table Browser (50) and the website of Gencode (51). The expression values per Gencode transcripts were obtained from the website of Cancer Cell Line Encyclopedia (CCLE; https://portals.broadinstitute.org/ccle) (27).
We also compared the methylation of each CpG site between the two methods. Pearson's correlation between them was 0.95 and 0.92 in MB231 and BT474, respectively (Figure 1C). We found a highly consistent methylation rate of CpG commonly covered by EMseq and WGBS around the CGI, WGBS generally showed significantly higher methylation rates than did EMseq at CGIs, and found higher methylation rates at CGIs, an artefact of biased coverage in favor of highly methylated CGIs, than on their neighboring regions (Figure 1D and Supplementary Figure S5). These results suggests that lowly methylated CGIs are under-represented in WGBS (Supplementary Figure S3C). Previous studies have suggested a correlation between the gene expression level and the CpG methylation level, indicated by a positive correlation at the gene body and a negative correlation around the transcriptional start sites (TSSs) (1,26). Using the RNA-seq datasets of BT474 and MB231 in the Cancer Cell Line Encyclopedia (CCLE) (27), we selected highly expressed transcripts, which we defined as those having an expression value of more than 10 transcript per million (TPM). We plotted the average methylation rate of the CpG sites at gene bodies and the transcription start sites for these highly expressed genes. We observed that the methylation rate was generally high for the gene body and low for the TSS (Figure 1E). WGBS also had significantly higher methylation rates than did EMseq around the TSS where CGIs are located. This is consistent with previous concerns that an unmethylated C-rich sequence is likely to be preferentially degraded during the bisulfite reaction (5).
A total of 181 670 and 75 471 CpH sites were detected to be methylated (≥90%) by EMseq on BT474 and MB231, respectively (Supplementary Figure S2B). Most of those methylated CpH sites were also found to be methylated by WGBS (Supplementary Figure S2C). Previous studies showed that methylated CpH sites were preferentially located within gene bodies (3,28,29). We examined and found that more than half of the methylated CpH sites detected by each method were also located within gene regions (Supplementary Figure S2B), thus should precisely represent the CpH methylation sites. From these results, we concluded that EMseq should be highly compatible with WGBS with a less biased representation than should WGBS (6).
NanoEM; Long-read sequencing of the EMseq library
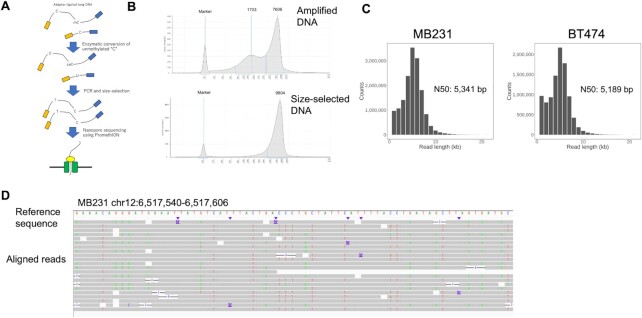
Then, we attempted to develop a new procedure for library construction, by which the EM-converted DNA templates should be subjected to long-read sequencing using a nanopore sequencer, PromethION of Oxford Nanopore Technologies (Figure 2A). To construct the sequence library, 50 ng gDNA of MB231 and BT474 at the average length of ∼10 kb, which should be a reasonable parameter for many clinical cancer specimens, was prepared. End repair, adapter ligation, and mC oxidation using the TET enzyme for protection were performed, followed by the deamination of C using APOBEC. For the subsequent PCR, we employed KOD, because KOD showed the highest capacity for amplification of longer DNA fragments among the tested PCR enzymes (Supplementary Figure S6). The size of the amplified DNA ranged between 200 and 15 000 bp (Figure 2B and Supplementary Figure S6). To eliminate short DNA fragments, size fractionation was performed using the ProNEX Size-Selective Purification System (ProNEX) (Supplementary Figure S7 for the condition optimization). After size fractionation using ProNEX, base-converted long DNA (∼600 ng) was used for the library preparation using the PromethION sequencer. Further details are shown in the Materials and Methods section. For further reduction of the input DNA amount, we also prepared a base-converted amplicon from 1 and 10 ng of DNA prepared from MB231. Enough DNA (200–400 ng) was obtained for preparation of the nanopore libraries (Supplementary Figure S8).
Figure 2.
Development of whole-genome nanoEM. (A) A schematic view of nanoEM. (B) Electropherograms of long amplicon of MB231 prepared by the Enzymatic Methyl-seq Kit and KOD Multi & Epi before and after size selection. Quantification was performed using a Genomic DNA Kit with 2200 TapeStation. (C) Sequence length distributions of nanoEM of 1D pass reads. N50 length of each datasets were shown in graphs. (D) The 1D pass reads of nanoEM on MB231 were aligned to the reference human genome using minimap2 (23). Base mismatch is shown for each read. The reads are shown in the Integrative Genomics Viewer (IGV) (52).
We conducted two runs and one run of nanoEM using 50 ng DNA for the MB231 and BT474 cells, respectively. The 1D pass reads of 15.9 and 9.9 M or 73 and 42 Gb (x24 and 14 of human genome) in total read bases, were obtained, respectively. The N50 read lengths were 5.3 and 5.2 kb, respectively (Figure 2C and Supplementary Table S2). To evaluate whether the base conversion was correctly detected by nanoEM, the reads were aligned to the human genome first by minimap2 (Figure 2D). Generally, C-to-T or G-to-A (in the reverse strand) substitutions were observed as expected.
Developing an analytical pipeline for nanoEM
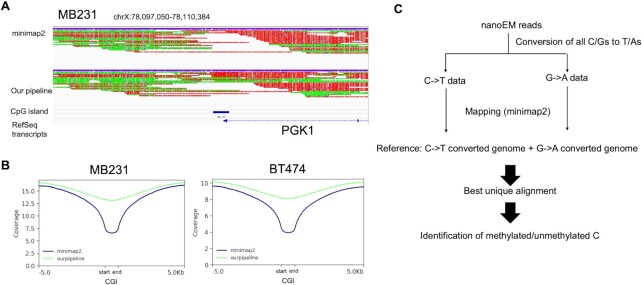
To analyze the obtained sequence data, a specially designed bioinformatics pipeline was needed. Owing to the base conversion, nanoEM reads should contain a C-to-T or G-to-A substitution for most of the bases in the original strand and complementary strand, respectively. This was not expected for the usual alignment tools for long-read sequencing, where the base call was originally error prone (Figure 3A). In particular, it was difficult to align the nanoEM reads at C-rich regions, such as the CGIs (Figure 3A and B), where the above two problems should be merged. In fact, when the reads were aligned to the human reference genome using minimap2 (23) as the default setting, a number of uncovered regions appeared. Although 81%–84% of reads were aligned, the coverage at the CGIs were frequently low (Figure 3A and B and Supplementary Table S2).
Figure 3.
Development of pipeline for analysis of long-read sequencing of base conversion. (A) Alignment result around PGK1 genes using minimap2 and our pipeline. Red and yellow colors on aligned reads represented substitution to T and A, respectively. (B) Coverage depth around CGIs when using minimap2 or our pipeline is shown. These graphs were prepared using deeptools (49). (C) Schematic view of the data analysis pipeline developed for this study.
To overcome this difficulty, we employed an algorithm in which the sequence reads should be aligned to the reference genome, which are treated with a C-to-T or G-to-A conversion (Supplementary Figure S9). As this strategy is employed by most of the standard analytical pipelines of WGBS, such as Bismark (22), we adopted the Bismark strategy for minimap2 (Figure 3C). The reference genome was firstly processed with C-to-T or G-to-A conversions. A new reference genome was prepared by combining the two reference genomes with C-to-T or G-to-A conversions. The reads with C-to-T or G-to-A conversions were aligned to the new reference genome. When both C-to-T reads and a G-to-A reads derived from the same read were aligned, we judged this as multi-mapping, and selected the best alignment according to mapping score. Those cases that gave the highest mapping quality values were selected. For details, see the Materials and Methods section. C-to-T or G-to-A SNPs in CpG sites should cause a miss-calling of methylation status. However, the frequency of miss-calling caused by these SNPs was quite limited on MB231 and BT474; only 1.0 and 1.2% of the CpG sites were overlapped with the corresponding SNPs. Therefore, we considered that the effect of SNPs on methylation calling should be limited (Supplementary Table S3).
Using the developed pipeline, 90% and 89% of nanoEM reads on MB231 and BT474, respectively, were aligned (Supplementary Table S2). The average aligned length of the reads from MB231 and BT474 were 4.6 and 4.3 kb, respectively. The mapping rate increased by 6–8% compared with that of the original minimap2 (Supplementary Table S2 and Figure 3A). Although these numbers may not be large, the recovered regions included many of biologically important regions, such as CGIs. In fact, by using this approach, a higher mapping rate and coverage of the CGI were achieved than those achieved solely using minimap2 (Figure 3B; also note that this approach may be useful for long-read WGBS sequencing using a PacBio sequencer, SMRT-BS (30)).
Evaluation of nanoEM
To further evaluate the performance of nanoEM, we compared the obtained results with those obtained from short-read WGBS and EMseq using the Illumina sequencer. The detected overall methylation rate of CpG was almost consistent between the long-read (45.7% and 79.2% on MB231 and BT474, respectively; Figure 1A) and short-read methods, although that of nanoEM in MB231 was somewhat lower than that of the short-read methods. The methylation rate of CpH in nanoEM was ∼1%, which showed that unmethylated C was nearly completely converted even when long DNA fragments were applied to enzymatic conversion, considering the error rate of nanopore sequencing. The number of CpG sites that were covered by at least five reads were 23 000 293 (39.4%) and 8 091 388 (13.9%) for MB231 and BT474, respectively, out of 58 351 766 CpG sites in the human genome (Figure 1B and Supplementary Figure S10). In MB231, for which the sequencing depth of nanoEM and WGBS was almost equal (Supplementary Tables S1 and S2), nanoEM showed slightly higher CpG coverage than did WGBS (Figure 1B). To further compare the performance of each of the methods, we examined how many of the CpG sites which overlapped repetitive elements were uniquely covered by each of the methods (Supplementary Table S4). In previous studies, it was shown that long-read sequencing can cover repetitive elements more efficiently than short read sequencing (31). NanoEM had a higher overlapping rate with long repetitive elements, such as LINE and SVA than other methods. The overall difference may also reflect the fact that nanoEM covered long repetitive regions, in which short reads had poorer coverage, due to the multiple mapping. In a comparison of coverage and GC content, although WGBS showed the most biased distribution as mentioned above, nanoEM and EMseq also showed moderate bias (Supplementary Figure S4). We found that nanoEM and EMseq showed the lowest bias among PCR-based methods. The observed overall methylation rates of the CpG sites were highly consistent between nanoEM and short-read methods (WGBS: R = 0.91 and 0.87; EMseq: 0.94 and 0.90 on MB231 and BT474, respectively) (Figure 1C). When we focused on the methylation rate around the CGIs and gene regions, we found similar methylation patterns of CpG, which were commonly covered by nanoEM, WGBS, and EMseq, for all methods (Figure 1D). As for the methylation rate of all CpG within CGIs and around TSS, nanoEM as well as EMseq showed, to some extent, a lower methylation rate than did WGBS, which may suggest that nanoEM and short-read EMseq were able to cover low methylated CGIs with higher efficiency than was WGBS able to.
Reducing the starting material
To reduce the amount of starting material, nanoEM libraries were prepared from 1 and 10 ng gDNA and were subjected to sequencing analysis. Although the number of CpG sites covered by at least five reads were comparable, between 10 and 1 ng input (11 370 815 [19.5%] and 10 988 803 [18.8%]), 1 ng input showed some biased distribution on CpG coverage (Supplementary Figures S10 and S11A). To assess the difference in PCR artifacts by the amount of starting material, we estimated the PCR duplicate rate by each condition according to the amount of starting material (Supplementary Figure S12). Starting material of 50 ng and 10 ng showed low (1.1–1.9%) and moderate (13.5%) duplicate rates, respectively, and that of 1 ng showed a high duplicate rate (46.0%). The number of PCR duplicates, which possibly originated from the same molecule was about two copies on average, was detected for 50 ng condition. It increased in proportion with a decreasing amount of the starting DNA. We considered that starting with ∼50 ng DNA, the effects of the PCR duplicates should be very small. However, with even lower input amounts, removing duplicates would be more important to provide accurate estimates of methylation rate. The observed overall methylation rates of the CpG sites with 10 ng input were highly consistent with short-read WGBS (R = 0.87), in contrast to 1 ng (R = 0.78) (Supplementary Figure S11B). Although CGI-specific low methylation within CGIs could be confirmed in both datasets, numerous PCR duplicate reads were generated in 1 ng (Supplementary Figure S11C). On the basis of these results, we concluded that nanoEM can be carried out from 1 ng of starting DNA, but >10 ng is preferable to avoid amplification bias. Vaisvila et al. described that 100 pg of genomic DNA was the minimum input for EMseq (6). But they also showed that coverage of CpG sites decreased by reducing the input amount of genomic DNA from 10 ng, and the number of CpG sites covered by at least five reads is almost zero in libraries prepared from 100 pg, of which sequencing depth was probably saturated (59–92 Gb) (6). Therefore, we considered the practical input amount for EMseq should be 10 ng, which is at the same level as nanoEM (Supplementary Figures S10–S12). As a result, we believe that even we started with a similar requirement for the material, far richer information was represented by 3–7 kb read length rather than 300–500 bp fragment (from nanoEM and EM-seq, respectively), as described later.
Comparison with the direct methylation call
We compared the results obtained from the direct methylation call from nanopore sequencing using nanopolish (13). We performed non-PCR nanopore WGS of the cell lines using PromethION. From MB231 and BT474, 5.9 and 11 M of reads, which were 51 and 101 Gb (x17 and 33 of human genome), were obtained at the N50 read length of 23 and 21 kb, respectively (Supplementary Table S5 and Supplementary Figure S13A). As expected, the length of the nanoEM reads was a quarter of that of nanopore WGS. For direct methylation calling, nanopolish, which distinguishes the electrical signals between the methylated and unmethylated C in the raw sequence data, called ‘squiggle,’ (13) was used. Nanopolish utilizes a 6-mer model of CpG motifs trained using a hidden Markov model. In this case, we considered the CpG sites giving a positive value in the log-likelihood ratio by nanopolish to be putative methylated sites. Using nanopolish analysis, the methylation rates of CpG appeared as 55.5 and 79.8% on MB231 and BT474, respectively (Supplementary Figure S13B). These numbers were quite similar to those obtained using short-read methods (Figure 1A). The number of CpG sites that covered at least five reads by nanopolish (24 174 358 [41.4%] and 44 260 634 [75.9%] on MB231 and BT474, respectively) was higher than that of nanoEM (Supplementary Figures S10 and S13C), which reflects an advantage over the PCR-free procedure. In a comparison of coverage and GC content, nanopore WGS produced the most uniform coverage by GC content as expected (Supplementary Figure S4). The correlation of the detected methylation rate of CpG sites was generally high between short-read methods and nanopolish (WGBS: R = 0.90 and 0.84; EMseq: 0.93 and 0.90 on MB231 and BT474, respectively) (Supplementary Figure S13D–F). NanoEM showed slightly higher correlation with WGBS than nanopolish (WGBS: R = 0.91 and 0.87; EMseq: 0.94 and 0.90 on MB231 and BT474; Figure 1C). High correlation was also observed between nanoEM and nanopolish (R = 0.89 and 0.84 for MB231 and BT474, respectively; Supplementary Figure S14). For MB231, nanoEM data represented 23 000 293 (39.4%) of CpG sites covered by at least five reads from the 73 Gb of the sequencing yield (Figure 1B and Supplementary Table S2). On the other hand, the nanopolish data represented 24 174 358 (41.4%) of CpG and covered by at least five reads from the 53 Gb of the sequencing yield (Supplementary Figure S13C and Supplementary Table S5). We considered that the fact that the 1.4-fold more sequencing depth (73 Gb versus 53Gb) yielded a similar coverage (39.4% and 41.4%, respectively for nanoEM and nanopolish) was derived from the PCR bias (Supplementary Figures S4 and S12). The results from the sequencing analysis of the libraries, which were constructed by varying PCR cycles, are shown in Supplementary Figures S12. Also, note that the degree of the PCR bias varied between samples, depending on not only PCR cycles but also the conditions of the initial samples and/or the overall methylation status of the samples. Importantly, for nanopolish, it is difficult to increase the coverage by increasing the sequencing depth. It requires 0.5–1 μg of gDNA per run, while nanoEM can repeat the sequencing analysis using the same amount of genomic DNA.
To further evaluate nanoEM, we calculated the overall precision and recall for three categories regarding the methylation rate, low level (<25%), high level (>75%), and middle level (Supplementary Table S6). In all categories of MB231 and BT474 data, precisions and recalls of the nanoEM datasets were comparable to or slightly higher than those of the nanopolish datasets. In addition, we calculated the per-read accuracy of methylation calling on 10 000 reads extracted randomly from the nanoEM and the nanopolish datasets against the WGBS dataset (Supplementary Figure S15). Both datasets had high accuracy, from 80% to 100%, and the nanoEM datasets were comparable to the nanopolish datasets. Overall, the results are generally highly consistent. Although the overall cover rate of the CpG sites in nanoEM was lower, it was probably caused by PCR bias. However, we consider that its influence is at an acceptable level in considering that the analysis was made from a limited amount of DNA material.
Viewing the long methylation data
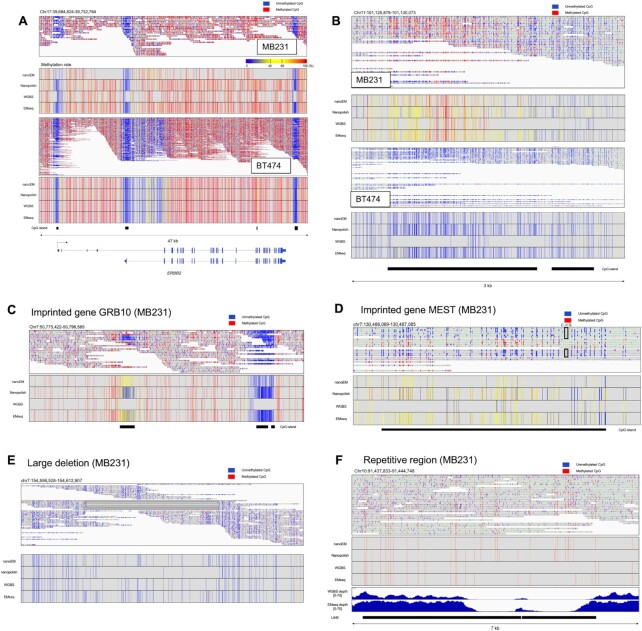
We visualized CpG methylation patterns and observed them for specific cases. First, for highly representative cases of key cancer-related genes, the cases of the HER2 gene and PGR genes are shown in Figure 4A and B, respectively. Note that, in both cases, the entire region of CGI is covered by single reads. BT474 harboring HER2 gene amplification (32) showed higher coverage on the HER2 gene locus than did MB231 (Figure 4A). A high methylation rate of the CGI in MB231 and low methylation of that in BT474 were detected by nanoEM, which were consistent with that of other methods (Supplementary Figure S3C and Figure 4B). For the second examples, the cases of the GRB10 and MEST genes are shown (Figure 4C and D). These two genes are known as so-called imprinted genes (33,34). For each case, a clear allele-specific methylation pattern was observed. This information was not resolved as an ‘intermediate’ methylation when the short-read data was analyzed. Moreover, in MB231, a heterozygous SNP (C/G) was detected in the CGI of genes, and the only C allele was detected as methylated. Because MEST is known as a paternal expressed gene, it is assumed that the C allele and G allele are located on a maternal chromosome and a paternal chromosome, respectively. For the third example, we attempted to show whether the methylation pattern on regions with structural variations (SVs) and repetitive regions can be detected by nanoEM. Owing to the general difficulty of aligning short reads to SVs, the methylation patterns of their surrounding regions have not been well characterized (35). We found that a heterozygous large deletion (∼8 kb) within an intron of the DPP6 gene, which was also identified in a previous study (36), was covered by nine nanoEM reads (Figure 4E) in MB231. For the last example, we analyzed the nanoEM reads that were aligned to repetitive sequences. In an intronic region of the HECTD2 gene resided two tandem LINE1 elements (Figure 4F). Although neither EMseq nor WGBS could completely cover this region, nanoEM was able to analyze this region. Therefore, nanoEM is a unique method that can be used to detect methylation patterns around SV and repetitive sequences.
Figure 4.
Methylation patterns of several genes and regions. (A) The nanoEM reads aligned to the HER2 gene on MB231 and BT474 are shown in the IGV in bisulfite mode in the top panel (52). Heatmap of the methylation rate, measured by nanoEM, nanopolish, short-read WGBS, and EMseq, is shown in the middle panel. Color bar of the methylation rate. The CGIs and the RefSeq transcripts are shown in the bottom panel. (B) The nanoEM reads of MB231 and BT474 aligned to the CGIs of the PGR gene are shown in the top panel. The location of CGIs is shown in the bottom panel. (C) The nanoEM reads aligned to the GRB10 gene are shown. Heatmap of the methylation rate, measured by nanoEM, nanopolish, short-read WGBS, and EMseq, is shown in the middle panel. The CGIs are shown in the bottom panel. (D) The nanoEM reads aligned to the MEST gene are shown. The heterozygous SNP (C to G) is shown in the box. Heatmap of the methylation rate, measured by nanoEM, nanopolish, short-read WGBS, and EMseq, is shown in the middle panel. The CGIs are shown in the bottom panel. (E) The nanoEM reads of MB231 mapped to the large deletion that was reported in a previous study (36). Split alignments of single reads are linked by lines. (F) The nanoEM reads of BT474 aligned to LINE1 located in HECTD2 intron is shown in the top panel. The read coverage by EMseq and WGBS is shown in the bottom panel.
Application to clinical samples
To evaluate the feasibility of nanoEM for clinical specimens, we applied nanoEM to two pairs of tumor and matched normal tissues of breast cancer specimens. Cases 7 and 8 were ‘ER-negative, PGR-negative, and HER2-positive’ and ‘ER-positive, PGR-positive, and HER2-negative’ breast cancer, respectively, as diagnosed from a histopathological viewpoint (Supplementary Figure S16). Case 7 was obtained from our previous study on spatial transcriptome analysis (19). We prepared the converted DNA from 25 or 100 ng of gDNA and sequenced it using PromethION. We obtained 5 924 844–20 134 343 of 1D pass reads (Supplementary Table S2). The N50 read length was 5.8–7.6 kb except for the tumor tissues of Case 8 (3.4 kb), for which the DIN before fragmentation was quite low (6.6) compared with that of other tissues (∼9.0). A number of short DNA fragments was retained even after size selection, which was attributed to degradation causing a short DNA fragment. For Cases 7, we performed WGS using PromethION and called methylation by nanopolish for the purpose of validation (Supplementary Table S5). For Case 8, only 25 ng of genomic DNA was available after dissecting the tumor part of the tissue and removing short fragments (Supplementary Table S2). Therefore, we could not employ nanopolish in the first place (For another example of the case for which sufficient amount of DNA was not available see Supplementary Figures S16 and S17).
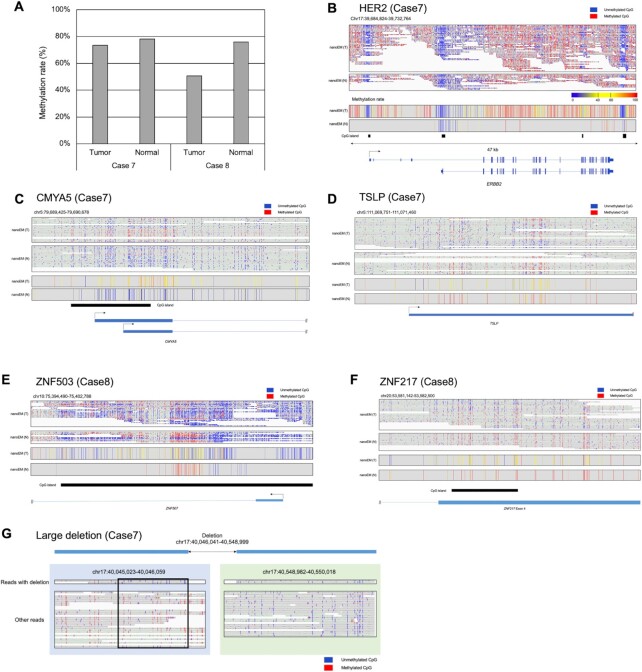
We similarly called methylation from nanoEM reads using our pipeline. The detected methylation rates of CpG were 50.7–73.4% and 76.0–78.0% for tumor and normal tissues, respectively (Figure 5A). Although similar methylation rates between the tumor and the normal samples were detected for Cases 7, the lowest methylation rate (50.7%) was detected for the tumor tissue of Case 8, in which a higher proportion of cancer cells was shown, compared with that of the tumor tissue of Case 7. Because of the high contents of hypomethylated cancer cells, the tissue might show a low methylation rate (Supplementary Figure S16). In the HER2-positive tumor tissue of Case 7, a higher read coverage of the HER2 locus was observed compared with that of the matched normal tissue (Figure 5B). We detected differential methylated regions (DMRs) between normal and tumor samples for Cases 7 and 8 using metilene (37). Then, 14 and 173 of DMRs were detected in Cases 7 and 8, respectively (Supplementary Table S7). For example, there was a higher methylation rate of CMYA5 in the tumor tissue of Case 7 (Figure 5C). CMYA5 was predicted to be an oncogene and potential prognosis indicator of the overall survival rate in breast cancer (38,39) and might be repressed in the tumor tissue of Case 7 via enhanced methylation. The TSLP gene showed a lower methylation rate in the tumor tissue of Case 7 than that in the normal counterpart (Figure 5D). It is known that TSLP promotes tumor cell survival and is important for metastasis in breast cancer (40). CGIs of ZNF503 and ZNF217 were less methylated in the tumor tissue of Case 8, compared with that in the normal counterpart (Figure 5E and F). It is shown that ZNF503 promotes proliferation and metastasis of breast cancer cells by repressing GATA3 (41). ZNF217 was shown to promote epithelial–mesenchymal transition and invasion in human mammary epithelial cells and is considered to be a biomarker of poor prognosis in breast cancer (42). Further intensive studies on the methylation of these gene, particularly with their chromosome background, would deepen the molecular etiology of these cancers. Last, we also attempted to detect the methylation of SV in tumor tissues in Case 7. The large deletion was detected by nanoEM in the tumor tissue of Case 7 (Figure 5G). Interestingly, CpG upstream of the large deletion was unmethylated, although the allele without large deletion was mostly methylated. That could be confirmed by nanopore WGS (Supplementary Figure S18). It might be possible that low methylation was caused by the deletion or vice versa.
Figure 5.
Applications to clinical samples. (A) The average cytosine methylation rate by CpG, measured by nanoEM. Cytosines covered by at least five reads were used for the calculation. (B–F) Examples of DMRs between tumor and normal tissues of Case 7 (B and C) and Case 8 (D and F). nanoEM reads of tumor and normal tissues aligned to each region are shown in the top panel. The location of transcript models and CGIs is shown in the bottom panel. Average methylation rate of CpG covered by at least five reads is shown in the middle panel. (G) Methylation patterns detected by nanoEM visualized using the IGV (52). Reads with and without deletion are shown separately. The unmethylated region specific to reads with deletion is shown in the box.
CONCLUSIONS
Here, we present nanoEM, a new method for long-read whole-genome methylation analysis. NanoEM is a combination of previous methods. Therefore, the methodological novelty of this method itself may not be at the highest rank. Nevertheless, we believe the practical impact that this method will give to various applications should be substantial. We summarized advantages and disadvantages of methods for whole-genome methylation analysis in Supplementary Table S8. We combined nanopore long-read sequencing and enzymatic base conversion using APOBEC, which can be implemented from a small amount of DNA (Supplementary Tables S8 and S9). Initially, on the basis of the comparison between WGBS and EMseq using an Illumina sequencer, EMseq showed higher CpG coverage and less bias than did WGBS. Using 1–100 ng of gDNA, we prepared long EMseq-converted DNA by optimizing a protocol for long PCR and DNA size selection from breast cancer cell lines and clinical specimens. The converted DNA was sequenced using a nanopore sequencer, PromethION. Then, 5.9–20.1 M of 1D pass reads of 3.4–7.6 kb N50 length was generated by using single flow cells of PromethION. We constructed a new pipeline for methylation analysis utilizing base-converted long reads. Our pipeline analyzes the obtained reads more efficiently, particularly at CGIs, than did a previous approach solely by using minimap2. The methylation rate detected by nanoEM was comparable with that by EMseq and nanopore WGS. Using cell lines, we demonstrated the methylation status and chromosome context of two breast cancer-related genes, HER2 and PGR, and two imprinted genes, GRB10 and MEST, were precisely analyzed. We also showed that nanoEM can detect methylation patterns around SV and repetitive regions, in which short reads are difficult to align. Using clinical samples, we detected the tumor-specific methylation status of four breast cancer-related genes, TSLP, CMYA5, ZNF503 and ZNF217. We also detected a tumor-specific large deletion and the differential methylation pattern between the alleles with and without the large deletion. It is unknown whether methylation is controlled at the chromosome level in most cancers, particularly at the site of chromosome rearrangements, including gene fusions.
Long-read methylation analysis would determine the methylation patterns on long single DNA molecules. By the obtained long read data, allele-biased methylation patterns will be more clearly detected. In fact, by the long-read methylation analysis using nanoEM methods, we could dissect the methylation pattern of imprinting genes and structural variations in an allele-sensitive manner (Figures 4C–E and 5G). We were also able to detect the methylation within repetitive sequences (Figure 4F). These pieces of information may have been overlooked by the short-read sequencing method. We believe this information should be vital because it is known that aberration of methylation in imprinting regions and long repetitive elements is critical for cancer (43). For cancerous structural variations, they are also known to cause carcinogenesis occasionally. Due to the difficulty in the sequence alignments of short-read sequences (44), the methylation status of these regions is still primarily elusive.
While direct calling from nanopore WGS has the potential to distinguish between mC and hmC on the same DNA molecule, nanoEM cannot distinguish between them (6,11). By adapting modified protocol of EMseq without TET oxidation of mC for protection from APOBEC deamination (8), nanoEM should be able to detect hmC. NanoEM can use as little as 10ng of DNA and produce reads in the 3.4–7.6 kb N50 lengths range (Supplementary Table S2). In contrast, nanopolish, which reads methylation from native DNA without the need for costly enzymatic or chemical conversion processes, obtains N50 read lengths that were ∼4× longer but requires ∼1 ug of DNA (Supplementary Table S5). Nanopore sequencing requires 150 fmol of DNA, corresponding to around 500 ng at 5 kb, as input for sample preparation. If 500 ng of DNA is available, it might be more advantageous to choose direct calling from nanopore WGS which does not require pretreatment for long-read methylation analysis. EM-seq using short-read sequencer produce higher coverage than other PCR-based methods, except for the regions, in which it is difficult to map short-reads, such as repetitive regions and SV. Although short-read sequencing has a cost advantage, the cost of long read sequencing is going down (45). In the near future, we expect the cost of long-read sequencing will be comparable to that of short-read sequencing.
For tissues with low tumor cellularity, the sequence reads originating from the cancer genome was usually embedded among the reads from the normal genome of the surrounding normal cells (Supplementary Figures S16 and S17). As shown in Supplementary Figure S17, if sequencing depth was enough, the aberrantly methylated reads derived from cancer cells can be detected even from the tumor with low tumor cellularity, and it is difficult to find them from short reads. If more than 100 ng of gDNA is available, it is possible to increase the coverage of sequencing by repeating the sequencing analysis of nanoEM, which would not be possible for nanopore WGS (nanopolish). In fact, the tumor cell-enrichment of a clinical specimen by microdissection typically leads to the DNA yield ranging from 50 to 300 ng (46). This amount of gDNA can be used for nanoEM but not for nanopolish. More generally, the surgically dissected samples are not always large enough to extract 1 μg of genomic DNA. If the most optimistic case were assumed where the tumors are packed with cancer cells, ∼1 mm3 human tissue would be needed to extract 1 μg of gDNA (47). Among a cohort of Stage I HER2-positive breast cancers, 7% of patients were diagnosed as T1mi tumors (48). For these cases, the maximum diameter is less than 1 mm. Even if assuming all the procedures were conducted ideally, without any loss, only 10 ng of gDNA would be extracted from 0.01 mm3 (a usually expected size) of the tissue (Supplementary Table S9). Therefore, an attempt to analyze most T1mi tumors using the nanopolish method would be theoretically impossible, which would leave our knowledge of early-stage cancers blank. Even for larger tumors than T1mi, a large part of the tumor tissue should be usually used for pathological and other diagnoses to benefit patients. For the research purpose, a small remaining part should be used, limiting the available amount of the starting material. We believe that long-read methylation analysis using nanoEM of a wide range of specimens from which is difficult to prepare an adequate amount of DNA, including very small samples (smaller than 1 mm3) of early-stage cancer tissues and biopsies for various cancer types, would deepen our understanding of epigenomic regulation and its disturbance in cancers. In addition to cancers, nanoEM may enable long-read methylation analysis of rare cells, such as oocytes and early embryos.
DATA AVAILABILITY
The scripts and the detailed explanations of our pipeline were used for data analyses of nanoEM is available in a GitHub repository at https://github.com/yos-sk/nanoEM.
With regard to cell lines, sequencing data and output files of methylation called by nanopolish were deposited to the DDBJ Sequence Read Archive (DRA) and Gene Expression Archive (GEA) under accession numbers DRA011237 and E-GEAD-408, respectively. The clinical samples were deposited to the Japanese Genotype-phenotype Archive (JGA) under accession number JGAS000265.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Terumi Horiuchi, Yuta Kuze and Erina Ishikawa for assistance with data processing and Kazumi Abe, Kiyomi Imamura, Mari Tsubaki, Yuni Ishikawa, Megumi Kombu and Etsuko Kobayashi for assistance with the experiments. The supercomputing resource was provided by the Human Genome Center of the University of Tokyo (http://sc.hgc.jp/shirokane.html). The authors would like to thank Enago (https://www.enago.jp/) for the English language review.
Contributor Information
Yoshitaka Sakamoto, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba, Japan.
Suzuko Zaha, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba, Japan.
Satoi Nagasawa, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba, Japan.
Shuhei Miyake, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba, Japan.
Yasuyuki Kojima, Division of Breast and Endocrine Surgery, Department of Surgery, St. Marianna University School of Medicine, Kawasaki, Kanagawa, Japan.
Ayako Suzuki, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba, Japan.
Yutaka Suzuki, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba, Japan.
Masahide Seki, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS KAKENHI [JP21K15074, JP19K16108]; MEXT KAKENHI [JP16H06279 (PAGS), JP17H06306, JP20H05906]; JSPS Fujita Memorial Fund for Medical Research; National Cancer Center Research and Development Fund (29-A-6). Funding for open access charge: JSPS KAKENHI [JP21K15074].
Conflict of interest statement. None declared.
REFERENCES
- 1.Greenberg M.V.C., Bourc’his D.. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019; 20:590–607. [DOI] [PubMed] [Google Scholar]
- 2.Ehrlich M.DNA methylation in cancer: too much, but also too little. Oncogene. 2002; 21:5400–5413. [DOI] [PubMed] [Google Scholar]
- 3.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.-M.et al.. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka K., Okamoto A.. Degradation of DNA by bisulfite treatment. Bioorg. Med. Chem. Lett. 2007; 17:1912–1915. [DOI] [PubMed] [Google Scholar]
- 5.Olova N., Krueger F., Andrews S., Oxley D., Berrens R. V., Branco M.R., Reik W.. Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biol. 2018; 19:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaisvila R., Chaithanya Ponnaluri V.K., Sun Z., Langhorst B.W., Saleh L., Guan S., Dai N., Campbell M.A., Sexton B., Marks K.et al.. EM-seq: detection of DNA methylation at single base resolution from picograms of DNA. 2019; bioRxiv doi:16 May 2020, preprint: not peer reviewed 10.1101/2019.12.20.884692. [DOI] [PMC free article] [PubMed]
- 7.Liu Y., Siejka-Zielińska P., Velikova G., Bi Y., Yuan F., Tomkova M., Bai C., Chen L., Schuster-Böckler B., Song C.-X.. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat. Biotechnol. 2019; 37:424–429. [DOI] [PubMed] [Google Scholar]
- 8.Sun Z., Vaisvila R., Hussong L.-M., Yan B., Baum C., Saleh L., Samaranayake M., Guan S., Dai N., Corrêa I.R. Jret al.. Nondestructive enzymatic deamination enables single-molecule long-read amplicon sequencing for the determination of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Genome Res. 2021; 31:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booth M.J., Ost T.W.B., Beraldi D., Bell N.M., Branco M.R., Reik W., Balasubramanian S.. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 2013; 8:1841–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakamoto Y., Xu L., Seki M., Yokoyama T.T., Kasahara M., Kashima Y., Ohashi A., Shimada Y., Motoi N., Tsuchihara K.et al.. Long-read sequencing for non-small-cell lung cancer genomes. Genome Res. 2020; 30:1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rand A.C., Jain M., Eizenga J.M., Musselman-Brown A., Olsen H.E., Akeson M., Paten B.. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods. 2017; 14:411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garalde D.R., Snell E.A., Jachimowicz D., Sipos B., Lloyd J.H., Bruce M., Pantic N., Admassu T., James P., Warland A.et al.. Highly parallel direct RN A sequencing on an array of nanopores. Nat. Methods. 2018; 15:201–206. [DOI] [PubMed] [Google Scholar]
- 13.Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W.. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods. 2017; 14:407–410. [DOI] [PubMed] [Google Scholar]
- 14.Ni P., Huang N., Zhang Z., Wang D.-P., Liang F., Miao Y., Xiao C.-L., Luo F., Wang J.. DeepSignal: detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics. 2019; 35:4586–4595. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q., Fang L., Yu G., Wang D., Xiao C.-L., Wang K. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 2019; 10:2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Cheng J., Siejka-Zielińska P., Weldon C., Roberts H., Lopopolo M., Magri A., D’Arienzo V., Harris J.M., McKeating J.A.et al.. Accurate targeted long-read DNA methylation and hydroxymethylation sequencing with TAPS. Genome Biol. 2020; 21:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cailleau R., Young R., Olivé M., Reeves W.J.. Breast tumor cell lines from pleural effusions. J. Natl. Cancer Inst. 1974; 53:661–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasfargues E.Y., Coutinho W.G., Redfield E.S.. Isolation of two human tumor epithelial cell lines from solid breast carcinomas. J. Natl. Cancer Inst. 1978; 61:967–978. [PubMed] [Google Scholar]
- 19.Nagasawa S., Kuze Y., Maeda I., Kojima Y., Motoyoshi A., Onishi T., Iwatani T., Yokoe T., Koike J., Chosokabe M.et al.. Genomic profiling reveals heterogeneous populations of ductal carcinoma in situ of the breast. Commun. Biol. 2021; 4:438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G. V., Sonkin D.et al.. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H.Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013; arXiv doi:26 May 2013, preprint: not peer reviewedhttps://arxiv.org/abs/1303.3997v2.
- 22.Krueger F., Andrews S.R.. Bismark: a flexible aligner and methylation caller for bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H.Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P.. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015; 31:2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y., Ecker J.R.. Non-CG methylation in the human genome. Annu. Rev. Genomics Hum. Genet. 2015; 16:55–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ando M., Saito Y., Xu G., Bui N.Q., Medetgul-Ernar K., Pu M., Fisch K., Ren S., Sakai A., Fukusumi T.et al.. Chromatin dysregulation and DNA methylation at transcription start sites associated with transcriptional repression in cancers. Nat. Commun. 2019; 10:2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G. V., Lo C.C., McDonald E.R., Barretina J., Gelfand E.T., Bielski C.M., Li H.et al.. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019; 569:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirane K., Toh H., Kobayashi H., Miura F., Chiba H., Ito T., Kono T., Sasaki H.. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLos Genet. 2013; 9:e1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J.H., Park S.J., Nakai K.. Differential landscape of non-CpG methylation in embryonic stem cells and neurons caused by DNMT3s. Sci. Rep. 2017; 7:11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y., Sebra R., Pullman B.S., Qiao W., Peter I., Desnick R.J., Geyer C.R., DeCoteau J.F., Scott S.A.. Quantitative and multiplexed DNA methylation analysis using long-read single-molecule real-time bisulfite sequencing (SMRT-BS). BMC Genomics. 2015; 16:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goerner-Potvin P., Bourque G.. Computational tools to unmask transposable elements. Nat. Rev. Genet. 2018; 19:688–704. [DOI] [PubMed] [Google Scholar]
- 32.Miller S.J., Suen T.C., Sexton T.B., Hung M.C.. Mechanisms of deregulated HER2/neu expression in breast cancer cell lines. Int. J. Oncol. 1994; 4:599–608. [DOI] [PubMed] [Google Scholar]
- 33.Blagitko N., Mergenthaler S., Schulz U., Wollmann H.A., Craigen W., Eggermann T., Ropers H.H., Kalscheuer V.M.. Human GRB10 is imprinted and expressed from the paternal and maternal allele in a highly tissue- and isoform-specific fashion. Hum. Mol. Genet. 2000; 9:1587–1595. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi S., Kohda T., Miyoshi N., Kuroiwa Y., Aisaka K., Tsutsumi O., Kaneko-Ishino T., Ishino F.. Human PEG1/MEST, an imprinted gene on chromosome 7. Hum. Mol. Genet. 1997; 6:781–786. [DOI] [PubMed] [Google Scholar]
- 35.Treangen T.J., Salzberg S.L.. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat. Rev. Genet. 2012; 13:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilpatrick T., Lee I., Graham J.E., Raimondeau E., Bowen R., Heron A., Downs B., Sukumar S., Sedlazeck F.J., Timp W.. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 2020; 38:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jühling F., Kretzmer H., Bernhart S.H., Otto C., Stadler P.F., Hoffmann S.. Metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016; 26:256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colaprico A., Olsen C., Bailey M.H., Odom G.J., Terkelsen T., Silva T.C., Olsen A. V., Cantini L., Zinovyev A., Barillot E.et al.. Interpreting pathways to discover cancer driver genes with Moonlight. Nat. Commun. 2020; 11:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao B., Hang J., Lei T., He Y., Kuang Z., Wang L., Chen L., He J., Zhang W., Liao Y.et al.. Identification of key genes relevant to the prognosis of ER-positive and ER-negative breast cancer based on a prognostic prediction system. Mol. Biol. Rep. 2019; 46:2111–2119. [DOI] [PubMed] [Google Scholar]
- 40.Kuan E.L., Ziegler S.F.. A tumor-myeloid cell axis, mediated via the cytokines IL-1α and TSLP, promotes the progression of breast cancer. Nat. Immunol. 2018; 19:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahi P., Wang C.Y., Lawson D.A., Slorach E.M., Lu A., Yu Y., Lai M.D., Velozo H.G., Werb Z.. ZNF503/Zpo2 drives aggressive breast cancer progression by down-regulation of GATA3 expression. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:3169–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vendrell J.A., Thollet A., Nguyen N.T., Ghayad S.E., Vinot S., Bièche I., Grisard E., Josserand V., Coll J.L., Roux P.et al.. ZNF217 is a marker of poor prognosis in breast cancer that drives epithelial-mesenchymal transition and invasion. Cancer Res. 2012; 72:3593–3606. [DOI] [PubMed] [Google Scholar]
- 43.Esteller M.Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007; 8:286–298. [DOI] [PubMed] [Google Scholar]
- 44.Mahmoud M., Gobet N., Cruz-Dávalos D.I., Mounier N., Dessimoz C., Sedlazeck F.J.. Structural variant calling: the long and the short of it. Genome Biol. 2019; 20:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Logsdon G.A., Vollger M.R., Eichler E.E.. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020; 21:597–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H., McDowell T.L., Hanson N.E., Tang X., Fujimoto J., Rodriguez-Canales J.. Laser capture microdissection for the investigative pathologist. Vet. Pathol. 2014; 51:257–269. [DOI] [PubMed] [Google Scholar]
- 47.Austin M.C., Smith C., Pritchard C.C., Tait J.F.. DNA yield from tissue samples in surgical pathology and minimum tissue requirements for molecular testing. Arch. Pathol. Lab. Med. 2016; 140:130–133. [DOI] [PubMed] [Google Scholar]
- 48.Parsons B.M., Uprety D., Smith A.L., Borgert A.J., Dietrich L.L.. A US registry-based assessment of use and impact of chemotherapy in stage I HER2-positive breast cancer. JNCCN J. Natl. Compr. Cancer Netw. 2018; 16:1311–1320. [DOI] [PubMed] [Google Scholar]
- 49.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karolchik D., Hinricks A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J.. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004; 32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frankish A., Diekhans M., Ferreira A.-M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J.et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scripts and the detailed explanations of our pipeline were used for data analyses of nanoEM is available in a GitHub repository at https://github.com/yos-sk/nanoEM.
With regard to cell lines, sequencing data and output files of methylation called by nanopolish were deposited to the DDBJ Sequence Read Archive (DRA) and Gene Expression Archive (GEA) under accession numbers DRA011237 and E-GEAD-408, respectively. The clinical samples were deposited to the Japanese Genotype-phenotype Archive (JGA) under accession number JGAS000265.