Abstract
Cancer and neurodegenerative diseases are caused by genetic and environmental factors. Expression of tumour suppressor genes is suppressed by mutations or epigenetic silencing, whereas for neurodegenerative disease-related genes, nucleic acid-based effects may be presented through loss of protein function due to erroneous protein sequences or gain of toxic function from extended repeat transcripts or toxic peptide production. These diseases are triggered by damaged genes and proteins due to lifestyle and exposure to radiation. Recent studies have indicated that transient, non-canonical structural changes in nucleic acids in response to the environment can regulate the expression of disease-related genes. Non-canonical structures are involved in many cellular functions, such as regulation of gene expression through transcription and translation, epigenetic regulation of chromatin, and DNA recombination. Transcripts generated from repeat sequences of neurodegenerative disease-related genes form non-canonical structures that are involved in protein transport and toxic aggregate formation. Intracellular phase separation promotes transcription and protein assembly, which are controlled by the nucleic acid structure and can influence cancer and neurodegenerative disease progression. These findings may aid in elucidating the underlying disease mechanisms. Here, we review the influence of non-canonical nucleic acid structures in disease-related genes on disease onset and progression.
INTRODUCTION
Under normal conditions, the number of cells in the body remains constant via the tight regulation of cell division and proliferation. However, genetic mutations can cause a cell population to grow uncontrollably. When this cell population grows invasively beyond the boundaries of normal tissues and/or shows metastasis, it forms malignant tumours (1). Mutations of genes are often observed in malignant tumour cells. According to the central dogma of biology, genetic information in DNA is transferred to RNA via transcription and then translated into proteins. Producing the appropriate amount of protein within cells is essential to maintain normal cell conditions. However, genetic mutations interfere with the proper production of proteins. External environmental factors such as chemical substances dispersed in the abiotic and biotic environments, lifestyle choices such as eating, drinking and smoking habits, and psychological factors like stress have been reported to induce gene mutations (2–4). Further, the structure of nucleic acids plays a role in cancer development by perturbing the processes of the central dogma. For example, in many human cancers, c-MYC transcriptional abnormalities are observed; however, the mechanism underlying c-MYC oncogene regulation of gene expression remains to be determined (5). Non-canonical structure formation in the c-MYC promoter and coding regions regulates transcription activity (6). In addition, the formation of non-canonical structures in several oncogenes inhibits transcription and translation. Interestingly, formation of such non-canonical structures strongly depends on the surrounding environment (7). Thus, the roles of nucleic acid structures in cancer cells have also attracted substantial research attention. Novel mechanisms for transcriptional dysregulation by c-MYC and other oncogenes have been illustrated by the discovery of phase separation with transcription factors. Indeed, phase separation formation may be critical for the regulation of transcription in cancer cells (8). Biomolecules related to cancer progression frequently undergo phase separation in cells and organisms and are thought to be involved in many complex human cancers (9).
Phase separations are important not only for cancers, but also for the progression of neurodegenerative diseases (10), which involve a gradual damage to specific nerve cells that are subsequently eliminated from the brain and the spinal cord. Neurodegenerative disease is a disorder that occurs in the central nervous system and is characterised by a decrease in the number of certain neurons and the accumulation and aggregation of fibrous material inside and outside the neurons (11). Accumulation and aggregation are thought to be caused by aberrant phase separation of protein–protein or protein–RNA interactions, abnormalities in protein folding, or the dysfunction of key proteolytic mechanisms in the cell. Abnormal aggregation and aggregates containing these proteins are thought to be the main cause of dysfunction and death in neurons, which are hallmarks of neurodegenerative diseases (12,13). Diseases with these characteristics are often referred to as ‘proteopathies’ and include diseases such as Parkinson's disease (14), amyotrophic lateral sclerosis (ALS) (15,16), and Huntington's disease (HD) (17–20). Examples of aggregation of peptides or proteins via solid–liquid phase separations include the cytoplasmic inclusions of the RNA-binding protein, TAR-DNA-binding protein and fused in sarcoma (FUS) in ALS and frontotemporal dementia (FTD), and polyglutamine aggregates in HD (21). Recently, the RNA transcripts of these neurodegenerative disease-related genes were demonstrated to form non-canonical structures and induce liquid–liquid phase separation (10). Thus, evidence for the association of aberrant phase separation with various diseases such as cancer and neurodegeneration has been accumulating. It is noteworthy that the speed and efficiency of phase separation depends on the higher-order structure of the RNA, such as hairpin or G-quadruplexes (22,23), suggesting that the structure of nucleic acids may play an important role in cancer and neurodegenerative diseases.
This review describes the non-canonical structures of nucleic acids and their functions in gene expression during disease progression and how nucleic acid structures are associated with phase separation along with their roles in the onset and progression of cancer and neurodegeneration.
Structural polymorphisms of nucleic acids
The canonical structure of nucleic acids is a duplex (24), which comprises two strands of nucleic acids linked in an antiparallel formation via Watson–Crick (W–C) base pairs between A and T (or U) and between G and C (Figure 1A). Interestingly, nucleic acids can also form non-canonical structures such as triplexes (25,26), tetraplexes of G-quadruplexes (27) and i-motifs (28), and junctions. Triplexes and G-quadruplexes are most commonly observed in disease-related genes and are considered to play important roles in disease onset and progression (29).
Figure 1.

Nucleic acids structures of (A) B-form duplex and Watson–Crick base pairs, (B) triplex and triads, and (C) G-quadruplex and G-quartet. Hydrogen bonds are shown in dashed line (green). The bases of A, C, G and T in the duplex, triplex and quadruplex structures are shown in red, pink, green and light blue, respectively.
Triplexes are formed by the binding of a third strand, the triplex-forming oligonucleotide (TFO), to a duplex from the major groove side via Hoogsteen (H) base pairs (Figure 1B). TFOs are composed of a pyrimidine base and a purine base bound in antiparallel and parallel to the duplex (30). Thus, the triplex structure consists of triads in which pyrimidine (Y) bases are bound to W–C base pairs of pyrimidine and purine (R) via H base pairs (Y-R*Y, where dash and star denote Watson–Crick and Hoogsteen base pairs, respectively), or triads in which purine bases are bound to Y-R base pairs (Y-R*R) (Figure 1B) (31). The TFO also interacts with the neighbouring bases via stacking interactions and stabilize the structures. Hence, the formation of triplexes generally requires a continuous homopurine-homopyrimidine run of 10 base pairs or more (32,33). Another characteristic of triplexes is that, unlike duplexes, their stability varies greatly with pH in solution. The width of the major groove in the duplex favours binding to the pyrimidine strand over the purine strand. Hence, the parallel triplex with three triads of C-G*C+ and T-A*T is more stable than the antiparallel triplex with three triads of C-G*G and T-A*A even under normal conditions (34,35). However, the Y-R*Y triplex is formed mainly under acidic conditions because protonation of N3 of cytosine stabilises the C-G*C+ triad in TFO (36).
G-quadruplexes are formed by hydrogen bonding of the four guanines of G-rich strands (H-bond) (Figure 1C). When four guanines are joined by hydrogen bonds between the H1 of the nitrogen and O6 of the carbonyl group and between the H2 of amine and N7, the four guanines are called G-quartet (Figure 1C) (37–39). Two G-quartets stacked with a cation in the middle, which is co-ordinated by O6 oxygen molecules in eight guanines between two G-quartets, rendering the G-quadruplexes highly stable. The order of the ability of monovalent cations to stabilise G-quadruplexes is K+ > NH4+ > Na+ > Li+ (40,41), as the stability of G-quartets is changed depending on the ionic radius and dehydration of the cation. G-quadruplex-forming sequences can be retrieved by the algorithm d(G3+N1–7G3+N1–7G3+N1–7G3+), wherein N refers to any base that may form G-quadruplexes (42,43). This algorithm can identify stable G-quadruplex sequences with three or more G-quartets. However, recent studies have shown that even sequences such as those forming two G-quartets or long loops (N number > 7) can form a G-quadruplex (44). An assay based on genome-wide sequences showed that there are more than 700,000 sites with G-quadruplex-forming sequences in the human genome (44). It was also reported that only 10 000 of the 700 000 G-quadruplex-forming sequences have been discovered by chromatin immunoprecipitation-sequencing using a G-quadruplex-binding antibody (45,46).
Nucleic acid structural changes based on the surrounding environment
The intracellular environment comprises diverse biomolecules that are intermingled (Figure 2). For example, in the nucleus, DNA is packed by chromatin with high density, while the cytoplasm contains macromolecules such as organelles, cytoskeleton, soluble and insoluble proteins, and low-molecular weight compounds such as osmoregulatory molecules and metabolites (Figure 2). These biomolecules occupy 20–40% of the cell volume, and their concentration can reach 300–400 mg/ml, thereby crowding the intracellular environment, which is referred to as molecular crowding (Figure 2). Meanwhile, an aqueous solution containing 100–150 mM NaCl or KCl has been used in vitro experiments to analyse the behaviour and function of biomolecules in cells. However, such standard solutions often do not reflect cell physiology. Therefore, the molecular environment in the cell is important for determining the behaviour and function of biomolecules.
Figure 2.

Image of the molecular crowding condition in cells. A cell exhibits a crowded environment comprising various organelles such as the nucleus, endoplasmic reticulum, mitochondria and Golgi apparatus, along with soluble and insoluble proteins, mRNAs, cytoskeletons and sugars filling the spaces between the organelles.
To evaluate the effect of the molecular environment in the cell, cell-mimicking solutions have been constructed in vitro to reproduce the molecular crowding environment by the addition of co-solutes to a standard aqueous solution. Molecular crowding studies use large or small molecules as co-solutes. For example, polyethylene glycol, polymers of sugars (ex. dextran, ficoll), proteins such as bovine serum albumin, and nucleic acids such as tRNA are used as crowding agents to mimic molecular crowding conditions. In solutions with such high-molecular-weight crowding molecules, the volume occupied by the biomolecules is reduced owing to the steric hindrance created by the crowding molecules, and hence, molecular motion and structure are restricted (47). In addition, the activity of the biomolecules increases owing to the limitation of the volume in which they exist. Such molecular crowding conditions change the chemical potential and activity coefficients of the biomolecules (48). Furthermore, the physical properties (ex. osmotic pressure, dielectric constant, surface tension, viscosity, etc.) of crowded solutions are very different from those of dilute solutions (7). These physical properties changes also affect the behaviour of biomolecules (7). For example, Minton et al. found that amyloid fibrillation and fibrinogen aggregation were enhanced in a molecular crowding environment (47,48). Furthermore, Winter et al. found that tubulin multimerisation was enhanced by the addition of ficoll (49). Bloomfield et al. had analysed the effect of molecular crowding on enzymatic reactions as well as aggregation and reported that in a molecular crowding environment, the affinity between the restriction enzyme EcoRV and DNA is increased, while the cleavage rate is decreased (50).
Focusing on nucleic acids, the stability and structure of nucleic acids are determined by five key interactions for hydrogen bonding, base stacking, conformational entropy, hydration, and cation binding (51–54). The hydration and cation binding are particularly important under molecular crowding conditions (55–60).
The excluded volume effect of the addition of a large co-solute has a small effect on the thermal stability of the structure formed by the short oligonucleotide (61). The reason for this is that the formation of nucleic acid structures involves binding with water molecules; hence, a reduction in water activity due to the addition of co-solutes has a pronounced effect on the stability formed by short oligonucleotides (7). The decrease in water activity is unfavourable for the formation of nucleic acid structures with hydration but favourable for the formation of nucleic acid structures with dehydration. The hydration and dehydration during structure formation of nucleic acids can be calculated by the osmotic pressure method. Duplex formation results in the uptake or a slight release of water molecules, thereby destabilising the structure under molecular crowding conditions (61–63). On the other hand, the formation of non-canonical structures such as triplexes, G-quadruplexes and cruciforms involves the release of water thereby stabilizing the structures under crowding conditions (57,64–66). Chalikian et al. used volumetric measurements to analyse the hydration structure of nucleic acids. Volumetric measurements have suggested that duplex formation is associated with changes in hydration around duplex structures (67). Molecular dynamic simulations have also shown that the ordered water network around a DNA duplex is disrupted in a solution containing co-solutes (68). Moreover, G-quadruplexes release water molecules (69,70). This change in the stability of the non-canonical structure anticipates that it may play a key role in gene regulation in response to disease-induced environmental changes.
Nucleic acid structures may control the formation of a phase separation
Phase separation is among the most important intracellular phenomena (71,72). These phase separations form membrane-less compartments in cells that are often termed as membrane-less organelles, condensates, granules, or droplets to distinguish them from compartments of organelles that are surrounded by membranes such as the nucleus and Golgi apparatus. For example, membrane-less compartments are the nucleolus (73) and Cajal body in the nucleus and centrosomes (74), and P-bodies and germ granules in the cytoplasm. These membrane-less organelles are formed by the phase separation of specific groups of molecules such as RNA and proteins with low-complexity regions. Importantly, the membrane-less organelles can selectively enrich certain biomolecules while excluding others and can regulate biological reactions (75) because of the diffusion property of the molecules in the membrane-less organelles. Various other functions have been proposed for membrane-less organelles, including the formation of molecular filters that can dissociate DNA duplexes (76), suggesting that these molecular filters may be associated with gene regulation. Moreover, the evolutionarily tuned response of stress-triggered phase separation (77) forms the stress granule, which marks stress onset. The stress granule is transiently formed in the cytoplasm in response to stresses such as heat, oxidation, hyperosmotic pressure and UV irradiation. The stress granule is involved in translation and RNA metabolism and is linked to diseases, including neurodevelopmental disorders and cancer (78). Moreover, the condensates of signalling molecules promote T cell receptor signal transduction (79), and the competing protein–RNA interaction networks control multiphase intracellular organisation (80).
The diverse growth factor signalling pathways are hyperactive in cancer cells (81,82), although it remains unknown whether these signalling pathways are constitutively active. Recently, the role of droplet formation in regulating cellular signalling was reported. Cyclic GMP-AMP synthase (cGAS), a DNA sensing and binding protein of an exogenous DNA in the cytoplasm that activates a type-I interferon response, induces droplet formation and the innate immune response (83). Thus, phase separation plays an important role in cancer through signalling and other mechanisms.
DNA in the nucleus is under a molecular crowding environment densely folded into histone proteins. The surrounding environment can drastically change. Transcription factors that control transcription initiation form densely concentrated droplets, which when containing DNA, lead to an increase in the transcriptional activity (84). The FUS protein induces ALS by forming rigid amyloid fibres; notably, the FUS protein forms droplets and is concentrated to form fibres (85). It has been reported that when the reversibility of the droplets is lost, precipitation occurs as amyloid fibres (85). The droplets have high reversibility and fluidity, and hence, they can move inside the cell while concentrating liquid inclusions. Therefore, in an intracellular molecular crowding environment, droplets may accumulate transcription factors and amyloid-forming proteins and host several reactions. For example, the intrinsically disordered domain of prions prevents abnormal protein aggregation (irreversible denaturation) by forming droplets (86) mitigated by RNA (87). The primary component of droplets has been thought to be proteins (or peptides); however, it has been recently reported that RNA molecules transcribed from genes of triplet repeat disorders also form droplets and are found in droplets. Recently, there have been some important reports on RNA structures that affect droplet formation (10). Droplet properties change depending on the structure of the RNAs. The rCAG and rCUG repeats that form hairpins were reported to form spherical droplets. In contrast, the rGGGCC repeats in the C9orf72, which is the most frequent mutation observed in ALS/FTD, form a G-quadruplex structure, and were shown to form cohesive droplets, with little or no RNA movement within them (10). Other studies have also shown that the formation of RNA G-quadruplexes promotes droplet formation (22,23). As these RNAs also interact with peptides that are cytotoxic in neurodegenerative diseases, the relationship between RNA phase separation and cytotoxicity has attracted attention (10,88–89). Thus, phase separation might be regulated by the RNA structure, which influences disease progression.
The mechanism of phase separation can be explained by intermolecular force interactions (e.g. dipole–dipole, ion–ion and hydrogen bonds) that determine the solvent properties of the coexisting phases. (90). As described earlier, the molecular crowding in cells significantly influences the regulation of the behaviour and function of biomolecules. In recent years, liquid–liquid phase separation has been considered to play a role in reducing fluctuations in protein concentration because of the constraints on concentration inside and outside the droplet (90). If the total concentration of protein changes, the droplet number and size will also change; however, the concentration outside the droplets may be unaffected by these changes (91). Concentration noise of proteins in cells is reduced in the presence of droplets; therefore, the phase separation may also affect the structure of nucleic acids by controlling molecular crowding.
Nucleic acids involved in cancer mechanisms
In normal cells, tumour suppressor genes maintain normal functions. Tumour suppressor genes encode proteins that suppress the development of cancer (tumour suppressor proteins) (92). A few well-known tumour suppressor genes include P53, Rb and BRCA1. Oncogenesis is thought to occur when tumour suppressor proteins are not produced, or when abnormal tumour suppressor proteins from damaged genes interfere with the function of normal tumour suppressor proteins.
In contrast, an oncogene is a normal gene that has been modified to cause abnormalities in protein expression, structure, and function, resulting in the transformation of normal cells to cancerous cells (93). In this case, the gene before modification is referred to as the proto-oncogene. Cancer cells can proliferate indefinitely and exhibit abnormalities in apoptosis control mechanisms. Additionally, the cells can invade and metastasise (93). Oncogenes include genes that regulate cell proliferation and growth, such as platelet-derived growth factor (PDGF), and the vascular endothelial growth factor receptor (VEGF). Low-molecular weight G proteins regulate cell proliferation, differentiation, gene expression and cell adhesion, and are encoded by the HRAS, KRAS and NRAS genes. Genes such as those of the MYC family encode transcription factors that activate cancer-related genes, and many malignant cancers overexpress MYC family genes (94). A potent oncoprotein encoded by BCL-2 has functions as an inhibitor of cell apoptosis and has been found to be aberrantly overexpressed in a wide range of human tumours (93). Hence, controlling the activation or inactivation of these cancer-related genes is an important factor in determining cancer progression. Moreover, the telomere synthase enzyme telomerase is activated in cancer cells and continues to elongate the telomeres. When telomeres are shortened, cells do not divide; however, in cancer cells, the length of the telomere chain is maintained, enabling cancer cells to divide indefinitely (39).
Non-canonical DNA and RNA structures perturb the expression of cancer-related genes, and these changes in expression may influence cancer progression
In recent years, the relationship between nucleic acid structure and cancer has attracted attention. Several examples implicating non-canonical DNA structures such as triplexes and G-quadruplexes have been described in the transcription of cancer-related genes (Figure 3 and Table 1). Sequences capable of forming intramolecular triplexes and G-quadruplexes are abundant in mammalian genomes. These sequences often correspond to sites of increased susceptibility to chromosomal translocations such as translocations involving the c-MYC oncogene. For example, a DNA triplex formation in the c-MYC promoter interferes with transcription (95) and induces anti-proliferative activity in breast cancer (5). More recently, the unexpected stability of certain long non-coding RNAs (lncRNA) involved in cancer, including metastasis-associated lung adenocarcinoma transcript 1 (96) and multiple endocrine neoplasia-β RNAs (97), was found to be the result of intramolecular RNA triplex formation near their 3′ ends. Furthermore, RNA in nuclear can bind to specific promoters of DNA duplex to form triplex structures (RNA/DNA triplex), which can inhibit transcription. For example, the lncRNA of MIR100HG binds to the p27 gene to form RNA/DNA triplex structures (98). The RNA/DNA triplex structures are also involved in cancer, targeting specific sequences in DNA and regulating transcription (98). Moreover, the lncRNA PARTICLE has been shown to repress the tumour suppressor gene MAT2A via RNA/DNA triplex formation (99). The lncRNA MEG3 binds to GA-rich sequences in the chromatin through RNA-DNA triplex formation and regulates the TGF-beta pathway (100). Thus, triplex structures in the gene can downregulate gene expression in cancer.
Figure 3.
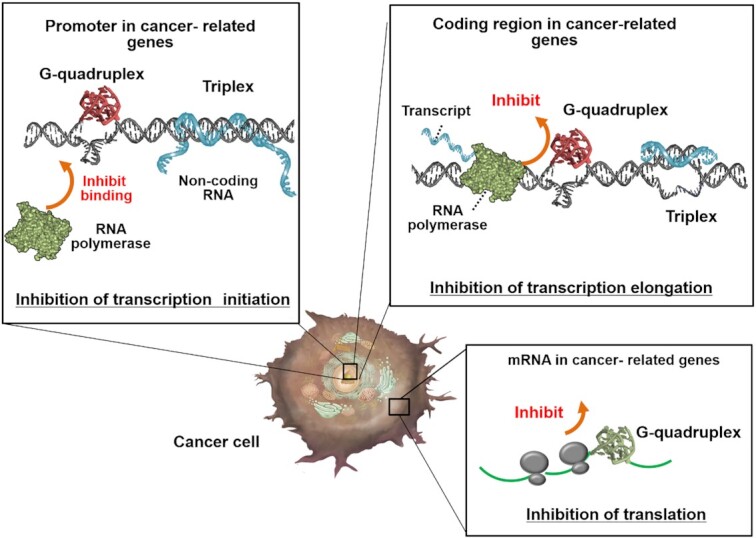
Roles of non-canonical structures in cancer cells. Non-canonical structures control gene expression. Formation of a non-canonical structure in the promoter region inhibits polymerase binding. Formation of non-canonical structures in the coding regions of DNA and mRNA inhibits transcription and translation, respectively.
Table 1.
Endogenous non-canonical structures related cancer and their roles
| Target gene (or RNA) | Where | Expected structure | Roles during cancer progression | Refs. |
|---|---|---|---|---|
| c–MYC | Promoter in c-MYC | DNA Triplex | Inhibition of expression of mRNA c-Myc Promotion of anti-proliferative activity in breast cancer | (95) |
| Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) | MALAT1 (lncRNA) | RNA Triplex | Increase the levels of an intronless β-globin reporter RNA via stabilization of MALAT1 | (96) |
| Multiple endocrine neoplasia-β (MENβ) RNA | MENβ RNA | RNA Triplex | Increase the levels of an intronless β-globin reporter RNA via stabilization of MENβ RNA | (97) |
| p27 | Promoter in p27 and lncRNA (MIR100HG) | RNA/DNA Triplex | Change for expression of p27 gene and the cell cycle, the progression of triple-negative breast cancer | (98) |
| MAT2A | Promoter in MAT2A and lncRNA (PARTICLE) | RNA/DNA Triplex | the ability of MALAT1 and MENβ sequences containing the predicted nuclear retention element and A-rich tract to increase the levels of an intronless β-globin reporter RNA | (99) |
| TGF-beta pathway | Promoter p27 and lncRNA (MEG3) | RNA/DNA Triplex | Regulation of thGF-beta pathway | (100) |
| c–MYC | Promoter in c-MYC | Antiparallel stranded DNA G-quadruplex | Decrease in transcript Promotion of cell migration | (102) |
| c-KIT | Promoter in c-KIT | Parallel stranded DNA G-quadruplex | Decrease in transcript Promotion of cell proliferation. | (103,104) |
| KRAS | Promoter in KRAS | Parallel stranded DNA G-quadruplex | Promotion of signal transduction pathways | (105) |
| VEGF | Promoter in VEGF | Parallel stranded DNA G-quadruplex | Promotion of tumor survival, growth and metastasis | (106,111) |
| PDGF-A | Promoter in PDGF-A | Parallel stranded DNA DNA G-quadruplex | Transcriptional regulation | (107) |
| BCL-2 | Promoter in BCL-2 | Parallel stranded DNA DNA G-quadruplex | Transcriptional regulation | (108) |
| insulin | Upstream of the insulin gene | DNA G-quadruplex | Increase in transcript levels | (109) |
| c-MYC | Coding region in c-MYC | Parallel stranded DNA G-quadruplex | Increase and decrease in transcript levels in response to G-quadruplex stability | (115) |
| Human Zic-1 zinc-finger protein | 5′-UTR of transcript | Parallel stranded RNA G-quadruplex | Promotion of cell migration, invasion, cellular development, and immunomodulation | (127) |
| Transforming growth factor β2 (TGFβ2) | 5′-UTR of transcript | Parallel stranded RNA G-quadruplex | Promotion of transforming growth factor βCell migration, invasion, cellular development, and immunomodulation | (128) |
The formation of G-quadruplexes in cancer related genes is considered to play important roles in cancer progression by altering the transcription and translation of cancer-related genes. The promoter region of the gene within 1 kb upstream of the transcription start site is significantly richer in G-quadruplex-forming sequences than the remainder of the genome (101). The structures of these G-quadruplex-forming sequences have been previously identified. For example, several G-quadruplex forming sequences such as those in c-MYC (102), c-KIT (103,104), KRAS (105), VEGF (106), PDGF-A (107) and BCL-2 (108) have been reported to form G-quadruplexes. As described above, these genes promote the progression of cancer, and the G-quadruplexes in their promoters may be involved in gene regulation.
The effect of non-canonical structures, especially of G-quadruplexes, on gene expression have been previously examined (Table 1). It has been reported that the G-quadruplex formation in the insulin-binding polymorphic region at upstream of the insulin gene promotes transcription (109). However, most G-quadruplexes inhibit transcription activity. For example, expression of the oncogene c-MYC which leads to enhanced cell proliferation (110), c-KIT which encodes a receptor tyrosine kinase, that when activated triggers signals (103,104). Interestingly, the G-quadruplex formations in the c-MYC and c-KIT promoter inhibit their transcriptional activation (102–104).
Recently, the role of G-quadruplex in response to changes in the chemical environment during tumour progression was elucidated in living cancer cells. The formation of the G-quadruplex is greatly influenced by the surrounding environment, especially K+ concentration (111). Because of overexpression of K+ channel, K+ concentration is decreased probably in malignant cancer cells compared with that in normal cells (112–114). In normal cells, the production of RNA transcribed from template DNA with G-quadruplexes is suppressed because the G-quadruplexes are stable. More transcripts are produced from the template with G-quadruplex ability in highly metastatic breast cancer cells than in milder cancer cells. This suggests that in normal cells, K+ ions repress the transcription of certain oncogenes by stabilising the G-quadruplex structure (115). In addition, tRNAs are highly expressed in malignant cancer cells. The 5′ untranslated region (UTR) mRNAs of oncogenes containing G-quadruplex-forming sequences are structurally altered by tRNAs from G-quadruplex to hairpin structure, and such alterations promote translation (116).
Conversely, transcriptional activity is controlled by the phase separation of transcription factors and DNA (117). The efficiency of formation of phase separation was determined using the highly ordered structures of nucleic acids (22,23). As the binding affinity of proteins with nucleic acids is determined by the structures of the nucleic acids, the structures of cancer-related genes may be involved in the formation of phase separation. Therefore, DNA non-canonical structures may regulate oncogene expression either directly or indirectly through phase separation.
G-quadruplexes are also present in transcribed RNA (37); for example, there are 4141 G-quadruplex-forming sequences in the 5′-UTR of human cancer-related genes (118,119) such as FGF-2 (120), NRAS (121), zinc-finger protein of the cerebellum 1 (Zic-1) (122), BCL2 (123), TRF-2 (124), VEGF (125) and ADAM-10 (126). Such G-quadruplexes of RNA in the 5′-UTR repress gene expression at the translational level. For example, the RNA G-quadruplex in Zic-1, which is overexpressed in medulloblastoma and is involved in cerebellar development, is reported to regulate translation (127). The G-quadruplex-forming sequence in the UTR was reported to reduce Zic-1 expression (122). Although it has been shown that some G-quadruplexes can augment translation, most of the well-characterised G-quadruplexes act as inhibitors. In vitro studies have demonstrated that 5′-UTR G-quadruplexes have regulatory functions, while in vivo studies have demonstrated the existence of G-quadruplex structures and their alteration in cancer tissues The translation of transforming growth factor β2 (TGFβ2) regulates cytokines and plays an important role in cell migration, invasion, and development and immune regulation (128). The formation of G-quadruplexes in the 5′-UTR of TGFβ2 mRNA enhances translational activity (128). Moreover, it has been reported that loss of the RNA G-quadruplex helicases DHX36 and DHX9 reduces translation of the coding regions of transcripts comprising proto-oncogenes, transcription factors, and epigenetic regulators through interaction between the helicases and the RNA G-quadruplex (129). Cancer is considered a multifactorial disease caused by a combination of genetic and environmental factors. Recently, whole genome sequencing of cancers has been used to identify mutations in cancer patients that overlap with those in potential RNA G-quadruplexes in the 5′-UTR. The results suggest that mutations in the G-quadruplex may act as cancer-promoting or suppressive mutations (130).
Recent studies have suggested that phase separation is critical for the regulation of transcription in an oncogene. For instance, the carboxy-terminal domain (CTD) in RNA polymerase II induces phase separation, and the RNA polymerase II forms clusters at the active genes via the interactions between the CTDs and activators (117). It is also reported that transcription factors activate genes via the phase-separation of mediator and activation domains (131). The transcription factors and RNA polymerase II are co-localised in stable clusters, which are associated with chromatins, have phase-separated condensate properties, and are sensitive to transcriptional inhibitors (132). Moreover, transcriptional coactivator proteins form droplets at super enhancers, which regulate genes, thereby influencing cell identity and function (133), especially in cancer-related genes including those related to diffuse large B cell lymphoma (134,135). The in vitro phase separation reconstitutes the super enhancer-enriched key components of the transcriptional machinery such as the mediator complex, suggesting that condensate formation near super enhancers may be associated with gene activation. Furthermore, malignancies occur due to chromosomal translocations resulting in new fusion proteins in which a DNA- or chromatin-binding domain is connected to the intrinsically disordered regions of a different protein, such as the fusion between the intrinsically disordered regions of the proteins in Ewing's sarcoma (136) or between the IDR of FUS and DNA-binding domains in certain types of liposarcoma (137). Phase separation drives aberrant expression programs of cancer-related genes. The relationship between nucleic acid structure and phase separation during cancer progression remains unclear; however, nucleic acid structures are also involved in biological reactions, hence, may relate also to the formation or dissociation of phase separations.
Nucleic acids involved in neurodegenerative diseases
Expansion of tandem repeats in cording or non-cording regions is associated with several neurodegenerative diseases. A typical hereditary neurodegenerative disease is the triplet repeat disease (138), which is caused by an abnormal extension of the triplet repeats. The association between repeat region expansion and neurodegenerative diseases was first reported in 1991 (139). The expansion of repeat length in triplet diseases leads to the expression of extended tandem repeat RNAs (140) and such repeat RNA leads to pathological conditions by loss of protein and RNA function. Moreover, protein and peptide aggregation translated from the expanded tandem repeats due to solid–liquid phase separation is also a major underlying cause of several neurodegenerative diseases (138).
As a typical example of repeat expansion disorders, the number of dCAG repeats in healthy individuals is ∼20, whereas HD patients have >40 repeats (Table 2) (141,142). The dCAG repeats produce a large number of abnormally elongated glutamine chains, called polyglutamine (poly-Q), and when poly-Q accumulates in neurons, it causes neuronal dysfunction and cell death (Figure 4) (143). In poly-Q related disease, with each generation, the repeat sequences are duplicated and lengthened. As the severity of the disease is usually proportional to the length of the repeat expansion, HD usually develops in middle-aged and older adults. Poly-Q disease can also induce abnormal gene expression, generally due to the loss of function of proteins that bind to aggregates and regulate transcription and translation. When huntingtin protein, the gene product responsible for HD, interacts with abnormally elongated poly-Q chains, fibres and aggregates are formed. Oculopharyngeal muscular dystrophy is caused by dGCG expansions in the PABP2 gene (144). Pathological expansion of polyalanine may result in accumulation of mutated PABP2 oligomers as filamentous inclusions in the nucleus (144). Huntington's disease-like 2 (HDL2), a neurodegenerative disease caused by the elongation of dCTG repeat sequences, involves symptoms that are very similar to those found in HD patients. JPH3, the gene responsible for HDL2, produces RNA with rCUG repeats and proteins with polyalanine and polyleucine (140). These RNAs are thought to be cytotoxic because of the abnormal aggregation of peptides encoded by them.
Table 2.
Repeat expansions involved in neurodegenerative diseases and their roles
| Related gene | Where | Repeat sequence | Expected DNA structure | Translation product | Objective | Disease | Refs. |
|---|---|---|---|---|---|---|---|
| ATN1 | DRPLA | (141), | |||||
| ATXN1 | CDS | dCAG | Hairpin | Poly-Q | Aggregation of peptide and protein | SCA1 | (143) |
| ATXN2 | rCAG | Cruciform | SCA 2 | ||||
| ATXN3 | SCA3(MJD) | ||||||
| CACNA1A | SCA6 | ||||||
| ATXN7 | SCA 7 | ||||||
| ATXN8 | SCA 8 | ||||||
| AR | SBMA | ||||||
| DRPLA | DRPLA | ||||||
| HTT | HD | ||||||
| PTB | SCA17 | ||||||
| PABP2 | CDS | dGCG | Hairpin | Poly-A | Aggregation of peptide and protein | OPMD | (144) |
| rGCG | |||||||
| JPH3 | CDS | dCTG | Hairpin | Poly-L | Aggregation of RNA (rCUG repeats), peptide and protein | HDL2 | (152) |
| rCUG | |||||||
| FMR1 | 5′UTR | dCGG | Hairpin | Transcription inhibition | FXS FXTAS | (155) | |
| rCGG | G-quadruplex | Loss of gene function | |||||
| PPP2R2B | 5′UTR | dCAG | Hairpin | Transcription inhibition | SCA12 | (143) | |
| rCAG | Loss of gene function | ||||||
| FXN | intron | dGAA | Triplex | Transcription inhibition | FRDA | (153) | |
| rGAA | Hairpin | Loss of gene function | |||||
| SCA10 | intron | dATTCT | Triplex | RNA foci increases stress in the nucleus | SCA10 | (156) | |
| rAUUCU | |||||||
| C9orf72 | intron | dGGGGCC | Hairpin | (RAN translation) | Aggregation of RNA (rGGGGCC repeats), peptide and protein | ALS/FTD | (147) |
| rGGGGCC | G-quadruplex | Poly-GA | |||||
| Poly-GP | |||||||
| Poly-GR | |||||||
| Poly-PA | |||||||
| Poly-PR | |||||||
| DMPK | 3′UTR | dCTG | Hairpin | Induce the multisystemic dominantly inherited disorder | DM1 | (158) | |
| rCUG | |||||||
| CNBP | intron | dCCTG | Aggregation of RNA | DM2 | (159) | ||
| rCCUG |
Figure 4.
Roles of non-canonical structures in neurodegenerative cells. Non-canonical structures control repeat expansion, gene expression, gene methylation, local translation and toxic accumulation.
In the case of repetition expansion disorder, when the DNA of a repetitive sequence forms a stable hairpin, the DNA strand ‘slips’ during replication, repair, and recombination, causing the repetitive sequence to elongate. Once the repeat sequence expands, the repeat sequence forms abnormal structures in multiple locations. RNA transcribed from the repeat region also forms an unusual structure. In these structure changes of DNA and RNA in the repeat region cause the disease to gradually increase with age and in subsequent generations (145).
Non-canonical structures of DNA and RNA in genes associated with neurodegenerative diseases
As shown above, the expansion of tandem repeats is closely related to neurodegenerative diseases (138). The detailed sequences related to neurodegenerative diseases are shown in Table 2. In repeat diseases, not only triplet repeats but also diseases involving sequences of five or six repeats have been reported (11,146). For example, the expansion of hexanucleotide dGGGGCC repeats in the first intron of C9ORF72 is the most common genetic cause of ALS and frontotemporal dementia (C9-ALS/FTD) (147). These repeat sequences form not only standard duplexes, but also hairpin, triplex, and G-quadruplex structures (Figure 5), such as the formation of hairpin structures, which are prevalent in dCNG, dGNC, dANT or dTNA repeat sequences (N = dA, dC, dG or dT) (148). The stem in the hairpin structures is composed of Watson–Crick base pairs and N–N mismatches (149). The dGAA repeats form a triplex (150) and dAGG and dTGG repeats form G-quadruplexes (151). Such non-canonical structures may cause repeat regions to elongate during DNA replication. For example, the dCTG triplet repeat sequence has been shown to self-expand during DNA replication when dCTG triplet repeat sequence forms hairpin structures (Figure 4) (152).
Figure 5.
Structures of repeat expansion DNAs involved in neurodegenerative diseases. Repeat sequences of dCAT, dATG, dCTG, dCTA, dTAG, dCAG, dTTA, dCGG or dCGG form hairpins, dGAA forms triplexes, and dAGG or dTGG form G-quadruplexes.
Triplet repeat diseases exhibit occasionally different pathogenic mechanisms depending on the location of the repeat region in the gene sequence (Table 2 and Figures 5 and 6). If this repeat region is in an open reading frame (ORF), the polypeptide is translated. These polypeptides accumulate in the cell as a collection of toxic peptides and proteins, resulting in gain-of-function mutations. However, the expansion of these triplet repeats can also occur in regions other than the ORF (untranslated regions). As these regions generally do not produce proteins, triplet repeat diseases caused by repeats in untranslated regions are different from those caused by repeats in the ORF, such as poly-Q disease.
Figure 6.
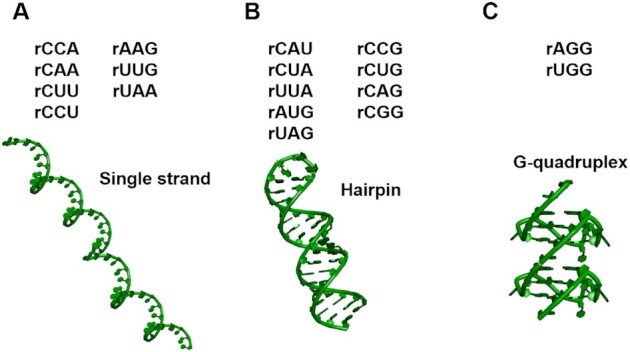
Structures of repeat expansion RNAs involved in neurodegenerative diseases. Among the repeats, rCCA, rAAG, rCAA, rUUG, rCUU, rUAA and rCCU do not form any higher-order structures; rCAU, rCCG, rCUA, rCUG, rUUA, rCAG, rAUG, rCGG and rUAG form hairpins; rAGG and rUGG repeats form G-quadruplexes.
Meanwhile, repeat sequences in introns reduce the expression levels of mRNAs. Such a decrease in the expression levels of mRNAs leads to loss-of-function type mutations in proteins. In addition, repeat expansion in non-coding regions such as introns and UTRs can also lead to gain-of-function of the repeated RNA, forming toxic foci in the cell. In recessive hereditary fragile X syndrome and Friedreich schizophrenia, an increased number of rCGG repeats in the 5′-UTR and rGAA repeats in the intron form a hairpin and triplex structure, respectively (Figure 5) (153), and consequently, inhibit transcription, thereby significantly decreasing the levels of gene products (loss of function) (154). Fragile X-associated tremor and ataxia syndrome (FXTAS) is a genetic disorder characterised by cerebellar ataxia and cognitive dysfunction. The extended rCGG repeat of the fragile X syndrome gene (FMR1) induces RAN translation, which is a translation reaction independent of AUG-initiation. This RAN translation has been shown to generate polyglycine (polyG) FMR1 protein, which accumulates in ubiquitin-positive inclusion bodies and causes toxicity. In myotonic dystrophy type 1 (DM1), there is an extended repeat in the 3′ UTR of the transcript, resulting in an abnormally long mRNA length. As this expansion becomes progressively longer in one allele, the pathogenesis of the disease also becomes more severe. The gain-of-function form of DM1 is unable to perform its normal function of splicing a particular gene family, as a result of inhibition of the activity of the rCUG-binding protein induced by the RNA repeats (155). Spinocerebellar ataxia type 10 (SCA10), which is an autosomal dominant genetic disorder, causes cerebellar ataxia and seizures. SCA10 patients exhibit expansion of a pentanucleotide (dATTCT) repeat in intron 9 of the ATXN10 gene. However, expanded dATTCT repeats are not passed from parent to child. Therefore, an inverse correlation between the length of dATTCT repeats and age of onset has been observed, which is unusual for a repeat disease (156). A dCTG expansion located in the 3′-UTR of a kinase gene causes DM1 (157). In addition, expansion of dCCTG causes a disease state similar to myotonic dystrophy type 2 (DM2). The behaviour of dCTC and dCCCTG expansions has shown that both DM1 and DM2 mutations are pathogenic at the RNA level (158). The finding that the accumulation of RNAs and peptides caused by repeat expansion of RNAs alters cellular functions indicates the existence of a new category of diseases.
In other genes associated with neurological diseases, the importance of non-canonical structures has been suggested. The transcriptional regulator, alpha-thalassemia/mental retardation syndrome X-linked (ATRX), binds G-rich tandem repeat sequences in both telomeres and euchromatin, and the binding affects nuclear processes (159). Moreover, ATRX also binds to the G-quadruplex of the CpG island of the Xlr3b gene and enhances its expression by recruiting DNA methyltransferases (160) (Figure 4). Specific neuronal mRNAs are transported to axons in nerve cells and dendrites and locally translated at synapses. Interestingly, bioinformatics analysis has shown that approximately 30% of the dendritic mRNAs have G-quadruplex-forming sequences in the 3′-UTR (161). Fragile X mental retardation protein uses its arginine–glycine–glycine (RGG) box to bind to the G-quadruplex located at the 5′-UTRs of the MAP1B, APP and PP2Ac mRNAs, and the binding induces translational repression (162) (Figure 4). The protein level correlates with the stability of G-quadruplexes in an mRNA (163). Such accumulating evidence illustrates the importance of the structure of nucleic acids, indicating that transcript RNAs may be important contributors to neurodegenerative diseases.
RNA structures affect cytotoxicity via phase separation in neurodegenerative diseases
How repeat expansion sequences that do not produce abnormal proteins or peptides can cause disease development is still unknown. Past studies have treated DNA structural abnormalities in repeat extension sequences and toxicity from expression of mutated proteins as the primary causes of neurodegeneration. However, the breakthrough in this field came from an RNA pathophysiology study in myotonic dystrophy (DM), wherein it was demonstrated that the extended dCTG repeat transcribes the rCUG RNA repeat, but the rCUG RNA repeat is not translated (164). The rCUG repeats undergo toxic gain-of-function and form droplets called rCUG RNA foci, which cause protein dysregulation and gene silencing in the nucleus (164). Such RNA foci are characteristically observed in DM (165), and the accumulation of RNA foci has been observed not only in cells but also in tissues. Moreover, transcripts of the extended dATTCT sequence were observed in a SCA10 patient (166). Furthermore, rAUUCU forms RNA foci in SCA10 lymphoblasts as well as in DM. Localisation of the foci in the nucleus and their co-localisation with some RNA-binding proteins were also clarified. In general, when RNA foci are formed, stress in the nucleus is increased (167,168), and RNA binding proteins are sequestered (169). As a result, nucleocytoplasmic transport is inhibited (167), which is thought to cause a variety of neurodegenerative symptoms. The repeat RNA sequences form several unique structures. The detail structures of repeat RNA sequences have been investigated because RNA structures are important in the formation of foci and binding to proteins. For example, the RNA repeat sequence of rCMG, rGMC, rAMU or rUMA (where M = rA, rC, rG or rU) also form hairpins. The rAGG and rUGG repeats fold into G-quadruplexes (Figure 6) (170). Moreover, rGGGGCC repeats can form the both of hairpins and parallel G-quadruplexes responsive to the solution conditions (171). Recently, the transcripts from repeat expansions have been reported to form RNA gels using phase-separation analysis (10). The rCAG repeats exhibit liquid-like properties in the solution containing polyvalent cations, whereas when the rGGGGCC repeats form predominantly G-quadruplexes, the repeats exhibit gel-like properties. These findings suggest that the secondary structure of the repeat RNA sequences during RNA gelation be one of a key factor in the pathogenesis because phase separation of the repeat RNA sequences may affect aggregate formation of cytotoxic peptides and proteins.
Nevertheless, the specific reason for RNA repeat toxicity has yet to be explained. The RNA repeat sequences form condensed liquid or gel-like states (droplets) or aggregates as previously described (Figure 4). The rGGGGCC repeats are phase-separated depending on the repeat length and the structure of the repeat region (89). The length of rGGGGCC repeat has been associated with the aetiology of C9-ALS/FTD (147), and cells from some C9-ALS/FTD patients are positive for cytoplasmic foci of rGGGGGCC (172). Importantly, the gelation of these RNAs is regulated by the RNA structures; RNA G-quadruplexes facilitate RNA accumulation in G-rich repeat expansions (22). The effects of RNA toxicity are thought to have mechanisms similar to those of protein-induced toxicity. For example, there may be changes in transcript levels, changes in the interactions of proteins that bind to mRNA, abnormalities in mRNA splicing and processing, abnormalities in translation or product mutation, abnormalities in protein quality control pathways, and abnormalities in signalling cascades (173). Overall, aberrant phase separations are associated with neurodegenerative diseases (72).
CONCLUSIONS, CHALLENGES AND PERSPECTIVES
Herein, we described the effects of nucleic acid structures on cancer and neurodegenerative diseases. Cancer and neurodegenerative diseases are completely different entities, but have some overlap, in which aberrant nucleic acid structures induce a topology change to control gene expression and biological reactions that contribute to disease onset (Figure 7). Specifically, transcription and translation are inhibited or promoted when non-canonical structures are formed from the canonical duplex structure in the gene and the mRNA (29). Moreover, various types of droplets and aggregates are formed in the cytoplasm and the nucleus by phase separation in the cells of patients with cancer and neurological diseases owing to liquid–liquid phase separation and liquid–solid phase separation. These droplets and aggregates encapsulate intracellular nucleic acids and proteins and promote or suppress various biological reactions, contributing to the progression of cancer and neurodegenerative diseases. Importantly, the non-canonical structures change the efficiency of liquid–liquid phase separation and liquid–solid phase separation. For example, droplets are triggered by the naturally denatured domains of proteins and the π–π stacking and cation-π interactions of nucleic acids (174). Droplet formation is also promoted by G-quadruplexes with an exposed π-plane, as this structure is favourable for interacting with droplet-forming proteins and peptides (22,23). In contrast, the structures of duplexes render them unfavourable for uptake within droplets (175). Thus, even for nucleic acids having the same sequences, their interactions with droplet-constituting biomolecules differ depending on their structures.
Figure 7.
Possibilities for effects of structural changes in nucleic acids on disease onset and progression. First, the structures of DNA and RNA change in response to environmental changes in the cell. These structural changes repress or promote related gene expression and, therefore, subsequent transcription and translation. Thus, the progression of cancer and neurodegenerative diseases may be altered.
From a physicochemical point of view, the mechanism of nucleic acid structure formation is primarily regulated by binding with cations and water, leading to the formation of hydrogen bonds and stacking interactions. The nucleic acid structures can be predicted by considering the interactions between the surrounding environment and the nucleic acids, as the mechanism is the same in cells during any disease; precisely, nucleic acids bind to cations to reduce electrostatic repulsion due to the negative charge of the phosphate groups. In particular, non-canonical structures are stabilised by the binding of specific ions; for example, increasing the H+ and Mg2+ content in a solution stabilises the triplexes (31,36). In addition, increasing the number of K+ ions specifically stabilises G-quadruplexes. Interestingly, in diseased cells such as those of cancer and neurodegenerative diseases, the concentration of ions (e.g. K+, H+, Na+, Mg2+) is typically changed by an overexpression (or inactivation) of disease-specific ‘ion channel’ proteins. Moreover, the overexpression of certain proteins due to a disease may reduce the water activity of the solution, which in turn, stabilises the non-canonical structures. Thus, clarification of the mechanisms underlying the control of biological reactions by nucleic acid structures can provide new perspectives and insights into the mechanisms underlying disease onset.
It is important to develop methods to regulate the structures and stabilities of nucleic acids in disease-associated genes for therapeutic applications (176,177). As the formation of the G-quadruplex regulates the expression of oncogenes, small molecules that stabilise the G-quadruplex structure have been evaluated for their effectiveness as anticancer agents (178). The G-quadruplex has a structure in which the large π-plane of the G-quartet is exposed, and many small molecules that bind specifically to the G-quadruplex recognize this π-plane and bind to the G-quadruplex through stacking interactions (179). Recently, small molecules with structural specificity for the RNA G-quadruplex of rGGGGCC repeats have been developed to tackle C9FTD/ALS. TMPyP4 (5,10,15,20-tetra(N-methyl-4-pyridyl) porphyrin), is a cationic porphyrin known to bind to the G-quadruplex. When TMPyP4 binds to the rGGGGCC repeats of C9orf72, it distorts the structure of the repeats and interferes with their interactions with hnRNPA1 or ASF/SF2. As a result, either the sequestration of proteins and/or their translation into toxic dipeptides is inhibited (180). Similarly, a small molecule was identified and shown to reduce RAN translation of C9orf72 and RNA aggregation in both cells and neurons from patients with expanded repeats (181). Moreover, antisense oligonucleotides (ASOs) targeting the G-quadruplex-forming rGGGGCC repeats in C9orf72 RNA have been reported to reduce toxicity (182). ASO has been utilized to normalize proteins that exhibit cytotoxicity and loss of function in other neurodegenerative diseases characterized by gain-of-function mechanisms that do not involve the G-quadruplex (183,184). These studies indicate that targeting RNA repeats using small molecules and oligonucleotides is an effective therapeutic strategy. Thus, the physicochemical insights into nucleic acid structures involved in cancer and neurodegenerative diseases summarised in this review are essential for the rational design of therapeutic materials.
DATA AVAILABILITY
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
ACKNOWLEDGEMENTS
We would like to acknowledge Professor Keiko Kawauchi of Konan University and Professor Eri Chatani of Kobe University for their inputs on this manuscript based on their expertise in the cancer and neurodegeneration fields.
Contributor Information
Hisae Tateishi-Karimata, Frontier Institute for Biomolecular Engineering Research (FIBER), Konan University, 7-1-20 Minatojima-minamimachi, Chuo-ku, Kobe 650-0047, Japan.
Naoki Sugimoto, Frontier Institute for Biomolecular Engineering Research (FIBER), Konan University, 7-1-20 Minatojima-minamimachi, Chuo-ku, Kobe 650-0047, Japan; Graduate School of Frontiers of Innovative Research in Science and Technology (FIRST), Konan University, 7-1-20 Minatojima-minamimachi, Chuo-ku, Kobe 650-0047, Japan.
FUNDING
Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT); Grant-in-Aid for Scientific Research on Innovative Areas of ‘Chemistry for Multimolecular Crowding in Biosystems’, JSPS KAKENHI [JP17H06351, JP19H00928, JP18KK0164]; MEXT-Supported Program for the Strategic Research Foundation at Private Universities (2014–2019), Japan; Hirao Taro Foundation of Konan Gakuen for Academic Research; Chubei Itoh Foundation; The Naito Foundation. Funding for open access charge: Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Conflict of interest statement. None declared.
REFERENCES
- 1.Weinberg R. A.2nd edn. The Biology of Cancer. 2013; Garland Science; 31–70. [Google Scholar]
- 2.Singletary K.W., Gapstur S.M.. Alcohol and breast cancer: review of epidemiologic and experimental evidence and potential mechanisms. JAMA. 2001; 286:2143–2151. [DOI] [PubMed] [Google Scholar]
- 3.Sasco A.J., Secretan M.B., Straif K.. Tobacco smoking and cancer: a brief review of recent epidemiological evidence. Lung Cancer. 2004; 45:S3–S9. [DOI] [PubMed] [Google Scholar]
- 4.Hopkinson J.B., Wright D.N.M., McDonald J.W., Corner J.L.. The prevalence of concern about weight loss and change in eating habits in people with advanced cancer. J. Pain Symptom Manag. 2006; 32:322–331. [DOI] [PubMed] [Google Scholar]
- 5.Kleine-Kohlbrecher D., Adhikary S., Eilers M.. Mechanisms of transcriptional repression by Myc. Curr. Top. Microbiol. Immunol. 2006; 302:51–62. [DOI] [PubMed] [Google Scholar]
- 6.Dutta D., Debnath M., Muller D., Paul R., Das T., Bessi I., Schwalbe H., Dash J.. Cell penetrating thiazole peptides inhibit c-MYC expression via site-specific targeting of c-MYC G-quadruplex. Nucleic Acids Res. 2018; 46:5355–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano S., Miyoshi D., Sugimoto N.. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014; 114:2733–2758. [DOI] [PubMed] [Google Scholar]
- 8.Han X., Yu D., Gu R., Jia Y., Wang Q., Jaganathan A., Yang X., Yu M., Babault N., Zhao C.et al.. Roles of the BRD4 short isoform in phase separation and active gene transcription. Nat. Struct. Mol. Biol. 2020; 27:333–341. [DOI] [PubMed] [Google Scholar]
- 9.Nozawa R.S., Yamamoto T., Takahashi M., Tachiwana H., Maruyama R., Hirota T., Saitoh N.. Nuclear microenvironment in cancer: control through liquid-liquid phase separation. Cancer Sci. 2020; 111:3155–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain A., Vale R.D.. RNA phase transitions in repeat expansion disorders. Nature. 2017; 546:243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balendra R., Isaacs A.M.. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol. 2018; 14:544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross C.A., Poirier M.A.. Protein aggregation and neurodegenerative disease. Nat. Med. 2004; 10:S10–S17. [DOI] [PubMed] [Google Scholar]
- 13.Pedersen J.T., Heegaard N.H.. Analysis of protein aggregation in neurodegenerative disease. Anal. Chem. 2013; 85:4215–4227. [DOI] [PubMed] [Google Scholar]
- 14.DeJesus-Hernandez M., Rayaprolu S., Soto-Ortolaza A.I., Rutherford N.J., Heckman M.G., Traynor S., Strongosky A., Graff-Radford N., Van Gerpen J., Uitti R.J.et al.. Analysis of the C9orf72 repeat in Parkinson's disease, essential tremor and restless legs syndrome. Parkinsonism Relat. Disord. 2013; 19:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J.et al.. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011; 72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L.et al.. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-Linked ALS-FTD. Neuron. 2011; 72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shelbourne P.F., Killeen N., Hevner R.F., Johnston H.M., Tecott L., Lewandoski M., Ennis M., Ramirez L., Li Z., Iannicola C.et al.. A Huntington's disease CAG expansion at the murine Hdh locus is unstable and associated with behavioural abnormalities in mice. Hum. Mol. Genet. 1999; 8:763–774. [DOI] [PubMed] [Google Scholar]
- 18.Rudnicki D.D., Holmes S.E., Lin M.W., Thornton C.A., Ross C.A., Margolis R.L.. Huntington's disease-like 2 is associated with CUG repeat-containing RNA foci. Ann. Neurol. 2007; 61:272–282. [DOI] [PubMed] [Google Scholar]
- 19.Labbadia J., Morimoto R.I.. Huntington's disease: underlying molecular mechanisms and emerging concepts. Trends Biochem. Sci. 2013; 38:378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aziz N.A., van Belzen M.J., Roos R.A.. Intergenerational CAG repeat instability is highly heritable in Huntington's disease. J. Med. Genet. 2008; 45:766. [DOI] [PubMed] [Google Scholar]
- 21.Gan L., Cookson M.R., Petrucelli L., La Spada A.R.. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 2018; 21:1300–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng Y., Tateishi-Karimata H., Sugimoto N.. RNA G-quadruplexes facilitate RNA accumulation in G-rich repeat expansions. Biochemistry. 2020; 59:1972–1980. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., Yang M., Duncan S., Yang X., Abdelhamid M.A.S., Huang L., Zhang H., Benfey P.N., Waller Z.A.E., Ding Y.. G-quadruplex structures trigger RNA phase separation. Nucleic Acids Res. 2019; 47:11746–11754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson J.D., Crick F.H.. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953; 171:737–738. [DOI] [PubMed] [Google Scholar]
- 25.Asensio J.L., Brown T., Lane A.N.. Solution conformation of a parallel DNA triple helix with 5′ and 3′ triplex-duplex junctions. Structure. 1999; 7:1–11. [DOI] [PubMed] [Google Scholar]
- 26.Felsenfeld G., Rich A.. Studies on the formation of two- and three-stranded polyribonucleotides. Biochim. Biophys. Acta. 1957; 26:457–468. [DOI] [PubMed] [Google Scholar]
- 27.Sen D., Gilbert W.. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990; 344:410–444. [DOI] [PubMed] [Google Scholar]
- 28.Phan A.T., Mergny J.L.. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002; 30:4618–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robles J., Grandas A., Pedroso E., Luque F.J., Eritja R., Orozco M.. Nucleic acid triple helices: stability effects of nucleobase modifications. Curr. Org. Chem. 2002; 6:1333–1368. [Google Scholar]
- 31.Lyamichev V.I., Voloshin O.N., Frank-Kamenetskii M.D., Soyfer V.N.. Photofootprinting of DNA triplexes. Nucleic Acids Res. 1991; 19:1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng A.J., Van Dyke M.W.. Oligodeoxyribonucleotide length and sequence effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1994; 22:4742–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gowers D.M., Fox K.R.. DNA triple helix formation at oligopurine sites containing multiple contiguous pyrimidines. Nucleic Acids Res. 1997; 25:3787–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scaria P.V., Shafer R.H.. Calorimetric analysis of triple helices targeted to the d(G3A4G3).d(C3T4C3) duplex. Biochemistry. 1996; 35:10985–10994. [DOI] [PubMed] [Google Scholar]
- 35.Chandler S.P., Fox K.R.. Specificity of antiparallel DNA triple helix formation. Biochemistry. 1996; 35:15038–15048. [DOI] [PubMed] [Google Scholar]
- 36.Lee J.S., Johnson D.A., Morgan A.R.. Complexes formed by (pyrimidine)n. (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979; 6:3073–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen D., Gilbert W.. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. [DOI] [PubMed] [Google Scholar]
- 38.Sundquist W.I., Klug A.. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989; 342:825–829. [DOI] [PubMed] [Google Scholar]
- 39.Williamson J.R., Raghuraman M.K., Cech T.R.. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989; 59:871–880. [DOI] [PubMed] [Google Scholar]
- 40.Raghuraman M.K., Cech T.R.. Effect of monovalent cation-induced telomeric DNA structure on the binding of Oxytricha telomeric protein. Nucleic Acids Res. 1990; 18:4543–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kankia B.I., Marky L.A.. Folding of the thrombin aptamer into a G-quadruplex with Sr2+: Stability, heat, and hydration. J. Am. Chem. Soc. 2001; 123:10799–10804. [DOI] [PubMed] [Google Scholar]
- 42.Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todd A.K., Johnston M., Neidle S.. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005; 33:2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 45.Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M.et al.. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 46.Kouzine F., Wojtowicz D., Baranello L., Yamane A., Nelson S., Resch W., Kieffer-Kwon K.R., Benham C.J., Casellas R., Przytycka T.M.et al.. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 2017; 4:344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilf J., Minton A.P.. Evidence for protein self-association induced by excluded volume. Myoglobin in the presence of globular proteins. Biochim. Biophys. Acta. 1981; 670:316–322. [DOI] [PubMed] [Google Scholar]
- 48.Zimmerman S.B., Minton A.P.. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 1993; 22:27–65. [DOI] [PubMed] [Google Scholar]
- 49.Schummel P.H., Gao M., Winter R.. Modulation of the polymerization kinetics of alpha/beta-tubulin by osmolytes and macromolecular crowding. ChemPhysChem. 2017; 18:189–197. [DOI] [PubMed] [Google Scholar]
- 50.Wenner J.R., Bloomfield V.A.. Crowding effects on EcoRV kinetics and binding. Biophys. J. 1999; 77:3234–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breslauer K.J., Frank R., Blocker H., Marky L.A.. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Auffinger P., Westhof E.. Melting of the solvent structure around a RNA duplex: a molecular dynamics simulation study. Biophys. Chem. 2002; 95:203–210. [DOI] [PubMed] [Google Scholar]
- 53.Auffinger P., Westhof E.. Hydrophobic groups stabilize the hydration shell of 2′-O-methylated RNA duplexes. Angew. Chem. Int. Ed. Engl. 2001; 40:4648–4650. [DOI] [PubMed] [Google Scholar]
- 54.Anderson C.F., Record M.T. Jr. Salt-nucleic acid interactions. Annu. Rev. Phys. Chem. 1995; 46:657–700. [DOI] [PubMed] [Google Scholar]
- 55.Feig M., Pettitt B.M.. Sodium and chlorine ions as part of the DNA solvation shell. Biophys. J. 1999; 77:1769–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feig M., Pettitt B.M.. A molecular simulation picture of DNA hydration around A- and B-DNA. Biopolymers. 1998; 48:199–209. [DOI] [PubMed] [Google Scholar]
- 57.Spink C.H., Chaires J.B.. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry. 1999; 38:496–508. [DOI] [PubMed] [Google Scholar]
- 58.Nordstrom L.J., Clark C.A., Andersen B., Champlin S.M., Schwinefus J.J.. Effect of ethylene glycol, urea, and N-methylated glycines on DNA thermal stability: the role of DNA base pair composition and hydration. Biochemistry. 2006; 45:9604–9614. [DOI] [PubMed] [Google Scholar]
- 59.Record M.T. Jr, Anderson C.F., Lohman T.M.. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys. 1978; 11:103–178. [DOI] [PubMed] [Google Scholar]
- 60.Rozners E., Moulder J.. Hydration of short DNA, RNA and 2′-OMe oligonucleotides determined by osmotic stressing. Nucleic Acids Res. 2004; 32:248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakano S., Karimata H., Ohmichi T., Kawakami J., Sugimoto N.. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 2004; 126:14330–14331. [DOI] [PubMed] [Google Scholar]
- 62.Spink C.H., Garbett N., Chaires J.B.. Enthalpies of DNA melting in the presence of osmolytes. Biophys. Chem. 2007; 126:176–185. [DOI] [PubMed] [Google Scholar]
- 63.Ghosh S., Takahashi S., Ohyama T., Endoh T., Tateishi-Karimata H., Sugimoto N.. Nearest-neighbor parameters for predicting DNA duplex stability in diverse molecular crowding conditions. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:14194–14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyoshi D., Karimata H., Sugimoto N.. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006; 128:7957–7963. [DOI] [PubMed] [Google Scholar]
- 65.Miyoshi D., Nakamura K., Tateishi-Karimata H., Ohmichi T., Sugimoto N.. Hydration of Watson-Crick base pairs and dehydration of Hoogsteen base pairs inducing structural polymorphism under molecular crowding conditions. J. Am. Chem. Soc. 2009; 131:3522–3531. [DOI] [PubMed] [Google Scholar]
- 66.Muhuri S., Mimura K., Miyoshi D., Sugimoto N.. Stabilization of three-way junctions of DNA under molecular crowding conditions. J. Am. Chem. Soc. 2009; 131:9268–9280. [DOI] [PubMed] [Google Scholar]
- 67.Son I., Shek Y.L., Dubins D.N., Chalikian T.V.. Hydration changes accompanying helix-to-coil DNA transitions. J. Am. Chem. Soc. 2014; 136:4040–4047. [DOI] [PubMed] [Google Scholar]
- 68.Nakano M., Tateishi-Karimata H., Tanaka S., Tama F., Miyashita O., Nakano S., Sugimoto N.. Thermodynamic properties of water molecules in the presence of cosolute depend on DNA structure: a study using grid inhomogeneous solvation theory. Nucleic Acids Res. 2015; 43:10114–101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan H.Y., Shek Y.L., Amiri A., Dubins D.N., Heerklotz H., Macgregor R.B. Jr, Chalikian T.V.. Volumetric characterization of sodium-induced G-quadruplex formation. J. Am. Chem. Soc. 2011; 133:4518–4526. [DOI] [PubMed] [Google Scholar]
- 70.Liu L., Kim B.G., Feroze U., Macgregor R.B. Jr, Chalikian T.V.. Probing the Ionic Atmosphere and Hydration of the c-MYC i-Motif. J. Am. Chem. Soc. 2018; 140:2229–2238. [DOI] [PubMed] [Google Scholar]
- 71.Alberti S., Gladfelter A., Mittag T.. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019; 176:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alberti S., Dormann D.. Liquid-liquid phase separation in disease. Annu. Rev. Genet. 2019; 53:171–194. [DOI] [PubMed] [Google Scholar]
- 73.Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P.. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016; 165:1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woodruff J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., Hyman A.A.. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell. 2017; 169:1066–1077. [DOI] [PubMed] [Google Scholar]
- 75.Riback J.A., Zhu L., Ferrolino M.C., Tolbert M., Mitrea D.M., Sanders D.W., Wei M.T., Kriwacki R.W., Brangwynne C.P.. Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020; 581:209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nott T.J., Craggs T.D., Baldwin A.J.. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat. Chem. 2016; 8:569–575. [DOI] [PubMed] [Google Scholar]
- 77.Riback J.A., Katanski C.D., Kear-Scott J.L., Pilipenko E.V., Rojek A.E., Sosnick T.R., Drummond D.A.. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017; 168:1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dumas L., Herviou P., Dassi E., Cammas A., Millevoi S.. G-Quadruplexes in RNA biology: recent advances and future directions. Trends Biochem. Sci. 2021; 46:270–283. [DOI] [PubMed] [Google Scholar]
- 79.Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., King D.S., Taunton J., Rosen M.K., Vale R.D.. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016; 352:595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanders D.W., Kedersha N., Lee D.S.W., Strom A.R., Drake V., Riback J.A., Bracha D., Eeftens J.M., Iwanicki A., Wang A.et al.. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell. 2020; 181:306–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hainaut P., Plymoth A.. Targeting the hallmarks of cancer: towards a rational approach to next-generation cancer therapy. Curr. Opin. Oncol. 2013; 25:50–51. [DOI] [PubMed] [Google Scholar]
- 82.Hanahan D., Weinberg R.A.. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 83.Du M., Chen Z.J.. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018; 361:704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X.et al.. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018; 361:aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rice A.M., Rosen M.K.. ATP controls the crowd. Science. 2017; 356:701–702. [DOI] [PubMed] [Google Scholar]
- 86.Franzmann T.M., Jahnel M., Pozniakovsky A., Mahamid J., Holehouse A.S., Nuske E., Richter D., Baumeister W., Grill S.W., Pappu R.V.et al.. Phase separation of a yeast prion protein promotes cellular fitness. Science. 2018; 359:eaao5654. [DOI] [PubMed] [Google Scholar]
- 87.Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillen-Boixet J., Franzmann T.M.et al.. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018; 360:918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R.et al.. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014; 507:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fay M.M., Anderson P.J., Ivanov P.. ALS/FTD-associated C9ORF72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 2017; 21:3573–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brangwynne C.P., Tompa P., Pappu R.V.. Polymer physics of intracellular phase transitions. Nat. Phys. 2015; 11:899–904. [Google Scholar]
- 91.Klosin A., Oltsch F., Harmon T., Honigmann A., Julicher F., Hyman A.A., Zechner C.. Phase separation provides a mechanism to reduce noise in cells. Science. 2020; 367:464–468. [DOI] [PubMed] [Google Scholar]
- 92.J.Levine A., Hu W., Feng Z. Chapter 3 - Tumor suppressor genes. The Molecular Basis of Cancer. 2008; 3rd edn31–38. [Google Scholar]
- 93.Kontomanolis E.N., Koutras A., Syllaios A., Schizas D., Mastoraki A., Garmpis N., Diakosavvas M., Angelou K., Tsatsaris G., Pagkalos A.et al.. Role of oncogenes and tumor-suppressor genes in carcinogenesis: a review. Anticancer Res. 2020; 40:6009–6015. [DOI] [PubMed] [Google Scholar]
- 94.Kawauchi K., Urano R., Kinoshita N., Kuwamoto S., Torii T., Hashimoto Y., Taniguchi S., Tsuruta M., Miyoshi D.. Photosensitizers based on G-quadruplex ligand for cancer photodynamic therapy. Genes (Basel). 2020; 11:1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Belotserkovskii B.P., De Silva E., Tornaletti S., Wang G., Vasquez K.M., Hanawalt P.C.. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007; 282:32433–32441. [DOI] [PubMed] [Google Scholar]
- 96.Li S., Ma F., Jiang K., Shan H., Shi M., Chen B.. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 promotes lung adenocarcinoma by directly interacting with specificity protein 1. Cancer Sci. 2018; 109:1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang Z., Zhou J.K., Peng Y., He W., Huang C.. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer. 2020; 19:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang S., Ke H., Zhang H., Ma Y., Ao L., Zou L., Yang Q., Zhu H., Nie J., Wu C.et al.. LncRNA MIR100HG promotes cell proliferation in triple-negative breast cancer through triplex formation with p27 loci. Cell Death. Dis. 2018; 9:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O’Leary V.B., Ovsepian S.V., Carrascosa L.G., Buske F.A., Radulovic V., Niyazi M., Moertl S., Trau M., Atkinson M.J., Anastasov N.. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015; 11:474–485. [DOI] [PubMed] [Google Scholar]
- 100.Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E.et al.. MEG3 longnoncoding RNA regulates the TGF-β pathwaygenes through formation of RNA-DNA triplex structures. Nat. Commun. 2015; 6:7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huppert J.L., Balasubramanian S.. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007; 35:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rankin S., Reszka A.P., Huppert J., Zloh M., Parkinson G.N., Todd A.K., Ladame S., Balasubramanian S., Neidle S.. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005; 127:10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fernando H., Reszka A.P., Huppert J., Ladame S., Rankin S., Venkitaraman A.R., Neidle S., Balasubramanian S.. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006; 45:7854–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cogoi S., Xodo L.E.. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006; 34:2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun D., Guo K., Rusche J.J., Hurley L.H.. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005; 33:6070–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qin Y., Rezler E.M., Gokhale V., Sun D., Hurley L.H.. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007; 35:7698–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dai J., Dexheimer T.S., Chen D., Carver M., Ambrus A., Jones R.A., Yang D.. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006; 128:1096–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Catasti P., Chen X., Moyzis R.K., Bradbury E.M., Gupta G.. Structure-function correlations of the insulin-linked polymorphic region. J. Mol. Biol. 1996; 264:534–545. [DOI] [PubMed] [Google Scholar]
- 110.Spencer C.A., Groudine M.. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer. Res. 1991; 56:1–48. [DOI] [PubMed] [Google Scholar]
- 111.Kim B.G., Long J., Dubins D.N., Chalikian T.V.. Ionic effects on VEGF G-quadruplex stability. J. Phys. Chem. B. 2016; 120:4963–4971. [DOI] [PubMed] [Google Scholar]
- 112.Spitzner M., Ousingsawat J., Scheidt K., Kunzelmann K., Schreiber R.. Voltage-gated K+ channels support proliferation of colonic carcinoma cells. FASEB J. 2007; 21:35–44. [DOI] [PubMed] [Google Scholar]
- 113.Ousingsawat J., Spitzner M., Puntheeranurak S., Terracciano L., Tornillo L., Bubendorf L., Kunzelmann K., Schreiber R.. Expression of voltage-gated potassium channels in human and mouse colonic carcinoma. Clin. Cancer Res. 2007; 13:824–831. [DOI] [PubMed] [Google Scholar]
- 114.Farias L.M., Ocana D.B., Diaz L., Larrea F., Avila-Chavez E., Cadena A., Hinojosa L.M., Lara G., Villanueva L.A., Vargas C.et al.. Ether a go-go potassium channels as human cervical cancer markers. Cancer Res. 2004; 64:6996–7001. [DOI] [PubMed] [Google Scholar]
- 115.Tateishi-Karimata H., Kawauchi K., Sugimoto N.. Destabilization of DNA G-quadruplexes by chemical environment changes during tumor progression facilitates transcription. J. Am. Chem. Soc. 2018; 140:642–651. [DOI] [PubMed] [Google Scholar]
- 116.Rode A.B., Endoh T., Sugimoto N.. tRNA shifts the G-quadruplex-hairpin conformational equilibrium in RNA towards the hairpin conformer. Angew. Chem. Int. Ed. Engl. 2016; 55:14315–14319. [DOI] [PubMed] [Google Scholar]
- 117.Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A.S., Yu T., Marie-Nelly H., McSwiggen D.T., Kokic G., Dailey G.M., Cramer P.et al.. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018; 25:833–840. [DOI] [PubMed] [Google Scholar]
- 118.Massague J.TGFbeta in Cancer. Cell. 2008; 134:215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kikin O., Zappala Z., D’Antonio L., Bagga P.S.. GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008; 36:D141–D148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bonnal S., Schaeffer C., Creancier L., Clamens S., Moine H., Prats A.C., Vagner S.. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003; 278:39330–39336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Balagurumoorthy P., Brahmachari S.K., Mohanty D., Bansal M., Sasisekharan V.. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992; 20:4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Naeem M.M., Maheshan R., Costford S.R., Wahedi A., Trajkovski M., Plavec J., Yatsunyk L.A., Ciesielski G.L., Kaufman B.A., Sondheimer N.. G-quadruplex-mediated reduction of a pathogenic mitochondrial heteroplasmy. Hum. Mol. Genet. 2019; 28:3163–3174. [DOI] [PubMed] [Google Scholar]
- 123.Shahid R., Bugaut A., Balasubramanian S.. The BCL-2 5′ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry. 2010; 49:8300–8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gomez D., Guedin A., Mergny J.L., Salles B., Riou J.F., Teulade-Fichou M.P., Calsou P.. A G-quadruplex structure within the 5′-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 2010; 38:7187–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morris M.J., Negishi Y., Pazsint C., Schonhoft J.D., Basu S.. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010; 132:17831–17839. [DOI] [PubMed] [Google Scholar]
- 126.Lammich S., Kamp F., Wagner J., Nuscher B., Zilow S., Ludwig A.K., Willem M., Haass C.. Translational repression of the disintegrin and metalloprotease ADAM10 by a stable G-quadruplex secondary structure in its 5′-untranslated region. J. Biol. Chem. 2011; 286:45063–45072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yokota N., Aruga J., Takai S., Yamada K., Hamazaki M., Iwase T., Sugimura H., Mikoshiba K.. Predominant expression of human zic in cerebellar granule cell lineage and medulloblastoma. Cancer Res. 1996; 56:377–383. [PubMed] [Google Scholar]
- 128.Agarwala P., Pandey S., Mapa K., Maiti S.. The G-quadruplex augments translation in the 5′ untranslated region of transforming growth factor beta2. Biochemistry. 2013; 52:1528–1538. [DOI] [PubMed] [Google Scholar]
- 129.Murat P., Marsico G., Herdy B., Ghanbarian A., Portella G., Balasubramanian S.. RNA G-quadruplexes at upstream open reading frames cause DHX36-and DHX9-dependent translation of human mRNAs. Genome Biol. 2018; 19:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zeraati M., Moye A.L., Wong J.W.H., Perera D., Cowley M.J., Christ D.U., Bryan T.M., Dinger M.E.. Cancer-associated noncoding mutations affect RNA G-quadruplex-mediated regulation of gene expression. Sci. Rep. 2017; 7:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M.et al.. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018; 175:1842–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cho W.K., Spille J.H., Hecht M., Lee C., Li C., Grube V., Cisse II.. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018; 361:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chapuy B., McKeown M.R., Lin C.Y., Monti S., Roemer M.G., Qi J., Rahl P.B., Sun H.H., Yeda K.T., Doench J.G.et al.. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013; 24:777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hnisz D., Schuijers J., Lin C.Y., Weintraub A.S., Abraham B.J., Lee T.I., Bradner J.E., Young R.A.. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell. 2015; 58:362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Boulay G., Sandoval G.J., Riggi N., Iyer S., Buisson R., Naigles B., Awad M.E., Rengarajan S., Volorio A., McBride M.J.et al.. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell. 2017; 171:163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Crozat A., Aman P., Mandahl N., Ron D.. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993; 363:640–644. [DOI] [PubMed] [Google Scholar]
- 138.Budworth H., McMurray C.T.. A brief history of triplet repeat diseases. Methods Mol. Biol. 2013; 1010:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P.et al.. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991; 65:905–914. [DOI] [PubMed] [Google Scholar]
- 140.Rohilla K.J., Gagnon K.T.. RNA biology of disease-associated microsatellite repeat expansions. Acta Neuropathol. Commun. 2017; 5:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ashizawa T., Wong L.J., Richards C.S., Caskey C.T., Jankovic J.. CAG repeat size and clinical presentation in Huntington's disease. Neurology. 1994; 44:1137–1143. [DOI] [PubMed] [Google Scholar]
- 142.Sugimoto N.Chemistry and Biology of Non-canonical Nucleic Acids. 2021; 1st ednWiley-VCH. [Google Scholar]
- 143.Zoghbi H.Y., Orr H.T.. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000; 23:217–247. [DOI] [PubMed] [Google Scholar]
- 144.Brais B., Bouchard J.P., Xie Y.G., Rochefort D.L., Chretien N., Tome F.M., Lafreniere R.G., Rommens J.M., Uyama E., Nohira O.et al.. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 1998; 18:164–167. [DOI] [PubMed] [Google Scholar]
- 145.Pearson C.E., Nichol Edamura K., Cleary J.D. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005; 6:729–742. [DOI] [PubMed] [Google Scholar]
- 146.Potaman V.N., Bissler J.J., Hashem V.I., Oussatcheva E.A., Lu L., Shlyakhtenko L.S., Lyubchenko Y.L., Matsuura T., Ashizawa T., Leffak M.et al.. Unpaired structures in SCA10 (ATTCT)n.(AGAAT)n repeats. J. Mol. Biol. 2003; 326:1095–1111. [DOI] [PubMed] [Google Scholar]
- 147.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J.et al.. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011; 72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.de Mezer M., Wojciechowska M., Napierala M., Sobczak K., Krzyzosiak W.J.. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011; 39:3852–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mitas M.Trinucleotide repeats associated with human disease. Nucleic Acids Res. 1997; 25:2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mariappan S.V., Catasti P., Silks L.A. 3rd, Bradbury E.M., Gupta G.. The high-resolution structure of the triplex formed by the GAA/TTC triplet repeat associated with Friedreich's ataxia. J. Mol. Biol. 1999; 285:2035–2052. [DOI] [PubMed] [Google Scholar]
- 151.Park Y., Kim K.T., Kim B.H.. G-Quadruplex formation using fluorescent oligonucleotides as a detection method for discriminating AGG trinucleotide repeats. Chem. Commun. (Camb.). 2016; 52:12757–12760. [DOI] [PubMed] [Google Scholar]
- 152.Chi L.M., Lam S.L.. Structural roles of CTG repeats in slippage expansion during DNA replication. Nucleic Acids Res. 2005; 33:1604–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A.et al.. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996; 271:1423–1427. [DOI] [PubMed] [Google Scholar]
- 154.Jain A., Ahmad F., Rajeswari M.R.. Structural studies on DNA triple helix formed by intronic GAA triplet repeat expansion in Friedreich's ataxia. Nucleosides. Nucleotides. Nucleic. Acids. 2003; 22:1517–1519. [DOI] [PubMed] [Google Scholar]
- 155.Singer R.H.Triplet-repeat transcripts: a role for DNA in disease. Science. 1998; 280:696–697. [DOI] [PubMed] [Google Scholar]
- 156.Matsuura T., Yamagata T., Burgess D.L., Rasmussen A., Grewal R.P., Watase K., Khajavi M., McCall A.E., Davis C.F., Zu L.et al.. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat. Genet. 2000; 26:191–194. [DOI] [PubMed] [Google Scholar]
- 157.Orengo J.P., Chambon P., Metzger D., Mosier D.R., Snipes G.J.. Expanded CTG and edgewise loops repeats within the DMPK 3' UTR causes severeskeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:2646–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ranum L.P., Day J.W.. Dominantly inherited, non-coding microsatellite expansion disorders. Curr. Opin. Genet. Dev. 2002; 12:266–271. [DOI] [PubMed] [Google Scholar]
- 159.Law M.J., Lower K.M., Voon H.P., Hughes J.R., Garrick D., Viprakasit V., Mitson M., De Gobbi M., Marra M., Morris A.et al.. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010; 143:367–378. [DOI] [PubMed] [Google Scholar]
- 160.Shioda N., Yabuki Y., Yamaguchi K., Onozato M., Li Y., Kurosawa K., Tanabe H., Okamoto N., Era T., Sugiyama H.et al.. Targeting G-quadruplex DNA as cognitive function therapy for ATR-X syndrome. Nat. Med. 2018; 24:802–813. [DOI] [PubMed] [Google Scholar]
- 161.Subramanian M., Rage F., Tabet R., Flatter E., Mandel J.L., Moine H.. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011; 12:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bole M., Menon L., Mihailescu M.R.. Fragile X mental retardation protein recognition of G quadruplex structure per se is sufficient for high affinity binding to RNA. Mol. Biosyst. 2008; 4:1212–1219. [DOI] [PubMed] [Google Scholar]
- 163.Bugaut A., Balasubramanian S.. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012; 40:4727–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Larkin K., Fardaei M.. Myotonic dystrophy–a multigene disorder. Brain Res. Bull. 2001; 56:389–395. [DOI] [PubMed] [Google Scholar]
- 165.Wojciechowska M., Krzyzosiak W.J.. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011; 20:3811–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liquori C.L., Ricker K., Moseley M.L., Jacobsen J.F., Kress W., Naylor S.L., Day J.W., Ranum L.P.. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001; 293:864–867. [DOI] [PubMed] [Google Scholar]
- 167.Zhang K., Donnelly C.J., Haeusler A.R., Grima J.C., Machamer J.B., Steinwald P., Daley E.L., Miller S.J., Cunningham K.M., Vidensky S.et al.. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015; 525:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kwon I., Xiang S.H., Kato M., Wu L., Theodoropoulos P., Wang T., Kim J., Yun J., Xie Y., McKnight S.L.. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014; 345:1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee Y.-B., Chen H.-J., Peres J.N., Gomez-Deza J., Attig J., Stalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C.et al.. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013; 5:1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang A.Y., Balasubramanian S.. The kinetics and folding pathways of intramolecular G-quadruplex nucleic acids. J. Am. Chem. Soc. 2012; 134:19297–19308. [DOI] [PubMed] [Google Scholar]
- 171.Blaszczyk L., Rypniewski W., Kiliszek A.. Structures of RNA repeats associated with neurological diseases. Wiley Interdiscip. Rev. RNA. 2017; 8:e1412. [DOI] [PubMed] [Google Scholar]
- 172.Donnelly C.J., Zhang P.W., Pham J.T., Haeusler A.R., Mistry N.A., Vidensky S., Daley E.L., Poth E.M., Hoover B., Fines D.M.et al.. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013; 80:415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Todd P.K., Paulson H.L.. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 2010; 67:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Uversky V.N.Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017; 44:18–30. [DOI] [PubMed] [Google Scholar]
- 175.Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D.et al.. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015; 57:936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mergny J.L., Helene C.. G-quadruplex DNA: a target for drug design. Nat. Med. 1998; 4:1366–1367. [DOI] [PubMed] [Google Scholar]
- 177.Simone R., Fratta P., Neidle S., Parkinson G.N., Isaacs A.M.. G-quadruplexes: emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015; 589:1653–1668. [DOI] [PubMed] [Google Scholar]
- 178.Tateishi-Karimata H., Sugimoto N.. Chemical biology of non-canonical structures of nucleic acids for therapeutic applications. Chem. Commun. (Camb.). 2020; 56:2379–2390. [DOI] [PubMed] [Google Scholar]
- 179.Monchaud D., Teulade-Fichou M.P.. A hitchhiker's guide to G-quadruplex ligands. Org. Biomol. Chem. 2008; 6:627–636. [DOI] [PubMed] [Google Scholar]
- 180.Zamiri B., Reddy K., Macgregor R.B. Jr, Pearson C.E.. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J. Biol. Chem. 2014; 289:4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Su Z., Zhang Y., Gendron T.F., Bauer P.O., Chew J., Yang W.Y., Fostvedt E., Jansen-West K., Belzil V.V., Desaro P.et al.. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014; 83:1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lagier-Tourenne C., Baughn M., Rigo F., Sun S., Liu P., Li H.R., Jiang J., Watt A.T., Chun S., Katz M.et al.. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E4530–E4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A.. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012; 488:111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lee J.E., Bennett C.F., Cooper T.A.. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.





